ABSTRACT
In a previous study, it was observed that cells infected with herpes simplex virus 2 (HSV-2) failed to accumulate stress granules (SGs) in response to oxidative stress induced by arsenite treatment. As a follow-up to this observation, we demonstrate here that disruption of arsenite-induced SG formation by HSV-2 is mediated by a virion component. Through studies on SG formation in cells infected with HSV-2 strains carrying defective forms of UL41, the gene that encodes vhs, we identify vhs as a virion component required for this disruption. Cells infected with HSV-2 strains producing defective forms of vhs form SGs spontaneously late in infection. In addition to core SG components, these spontaneous SGs contain the viral immediate early protein ICP27 as well as the viral serine/threonine kinase Us3. As part of these studies, we reexamined the frameshift mutation known to reside within the UL41 gene of HSV-2 strain HG52. We demonstrate that this mutation is unstable and can rapidly revert to restore wild-type UL41 following low-multiplicity passaging. Identification of the involvement of virion-associated vhs in the disruption of SG formation will enable mechanistic studies on how HSV-2 is able to counteract antiviral stress responses early in infection. In addition, the ability of Us3 to localize to stress granules may indicate novel roles for this viral kinase in the regulation of translation.
IMPORTANCE Eukaryotic cells respond to stress by rapidly shutting down protein synthesis and storing mRNAs in cytoplasmic stress granules (SGs). Stoppages in protein synthesis are problematic for all viruses as they rely on host cell machinery to synthesize viral proteins. Thus, many viruses target SGs for disruption or modification. Infection by herpes simplex virus 2 (HSV-2) was previously observed to disrupt SG formation induced by oxidative stress. In this follow-up study, we identify virion host shutoff protein (vhs) as a viral protein involved in this disruption. The identification of a specific viral protein involved in disrupting SG formation is a key step toward understanding how HSV-2 interacts with these antiviral structures. Additionally, this understanding may provide insights into the biology of SGs that may find application in studies on human motor neuron degenerative diseases, like amyotrophic lateral sclerosis (ALS), which may arise as a result of dysregulation of SG formation.
INTRODUCTION
Eukaryotic cells respond to environmental stresses such as nutrient depletion, oxidation, and heat shock by halting the synthesis of most proteins and shifting cellular resources toward synthesizing proteins required for coping with the stress. Following translational arrest, mRNAs contained within stalled 48S ribosomal preinitiation complexes accumulate in cytoplasmic structures known as stress granules (SGs). These structures assemble within minutes in response to stoppages in translation and, likewise, disassemble rapidly once stress is alleviated. As the mRNAs stored within SGs remain associated with translation initiation proteins, translation can quickly resume once SGs are disassembled. Thus, the dynamic cycle of SG assembly/disassembly can be viewed as a strategy to cope with disruptions in translation caused by environmental stress. In keeping with this view, cells with impairments in SG assembly show decreased ability to survive exposure to stressful stimuli (1–3).
The cellular response to environmental stress is primarily mediated by four stress-activated kinases, double-stranded RNA-dependent protein kinase (PKR), PKR-like endoplasmic reticulum kinase (PERK), heme-regulated inhibitor kinase (HRI), and general control nonrepressed kinase 2 (GCN2). Each of these kinases is activated by a distinct stress. For example, HRI responds to conditions of oxidative stress (4) and can be evoked experimentally by treating cultured cells with arsenite. Following activation, stress-activated kinases phosphorylate the alpha subunit of eukaryotic initiation factor 2 (eIF2α), leading to translational arrest and accumulation of mRNAs in stalled preinitiation complexes. Cellular RNA-binding proteins such as T cell internal antigen 1 (TIA-1) and Ras-GTPase-activating SH3 domain-binding protein (G3BP) bind to these mRNAs and to one another via prionlike domains to nucleate SG assembly (5, 6). After nucleation, other cellular proteins are recruited into the complex to complete SG formation. Recently, the kinase activity of the dual-specificity tyrosine-phosphorylation-regulated kinase 3 (DYRK3) was found to facilitate the cycle of SG assembly/disassembly and link this cycle to signaling through the mammalian target of rapamycin (mTOR) pathway (7). Following the induction of stress, the kinase-inactive form of DYRK3 partitions into SGs along with mTOR complex 1 (mTORC1) components. Once stress is alleviated, restoration of the kinase activity of DYRK3 promotes the disassembly of SGs and the release of mRNAs and mTORC1 components, thereby allowing the initiation of mRNA translation to resume.
Viral infection is known to activate both PKR and PERK (8, 9). The resulting translational arrest is problematic for viruses, as all viruses rely on the host cell protein synthesis machinery for production of their proteins. Consequently, viruses have evolved sophisticated strategies to cope with translational arrest, including manipulating or disrupting SGs (10–18). The varied impact of viral infection on SGs is presently a subject of intense investigation and the topic of several recent reviews (19–21). In addition to providing information on virus-host cell interactions, a deeper understanding of how viruses impact SGs has the potential to provide fundamental insights into the assembly/disassembly cycle of SGs. These insights may find application in studies on human motor neuron degenerative diseases like amyotrophic lateral sclerosis (ALS), which may arise as a result of dysregulation of the assembly/disassembly cycle of SGs (22).
We recently discovered that early after infection with HSV-2, cells fail to accumulate SGs in response to arsenite treatment (23). HSV-2-infected cells phosphorylate eIF2α in response to arsenite, suggesting that HSV-2 disrupts SG formation at a step that is downstream of eIF2α phosphorylation. We sought to better characterize the mechanism by which HSV-2 disrupts arsenite-induced SG formation by identifying viral proteins that mediate this disruption. The only herpesvirus protein connected with SGs thus far is the virion host shutoff protein (vhs), an endoribonuclease that promotes the decay of both cellular and viral mRNAs and that is encoded by the UL41 gene (24, 25). SGs were observed to accumulate at late times following infection with vhs-deficient mutants of HSV-1, whereas infection with wild-type (WT) viruses did not result in the accumulation of SGs (26, 27). These observations may be indicative of a role for vhs in preventing SG accumulation. In this report, we establish that disruption of arsenite-induced SG formation is mediated by a virion component and identify vhs as a virion component required for this disruption.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero), cervical carcinoma cells (HeLa), and transformed keratinocyte cells (HaCaT) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) in a 5% CO2 environment. HSV-2 strains were propagated in HaCaT or Vero cells and their titers were determined on Vero cells. Infection of HeLa cells for most SG analyses was carried out using a multiplicity of infection (MOI) of 10 to 20; infection of HeLa cells for SG analyses, including ΔUL41 virus, were carried out at an MOI of <0.5. Times postinfection, reported as hours postinfection (hpi) or minutes postinfection (mpi), refer to the time elapsed following medium replacement after a 1-h inoculation period. For phosphonoacetic acid (PAA)-treated infections, 200 μg/ml PAA (Sigma, St. Louis, MO) was applied 1 h prior to inoculation and cells were maintained in the continuous presence of 200 μg/ml PAA for the duration of the experiment. For actinomycin D (ActD)-treated infections, 2 μg/ml ActD (Sigma) was applied 30 min prior to inoculation and cells were maintained in the continuous presence of 2 μg/ml ActD for the duration of the experiment.
Construction of recombinant HSV-2 strains.
pYEbac373, the full-length infectious HSV-2 186 bacterial artificial chromosome (BAC), was constructed as described previously (28). The HSV-2 mutant lacking the UL41 gene (pYEbac373-ΔUL41) was constructed by the two-step Red-mediated mutagenesis procedure, using pYEbac373 in Escherichia coli GS1783 (29). Primers 5′-AGCTCTGTAGAGAGACCTATCCGCACCTACAATCGTGCCGGAATGGAGCTGGTGGAGCACAGGATGACGACGATAAGTAGGG-3′ and 5′-GATAAGCGATATGACGTACTTGACGTATCTGTGCTCCACCAGCTCCATTCCGGCACGATTCAACCAATTAACCAATTCTGATTAG-3′ were used to amplify a PCR product from pEP-Kan-S2, a kind gift of Klaus Osterrieder, Freie Universität Berlin, and used to remove most of the UL41-coding sequence, leaving only the carboxy-terminal 82 amino acids. To repair HSV-2 ΔUL41, the I-SceI-flanked kanamycin cassette portion of pEP-Kan-S2 was amplified using primers 5′-GATCGGCGCGCCCAAGGATGACGACGATAAGTAGGG-3′ and 5′-GATCGGCGCGCCCGAGGGGGCCGCCGGGCCTGGGCCGGCGGCCCGCGTTCCCCGCTCGAGCGGCCGCCAGTGTGATGG-3′, designed to introduce AscI sites (italicized) and the additional UL41-derived sequence (underlined) to facilitate recombination. This PCR product was then introduced at the AscI site of a UL41 expression vector (described below) to generate pRF87. Primers 5′-ACCTATACAGCTCTGTAGAGAGACCTATCCGCACCTACAATCGTGCCGGAATGGGTCTGTTTGGCATGATGAAGTTTGC-3′ and 5′-GATCGTCGACCTACTCGTCCCAGAATTTAGCCAGGACGTCC-3′ were used to amplify a PCR product from pRF87. This PCR product was then used in the two-step Red-mediated mutagenesis procedure to restore the complete UL41 gene (pYEbac373-ΔUL41Rep).
Restriction fragment length polymorphism analysis was used to confirm the integrity of each recombinant BAC clone compared to the WT BAC by digestion with EcoRI. Additionally, a PCR fragment that spanned the region of interest was amplified and sequenced to confirm the deletion or repair of UL41. Virus was reconstituted from BAC DNA as described previously (28).
Plasmids and transfections.
To construct expression plasmids containing WT UL41, the UL41 gene was amplified by PCR with forward primer (5′-GATCGAATTCGAATGGGTCTGTTTGGCATGATGAAGTTTGC-3′) and reverse primer (5′-GATCGTCGACCTACTCGTCCCAGAATTTAGCCAGGACGTCC-3′) using viral DNA templates purified from infected cells. The amplified DNA was digested with EcoRI and SalI and ligated into similarly digested pCI-neo vector (Promega, Madison, WI). All plasmids utilizing PCR in their construction were sequenced to ensure that no spurious mutations were introduced during their construction. Transfection of HeLa cells with UL41 expression plasmid or with pCI-neo vector was carried out using X-treme Gene HP DNA transfection reagent (Roche, Laval, QC, Canada) according to the manufacturer's protocols. The mCherry red fluorescent protein expression plasmid pJR70, a pEGFP-N1-based expression vector in which the enhanced green fluorescent protein (EGFP) gene was replaced with the mCherry gene (30), was included in the transfection mixes in order to identify transfected cells.
Immunological reagents.
Goat polyclonal antiserum against human TIA-1 (Santa Cruz Biotechnology, Santa Cruz, CA) was used for indirect immunofluorescence microscopy at a dilution of 1:500; mouse monoclonal antibody against human G3BP (BD Biosciences, Mississauga, ON, Canada) was used for indirect immunofluorescence microscopy at a dilution of 1:1,000; goat polyclonal antiserum against human poly(A)-binding protein (PABP; Santa Cruz Biotechnology) was used for indirect immunofluorescence microscopy at a dilution of 1:100; mouse monoclonal antibody against HSV ICP27 (Virusys, Taneytown, MD) was used for indirect immunofluorescence microscopy at a dilution of 1:1,000 and for Western blotting at a dilution of 1:500; mouse monoclonal antibody against HSV ICP8 (Virusys) was used for immunofluorescence microscopy at a dilution of 1:1,000; mouse monoclonal antibody against HSV gC (Virusys) was used for immunofluorescence microscopy at a dilution of 1:300; rat polyclonal antibody against Us3 (30) was used for immunofluorescence microscopy at a dilution of 1:1,000; rabbit polyclonal against vhs (31) was used for Western blotting at a dilution of 1:3,000; mouse monoclonal antibody against beta actin (Sigma) was used for Western blotting at a dilution of 1:2,000; Alexa Fluor 488-conjugated donkey anti-goat, Alexa Fluor 568-conjugated donkey anti-goat, Alexa Fluor 568-conjugated donkey anti-mouse, and Alexa Fluor 488-conjugated donkey anti-rat (Molecular Probes, Eugene, OR) were used for indirect immunofluorescence at a dilution of 1:500; horseradish peroxidase-conjugated goat anti-mouse and horseradish peroxidase-conjugated goat anti-rabbit (Sigma) were used for Western blotting at a dilution of 1:10,000.
Indirect immunofluorescence microscopy.
Cells for microscopic analyses were grown either on glass coverslips or on glass bottom dishes (MatTek, Ashland, MA). Cells to be stained for the presence of PABP were fixed with cold 100% methanol at 4°C for 20 min; otherwise, cells were fixed with 4% paraformaldehyde and stained as described previously (30). Images were captured using either a Nikon Eclipse TE200 inverted fluorescence microscope and Metamorph 7.1.2.0 software or an Olympus FV1000 laser scanning confocal microscope and Fluoview 1.7.3.0 software. Images from the inverted fluorescence microscope were captured using a 60× (1.40 numerical aperture [NA]) oil immersion objective. Images from the confocal microscope were captured using a 60× (1.42 NA) oil immersion objective. Relative fluorescence intensities in images captured by confocal microscopy were determined using Fluoview 1.7.3.0 software. Composites of representative images were prepared using Adobe Photoshop software. Unpaired t tests of data yielded from microscopic analyses were performed using GraphPad software.
RNA in situ hybridization.
Paraformaldehyde-fixed cells on glass bottom dishes were permeabilized by the addition of phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.01% Triton X-100 for 5 min at room temperature. Fixed and permeabilized cells were washed twice with PBS-BSA and twice with 2× SSC (1× SSC is 0.15 M sodium chloride and 0.015 M sodium citrate) and incubated overnight with 50 ng biotinylated oligo(dT) probe in a hybridization buffer composed of 50% formamide, 2× SSC, 10% dextran sulfate, 0.2% BSA, and 20 mM vanadyl ribonucleoside complex (New England BioLabs, Pickering, ON, Canada) in a chamber humidified with 2× SSC at 43°C. The following day, the hybridization solution was removed and cells were washed twice with 2× SSC and twice with PBS-BSA. Staining for TIA-1 was carried out as described previously (30) and streptavidin-conjugated Alexa Fluor 568 (Molecular Probes), reconstituted according to the manufacturer's protocols and diluted 1:3,000, was used to detect biotin bound to polyadenylated mRNA.
Preparation and analysis of whole-cell extracts.
To prepare whole-cell extracts of infected cells for Western blot analyses, cells were washed with cold PBS and then scraped into cold PBS containing protease inhibitors (Roche) plus 5 mM sodium fluoride (New England BioLabs) and 1 mM sodium orthovanadate (New England BioLabs) to inhibit phosphatases. Harvested cells were transferred to a 1.5-ml microcentrifuge tube containing 3× SDS-PAGE loading buffer. The lysate was repeatedly passed through a 28 1/2 gauge needle to reduce viscosity and then heated at 100°C for 5 min. For Western blot analysis, 10 to 20 μl of whole-cell extract was electrophoresed through SDS-PAGE gels. Separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) and probed with appropriate dilutions of primary antibody followed by appropriate dilutions of horseradish peroxidase-conjugated secondary antibody. The membranes were treated with Pierce ECL Western blotting Substrate (Thermo Scientific, Rockford, IL) and exposed to film.
RESULTS
Disruption of arsenite-induced SG formation is mediated by a virion component.
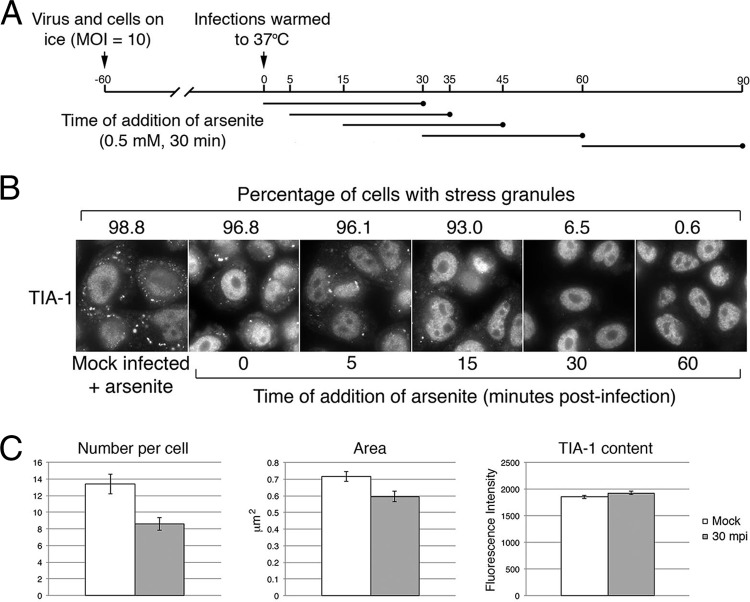
In a previous study, HSV-2 disruption of arsenite-induced SG formation was observed as early as 1 h after infection (23). To further investigate the kinetics of HSV-2 disruption of arsenite-induced SG formation, we determined the percentage of infected cells with SGs, the number of SGs per cell, and the size of the SGs formed following arsenite treatment at multiple early time points after infection. Prechilled HeLa cells were inoculated with HSV-2 strain HG52 at a multiplicity of infection (MOI) of 10 and held on ice to allow virions to bind to cells. After 1 h, warm medium was added to initiate virus infection. Under these infection conditions, greater than 95% of cells were infected (data not shown). Infected cells were treated with 0.5 mM arsenite for 30 min immediately after the temperature shift or at 5, 15, 30, or 60 min post-temperature shift (Fig. 1A). Decreases in the percentage of cells that were able to respond to arsenite treatment by forming SGs were observed over time, with a steep decrease noted between 15 and 30 min after arsenite addition (45 and 60 mpi, respectively [Fig. 1B]). Although the drop in the percentage of cells containing SGs when arsenite was added immediately after the temperature shift was negligible in comparison to the drop observed with arsenite-treated mock-infected cells, differences in the numbers of SGs per cell and in the size of the resulting SGs were detected (Fig. 1C). These results indicate that HSV-2 disruption of arsenite-induced SG formation begins as early as 30 mpi and suggest that this activity is mediated by a virion component. In keeping with this argument, disruption of arsenite-induced SG formation was not detected when virions were removed from the inoculum (23).
FIG 1.
Disruption of arsenite-induced SG formation is detectable 30 min after infection. (A) Timeline of experiments depicted in panels B and C. (B) HeLa cells were mock infected and then treated with 0.5 mM arsenite for 30 min or infected at high multiplicity with HSV-2 and treated with arsenite at various times postinfection as depicted in panel A. Cells were fixed and stained with goat polyclonal antiserum specific for TIA-1 followed by staining with Alexa Fluor 488-conjugated donkey anti-goat IgG. Stained cells were examined by wide-field fluorescence microscopy, and representative images are shown. Numeric values above the images indicate the fraction of cells containing at least one SG from 30 independent fields of view. (C) Cells stained as described for panel B were examined by confocal fluorescence microscopy, and images were captured using a 60× oil immersion lens with a zoom factor of 4. Mock-infected cells were compared with HSV-2-infected cells that received arsenite treatment immediately after the temperature shift. Numbers of SGs per cell were counted in images of 10 independent fields. From these images, all SGs in 22 cells were analyzed for mock-infected cells and all SGs in 24 cells were analyzed for HSV-2-infected cells. The numbers of SGs analyzed were 294 and 206 for mock-infected and HSV-2 infected cells, respectively. The area and fluorescence intensity of all resulting SGs were measured using Fluoview software. Error bars show standard errors of the means. Differences in the number of SGs per cell and the area of the SGs were statistically very significant (P = 0.0012 and 0.008, respectively), while differences in TIA-1 content were not statistically significant (P = 0.1118).
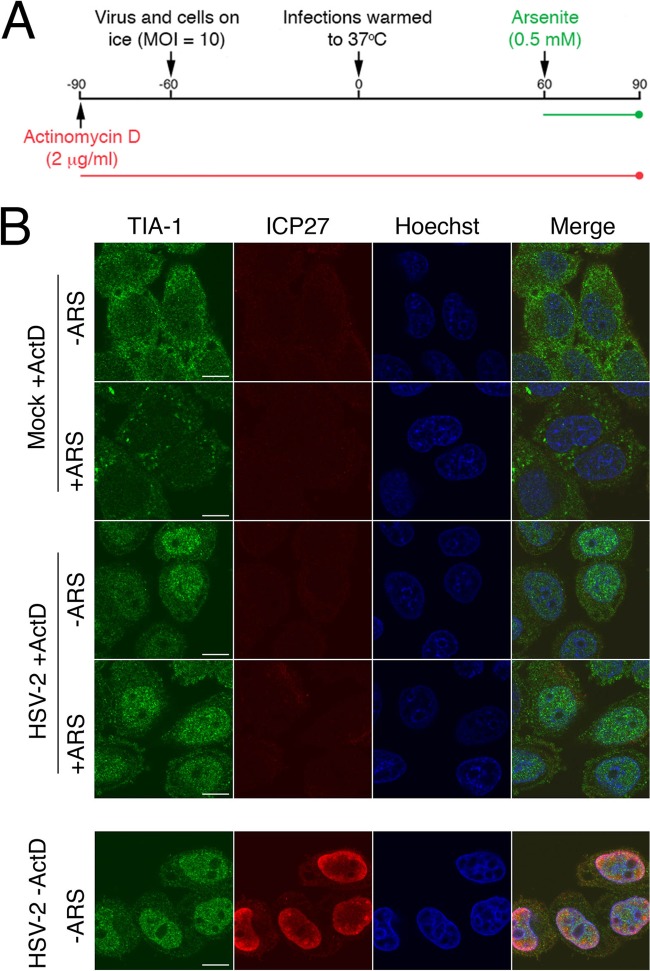
If disruption of arsenite-induced SG formation were mediated by a virion component, neither viral DNA replication nor viral gene transcription should be required for this activity. A previous study demonstrated that inhibition of viral DNA replication with either phosphonoacetic acid (PAA) or acyclovir had no effect on the ability of HSV-2 to disrupt SG accumulation (23). To examine the requirement for viral gene transcription, HeLa cells were pretreated with 2 μg/ml ActD, infected for 1 h, and then treated with arsenite in the continuous presence of ActD (Fig. 2A). Under these conditions, mock-infected HeLa cells formed SGs in response to arsenite treatment whereas HSV-2-infected cells did not (Fig. 2B). Similar results were observed in an independent experiment. As de novo viral gene expression is not required for disruption of arsenite-induced SG formation, we conclude that this activity is mediated by a component of the HSV-2 virion.
FIG 2.
Disruption of arsenite-induced SG formation is detectable in the presence of actinomycin D. (A) Timeline of the experiment depicted in the top portion of panel B. (B) HeLa cells were pretreated with actinomycin D (ActD) and then mock infected or infected at high multiplicity with HSV-2 and treated with arsenite (+ARS) in the continuous presence of ActD as depicted in panel A. Cells were fixed and stained with goat polyclonal antiserum specific for TIA-1 and mouse monoclonal antibody specific for HSV-2 ICP27 followed by staining with Alexa Fluor 488-conjugated donkey anti-goat IgG and Alexa Fluor 568-conjugated donkey anti-mouse IgG secondary antibodies. Nuclei were stained with Hoechst 33342. To demonstrate the effectiveness of the ActD treatment, untreated cells were infected at a high MOI, fixed, and stained in parallel (lower set of images). An example of HSV-2-infected cells treated with arsenite in the absence of ActD is shown in the rightmost panel of Fig. 1B. Stained cells were examined by confocal microscopy, and representative images are shown. Scale bars, 10 μm.
Investigation of virion host shutoff protein (vhs) in HSV-2 strain HG52.
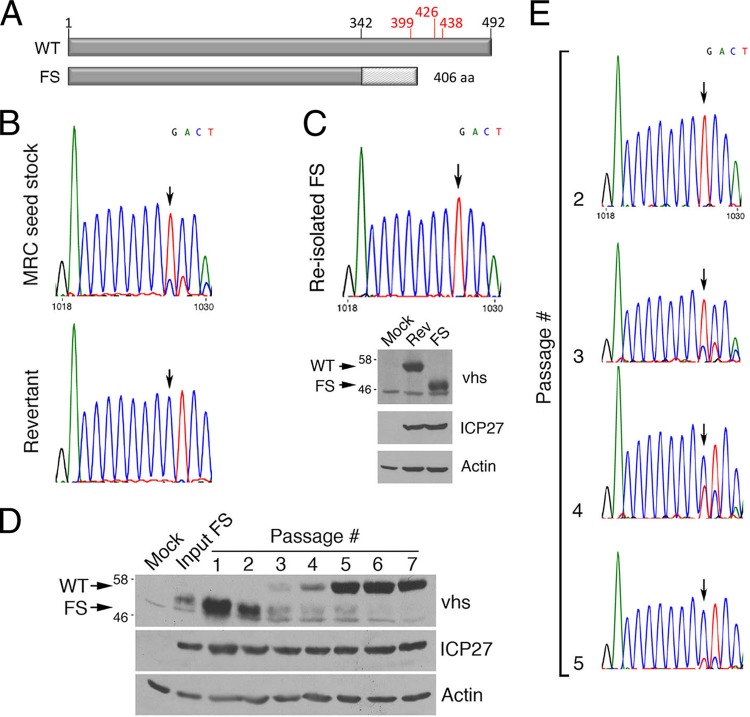
In considering virion components that could mediate the disruption of arsenite-induced SG formation, we initially dismissed the involvement of vhs, as much of our initial work was carried out with HSV-2 strain HG52, which was reported to carry a frameshift (FS) mutation within a homopolymeric tract of deoxycytidines in the UL41 gene that encodes vhs (Fig. 3A) (32). However, cells infected with HSV-1 bearing UL41 mutations accumulate SGs late in infection (26, 27), consistent with a role for vhs in preventing SG formation. This prompted us to reevaluate the status of UL41 and vhs in our stocks of HG52. Sequence analysis of populations of PCR products generated from viral DNA templates isolated from low-passage-number HG52 stocks revealed that while the majority of virus genomes contained the expected FS mutation, a subpopulation of viruses containing the full-length WT sequence was also present (Fig. 3B, top chromatograph). Sequence analysis of PCR products generated from a passaged stock, used in our previous studies, confirmed that the UL41 gene in this virus stock had reverted to the WT (Fig. 3B, bottom chromatograph). As our previous studies had utilized virus carrying WT UL41 as opposed to the defective FS reported for HG52, the contribution of vhs to arsenite-induced SG formation was further analyzed.
FIG 3.
Investigation of UL41 and vhs in HSV-2 strain HG52. (A) Linear diagram of HSV-2 vhs. Amino acids in red have been implicated in the endoribonuclease activity of vhs (35) or its ability to bind eIF4H in the case of amino acid 438 (43, 44). HG52 contains a frameshift (FS) after amino acid 342 (32), resulting in a vhs that is smaller than wild-type (WT) vhs. (B) UL41 sequence analysis. Shown are portions of the sequence chromatographs yielded from reactions performed on PCR products amplified from viral DNA isolated from the first passage of a seed stock of HG52 received from the MRC Virology Unit at the University of Glasgow (top) and the seventh passage of this stock in which the UL41 FS has reverted to WT (bottom). The vertical arrows indicate the positions of the FS. (C) Analysis of reisolated HG52 virus with FS in UL41. At the top is a portion of the sequence chromatograph yielded from reactions performed on PCR products amplified from viral DNA isolated from reisolated FS virus. The vertical arrow indicates the position of the FS. At the bottom is Western analysis of whole-cell lysates prepared from Vero cells infected with revertant virus (Rev) or FS virus or from mock-infected Vero cells at 9 hpi. Equal volumes of lysates were electrophoresed through 8% polyacrylamide gels and transferred to PVDF membranes. Membranes were probed with the antisera indicated at the right. Molecular masses in kDa and the migration position of WT and FS vhs are indicated at the left. (D, E) Serial passage analysis. HaCaT cells were infected at low multiplicity with FS virus (input) and then serially passaged in HaCaT cells at low multiplicity. (D) Whole-cell lysates were prepared from mock-infected cells, from input virus-infected cells, or from infected cells at each passage. Lysates were electrophoresed through 8% polyacrylamide gels and transferred to PVDF membranes. Membranes were probed with the antisera indicated on the right. Molecular masses in kDa and the migration positions of WT and FS vhs are indicated on the left. (E) Portions of the sequence chromatograph yielded from reactions performed on PCR products amplified from viral DNA isolated from several different passages are shown. The vertical arrows indicate the positions of the frameshifts.
To determine if viruses expressing the truncated, FS-derived vhs could disrupt arsenite-induced SG formation, we first reisolated HG52 virus that carries the UL41 FS mutation and confirmed that this virus stock produced a truncated form of vhs in comparison to revertant virus (Fig. 3C). Given the potential of virus with WT UL41 sequence to supplant virus with FS UL41, we examined the stability of the UL41 FS mutation. FS virus was serially passaged using a low MOI (≤0.01) seven times. After each passage, the vhs produced was analyzed by Western blotting (Fig. 3D) and the UL41 sequence carried by progeny virus was determined (Fig. 3E). Remarkably, reversion of FS UL41 to WT UL41 could be detected after just three passages, with the majority of viruses carrying the WT UL41 sequence and producing WT vhs after four passages. An independent passaging experiment yielded similar results (data not shown). These data demonstrate that the FS mutation in UL41 of HSV-2 strain HG52 is not stable and can readily revert to restore the WT UL41 sequence and may indicate that HG52 carrying WT UL41 has a selective advantage over virus carrying defective UL41.
HSV-2 carrying a defective vhs does not disrupt arsenite-induced SG formation.
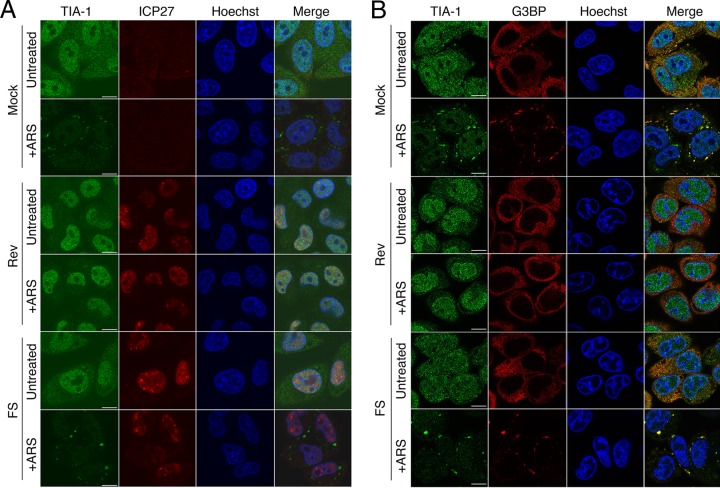
We compared the ability of HG52 viruses with WT UL41 sequence to that of viruses with FS UL41 sequence (designated Rev and FS, respectively, in Fig. 4) to disrupt SGs induced by arsenite treatment at 1 and 4 hpi. In keeping with our previous observations (23) and the data presented in Fig. 1 and 2, cells infected with Rev virus did not form SGs in response to arsenite treatment. By contrast, cells infected with FS virus formed SGs positive for both TIA-1 and G3BP (Fig. 4B) in response to arsenite treatment. These observations are consistent with the involvement of vhs in HSV-2-mediated disruption of arsenite-induced SG formation.
FIG 4.
HSV-2 carrying a defective vhs does not disrupt SG formation. HeLa cells were infected at high multiplicity with the indicated viruses or mock infected. One hour (A) or 4 h (B) later, 0.5 mM arsenite (+ARS) was applied for 30 min or cells were left untreated. Cells were fixed and stained as described for Fig. 2B. Stained cells were examined by confocal microscopy, and representative images are shown. Scale bars, 10 μm.
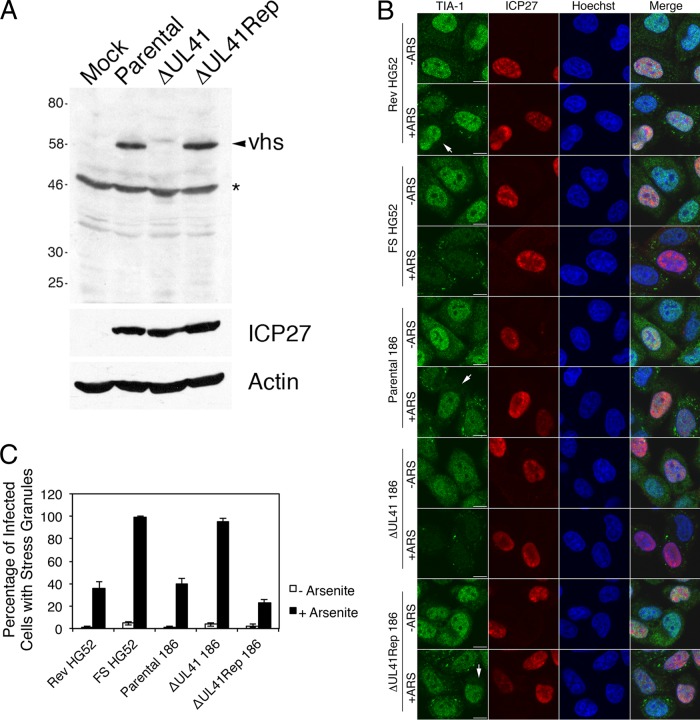
To further confirm the involvement of vhs in disruption of arsenite-induced SG formation, we utilized en passant mutagenesis to construct an HSV-2 strain 186 virus carrying a large deletion in UL41 (ΔUL41) and a virus carrying a repair of this mutation (ΔUL41Rep). The deletion in ΔUL41 removes all but the last 82 amino acids of vhs. Western blot analysis confirmed that vhs was not detectable in cells infected with ΔUL41 virus and that full-length vhs was produced in cells infected with ΔUL41Rep virus (Fig. 5A). We compared the ability of parental 186, ΔUL41, and ΔUL41Rep viruses to disrupt SGs induced by arsenite treatment. The FS and Rev viruses described above were also included in this analysis. Due to the low titers yielded by ΔUL41 virus, low MOIs (<0.5) were used for all viruses analyzed in this experiment to facilitate comparisons between infections. Rev, parental 186, and ΔUL41Rep viruses all disrupted SG formation in response to arsenite. While this disruption was detectable for all three viruses (indicated by the arrows in Fig. 5B), it was not complete (Fig. 5C), presumably due to the lower multiplicities of infection used in this experiment. Images shown in Fig. 5B depict both types of staining patterns observed in infections with viruses carrying WT UL41; quantitative analysis of the experiment depicted in Fig. 5B is shown in Fig. 5C. Both FS and Δ41 viruses failed to disrupt arsenite-induced SG formation. These data demonstrate that vhs is involved in HSV-2-mediated disruption of arsenite-induced SG formation, regardless of background strain, and indicate that vhs is a virion component required for this disruption. In keeping with the view that vhs is sufficient for this disruption, HeLa cells transfected with a plasmid expressing WT vhs and treated with arsenite showed an extremely significant decrease in the percentage of cells with SGs in comparison to cells transfected with vector alone and treated with arsenite (62.0% ± 18.6% versus 96.4% ± 4.6%, respectively, from 30 fields in experiment 1 and 78.9% ± 11.0% versus 98.7% ± 1.6%, respectively, from 30 fields in experiment 2; P = <0.0001 in both experiments).
FIG 5.
Recombinant HSV-2 carrying a large deletion in vhs does not disrupt SG formation. (A) Western analysis of recombinant HSV-2 generated from pYEbac373. Whole-cell lysates were prepared from HeLa cells infected at low multiplicity for 16 h with the indicated viruses or from mock-infected cells. An arrowhead indicates the migration position of WT vhs; an asterisk indicates a prominent background band detected in all samples. Equal volumes of lysates were electrophoresed through a 10% polyacrylamide gel and transferred to a PVDF membrane. The membrane was probed with the antisera indicated on the right. An arrowhead indicates the migration position of WT vhs; an asterisk indicates a prominent background band detected in all samples. Molecular masses in kDa are indicated on the left. (B) HeLa cells were infected at low multiplicity with the indicated viruses or mock infected. Four hours later, 0.5 mM arsenite (+ARS) was applied for 30 min or cells were left untreated. Cells were fixed and stained as described for Fig. 2B. Stained cells were examined by confocal microscopy. Note that SGs were not detected following arsenite treatment in the majority of cells infected with all three viruses carrying the WT UL41 sequence; examples are indicated by the arrows. Scale bars, 10 μm. (C) Quantitative analysis of SG formation following arsenite treatment of infected cells. The percentage of infected cells with at least one SG was scored in at least 100 infected cells in approximately 30 independent fields of view for each virus under both treatment conditions. Error bars show standard errors of the means.
Spontaneous SGs accumulate in cells infected with HSV-2 strains defective in vhs.
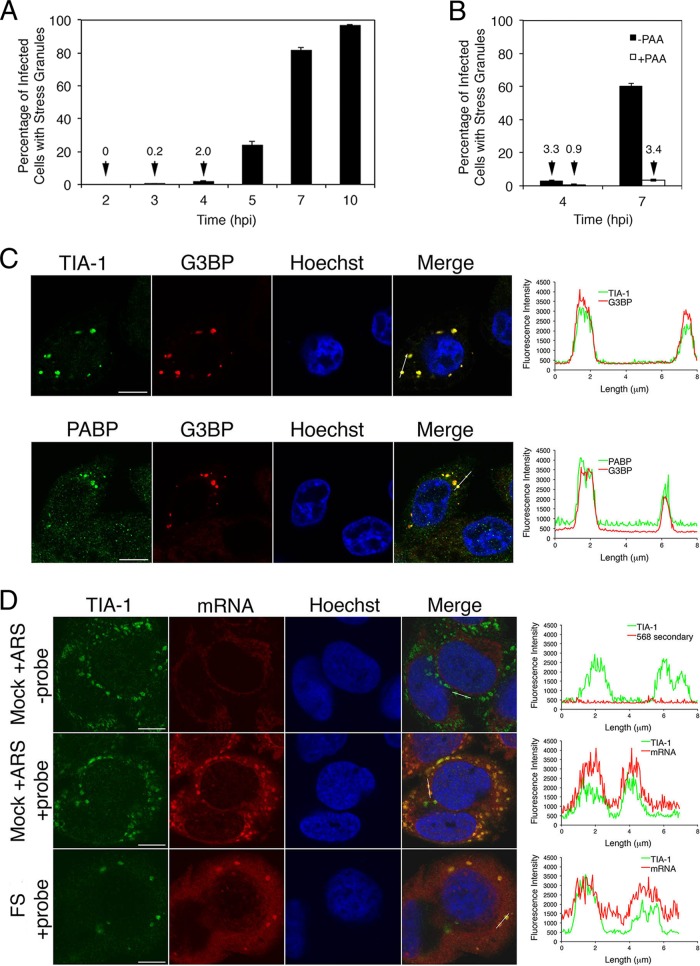
As cells infected with HSV-1 bearing UL41 mutations have been reported to accumulate SGs late in infection (26, 27), we wished to test whether this was also true for HSV-2. HeLa cells infected with FS at an MOI of 10 formed SGs starting at 4 hpi with the majority of infected cells containing SGs by 7 hpi (Fig. 6A). This is in sharp contrast to cells infected with HSV-2 carrying WT vhs, where SGs do not accumulate over time (23). Spontaneous SG formation was reduced when viral DNA replication was inhibited with PAA, indicating that late events in the infectious cycle are required for their formation (Fig. 6B). Spontaneous SGs were observed in 20.6% ± 17.6% and 43.6% ± 19.6% of infected cells at 10 and 13 hpi, respectively, in low-multiplicity infections of HeLa cells with ΔUL41 virus (data not shown).
FIG 6.
SGs accumulate spontaneously in infections with HSV-2 carrying a defective vhs. (A) HeLa cells were infected at high multiplicity with FS virus. At the indicated times postinfection, cells were fixed and stained as described for Fig. 2B, and percentages of infected cells with at least one SG were determined from 20 independent fields of view. Error bars show standard errors of the means. (B) HeLa cells were pretreated with phosphonoacetic acid (PAA) or left untreated and then infected at high MOI with FS virus in the continuous presence of PAA (+PAA) or in the absence of PAA (−PAA). At the indicated times postinfection, cells were fixed and stained as described for Fig. 2B, and percentages of infected cells with at least one SG were determined from 20 independent fields of view. Error bars show standard errors of the means. (C) HeLa cells were infected at high multiplicity with FS virus. Seven hours later, cells were fixed and stained with goat polyclonal antiserum specific for TIA-1 and mouse monoclonal antibody specific for G3BP (upper images) or goat polyclonal antiserum specific for PABP and mouse monoclonal antibody specific for G3BP (lower images), followed by staining with Alexa Fluor 488-conjugated donkey anti-goat IgG and Alexa Fluor 568-conjugated donkey anti-mouse IgG secondary antibodies. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy, and representative images are shown. Scale bars, 10 μm. The graphs on the right show the fluorescence intensity across an approximately 8-μm line indicated on the merged image panels. (D) HeLa cells were infected at high multiplicity with FS virus. Seven hours later, cells were fixed, permeabilized, and hybridized to a biotinylated oligo(dT) probe. As procedural controls, mock-infected cells treated with 0.5 mM arsenite for 30 min were hybridized to a biotinylated oligo(dT) probe or to a hybridization solution lacking probe; FS-infected cells incubated with hybridization solution lacking probe were also analyzed (data not shown). Following hybridization, cells were stained with goat polyclonal antiserum specific for TIA-1 followed by staining with Alexa Fluor 488-conjugated donkey anti-goat IgG and Alexa Fluor 568-conjugated streptavidin. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy, and representative images are shown. Scale bars, 10 μm. For comparative purposes, the confocal acquisition setting on the −probe samples was the same as that used for the +probe samples. The level of background staining in FS-infected cells without probe was comparable to that shown for mock-infected arsenite-treated cells without probe shown in the top panel of images. Similar results were observed in an independent fluorescent in situ hybridization experiment.
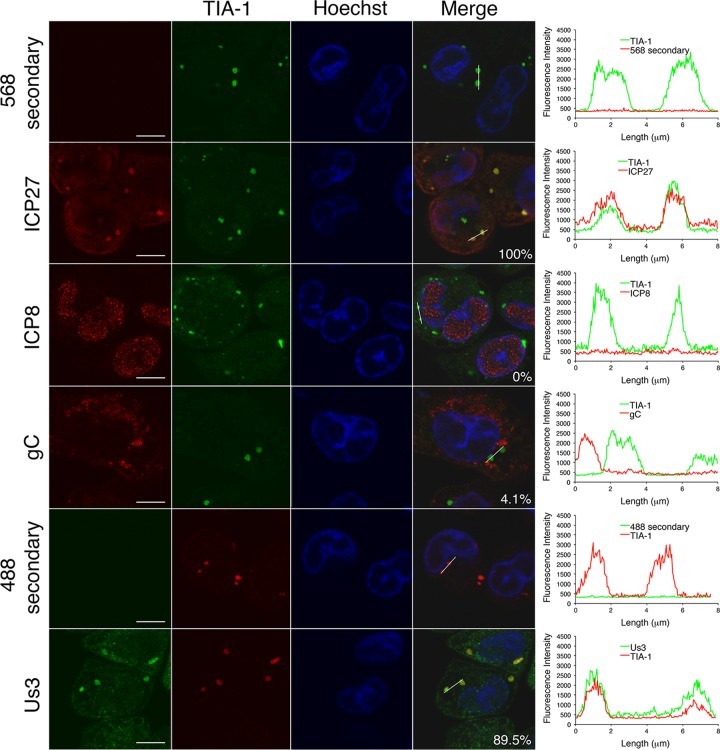
Spontaneous SGs contained the SG marker proteins TIA-1, G3BP, and poly(A) binding protein (PABP) as well as polyadenylated mRNA (Fig. 6C and D). Intriguingly, they also contained the viral proteins Us3 and ICP27 (Fig. 7), raising the possibility that these viral proteins contribute to the formation of these structures or that these viral proteins may become passively sequestered within these structures.
FIG 7.
Spontaneous SGs contain viral proteins. HeLa cells were infected at high MOI with FS virus. Seven hours later, cells were fixed and stained with goat polyclonal antiserum specific for TIA-1 and mouse monoclonal antibody specific for ICP27, ICP8, or gC or with rat polyclonal antiserum specific for Us3 followed by staining with Alexa Fluor 488- or Alexa Fluor 568-conjugated donkey anti-goat IgG and Alexa Fluor 568-conjugated donkey anti-mouse IgG secondary antibodies or Alexa Fluor 488-conjugated donkey anti-rat IgG antibodies. Nuclei were stained with Hoechst 33342. Stained cells were examined by confocal microscopy, and representative images are shown. The percentage of colocalization observed between TIA-1 and viral protein in approximately 200 spontaneous SGs is shown on the merged image panels. Scale bars, 10 μm. The graphs on the right show the fluorescence intensity across an approximately 8-μm line indicated on the merged image panels. For comparative purposes, the confocal acquisition settings used for the secondary-antibody-alone samples were the same as those used for the corresponding viral protein(s).
DISCUSSION
In our previous study, HSV-2 was found to impact SG accumulation (23), thereby adding HSV-2 to an increasing list of viruses that impact these cellular structures (20). We now demonstrate that HSV-2 virion components are sufficient to mediate the disruption of arsenite-induced SG formation and identify the tegument protein vhs as a necessary virion component. The conclusion that vhs is a required virion component is based on (i) the reduced ability of defective forms of vhs from two different HSV-2 strains to mediate the disruption of arsenite-induced SG formation, (ii) the recovery of this disruption when a large deletion within UL41 is specifically repaired, and (iii) the ability of vhs to mediate this disruption in the absence of other viral proteins.
It is well established that vhs is an endoribonuclease whose activity in vivo is directed toward cellular and viral mRNAs (24, 25, 33–35). Packaging of vhs into the tegument of virions allows this endoribonuclease to be delivered to the cytoplasm immediately upon fusion of viral and cellular membranes and to commence the degradation of cellular mRNAs well in advance of de novo viral gene expression. Degradation of cellular mRNAs in combination with disruptions in cellular mRNA biogenesis mediated by the immediate early viral protein ICP27 (36, 37) leads to considerable suppression of cellular protein synthesis. Later in infection, the association of newly synthesized vhs with the viral proteins VP16 and VP22 has been proposed to dampen vhs endoribonuclease activity, thereby permitting robust production of late viral mRNAs (38–41). Targeting of vhs activity toward mRNAs in vivo may be facilitated by its ability to associate with the cellular translation initiation factors eIF4A, eIF4B, and eIF4H (42–44). However, these associations may also be indicative of a direct role for vhs in translation. Indeed, enhanced translation of viral late mRNAs has recently been ascribed to vhs (26). As vhs may interact with mRNA in two distinct manners and has the ability to associate with multiple viral and cellular proteins, predicting how vhs is able to disrupt arsenite-induced SG formation is not straightforward. For example, the defective form of vhs synthesized by the FS virus used in this study is lacking C-terminal residues that, in the case of the highly related protein from HSV-1 (32), have been implicated in vhs endoribonuclease activity (amino acids 399, 426, and 438 [Fig. 3A]). While this may implicate the involvement of vhs endoribonuclease activity in the disruption of SG formation, one of these residues, amino acid 438, is also required for eIF4H binding (43). In addition, packaging defects have been noted with defective forms of HSV-1 vhs (45, 46), raising the possibility that decreases in the amount of vhs delivered to an infected cell as a result of a packaging defect could contribute to a lack of ability to disrupt SG formation. Future studies will take advantage of the wealth of knowledge about functional domains and residues within vhs to define which activities contribute to disruption of SG formation while taking possible packaging defects into consideration.
Our studies demonstrate that the homopolymer-based frameshift mutation (HFM) in the UL41 gene of HSV-2 strain HG52, identified by Everett and Fenwick nearly 25 years ago (32), is unstable and will rapidly revert to restore the WT UL41 sequence. As a practical consequence, stocks of HG52 that have undergone repeated low-multiplicity passaging will very likely carry WT UL41. The basis for the selective pressure that favors WT UL41 over the FS version found in HG52 is not known. We can routinely grow HG52 with FS UL41 to peak titers that are 5- to 10-fold lower than those of HG52 with WT UL41 in both HaCaT and Vero cells and have not observed differences in either size or morphology of plaques yielded from HG52 with FS UL41 on Vero cells in comparison to those yielded from HG52 with WT UL41. Despite this, the selection of WT UL41 over FS UL41 in HaCaT cells proceeds rapidly. It has been demonstrated that HSV-2 strain 333 carrying an internal deletion, or a point mutation (D215N), in vhs is hypersensitive to type I interferon (IFN), resulting in impaired viral replication (47, 48). As the type I IFN signaling pathway is present and functional in HaCaT cells (49, 50), it is possible that a growth advantage gained by acquiring vhs-mediated interference with type I IFN responses drives the selection of WT UL41. The dominance of HSV-1 strain KOS carrying WT UL41 over HSV-1 strain KOS carrying a defective UL41 (vhs1; T214I) was also observed following serial passaging of mixed infections (51). Regarding the restoration of the WT UL41 sequence, homopolymeric tracts of nucleotides such as that found within UL41 are known to be mutational hot spots. Two well-known examples of HSV open reading frames with HFMs that have functional consequences are UL23, which encodes thymidine kinase (52–55), and Us4, which encodes glycoprotein G (56, 57). Szpara and colleagues have recently identified many other examples of HFMs within the HSV genome in a comparative survey of the complete genomes of 26 different HSV-1 isolates (58). In this survey, instances of HFMs were found in one-third of the genomes analyzed, and so it seems very probable that a copying error by the viral DNA polymerase within the homopolymeric nucleotide tract of UL41 occurred to restore the WT sequence.
Finally, we confirm that cells infected with HSV-2 strains carrying a defective vhs form SGs spontaneously late in infection as has been observed for HSV-1 strains carrying defects in vhs (26, 27). The increased presence of spontaneous SGs at late times postinfection coupled with their decreased presence when viral DNA replication is inhibited argues that late events in the infectious cycle are required for their formation. Dauber et al. have proposed an “mRNA overload” model to explain their formation, whereby in the absence of the mRNA-destabilizing function of vhs, viral mRNAs eventually accumulate to levels that exceed the capacity of the host translational machinery, leading to SG formation (26, 59). We also demonstrate that these spontaneous SGs contain the viral proteins ICP27 and Us3. One of the many roles that ICP27 plays during viral infection is to facilitate the export of viral mRNAs via direct interactions with both mRNA and cellular RNA export proteins (60–62). ICP27 also has the ability to boost the translation of viral mRNAs (63, 64) and can associate with the cellular translation initiation factors PABP, eIF3, and eIF4G (65), all of which are core SG components (5). Thus, there are multiple ways that ICP27 could passively localize to SGs. The direct association of the viral serine/threonine kinase Us3 with mRNA, mRNA binding proteins, or other SG components has not been described. In yeast two-hybrid analyses performed using pseudorabies virus Us3 as bait, eIF4A (a core SG component), ribonucleoprotein L (an mRNA binding protein), and ribosomal protein S26 (a component of the small ribosomal subunit) were all identified as potential binding partners for Us3 (M. G. Lyman and B. W. Banfield, unpublished data). Association of Us3 with any of these proteins could allow it to passively localize to SGs. As both ICP27 and Us3 are multifunctional viral proteins, there is a possibility that they could play more-active roles in SG formation.
Future studies will aim to define which activities of vhs contribute to its ability to disrupt SG formation and to probe the relevance of this ability to viral infection. Further investigation into Us3's ability to localize to SGs has the potential to uncover novel roles for Us3 in the regulation of translation.
ACKNOWLEDGMENTS
This work was supported by Canadian Institutes of Health Research operating grant 93804, Natural Sciences and Engineering Council of Canada Discovery Grant 418719, Canada Foundation for Innovation award 16389, and an award from the Violet E. Powell Research Fund to B.W.B., by Canadian Institutes of Health Research operating grant 37955 to J.R.S, and by a postdoctoral fellowship from Alberta Innovates-Health Solutions to B.D. J.R.S. is a Canada Research Chair in Molecular Virology (Tier 1).
We thank D. W. Wilson, Albert Einstein College of Medicine, for vhs antisera, D. J. McGeoch, MRC Virology Unit, Glasgow, for HSV-2 strain HG52, and N. Osterriedder, Freie Universität Berlin, for providing plasmid pEP-Kan-S2.
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925–16933. 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 2.Eisinger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, Lannigan DA. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31:722–736. 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim B, Cooke HJ, Rhee K. 2012. DAZL is essential for stress granule formation implicated in germ cell survival upon heat stress. Development 139:568–578. 10.1242/dev.075846. [DOI] [PubMed] [Google Scholar]
- 4.Lu L, Han AP, Chen JJ. 2001. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol. Cell. Biol. 21:7971–7980. 10.1128/MCB.21.23.7971-7980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383–5398. 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152:791–805. 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. 2006. Inhibition of PKR by RNA and DNA viruses. Virus Res. 119:100–110. 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol. 81:3377–3390. 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 123:369–383. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez E, Castello A, Menendez-Arias L, Carrasco L. 2006. HIV protease cleaves poly(A)-binding protein. Biochem. J. 396:219–226. 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. 2011. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 85:6882–6892. 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046. 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989–12000. 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Legros S, Boxus M, Gatot JS, Van Lint C, Kruys V, Kettmann R, Twizere JC, Dequiedt F. 2011. The HTLV-1 Tax protein inhibits formation of stress granules by interacting with histone deacetylase 6. Oncogene 30:4050–4062. 10.1038/onc.2011.120. [DOI] [PubMed] [Google Scholar]
- 17.McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763. 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Q, Hastings C, Miller CL. 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 83:11090–11101. 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valiente-Echeverria F, Melnychuk L, Mouland AJ. 2012. Viral modulation of stress granules. Virus Res. 169:430–437. 10.1016/j.virusres.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reineke LC, Lloyd RE. 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436:255–267. 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montero H, Trujillo-Alonso V. 2011. Stress granules in the viral replication cycle. Viruses 3:2328–2338. 10.3390/v3112328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YR, King OD, Shorter J, Gitler AD. 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201:361–372. 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finnen RL, Pangka KR, Banfield BW. 2012. Herpes simplex virus 2 infection impacts stress granule accumulation. J. Virol. 86:8119–8130. 10.1128/JVI.00313-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063–1068. 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smiley JR, Elgadi MM, Saffran HA. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440–451. 10.1016/S0076-6879(01)42565-1. [DOI] [PubMed] [Google Scholar]
- 26.Dauber B, Pelletier J, Smiley JR. 2011. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 85:5363–5373. 10.1128/JVI.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esclatine A, Taddeo B, Roizman B. 2004. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J. Virol. 78:8582–8592. 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Sage V, Jung M, Alter JD, Wills EG, Johnston SM, Kawaguchi Y, Baines JD, Banfield BW. 2013. The herpes simplex virus 2 UL21 protein is essential for virus propagation. J. Virol. 87:5904–5915. 10.1128/JVI.03489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 30.Finnen RL, Roy BB, Zhang H, Banfield BW. 2010. Analysis of filamentous process induction and nuclear localization properties of the HSV-2 serine/threonine kinase Us3. Virology 397:23–33. 10.1016/j.virol.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee GE, Church GA, Wilson DW. 2003. A subpopulation of tegument protein vhs localizes to detergent-insoluble lipid rafts in herpes simplex virus-infected cells. J. Virol. 77:2038–2045. 10.1128/JVI.77.3.2038-2045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett RD, Fenwick ML. 1990. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J. Gen. Virol. 71(Part 6):1387–1390. 10.1099/0022-1317-71-6-1387. [DOI] [PubMed] [Google Scholar]
- 33.Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. U. S. A. 84:1926–1930. 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oroskar AA, Read GS. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everly DN, Jr, Read GS. 1999. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J. Virol. 73:9117–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy WR, Sandri-Goldin RM. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spencer CA, Dahmus ME, Rice SA. 1997. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 71:2031–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knez J, Bilan PT, Capone JP. 2003. A single amino acid substitution in herpes simplex virus type 1 VP16 inhibits binding to the virion host shutoff protein and is incompatible with virus growth. J. Virol. 77:2892–2902. 10.1128/JVI.77.5.2892-2902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sciortino MT, Taddeo B, Giuffre-Cuculletto M, Medici MA, Mastino A, Roizman B. 2007. Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 81:10924–10932. 10.1128/JVI.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam Q, Smibert CA, Koop KE, Lavery C, Capone JP, Weinheimer SP, Smiley JR. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 42.Doepker RC, Hsu WL, Saffran HA, Smiley JR. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 78:4684–4699. 10.1128/JVI.78.9.4684-4699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng P, Everly DN, Jr, Read GS. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651–9664. 10.1128/JVI.79.15.9651-9664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Page HG, Read GS. 2010. The virion host shutoff endonuclease (UL41) of herpes simplex virus interacts with the cellular cap-binding complex eIF4F. J. Virol. 84:6886–6890. 10.1128/JVI.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mbong EF, Woodley L, Dunkerley E, Schrimpf JE, Morrison LA, Duffy C. 2012. Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J. Virol. 86:12351–12361. 10.1128/JVI.01975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read GS, Karr BM, Knight K. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duerst RJ, Morrison LA. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322:158–167. 10.1016/j.virol.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 48.Wylie KM, Schrimpf JE, Morrison LA. 2009. Increased eIF2alpha phosphorylation attenuates replication of herpes simplex virus 2 vhs mutants in mouse embryonic fibroblasts and correlates with reduced accumulation of the PKR antagonist ICP34.5. J. Virol. 83:9151–9162. 10.1128/JVI.00886-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Fits L, van der Wel LI, Laman JD, Prens EP, Verschuren MC. 2003. Psoriatic lesional skin exhibits an aberrant expression pattern of interferon regulatory factor-2 (IRF-2). J. Pathol. 199:107–114. 10.1002/path.1263. [DOI] [PubMed] [Google Scholar]
- 50.Gianni T, Leoni V, Campadelli-Fiume G. 2013. Type I interferon and NF-kappaB activation elicited by herpes simplex virus gH/gL via alphavbeta3 integrin in epithelial and neuronal cell lines. J. Virol. 87:13911–13916. 10.1128/JVI.01894-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwong AD, Kruper JA, Frenkel N. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K, Mahalingam G, Hoover SE, Mont EK, Holland SM, Cohen JI, Straus SE. 2007. Diverse herpes simplex virus type 1 thymidine kinase mutants in individual human neurons and ganglia. J. Virol. 81:6817–6826. 10.1128/JVI.00166-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasadeusz JJ, Tufaro F, Safrin S, Schubert K, Hubinette MM, Cheung PK, Sacks SL. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grey F, Sowa M, Collins P, Fenton RJ, Harris W, Snowden W, Efstathiou S, Darby G. 2003. Characterization of a neurovirulent aciclovir-resistant variant of herpes simplex virus. J. Gen. Virol. 84:1403–1410. 10.1099/vir.0.18881-0. [DOI] [PubMed] [Google Scholar]
- 55.Sauerbrei A, Deinhardt S, Zell R, Wutzler P. 2010. Phenotypic and genotypic characterization of acyclovir-resistant clinical isolates of herpes simplex virus. Antiviral Res. 86:246–252. 10.1016/j.antiviral.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Liljeqvist JA, Svennerholm B, Bergstrom T. 1999. Herpes simplex virus type 2 glycoprotein G-negative clinical isolates are generated by single frameshift mutations. J. Virol. 73:9796–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rekabdar E, Tunback P, Liljeqvist JA, Lindh M, Bergstrom T. 2002. Dichotomy of glycoprotein g gene in herpes simplex virus type 1 isolates. J. Clin. Microbiol. 40:3245–3251. 10.1128/JCM.40.9.3245-3251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J. Virol. 88:1209–1227. 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dauber B, Saffran HA, Smiley JR. 2014. The HSV-1 virion host shutoff protein enhances translation of viral late mRNAs by preventing mRNA overload. J. Virol. 88:9624–9632. 10.1128/JVI.01350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson LA, Sandri-Goldin RM. 2009. Efficient nuclear export of herpes simplex virus 1 transcripts requires both RNA binding by ICP27 and ICP27 interaction with TAP/NXF1. J. Virol. 83:1184–1192. 10.1128/JVI.02010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandri-Goldin RM. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868–879. 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian X, Devi-Rao G, Golovanov AP, Sandri-Goldin RM. 2013. The interaction of the cellular export adaptor protein Aly/REF with ICP27 contributes to the efficiency of herpes simplex virus 1 mRNA export. J. Virol. 87:7210–7217. 10.1128/JVI.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fontaine-Rodriguez EC, Knipe DM. 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 82:3538–3545. 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellison KS, Maranchuk RA, Mottet KL, Smiley JR. 2005. Control of VP16 translation by the herpes simplex virus type 1 immediate-early protein ICP27. J. Virol. 79:4120–4131. 10.1128/JVI.79.7.4120-4131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fontaine-Rodriguez EC, Taylor TJ, Olesky M, Knipe DM. 2004. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology 330:487–492. 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]