ABSTRACT
Replication and packaging of the rotavirus genome occur in cytoplasmic compartments called viroplasms, which form during virus infection. These processes are orchestrated by yet-to-be-understood complex networks of interactions involving nonstructural proteins (NSPs) 2, 5, and 6 and structural proteins (VPs) 1, 2, 3, and 6. The multifunctional enzyme NSP2, an octamer with RNA binding activity, is critical for viroplasm formation with its binding partner, NSP5, and for genome replication/packaging through its interactions with replicating RNA, the viral polymerase VP1, and the inner core protein VP2. Using isothermal calorimetry, biolayer interferometry, and peptide array screening, we examined the interactions between NSP2, VP1, VP2, NSP5, and NSP6. These studies provide the first evidence that NSP2 can directly bind to VP1, VP2, and NSP6, in addition to the previously known binding to NSP5. The interacting sites identified from reciprocal peptide arrays were found to be in close proximity to the RNA template entry and double-stranded RNA (dsRNA) exit tunnels of VP1 and near the catalytic cleft and RNA-binding grooves of NSP2; these sites are consistent with the proposed role of NSP2 in facilitating dsRNA synthesis by VP1. Peptide screening of VP2 identified NSP2-binding sites in the regions close to the intersubunit junctions, suggesting that NSP2 binding could be a regulatory mechanism for preventing the premature self-assembly of VP2. The binding sites on NSP2 for NSP6 were found to overlap that of VP1, and the NSP5-binding sites overlap those of VP2 and VP1, suggesting that interaction of these proteins with NSP2 is likely spatially and/or temporally regulated.
IMPORTANCE Replication and packaging of the rotavirus genome occur in cytoplasmic compartments called viroplasms that form during virus infection and are orchestrated by complex networks of interactions involving nonstructural proteins (NSPs) and structural proteins (VPs). A multifunctional RNA-binding NSP2 octamer with nucleotidyl phosphatase activity is central to viroplasm formation and RNA replication. Here we provide the first evidence that NSP2 can directly bind to VP1, VP2, and NSP6, in addition to the previously known binding to NSP5. The interacting sites identified from peptide arrays are consistent with the proposed role of NSP2 in facilitating dsRNA synthesis by VP1 and also point to NSP2's possible role in preventing the premature self-assembly of VP2 cores. Our findings lead us to propose that the NSP2 octamer with multiple enzymatic activities is a principal regulator of viroplasm formation, recruitment of viral proteins into the viroplasms, and possibly genome replication.
INTRODUCTION
Rotavirus, a member of the family Reoviridae, is a large icosahedral virus, with a genome consisting of 11 segments of double-stranded RNA (dsRNA) encoding six structural proteins (VPs) and six nonstructural proteins (NSPs). The genome is enclosed within a triple-layered capsid, composed of three concentric protein layers: (i) an outer capsid layer of VP7 and VP4; (ii) an intermediate layer of VP6; (iii) an inner layer composed of VP2, with VP1, the RNA-dependent RNA polymerase (RdRP), and VP3, the viral methyltransferase and guanylyltransferase, positioned on the inside surface of this inner layer at each of the five-fold icosahedral axes (1, 2). The outer capsid layer is lost during cell entry, resulting in activation of endogenous transcription within the double-layered particle (DLP). Transcription yields capped, nonpolyadenylated, positive-strand RNA [(+)RNA] molecules that are extruded into the cytoplasm of the infected cell, where they serve as mRNAs for translation of viral proteins and as the templates for negative-strand RNA [(–)RNA] synthesis (2, 3).
Genome replication, packaging, and DLP assembly occur in cytoplasmic structures called viroplasms (4–8) that form early during infection. These processes are regulated by a network of interactions involving NSPs 2, 5, and 6 and VPs 1, 2, 3, and 6 (9, 10). NSP2, a multifunctional protein that exhibits nucleoside triphosphatase (NTPase), RNA triphosphatase (RTPase), and helix-destabilizing activities (10–14), is critical for both viroplasm formation with its binding partner NSP5 (15, 16) and for viral replication and packaging via its interactions with RNA, VP1, and VP2 (10, 12, 17, 18). Strains expressing temperature-sensitive mutants of NSP2 lose their ability to synthesize dsRNA and produce empty virus particles at nonpermissive temperatures (19, 20). NSP2's role in replication is further supported by the fact that NSP2 is a component of early replication intermediates (RIs) (18, 21). The crystal structure of NSP2 revealed a donut-shaped octamer with a 35-Å central hole and deep grooves positioned diagonally along the two-fold axis. The grooves are lined by positively charged residues and are the single-stranded RNA (ssRNA)-binding sites on the octamer (10, 22–24). They are also the binding sites for NSP5, an O-glycosylated phosphoprotein that has RNA-binding (11, 12, 25–27), VP1-binding (28), VP2-binding (29), and NSP6-binding (30) activities. Even though the exact function of NSP5 in the infection process is unresolved, it can be speculated that the protein is involved in genome replication and packaging. The localization of NSP5 in viroplasms and its activity are correlated with the phosphorylation state of the protein, which in turn is dependent on its interaction with NSP2 (15, 31, 32).
Another nonstructural protein found in the viroplasms is NSP6. NSP6 is encoded by the alternative open reading frame (ORF) of segment 11 of most viral strains and, together with NSP2 and NSP5, it accumulates in the viroplasms (7, 27, 30, 33). NSP6 is known to interact with NSP5 and is thought to play a regulatory role in multimerization and hyperphosphorylation of NSP5 (30). It is also a sequence-independent nucleic acid-binding protein, with similar affinities for ssRNA and dsRNA (34). In contrast to NSP2 and NSP5, very little is known about the role NSP6 plays in rotavirus infection.
Inside the viroplasms, replication is thought to be initiated when the viral polymerase VP1, in complex with VP3 and the (+)RNA, comes into contact with the core protein VP2. Upon binding to VP2, VP1 undergoes a structural reorganization leading to activation of the RdRP and initiation of dsRNA synthesis (21, 35–39). The N-terminal domain of VP2 protrudes inside the core at the five-fold axis, is involved in VP2-RNA interactions, and serves as a scaffold for the viral transcription complex in viral particles (39–42). It is suggested that the N terminus of VP2 (amino acids [aa] ∼1 to 100) plays a regulatory role in VP1 polymerase activation by possibly forming tethers that support the interaction of VP1 with the principal scaffold domain of VP2 (aa ∼101 to 880) (43, 44). NSP2, as a component of replication intermediates, interacts with VP1 (17, 45); however, whether this interaction involves direct binding of the proteins is not known. The N-terminal domain of VP2 is required for activation of VP1, and the core assembly is thought to happen concurrently with dsRNA synthesis; therefore, it is reasonable to suppose that, in addition to VP1 and RNA, NSP2 interacts with VP2 (specifically, the N-terminal region) as well.
In this study, we examined the interactions of NSP2 with viral proteins that are known constituents of viroplasm and RIs, specifically, VP1, VP2, and NSP6, by using isothermal titration calorimetry (ITC) and biolayer interferometry (BLI), which clearly demonstrated that NSP2 directly interacts with VP1, VP2, and NSP6. ITC and BLI are standard techniques for determining binding affinities (KD) without relying on radioactive or fluorescent labels. Further, using peptide arrays, we identified possible specific binding sites involved in these interactions, both on NSP2 and on its binding partners. Peptide arrays offer a rapid means of screening and identifying peptides that interact with target molecules by synthesizing a large array of specifically designed peptides on cellulose membranes, which can then be probed using a protein of choice (46, 47). Peptide array experiments have been effectively used to identify peptide inhibitors of β-lactamases (48), novel binding sites for the tumor suppressor protein p53 on its major regulator HDM2 (49), and nuclear carrier binding sites on nucleoporins (Nups) (50) and to develop antibodies with high therapeutic potential against viral infections (51, 52). In every tested case, array-identified interactions have been confirmed by solution-based binding studies (49, 51), X-ray crystallography (53, 54), and site-directed mutagenesis experiments (50). In our experiments, arrays spanning the full length of NSP2 were screened for sequences that bound VP1, the N terminus of VP2, and NSP6. This allowed us to map the regions in these proteins that participate in the interactions. Our results suggest that the NSP2 octamer provides a suitable platform to coordinate a variety of interactions and plays a significant role in the formation of RIs and initiation of viral replication.
MATERIALS AND METHODS
Protein expression and purification.
Baculovirus expressing C-terminally His-tagged VP1 (accession number O37061) from simian rotavirus strain SA11 was obtained from J. Patton, NIH, Bethesda, MD (37). SF9 insect cells were infected at a multiplicity of infection (MOI) of 5 for 96 h at 27°C at the Baculovirus Core Facility, Baylor College of Medicine. The cells were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended in the lysis buffer (25 mM HEPES [pH 7.8], 100 mM NaCl) containing EDTA-free protease inhibitors. The cells were lysed in a microfluidizer (Microfluidics, Newton, MA) and applied to Talon metal affinity resin (Clontech) preequilibrated with lysis buffer. VP1 was eluted using a gradient of imidazole from 0.1 M to 1 M. Protein peak fractions were pooled, bound to a HiTrap FF heparin HP column (GE Healthcare), and eluted using a salt gradient from 0.1 M to 1 M. Purified VP1 was dialyzed into a final buffer (25 mM HEPES [pH 7.8], 100 mM NaCl) and used for further studies. SA11 (group A) NSP2 (accession number Q03242) and Bristol (group C) NSP2 (accession number CAB52753) were cloned into the bacterial expression vector pQE60 (Qiagen) with a C-terminal 6×His tag preceded by a thrombin cleavage site (LVPRGS), as described by Hu et al. and Taraporewala et al., respectively (10, 23). Both proteins were expressed in Escherichia coli SG13009 cells and purified using Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography (Qiagen), and HiTrap SP HP column (GE Healthcare) as described above, followed by gel filtration chromatography (HiLoad 16/60 Superdex 200; GE Healthcare). Gel filtration confirmed that both SA11 and Bristol NSP2 form an octamer in solution. Purified NSP2 was dialyzed into a final buffer containing 10 mM Tris-HCl (pH 7.8) and 100 mM NaCl. The SA11 VP2 (accession number AAA47349.1) construct corresponding to residues 1 to 134 was cloned into a pET46 vector (Invitrogen) with an N-terminal 6×His tag followed by a thrombin cleavage site (LVPRGS). Protein was expressed in E. coli BL21 cells and purified as described above. Purified VP21-134 was dialyzed into 10 mM Tris-HCl (pH 7.8), 100 mM NaCl. The full-length SA11 NSP6 gene (accession number AF306493.1) was cloned into a pET32 expression vector (Invitrogen) with an N-terminal 6×His tag followed by a thrombin cleavage site (LVPRGS). Protein was expressed in the E. coli BL21 cells, extracted from the insoluble fraction in a buffer containing 50 mM n-dodecyl-β-d-maltoside (DDM) and 10 mM imidazole, 300 mM NaCl, 50 mM Tris (pH 7.2), and purified using a Ni-NTA affinity chromatography (Qiagen) followed by gel filtration chromatography (HiLoad 16/60 Superdex 200; GE Healthcare). Purified NSP6 was dialyzed into a final buffer containing 10 mM Tris-HCl (pH 7.8), 100 mM NaCl, and 0.005% DDM. For the binding analyses, the affinity tags on NSP2, VP2, and NSP6 were cleaved off by using thrombin (Hematologic Tech).
ITC and BLI.
ITC analysis was carried out using a MicroCal Auto-iTC200 instrument (GE Healthcare). NSP2 was used at 20 μM in ITC buffer (10 mM Tris [pH 7.8], 150 mM NaCl). VP21-134 was used at 200 μM in the same buffer, and NSP6 was used at 200 μM in the same buffer supplemented with 0.05% DDM. For each experiment, 40 and 50 titrations were carried out at 4°C and at 10°C, respectively. Experiments were performed in triplicates. Each titration was fitted into a one-site binding model by using the Origin software provided with the instrument.
BLI was carried out using an Octet RED96 instrument (ForteBio). Biotinylation of proteins for loading onto streptavidin-coated biosensors (ForteBio) was carried out using EZ-link NHC-LC-LC-biotin (catalog number 21343; Thermo Scientific) following the instructions of the manufacturer. For NSP2-VP1-binding analysis, biotinylated VP1 was loaded onto streptavidin biosensors at a concentration of 1.25 μg/ml in BLI running buffer (20 mM HEPES [pH 7.8], 150 mM NaCl, 0.05% surfactant P20, and 2 mg/ml bovine serum albumin) for 600 s, resulting in capture levels of 0.8 to 1.0 nm within a row of eight tips. NSP2-VP1 association and dissociation curves were obtained through serial dilutions of NSP2 (0.5, 0.8, 1.5, 3, 5, 7, and 10 μM) plus buffer blanks and using the Octet acquisition software. For NSP2-VP21-134 analysis, biotinylated NSP2 (1.25 μg/ml) was immobilized on streptavidin biosensors, and association and dissociation curves were obtained through serial dilutions of VP21-134 (18, 56, and 112 μM). For the analysis of NSP2 interactions with NSP6, biotinylated NSP6 (18 μg/ml) was immobilized onto a streptavidin biosensor, and the association and dissociation curves were obtained through serial dilutions of NSP2 (2.5, 5, 10, 20, and 40 μM). The binding data in each case were fitted using the Octet analysis software.
Peptide array synthesis.
Peptide arrays of 17-mer peptides corresponding to the each of the rotavirus protein VP1, VP2, NSP5, NSP6, and NSP2 (SA11 and Bristol) sequences were synthesized directly onto cellulose membranes (amino-PEG500-UC450; Intavis AG) by using an Auto Spot Robot ASP 222 peptide synthesizer (Intavis AG) and the autospot program (55) with 9-fluorenylmethoxy carbonyl (Fmoc)-protected amino acids (Intavis AG) that were prepared according to the manufacturer's instructions. Each peptide array started at the N terminus and spanned the entire length of the protein sequence, with each successive spot containing 17 amino acids along the sequence shifted by 3 amino acids toward the C terminus, i.e., each spot in the array had a 14-residue overlap with the previous spot. Prior to the first round of synthesis, cellulose membranes were prepared by washing three times for 2 min in N,N-dimethyl formamide (DMF), then washing 2 times in ethanol, and finally drying the sheets for 10 min. Fmoc-protected amino acids were activated with 0.2 M diisopropylcarbodiimide (DIC) and 0.13 M 1-hydroxybenzotriazole (HOBt) in N-methylpyrrolidone (NMP). After the last round of synthesis, membranes were washed with 20% piperidine in DMF for 30 s, followed by two 2-min washes in H2O. To deprotect the side chains that had been acetylated, membranes were incubated in 20 ml of side chain deprotection solution (95% trifluoroacetic acid [TFA], and 3% tri-isopropyl saline [TIS]) for 1 h. After deprotection, membranes were washed with 20 ml DCM, followed by four 2-min washes with ethanol. Finished membranes were dried overnight in a chemical hood (50).
Peptide array screening.
Cellulose membranes containing synthesized VP1 (SA11 and Bristol strains), VP2, NSP5, and NSP6 peptide arrays were screened to identify peptides that bound SA11 and Bristol NSP2. In reciprocal experiments, membranes containing synthesized NSP2 peptides were screened to identify peptides that bound SA11 VP1, VP21-134, and NSP6. Briefly, prepared cellulose membranes were washed three times for 5 min in Millipore H2O and blocked overnight at 4°C with 3% (wt/vol) milk–PBS (MPBS). Purified proteins (15 μM) were incubated with the membranes in MPBS for 3 h at room temperature with gentle agitation. For negative controls, membranes were prepared and treated as above but incubated with 3% MPBS and no protein. After incubation, membranes were washed four times for 5 min with PBS supplemented with 1% Tween 20 (PBST). The peptide-bound proteins were detected by incubating the membranes with either anti-His tag (VP2, VP1, and NSP2) or anti-S tag (NSP6)–horseradish peroxidase (HRP)–conjugated antibodies (2 μg/ml; Abcam) in MPBS for 2 h at 4°C with gentle agitation. The membranes were then washed three times for 5 min with PBST, incubated in 5 ml of the enhanced chemiluminescence (ECL) developing solution, and exposed to X-ray films in the ECL system (Amersham Biosciences) (50). Autoradiograms were scanned into the Typhoon TRIO variable mode imager (GE Healthcare), and the Quantity One program was used to measure intensities of each spot. No oversaturation of the spots was detected. For each peptide array, after subtracting the background, as measured from a negative-control membrane, spot intensities were plotted relative to the spot that showed the highest intensity, which was set as 100% binding. Spots with intensities higher than a cutoff of 60%, based on the NSP5 peptide array analysis (see Results), were considered to represent positive binding. To take into consideration that some positive signals could represent nonspecific binding, only those residues that were present in at least two spots that showed a binding intensity above a more stringent 80% cutoff were considered significant for mapping onto the crystal structures when available, such as that of NSP2, VP2, and VP1. Each peptide array experiment was carried out at least two times to ensure reproducibility.
Mutagenesis of the identified VP1- and NSP6-binding sites on full-length NSP2.
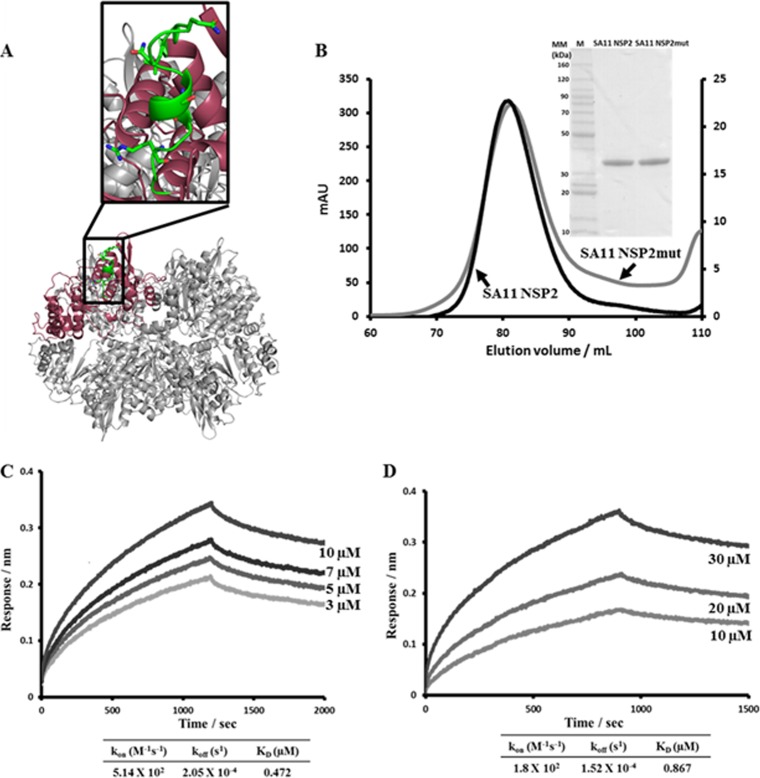
To validate the observations from the peptide array analysis, in the context of full-length proteins, the SA11 NSP2 peptide LKVTQANVSNVLSRVVS (aa 91 to 107), identified by peptide array analysis, that showed binding to VP1 as well as NSP6, was chosen for site-directed mutagenesis experiments. In the full-length NSP2, residues 91, 95, 97, 100, and 104 (shown in bold) were replaced with alanine by using Epoch (Epoch Life Science, Inc., Missouri City, TX). A full-length SA11 NSP2 construct containing all five mutations (NSP2mut) was cloned into the bacterial expression vector pQE60 (Qiagen) with a C-terminal His tag preceded by a thrombin cleavage site. The protein was expressed in E. coli SG13009 cells and purified using Ni-NTA affinity chromatography (Qiagen) and gel filtration chromatography (HiLoad 16/60 Superdex 200; GE Healthcare) as described above. Gel filtration confirmed that NSP2mut still formed an octamer in solution, similar to wild-type SA11 NSP2. Purified NSP2mut was dialyzed into a final buffer (10 mM Tris-HCl [pH 7.8], 100 mM NaCl), and its ability to bind VP1 and NSP6 was analyzed using BLI following the protocol described above.
RESULTS
Interaction of NSP2 with VP1.
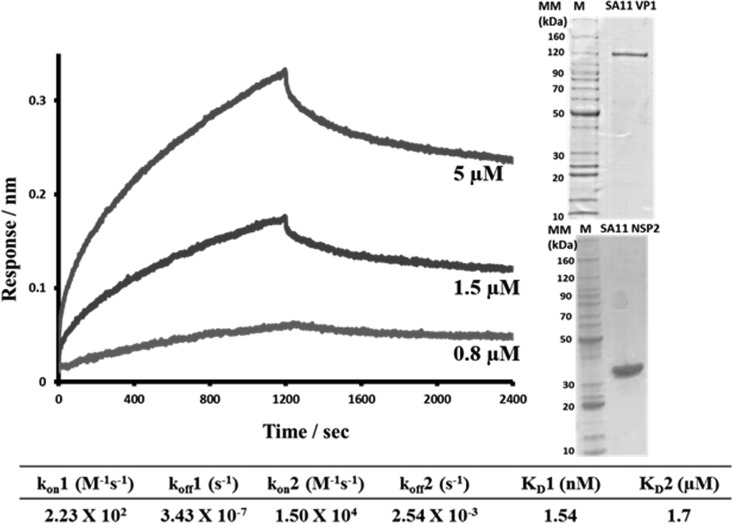
The RTPase activity of NSP2 is proposed to aid in genome circularization and spooling around the VP1/VP3 enzyme complex by specifically recognizing the 5′ consensus sequence of (–)RNA as soon as the dsRNA emerges from VP1 and allowing it to be recaptured by VP1 after the hydrolysis is complete (10, 56). Furthermore, the NSP2 octamer with electropositive RNA-binding grooves is suggested to play a role in relaxing the mRNA in preparation for dsRNA synthesis by VP1 (13). Based on these findings and biochemical studies that strongly suggest an interaction between NSP2 and VP1 (17, 28), we hypothesized that NSP2 directly binds VP1 during replication and/or core assembly. To analyze the interaction between NSP2 and VP1, we examined the rates of association and dissociation of the proteins by using BLI. For NSP2-binding analysis, we used a biotinylated VP1 immobilized on the streptavidin biosensor that was dipped into wells containing serial dilutions of NSP2. The sensograms obtained for NSP2 binding to VP1 were best fitted by a global two-site binding model with the χ2/degrees of freedom (DoF) value of 0.00115 for the fit (Fig. 1). Data analysis showed the binding affinity between the two proteins to be 1.54 nM for site one and 1.7 μM for site two, indicating a strong direct interaction between NSP2 and VP1.
FIG 1.
BLI analysis of NSP2-VP1 interactions. VP1-NSP2 association and dissociation curves were obtained through serial dilutions of NSP2 (0.5, 0.8, 1.5, 3, 5, 7, and 10 μM) plus buffer blanks and using the Octet acquisition software. Representative sensograms for three of the concentrations are shown. Sensograms were fitted with a global two-site binding model. The KD1, KD2, kon, and koff values for two sites are shown below the graph. The insert shows Coomassie blue-stained SDS-PAGE gels of SA11 VP1 and SA11 NSP2 proteins.
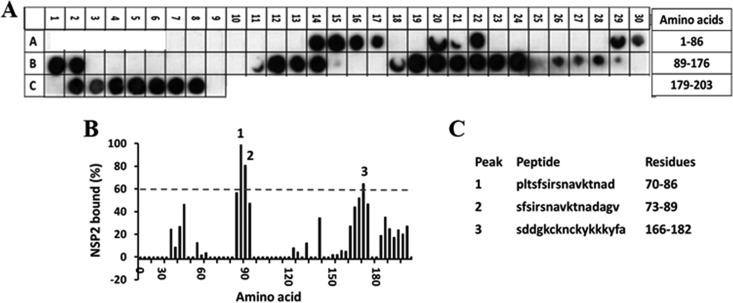
To identify the possible regions on the proteins that participate in the interaction, we synthesized peptide arrays encompassing full-length VP1 or NSP2 and screened them for peptides that bound either NSP2 or VP1, respectively. Before proceeding with the VP1 peptide array analysis, we used an NSP5 peptide array as a positive control, since NSP5 is a known NSP2-binding protein and some of the NSP2-interacting regions on NSP5 have been described previously (15, 16, 31, 32). We synthesized a peptide array encompassing the full-length NSP5 sequence and probed it for binding to NSP2. The array indicated two major sites for NSP2 binding (Fig. 2). The first site (peaks 1 and 2, aa 70 to 89) was previously unidentified and located in the N-terminal domain of NSP5. The second site (peak 3, aa 166 to 182) was consistent with previous studies, which showed that this region in NSP5 is essential for binding to NSP2 (15, 31). Based on an ∼60% intensity level of the peptide (SDDGKCKNCKYKKKYFA) in this region, a 60% cutoff was considered to represent positive binding in the subsequent peptide array analysis.
FIG 2.
Binding of the full-length SA11 NSP2 to the peptide array of the SA11 NSP5 sequence. (A) Autoradiograph of the NSP5 peptide array probed with full-length NSP2. The peptide array consists of spots of 17-residue peptides in the protein sequence, starting from the N terminus (spot A7, upper left corner of the array) and ending with the C-terminal peptide (spot C9, lower right corner of the array), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the protein sequence. Dark spots on the autoradiograms indicate peptides with high binding specificities, whereas the light spots indicate peptides with weak or no binding. (B) Graph showing relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). The tick marks on the x axis correspond to spots A7 through C8. In this graph, the spot intensities, after subtracting the background (a negative control), were plotted relative to the spot with the highest intensity, which was set as 100% binding. Spots that show intensities higher than the 60% cutoff are numbered. (C) The peptide sequences corresponding to the three spots (1 to 3) in panel B.
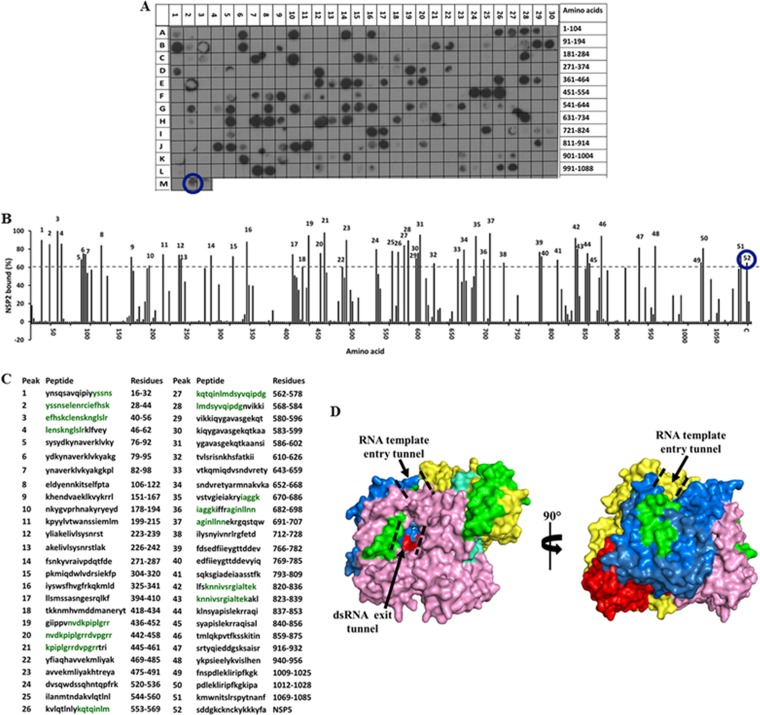
For the VP1-NSP2 peptide arrays, the intensity of each binding spot was calculated and plotted against 100% binding (Fig. 3 and 4). On the VP1 array, 52 spots showed binding to NSP2 with at least a 60% cutoff intensity (Fig. 3A to C), with 20 spots showing ≥80% cutoff intensities. Residues that were present in at least two peptides that bound NSP2 with ≥80% intensity were considered significant and were mapped onto the crystal structure of VP1 (Fig. 3D, shown in green). NSP2-binding sites localized to the N-terminal domain (aa 28 to 56), the polymerase fingers subdomain (aa 442 to 458 and 562 to 578), thumb subdomain (aa 682 to 698), and the C-terminal “bracelet” domain (aa 823 to 836) of VP1 (37). All of the residues mapped to the surface-exposed regions of VP1 with the exception of amino acids 682 to 698, which mapped to inside the catalytic core of VP1 and are unlikely to participate in the interaction with NSP2. Residues 442 to 458 and 562 to 578 line one side of the RNA template entry tunnel of VP1. Binding of NSP2 near the tunnel is in agreement with its helix-destabilizing activity, which has been suggested to relax the secondary structures on mRNA templates in preparation for dsRNA synthesis (13). Residues 823 to 836 line the side of the dsRNA product exit tunnel of VP1, and binding of NSP2 at these sites would bring it into close proximity with the 5′ (−)RNAs, leading to more efficient γ-phosphate hydrolysis.
FIG 3.
Binding of the full-length SA11 NSP2 to the peptide array of the SA11 VP1 sequence. (A) Autoradiograph of the VP1 peptide array probed by full-length NSP2. The peptide array consists of spots of 17-residue peptides in the VP1 sequence starting from the N terminus (spot A1) and ending with the C-terminal peptide (spot L27), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the VP1 sequence. (B) Graph showing the relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). The spot with the highest intensity was set as 100% binding. The spots with intensities higher than the 60% cutoff are numbered 1 to 52. (C) The peptide sequences corresponding to the peaks (1 to 52) in panel B. VP1 residues present in at least two spots that showed ≥80% binding intensity (green) were mapped onto the crystal structure of VP1 (pdb 2R7R). (D) Surface representation of the VP1 structure with the N-terminal domain (yellow), C-terminal domain (pink), and the polymerase fingers (blue), palm (red), and thumb (cyan) subdomains labeled. NSP2-binding regions are shown in green. The RNA template entry tunnel and dsRNA exit tunnel are shown by dashed black lines. In panels A and B, the NSP5 peptide is marked by a blue circle.
FIG 4.
Binding of the full-length SA11 VP1 to the peptide array of the SA11 NSP2 sequence. (A) Autoradiograph of the NSP2 peptide array probed by full-length VP1. The peptide array consists of spots of 17-residue peptides in the NSP2 sequence, starting from the N terminus (spot A1) and ending with the C-terminal peptide (D11), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the NSP2 sequence. (B) Graph showing the relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). (C) The peptide sequences corresponding to the spots denoted 1 to 18 in panel B that showed intensities higher than the 60% cutoff. NSP2 residues present in at least two peptides that showed a ≥80% binding intensity (green) were mapped onto the known structure of NSP2 (pdb 1L9V). (D) Surface model of the NSP2 octamer (gray) with VP1-binding residues shown in green on monomer A (pink). The RNA-binding groove is shown by dashed black lines, and the catalytic cleft is indicated by a black arrow.
On the NSP2 array, 18 spots showed binding to VP1 with at least 60% intensity (Fig. 4A to C), with 5 spots showing ≥80% binding. Residues that were present in at least two of the spots that showed 80% binding or higher were mapped onto a known structure of NSP2 (Fig. 4D, shown in green). Consistent with the VP1 array results, VP1-binding sites on NSP2 (aa 22 to 32, 67 to 74, and 91 to 104) clustered around the catalytic cleft of the NSP2 monomer, which is the site of the ssRNA-binding and RTPase activity of NSP2 (10) (Fig. 4D).
Interaction of NSP2 with the N-terminal domain of VP2.
The structure of VP2, which forms the innermost shell, has been determined in the context of the virion architecture by cryo-electron microscopy (EM) and X-ray crystallography (39, 57–59). Most of VP2 folds into a broad, flat sheet. In the shell, composed of 120 VP2 subunits, five VP2 dimers come together at each of the five-fold axes, with their N-terminal domains (aa 1 to 100) projecting inward (39). This highly flexible N-terminal region of the protein is critical for RNA binding, encapsidation of VP1 and VP3, and activation of RdRP for genome replication (35, 39–42). Direct interaction of NSP2 and VP2, although suspected, has not been demonstrated previously. Previous studies showing that high levels of NSP2 are associated with RIs recovered from infected cells that specifically contained VP2 (21) and more recent studies showing that VP2 is pulled down by NSP2 from the lysates of rotavirus-infected cells strongly suggest a possible interaction between the two proteins (60). We hypothesized that in addition to VP1, NSP2 directly binds VP2, possibly coordinating the VP2-VP1 interaction for initiation of replication. Overexpression of full-length VP2 leads to the autoassembly of core-like particles (61), limiting our ability to analyze its binding to NSP2. Therefore, we cloned, expressed, and purified a VP2 construct corresponding to residues 1 to 134, the N-terminal domain, and analyzed its binding to NSP2 by ITC and BLI.
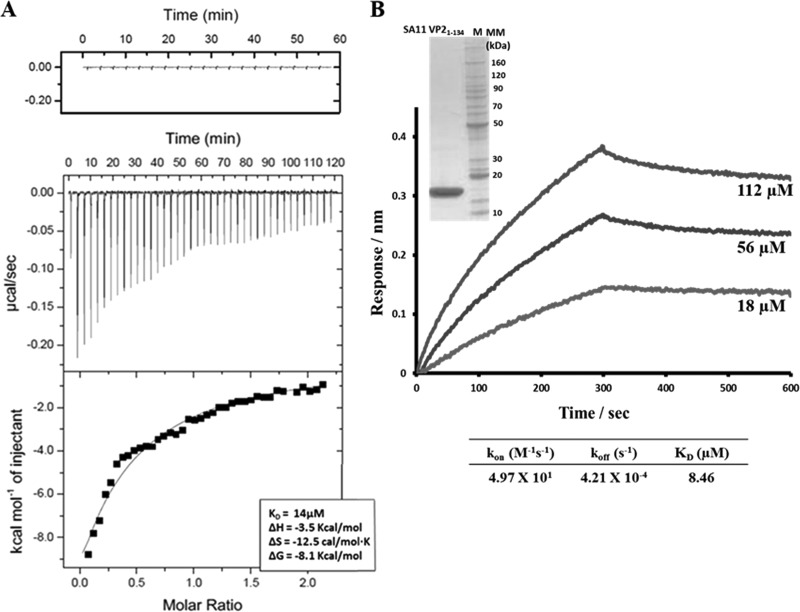
For ITC analysis, NSP2-VP21-134 titrations were carried out in triplicate at 4°C. The ΔH of VP21-134 titrated into buffer alone was negligible and subtracted as background. Apparent exothermic heat change was detected as VP21-134 bound to NSP2. Upon analysis of the resulting isotherm, the binding constant was calculated to be 14 μM (Fig. 5A). BLI analysis was also carried out by immobilizing biotinylated NSP2 onto a streptavidin biosensor and dipping it into wells containing serial dilutions of VP21-134. The binding sensograms were fitted using a global one-site binding model with χ2/DoF of 0.000123, which showed a binding constant (KD) 8.5 μM, similar to ITC results (Fig. 5B).
FIG 5.
ITC and BLI analyses of NSP2-VP21-134 interactions. (A) ITC measurement of NSP2 binding to VP21-134. The original raw data (middle graph) and the fit after integration (lower graph) are shown. The control titration is shown on top. (B) BLI measurement of NSP2 binding to VP21-134. NSP2-VP21-134 association and dissociation curves were obtained through serial dilutions of VP21-134 (18, 56, and 112 μM). Sensograms were fitted with a global one-site binding model. The KD, kon, and koff values are shown. The insert shows the Coomassie blue-stained SDS-PAGE gel of the SA11 VP21-134 protein.
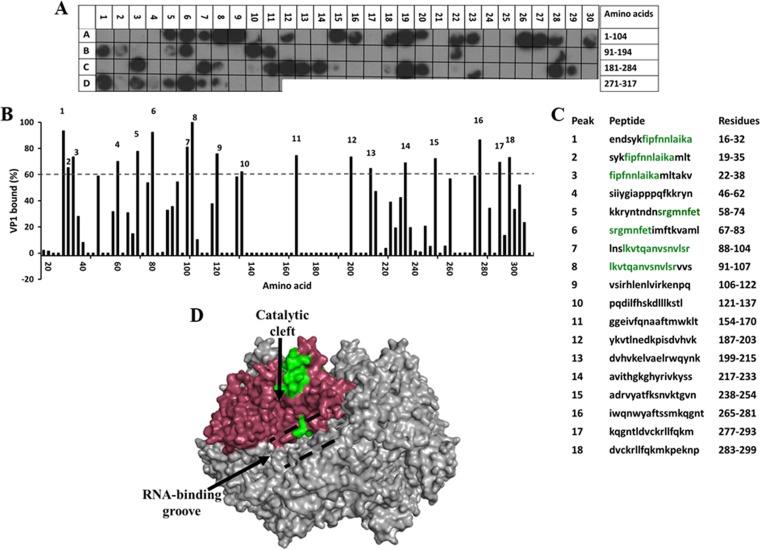
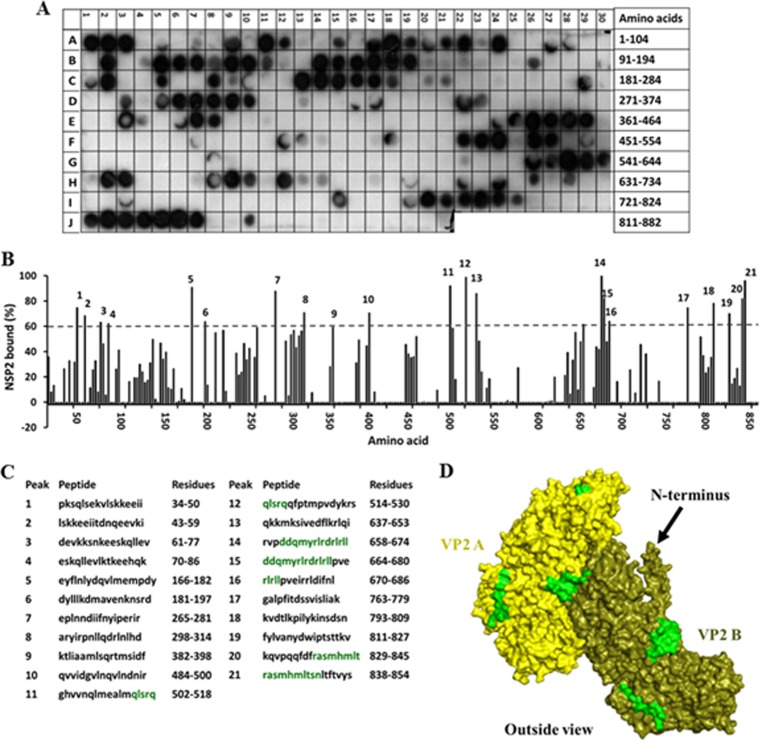
To further understand NSP2 binding to VP2 and to determine if other regions on VP2, in addition to the N-terminal domain, also participate in the binding, we synthesized a peptide array spanning the complete VP2 sequence and screened it with purified NSP2 to identify the NSP2-binding peptides (Fig. 6). In the VP2 peptide array, 21 spots showed binding to NSP2 with at least a 60% cutoff intensity (Fig. 6A to C). These included peptides corresponding to amino acids 34 to 86 (Fig. 6B and C, peaks 1 to 4), consistent with our ITC and BLI results, which indicated a direct interaction between NSP2 and the N terminus of VP2. Of the 21 spots, 9 showed ≥80% binding compared to the control, and the residues present in at least two of these spots were mapped onto the VP2 structure. These residues mapped primarily near the intersubunit-interacting regions of VP2 (aa 514 to 518, 664 to 674, and 838 to 845) (Fig. 6D, shown in green). At high concentrations, VP2 self-assembles into core-like particles (61), a characteristic that potentially could result in inefficient genome packaging and the production of empty particles. Rotavirus must employ a mechanism to prevent premature oligomerization of VP2 and to ensure that the core assembly happens in a timely and controlled manner. Binding of NSP2 near these sites may serve to prevent premature VP2 autoassembly into core-like particles.
FIG 6.
Binding of the full-length SA11 NSP2 to the peptide array of the SA11 VP2 sequence. (A) Autoradiograph of the VP2 peptide array probed by full-length NSP2. The peptide array consists of spots of 17-residue peptides in the protein sequence, starting from the N terminus (spot A1) and ending with the C-terminal peptide (J21), with the N-terminal residue of the peptide in each spot shifted by 2 residues from the previous spot along the protein sequence. (B) Graph showing relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). (C) The peptide sequences corresponding to the spots denoted 1 to 21 in panel B that showed intensities higher than the 60% cutoff. VP2 residues present in at least two peptides that showed ≥80% binding intensity (green) were mapped onto the known structure of VP2 (pdb 3KZ4). (D) Surface model of the VP2 dimer, with NSP2-binding sites (green) labeled on monomer A (yellow) and monomer B (olive). The N-terminal domain is indicated by a black arrow.
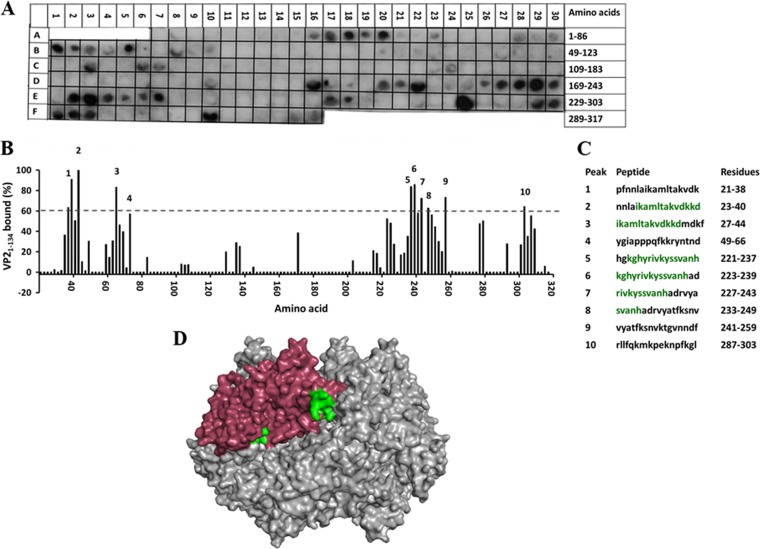
In a reciprocal experiment, to identify the VP21-134-binding sites on NSP2, we synthesized a peptide array spanning the complete NSP2 sequence and screened it with VP21-134. The NSP2 array showed 10 spots with at least 60% binding to NSP2 compared to the control (Fig. 7A to C), with 5 spots showing ≥80% binding. When the residues that showed significant binding were mapped onto the NSP2 octamer, they formed two distinct sites on NSP2 (aa 27 to 40 and 223 to 237) (Fig. 7D, shown in green). The first site is surface exposed and falls right at the two-fold symmetric axis of the octamer. The second site, on the other hand, is located deep inside the catalytic cleft of the NSP2 octamer and would not be readily available for interaction with other proteins (Fig. 7D). However, it is important to remember that this membrane was probed against the N-terminal domain of VP2, consisting mostly of coil and flexible loops. Thus, in the context of the full-length VP2 protein, this region could certainly extend into the cleft to reach this particular binding site.
FIG 7.
Binding of the SA11 VP21-134 to the peptide array of the SA11 NSP2 sequence. (A) Autoradiograph of the NSP2 peptide array probed by full-length VP21-134. The peptide array consists of spots of 17-residue peptides in the protein sequence, starting from the N terminus (spot A7) and ending with the C-terminal peptide (F16), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the protein sequence. (B) Graph showing relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). (C) The peptide sequences corresponding to the spots denoted 1 to 10 in panel B that showed intensities higher than the 60% cutoff. NSP2 residues present in at least two peptides that showed ≥80% binding intensity (green) were mapped onto the known structure of NSP2 (pdb 1L9V). (D) Surface model of the NSP2 octamer (gray) with VP21-134-binding residues shown in green on monomer A (pink).
Interaction of NSP2 with NSP6.
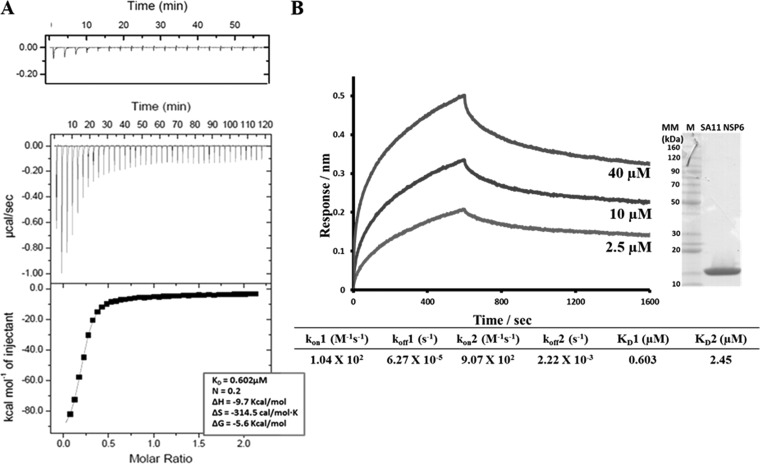
NSP6 is the least-studied rotavirus protein. It is encoded by the alternative ORF of segment 11 (33, 34) and is known to interact with NSP5 (30) and accumulate in the viroplasms (7, 27, 62). Based on this and the fact that NSP6 is a sequence-independent nucleic acid-binding protein, with similar affinities for ssRNA and dsRNA (34), we hypothesized that NSP6, via its interaction with NSP5 and possibly NSP2, plays a regulatory role in the viral life cycle, possibly in viroplasm formation. To determine if NSP6 interacts with NSP2, we carried out binding experiments using ITC and BLI. The ITC binding isotherm for NSP2-NSP6 binding was obtained at 10°C. Fitting analysis indicated that NSP6 bound NSP2 with an n = 0.23 binding stoichiometry, suggesting that one molecule of NSP6 bound four molecules of NSP2, or in other words, two molecules of NSP6 bound to one NSP2 octamer with a calculated KD of 0.60 μM (Fig. 8A). For the BLI analysis, we immobilized biotinylated NSP2 onto streptavidin biosensors and dipped them into wells containing serial dilutions of NSP6. The sensograms of NSP6 binding with NSP2 were best fitted with the global two-site binding model, with a χ2/DoF value of 0.00002, and the calculated KD of the reaction was 0.603 μM for site one, consistent with ITC results, and 2.46 μM for the second site (Fig. 8B).
FIG 8.
ITC and BLI analyses of NSP2-NSP6 interactions. (A) ITC measurement of NSP2 binding to NSP6. The middle graph shows the original raw data, and the fit after integration is shown in the bottom graph (KD = 0.602 μM). The control titration is shown on top. (B) BLI measurement of NSP2 binding to NSP6. NSP2-NSP6 association and dissociation curves were obtained through serial dilutions of NSP2 (2.5, 5, 10, 20, and 40 μM). Representative sensograms for three of the concentrations are shown. Sensograms were fitted with a global two-site binding model. The KD1, KD2, kon, and koff values for two sites are shown. The insert shows the Coomassie blue-stained SDS-PAGE gel of the SA11 NSP6 protein.
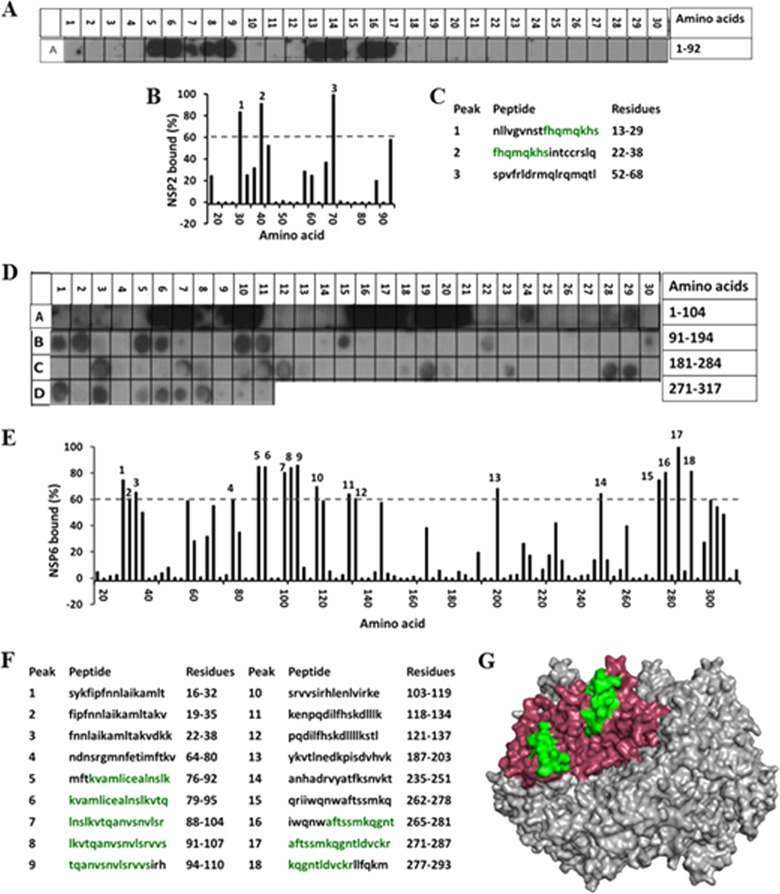
To shed further light on the NSP2-NSP6 interaction, we sought to identify the sites on the two proteins that participate in the interaction by using peptide arrays. The NSP6 peptide array probed by NSP2 showed two distinct regions (aa 22 to 29 and 52 to 68) in NSP6 with strong NSP2-binding signals (Fig. 9A to C). These regions fall within the central part of the NSP6 protein. Further experiments are necessary to determine if these sites overlap the NSP5- or RNA-binding sites on NSP6.
FIG 9.
Mapping the NSP2-NSP6 interaction sites. (A) Autoradiograph of the NSP6 peptide array probed by full-length NSP2. The peptide array consists of spots of 17-residue peptides in the protein sequence, starting from the N terminus (spot A1) and ending with the C-terminal peptide (A30), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the protein sequence. (B) Graph showing the relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). (C) The peptide sequences corresponding to the spots denoted 1 to 3 in panel B, which showed intensities higher than the 60% cutoff. (D) Autoradiograph of the NSP2 peptide array probed by full-length NSP6. The peptide array consists of spots of 17-residue peptides in the protein sequence starting from the N terminus (spot A1) and ending with the C-terminal peptide (D11), with the N-terminal residue of the peptide in each spot shifted by 3 residues from the previous spot along the protein sequence. (E) Graph showing relative intensity (y axis) of each spot (black bars) in the array with its position relative to the protein sequence (x axis). (F) The peptide sequences corresponding to the spots denoted 1 to 18 in panel B, which showed intensities higher than the 60% cutoff. NSP2 residues present in at least two peptides that showed ≥80% binding intensity (green) were mapped onto the known structure of NSP2 (pdb 1L9V). (G) Surface model of the NSP2 octamer (gray) with NSP6-binding residues shown in green on monomer A (pink).
In a reciprocal experiment, the NSP2 peptide array probed by NSP6 showed 18 NSP2 peptides binding to NSP6 with at least a 60% cutoff intensity (Fig. 9D to F), with 6 of them showing ≥80% binding. Mapping of the corresponding significant residues onto the NSP2 structure showed that they clustered primarily in the N terminus (aa 79 to 107) and the C terminus (aa 271 to 287) of the protein (Fig. 9G, shown in green). Interestingly, residues 79 to 107 overlap entirely with the VP1-binding site on NSP2 (aa 91 to 104), suggesting that VP1 and NSP6 may interact with NSP2 at different times during infection.
Validation of the identified NSP2-VP1 and NSP2-NSP6 interaction interfaces in the context of full-length proteins.
To validate the biological significance of the binding sites identified by peptide arrays, in the context of full-length proteins, we mutated a selected region in NSP2 based on the criteria that the mutation(s) did not adversely affect the expression level or the oligomeric state of NSP2 and that it is implicated in binding to both VP1 and NSP6. The peptide LKVTQANVSNVLSRVVS (aa 91 to 107) was chosen for the site-directed mutagenesis experiments because it was the strongest VP1-binding peptide on NSP2 (Fig. 4C, peptide 8) and also showed high specificity to NSP6 (Fig. 9F, peptide 8). In the full-length NSP2, amino acids 91, 95, 97, 100, and 104 (shown in bold) were mutated to alanine, and the resulting NSP2mut, which expressed well and showed the same octameric state as the wild type, was tested for binding to VP1 and NSP6 by BLI (Fig. 10A and B). The obtained NSP2mut-VP1 sensograms were best fitted using a global one-site binding model with the χ2/DoF value of 0.0003 for the fit (Fig. 10C). The estimated KD of 0.47 μM for the NSP2mut was substantially higher than the KD of 1.54 nM for wild-type NSP2, indicating a significant loss of affinity of the mutant toward VP1 and thus confirming our peptide array interpretation for this site as a significant binding interface between NSP2 and VP1. Similarly, NSP2mut also showed decreased binding with NSP6 (KD of 0.87 μM versus 0.60 μM for wild-type NSP2), although the association rate constant (kon) was similar (1.04 × 102 M−1 s−1 for wild-type NSP2 versus 1.8 × 102 M−1 s−1 for NSP2mut), the dissociation constant (koff) increased significantly (6.3 × 10−5 s−1 for wild-type NSP2 versus 1.5 × 10−4 s−1 for NSP2mut), suggesting that the mutation destabilizes the interaction (Fig. 10D).
FIG 10.
BLI analysis of SA11 NSP2mut-VP1 and NSP2mut-NSP6 interactions. (A) Ribbon diagram of the NSP2 octamer (gray) with the peptide LKVTQANVSNVLSRVVS (aa 91 to 107) chosen for the site-directed mutagenesis experiments shown in green on one of the subunits (pink) in the octamer. Amino acids 91, 95, 97, 100, and 104 (shown as sticks) were mutated to alanine, and the resulting NSP2mut was tested for binding to VP1 and NSP6 by BLI. (B) Gel filtration profiles of the SA11 strain wild-type NSP2 and NSP2mut. (Insert) Coomassie blue-stained SDS-PAGE gels of the wild-type SA11 NSP2 and SA11 NSP2mut proteins. (C) NSP2mut-VP1 association and dissociation curves were obtained through serial dilutions of NSP2mut (0.8, 1.5, 3, 5, 7, and 10 μM). Representative sensograms for four of the concentrations are shown. (D) NSP2mut-NSP6 association and dissociation curves were obtained through serial dilutions of NSP2mut (1.25, 2.5, 5, 10, 20, and 30 μM). Representative sensograms for three of the concentrations are shown. Sensograms were fitted with global one-site binding model. The KD, kon, and koff values for the sites are shown.
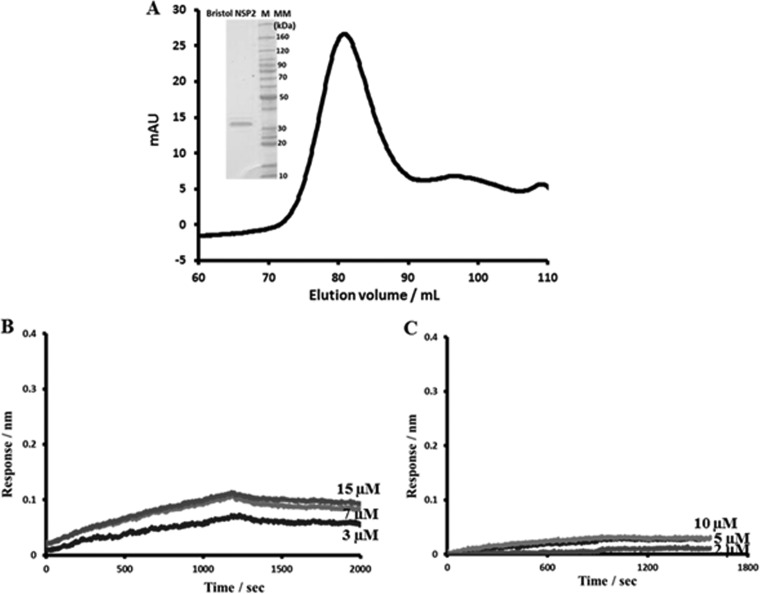
To further validate the NSP2-binding sites with VP1 and NSP6, we expressed and purified NSP2 from a group C rotavirus strain (Bristol) and tested its ability to bind VP1 and NSP6 of the SA11 rotavirus strain (group A). The NSP2 sequence varies between rotavirus groups A, B, and C, with group C viruses being the most divergent. Previous studies with VP2 and VP1 showed that proteins belonging to different rotavirus groups, particularly from group A strain SA11 and group C strain Bristol, do not functionally replace one another in vitro (43). Despite sequence conservation of only 33%, the Bristol NSP2 forms a donut-shaped octamer, almost identical to that of the SA11 NSP2 (23). The SA11 VP1- and NSP6-binding sites identified by our peptide array of SA11 NSP2 are not conserved in the Bristol NSP2 sequence, indicating that Bristol NSP2 should not bind to SA11 VP1 or NSP6. Accordingly, in the BLI experiments, Bristol NSP2 showed no binding with either SA11 VP1 or NSP6 (Fig. 11). Next, we synthesized a peptide membrane containing full-length SA11 VP1 sequence and probed it for binding to the Bristol NSP2. Consistent with the BLI results, the peptide array showed no binding between Bristol NSP2 and SA11 VP1 peptides (data not shown).
FIG 11.
BLI analysis of the Bristol NSP2-SA11 VP1 and Bristol NSP2-SA11 NSP6 interactions. (A) Gel filtration profile of the Bristol strain NSP2. (Insert) Coomassie blue-stained SDS-PAGE gel of the Bristol NSP2 protein. (B) BLI analysis of the Bristol NSP2-SA11 VP1 interaction. Association and dissociation curves were obtained through serial dilutions of Bristol NSP2 (0.5, 0.8, 1.5, 3, 5, 7, and 10 μM). Representative sensograms for three of the concentrations are shown. (C) BLI analysis of the Bristol NSP2-SA11 NSP6 interaction. Association and dissociation curves were obtained through serial dilutions of Bristol NSP2 (1.25, 2.5, 5, 10, and 20 μM). Representative sensograms for three of the concentrations are shown. In contrast to SA11 NSP2, only minimal signals, comparable to those of the negative control, were detected, indicating no binding between the proteins.
DISCUSSION
NSP2 is a multifunctional enzyme that participates in a variety of activities during rotavirus infection, specifically, viroplasm formation, genome replication, and assembly of the new viral particles (39–42). To test if NSP2 directly interacts with other known proteins of the viroplasms or the RIs, we performed solution-based binding experiments with NSP2 and VP1, NSP2 and VP21-134, and NSP2 and NSP6. ITC and BLI experiments clearly confirmed NSP2's binding to VP1, NSP6, and VP21-134. Furthermore, using peptide arrays, we identified the sites on VP1, VP2, NSP6, and NSP2 that participate in the binding.
NSP2 exhibits high-affinity binding to the viral polymerase VP1.
Using BLI, we have shown here a direct and high-affinity binding between NSP2 and VP1, and by using peptide arrays we have identified potential binding sites on these proteins. Although previous biochemical studies suggested a possible interaction between NSP2 and VP1 (17, 28), it was unclear whether this interaction involved direct binding or was mediated by RNA or other viral proteins. The interacting sites on NSP2 and VP1, as identified by reciprocal peptide array analysis and then mapped onto their respective structures, are consistent with the known functional and biochemical properties of these two proteins. The four NSP2-binding sites on VP1 are in close proximity to the RNA template entry and dsRNA exit tunnels of VP1 (35, 37), whereas the VP1-binding sites on NSP2 map within the regions lining the catalytic cleft and RNA-binding grooves of the NSP2 octamer (10). The binding site of NSP2 near the template entry site on VP1 is consistent with the helix-destabilizing activity of the donut-shaped NSP2 octamer with electropositive RNA-binding grooves on its sides being involved in relaxing the mRNA template in preparation for the initiation of dsRNA synthesis by VP1 (13). The NSP2-binding region in the C-terminal domain immediately adjacent to the dsRNA exit tunnel of VP1 (37) is in line with the demonstrated RTPase activity of NSP2, i.e., to hydrolyze γ-phosphates on the 5′ (–)RNAs of the newly synthesized dsRNA in vitro (56). This activity is suggested to play a role in packaging the newly synthesized dsRNA around the VP1/VP3 complex inside the cores by specifically recognizing the 5′ consensus sequence of (–)RNA as soon as the dsRNA emerges from the VP1 active site and allowing it to be recaptured by VP1 after the γ-phosphate hydrolysis is complete (10). Accordingly, as previously mentioned, VP1 binding on NSP2 maps within the regions flanking the catalytic cleft, a site for specific ssRNA binding and RTPase activity of NSP2 (9).
The unexpected observation from our studies is that NSP2 binds VP1 with remarkably high affinity. As a nonstructural protein, NSP2 is not incorporated into the newly forming viral particles, leaving a question as to how it is able to dissociate from VP1 during particle formation in the viroplasms. Rotavirus must employ a mechanism by which NSP2 is released from the replication complex before replication is complete and the cores are fully assembled.
NSP2 interacts with the inner core protein VP2.
BLI and ITC analysis of NSP2 and the N-terminal domain of VP2 (aa 1 to 134) showed a direct interaction between the two proteins. The NSP2 peptide array probed against VP21-134 resolved two possible VP2-binding sites on NSP2 that map near the two-fold symmetry axis of the octamer extending into the catalytic cleft. Reciprocal peptide array screening with NSP2 sequence and binding analysis using a full-length VP2, similar to what was done with VP1-NSP2 binding, was not possible, as the full-length VP2 exhibits an intrinsic ability to readily aggregate into larger oligomers leading to core assembly, a process that cannot be easily controlled. However, screening of the VP2 peptide array identified NSP2-binding sites dispersed throughout the length of the protein with all of the sites mapping near the intersubunit-interacting regions of VP2. Several of these sites are conserved among the VP2 sequences of group A rotaviruses. Although N-terminal regions of VP2 show some variation among group A strains, the VP2 peptides that showed strong binding (>80%) are well conserved. Binding of NSP2 to these sites could interfere with the assembly of VP2 into core structures. Recently, a new cytoplasmic form of NSP2 has been characterized that interacts with cellular and viral proteins, including VP2 (60). Based on our results and the previous observations, which showed that purified NSP2 impedes viral replication in vitro by interfering with the role of VP2 in forming the minus-strand initiation complex (63), a reasonable hypothesis is that NSP2-VP2 interactions, among other functions, play a role in preventing the premature assembly of VP2 cores in the cytoplasm, prior to VP2 being shuttled into the viroplasms. This hypothesis is in line with the proposal that the pentamers of VP2 dimers, together with VP1/VP3, and a dsRNA segment constitute an initial building unit for RNA replication and core assembly (36, 64). This could be a mechanism by which rotavirus ensures that both replication and viral particle assembly occur concurrently inside viroplasms. In the viroplasms, NSP2 is released from VP2, allowing pentamers of VP2 dimers, together with VP1 and newly synthesized dsRNA segments, to begin core assembly. It is possible that the dynamic process of VP1 activation, dsRNA synthesis, incorporation of VP3, and VP2 assembly leads to disassociation of NSP2.
NSP2's interactions with NSP6 and NSP5.
Both ITC and BLI clearly showed a direct interaction between NSP6 and NSP2, with peptide array studies identifying two NSP2-binding sites in the central portion of NSP6 and two NSP6-binding regions mapping to the N terminus and the C terminus of NSP2. The precise function of NSP6 is unknown, and more experiments are needed to identify the exact role this particular interaction plays in the rotavirus life cycle. Notable was the fact that one of the two NSP6-binding sites on NSP2 overlaps with the VP1-binding site on the protein, suggesting that these proteins interact with NSP2 at different times during infection. Since NSP6 is only expressed transiently early during the infection, it is likely that NSP6 plays a role in either viroplasm formation or possibly suppressing the RNA-activated innate cellular immune response.
Finally, although the NSP5 peptide array was used as a control, it is noteworthy that the peptide array of NSP5 resolved two major sites for NSP2 binding. The first site in the C-terminal domain is consistent with previous studies, which showed that this region in NSP5 is critical for binding to NSP2 (15, 31). In addition, the second site, previously unidentified and located in the central region of the protein, showed strong binding to NSP2. The structure of NSP2 in complex with the NSP5 construct from aa 66 to 188 has been determined by cryo-EM (24). In this structure, the NSP5 construct binds to the same region (aa 64 to 68, 179 to 183, 232 to 251, and 291 to 302) in the NSP2 octamer as the RNA, across the groves lined by positively charged residues (24). Our findings show that some of these sites also overlap the VP2- and VP1-binding sites on NSP2, suggesting that, like NSP6, NSP5's binding to NSP2 is spatially and/or temporally separated from NSP2's interactions during the replication process.
In conclusion, our studies provide new insights into NSP2's interactions with the proteins involved in viroplasm formation and genome replication. We present here unequivocal evidence of direct binding between NSP2 and VP1, NSP2 and NSP6, and NSP2 and the N-terminal domain of VP2. In addition, we were able to map the sites on these proteins that participate in the binding and validate some of the sites in the context of the full-length proteins by mutational analysis, thus providing further insight into the complex network of interactions that are involved in genome replication. Our findings lead us to propose that the NSP2 octamer with multiple enzymatic activities is a principal regulator of viroplasm formation, recruitment of viral proteins into the viroplasms, and possibly genome replication. Further studies are required to understand how NSP2 uses its enzymatic and protein-binding activities to coordinate the multiple protein-protein and protein-RNA interactions required for viroplasm formation, genome replication, and packaging of the viral particles.
ACKNOWLEDGMENTS
We acknowledge support from NIH grants R37 AI36040 (B.V.V.P.) and R01 AI080656 and P30 DK56338 (M.K.E.), NLM Training Program in Biomedical Informatics T15LM007093-21 (A.M.H.), and the Robert Welch Foundation (Q1279; to B.V.V.P.). We acknowledge the Monoclonal Antibody/Recombinant Protein Expression Shared Resource at the Baylor College of Medicine, supported by NIH grant P30 CA125123.
Footnotes
Published ahead of print 27 August 2014
REFERENCES
- 1.Chen D, Luongo CL, Nibert ML, Patton JT. 1999. Rotavirus open cores catalyze 5′-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology 265:120–130. 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- 2.Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471–473. 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 3.Jayaram H, Estes MK, Prasad BV. 2004. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 101:67–81. 10.1016/j.virusres.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Silvestri LS, Taraporewala ZF, Patton JT. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763–7774. 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altenburg BC, Graham DY, Estes MK. 1980. Ultrastructural study of rotavirus replication in cultured cells. J. Gen. Virol. 46:75–85. 10.1099/0022-1317-46-1-75. [DOI] [PubMed] [Google Scholar]
- 6.Lopez T, Rojas M, Ayala-Breton C, Lopez S, Arias CF. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 86:1609–1617. 10.1099/vir.0.80827-0. [DOI] [PubMed] [Google Scholar]
- 7.Petrie BL, Greenberg HB, Graham DY, Estes MK. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133–152. 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez RA, Espinosa R, Romero P, Lopez S, Arias CF. 2000. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol. 145:1963–1973. 10.1007/s007050070069. [DOI] [PubMed] [Google Scholar]
- 9.Trask SD, McDonald SM, Patton JT. 2012. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 10:165–177. 10.1038/nrmicro2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu L, Chow DC, Patton JT, Palzkill T, Estes MK, Prasad BV. 2012. Crystallographic analysis of rotavirus NSP2-RNA complex reveals specific recognition of 5′ GG sequence for RTPase activity. J. Virol. 86:10547–10557. 10.1128/JVI.01201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton JT, Salter-Cid L, Kalbach A, Mansell EA, Kattoura M. 1993. Nucleotide and amino acid sequence analysis of the rotavirus nonstructural RNA-binding protein NS35. Virology 192:438–446. 10.1006/viro.1993.1059. [DOI] [PubMed] [Google Scholar]
- 12.Kattoura MD, Clapp LL, Patton JT. 1992. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology 191:698–708. 10.1016/0042-6822(92)90245-K. [DOI] [PubMed] [Google Scholar]
- 13.Taraporewala ZF, Patton JT. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519–4527. 10.1128/JVI.75.10.4519-4527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaram H, Taraporewala Z, Patton JT, Prasad BV. 2002. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 417:311–315. 10.1038/417311a. [DOI] [PubMed] [Google Scholar]
- 15.Eichwald C, Rodriguez JF, Burrone OR. 2004. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 85:625–634. 10.1099/vir.0.19611-0. [DOI] [PubMed] [Google Scholar]
- 16.Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333–339. [DOI] [PubMed] [Google Scholar]
- 17.Kattoura MD, Chen X, Patton JT. 1994. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803–813. 10.1006/viro.1994.1402. [DOI] [PubMed] [Google Scholar]
- 18.Aponte C, Poncet D, Cohen J. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J. Virol. 70:985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Gombold JL, Ramig RF. 1990. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology 178:143–151. 10.1016/0042-6822(90)90387-7. [DOI] [PubMed] [Google Scholar]
- 20.Ramig RF, Petrie BL. 1984. Characterization of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J. Virol. 49:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallegos CO, Patton JT. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616–627. 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 22.Kumar M, Jayaram H, Vasquez-Del Carpio R, Jiang X, Taraporewala ZF, Jacobson RH, Patton JT, Prasad BV. 2007. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals a nucleoside diphosphate kinase-like activity. J. Virol. 81:12272–12284. 10.1128/JVI.00984-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taraporewala ZF, Jiang X, Vasquez-Del Carpio R, Jayaram H, Prasad BV, Patton JT. 2006. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J. Virol. 80:7984–7994. 10.1128/JVI.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X, Jayaram H, Kumar M, Ludtke SJ, Estes MK, Prasad BV. 2006. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J. Virol. 80:10829–10835. 10.1128/JVI.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welch SK, Crawford SE, Estes MK. 1989. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J. Virol. 63:3974–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vende P, Taraporewala ZF, Patton JT. 2002. RNA-binding activity of the rotavirus phosphoprotein NSP5 includes affinity for double-stranded RNA. J. Virol. 76:5291–5299. 10.1128/JVI.76.10.5291-5299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez SA, Burrone OR. 1991. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology 182:8–16. 10.1016/0042-6822(91)90642-O. [DOI] [PubMed] [Google Scholar]
- 28.Arnoldi F, Campagna M, Eichwald C, Desselberger U, Burrone OR. 2007. Interaction of rotavirus polymerase VP1 with nonstructural protein NSP5 is stronger than that with NSP2. J. Virol. 81:2128–2137. 10.1128/JVI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berois M, Sapin C, Erk I, Poncet D, Cohen J. 2003. Rotavirus nonstructural protein NSP5 interacts with major core protein VP2. J. Virol. 77:1757–1763. 10.1128/JVI.77.3.1757-1763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres-Vega MA, Gonzalez RA, Duarte M, Poncet D, Lopez S, Arias CF. 2000. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J. Gen. Virol. 81:821–830. [DOI] [PubMed] [Google Scholar]
- 31.Afrikanova I, Fabbretti E, Miozzo MC, Burrone OR. 1998. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 79:2679–2686. [DOI] [PubMed] [Google Scholar]
- 32.Poncet D, Lindenbaum P, L'Haridon R, Cohen J. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 71:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattion NM, Mitchell DB, Both GW, Estes MK. 1991. Expression of rotavirus proteins encoded by alternative open reading frames of genome segment 11. Virology 181:295–304. 10.1016/0042-6822(91)90495-W. [DOI] [PubMed] [Google Scholar]
- 34.Rainsford EW, McCrae MA. 2007. Characterization of the NSP6 protein product of rotavirus gene 11. Virus Res. 130:193–201. 10.1016/j.virusres.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Valenzuela S, Pizarro J, Sandino AM, Vasquez M, Fernandez J, Hernandez O, Patton J, Spencer E. 1991. Photoaffinity labeling of rotavirus VP1 with 8-azido-ATP: identification of the viral RNA polymerase. J. Virol. 65:3964–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton JT, Gallegos CO. 1990. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J. Gen. Virol. 71:1087–1094. 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC. 2008. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16:1678–1688. 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald SM, Aguayo D, Gonzalez-Nilo FD, Patton JT. 2009. Shared and group-specific features of the rotavirus RNA polymerase reveal potential determinants of gene reassortment restriction. J. Virol. 83:6135–6148. 10.1128/JVI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. 2010. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J. Mol. Biol. 397:587–599. 10.1016/j.jmb.2010.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labbe M, Baudoux P, Charpilienne A, Poncet D, Cohen J. 1994. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J. Gen. Virol. 75:3423–3430. 10.1099/0022-1317-75-12-3423. [DOI] [PubMed] [Google Scholar]
- 41.Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J. 1997. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71:9618–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng CQ, Wentz MJ, Cohen J, Estes MK, Ramig RF. 1996. Characterization and replicase activity of double-layered and single-layered rotavirus-like particles expressed from baculovirus recombinants. J. Virol. 70:2736–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald SM, Patton JT. 2011. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 85:3095–3105. 10.1128/JVI.02360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boudreaux C, Vile D, Gilmore B, Tanner J, Kelly D, McDonald S. 2013. Rotavirus core shell subdomains involved in polymerase encapsidation into virus-like particles. J. Gen. Virol. 94:1818–1826. 10.1099/vir.0.052951-0. [DOI] [PubMed] [Google Scholar]
- 45.Taraporewala ZF, Patton JT. 2004. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 101:57–66. 10.1016/j.virusres.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 46.Reineke U, Volkmer-Engert R, Schneider-Mergener J. 2001. Applications of peptide arrays prepared by the SPOT-technology. Curr. Opin. Biotechnol. 12:59–64. 10.1016/S0958-1669(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 47.Reimer U, Reineke U, Schneider-Mergener J. 2002. Peptide arrays: from macro to micro. Curr. Opin. Biotechnol. 13:315–320. 10.1016/S0958-1669(02)00339-7. [DOI] [PubMed] [Google Scholar]
- 48.Huang W, Beharry Z, Zhang Z, Palzkill T. 2003. A broad-spectrum peptide inhibitor of beta-lactamase identified using phage display and peptide arrays. Protein Eng. 16:853–860. 10.1093/protein/gzg108. [DOI] [PubMed] [Google Scholar]
- 49.Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. 2006. The central region of HDM2 provides a second binding site for p53. Proc. Natl. Acad. Sci. U. S. A. 103:1227–1232. 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cushman I, Palzkill T, Moore MS. 2006. Using peptide arrays to define nuclear carrier binding sites on nucleoporins. Methods 39:329–341. 10.1016/j.ymeth.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Li PC, Liao MY, Cheng PC, Liang JJ, Liu IJ, Chiu CY, Lin YL, Chang GJ, Wu HC. 2012. Development of a humanized antibody with high therapeutic potential against dengue virus type 2. PLoS Negl. Trop. Dis. 6:e1636. 10.1371/journal.pntd.0001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D, Mao Y, Cao Q, Xiong L, Wen J, Chen R, Zhu J. 2013. Generation and characterization of a novel recombinant antibody against LMP1-TES1 of Epstein-Barr virus isolated by phage display. Viruses 5:1131–1142. 10.3390/v5041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. 2002. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277:50597–50606. 10.1074/jbc.M209037200. [DOI] [PubMed] [Google Scholar]
- 54.Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M. 2002. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21:2843–2853. 10.1093/emboj/cdf305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank R, Overwin H. 1996. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol. Biol. 66:149–169. [DOI] [PubMed] [Google Scholar]
- 56.Vasquez-Del Carpio R, Gonzalez-Nilo FD, Riadi G, Taraporewala ZF, Patton JT. 2006. Histidine triad-like motif of the rotavirus NSP2 octamer mediates both RTPase and NTPase activities. J. Mol. Biol. 362:539–554. 10.1016/j.jmb.2006.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawton JA, Zeng CQ, Mukherjee SK, Cohen J, Estes MK, Prasad BV. 1997. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 71:7353–7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. 2008. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl. Acad. Sci. U. S. A. 105:1867–1872. 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. 2009. Rotavirus architecture at subnanometer resolution. J. Virol. 83:1754–1766. 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Criglar JM, Hu L, Crawford SE, Hyser JM, Broughman JR, Prasad BV, Estes MK. 2013. A novel form of rotavirus NSP2 and phosphorylation-dependent NSP2-NSP5 interactions are associated with viroplasm assembly. J. Virol. 88:786–798. 10.1128/JVI.03022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng CQ, Labbe M, Cohen J, Prasad BV, Chen D, Ramig RF, Estes MK. 1994. Characterization of rotavirus VP2 particles. Virology 201:55–65. 10.1006/viro.1994.1265. [DOI] [PubMed] [Google Scholar]
- 62.Tian P, Ball JM, Zeng CQ, Estes MK. 1996. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J. Virol. 70:6973–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vende P, Tortorici MA, Taraporewala ZF, Patton JT. 2003. Rotavirus NSP2 interferes with the core lattice protein VP2 in initiation of minus-strand synthesis. Virology 313:261–273. 10.1016/S0042-6822(03)00302-7. [DOI] [PubMed] [Google Scholar]
- 64.Tortorici MA, Broering TJ, Nibert ML, Patton JT. 2003. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J. Biol. Chem. 278:32673–32682. 10.1074/jbc.M305358200. [DOI] [PubMed] [Google Scholar]