ABSTRACT
The unfolded-protein response (UPR) is a signal transduction cascade triggered by perturbation of the homeostasis of the endoplasmic reticulum (ER). UPR resolves ER stress by activating a cascade of cellular responses, including the induction of molecular chaperones, translational attenuation, ER-associated degradation, and other mechanisms. Under prolonged and irremediable ER stress, however, the UPR can also trigger apoptosis. Here, we report that in cells infected with the avian coronavirus infectious bronchitis virus (IBV), ER stress was induced and the IRE1α-XBP1 pathway of UPR was activated. Knockdown and overexpression experiments demonstrated that IRE1α protects infected cells from IBV-induced apoptosis, which required both its kinase and RNase activities. Our data also suggest that splicing of XBP1 mRNA by IRE1α appears to convert XBP1 from a proapoptotic XBP1u protein to a prosurvival XBP1s protein. Moreover, IRE1α antagonized IBV-induced apoptosis by modulating the phosphorylation status of the proapoptotic c-Jun N-terminal kinase (JNK) and the prosurvival RAC–alpha serine/threonine-protein kinase (Akt). Taken together, the data indicate that the ER stress sensor IRE1α is activated in IBV-infected cells and serves as a survival factor during coronavirus infection.
IMPORTANCE Animal coronaviruses are important veterinary viruses, which could cross the species barrier, becoming severe human pathogens. Molecular characterization of the interactions between coronaviruses and host cells is pivotal to understanding the pathogenicity and species specificity of coronavirus infection. It has been well established that the endoplasmic reticulum (ER) is closely associated with coronavirus replication. Here, we report that inositol-requiring protein 1 alpha (IRE1α), a key sensor of ER stress, is activated in cells infected with the avian coronavirus infectious bronchitis virus (IBV). Moreover, IRE1α is shown to protect the infected cells from apoptosis by modulating the unfolded-protein response (UPR) and two kinases related to cell survival. This study demonstrates that UPR activation constitutes a major aspect of coronavirus-host interactions. Manipulations of the coronavirus-induced UPR may provide novel therapeutic targets for the control of coronavirus infection and pathogenesis.
INTRODUCTION
In eukaryotic cells, the endoplasmic reticulum (ER) is the major site where secreted and transmembrane proteins are synthesized and folded. When excessive amounts of protein enter the ER, unfolded proteins accumulate in the ER lumen and cause ER stress. To maintain homeostasis, signaling pathways collectively known as the unfolded-protein response (UPR) are activated (1). To date, three UPR sensors have been identified, namely, PKR-like ER protein kinase (PERK), activating transcriptional factor 6 (ATF6), and inositol-requiring protein 1 alpha (IRE1α). Activated PERK phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α) and results in a global shutdown of protein synthesis to reduce the protein flux into the ER (2). Activated ATF6 is cleaved twice to release a cytosolic fragment, which translocates to the nucleus and transactivates ER protein chaperones that enhance the ER folding capacity (3, 4).
The IRE1α-XBP1 branch of the UPR is evolutionarily conserved from yeast to humans. In response to unfolded proteins, IRE1α dissociates from ER protein chaperones and undergoes oligomerization (5). This results in the autophosphorylation of the kinase domain and activation of the RNase domain. The best-characterized substrate for the RNase domain is mRNA of X box binding protein 1 (XBP1) (6, 7). IRE1α removes a 26-nucleotide (nt) intron from XBP1 mRNA to form a frameshift transcript, the spliced XBP1 (XBP1s). While the unspliced XBP1 (XBP1u) mRNA encodes an inhibitor of the UPR, XBP1s mRNA encodes a potent transcription activator, which translocates to the nucleus and enhances the expression of many UPR genes, including those encoding molecular chaperones and proteins contributing to ER-associated degradation (ERAD) (8, 9).
If ER homeostasis is not reestablished, the UPR can induce apoptosis to eliminate the overly stressed cells. Apoptosis is a highly controlled mode of cell death characterized by cell shrinkage, plasma membrane blebbing, and nuclear fragmentation (10). Previously, ER stress-induced apoptosis was mainly attributed to the induction of C/EBP homologous protein (CHOP) (11). Recently, it has been demonstrated that the IRE1α branch is also involved in regulation of ER stress-induced apoptosis. Activated IRE1α has been found to be associated with tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2). This complex further recruits apoptosis signal-regulating kinase 1 (ASK1), which induces apoptosis by activating the mitogen-activated protein (MAP) kinase c-Jun N-terminal kinase (JNK) (12). In another study, IRE1α has been shown to promote clustering and activation of procaspase 12, which subsequently cleaves caspase 3 and induces apoptosis (13, 14).
Coronaviruses are enveloped viruses with a large, single-stranded, positive-sense RNA genome. Infectious bronchitis virus (IBV) is an avian gammacoronavirus that causes respiratory disease in chickens, resulting in a major economic burden to the poultry industry worldwide. During coronavirus infection, tremendous amounts of viral proteins are synthesized in the ER. Moreover, the replication and transcription complexes (RTCs), where coronavirus RNA synthesis occurs, originate from a reticular network of modified ER membranes (15, 16). The overloading of the ER folding capacity and extensive rearrangement of the ER membrane may cause ER stress and induce the UPR, as previously demonstrated in cells infected with mouse hepatitis virus (MHV) (17). Moreover, the envelope protein of severe acute respiratory syndrome coronavirus (SARS-CoV) has been shown to counteract the IRE1α-XBP1 pathway of UPR and inhibit SARS-CoV-induced apoptosis (18). However, the significance of the UPR in coronavirus-host interaction remains largely unexplored.
Previously, we showed that IBV induces apoptosis in late-stage infected cells and identified two Bcl-2 family proteins that modulate IBV-induced apoptosis (19–22). However, the mechanisms regulating this process remain largely unexplored. In this study, we focus on the UPR sensor IRE1α and its function in IBV-induced apoptosis. It was found that IBV induced ER stress in infected cells and activated the IRE1α-XBP1 pathway at a late stage of infection. Knockdown and overexpression studies showed that IRE1α protected infected cells from IBV-induced apoptosis, which required both the kinase and RNase domains of IRE1α. Splicing of XBP1 by IRE1α appears to convert it from a proapoptotic unspliced form to an antiapoptotic spliced form. Moreover, phosphorylation of the proapoptotic kinase JNK and the prosurvival kinase Akt was also modulated by IRE1α to promote cell survival during IBV infection. Taken together, our data demonstrate the important prosurvival function of the UPR sensor IRE1α during coronavirus infection.
MATERIALS AND METHODS
Virus and cell lines.
The egg-adapted Beaudette strain of IBV (ATCC VR-22) was obtained from the American Type Culture Collection (ATCC) and adapted to Vero cells as described previously (23). To prepare virus stock, monolayers of Vero cells were infected at a multiplicity of infection (MOI) of approximately 0.1 and cultured in plain Dulbecco modified Eagle medium (DMEM) at 37°C for 24 h. After three freeze-thaw cycles, the cell lysate was clarified by centrifugation at 1,500 × g at 4°C for 30 min. The supernatant was aliquoted and stored at −80°C as virus stock. The titer of the virus preparation was determined by plaque assays. Mock cell lysate was prepared by the same treatment of uninfected Vero cells.
Inactivation of IBV was performed by exposing the virus stock to 120,000 mJ/cm2 of 254-nm shortwave UV radiation for 15 min with a CL-1000 cross-linker (UVP) (24) (UV-inactivated IBV [UV-IBV]). To demonstrate that IBV had been inactivated, Vero cells were incubated with UV-IBV, and the cell lysates were analyzed by Western blotting to confirm that no viral proteins could be detected.
H1299 cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco). All cells were grown in a 37°C incubator supplied with 5% CO2. In all the experiments, cells were washed twice with phosphate-buffered saline (PBS) before being infected with IBV at an MOI of approximately 2 or incubated with an equal volume of UV-IBV in serum-free medium. After 2 h of adsorption, the cells were washed twice with serum-free medium and kept incubating at 37°C before being harvested.
Antibodies, chemicals, and reagents.
The antibodies against IRE1α (number 3294), PARP (number 9532), caspase 3 (number 9662), caspase 8 (number 9746), caspase 9 (number 9502), JNK (number 9258), phosphorylated JNK (phosphor-JNK) (number 4668), Akt (number 4691), and phosphor-Akt (number 4060) were purchased from Cell Signaling Technology. The antibody against enhanced green fluorescent protein (EGFP) was from Sigma. The antibody against β-actin (sc-1616) was from Santa Cruz Biotechnology. The antisera against IBV S protein and N protein were from rabbits immunized with bacterial expressed fusion proteins, as previously described (25, 26).
Dithiothreitol (DTT) was purchased from Sigma. The 1 M DTT stock was prepared by dissolving in autoclaved water and stored at −20°C. To induce ER stress, cells were treated with 2 mM DTT for 2 h before being harvested for RNA extraction.
Plasmid constructions and transfection.
The cDNA of human IRE1α (RefSeq NM_001433.3) was amplified from total RNA of H1299 cells by reverse transcriptase PCR (RT-PCR) using IRE1α-specific primers (forward primer, 5′-CGGGAATTCGGCCGAGTCCTCGCCATG-3′; reverse primer, 5′-CAAGCGGCCGCCTTTCCCAACTATCACCACGCT-3′). The PCR product was digested with EcoRI and NotI and inserted into pHA-C, which has a hemagglutinin (HA) tag coding sequence inserted between the NotI and XbaI sites in the parental construct, pcDNA3.1 (Invitrogen). The resulting plasmid was named pcDNA3.1-IRE1α-HA. The kinase-dead mutant K599A was generated using site-directed mutagenesis (forward primer, 5′-GACGTGGCCGTGGCGAGGATCCTCCCC-3′; reverse primer, 5′-GGGGAGGATCCTCGCCACGGCCACGTC-3′; mutated nucleotides are underlined). The mutant with RNase deleted was generated by amplifying a DNA fragment from pcDNA3.1-IRE1α-HA using specific primers (forward primer, 5′-CGGGAATTCGGCCGAGTCCTCGCCATG-3′; reverse primer, 5′-CAAGCGGCCGCCTTTCCCAACTATCACCACGCT-3′) and ligating it into the same sites. The cDNA of human XBP1, unspliced isoform (RefSeq NM_005080.3), was amplified from total RNA of H1299 cells by RT-PCR using XBP1u-specific primers (forward primer, 5′-GGAAGATCTGGAGCTATGGTGGTG-3′; reverse primer, 5′-CGGGGTACCTTAGTTCATTAATGGCTTCCAGC-3′). The cDNA of human XBP1, spliced isoform (RefSeq NM_001079539.1), was amplified from total RNA of H1299 cells treated with 2 mM DTT for 2 h, using XBP1s-specific primers (forward primer, 5′-GGAAGATCTGGAGCTATGGTGGTG-3′; reverse primer, 5′-CGGGGTACCTTAGACACTAATCAGCTGGGG-3′). The PCR products of both XBP1u and XBP1s were digested with BglII and KpnI and inserted into pEGFP-C1 (Clontech). The forward primer 5′-GGAAGATCTGGAGCTATGGTGGTG-3′ and the reverse primer, 5′-CGGGGTACCTTATACCGCCAGAATCCATGGGGAGATG-3′ were used to amplify the coding sequence of the dominant-negative form of XBP1 (XBP1-DN), which was inserted between the BamHI and KpnI sites in the vector pXJ40-FLAG. The expression plasmid for constitutively active Akt, pcDNA-myr-AKT1, was a generous gift from Jean-Ehrland Ricci, as described previously (27).
Transfection of plasmid DNA into H1299 cells was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Briefly, H1299 cells were plated on a 12-well plate the day before transfection. For each well, 0.8 μg plasmid DNA and 2 μl Lipofectamine 2000 were each diluted with 100 μl RPMI and incubated for 5 min. Then, the diluted plasmid and transfection reagent were mixed by brief vortexing and incubated for another 20 min. The culture medium was exchanged with 800 μl RPMI containing 5% FBS, and the transfection mixture was added dropwise to each well. The cells were incubated at 37°C for 6 to 8 h before the medium was replaced with complete medium. At 24 h posttransfection, the cells were infected with IBV at an MOI of 2 or mock infected, and incubation was continued until cells were harvested for protein and/or RNA analysis at the indicated time points.
RNA interference.
IRE1α siRNA (+) (5′-GGACGUGAGCGACAGAAUAdTdT-3′), XBP1 siRNA (+) (5′-ACAGCAAGUGGUAGAUUUAdTdT-3′), JNK1/2 siRNA (+) (5′-AAAGAAUGUCCUACCUUCUdTdT-3′), and control EGFP siRNA (+) (5′-GCUGACCCUGAAGUUCAUCdTdT-3′) were purchased from Sigma (28, 29). Transfection of small interfering RNA (siRNA) was performed using DharmaFECT2 transfection reagent (Dharmacon Thermo Fisher Scientific) according to the manufacturer's instructions. At 48 h posttransfection, cells were infected with IBV at an MOI of 2 or mock infected, and incubation was continued until cells were harvested for protein and/or RNA analysis at the indicated time points.
RNA extraction and RT-PCR analysis.
Total RNA from cultured cells was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Briefly, cells were lysed with 1 ml TRIzol per 10-cm2 effective growth area, and the lysates were mixed with one-fifth volume of chloroform. After centrifugation at 12,000 × g at 4°C for 15 min, the aqueous phase was mixed with an equal volume of isopropanol. RNA was pelleted by centrifugation at 12,000 × g at 4°C for 15 min, washed with 70% ethanol twice, and dissolved in RNase-free H2O. The concentration of the total RNA was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific).
cDNA was reverse transcribed from total RNA using oligo(dT) and the ImProm-II Reverse Transcription System (Promega) according to the manufacturer's instructions. The following primers (forward and reverse) were used for PCR: IRE1α, GAAAAGGAATCCCTGGATGG and TCAGAGGGCGTCTGGAGTC; XBP1, CAGCGCTTGGGGATGGATGC and GGGCTTGGTATATATGTGG; ERdj4, GATACACTTGGACACAGTGC and CTACTGTCCTGAACAGTCAG; EDEM1, CTACCAGGCAACCAAGAATC and CCAACCATCTGGTCAATCTG; P58IPK, GGCTCGGTATTCCCCTTCCT and AGTAGCCCTCCGATAATAAGCAA; HERPUD1, GGACCTATTCAGCAGCTACA and ATCAGTTTGCGATGGCTGGG; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), GGGCTCATCTGAAGGGTGGTGC and GGACGCTGGGATGATGTTCTGG; IBV genomic RNA (gRNA), GAGTAACATAATGGACCTGT and TGCTGTACCCTCGATCGTAC; and IBV subgenomic RNA2 (sgRNA2), CTATTACACTAGCCTTGCGCTAGA and CTCTGGATCCAATAACCTAC. The PCR products were resolved using a 1% agarose gel prestained with ethidium bromide and visualized under UV shadowing. The band intensities of specific genes were determined by densitometry using ImageJ software and normalized to the intensities of the corresponding GAPDH bands. To resolve the unspliced and spliced forms of XBP1, the PCR products were resolved using a 4% agarose gel. The percentage of XBP1 splicing was calculated as the intensity of XBP1s divided by the total intensities of XBP1u and XBP1s.
Real-time RT-PCR was performed using a SYBR Select PCR kit (Life Technologies) in an Applied Biosystems 7500 real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. The mRNA levels of specific genes were calculated using GAPDH as an internal reference and normalized to 0-h samples (in time course experiments) or siEGFP-transfected samples (in siRNA knockdown experiments). The following real-time PCR primers (forward and reverse) were used: IRE1α, CGGGAGAACATCACTGTCCC and CCCGGTAGTGGTGCTTCTTA; total XBP1, TTGTCACCCCTCCAGAACATC and TCCAGAATGCCCAACAGGAT; spliced XBP1, TGCTGAGTCCGCAGCAGGTG and GCTGGCAGGCTCTGGGGAAG; ERdj4, TCTTAGGTGTGCCAAAATCGG and TGTCAGGGTGGTACTTCATGG; EDEM1, CGGACGAGTACGAGAAGCG and CGTAGCCAAAGACGAACATGC; P58IPK, GGCTCGGTATTCCCCTTCCT and AGTAGCCCTCCGATAATAAGCAA; and GAPDH, CCACTCCTCCACCTTTGAC and ACCCTGTTGCTGTAGCCA.
SDS-PAGE and Western blot analysis.
Cells were infected with IBV and harvested at the indicated time points using cell scrapers (Corning). After centrifugation at 16,000 × g for 1 min, the supernatant was discarded and the pellets were lysed in 1× RIPA buffer. After clarification by centrifugation and determination of the protein concentration with a spectrophotometer, the cell lysates were mixed with Laemmli sample buffer containing 100 mM dithiothreitol (30). The protein samples were boiled at 90°C for 5 min and centrifuged at 16,000 × g for 5 min. Equal amounts of protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to 0.2-μm nitrocellulose membranes (Bio-Rad). After the nonspecific antibody binding sites were blocked with 5% skim milk in Tris-buffered saline (20 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing 0.1% Tween 20, the membranes were incubated with 1 μg/ml primary antibodies at 4°C overnight. After washing with Tris-buffered saline, the membranes were incubated with 1:2,000-diluted anti-mouse or anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Dako) at room temperature for 2 h. The membranes were washed, and the proteins were detected with a chemiluminescence detection kit (Amersham Biosciences) and medical X-ray films (Fujifilm) according to the manufacturer's instructions. The films were scanned as grayscale 8-bit images, and the densities of the bands were determined with the NIH software ImageJ.
Virus titration.
Cell-free supernatants of IBV-infected cells collected at different time points were clarified by centrifugation and 10-fold serially diluted using serum-free DMEM. The viral titers were determined by plaque assay. Briefly, 250 μl of diluted supernatants was applied to confluent monolayers of Vero cells in 6-well plates. The plates were agitated every 10 min to ensure proper coverage of the monolayers. After 2 h of adsorption, unbound viruses were removed, and the cells were washed twice with DMEM. Overlay medium (2 ml; 0.4% agarose in DMEM) was added to each well, and the plates were incubated at 37°C for 2 days before plaques formed. The agarose overlay was removed, and the cells were fixed with 4% formaldehyde before staining with crystal violet. Finally, the plaques were counted, and the titers of individual samples were expressed as the logarithm of plaque-forming units (PFU) per milliliter. Each sample was titrated in triplicate in each experiment.
RESULTS
IBV infection activates the IRE1α-XBP1 pathway.
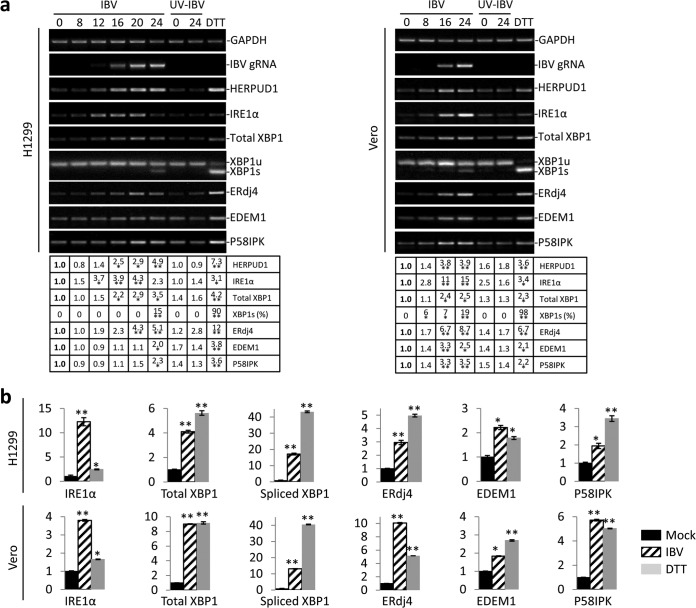
Our previous studies have shown that the PERK branch of the ER stress response is activated in cells infected with IBV at an early stage of infection (31, 32). To determine whether IBV infection also modulates the IRE1α pathway, H1299 cells were either infected with IBV at an MOI of ∼2 or incubated with UV-IBV. Cells treated with DTT, a strong ER stress inducer, were included as a positive control. Total RNAs were extracted and subjected to semiquantitative RT-PCR analysis. The IBV genomic RNA could be detected from 12 h postinfection (p.i.) and accumulated till the end of infection (Fig. 1a). RT-PCR analysis of HERPUD1, a component of the ERAD pathway and a commonly used ER stress marker (33, 34), showed significant increase at the mRNA level in cells infected with IBV and treated with DTT, but not in cells incubated with UV-IBV (Fig. 1a). Therefore, similar to other coronaviruses studied, IBV infection induces potent ER stress. Determination of the mRNA levels of components in the IRE1α-XBP1 pathway of the UPR showed stable increase of IRE1α through the course of infection with slight reduction at 24 h p.i., possibly due to extensive cell death at a late stage of infection (Fig. 1a). The total amount of XBP1 mRNA also gradually accumulated over time, whereas both IRE1α and XBP1 mRNA remained relatively unchanged in cells incubated with UV-IBV (Fig. 1a).
FIG 1.
Activation of the IRE1α-XBP1 pathway by IBV infection. (a) IBV infection causes ER stress and partially activates the IRE1α-XBP1 pathway in H1299 cells and Vero cells. H1299 cells (left) or Vero cells (right) were infected with IBV (MOI, ∼2) or incubated with UV-IBV and harvested at the time points indicated at the top (hours). As a positive control, H1299 cells were treated with 2 mM DTT for 2 h. Total RNA was extracted and subjected to RT-PCR using primer pairs specific for the indicated genes. The PCR products were resolved using 1% agarose gel electrophoresis, except for XBP1u/XBP1s, where a 4% agarose gel was used. The band intensities of HERPUD1, IRE1α, total XBP1, EDEM1, and ERdj4 were determined by densitometry and normalized to the intensities of the corresponding GAPDH bands. The percentage of XBP1 splicing [XBP1s (%)] was calculated as the intensity of XBP1s divided by the total intensities of XBP1u and XBP1s. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the 0-h p.i. sample (*, P < 0.05; **, P < 0.01). (b) H1299 cells and Vero cells were infected with IBV, mock infected for 20 h, or treated with DTT as for panel a. Total RNA was extracted and subjected to real-time RT-PCR analysis. The fold inductions of specific genes were calculated using GAPDH as an internal reference and normalized to the mock-infected samples. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the mock-treated sample (*, P < 0.05; **, P < 0.01). The error bars indicate standard deviations.
During ER stress, the activated IRE1α mediates splicing of the XBP1 mRNA by removing a 26-nt intron, as previously described (6). A moderate degree of XBP1 splicing (∼15%) was detected in IBV-infected H1299 cells at 24 h p.i., compared to ∼90% splicing in cells treated with DTT (Fig. 1a). No XBP1 splicing was observed in cells incubated with UV-IBV (Fig. 1a). ERdj4, EDEM1, and P58IPK are known to be specifically induced by the spliced form of XBP1 (9). Significant induction of all three genes was observed in cells treated with DTT. Considerable increase of ERdj4 mRNA was detected in IBV-infected cells, but only moderate induction of P58IPK and EDEM1 by IBV infection was observed (Fig. 1a). Since ERdj4 has been shown to be specifically induced by XBP1s, it was interesting that ERdj4 was induced in the IBV-infected cells in the absence of detectable spliced XBP1 at early time points (8 to 20 h). The same time course experiment was performed in Vero cells. Similarly, the mRNA levels of HERPUD1, IRE1α, XBP1, EDEM1, P58IPK, and ERdj4 were upregulated in IBV-infected Vero cells, and a moderate level of XBP1 splicing was observed at 24 h p.i. (Fig. 1a, right).
To confirm the semiquantitative RT-PCR results, real-time RT-PCR was performed. As shown in Fig. 1b, IBV infection induced an ∼12-fold increase in IRE1α mRNA and an ∼4-fold increase in total XBP1 mRNA in H1299 cells at 20 h p.i. The mRNA level of XBP1s was determined using a pair of primers previously validated to amplify only the spliced variant (35). Compared with a mock-infected control, IBV infection induced an ∼18-fold increase in XBP1s mRNA. Consistently, ERdj4, EDEM1, and P58IPK mRNA levels increased by ∼2- to 3-fold in IBV-infected cells compared with the mock-infected control (Fig. 1b). The same pattern was also observed in Vero cells, where IBV infection induced significant upregulation of IRE1α, total XBP1, XBP1s, ERdj4, and P58IPK (Fig. 1b, bottom). Taken together, the data indicate that IBV infection causes ER stress in the infected cells and activates the IRE1α-XBP1 pathway.
Knockdown of IRE1α and XBP1 attenuates IBV-induced activation of the IRE1α-XBP1 pathway.
To confirm that the IRE1α-XBP1 pathway is induced in IBV-infected cells, we used siRNA to specifically knock down IRE1α or XBP1 in H1299 cells. Vero cells were not chosen for the knockdown experiments due to their very low transfection efficiency. As shown in Fig. 2a, H1299 cells were transfected with siRNAs targeting IRE1α, XBP1, or EGFP (as a negative control) before being treated with DTT. As expected, the mRNA levels of IRE1α, total XBP1, XBP1s, and ERdj4 significantly increased in the siEGFP-transfected DTT-treated cells compared with a mock-treated control. Transfection of siIRE1α significantly reduced DTT-induced upregulation of IRE1α and XBP1 splicing by ∼50%. On the other hand, transfection of siXBP1 drastically reduced the mRNA level of total XBP1 and spliced XBP1 induced by DTT by ∼70%. Moreover, knockdown of either IRE1α or XBP1 also reduced DTT-induced ERdj4 upregulation by 30 to 40%. This suggested that transfection of the siRNAs specifically reduced the mRNA levels of target genes and downregulated the activation of the IRE1α-XBP1 pathway triggered by DTT.
FIG 2.
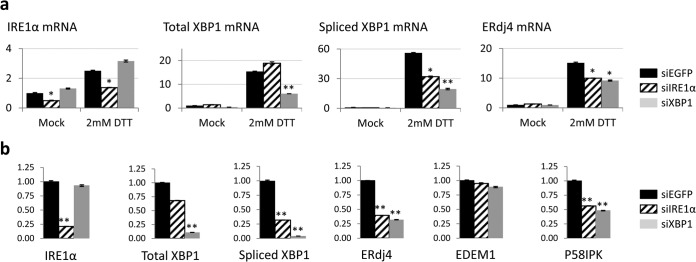
Effects of IRE1α and XBP1 knockdown on the activation of the IRE1α-XBP1 pathway. (a) H1299 cells were transfected with siEGFP, siIRE1α, or siXBP1 before being treated with 2 mM DTT or mock treated with the same volume of solvent control for 2 h. Total RNA was extracted and subjected to real-time RT-PCR analysis. The fold induction of specific genes was calculated using GAPDH as an internal reference and normalized to the siEGFP-transfected mock-treated sample. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the siEGFP-transfected samples for the same treatment (*, P < 0.05; **, P < 0.01). (b) H1299 cells were transfected with siRNAs before being infected with IBV (MOI, ∼2) for 20 h. Total-RNA extraction, real-time RT-PCR, and data analysis were performed as for panel a. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the siEGFP-transfected sample (**, P < 0.01). The error bars indicate standard deviations.
We then infected the siRNA-transfected cells with IBV, harvested total RNA at 20 h p.i., and compared the mRNA levels of genes related to the IRE1α-XBP1 pathway using RT-quantitative PCR (qPCR). As shown in Fig. 2b, transfection with siIRE1α and siXBP1 resulted in ∼75% and ∼90% knockdown efficiencies in the endogenous IRE1α and total XBP1 mRNA levels, respectively. Compared with the DTT treatment experiment, the higher knockdown efficiency could be a result of the longer incubation time after siRNA transfection before cell harvest. Notably, whereas siXBP1 nearly completely abolished the IBV-induced upregulation of XBP1s, siIRE1α also reduced the level of XBP1s mRNA by ∼75% (Fig. 2b). This is not surprising, because IRE1α is required for efficient splicing of XBP1 in cells under ER stress. Knockdown of IRE1α or XBP1 significantly reduced IBV-induced upregulation of ERdj4 and P58IPK by more than 50%, but the mRNA level of EDEM1 was only minimally affected (Fig. 2b). Therefore, IBV infection indeed activates the IRE1α-XBP1 pathway, and the IBV-induced upregulation of downstream ERdj4 and P58IPK genes was dependent on both IRE1α and XBP1.
Knockdown of IRE1α potentiates IBV-induced apoptosis in infected cells.
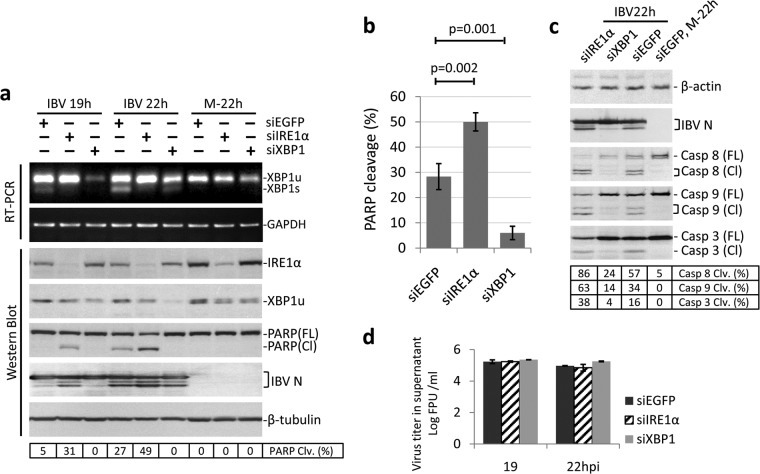
One outcome for cells under ER stress is the activation of caspase-dependent apoptosis, and IRE1α has been previously demonstrated to mediate ER stress-induced apoptosis (12–14). We next looked at the roles of IRE1α and XBP1 in IBV-induced apoptosis. As shown in Fig. 3a, H1299 cells were transfected with siIRE1α, siXBP1, and siEGFP before being infected with IBV or mock infected. The knockdown efficiencies were determined by Western blotting, although we were able to detect only the unspliced form and not the spliced form of XBP1. Successful knockdown of the two genes was also reflected by the effects on XBP1 splicing determined by RT-PCR. Consistent with the real-time PCR results (Fig. 2b), whereas knockdown of IRE1α reduced the mRNA level of spliced XBP1, knockdown of XBP1 depleted the mRNAs of both XBP1u and XBP1s. Poly(ADP-ribose) polymerase (PARP), a well-characterized apoptosis marker and a cleavage target of caspase 3, was used to monitor apoptosis. In cells transfected with siEGFP, significant PARP cleavage (∼27%) was detected at 22 h p.i. Interestingly, in IRE1α knockdown cells, PARP cleavage could be detected at an earlier time point (∼19 h p.i.). Moreover, at 22 h p.i., more prominent PARP cleavage (∼49%) was observed in IBV-infected IRE1α knockdown cells than with the siEGFP control (Fig. 3a). Surprisingly, knockdown of XBP1 did not result in a similar phenotype. Indeed, in XBP1 knockdown cells, no significant PARP cleavage could be detected throughout the course of infection (Fig. 3a). The same experiment was performed multiple times, and the observed effects of gene knockdown on IBV-induced PARP cleavage were reproducible and statistically significant (Fig. 3b).
FIG 3.
Effects of IRE1α and XBP1 knockdown on IBV-induced apoptosis. (a) Effects of IRE1α and XBP1 knockdown on IBV-induced PARP cleavage. H1299 cells in duplicate were transfected with siIRE1α, siXBP1, or siEGFP. At 48 h posttransfection, the cells were infected with IBV (MOI, ∼2) or mock infected. One set of cells was harvested at the indicated time points and subjected to Western blot analysis using antibodies against IRE1α, XBP1, IBV N, and PARP. β-Tubulin was included as a loading control. The percentage of PARP cleavage [PARP Clv. (%)] was calculated as the intensity of cleaved PARP [PARP(Cl)] divided by the total intensities of full-length PARP [PARP(FL)] and PARP(Cl). In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for XBP1 and GAPDH. (b) Quantification of PARP cleavage in siRNA-transfected cells infected with IBV. The percentage of PARP cleavage in cells transfected with siIRE1α, siXBP1, or siEGFP and infected with IBV for 22 h was determined as for panel a. The bar chart shows the results from three independent experiments with standard deviations and P values. (c) Effects of IRE1α and XBP1 knockdown on IBV-induced caspase activation. The IBV-infected, 22-h p.i. protein samples and the siEGFP-transfected mock-infected protein sample from panel a were subjected to Western blot analysis using antibodies against IBV N, caspase 8, caspase 3, and caspase 9. β-Actin was included as a loading control. The percentage of caspase cleavage was calculated as the intensity of cleaved caspase divided by the total intensities of full-length and cleaved caspase. (d) The culture supernatants from IBV-infected samples in panel a were clarified by centrifugation and subjected to plaque assay analysis using a confluent monolayer of Vero cells. Virus titers are expressed as the logarithm of PFU per milliliter of supernatants. The experiment was repeated three times with similar results, and the result of one representative experiment is shown.
To further validate the results, we also determined the activation of caspases (caspases 3, 8, and 9) during IBV infection in the knockdown cells. Samples collected at 22 h p.i. and the siEGFP-transfected mock-infected sample shown in Fig. 3a were further probed with individual caspase antibodies. As shown in Fig. 3c, consistent with the PARP cleavage pattern, the percentages of cleavage of caspases 3, 8, and 9 were significantly higher in IRE1α knockdown cells and lower in XBP1 knockdown cells than in the siEGFP control. The knockdown of IRE1α or XBP1 did not affect IBV replication, as determined from the similar N protein levels (Fig. 3a) and the similar virus titers in the supernatant (Fig. 3d). Taken together, these data suggest that although IRE1α is not essential for IBV replication, it protects the infected cells from IBV-induced apoptosis.
The seemingly opposite effect of XBP1 knockdown on apoptosis was unexpected, since XBP1 is the main target of IRE1α-mediated splicing. However, it is well known that whereas XBP1s serves as a potent activator of downstream UPR genes, XBP1u is actually a negative regulator of UPR (36). Therefore, it is possible that XBP1u and XBP1s may also demonstrate opposite effects on IBV-induced apoptosis. Because the siXBP1 used in the experiments targets both XBP1u and XBP1s, it is difficult to elucidate the functions of individual isoforms. As shown below, when we shifted to the overexpression approach using wild-type and dominant-negative XBP1, it became apparent that XBP1u and XBP1s indeed exert opposite effects on IBV-induced apoptosis.
Overexpression of full-length IRE1α protects cells from IBV-induced apoptosis.
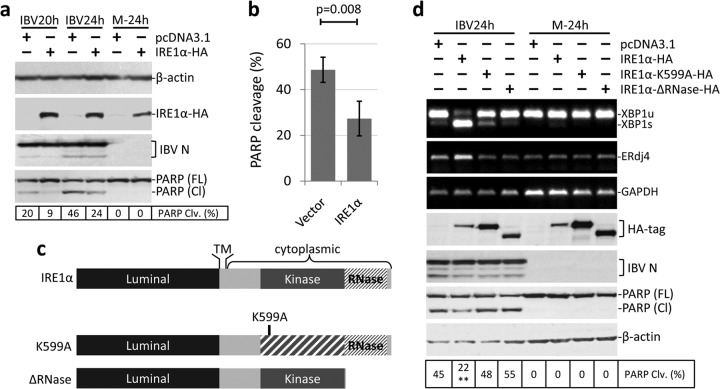
We next adopted the transient-overexpression approach to study the antiapoptotic activity of IRE1α. A plasmid encoding the full-length human IRE1α with an HA tag at the C terminus was constructed. H1299 cells were transfected with the construct or a vector control (pcDNA3.1) before being infected with IBV at an MOI of ∼2 or mock infected. As shown in Fig. 4a, the expression of IRE1α-HA was detected by Western blotting using antibodies against the HA tag. Transfection of IRE1α did not significantly affect the replication of IBV, as indicated by the similar level of N protein to that of the control (Fig. 4a). In the vector control, IBV induced prominent PARP cleavage at 20 h p.i. (∼20%) and 24 h p.i. (∼46%). However, in cells transfected with IRE1α-HA, IBV-induced PARP cleavage was partially reduced to ∼9% at 20 h p.i. and ∼24% at 24 h p.i. The experiment was performed multiple times, and the reduction of PARP cleavage by transfection of full-length IRE1α was reproducible and statistically significant (Fig. 4b). Thus, ectopic expression of IRE1α could indeed protect cells from IBV-induced apoptosis.
FIG 4.
Overexpression of full-length IRE1α protects cells from IBV-induced apoptosis. (a) H1299 cells were transfected with pcDNA3.1-IRE1α-HA or pcDNA3.1. At 24 h posttransfection, the cells were infected with IBV (MOI, ∼2) or mock infected. Cells were harvested at the indicated time points and subjected to Western blot analysis using antibodies against HA tag, IBV N, and PARP. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. (b) Quantification of PARP cleavage in plasmid-transfected cells infected with IBV. The percentage of PARP cleavage in cells transfected with pcDNA3.1 or pcDNA3.1-IRE1α-HA and infected with IBV for 24 h was determined as for panel a. The bar chart shows results from three independent experiments with standard deviations and P values. (c) Schematic diagrams showing the functional domains of IRE1 protein. A lysine-to-alanine mutation at K599 results in loss of IRE1 kinase activity. The RNase domain of IRE1 was deleted to generate the ΔRNase mutant. TM, transmembrane domain. (d) H1299 cells were transfected with pcDNA3.1-IRE1α-HA, pcDNA3.1-IRE1α-K599A-HA, pcDNA3.1-IRE1α-ΔRNase-HA, or pcDNA3.1 in duplicate. At 24 h posttransfection, cells were infected or mock infected as for panel a. In one set of cells, Western blot analysis was performed as for panel a. In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for XBP1, ERdj4, and GAPDH. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the pcDNA3.1-transfected 24-h p.i. sample (**, P < 0.01).
The IRE1α protein contains a lumenal domain that recognizes unfolded protein, a kinase domain that triggers autophosphorylation and dimerization, and an RNase domain that mediates XBP1 mRNA splicing (Fig. 4c). To determine which domain of IRE1α is required for its antiapoptotic activity, a kinase-dead mutant (K599A) and an RNase deletion mutant (ΔRNase) were generated (Fig. 4c). H1299 cells were transfected with vector, wild-type IRE1α, or the two mutants before being infected with IBV or mock infected for 24 h. As shown in Fig. 4d, the expression of transfected plasmids was determined by Western blotting. The protein level of wild-type IRE1α was lower than those of the two mutants, possibly because removal of the kinase or RNase activity reduced the basal activation and subsequent degradation of the protein, rendering it more stable. Transfection of wild-type IRE1α, but not the kinase-dead or RNase deletion mutant, significantly increased the IBV-induced XBP1 splicing and upregulation of ERdj4 compared with the vector control (Fig. 4d). As expected, overexpression of wild-type IRE1α partially reduced IBV-induced PARP cleavage. However, in IBV-infected cells transfected with the IRE1α mutants, PARP cleavage was indeed slightly higher than that of the vector control (Fig. 4d). IBV replication is not significantly affected by transfection of the constructs, as indicated by similar IBV N levels. Therefore, the results suggested that both the kinase and RNase activities of IRE1α are required for its antiapoptotic activity during IBV infection.
Overexpression of XBP1s, but not XBP1u, protects cells from IBV-induced apoptosis.
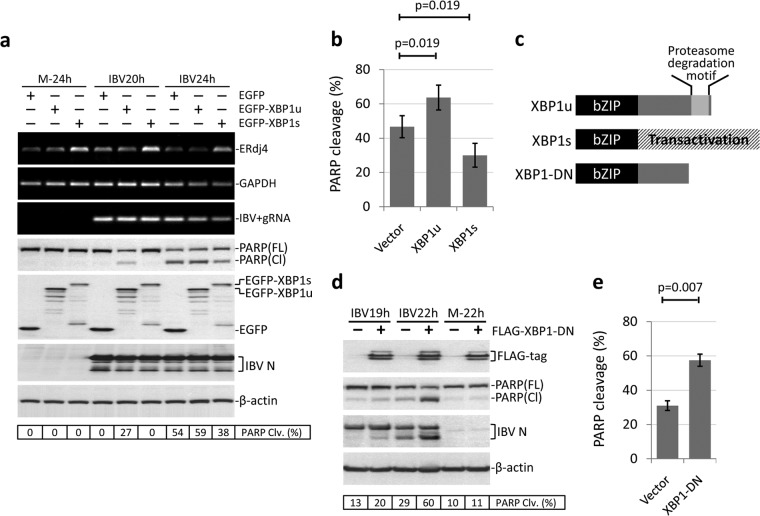
Because siRNA for XBP1 targets both the unspliced and the spliced forms, it is difficult to attribute the observed phenotype to individual isoforms. Therefore, we also adopted the overexpression approach to study the functions of XBP1u and XBP1s during IBV infection. Initially, the coding sequence of XBP1u or XBP1s was inserted into the C-terminal HA-tagged pcDNA3.1 vector as for the IRE1α constructs described above. However, when transfected into H1299 cells, the protein expression levels were very low for both XBP1u-HA and XBP1s-HA (data not shown). To enhance the expression level and monitor transfection efficiency, XBP1u and XBP1s were fused with an N-terminal EGFP tag (37). As shown in Fig. 5a, H1299 cells were transfected with EGFP-XBP1u, EGFP-XBP1s, or vector plasmid before being infected with IBV or mock infected. Using anti-EGFP antibody, the expression of both fusion proteins was clearly detectable. Many degradation bands could be detected for XBP1u, which was probably due to the proteosome degradation motif in the C-terminal region, as described previously (36). Transfection of EGFP-XBP1u or EGFP-XBP1s did not significantly affect the replication of IBV, as determined by the similar level of IBV N protein to that of the control (Fig. 5a). It was noted that the levels of IBV positive-stranded gRNA at 24 h p.i. in the EGFP-XBP1u- or EGFP-XBP1s-transfected cells seemed to be lower than that of the control, but the intensity ratios were actually very similar after being normalized to the GAPDH bands. Detection of similar amounts of IBV positive-stranded gRNA in all transfected cells at the same time point was also confirmed by real-time PCR analysis. Therefore, IBV replication was not affected by the overexpression of EGFP-XBP1u or EGFP-XBP1s. As shown in Fig. 5a, transfection of EGFP-XBP1s, but not EGFP-XBP1u, significantly enhanced the IBV-induced upregulation of ERdj4, indicating that the ectopically expressed XBP1s had normal function as a potent UPR transcription factor. Compared with the vector control, in cells transfected with EGFP-XBP1u, a low level of PARP cleavage could be detected at an early time point (20 h p.i.), and a slightly higher level of PARP cleavage was also observed at 24 h p.i. This suggested that XBP1u might function as a weak proapoptotic protein during IBV infection. In contrast, in cells transfected with EGFP-XBP1s, the level of IBV-induced PARP cleavage was lower than in the control at 24 h p.i. Although the difference was not great, it was observed in multiple experiments and was statistically significant (Fig. 5b). Therefore, unlike XBP1u, the XBP1s protein demonstrated antiapoptotic characteristics during IBV infection.
FIG 5.
Overexpression of XBP1s, but not XBP1u, protects cells from IBV-induced apoptosis. (a) H1299 cells were transfected with pEGFP-C1, pEGFP-XBP1u, or pEGFP-XBP1s in duplicate. At 24 h posttransfection, the cells were infected with IBV (MOI, ∼2) or mock infected. Cells were harvested at the indicated time points and subjected to Western blot analysis using antibodies against EGFP, IBV N, and PARP. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. In the second set of cells, total RNA was extracted and subjected to RT-PCR using primers specific for ERdj4, IBV genomic RNA, and GAPDH. (b) Quantification of PARP cleavage in plasmid-transfected cells infected with IBV. The percentage of PARP cleavage in cells transfected with pEGFP-C1, pEGFP-XBP1u, or pEGFP-XBP1s and infected with IBV for 24 h was determined as for panel a. The bar chart shows the results from three independent experiments with standard deviations and P values. (c) Schematic diagrams showing the functional domains of XBP1u, XBP1s, and XBP1-DN. All three proteins contain the bZIP DNA binding domain. The proteasome degradation motif in XBP1u was deleted to generate the dominant-negative XBP1-DN. (d) H1299 cells were transfected with pXJ40-FLAG or pXJ40-FLAG-XBP1-DN. At 24 h posttransfection, the cells were infected with IBV (MOI, ∼2) or mock infected. Cells were harvested at the indicated time points and subjected to Western blot analysis using antibodies against FLAG tag, IBV N, and PARP. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. (e) Quantification of PARP cleavage in plasmid-transfected cells infected with IBV. The percentage of PARP cleavage in cells transfected with pXJ40-FLAG or pXJ40-FLAG-XBP1-DN and infected with IBV for 24 h was determined as for panel d. The bar chart shows the results from three independent experiments with standard deviations and P values.
Both XBP1u and XBP1s contain the bZIP DNA binding domain in the N terminus (Fig. 5c). In XBP1u, the presence of a proteasome degradation motif renders it highly unstable in the cells. On the other hand, due to the frameshifting resulting from IRE1α-mediated splicing, XBP1s encodes a transactivation domain at the C terminus that accounts for its ability to induce downstream UPR genes. Previous studies have established that when the proteasome degradation motif of XBP1u is deleted, the resulting protein is stabilized and serves as a dominant-negative inhibitor of XBP1s (38) (Fig. 5c). Therefore, we decided to investigate the effect of this dominant-negative XBP1 (XBP1-DN) on IBV-induced apoptosis. As shown in Fig. 5d, the expression of FLAG-tag XBP1-DN was clearly detected. Multiple bands were observed, possibly due to posttranslational modifications of the protein. Compared with the vector control, overexpression of XBP1-DN significantly enhanced IBV-induced PARP cleavage, especially at the late time point, 22 h p.i. The proapoptotic effect of XBP1-DN was also reflected in the much stronger cleavage of the IBV N protein, because coronavirus N protein has been shown to be a substrate of activated caspases (39, 40). The observation was reproducible and statistically significant (Fig. 5e), suggesting that inhibition of XBP1s by overexpressing XBP1-DN markedly potentiates IBV-induced apoptosis. Taken together, the data indicate that the two isoforms of XBP1 display distinct properties during IBV infection: XBP1u is a weak proapoptotic protein, whereas XBPs is an antiapoptotic protein.
The kinases JNK and Akt are involved in the antiapoptotic function of IRE1α.
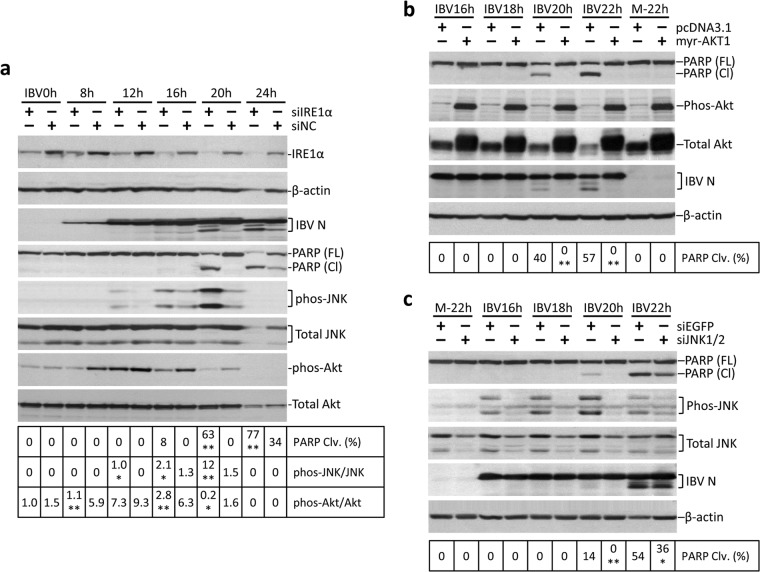
Since the MAP kinase JNK has been implicated in ER stress-induced apoptosis mediated by IRE1α (12), we proceeded to investigate the involvement of JNK in IBV-induced apoptosis in H1299 cells. As shown in Fig. 6a, a time course IBV infection experiment was performed in H1299 cells. As previously described, cells with IRE1α knocked down had an earlier onset and a significantly higher level of PARP cleavage than negative-control samples at the same time point. Interestingly, we also observed significant differences in the phosphorylation levels of two kinases: the proapoptotic kinase JNK and the prosurvival kinase Akt. In the negative control, relatively weak JNK phosphorylation was detected at 16 and 20 h p.i. In contrast, in IRE1α knockdown cells, JNK phosphorylation could be detected earlier (12 h p.i.) and at a significantly higher level than with the control samples at the same time point (Fig. 6a). On the other hand, phosphorylation of Akt was significantly induced at 8 h p.i. and sustained to 20 h p.i. in the negative-control cells infected with IBV. However, in IRE1α knockdown cells, significant Akt phosphorylation could be observed only at 12 h p.i. and rapidly diminished at 16 h p.i. (Fig. 6a). Therefore, the hypophosphorylation of Akt and the hyperphosphorylation of JNK may contribute to the enhanced IBV-induced apoptosis observed in the IRE1α knockdown cells.
FIG 6.
JNK and Akt are involved in the antiapoptotic function of IRE1α. (a) JNK hyperphosphorylation and Akt hypophosphorylation in IRE1 knockdown cells infected with IBV. H1299 cells were transfected with siIRE1 or nontarget siRNA before being infected with IBV and harvested at the indicated time points. Western blot analysis was performed using antibodies against IRE1α, IBV N, PARP, phosphor-JNK, total JNK, phosphor-Akt, and total Akt. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. The percentage of JNK or Akt phosphorylation was calculated as the band intensity of phosphorylated JNK or phosphorylated Akt divided by the band intensity of the corresponding total JNK or total Akt, respectively. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the siNC-transfected samples at the same time point (*, P < 0.05; **, P < 0.01). (b) Akt protects cells from IBV-induced apoptosis. H1299 cells were transfected with pcDNA3.1 or pcDNA3.1-myr-AKT1. At 24 h posttransfection, the cells were infected with IBV (MOI, ∼2) or mock infected. Cells were harvested at the indicated time points and subjected to Western blot analysis using antibodies against IBV N, PARP, phosphor-Akt, and total Akt. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the pcDNA3.1-transfected samples at the same time point (**, P < 0.01). (c) JNK is required for IBV-induced apoptosis. H1299 cells were transfected with siJNK1/2 or siEGFP before being infected with IBV or mock infected. Cells were harvested at the indicated time points and subjected to Western blot analysis using antibodies against IBV N, PARP, phosphor-JNK, and total JNK. β-Actin was included as a loading control. The percentage of PARP cleavage was calculated as for Fig. 3a. The experiment was repeated three times with similar results, and the result of one representative experiment is shown. The asterisks indicate significant differences between the indicated samples and the siEGFP-transfected samples at the same time point (*, P < 0.05; **, P < 0.01).
Next, we performed further experiments to confirm the involvement of Akt and JNK in IBV-induced apoptosis. As shown in Fig. 6b, H1299 cells were transfected with a constitutive-active form of Akt (myr-AKT1) (27) or the vector control before being infected with IBV. The expression of the transfected Akt could be determined from the significantly higher level of total Akt protein, as well as the highly intense phosphor-Akt bands in the transfected samples. Transfection of constitutively active Akt did not affect IBV replication, as indicated by the similar levels of IBV N protein. In the vector control, IBV induced PARP cleavage at 20 and 22 h p.i. (Fig. 6b). In contrast, PARP cleavage was completely abolished in cells transfected with myr-AKT1. Therefore, Akt is indeed a very strong antiapoptotic protein during IBV infection.
In another experiment, H1299 cells were transfected with siRNA targeting JNK1/2 or EGFP before being infected with IBV or mock infected (Fig. 6c). The knockdown of JNK was determined from the lower levels of both phosphor-JNK and total JNK in the siJNK1/2-transfected cells. JNK knockdown did not significantly affect IBV replication, as indicated by the similar levels of IBV N protein. In cells transfected with siJNK1/2, IBV-induced PARP cleavage was significantly reduced compared with the negative samples at the same time points (Fig. 6c). Thus, it is quite likely that JNK plays a proapoptotic role during IBV infection.
DISCUSSION
As the primary site of protein synthesis and folding, the ER is a front line in the battle between virus and host cell. Infection by many RNA viruses is known to induce modification of the ER membrane and to cause ER stress (17, 28, 37, 41). The replication and maturation of coronavirus are intimately associated with the ER, and ER stress is possibly induced through multiple mechanisms (42): first, massive synthesis of the highly glycosylated spike protein, which imposes a significant burden on the ER (33, 43); second, rearrangement of the ER membrane structure for the formation of double membrane vesicles (16); and third, extensive use of membranes from the ER-Golgi intermediate compartment (ERGIC) for virus morphogenesis and budding. Moreover, the envelope proteins of SARS-CoV and other coronaviruses have been demonstrated to have membrane permeabilization and ion channel activities (44–46). The envelope protein of SARS-CoV has also been demonstrated to suppress the IRE1-XBP1 pathway of the UPR and inhibit virus-induced apoptotic cell death. In this study, we found that IBV infection induced ER stress and activated the IRE1α-XBP1 pathway of UPR, as indicated by the upregulation of HERPUD1, IRE1α, total XBP1, XBP1s, ERdj4, and P58IPK mRNAs in IBV-infected cells. IBV infection also increased the mRNA and protein levels of GRP78 and GRP94 (data not shown), two protein chaperones commonly used as markers of ER stress. Together with the recent publications on MHV and SARS-CoV, these findings demonstrate that induction of ER stress and the UPR is a general host response during infection with coronaviruses (17, 33).
In the previous study on MHV, up to 75% XBP1 splicing was observed in MHV-infected cells, although the protein level of XBP1s was found to be only slightly increased (17). Similarly, significant XBP1 splicing was detected in IBV-infected cells at a late stage of infection. Although we were not able to detect a specific band of XBP1s using Western blotting, significant induction of downstream UPR genes, such as the ERdj4 and P58IPK genes, was observed in IBV-infected cells. Moreover, when the endogenous level of IRE1α or XBP1was depleted by siRNA, the mRNA level of ERdj4 and P58IPK was significantly reduced compared with the control. Also, when IRE1α or XBP1s was overexpressed in the cells, the mRNA level of ERdj4 was markedly increased. These results suggested that IBV infection indeed activated the IRE1α-XBP1 pathway and that the induction of UPR genes required both IRE1α-mediated XBP1 splicing and the action of the XBP1s protein. On the other hand, it is important to note that coronavirus may employ various strategies to suppress UPR activation. For example, the E protein of SARS-CoV has been shown to inhibit XBP1 splicing and suppress ER stress induced by infection with SARS-CoV and respiratory syncytial virus (18), whereas in MHV-infected cells, sustained translation shutdown via eIF2α phosphorylation has been attributed to the low level of XBP1s synthesis and the failure to induce UPR downstream genes (17). Whether similar mechanisms apply to IBV infection remain to be investigated.
Apoptosis is one possible outcome for cells infected with coronaviruses. Programmed demolition of infected cells can eliminate viruses before infectious progeny are formed. However, if it occurs at a later stage, apoptosis can also facilitate virus spread. When neighboring cells engulf apoptotic bodies containing infectious virions, the viruses gain access to new host cells without an extracellular stage and subvert recognition by the immune system. Previous studies have demonstrated that coronaviruses induce caspase-dependent and p53-independent apoptosis in infected cells (19, 20, 47). In this study, we investigated the involvement of the IRE1α-XBP1 pathway of UPR in IBV-induced apoptosis. The antiapoptotic activity of IRE1α was clearly demonstrated by the potentiated cleavage of PARP and caspases in IRE1α knockdown cells infected with IBV and the partially reduced apoptosis when cells overexpressed wild-type IRE1α, but not its mutants. To our surprise, contrary to that of IRE1α, knockdown of XBP1 seemed to inhibit IBV-induced apoptosis. However, since siXBP1 targeted both XBP1u and XBP1s, the contributions of individual isoforms could not be determined. Further experiments using the overexpression approach demonstrated that whereas unspliced XBP1 was proapoptotic, the spliced form is antiapoptotic. This was further confirmed when a stabilized, dominant-negative form of XBP1 was transfected and IBV-induced apoptosis was significantly potentiated. Considering the fact that IBV induced a relatively low level of XBP1 splicing compared to DTT treatment and that only XBP1u, but not XBP1s, could be detected using Western blotting, it is likely that the predominant isoform of XBP1 in the infected cells was the unspliced form. This may explain why a lower level of IBV-induced apoptosis was observed when XBP1 was knocked down using siRNA. In that case, the antiapoptotic function of IRE1α may be mediated, at least in part, via the conversion of proapoptotic XBP1u to antiapoptotic XBP1s.
Previous studies have shown that under prolonged ER stress, the MAP kinase JNK is phosphorylated by IRE1α to induce apoptosis (12). Activation of JNK could be detected in cells infected with SARS-CoV (48), MHV-A59 (49), or IBV. If JNK phosphorylation was mediated by IRE1α in coronavirus-infected cells, a reduced level of phosphor-JNK would be detected in IRE1α-deficient cells. However, the enhanced IBV-induced apoptosis observed in IRE1α knockdown cells was associated with hyperphosphorylation of JNK. In fact, knockdown of JNK partially reduced IBV-induced PARP cleavage, suggesting that JNK was proapoptotic in nature. Therefore, it is likely that during IBV infection JNK was activated by other upstream kinases (such as MKK4/MKK7) to promote apoptosis, which was modulated by the uncharacterized activity of IRE1α.
On the other hand, the Akt kinase has been demonstrated to play an important role in cell survival under ER stress (50). Previous studies on SARS-CoV demonstrated that limited activation of Akt was observed at an early stage of infection but could not prevent virus-induced apoptosis (51). In the current study, we found that enhanced IBV-induced apoptosis in IRE1α knockdown cells was associated with hypophosphorylation of Akt. So far, there has been no report of direct phosphorylation of Akt by the kinase activity of IRE1α. Of note, it has been shown that inhibition of Akt phosphorylation actually promotes the activity of IRE1α and IRE1α-mediated JNK phosphorylation (52). Therefore, the signaling between Akt and IRE1α seems to be complicated and may differ in cells under various stress conditions and may involve the actions of other regulatory factors. The signaling cross talk between Akt and JNK may also be complicated. As a survival kinase, Akt has been known to decrease the kinase activity of ASK1, which is the upstream MAP kinase kinase kinase of JNK (53). Moreover, Akt also interacts with JNK-interacting protein 1 (JIP1) and prevents its association with JNK to form active signaling complexes (54). On the other hand, it has been shown that JNK can inhibit the survival signals of Akt by phosphorylating the 14-3-3 protein (55). It is possible that the phosphorylation status and antagonizing actions of Akt and JNK have important functions in determining cell death/survival at the late stage of IBV infection, and as an ER stress sensor, IRE1α modulates the phosphorylation of Akt and JNK to promote cell survival.
To summarize the findings in the current study, we propose a working model (Fig. 7). IBV infection induces ER stress and the activation of IRE1α. IRE1α mediates splicing of XBP1, and downstream UPR genes (such as the ERdj4 and P58IPK genes) are upregulated to restore ER homeostasis. Whereas the unspliced form of XBP1 is proapoptotic, the spliced form is antiapoptotic. IRE1α protects cells from IBV-induced apoptosis, possibly by mediating the splicing of XBP1 and converting it from the proapoptotic XBP1u to the antiapoptotic XBP1s. IRE1α also modulates the phosphorylation status of the kinases Akt and JNK during IBV infection. The activation of the prosurvival kinase Akt seems to be promoted by IRE1α, whereas the phosphorylation of the proapoptotic kinase JNK seems to be negatively regulated by IRE1α (Fig. 7).
FIG 7.
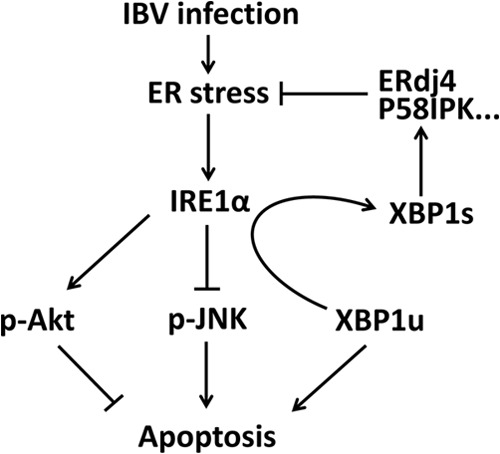
Working model. IBV infection induces ER stress and activation of the IRE1α-XBP1 pathway of the UPR. Downstream UPR genes, such as the ERdj4 and P58IPK genes, were induced to resolve the ER stress. IRE1 protects IBV-infected cells from apoptosis by (i) converting the proapoptotic XBP1u to antiapoptotic XBP1s, (ii) suppressing the proapoptotic kinase JNK, and (iii) promoting the antiapoptotic kinase Akt.
Previously, we demonstrated that the PERK branch of the UPR and the double-stranded RNA (dsRNA)-dependent kinase PKR are activated at the early stage of IBV infection (31). This leads to phosphorylation of eIF2α and upregulation of the growth arrest- and DNA damage-inducible protein 153 (GADD153), which promotes IBV-induced apoptosis by suppressing the prosurvival extracellular signal-related kinase (ERK) (31). Among the three UPR sensors, PERK is generally believed to be activated first in response to ER stress, followed by ATF6 and IRE1α (1, 56). It is interesting to consider the temporal control of UPR activation and its implication in coronavirus infection. From the host perspective, early activation of the PERK pathway and eIF2α phosphorylation induces translation attenuation, which serves as an effective antiviral defense mechanism. The induction of apoptosis through the eIF2α-ATF4-GADD153 pathway may also restrict virus replication. On the other hand, activation of IRE1α at the late stage of infection seems to promote the survival of infected cells. This may allow more virions to be assembled and released before the infected cells succumb to apoptotic cell death. Moreover, the cross talk between the UPR and the innate immune response may also constitute an important aspect of antiviral response during coronavirus infection (42).
In conclusion, the current study demonstrates that IRE1α, a major ER stress transducer, modulates apoptosis signaling during coronavirus infection. This work reveals the antiapoptotic signaling of the UPR in cells infected with IBV and provides new insights into the intricate signaling networks in IBV-induced apoptosis.
ACKNOWLEDGMENTS
This work was partially supported by a Competitive Research Programme (CRP) grant (R-154-000-529-281) from the National Research Foundation, Singapore, and an Academic Research Fund (AcRF) Tier 1 grant (RGT17/13) from Nanyang Technological University and the Ministry of Education, Singapore.
Footnotes
Published ahead of print 20 August 2014
REFERENCES
- 1.Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8:519–529. 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355–1364. 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013–27020. 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 5.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326–332. 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 6.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891. 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 8.Ng DT, Spear ED, Walter P. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77–88. 10.1083/jcb.150.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell. Biol. 9:231–241. 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 11.Oyadomari S, Mori M. 2004. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11:381–389. 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 12.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664–666. 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 13.Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. 2001. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J. Biol. Chem. 276:13935–13940. 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- 14.Hitomi J, Katayama T, Taniguchi M, Honda A, Imaizumi K, Tohyama M. 2004. Apoptosis induced by endoplasmic reticulum stress depends on activation of caspase-3 via caspase-12. Neurosci. Lett. 357:127–130. 10.1016/j.neulet.2003.12.080. [DOI] [PubMed] [Google Scholar]
- 15.Snijder EJ, Van Der Meer Y, Zevenhoven-Dobbe J, Onderwater JJM, Van Der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927–5940. 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoops K, Kikkert M, Van Den Worm, SHE. Zevenhoven-Dobbe JC, Van Der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS biology 6:e226. 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechill J, Chen Z, Brewer JW, Baker SC. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 82:4492–4501. 10.1128/JVI.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Regla-Nava JA, Álvarez E, Oliveros JC, Zhao J, Fett C, Perlman S, Enjuanes L. 2011. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 7:e1002315. 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Xu HY, Liu DX. 2001. Induction of caspase-dependent apoptosis in cultured cells by the avian coronavirus infectious bronchitis virus. J. Virol. 75:6402–6409. 10.1128/JVI.75.14.6402-6409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li FQ, Tam JP, Liu DX. 2007. Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology 365:435–445. 10.1016/j.virol.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong Y, Liao Y, Fang S, Tam JP, Liu DX. 2012. Up-regulation of mcl-1 and bak by coronavirus infection of human, avian and animal cells modulates apoptosis and viral replication. PLoS One 7:e30191. 10.1371/journal.pone.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Y, Tan YW, Liu DX. 2012. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses 4:980–1010. 10.3390/v4060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng LF, Liu DX. 1998. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology 243:388–395. 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu LH, Huang M, Fang SG, Liu DX. 2011. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase δ. J. Biol. Chem. 286:39546–39559. 10.1074/jbc.M111.242206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu DX, Inglis SC. 1991. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology 185:911–917. 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li FQ, Xiao H, Tam JP, Liu DX. 2005. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome coronavirus. FEBS Lett. 579:2387–2396. 10.1016/j.febslet.2005.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacquin M, Chiche J, Zunino B, Bénéteau M, Meynet O, Pradelli L, Marchetti S, Cornille A, Carles M, Ricci J. 2013. GAPDH binds to active Akt, leading to Bcl-xL increase and escape from caspase-independent cell death. Cell Death Differ. 20:1043–1054. 10.1038/cdd.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang ZM, Tan T, Yoshida H, Mori K, Ma Y, Yen TS. 2005. Activation of hepatitis B virus S promoter by a cell type-restricted IRE1-dependent pathway induced by endoplasmic reticulum stress. Mol. Cell. Biol. 25:7522–7533. 10.1128/MCB.25.17.7522-7533.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964–6972. 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Liao Y, Fung TS, Huang M, Fang SG, Zhong Y, Liu DX. 2013. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 87:8124–8134. 10.1128/JVI.00626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J. Virol. 83:12462–12472. 10.1128/JVI.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Versteeg GA, Van De Nes PS, Bredenbeek PJ, Spaan WJM. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 81:10981–10990. 10.1128/JVI.01033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meir O, Dvash E, Werman A, Rubinstein M. 2010. C/EBP-β regulates endoplasmic reticulum stress-triggered cell death in mouse and human models. PLoS One 5:e9516. 10.1371/journal.pone.0009516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirota M, Kitagaki M, Itagaki H, Aiba S. 2006. Quantitative measurement of spliced XBP1 mRNA as an indicator of endoplasmic reticulum stress. J. Toxicol. Sci. 31:149–156. 10.2131/jts.31.149. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida H, Oku M, Suzuki M, Mori K. 2006. pXBP1 (U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1 (S) in mammalian ER stress response. J. Cell Biol. 172:565–575. 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu CY, Hsu YW, Liao CL, Lin YL. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 80:11868–11880. 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. 2006. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281:5852–5860. 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- 39.Eléouët JF, Slee EA, Saurini F, Castagné N, Poncet D, Garrido C, Solary E, Martin SJ. 2000. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 74:3975–3983. 10.1128/JVI.74.9.3975-3983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diemer C, Schneider M, Seebach J, Quaas J, Frösner G, Schätzl HM, Gilch S. 2008. Cell type-specific cleavage of nucleocapsid protein by effector caspases during SARS coronavirus infection. J. Mol. Biol. 376:23–34. 10.1016/j.jmb.2007.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pena J, Harris E. 2011. Dengue virus modulates the unfolded protein response in a time-dependent manner. J. Biol. Chem. 286:14226–14236. 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fung TS, Liu DX. 2014. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 5:296. 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan CP, Siu KL, Chin KT, Yuen KY, Zheng B, Jin DY. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 80:9279–9287. 10.1128/JVI.00659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Lescar J, Tam J, Liu D. 2004. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 325:374–380. 10.1016/j.bbrc.2004.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y, Yuan Q, Torres J, Tam J, Liu D. 2006. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349:264–275. 10.1016/j.virol.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verdiá-Báguena C, Nieto-Torres JL, Alcaraz A, DeDiego ML, Torres J, Aguilella VM, Enjuanes L. 2012. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology 432:485–494. 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eleouet JF, Chilmonczyk S, Besnardeau L, Laude H. 1998. Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J. Virol. 72:4918–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. 2005. JNK and PI3k/Akt signaling pathways are required for establishing persistent SARS-CoV infection in Vero E6 cells. Biochim. Biophys. Acta 1741:4–10. 10.1016/j.bbadis.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D, Zhu H, Liu Y, Cao J, Zhang X. 2009. Regulation of proinflammatory cytokine expression in primary mouse astrocytes by coronavirus infection. J. Virol. 83:12204–12214. 10.1128/JVI.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu P, Han Z, Couvillon AD, Exton JH. 2004. Critical role of endogenous Akt/IAPs and MEK1/ERK pathways in counteracting endoplasmic reticulum stress-induced cell death. J. Biol. Chem. 279:49420–49429. 10.1074/jbc.M407700200. [DOI] [PubMed] [Google Scholar]
- 51.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. 2004. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology 327:169–174. 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. 2012. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1–JNK pathway. Cell Death Differ. 19:310–320. 10.1038/cdd.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim AH, Khursigara G, Sun X, Franke TF, Chao MV. 2001. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol. Cell. Biol. 21:893–901. 10.1128/MCB.21.3.893-901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim AH, Yano H, Cho H, Meyer D, Monks B, Margolis B, Birnbaum MJ, Chao MV. 2002. Akt1 regulates a JNK scaffold during excitotoxic apoptosis. Neuron 35:697–709. 10.1016/S0896-6273(02)00821-8. [DOI] [PubMed] [Google Scholar]
- 55.Sunayama J, Tsuruta F, Masuyama N, Gotoh Y. 2005. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 170:295–304. 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szegezdi E, Logue SE, Gorman AM, Samali A. 2006. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7:880–885. 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]