ABSTRACT
Superoxide dismutases (SODs) are metalloproteins that protect organisms from toxic reactive oxygen species by catalyzing the conversion of superoxide anion to hydrogen peroxide and molecular oxygen. Chlorovirus PBCV-1 encodes a 187-amino-acid protein that resembles a Cu-Zn SOD with all of the conserved amino acid residues for binding copper and zinc (named cvSOD). cvSOD has an internal Met that results in a 165-amino-acid protein (named tcvSOD). Both cvSOD and tcvSOD recombinant proteins inhibited nitroblue tetrazolium reduction of superoxide anion generated in a xanthine-xanthine oxidase system in solution. tcvSOD was chosen for further characterization because it was easier to produce. Recombinant tcvSOD also inhibited a riboflavin photochemical reduction system in a polyacrylamide gel assay, which was blocked by the Cu-Zn SOD inhibitor cyanide but not by azide, which inhibits Fe and Mn SODs. A kcat/Km value for cvSOD was determined by stop-flow spectrophotometry as 1.28 × 108 M−1 s−1, suggesting that cvSOD-catalyzed O2− dismutation was not a diffusion controlled encounter. The cvsod gene was expressed as a late gene, and cvSOD activity was detected in purified virions. Superoxide accumulated rapidly during virus infection, and circumstantial evidence indicates that cvSOD aids its decomposition to benefit virus replication. Cu-Zn SOD homologs have been described to occur in 3 other families of large DNA viruses, poxviruses, baculoviruses, and mimiviruses, which group as a clade. Interestingly, cvSOD does not group in the same clade as the other virus SODs but instead groups in an expanded clade that includes Cu-Zn SODs from many cellular organisms.
IMPORTANCE Virus infection often leads to an increase in toxic reactive oxygen species in the host, which can be detrimental to virus replication. Viruses have developed various ways to overcome this barrier. As reported in this article, the chloroviruses often encode and package a functional Cu-Zn superoxide dismutase in the virion that presumably lowers the concentration of reactive oxygen induced early during virus infection.
INTRODUCTION
Reactive oxygen species (ROS) are generated both through normal metabolic activity and under stress situations in aerobic organisms. Physiological concentrations of ROS are beneficial and are involved in cell signaling pathways and protection of hosts from pathogen invasion. However, higher levels of ROS can damage macromolecules and cell structure (1, 2, 3). Cell defense systems can minimize these deleterious effects, with superoxide dismutases (SODs) providing the first line of defense against ROS (4, 5). SODs are metalloproteins that protect organisms from toxic ROS by catalyzing the conversion of superoxide anion to hydrogen peroxide and molecular oxygen (6). Depending on the associated metal cofactors required for the superoxide ion scavenging function, SODs are classified into copper-zinc SODs (Cu-Zn SODs), iron SODs (Fe SOD), manganese SODs (Mn SOD), and nickel SODs (Ni SOD). In mammalian cells, Cu-Zn SODs are located in the cytoplasm (named SOD1) and/or in extracellular spaces (SOD3 or EC-SOD); Mn SODs (named SOD2) are located in the mitochondria (7). Higher plants usually have mitochondrial Mn SODs, chloroplast-localized Fe SODs and multiple isoforms of Cu-Zn SODs located in various cellular compartments, including the cytoplasm, chloroplast, and peroxisome (8). Eukaryotic algae and protozoa typically contain Mn SODs and Fe SODs but lack Cu-Zn SODs; prokaryotic organisms have Mn SODs and/or Fe SODs (9), although many Gram-negative bacteria have Cu-Zn SODs (10). Ni SODs exist only in a few Streptomyces species (11).
It is rare for viruses to encode a functional SOD, but a Cu-Zn SOD-like protein-encoding gene is present in several large DNA viruses, including poxviruses, baculoviruses, mimiviruses, and phycodnaviruses. Interestingly, with one exception, the poxvirus-encoded homologs are not functional as SODs because they lack a copper-binding domain (12, 13, 14). Furthermore, the genes encoding the Cu-Zn SOD homologs in the poxviruses can be disrupted or deleted without having much effect on virus replication. The single poxvirus exception is Amsacta moori entomopoxvirus (AmEPV), which encodes a functional Cu-Zn SOD (15). However, like for other poxviruses, the gene is not essential for virus replication. It is interesting that the Cu-Zn SOD-like protein encoded by two leporipoxviruses can bind Zn, but the Cu binding amino acid residues are absent. Recombinant leporipoxvirus SODs form a stable heterodimer with a cellular copper chaperone protein associated with the host Cu-Zn SODs, leading to the hypothesis that this interaction reduces host Cu-Zn SOD activity by sequestering Cu, thus keeping Cu from the host enzyme (16). Gene knockout experiments indicated that the viral Cu-Zn SOD homolog was involved in upregulating the intracellular ROS level, which inhibited the programmed cell death response; this upregulation was proposed to benefit virus replication (17).
Most baculoviruses encode a Cu-Zn SOD homolog (18) that has the typical Cu-Zn binding domains in the protein (e.g., see references 18 and 19). However, the role of the SOD in virus replication is unclear. A Cu-Zn SOD-encoding gene was identified in Autographa californica nuclear polyhedrosis virus (AcMNPV) many years ago (20). The putative protein contained metal binding domains, and the protein was predicted to have enzyme activity. However, attempts to detect Cu-Zn SOD activity in lysates from virus-infected cells were negative. Furthermore, the AcNMPV gene could be deleted without any obvious effect on virus replication in cell culture or in insects (20). To our knowledge, a recombinant baculovirus-encoded Cu-Zn SOD has not been produced and tested for activity.
Some of the huge viruses included in the Mimiviridae family also encode a Cu-Zn SOD homolog. The Megavirus-encoded Cu-Zn SOD has all the relevant metal binding domains, and a recombinant protein has been crystallized; its structure resembles that of functional Cu-Zn SODs (21). However, the recombinant protein has not been characterized enzymatically.
The other family of viruses that encode Cu-Zn SOD homologs is the Phycodnaviridae family, including the genus Chlorovirus. Chloroviruses are large icosahedral, plaque-forming, double-stranded DNA (dsDNA) viruses that infect certain eukaryotic, chlorella-like, symbiotic green algae (22, 23). The chloroviruses are proposed to have a common evolutionary ancestor with some other large DNA viruses, including the poxviruses, asfarviruses, iridoviruses, ascoviruses, mimiviruses, and marseilleviruses, and collectively are often referred to as nucleocytoplasmic large DNA viruses (NCLDVs) (24, 25, 26). The 331-kb linear genome of the chlorovirus type species, Paramecium bursaria chlorella virus 1 (PBCV-1), has 416 predicted protein coding sequences (CDSs) (27). About 40% of the predicted CDSs resemble those for known proteins (23), including a gene, a245r, which encodes a putative Cu-Zn SOD. We report here that the PBCV-1 a245r-encoded SOD recombinant protein has all the biochemical properties expected of a Cu-Zn SOD.
MATERIALS AND METHODS
Viruses and host strains.
Growth of Chlorella variabilis NC64A on modified Bold's basal medium, the plaque assay, the production of the viruses, and the isolation of virus DNAs have been described previously (28, 29, 30).
a245r gene cloning and expression.
A 564-bp putative Cu-Zn SOD-encoding gene, a245r (cvsod), and its truncated version, which starts from an internal ATG codon and is 63 bp shorter, tcvsod, were amplified with primer pairs cvsod Frw/cvsod Rev and tcvsod Frw/cvsod Rev (see Table S1 in the supplemental material) from PBCV-1 genomic DNA. The cvsod and tcvsod PCR products were digested with BamHI and EcoRI and inserted into pGEX-2T (GE Healthcare Life Science) to generate two recombinant plasmids, pGEXcvsod and pGEXtcvsod. Plasmids were transformed into Escherichia coli BL21expression cells (Novagen); single colonies were picked and cultured in 5 ml of 2× YT medium (16 g of tryptone, 10 g of yeast extract, and 5 g NaCl in 1 liter [pH 7.0]) overnight at 37°C. One hundred milliliters of fresh 2× YT medium supplemented with 0.5 mM CuCl2 and 0.1 mM ZnSO4 (15) and with 5% inoculum were incubated at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.8. The culture was cooled to 25°C, and isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 0.1 mM to induce protein expression; the cells were incubated for 3 h. Cells were collected by centrifugation for 5 min at 11,000 × g and 4°C, suspended in 5 ml of modified phosphate-buffered saline (PBS) solution (2× PBS, 280 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]), and sonicated with a Tekmar sonic disruptor for 2 min with 5-s pulses at the 50% energy level. The cell lysate was centrifuged for 30 min at 28,500 × g and 4°C to separate the supernatant and particulate material.
To isolate recombinant cvSOD and tcvSOD proteins, 0.4 ml of glutathione Sepharose 4B (GE Healthcare) resin was loaded in a 5-ml self-packing column, washed, and equilibrated with 15 ml of 2× PBS. The cell lysate was applied to the column; the flowthrough was collected and reloaded a second time. The column was washed with another 15 ml of 2× PBS to remove unbound proteins. The glutathione-tagged SOD proteins were either eluted from the column with elution buffer (100 mM Tris-HCl, 30 mM reduced glutathione [pH 8.5]) or digested with 25 μl of thrombin while the protein was on the column. Bovine thrombin powder was purchased from MP Biochemicals and prepared as 5 U/μl in 2× PBS. After 16 h of digestion at 4°C, recombinant SODs were collected from the column and passed through a Hitrap benzamidine FF column (GE Healthcare) to remove the thrombin. Microliter protein samples were collected at every purification step and electrophoresed on precast 4 to 20% Tris-glycine PAGEr Gold (Lanza) polyacrylamide gels (PAG) to monitor protein purification. Eight-microliter protein samples were mixed with 2 μl of 5-fold loading buffer (250 mM Tris-Cl [pH 6.8], 500 mM dithiothreitol [DTT], 10% SDS, 50% glycerol, 0.5% bromophenol blue), heated at 100°C for 10 min, and loaded and electrophoresed in a buffer system containing 25 mM Tris, 192 mM glycine, and 0.1% SDS (pH 8.3) for 1 h at 200 V in a Bio-Rad vertical mini-gel electrophoresis system. The gel was stained in 40% methanol, 10% acetic acid, 50% water, and 0.1% Coomassie brilliant blue R250 for 3 to 10 min; destaining occurred in the same solution minus the dye.
SOD activity assay in solution.
In an indirect assay, the SOD-associated inhibition of nitroblue tetrazolium (NBT) reduction in the presence of superoxide anion (O2−) generated by xanthine/xanthine oxidase was monitored by measuring the absorbance change at 560 nm (31). The 900-μl reaction mixture contained 50 mM phosphate buffer (pH 7.8), 1 mM diethylenetriamine pentaacetic acid, 1 U of catalase, 56 μM NBT, 100 μM xanthine, and 0 to 10 μg of SOD in 100 μl of 1× PBS. The absorbance at 560 nm was monitored after adding 100 μl of 0.2-U/ml xanthine oxidase using a Beckman DU530 UV-visible (UV-Vis) spectrophotometer.
SOD activity in situ assay.
One microgram of SOD protein was mixed with an appropriate amount of 5× loading buffer without DTT, applied to a precast PAGEr Gold gel, and electrophoresed as described above without further denaturing. For the in situ assay of recombinant SOD protein activity, the gel was stained by following the procedure of Beauchamp and Fridovich (32), with a slight modification (15). The gel was washed 3 times in deionized water, 10 min each time, then soaked in 2.45 mM NBT solution for 20 min in the dark, rinsed briefly with water, and incubated in 28 μM riboflavin and putrescine at 4.5 g/liter in 50 mM KPO4 (pH 7.8) under a fluorescent light until the transparent band completely developed. To determine the metal cofactors associated with the recombinant tcvSOD, 50 mM NaCN or 50 mM NaN3 was mixed with both NBT and riboflavin solutions (33).
SOD activity in virion particles.
To test for SOD activity in virus particles, 6 × 1011 PFU of either chlorovirus PBCV-1 or NY-2A was suspended in 100 μl of 50 mM Tris-HCl (pH 7.8) and sonicated at 20% amplitude with 5-s pulses for 4 min. The sonicated products were centrifuged for 10 min at 16,800 × g and 4°C. The supernatant fractions were assayed for SOD activity by both xanthine/xanthine oxidase spectrophotometry and PAGE in situ staining.
Stopped-flow spectrophotometry.
Catalysis by tcvSOD was monitored by stopped-flow spectrophotometry (model SX-18 MV stopped-flow spectrophotometer; Applied Photophysics) in a sequential-mixing chamber in which KO2 in a solution of dimethyl sulfoxide and 18-crown-6-ether (34) was diluted (1:10) in two sequential rapid mixings (35). The final solution contained 0.25 μM enzyme, 25 μM EDTA, and 100 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES) buffer at pH 8.9 and 25°C, as well as 8.3% (by volume) dimethyl sulfoxide with various amounts of superoxide. The active tcvSOD concentration was adjusted by the percentage of Cu-saturated recombinant protein. The change in absorbance due to superoxide at 260 nm was monitored.
Dot blot hybridization.
DNA was isolated from C. variabilis and 44 viruses that infect C. variabilis, transferred to a nylon membrane (Osmonics), and cross-linked by a UV Stratalinker 2400 at the auto-cross-link level. The cvsod gene was amplified by PCR as a hybridization probe. The probe was labeled with a Random Primers DNA Labeling System (Invitrogen). The dot blot membrane was prehybridized in 6× SSC (1× SSC is 0.15 M NaCl and 15 mM sodium citrate), 5× Denhardt's reagent, 0.5% SDS, and denatured salmon sperm DNA at 68°C for 1 h. The denatured dsDNA probe, labeled with [32P]dATP, was added to the membrane and hybridized at 68°C for 1 h. The membrane was washed in 2× SSC–0.5% SDS at room temperature for 5 min, 2× SSC–0.1% SDS at room temperature for 15 min twice, and 0.1× SSC–0.5% SDS at 65°C for 2 h and, finally, subjected to signal detection with a Storm phosphorimager equipped with ImageQuant software (Molecular Dynamics).
Northern hybridization.
C. variabilis cells (3 × 109) were collected at various times after virus PBCV-1 infection at a multiplicity of infection (MOI) of 5, frozen in liquid nitrogen, and stored at −80°C. RNA was extracted with TRIzol reagent (Invitrogen), denatured with formaldehyde, separated on a 1.5% agarose gel, and then transferred to a nylon membrane. A [32P]dATP-labeled probe was prepared as in the dot blot hybridization experiment. The membrane was preincubated in 20 ml of Church's buffer (1 mM EDTA, 0.5 M NaPO4, 7% SDS [pH 8.0]) for 1 h at 65°C, hybridized in fresh Church's buffer and a denatured probe for 16 h. After hybridization, the membrane was washed twice with 0.1× SSC–0.1% SDS, first for 30 min and then for 15 min. The signal detection was the same as in the dot blot hybridization experiment.
Sedimentation equilibrium.
The oligomeric state of tcvSOD was determined at 20°C by sedimentation equilibrium using an Optima XL-I analytical ultracentrifuge (Beckman Coulter, Inc.). tcvSOD was dialyzed into 12 mM sodium phosphate buffer (pH 7.3) containing 2.7 mM KCl and 140 mM NaCl. The sample cells were loaded with 110 μl of tcvSOD (0.2-mg/ml and 0.4-mg/ml concentrations), and reference cells were filled with the dialysis buffer. Radial scans were collected at 270 nm after 22 h at rotor speeds of 16,000, 18,000, and 20,000 rpm. Data from the three different rotor speeds were fit by global analysis to an equation describing a single-species model using Origin 6.0. A partial specific volume for tcvSOD of 0.7352 was calculated by SedTerp and was used for the best-fit analysis. The solvent density of the buffer was calculated to be 1.0062 g/ml.
Superoxide anion detection by DHE.
C. variabilis cells (4 × 108/ml) were mixed with 1 μl of 5 mM dihydroethidium (DHE; Molecular Probes) to a final concentration of 5 μM and incubated for 1 h by rolling to let the dye penetrate the cells. (DHE forms a red fluorescent product when it reacts with ROS.) The cells were then infected with either PBCV-1 or NY-2A at an MOI of 10 and analyzed by a Becton Dickinson flow cytometer at 0, 1, 5, 10, 15, 20, 30, 60, 120, 180, 240, and 300 min postinfection (p.i.).
Phylogenetic analysis of cvSOD.
A BLAST analysis was conducted (http://www.ncbi.nlm.nih.gov/Blast.cgi) using PBCV-1 tcvSOD (GenBank accession number NP_048593.1; all accession numbers in this article are GenBank numbers unless otherwise specified) to acquire taxa for phylogenetic analysis. While some green algae encode a Cu-Zn superoxide dismutase, the PBCV-1 host, C. variabilis (accession number ADIC01003810.1), does not. Phylogenetic analyses were conducted at http://www.phylogeny.fr (36). ClustalW was used to align the sequences. Positions with gaps were removed during alignment curation. BioNJ (distance method), maximum likelihood, and maximum parsimony tree construction methods were used.
Other procedures.
The concentrations of either the recombinant tcvSOD protein or the virion-associated proteins were determined with a Pierce Coomassie (Bradford) protein assay kit (Thermo Scientific). The Cu and Zn concentrations in the recombinant cvSOD were determined by mixing the protein with concentrated nitric acid and digested on a block heater at 95°C for 4 to 6 h. If necessary, 30% H2O2 was added to help oxidize the organic matter. After digestion, the solution was evaporated to <1 ml and then diluted with 1% nitric acid before analysis using a GVI Platform XS inductively coupled plasma mass spectrometer (ICP-MS). The ICP-MS uses a helium-hydrogen hexapole collision cell for removal of polyatomics and was calibrated using matrix-matched standards.
RESULTS AND DISCUSSION
A245R cloning, expression, and purification.
The PBCV-1 CDS A245R amino acid sequence has all the conserved residues required for copper binding (His 76, 78, and 150), zinc binding (His 93 and 110 and Asp 113), and formation of disulfide bonds (Cys 87 and 176) (Fig. 1), suggesting that A245R is probably a functional Cu-Zn SOD. A245R consists of 187 amino acids, which is larger than most Cu-Zn SODs, which are about 35 amino acids smaller; e.g., human SOD1 is 152 amino acids, bovine SOD1 is 154 amino acids, and the only reported functional viral SOD, AMEPVSOD, is 152 amino acids (Fig. 1). The major difference between A245R and the other SODs is an extension at the amino-terminal end. However, an internal Met residue exists at position 22 in A245R. Therefore, both the entire a245r gene and its truncated version that starts from the internal Met residue were cloned into vector pGEX2T at the BamHI and EcoRI sites, which produced a glutathione S-transferase (GST)-tagged protein. When the genes were overexpressed in E. coli, the larger gene produced only a small amount of recombinant protein, whereas the truncated version produced much more soluble protein. Both the entire protein and the truncated protein have Cu-Zn SOD activity; i.e., they inhibited NBT reduction in the xanthine-xanthine oxidase system by 90% (results not shown). However, because higher quantities of the recombinant truncated protein could be produced, it was chosen for further studies and named tcvSOD. Searching databases with the N-terminal 30 amino acids by BLAST did not identify any domains for this region.
FIG 1.
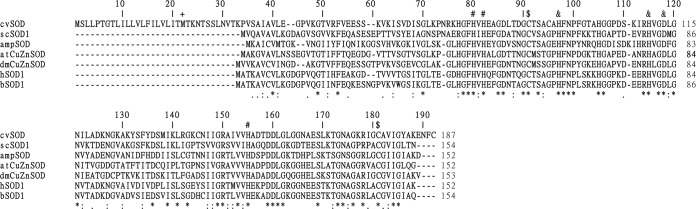
Sequence alignment of cvSOD with functional Cu-Zn SODs from different organisms. The sequences of bovine SOD1 (bSOD1; GenBank accession number NP_777040.1), human SOD1 (hSOD1; NP_000445.1), Drosophila melanogaster Cu-Zn SOD (dmCu-ZnSOD; CAA35210.1), Arabidopsis thaliana Cu-Zn SOD (atCuZnSOD; NP_172360.1), Saccharomyces cerevisiae SOD1 (scSOD1; NP_012638.1), Amsacta moorei entomopoxvirus (AMPSOD; NP_065037.1), and PBCV-1 SOD (cvSOD; NP_048593.1) were aligned with multiple-sequence alignment tool Clustal Omega from EMBL-EBI. Asterisks indicate positions which have a fully conserved residue. A colon indicates conservation between amino acids with strongly similar properties. A period indicates conservation between amino acids with less similar properties. Copper and zinc binding sites and intramolecular disulfide bonds are labeled with the symbols #, &, and $, respectively. The internal Met residue in PBCV-1 cvSOD is labeled with a plus (+).
The GST-tagged recombinant tcvSOD protein distributed about equally into the soluble and insoluble fractions (see Fig. S1 in the supplemental material). The tagged protein was eluted with glutathione elution buffer and cleaved with bovine thrombin at the Gly-Ser-Pro-Gly site to produce a 37-kDa thrombin, a 26-kDa GST, and a 17-kDa tcvSOD band on PAGE. Alternatively, tcvSOD was digested on the column with thrombin. Extra thrombin was removed by an affinity benzamidine FF column, and purified tcvSOD was collected for characterization (see Fig. S1, lane 6). Cu and Zn concentrations were determined in the recombinant tcvSOD by coupled plasma mass spectrometry; 54% of the subunits had Cu, and 70% of the subunits had Zn.
Recombinant tcvSOD is active.
Recombinant tcvSOD was tested for enzyme activity by 3 procedures, described below.
(i) Assay in free solution.
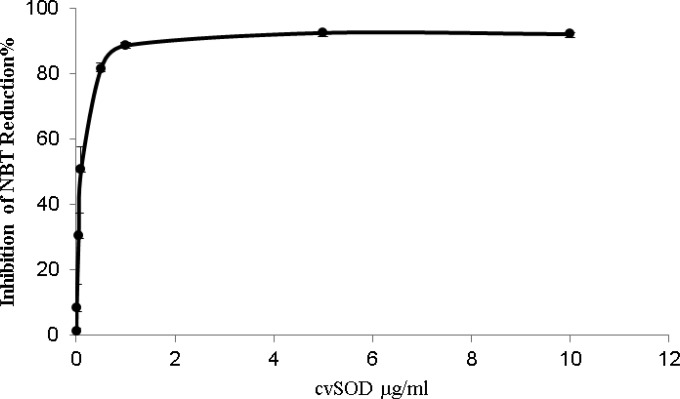
A xanthine-xanthine oxidase system was used to generate a reproducible flux of superoxide anion (O2−) to reduce NBT; tcvSOD activity was measured by its ability to inhibit this reduction. SOD activity is usually expressed as units per mg protein, and 1 U of activity is the amount of protein that gives half-maximal inhibition (32). The tcvSOD exhibited a typical inhibition curve and reached ∼90% inhibition of NBT reduction in the xanthine-xanthine oxidase/NBT assay system (Fig. 2). Inhibitors were used to distinguish the different types of SODs. Cyanide inhibits Cu-Zn SODs but not Fe and Mn SODs. Hydrogen peroxide inactivates Cu-Zn SODs and Fe SODs but not Mn SOD. Finally, both Fe and Mn SODs are sensitive to azide, and Cu-Zn SODs are not (37, 38). cvSOD activity was completely inhibited by 50 mM NaCN and was insensitive to NaN3, indicating that it is an authentic Cu-Zn SOD (results not shown).
FIG 2.
tcvSOD activity measurement using NBT as an indicator. Inhibition of NBT reduction in the presence of superoxide anion (O2−) generated by xanthine/xanthine oxidase was monitored by measuring the change in absorbance at 560 nm.
(ii) Assay on an acrylamide gel.
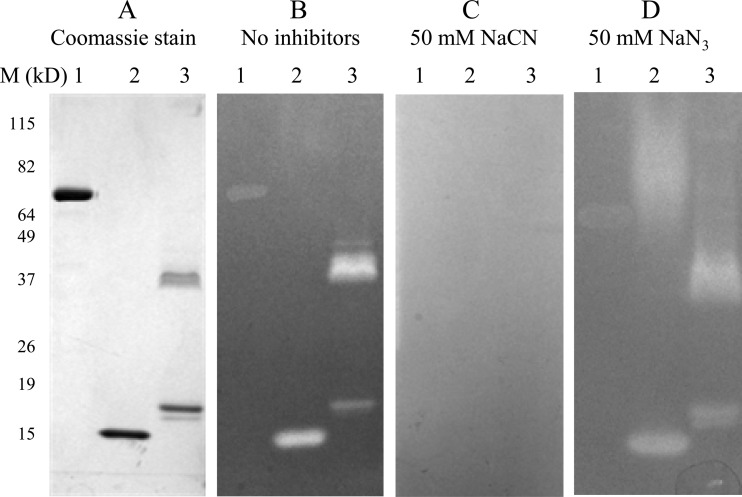
To confirm the tcvSOD activity by inhibition of NBT reduction, 1 μg of tcvSOD protein was loaded on a 4 to 20% Tris-glycine PAG, with the same amount of bovine serum albumin (BSA) and commercial bovine Cu-Zn SOD serving as negative and positive controls, respectively, and was electrophoresed in the presence of 0.1% SDS. After staining with NBT in the dark and exposure of the gel to light in the presence of riboflavin, clear bands developed in the positions where tcvSOD and bovine Cu-Zn SOD localized in the blue background generated by NBT photoreduction (Fig. 3A and B). The addition of SDS in loading and running buffers resulted in bovine SOD1 appearing as a monomer; however, tcvSOD migrated as both monomeric and dimeric bands on the gel. When SDS was washed away, the monomers were still active on the gel, indicating that after removal of the SDS (39), they can be restored to functional dimers. Addition of 50 mM NaCN to both the NBT and riboflavin solutions during the staining prevented the appearance of the tcvSOD and bovine SOD bands (Fig. 3C); 50 mM NaN3 did not inhibit the development of the clear bands (Fig. 3D). The results of this in situ gel assay agreed with those obtained in the xanthine-xanthine oxidase assay.
FIG 3.
tcvSOD activity gel in situ assay. One microgram of tcvSOD was mixed with 5× loading buffer without DTT and separated by PAGE with buffer that contained SDS. Lanes 1, BSA; lanes 2, bovine SOD1; lanes 3, recombinant tcvSOD. (A) Gel stained with Coomassie brilliant blue. (B) Gel stained with NBT and riboflavin. (C and D) Gel stained with NBT and riboflavin plus 50 mM NaCN (C) and NaN3 (D).
(iii) Stopped-flow spectrometry.
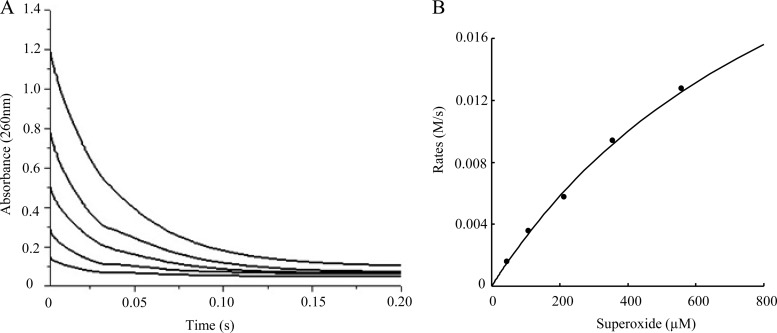
A decrease in O2− absorbance at 260 nm was measured by stopped-flow spectrophotometry after mixing KO2 and Cu-Zn SOD in solutions containing different amounts of KO2. The first 10 to 15% of each scan was used to derive the initial velocity of the reaction. The rate of the uncatalyzed dismutation of superoxide was negligible compared with the catalyzed rates. Saturation of catalytic activity was not observed by increasing superoxide, consistent with previous results with bovine and human SODs (40, 41), and the tcvSOD-catalyzed reaction was not a diffusion-controlled encounter. tcvSOD had a kcat/Km value of 1.28 × 108 ± 0.05 × 108 M−1s−1 at pH 8.9 (Fig. 4). The kcat/Km value is about 20- and 50-fold lower than those of bovine Cu-Zn SOD (kcat/Km values, 2.3 × 109 M−1 s−1 at pH 7 and 1.9 × 109 M−1 s−1 at pH 9.5) (41) and Photobacterium leiognathi Cu-Zn SOD (kcat/Km values, 5 × 109 M−1 s−1 at pH 7) (42), respectively, which are close to the diffusion limit. However, the tcvSOD kcat/Km value is slightly higher than that of the poxvirus SOD AMEPV-SOD (kcat/Km values, 8.5 × 107 M−1 s−1 at pH 8 and 4.1 × 107 M−1 s−1 at pH 9.5) (15) (Table 1). (Note that only 54% of the tcvSOD subunits were saturated with Cu and so we assumed that only half of the tcvSOD protein was enzymatically active. This assumption was included in the kcat/Km calculations.)
FIG 4.
Rate constant kcat/Km measurement by stopped-flow spectrophotometry. (A) The decrease in O2− absorbance at 260 nm measured after adding superoxide. Scans were recorded with decreasing O2− concentrations (from top to bottom, the concentrations were 611, 404, 261, 153, and 83.7 μM, respectively). (B) The first 10 to 15% of each scan from panel A was used to get the initial velocity of the reaction. The solid line is a least-squares fit of the Michaelis-Menten equation to the data resulting in a kcat of 7.8 × 104 ± 0.7 × 104 s−1 and kcat/Km of 1.28 × 108 ± 0.05 × 108 M−1 s−1.
TABLE 1.
Catalytic rate constant (kcat/Km) values for several Cu-Zn SODs
Like other Cu-Zn SODs, tcvSOD behaved as a homodimer in sedimentation equilibrium experiments. A best-fit value of 35,519 Da was determined for the molecular mass of tcvSOD, which agrees with the predicted molecular mass of a dimeric species (35,252 Da). Therefore, all of our experimental results indicate that tcvSOD is an authentic Cu-Zn SOD, and to our knowledge, this is only the second virus-encoded Cu-Zn SOD demonstrated to be enzymatically active.
cvSOD is a late gene product.
Total RNAs were isolated from uninfected and the PBCV-1-infected host C. variabilis at several times after infection. The RNAs were denatured, displayed on a 1.5% agarose gel, transferred to a nylon membrane, and hybridized with a [32P]ATP-labeled, tcvsod gene probe. Hybridization signals first appeared at 60 min p.i., reached a peak at 180 min, and then decreased (see Fig. S2 in the supplemental material). Since the synthesis of PBCV-1 DNA begins at 60 to 90 min p.i. (43), the cvSOD-encoding gene a245r was expressed as a late gene. A microarray experiment also indicated that the a245r gene was expressed as a late gene (44). The hybridized RNA band was ∼1.3 kb in size (see Fig. S2), which was larger than expected for a gene encoding a 187-amino-acid peptide. Larger gene transcripts are not unusual for PBCV-1 genes, and some of them may represent polycistronic mRNAs in which adjacent genes are present in the same transcript (e.g., see reference 45).
cvSOD is packaged in the PBCV-1 virion.
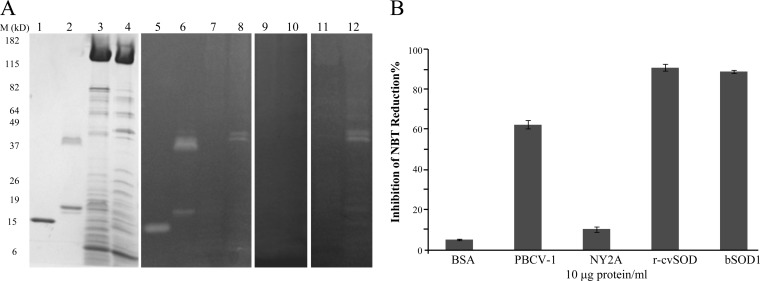
Late gene-encoded proteins are often packaged in virus particles, and so we examined purified PBCV-1 virions for SOD activity. This experiment also included chlorovirus NY-2A; NY-2A infects the same host as PBCV-1 but lacks a SOD-encoding gene. The purified viruses were suspended in 50 mM Tris-Cl, pH 7.8, and disrupted by sonication. After centrifugation, the supernatants were assayed for SOD activity by the PAGE assay. Ten micrograms of soluble virion protein was subjected to PAGE with BSA and recombinant tcvSOD serving as negative and positive controls, respectively. After NBT and riboflavin staining, the PBCV-1 sample generated two clear bands that migrated slightly more slowly than tcvSOD, suggesting that the native cvSOD has a slightly higher molecular mass (compare lanes 6 and 8 in Fig. 5A). One explanation is that the tcvSOD is a truncated protein lacking 22 amino acid residues at the N terminus, whereas the native protein consists of the entire 187 amino acid residues. Another possibility is that cvSOD might undergo posttranslational modification. Why the native cvSOD and the recombinant tcvSOD consistently produced two active bands on the gels is unknown, but the simplest explanation is that the higher-molecular-mass band is the dimeric form of the enzyme. No activity was detected in virus NY-2A, as expected.
FIG 5.
cvSOD activity detected in virion-associated proteins. (A) One microgram of virion protein was subjected to PAGE and stained with Coomassie brilliant blue or NBT with or without inhibitors. Lanes 1, 2, 3, and 4 are bovine SOD1, tcvSOD, NY-2A virion protein, and PBCV-1 virion protein, respectively; samples were stained with Coomassie brilliant blue. Lanes 5, 6, 7, and 8 are the same samples stained with NBT. Lanes 9 and 10 are NY-2A and PBCV-1 virion protein stained with NBT plus 50 mM NaCN, and lanes 11 and 12 are NY-2A and PBCV-1 virion proteins stained with NBT plus 50 mM NaN3. (B) cvSOD activity of virion protein measured in solution by the xanthine/xanthine oxidase assay.
The SOD activity observed on the gels was due to the PBCV-1-encoded enzyme and not a host enzyme for the following reasons. (i) The genome of C. variabilis, the host for both viruses, lacks a Cu-Zn SOD-encoding gene (46). (ii) The two activity bands were not the result of a host-encoded Fe or Mn SOD because of their sizes. (iii) The viral packaged cvSOD exhibited the same sensitivity to inhibitors as the recombinant protein; i.e., activity was inhibited by 50 mM NaCN (Fig. 5A, lane 10) but not by 50 mM NaN3 (Fig. 5A, lane 12). (iv) The NY-2A virion protein sample lacked SOD activity, which provides indirect evidence that the PBCV-1 activity was virus encoded. (v) Finally, a PBCV-1 proteome study reported that the cvSOD protein was packaged in the virion (27). Therefore, the SOD activity detected in the PBCV-1 virion is the virus-encoded protein and not from the host.
The virion particle-associated SOD activity was also measured by the xanthine-xanthine oxidase assay with BSA as a negative control and tcvSOD and bovine Cu-Zn SOD as positive controls. Compared to over 70% inhibition of NBT reduction by SODs from tissue homogenates (32), the PBCV-1 virion-associated Cu-Zn SOD produced only 60% inhibition, possibly due to the low number of cvSOD molecules packaged in the virion. The inhibition of NBT reduction by both tcvSOD and bovine Cu-Zn SOD was over 90%, while NY-2A virion-associated activity was similar to that of the BSA negative control (Fig. 5B).
cvSOD may be involved in scavenging host-generated superoxide.
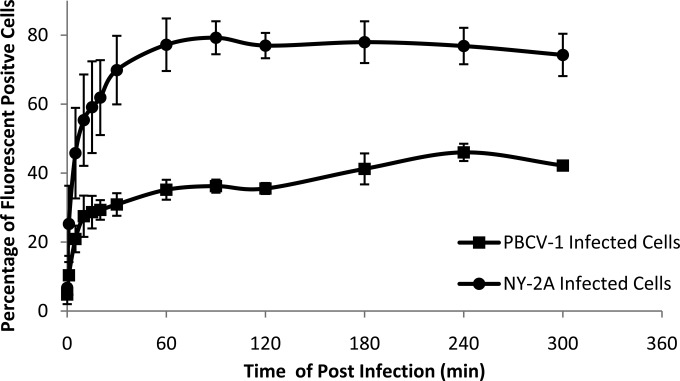
The discovery that active cvSOD is packaged in the virion suggests that it performs some early role(s) during virus infection. This assumption prompted us to examine intracellular superoxide levels during PBCV-1 infection. Chlorovirus NY-2A was chosen for comparison because it lacks a SOD gene. C. variabilis cells were incubated with 5 μM DHE for 1 h and then infected with viruses at an MOI of 10. DHE is a cell-permeative superoxide indicator that forms DNA-binding fluorophore ethidium bromide or a structurally similar product after oxidation by superoxide that can be monitored by flow cytometry (1). Superoxide accumulated rapidly in both PBCV-1- and NY-2A-infected cells in the first 30 min p.i., but only about 25 to 35% of the PBCV-1-infected cells fluoresced, and this number increased to 35 to 45% at 2 h p.i. (Fig. 6), possibly due to disruption of photosynthesis and damage to cellular organelles that resulted in higher superoxide accumulation (47). In contrast, 70 to 80% of NY-2A-infected cells were fluorescent at 2 h p.i., and this level remained stable until the end of the experiment (5 h). The lower percentage of fluorescent cells infected by PBCV-1 suggests that virion-associated cvSOD enters the host cell together with viral genomic DNA and other virion-associated proteins and scavenges intracellular superoxide anions to benefit virus replication. Due to the absence of cvSOD, NY-2A may encounter difficulties in its replication, and this might be one of the reasons why NY-2A has a smaller burst size and a 15- to 18-h replication cycle, 2.5 times longer than that of PBCV-1 (48).
FIG 6.
Measurement of superoxide levels in PBCV-1- and NY-2A virus-infected C. variabilis cells. C. variabilis cells (4 × 108/ml) were mixed with 1 μl of 5 mM DHE to a final concentration of 5 μM and incubated for 1 h by rolling to let the dye penetrate the cells. (DHE forms a red fluorescent product when it reacts with superoxide anions.) The cells were then infected with either PBCV-1 or NY-2A at an MOI of 10 and analyzed by flow cytometry. cvSOD is encoded by PBCV-1 and packaged in its capsid, while NY-2A does not encode a cvSOD.
These results are consistent with the hypothesis that PBCV-1 cvSOD is transferred into the host, together with viral genomic DNA and several other virion proteins, during virus infection; the cvSOD could then begin to scavenge ROS as infection begins. PBCV-1 also encodes other enzymes that may be involved in reducing ROS concentrations, including a thioredoxin, glutaredoxin, thiol oxidoreductase, and a protein disulfide isomerase. The thioredoxin, glutaredoxin, and protein disulfide isomerase are also packaged in the virion (27). In contrast to PBCV-1, chlorovirus NY-2A, which lacks an SOD protein, is presumably confronted with higher concentrations of superoxide anions. Unfortunately, we currently do not have the technology to specifically remove or inactivate the PBCV-1 cvsod gene to directly test this hypothesis.
Cu-Zn SOD-encoding genes exist in additional viruses that infect C. variabilis.
To determine whether the cvSOD gene is common in the chloroviruses, viral genomic DNAs were prepared from 44 different chloroviruses that infect C. variabilis. The chloroviruses were isolated from diverse geographical locations worldwide (49) and hybridized with the 32P-labeled PBCV-1 tcvsod gene probe. The probe hybridized strongly to 37 of the 44 viruses (see Fig. S3 in the supplemental material). DNA from 7 viruses (Ar158, KS-1B, MA-1D, NY-2A, NY-2B, NYs-1, and XZ-3A) did not hybridize at all or hybridized poorly with the cvsod probe, suggesting that the gene is absent in their genomes. Six of these 7 viral genomes have been sequenced and annotated recently, and they all lack a cvsod gene (50, 51). Interestingly, all 7 of these viruses form small plaques, 0.5 to 1.0 mm, compared to the 3-mm plaques of PBCV-1 and many other viruses that infect C. variabilis (52). Presumably, like NY-2A, these viruses have smaller burst sizes and/or longer replication cycles than PBCV-1, suggesting that these viruses might encounter difficulties during replication; inefficient suppression of the elevated ROS levels in the host intracellular environment might be due to the absence of a cvSOD.
We recently sequenced 28 additional chloroviruses that infect two hosts other than those infected by PBCV-1 (51). Ten of the 13 viruses that infect Chlorella heliozoae have a Cu-Zn SOD gene, and 14 of the 15 viruses that infect Micractinium conductrix also have the gene. Thus, in total, 61 of the 72 chloroviruses examined contain a cvsod gene, including members classified in the genus Chlorovirus that infect all three algal species, suggesting that cvSOD presumably benefits virus replication, even if a few NC64A viruses replicate without the gene.
Thirty-five of the 61 viral genomes encoding a Cu-Zn SOD have been sequenced, and the sizes of the predicted Cu-Zn SOD proteins range from 169 amino acid residues to 187 residues, all of which are larger than typical Cu-Zn SODs (∼152 amino acids). (In total, 85 amino acid residues are conserved among the 35 cvSODs.) Each of these 35 cvSODs has an internal Met at its N terminus; using this Met as the starting codon results in proteins of 154 to 167 amino acids. Since all of the chlorovirus encoded Cu-Zn SODs have this ∼20-amino-acid extension at the N-terminal end, this domain may perform some unknown function.
As noted in the introduction, Chloroviruses members comprise one genus of viruses in the family Phycodnaviridae. The family includes 6 genera classified primarily by host range, which is supported by sequence comparisons. Members in the Chlorovirus genus infect freshwater algae, while most of the viruses in the other 5 genera infect marine eukaryotic algae (53). Other phycodnaviruses, including Micromonas pusilla virus and Emiliania huxleyi virus, also contain putative Cu-Zn SOD-encoding genes.
Phylogenetic analysis of cvSOD.
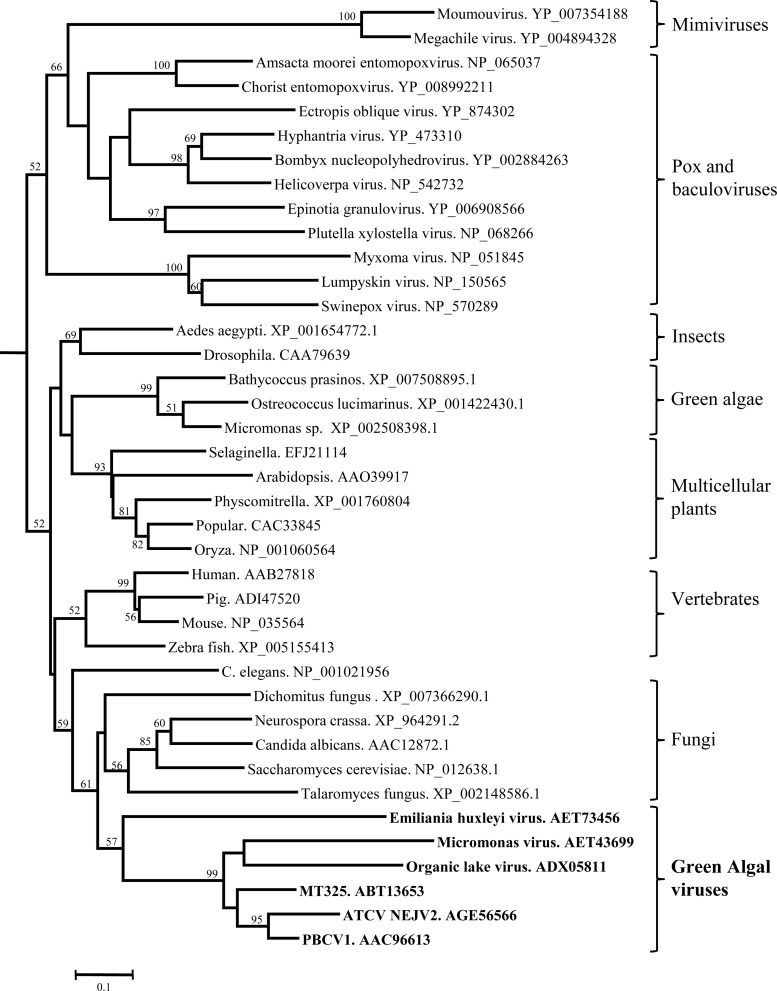
Phylogenetic analyses produced a surprise in that the Cu-Zn SODs from the phycodnaviruses formed a distinct clade from the Cu-Zn SOD-like proteins encoded by the other large dsDNA viruses (Fig. 7). BioNJ (distance method), maximum likelihood, and maximum parsimony tree construction methods were used. BioNJ and maximum likelihood methods produced nearly identical tree topologies. Maximum parsimony produced a polytomy, but the clade groupings bracketed on the right in Fig. 7 were the same. The Cu-Zn SOD homologs from the baculoviruses, poxviruses, and mimiviruses formed their own clade, whereas the phycodnaviruses, including the chloroviruses, Micromonas viruses, and the E. huxleyi virus, were in a separate clade. The group phylogenetically closest to the phycodnaviruses was from fungi. Indeed, when the cvSOD sequence from PBCV-1 (NP_048593.1) was blasted against the nonredundant database that excluded all viruses (http://www.ncbi.nlm.nih.gov/Blast.cgi), the best hits were from fungi. The Cu-Zn SOD sequence from PBCV-1 is 187 amino acids in length, similar to those in some fungi, but other fungi such as Aspergillus and Neurospora lack the 30+ amino acids at the N terminus. These results indicate a phylogenetic relationship between fungi and the phycodnaviruses and suggest that an ancient horizontal gene transfer event occurred. Indeed, we have long suspected that PBCV-1 has had, or may still have, a host(s) in nature other than just the green alga C. variabilis.
FIG 7.
Phylogram of Cu/Zn superoxide dismutases. A BLAST analysis was conducted (http://www.ncbi.nlm.nih.gov/Blast.cgi) using cvSOD from PBCV-1 (accession number NP_048593.1). While some green algae have Cu-Zn SODs, the PBCV-1 host, C. variabilis (ADIC01003810.1), does not. Excluding viruses, the best BLAST hits to cvSOD were from fungi. Phylogenetic analyses were conducted at http://www.phylogeny.fr (36). ClustalW was used to align the sequences. Positions with gaps were removed during alignment curation. The figure was produced by BioNJ (distance method). BioNJ and maximum likelihood methods produced nearly identical tree topologies. Maximum parsimony produced a polytomy, but the clade groupings bracketed on the right were the same. The values on the branches are the percentages of bootstrap support (200 replicates). Only bootstrap values >50% are shown.
Conclusions.
Many, but not all, chloroviruses encode a Cu-Zn SOD; however, viruses that encode the enzyme have an extra ∼35 amino acids located at the N-terminal end of the protein compared to most Cu-Zn SODs. The chlorovirus PBCV-1 protein has an internal Met located at residue 22, and this site was used to produce a recombinant Cu-Zn SOD that has all of the enzymatic properties of Cu-Zn SOD enzymes from cellular organisms. To our knowledge, this is only the second virus-encoded Cu-Zn SOD to be demonstrated to be enzymatically active. The fact that the PBCV-1 enzyme is active is probably not surprising because phylogenetically, the enzyme groups with Cu-Zn SODs from most cellular organisms. In contrast, the Cu-Zn SOD-like homologs encoded by the other 3 groups of large dsDNA viruses, poxviruses, baculoviruses, and mimiviruses, group into a separate clade. The PBCV-1 enzyme is packaged in the virion and is very likely involved in reducing ROS during the early phases of virus infection because chlorovirus NY-2A, which lacks a Cu-Zn SOD, induces a much higher level of ROS during the early stages of infection. We propose that as a consequence, NY-2A has a longer replication cycle than PBCV-1 (6 to 8 h versus 18 h or more) and also a smaller burst size.
Supplementary Material
ACKNOWLEDGMENTS
This investigation was supported in part by National Institutes of Health grants GM32241 (J.L.V.E.), GM079393 (D.F.B.), and P30GM103335 and grant P20-RR15635 from the COBRE program of the National Center for Research Resources (J.L.V.E.), as well as grants from the National Center for Research Resources (5P20RR016469) and the National Institute for General Medical Science (NIGMS) (8P20GM103427) (G.A.D.). This work was also supported by National Science Foundation grant DBI-0619764 (D.F.B.).
Special thanks are due to David Silverman at the University of Florida for help with the stopped-flow spectrophotometry experiments and Daniel Snow at the University of Nebraska—Lincoln Water Science Center for the Cu and Zn measurements of cvSOD samples.
Footnotes
Published ahead of print 20 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02031-14.
REFERENCES
- 1.Tarpey MM, Wink DA, Grisham MB. 2004. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am. J. Physiol. Regul. Integr. Comp. Physiol. 286:R431–R444. 10.1152/ajpregu.00361.2003. [DOI] [PubMed] [Google Scholar]
- 2.Miller AF. 2012. Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 586:585–595. 10.1016/j.febslet.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buettner GR. 2011. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med. Chem. 11:341–346. 10.2174/187152011795677544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apel K, Hirt H. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373–399. 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 5.Miao L, Clair DKS. 2009. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med. 47:344–356. 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridovich I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64:97–112. 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 7.Zelko IN, Mariani TJ, Folz RJ. 2002. Superoxide dismutase multigene family: a comparison of the Cu/Zn SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33:337–349. 10.1016/S0891-5849(02)00905-X. [DOI] [PubMed] [Google Scholar]
- 8.Huang CH, Kuo WY, Weiss C, Jinn TL. 2012. Copper chaperone-dependent and independent activation of three copper-zinc superoxide dismutase homologs localized in different cellular compartments in Arabidopsis. Plant Physiol. 158:737–746. 10.1104/pp.111.190223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowler C, Van Montagu M, Inze D. 1992. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43:83–116. [Google Scholar]
- 10.Fang FC, DeGroote MA, Foster JW, Baumler AJ, Ochsner U, Testerman T, Bearson S, Giard JC, Xu Y, Campbell G, Laessig T. 1999. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. U. S. A. 96:7502–7507. 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclere V, Boiron P, Blondeau R. 1999. Diversity of superoxide dismutases among clinical and soil isolates of Streptomyces species. Curr. Microbiol. 39:365–368. 10.1007/s002849900473. [DOI] [PubMed] [Google Scholar]
- 12.Almazán F, Tscharke DC, Smith GL. 2001. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J. Virol. 75:7018–7029. 10.1128/JVI.75.15.7018-7029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao JX, Teoh ML, Moon M, McFadden G, Evans DH. 2002. Leporipoxvirus Cu-Zn superoxide dismutase homologs inhibit cellular superoxide dismutase, but are not essential for virus replication or virulence. Virology 296:125–135. 10.1006/viro.2002.1383. [DOI] [PubMed] [Google Scholar]
- 14.Smith GL, Chan YS, Howard ST. 1991. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J. Gen. Virol. 72:1349–1376. 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 15.Becker MN, Greenleaf WB, Ostrov DA, Moyer RW. 2004. Amsacta moorei entomopoxvirus expresses an active superoxide dismutase. J. Virol. 78:10265–10275. 10.1128/JVI.78.19.10265-10275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teoh MLT, Walasek PJ, Evans DH. 2003. Leporipoxvirus Cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J. Biol. Chem. 278:33175–33184. 10.1074/jbc.M300644200. [DOI] [PubMed] [Google Scholar]
- 17.Teoh MLT, Turner PV, Evans DH. 2005. Tumorigenic poxviruses up-regulate intracellular superoxide to inhibit apoptosis and promote cell proliferation. J. Virol. 79:5799–5811. 10.1128/JVI.79.9.5799-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohrmann GF. 2013. The AcMNPV genome: gene content, conservation, and function. In Rohrmann GF. (ed), Baculovirus biology, 3rd ed. National Center for Biotechnology Information, Bethesda, MD: http://www.ncbi.nlm.nih.gov/pubmed/24479205. [Google Scholar]
- 19.Hu ZH, Arif BM, Sun JS, Chen XW, Zuidema D, Goldbach RW, Vlak JM. 1998. Genetic organization of the HindIII-I region of the single-nucleocapsid nucleopolyhedrovirus of Buzura suppressaria. Virus Res. 55:71–82. 10.1016/S0168-1702(98)00029-X. [DOI] [PubMed] [Google Scholar]
- 20.Tomalski MD, Eldridge R, Miller LK. 1991. A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology 184:149–161. 10.1016/0042-6822(91)90831-U. [DOI] [PubMed] [Google Scholar]
- 21.Lartigue A, Philippe N, Jeudy S, Abergel C. 2012. Preliminary crystallographic analysis of the Megavirus superoxide dismutase. Acta Crystallogr. F 68:1557–1559. 10.1107/S174430911204657X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang M, Graves M, Mehmel M, Moroni A, Gazzarrini S, Thiel G, Gurnon JR, Van Etten JL. 2004. Genetic diversity in chlorella viruses flanking kcv, a gene that encodes a potassium ion channel protein. Virology 326:150–159. 10.1016/j.virol.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Van Etten JL, Dunigan DD. 2012. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 17:1–8. 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734. 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleocytoplasmic large DNA viruses. Virus Res. 117:156–184. 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223. 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunigan DD, Cerny RL, Bauman AT, Roach JC, Lane LC, Agarkova K, Wulser K, Yanai-Balser GM, Gurnon JR, Vitek JC, Kronschnabel BJ, Jeannard A, Blanc G, Upton C, Duncan GA, McClung OW, Ma F, Van Etten JL. 2012. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J. Virol. 86:8821–8834. 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Etten JL, Meints RH, Burbank DE, Kuczmarski D, Cupples DA, Lane LC. 1981. Isolation and characterization of a virus from the intracellular green algae symbiotic with Hydra viridis. Virology 113:704–711. 10.1016/0042-6822(81)90199-9. [DOI] [PubMed] [Google Scholar]
- 29.Van Etten JL, Burbank DE, Kuczmarski D, Meints RH. 1983. Virus infection of culturable chlorella-like algae and development of a plaque assay. Science 219:994–996. 10.1126/science.219.4587.994. [DOI] [PubMed] [Google Scholar]
- 30.Van Etten JL, Burbank DE, Xia Y, Meints RH. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126:117–125. [DOI] [PubMed] [Google Scholar]
- 31.Oberley LW, Spitz DR. 1985. Quantitation of SOD, p 211–220 In Greenwald RA. (ed), CRC handbook of methods for oxygen radical species. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 32.Beauchamp C, Fridovich I. 1971. Superoxide dismutase: improved assays and assays applicable to acrylamide gels. Anal. Biochem. 44:276–287. 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 33.Regelsberger G, Laaha U, Dietmann D, Ruker F, Canini A, Grilli-Caiola M, Furtmuler PG, Jakopitsch C, Peschek GA, Obinger C. 2004. The iron superoxide dismutase from the filamentous cyanobacterium Nostoc PCC 7120. J. Biol. Chem. 279:44384–44393. 10.1074/jbc.M406254200. [DOI] [PubMed] [Google Scholar]
- 34.Valentine JS, Curtis ABA. 1975. Convenient preparation of solutions of superoxide anion and the reaction of superoxide anion with a copper(I1) complex. J. Am. Chem. Soc. 97:224–226. 10.1021/ja00834a058. [DOI] [PubMed] [Google Scholar]
- 35.Greenleaf WB, Perry JJ, Hearn AS, Cabelli DE, Lepock JR, Stroupe ME, Tainer JA, Nick HS, Silverman DN. 2004. Role of hydrogen bonding in the active site of human manganese superoxide dismutase. Biochemistry 43:7038–7045. 10.1021/bi049888k. [DOI] [PubMed] [Google Scholar]
- 36.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borders CL, Fridovich I. 1985. A comparison of the effects of cyanide, hydrogen peroxide, and phenylglyoxal on eukaryotic and prokaryotic Cu, Zn superoxide dismutase. Arch. Biochem. Biophys. 241:472–476. 10.1016/0003-9861(85)90572-7. [DOI] [PubMed] [Google Scholar]
- 38.Misra HP, Fridovich I. 1978. Inhibition of superoxide dismutase by azide. Arch. Biochem. Biophys. 189:317–322. 10.1016/0003-9861(78)90218-7. [DOI] [PubMed] [Google Scholar]
- 39.Chen JR, Weng CN, Ho TY, Cheng IC, Lai SS. 2000. Identification of the copper-zinc superoxide dismutase activity in Mycoplasma hyopneumoniae. Vet. Microbiol. 73:301–310. 10.1016/S0378-1135(99)00170-4. [DOI] [PubMed] [Google Scholar]
- 40.Goto JJ, Zhu H, Sanchez RJ, Nersissian A, Gralla EB, Valentine JS, Cabelli DE. 2000. Loss of in vitro metal ion binding specificity in mutant copper-zinc superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 275:1007–1014. 10.1074/jbc.275.2.1007. [DOI] [PubMed] [Google Scholar]
- 41.Klug D, Rabani J, Fridovich I. 1972. A direct demonstration of catalytic action of superoxide dismutase through use of pulse radiolysis. J. Biol. Chem. 247:4839–4842. [PubMed] [Google Scholar]
- 42.Stroppolo ME, Sette M, O'Neill P, Polizio F, Cambria MT, Desideri A. 1998. Cu,Zn superoxide dismutase from Photobacterium leiognathi is an hyperefficient enzyme. Biochemistry 37:12287–12292. 10.1021/bi980563b. [DOI] [PubMed] [Google Scholar]
- 43.Van Etten JL, Burbank DE, Joshi J, Meints RH. 1984. DNA synthesis in a chlorella-like alga following infection with the virus PBCV-1. Virology 134:443–449. 10.1016/0042-6822(84)90311-8. [DOI] [PubMed] [Google Scholar]
- 44.Yanai-Balser GM, Duncan GA, Eudy JD, Wang D, Li X, Agarkova IV, Dunigan DD, Van Etten JL. 2010. Microarray analysis of Paramecium bursaria chlorella virus 1 transcription. J. Virol. 84:532–542. 10.1128/JVI.01698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang M, Dunigan DD, Van Etten JL. 2005. Chlorovirus: a genus of Phycodnaviridae that infects certain chlorella-like green algae. Mol. Plant Pathol. 6:213–224. 10.1111/j.1364-3703.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- 46.Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL. 2010. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955. 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans C, Malin G, Mills GP, Wilson WH. 2006. Virus infection of Emiliania huxleyi (Prymnesiophyceae) leads to elevated production of reactive oxygen species. J. Phycol. 42:1040–1047. 10.1111/j.1529-8817.2006.00256.x. [DOI] [Google Scholar]
- 48.Van Etten JL, Schuster AM, Meints RH. 1988. Viruses of eukaryotic chlorella-like algae, p 411–428 In Koltin Y, Leibowitz MJ. (ed), Viruses of fungi and simple eukaryotes. Marcel Dekker Inc, New York, NY. [Google Scholar]
- 49.Sun L, Li Y, McCullough AK, Wood TG, Lloyd RS, Adams B, Gurnon JR, Van Etten JL. 2000. Intron conservation in a UV-specific DNA repair gene encoded by chlorella viruses. J. Mol. Evol. 50:82–92. 10.1007/s002399910009. [DOI] [PubMed] [Google Scholar]
- 50.Fitzgerald LA, Graves MV, Li X, Feldblyum T, Nierman WC, Van Etten JL. 2007. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 358:472–484. 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, Van Etten JL, Blanc G. 2013. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14:158. 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Etten JL, Lane LC, Meints RH. 1991. Viruses and virus-like particles of eukaryotic algae. Microbiol. Rev. 55:586–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson WH, Van Etten JL, Allen MJ. 2009. The Phycodnaviridae: the story of how tiny giants rule the world. Curr. Top. Microbiol. Immunol. 328:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.