ABSTRACT
The ubiquitin-proteasome system is targeted by many viruses that have evolved strategies to redirect host ubiquitination machinery. Members of the genus Chlorovirus are proposed to share an ancestral lineage with a broader group of related viruses, nucleo-cytoplasmic large DNA viruses (NCLDV). Chloroviruses encode an Skp1 homolog and ankyrin repeat (ANK) proteins. Several chlorovirus-encoded ANK repeats contain C-terminal domains characteristic of cellular F-boxes or related NCLDV chordopox PRANC (pox protein repeats of ankyrin at C-terminal) domains. These observations suggested that this unique combination of Skp1 and ANK repeat proteins might form complexes analogous to the cellular Skp1-Cul1-F-box (SCF) ubiquitin ligase complex. We identified two ANK proteins from the prototypic chlorovirus Paramecium bursaria chlorella virus-1 (PBCV-1) that functioned as binding partners for the virus-encoded Skp1, proteins A682L and A607R. These ANK proteins had a C-terminal Skp1 interactional motif that functioned similarly to cellular F-box domains. A C-terminal motif of ANK protein A682L binds Skp1 proteins from widely divergent species. Yeast two-hybrid analyses using serial domain deletion constructs confirmed the C-terminal localization of the Skp1 interactional motif in PBCV-1 A682L. ANK protein A607R represents an ANK family with one member present in all 41 sequenced chloroviruses. A comprehensive phylogenetic analysis of these related ANK and viral Skp1 proteins suggested partnered function tailored to the host alga or common ancestral heritage. Here, we show protein-protein interaction between corresponding family clusters of virus-encoded ANK and Skp1 proteins from three chlorovirus types. Collectively, our results indicate that chloroviruses have evolved complementing Skp1 and ANK proteins that mimic cellular SCF-associated proteins.
IMPORTANCE Viruses have evolved ways to direct ubiquitination events in order to create environments conducive to their replication. As reported in the manuscript, the large chloroviruses encode several components involved in the SCF ubiquitin ligase complex including a viral Skp1 homolog. Studies on how chloroviruses manipulate their host algal ubiquitination system will provide insights toward viral protein mimicry, substrate recognition, and key interactive domains controlling selective protein degradation. These findings may also further understanding of the evolution of other large DNA viruses, like poxviruses, that are reported to share the same monophyly lineage as chloroviruses.
INTRODUCTION
The ubiquitin-proteasome proteolytic pathway is an attractive target for viruses in their battle to create an intracellular environment conducive for their replication. In fact, targeting of ubiquitin-associated enzymes is a reoccurring theme in permissive viral infections (1). This eukaryotic regulatory system mediates a variety of biological processes, including protein turnover, DNA repair, trafficking, and signal transduction (2). Through sequential reactions of three enzyme types, covalent attachment of ubiquitin chains to the substrate is achieved, which then targets the substrate protein for proteasomal degradation. This cascade is initiated by the ubiquitin-activating enzyme (E1), which forms an ATP-dependent high-energy thioester bond with an ubiquitin moiety, enabling passage to a ubiquitin conjugase (E2). Substrate-specific ubiquitin ligases (E3) then catalyze the transfer of ubiquitin from the E2 enzyme to the target protein, creating an isopeptide bond most commonly between a lysine of the substrate and the C-terminal glycine of ubiquitin (Fig. 1) (3). In this system the E3 ubiquitin ligase is the most critical component in determining the selection of substrate proteins for ubiquitin modification, which in turn has pleiotropic cell-regulatory effects, particularly by inducing substrate protein degradation.
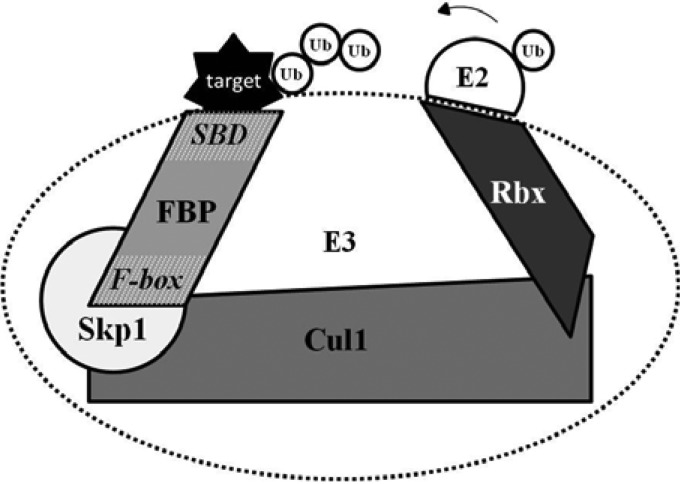
FIG 1.
The SCF ubiquitin ligase complex. Schematic representation of the cullin-based RING E3 ligase (CRL) SCF complex mediating the catalytic transfer of ubiquitin (Ub) from the recruiting thioester-bound E2 conjugating enzyme to the target substrate, forming a polyubiquitin chain. As the target recognition component, the F-box protein (FBP) harbors two domains: (i) the F-box domain which resides in the amino terminus of the protein where it binds linker protein Skp1, and (ii) the substrate-binding domain (SBD) responsible for bringing specific target proteins in close proximity to the catalytic core of the SCF complex.
Several ubiquitin-interfering viral proteins interact directly with E3 family enzymes, in particular the cullin-based RING (really interesting new gene) finger-type ubiquitin ligase (CRL) SCF (Skp1, cullin, F-box) complex (4, 5). The F-box protein is one of the four subunits of the SCF complex which mediates target recruitment for ubiquitination. F-box proteins are a large family of proteins present in all eukaryotes that are characterized by the presence of the F-box domain, a conserved sequence of approximately 50 amino acids that interacts with the Skp1 component of the SCF CRL complex (6). The highly conserved F-box motif is usually located at the N terminus and is required for interaction with the Skp1 element of the SCF complex. Most of the characterized cellular F-box proteins harbor substrate-binding repeat-containing motifs in the C-terminal region, such as leucine-rich repeats (LRR), WD40 repeats (WD), or Kelch repeats (7).
Virulence factors from bacteria and viruses have harnessed F-box proteins compatible with cellular SCF ligase components to mediate substrate recruitment and degradation for the benefit of the pathogen (8–13). Recent genomic analyses have identified a conserved C-terminal PRANC (pox protein repeats of ankyrin [ANK] at C-terminal) motif in chordopox ankyrin repeat-containing proteins that is involved in the hijacking of the host cell ubiquitin machinery (14, 15). These ANK/PRANC proteins play a critical role in reprogramming various cellular signaling cascades to regulate host cell tropism and permissiveness (16). ANK proteins participate in protein-protein interactions and are involved in many functions, such as cell cycle regulation, signal transduction, and intracellular trafficking (17, 18). Experimentally, it has been established that the PRANC domain present in ANK proteins of several poxvirus family members interacts with cellular SCF complexes to redirect ubiquitin ligase activity (9, 19–23). Interestingly, in mammals the ANK repeats are commonly the substrate recognition module in combination with SOCS boxes in another class of CRLs which contain cullin-5 and the elongin B and C complex in mammals (24). Although common in eukaryotes and bacteria, the 33-residue ANK motif is relatively rare among viruses, with the exception of poxviruses in the nucleo-cytoplasmic large DNA virus (NCLDV) family.
As members of the Phycodnaviridae family, chloroviruses are included in the same seven-member NCLDV monophyletic group as Poxviridae. Mimiviruses, another member of the NCLDV family, have several N-terminal F-box motifs with C-terminal ANK proteins. The NCLDVs infect animals and diverse unicellular eukaryotes and replicate either exclusively in the cytoplasm of their host cells or possess both nuclear and cytoplasmic stages in their life cycles. Members of this group contain genome sizes that range from 100 kb to 1.2 Mb. A comprehensive genome analysis of the NCLDVs identified approximately 50 conserved genes that mapped to the genome of a common ancestral virus while five genes were conserved in all NCLDV sequenced genomes (25). These conserved common genes are referred to as NCLDV core genes. Tracing the evolutionary lineage of NCLDVs may reveal shared ancestor genes designed to optimize the subversion of cellular E3 ubiquitin ligase activity.
Collectively referred to as chloroviruses, NC64A, SAG, and Pbi viruses infect one of three different symbiotic chlorella-like green algal species: the sequenced and annotated Chlorella variabilis NC64A, Chlorella sp. SAG 3.83 (renamed Chlorella heliozoae), and Chlorella sp. Pbi (renamed Micractinium conductrix), respectively (26). The prototype chlorovirus, Paramecium bursaria chlorella virus-1 (PBCV-1) is a 331-kb double-stranded DNA, plaque-forming virus that is predicted to encode 416 proteins and 11 tRNAs (27). The host of PBCV-1, the unicellular green alga C. variabilis, is a natural endosymbiont of the protozoan Paramecium bursaria. Chloroviruses encode many candidate proteins that have the potential to manipulate the SCF complex. For example, PBCV-1 encodes eight ANK proteins that appear to contain C-terminal domains with potential F-box-like function, imitating poxvirus ANK genes. The current manuscript addresses this issue.
PBCV-1 and 39 of the 41 annotated chloroviruses also encode a putative viral Skp1 protein (vSkp1PBCV-1). Cellular Skp1 serves as a critical subunit of the SCF complex which governs cell cycle regulation. As the linker subunit, cellular Skp1 bridges cullin-1 (Cul1) and F-box proteins to form the multisubunit E3 ligase (Fig. 1). The C. variabilis algal host encodes two Skp1 homologs (GenBank accession numbers EFN54040 and EFN51326); many organisms encode multiple Skp1 or Skp1-like genes (28). Of this pair of C. variabilis Skp1 genes, GenBank EFN54040 is more similar in both size and amino acid sequence to known Skp1 proteins. We hypothesize that chloroviruses use their vSkp1s and one or more ANK proteins to hijack the host alga SCF complex to favor viral infection. Our results demonstrate an interaction between cellular SCF proteins and virus-encoded proteins and provide the first evidence of a functioning vSkp1 with partnering virus and cellular genes. Furthermore, we identified the minimum viral ANK domain required for Skp1 binding. We predict that vSkp1PBCV-1 and its partnering ANK proteins are capable of mimicking the Skp1 and F-box components of the eukaryotic SCF ubiquitin ligase machinery, respectively.
MATERIALS AND METHODS
Cells and media.
Escherichia coli DH5α competent cells, prepared according to the method of Inoue et al.(29), and BL21-Gold(DE3)pLysS competent cells (Stratagene, La Jolla, CA) were grown in LB medium or super optimal broth (SOB) medium (2% [wt/vol] tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2) after transformation. Media were supplemented with proper antibiotics if needed. Saccharomyces cerevisiae MaV203 (Proquest Two-hybrid System; Invitrogen) served as the host yeast strain for the bait and prey plasmids. The yeast GAL4-based two-hybrid system includes deletions in the endogenous GAL4 and GAL80 genes. The strain also contains a set of nonreverting auxotrophic mutations (leu2 and trp1) for the selection of bait and prey fusion vectors. MaV203 competent cells were prepared according to the procedure of Gietz et al. (30), with modification. Cells were saturated by overnight shaking in YPAD (10 g of yeast extract, 20 g of peptone, 20 g of dextrose, and 30 mg of adenine sulfate in 1 liter) medium at 30°C and 250 rpm and diluted to an optical density at 600 nm (OD600) of 0.1 in fresh YPAD medium. Cells were then cultured for an additional 4 h to reach an OD600 of 0.4 to 0.6, pelleted by centrifugation at 4,000 × g for 5 min at room temperature, and washed in lithium acetate (LiAc) solution (100 mM LiAc, 10 mM Tris-Cl, 1 mM EDTA, pH 8.0) in a 1/2 volume of original culture. Finally, cells were suspended in LiAc solution in a 1/100 volume of original culture, dispersed into 100-μl aliquots for transformation or supplemented with 5% glycerol and 10% (vol/vol) dimethyl sulfoxide (DMSO), and stored at −80°C as frozen competent cells. Synthetic complete (SC) medium contained 6.7 g/liter yeast nitrogen base, 20 g/liter dextrose, and 1.4 g/liter amino acid dropout mix (Sigma, St. Louis, MO) omitting leucine and tryptophan (SC-Leu−-Trp−).
Gene cloning and plasmid construction.
Genes carried by the host and virus were synthesized with optimization according to the codon preference of S. cerevisiae by GenScript (Piscataway Township, NJ). Coding regions were amplified by PCR and incorporated into entry clone pENTR/D-TOPO based on Gateway cloning technology (Invitrogen, Carlsbad, CA) (data not shown). PCR products were cloned directionally by adding four bases to the forward primer (CACC) (data not shown). The entry clone was selected with 50 μg/ml kanamycin. The tailor-made entry clones flanked with attL1 and attL2 sites underwent LR recombination (Gateway technology; Invitrogen) into the destination vectors. As a result, the gene of interest was fused in frame with the GAL4 DNA-binding domain (DBD) or the activation domain (AD) in the yeast two-hybrid (Y2H) yeast expression vectors pDEST32 and pDEST22 containing the GAL4 DNA-binding domain and GAL4 DNA activation domain, respectively. The E. coli cells containing positive cloned plasmids were selected with 50 μg/ml gentamicin and 100 μg/ml ampicillin, respectively, for pDEST32 and pDEST22. The structure of each vector was sequence verified. Plasmids were isolated with a QIAprep Spin Miniprep kit (Valencia, CA) according to the manufacturer's instructions. For overexpression of the vSkp1 proteins in E. coli, a pET-23a(+) vector (Novagen Biosciences, San Diego, CA), which carries an N-terminal T7 tag sequence in addition to a C-terminal His6 sequence tag, was used. Total plasmid concentration was quantified with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Y2H screening.
Yeast strain MaV203 competent cells were transformed with bait and prey vectors according to a high-efficiency transformation protocol (30). Yeastmaker carrier DNA (salmon sperm DNA; Clontech, Mountain View, CA) was used to increase transformation efficiency. Selection criteria regarding yeast transformants containing bait and prey vectors were based on the ProQuest two-hybrid system (Invitrogen), according to the manufacturer's instructions. Nutritionally selective SC-Leu−-Trp− medium was used to test for positive transformation. To assess protein-protein interaction, induction of three unrelated GAL4-inducible reporter genes (HIS3, URA3, and lacZ) downstream at separate loci in the yeast genome enabled the measurement of interaction strength between bait and prey proteins. Reporter genes HIS3 and URA3 were tested on respective dropout selection media. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) colorimetric assays were used to test induction of the lacZ reporter gene measuring β-galactosidase activity. To select against positive interactions, medium was supplemented with 5-fluoroorotic acid (5FOA), which is toxic to yeast cells expressing URA3. This combination of reporter genes and sensitivity assays allowed the interpretation of four phenotypes to access true interactions. Clones containing bait and prey that induced the reporter genes without interacting in the two-hybrid system were defined as false positives. False positives in which bait and prey underwent self-activation were reduced by testing cotransformed yeast cells with bait or prey plasmid and an empty plasmid according to the manufacturer's protocol (Invitrogen). To further test for self-activation, the threshold of resistance to 3-amino-1,2,4-triazole, an enzyme encoded by HIS3 involved in histidine biosynthesis, was determined in a dose-dependent manner (10, 25, 50, and 100 mM). The transformants that contained empty bait and prey vectors pDEST32 and pDEST22 and the vector pairs of pEXP32/Krev1 and pEXP22/RalGDS-m2 were used as negative controls, whereas yeast strains which were cotransformed with the pEXP32/krev1 and pEXP22/RalGDS-wt vectors were used as positive controls (Invitrogen).
Viral lysate preparation.
C. variabilis cells (2 × 107 cells/ml) from an actively growing culture in modified Bold's basal medium (31) were inoculated with purified PBCV-1 at a multiplicity of infection (MOI) of 5 with agitation to ensure uniform infection. Infected cells were harvested by centrifugation (at 5,000 × g) for 5 min at 4°C at 0, 15, 30, 45, 60, 90, 120, 180, 240, and 360 min postinfection (p.i.). Cells were immediately harvested following virus addition at time zero. C. variabilis cells were also harvested without addition of virus as a control. The cells were resuspended in 500 μl of phosphate-buffered saline (PBS) buffer supplemented with 2 mM phenylmethanesulfonyl fluoride (PMSF). Glass beads (0.25 to 0.3 mm) were added to the infected cell suspension and vortexed for 5 min. Cells were then sonicated (5-s pulse at 30% amplitude using a Tekmar Sonic Disruptor TM-100) for 5 min on ice. The cell lysate was centrifuged (16,000 × g) for 10 min at 4°C, and supernatant was collected. For pulldown experiments, the supernatant was collected and diluted with 4 ml of PBS before it was applied to a vSkp1-bound column.
vSkp1PBCV-1 overexpression and candidate F-box protein pulldown by metal chelation chromatography.
BL21-Gold(DE3)pLysS cells were transformed with pET-23a(+) vSkp1PBCV-1, and transformants were selected by resistance to carbenicillin and chloramphenicol. Batch culture was grown to an OD600 of 0.6 at 37°C. Induction of protein overexpression was achieved with the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 2 h at 25°C. Cells were harvested by centrifugation (5,000 × g) for 5 min at 4°C and then disrupted by sonication twice in PBS buffer with 2 mM PMSF for 5 min on ice. Cell lysate was centrifuged (5,000 × g) for 5 min at 4°C, and the supernatant was applied to a Ni2+ column. Column resin and elution buffer were from a Ni-nitrilotriacetic acid (NTA) buffer kit (Novagen Biosciences, San Diego, CA). One milliliter of resuspended Ni-NTA agarose in 4 ml of PBS was loaded on a 5-ml self-packing column with a 45- to 90-μm-pore-size polyethylene filter (frit) (Life Science Products, Frederick, CO), and the resin was allowed to settle. After the resin was equilibrated with 8 ml of PBS, 8 ml of sample lysate containing overexpressed vSkp1PBCV-1 was loaded on the column. The Ni2+ column was washed four consecutive times using 4 ml of PBS buffer with 10 mM imidazole. Three lysates were applied individually to the vSkp1PBCV-1 column: (i) uninfected C. variabilis cells, (ii) C. variabilis cells at 30 min p.i., and (iii) C. variabilis cells at 180 min p.i. Protein was eluted from the column with 0.5 ml of Ni-NTA elution buffer four consecutive times. Collected protein was concentrated by a Microcon (YM-10) centrifugal filter (Millipore, Bedford, MA). The protein concentration was determined by a Coomassie Plus Bradford assay (Thermo Fisher Scientific, Waltham, MA). Eluted proteins were resolved by SDS-PAGE using PAGEr Gold precast gels (Lonza, Rockland, ME) with Coomassie brilliant blue staining solution. Bands were excised from the gel and analyzed for protein identification by mass spectrometry.
Protein identification by nano-LC-MS/MS.
The excised protein bands from the SDS-PAGE gel were washed with deionized water and destained by repeated cycles of dehydration (50 mM ammonium bicarbonate in 50:50 acetonitrile) and rehydration in 50 mM ammonium bicarbonate. Proteins were reduced with 10 mM dithiothreitol (DTT) in 50 mM ammonium bicarbonate buffer for 45 min. For alkylation, 40 mM iodoacetamide in ammonium bicarbonate was added, and the sample was incubated in the dark for an additional 30 min at room temperature. Following two cycles of dehydration/rehydration as described above, sequencing-grade trypsin in 50 mM ammonium bicarbonate was added to dried gel pieces, which were incubated overnight at 37°C. Peptides were extracted with 100% acetonitrile–0.1% formic acid (FA) and dried using a SpeedVac. The dried mixture of tryptic peptides was dissolved in 5% acetonitrile–0.1% FA and analyzed by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) using an UltiMate 3000 RSLCnano system (Dionex Corp., Sunnyvale, CA) coupled to an linear trap quadrupole (LTQ) Velos Pro dual-pressure linear ion trap mass spectrometer integrated with electron transfer dissociation (ETD) (Thermo Scientific, San Jose, CA). After injection, the peptides were concentrated on a monolithic C18 trap column (Pep Map; internal diameter [i.d.], 300 μm; length, 5 mm; pore size, 100 Å; particle size, 5 μm [Dionex Corp.]) and washed with 5% acetonitrile–0.1% FA. The enrichment column was then switched into the nanoflow path (200 nl/min), and the sample was further separated on a C18 Pep Map column (75-μm i.d. by 15-cm length; 3-μm particle size and 100-Å pore size [New Objective Inc., Woburn, MA]) by applying an acetonitrile gradient (acetonitrile plus 0.1% FA; 60 min) at a flow rate of 200 nl/min. Automated MS/MS spectra were acquired during the run-in data-dependent acquisition mode with selection of the six most abundant precursor ions. The LTQ Velos Pro mass spectrometer was operated with the following parameters: nanospray voltage of 2.0 kV, heated capillary temperature of 250°C, and full-scan m/z range of 400 to 2,000. The data acquisition was done with alternating collision-induced dissociation (CID), high-energy collision dissociation (HCD), and ETD; six MS/MS spectra were obtained for every full scan, and five microscans were averaged for full scans and MS/MS scans. The normalized collision energy was set to 35% for CID, with an isolation width of 3.0 and activation time of 10 ms. Supplemental activation was enabled for ETD; the activation time was set to 100 ms with an isolation width of 3.0. The signal threshold for triggering an MS/MS event was set to 500 counts. Extracted spectra were searched against NCBI and a custom-based viral protein database using the MASCOT search engine.
Antibody.
Polyclonal rabbit anti-vSkp1PBCV-1 was produced by Harlan Bioproducts for Science, Inc. (Indianapolis, IN). The antiserum was prepared using their standard 73-day protocol by immunizing one rabbit with 500 μg of vSkp1 peptide for three consecutive boosts.
Western blot analyses.
Samples were separated on SDS-PAGE gels and transferred to polyvinyl difluoride (PVDF) membranes (0.45-μm pore size) using standard procedures (32). Membranes were blocked for 1 h at room temperature with 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 (TBS-T). Blots were probed using primary polyclonal antibody rabbit anti-vSkp1PBCV-1 (Harlan) overnight with agitation at 4°C in blocking buffer. After five washes with TBS-T, membranes were probed with goat anti-rabbit conjugated to horseradish peroxidase (HRP) (A6154; Sigma) as the secondary antibody. After an additional five washes with TBS-T, the membranes were developed using ECL SuperSignal West Pico chemiluminescent substrate (ThermoFisher Scientific). Bands were visualized using a ChemiDoc MP Imaging System with Imagine Lab Software (Bio-Rad, Hercules, CA).
Protein modeling.
vSkp1PBCV-1 and PBCV-1 A682L structure predictions were constructed by PHYRE (protein homology/analogy recognition engine), version 2.0 (33). Molecular docking of viral proteins was performed using PatchDock (34), a molecular docking algorithm based on shape complementarity principles. Protein complexes were inputted into the molecular modeling system, Chimera, where three-dimensional protein models were constructed. Analysis of conformational ensembles of the Skp1-Skp2 complex (Protein Data Bank [PDB] accession number 1FQV) (35) and the vSkp1PBCV-1-A682L complex was completed using Chimera imagery.
Sequence identification and alignments.
Gene sequences were identified by a BLAST search using the National Center for Biotechnology Information nonredundant database. The C termini of the eight PBCV-1-encoded ANK proteins were aligned with the F-box consensus sequence adapted from Kipreos and Pagano (36) using ClustalW2 (37) running MEGA, version 3.1, with default settings (Gonnet matrix; gap opening penalty, 10; gap extension penalty, 0.1 for pairwise alignments and 10 and 0.2 for multiple alignments). Known residues of Skp2 that contact cellular Skp1 were reported by Sonnberg et al. (19). Sequence alignment of Skp1 homologs was created by the Phylogeny.fr platform (38, 39) using default settings (MUSCLE, BLOSUM62 substitution matrix, and Gblocks for alignment curation).
Phylogenetic analysis.
To construct a robust phylogenetic tree from chlorovirus proteins, sequence alignments were created using the Phylogeny.fr platform (38, 39) with default settings (MUSCLE, version 3.7, for multiple alignment; Gblocks, version 0.91b, for alignment curation; PhyML, version 3.0, approximate likelihood ratio test [aLRT], for phylogeny; and TreeDyn, version 198.3, for tree rendering). The following additional parameters were enforced: the minimum number of sequences for a flank position was 85%; all segments with contiguous nonconserved positions bigger than 8 were rejected; minimum block length was 10; no gaps were allowed in the final alignment. Phylogenetic trees were computed using the Whelan and Goldman (WAG) substitution model. We show only bootstrap values of <90%. Branches with bootstrap support of less than 50 were collapsed.
Identification of proteins used for analysis.
All viral genes investigated in this article correspond to their gene locus tag identifiers in the National Center for Biotechnology Information (NCBI) database. The following are PBCV-1 ANK proteins: A672R (GenBank accession number NP_049028), A005R (GenBank NP_048353), A682L (GenBank NP_049038), A247R (GenBank NP_049038), A607R (GenBank NP_048963), A330R (GenBank NP_048686), A7/8 (GenBank NP_048355), and A429L (GenBank NP_048786). The following vSkp1 proteins were investigated: A039L (PBCV-1; GenBank accession number NP_048387), Z339L (Acanthocystis turfacea chlorella virus-1 [vSkp1ATCV-1]; GenBank YP_001426820), and N799R (Paramecium bursaria chlorella virus FR483 [vSkp1FR483]; GenBank YP_001426431). Skp1-binding ANK proteins from ATCV-1 and FR483 are Z568R (GenBank accession number YP_001427049) and N710R (GenBank YP_001426342), respectively. Skp1 proteins correspond to GenBank accession numbers EFN54040 and EFN51326 (C. variabilis), P63208 (Homo sapiens), P52285 (Dictyostelium discoideum), and P52286 (S. cerevisiae). F-box proteins from C. variabilis were GenBank EFN59680, UniProt E1ZSC7, GenBank EFN54870, and GenBank EFN51283.
RESULTS
Chlorovirus ANK repeat proteins have a possible carboxyl-terminal Skp1 interactional motif.
The combination of a substrate-binding domain, often containing protein sequence repeats, and an F-box domain is responsible for the bipartite functionality of cellular F-box proteins. Chlorovirus ANK proteins are composed of similar domain elements. ANK regions, located toward the N-terminal region, are presumed to be substrate binding. Interestingly, alignment of the eight PBCV-1-encoded ANK proteins revealed a common motif in the C-terminal region (Fig. 2). Manual inspection of this region revealed a conserved domain with some key binding residues, including some with Leu-Pro residues characteristic of the 50-amino-acid cellular F-box motif. Although sharing some common residues with the F-box motif, the putative Skp1 interactional domains in the viral ANK proteins are truncated (30 to 40 amino acids) and are positioned at the opposite end of cellular F-boxes. However, they resemble the C-terminal location of the F-box-like PRANC domain of ANK proteins in chordopox viruses (23, 40).
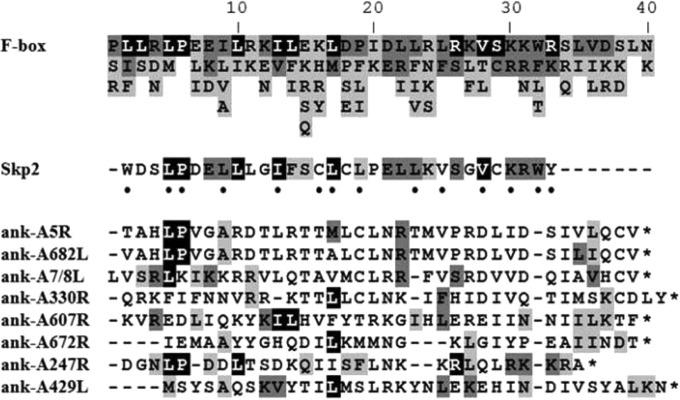
FIG 2.
PBCV-1 ANK repeat proteins contain putative carboxyl-terminal Skp1-binding domains. ClustalW2 was used to align the carboxyl-termini of the eight PBCV-1-encoded ANK proteins with the F-box consensus sequence adapted from Kipreos and Pagano (36) and the F-box domain from Skp2, a cellular F-box protein. Residues are shaded according to frequency in the F-box consensus sequence (top), which was derived from the alignment of 234 sequences used to create the Pfam F-box profile. Black signifies residues found in over 40% of the F-box sequences; dark gray signifies residues found in 20 to 40% of the F-boxes; light gray signifies residues found in 10 to 20% of the F-boxes. The dots indicate known residues of Skp2 that contact cellular Skp1 according to Sonnberg et al. (19). A dash indicates a space, and an asterisk indicates the end of the protein.
When aligned with a consensus F-box sequence (36) derived from the Pfam F-box profile, the C-terminal regions of the eight PBCV-1-encoded ANK proteins contain a limited number of residues that are present in characterized cellular F-boxes (Fig. 2). Among these shared residues are amino acids known to mediate contact between cellular proteins Skp1 and F-box protein Skp2 (19). Based on domain sequence similarity, PBCV-1 ANK proteins A5R, A682L, and A247R contain a C-terminal motif that most closely resembles a canonical F-box protein. Specifically, these three ANK proteins possess Leu-Pro residues common to most cellular F-box proteins. The viral ANK proteins are highly conserved in the chloroviruses. While their C-terminal sequences are somewhat divergent from those of cellular F-boxes, their locations in the protein are similar to the location of the PRANC domain from poxvirus-encoded ANK proteins (41). We hypothesize that the combination of the ANK domain and conserved C-terminal motif in chlorovirus ANK proteins may function as the substrate-binding and Skp1-binding domains, respectively. This hypothesis was supported by secondary and tertiary structure predictions of the carboxyl-terminal portion of the eight PBCV-1 ANK proteins, which indicated protein folding and exposed active sites for protein interactions similar to those that occur in F-boxes. The predicted structure of the interaction of the C terminus of PBCV-1 ANK protein A682R with the viral Skp1 protein compared with the crystal structure of human Skp1 interacting with Skp2 is reported in Fig. 3 and demonstrates the similarity of the Skp1 and F-box like interactions.
FIG 3.
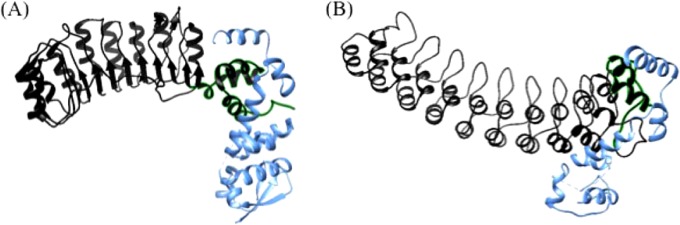
Protein modeling shows conformation similarities between SCF subunits and viral genes. (A) Crystal structure of the Skp1-Skp2 complex (blue and black, respectively) (PDB accession number 1FQV). The F-box domain of Skp1 is highlighted in green. (B) Predicted protein interaction model of vSkp1PBCV-1 (blue) and A682L (black). The vSkp1 interactional motif located at the carboxyl terminus of the viral ANK protein is highlighted in green. There is excellent similarity between the interaction of vSkp1 to the C terminus of A682L and the interaction of Skp1-Skp2 (F-box protein). This similarity supports the hypothesis that the C terminus of A682L can bind to Skp1 similarly to an F-box domain despite its weak amino acid similarity.
Highly conserved ANK repeat protein families among chloroviruses group into monophyletic clusters according to the host alga.
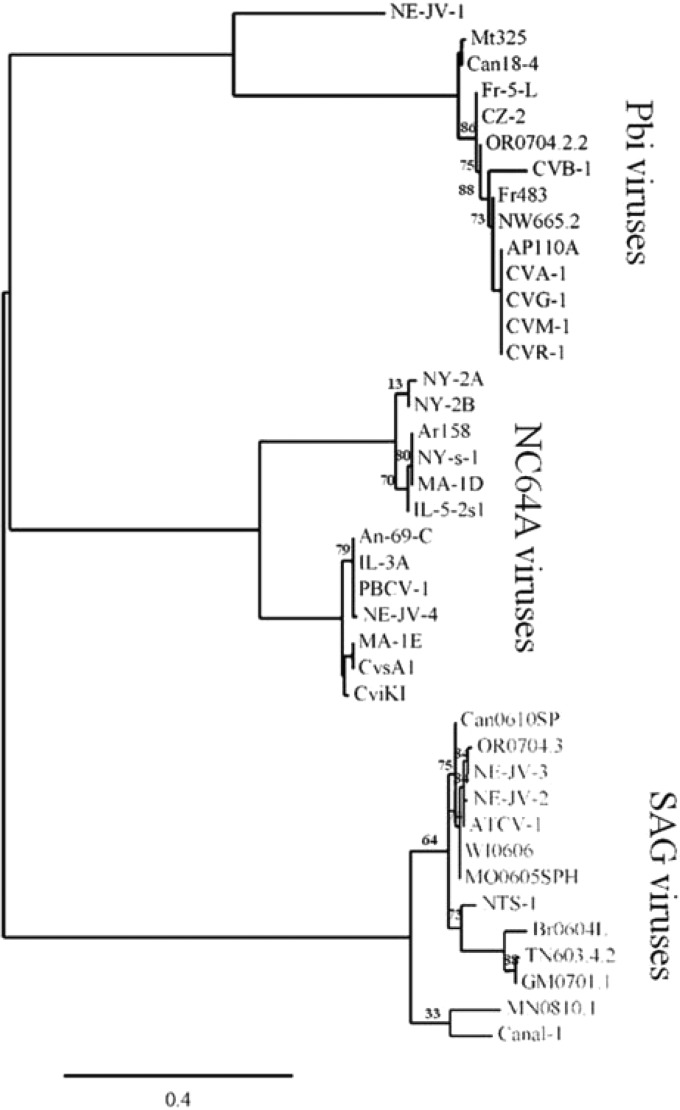
Annotated genomes of 41 chloroviruses, which are classified by their ability to infect one of three different symbiotic chlorella-like host algae, contain between 319 and 416 predicted protein-encoding genes per genome (57). Collectively, the 41 chloroviruses encoded 178 ANK-containing proteins. One ANK protein family contains 51 members and, after likely gene duplications are eliminated, all 41 annotated chloroviruses contain at least one member of this protein family. The PBCV-1 family member is A607R. This family of ANK proteins was analyzed by a maximum-likelihood reconstruction method in two ways: (i) only the ANK domains (Fig. 4A) and (ii) only the C-terminal 40 amino acids which contain the putative Skp1-binding domains (Fig. 4B). Sequences were aligned with MUSCLE (version 3.7), and phylogenetic trees were computed using the WAG substitution model. Phylogenetic analysis of the ANK domain from the 51-member ANK protein family revealed three strongly supported clades clustered by the virus host alga. Bootstrap branch supports of 100, 100, and 97 expected-likelihood weights (ELW) provide convincing evidence in support of the monophyly of the NC64A, SAG, and Pbi viruses, respectively (including Pbi virus NE-JV-1; 63 ELW). Surprisingly, the ANK domains from this protein family are well preserved among the chlorovirus families as several of these ANK domains have 100% sequence identity (Table 1). These ANK domains are most diverse when viruses that infect different host algae are compared (e.g., the highest degree of sequence identity of the ANK domains between any two virus types is less than 50%).
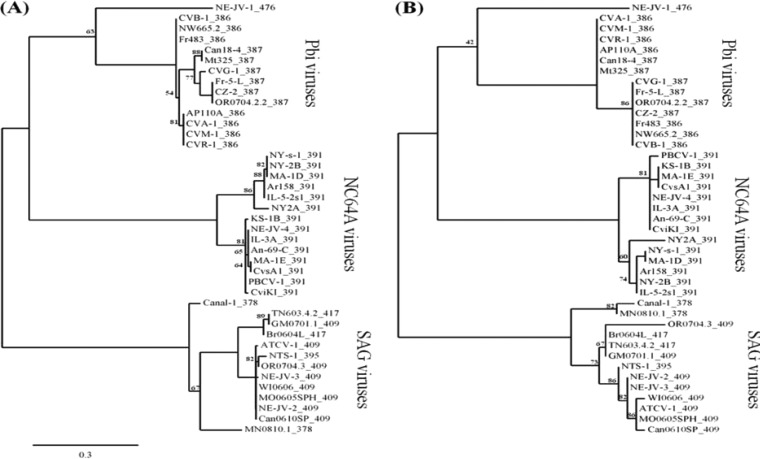
FIG 4.
Phylogenetic trees of the 51-member ANK repeat protein family coded by chloroviruses. (A) ANK domain of the ANK family proteins. (B) Putative Skp1-binding domain (last 40 residues of the C terminus) of the ANK family proteins. A maximum-likelihood reconstruction method was based on the separate alignments of the ANK and Skp1-binding domains from the ANK family proteins present in 41 chloroviruses that infect three algal hosts (10 gene duplicates were omitted from the analysis). The ANK and Skp1-binding domains are located in the amino and carboxyl termini of the ANK protein, respectively. The following chloroviruses are represented (corresponding geographical sampling locations): NC64A viruses NY-2A (NY, USA), NY-2B (NY, USA), Ar158 (Buenos Aires, Argentina), NY-s-1 (NY, USA), MA-1D (MA, USA), IL-5-2s1 (IL, USA), An-69-C (Canberra, Australia), IL-3A (IL, USA), PBCV-1 (NC, USA), NE-JV-4 (NE, USA), MA-1E (MA, USA), CvsA1 (Sawara, Japan), and CviKI (Kyoto, Japan); SAG viruses Can0610SP (British Columbia, Canada), OR0704.3 (OR, USA), NE-JV-3 (NE, USA), NE-JV-2 (NE, USA), ATCV-1 (Stuttgart, Germany), WI0606 (WI, USA), MO0605SPH (MO, USA), NTS-1 (NE, USA), Br6064L (Sao Paolo, Brazil), TN603.4.2 (TN, USA), GM0701.1 (Guatemala), MN0810.1 (MN, USA), and Canal-1 (NE, USA); Pbi viruses NE-JV-1 (NE, USA), Mt325 (MT, USA), Can18-4 (Canada), Fr-5-L (France), CZ-2 (Czech Republic), OR0704.2.2 (OR, USA), CVB-1 (Berlin, Germany), Fr483 (France), NW665.2 (Norway), AP110A (Unknown), CVA-1 (Amönau, Germany), CVG-1 (Göttingen, Germany), CVM-1 (Marburg, Germany), and CVR-1 (Rauschenberg, Germany). Numbers following virus names on the figure correspond to the total number of amino acid residues.
TABLE 1.
Protein sequence similarity of chlorovirus A607R ANK family and vSkp1 proteins
| Chlorovirus group | % sequence similarity |
||
|---|---|---|---|
| ANK domain | ANK protein C terminus | vSkp1 full-length protein | |
| NC64A | 72.7–100 | 62–100 | 64.9–100 |
| SAG | 60.7–100 | 62.5–100 | 74.5–100 |
| Pbia | 73.2–100 | 85–100 | 89–100 |
Percent protein sequence similarity of Pbi viruses was calculated excluding NE -JV-1.
Analysis of the last 40 residues of the C-terminal region (putative Skp1-binding domain) from the 51-member ANK family again showed three strongly supported clades grouped by virus host alga with equivalent bootstrap branch supports (Fig. 4B). The highest degree of sequence identity of the terminal region between any two virus types was only 35% while several of these C-terminal regions are identical among viruses that infect the same host (Table 1). As expected, the clustering of the C-terminal region from Pbi virus NE-JV-1 was a close outlier, as it was for the ANK domain (42 ELW).
Chloroviruses encode an Skp1.
Genome analysis revealed one vSkp1 protein in all sequenced chloroviruses except for NC64A virus KS-1B. Skp1 is a core component of the eukaryotic SCF complex that mediates proteosome-dependent protein degradation. Cellular Skp1 also functions in cell cycle progression, transcriptional regulation, and signal transduction (42). Multiple-sequence alignment of cellular Skp1 homologs, including ones from the host C. variabilis, were aligned with vSkp1 proteins from NC64A, SAG, and Pbi virus members PBCV-1, ATCV-1, and FR483, respectively (Fig. 5). Protein alignment showed 70% conserved amino acid composition among the cellular and viral proteins. Phylogenetic analysis of the 40 vSkp1 proteins revealed three strongly supported clades grouped by the virus host alga (Fig. 6). Bootstrap branch supports of 99, 100, and 99 ELW support the monophyly of the NC64A, SAG, and Pbi viruses, respectively (including Pbi virus NE-JV-1; 97 ELW). While several chloroviruses infecting the same host have identical Skp1 proteins, the highest sequence identity between Skp1 proteins from chloroviruses that infect different hosts is less than 40% (Table 1).
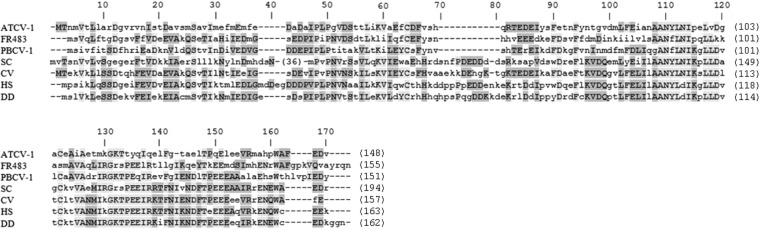
FIG 5.
Sequence alignment of Skp1 homologs. Amino acid identities are shaded as the most conserved (according to BLOSUM62). Average BLOSUM62 scores represented as a maximum of 3.0 (light gray) and a minimum of 0.5 (dark gray). S. cerevisiae Skp1 has a 36-amino-acid insertion of primarily N, D, and E residues in the N-terminal domain. SC, S. cerevisiae; CV, C. variabilis (GenBank accession number EFN54040); HS, H. sapiens; DD, D. discoideum. Protein alignment was created using Phylogeny.fr (38).
FIG 6.
Phylogenetic tree of vSkp1 homologs coded by chloroviruses. A maximum-likelihood reconstruction method was based on an alignment of chlorovirus-encoded vSkp1 genes native to 40 chloroviruses that infect three algal hosts. Sequences were aligned with MUSCLE (version 3.7) configured with default settings. The following parameters were enforced: the minimum number of sequences for a flank position was 85%; all segments with contiguous nonconserved positions bigger than 8 were rejected; the minimum block length was 10; no gaps were allowed in the final alignment. The phylogenetic tree was computed using the WAG substitution model. We show only bootstrap values of <90%. Branches with bootstrap support of less than 50 were collapsed.
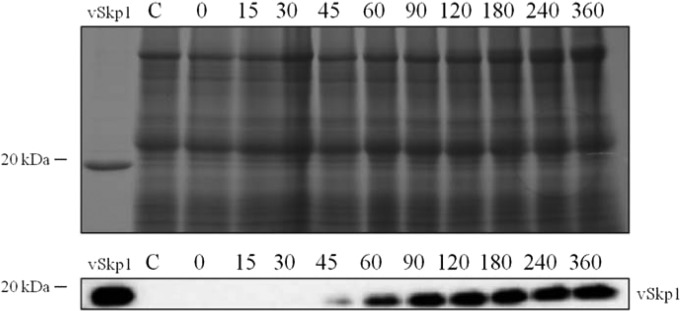
Neither the chlorovirus ANK proteins nor vSkp1 proteins were associated with the PBCV-1 particle proteome, which contains 148 unique virus-encoded proteins (about 35% of the virus coding capacity) and 1 host protein (43). The genes encoding these virion-associated proteins are dispersed throughout the virus genome, and most are transcribed late or early-late in the infection cycle. In contrast, the PBCV-1 ANK and vSkp1 genes are expressed as early genes (44, 45). Western blotting confirmed the appearance of the vSkp1PBCV-1 protein at 45 min p.i. (Fig. 7).
FIG 7.
Early translation of vSkp1PBCV-1 protein. Total protein was isolated from uninfected (control) and PBCV-1-infected C. variabilis cells at the indicated times (min). Time zero signifies cells that were immediately harvested after virus addition. Viral lysates were analyzed by SDS-PAGE (upper panel) and immunoblotting (lower panel). Detection of the vSkp1PBCV-1 protein is shown with increasing intensity with time. The protein was probed with antiserum against vSkp1PBCV-1.
Cellular Skp1 and vSkp1PBCV-1 proteins interact differently with two of eight ANK repeat proteins from PBCV-1.
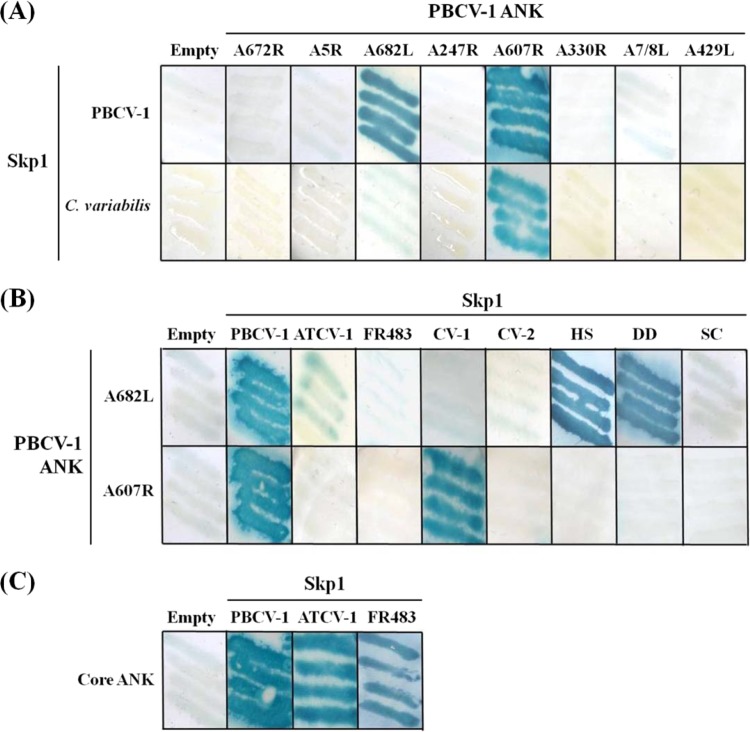
We performed Y2H analysis to test the hypothesis that PBCV-1 ANK proteins interacted with either cellular or vSkp1PBCV-1. Sequences encoding the ANK proteins and the Skp1 targets were yeast codon optimized and cloned into the pDEST32 bait and pDEST22 prey vectors to allow testing for interactions. The Y2H analyses revealed selective binding interactions between PBCV-1 ANK and Skp1 proteins. The results established that vSkp1PBCV-1 interacted strongly with two of the eight ANK proteins, PBCV-1 A682L and A607R (Fig. 8A). PBCV-1 A607R interacted with one of the two host C. variabilis Skp1 proteins (GenBank accession number EFN54040), which is the more highly conserved Skp1 member. The remaining six PBCV-1 ANK proteins did not interact with any of the Skp1 proteins. To further test the Skp1-binding potential of the ANK proteins A682L and A607R, we assayed these ANK proteins in the bait vector for binding with the cellular Skp1 proteins from human, Dictyostelium discoideum, and S. cerevisiae, as well as vSkp1ATCV-1 and vSkp1FR483 proteins in addition to vSkp1PBCV-1. ANK protein PBCV-1 A682L interacted with cellular Skp1 proteins from human and D. discoideum while weakly interacting with vSkp1ATCV-1 (Fig. 8B). PBCV-1 A682L did not interact with any additional Skp1 homologs. A summary of Y2H interactions involving ANK proteins PBCV-1 A682L and A607R is reported in Table 2.
FIG 8.
Interactions between PBCV-1 and cellular SCF-associated proteins detected by Y2H. (A) PBCV-1-encoded ANK proteins A682L and A607R interacted with vSkp1PBCV-1; however, only A607R interacted with Skp1 from host C. variabilis (GenBank accession number EFN54040). (B) PBCV-1 A682L interacted with cellular Skp1 proteins from human (HS) and D. discoideum (DD) while weakly interacting with vSkp1ATCV-1. Both PBCV-1 ANK proteins did not interact with the second Skp1 from host C. variabilis (CV-2) (GenBank EFN51326). (C) Similar to the binding between vSkp1PBCV-1 and A607R, vSkp1ATCV-1 and vSkp1FR483 proteins also interact with their partner ANK family proteins, Z568R and N710R, respectively. Interacting expression vectors induce the downstream colorimetric reporter gene lacZ (blue), indicating positive protein-protein interaction. The eight PBCV-1 ANK proteins are designated by gene locus tag.
TABLE 2.
Protein interactions between Skp1 and ANK tested via yeast two-hybrid assays
| Skp1 proteinb | Interaction with ANK proteina |
|
|---|---|---|
| A682L | A607R | |
| vSkp1PBCV-1 | + | + |
| vSkp1ATCV-1 | + | − |
| vSkp1FR483 | − | − |
| Skp1C. variabilis | − | + |
| Skp1H. sapiens | + | − |
| Skp1D. discoideum | + | − |
+, proteins Skp1 and ANK interacted; −, proteins Skp1 and ANK did not interact.
The origins of Skp1 homologs are identified by subscripts.
ANK protein PBCV-1 A682L interacts with Skp1 proteins via a carboxyl Skp1-interactional motif.
We constructed truncated versions of ANK protein PBCV-1 A682L by making a series of deletions from the N terminus (Fig. 9A) in order to identify the minimum domain required for binding to the Skp1 proteins. According to ExPASy's PROSITE motif scan (46), A682L has seven ANK domains, designated by the following residue numbers: 38 to 70, 71 to 103, 104 to 136, 137 to 169, 170 to 202, 203 to 235, and 236 to 268. We constructed six mutants with different combinations of ANK repeats and Skp1-binding region deletions. One mutant contained only the predicted Skp1-binding domain excluding all ANK repeat motifs while another construct contained all the ANK repeats without the C-terminal putative Skp1-binding domain. An additional double point mutation of A682L was constructed by replacing the highly conserved C-terminal dipeptide L335-P336 of the putative Skp1-binding motif with two alanine residues. Y2H screening was performed using the series of truncated A682L mutants with Skp1 proteins (Fig. 9B). The results showed that deletion of the Skp1-binding domain (ΔF-box) eliminated binding ability to the PBCV-1, human, and D. discoideum Skp1 proteins. The site-directed A682L mutant (L335A-P336A) did not bind to any Skp1 homologs, further supporting its role as a key binding dipeptide. Similar mutations in yeast F-box proteins also disrupt Skp1 protein interactions in vitro (47). Evidence of binding of the mutant lacking ANK domains 1 to 5 (ΔANKR1–5) demonstrated that at least two ANK domains adjacent to the C-terminal putative Skp1-binding region (approximately the last 40 residues) are sufficient for virus and heterogeneous Skp1 protein interaction. Thus, the interactive behavior of A682L requires the equivalent obligatory domains regardless of origin of the Skp1 binding partner.
FIG 9.
PBCV-1-encoded ANK protein A682L deletion mutant constructs interact in a putative Skp1 interactional motif-dependent manner. (A) Five truncated versions of the full-length PBCV-1 A682L were constructed by making a series of deletions from the N terminus (deletion ANK domains 1 to 3 [ΔANKR1–3], ΔANKR1–5, ΔANKR1–6, and ΔANKR1–7). Another mutant incorporated all the ANK domains without the C-terminal putative Skp1-binding domain (ΔF-box). ANK domains 1 to 7 in A682L are located at the following residue numbers: 38 to 70, 71 to 103, 104 to 136, 137 to 169, 170 to 202, 203 to 235, and 236 to 268, respectively. The putative Skp1-binding domain is shown in dark gray, the ANK domain is shown in black, and functional unknown domains are shown in light gray. (B) Growth of yeast strains harboring versions of the A682L protein as prey and Skp1 as bait assayed with X-Gal, together with their β-galactosidase activities, are shown. Y2H results indicate that truncated mutants missing the first three and five ANK domains (ΔANKR1–3 and ΔANKR1–5, respectively) retain their ability to interact with the Skp1 protein from PBCV-1 and slightly with Skp1 from human and D. discoideum. Deletion of the Skp1-binding domain (ΔF-box) from the viral ANK protein eliminated binding to all Skp1 members. The site-directed A682L double mutant (A682L-L335A-P336A) did not bind to any Skp1 homolog, further supporting thee role of these residues as key binding residues in cellular F-box proteins. Empty vectors were used as negative controls. Values are means ± standard deviations (n = 3).
Chlorovirus-encoded ANK repeat family members and vSkp1 proteins interact exclusively within corresponding monophyly clusters.
To test the interactive boundaries of the 51-member ANK protein family, we performed additional Y2H trials using ANK family members from the remaining two chlorovirus types established by their algal host: ATCV-1 Z568R (a SAG virus) and FR483 N710R (a Pbi virus). We also cloned vSkp1 proteins from chloroviruses ATCV-1 and FR483. The results indicated positive protein interactions between all vSkp1 proteins tested and their partner ANK family proteins (Fig. 8C). These data confirm the binding of vSkp1PBCV-1 and PBCV-1 A607R. To examine binding exclusivity, we tested the same ANK proteins against C. variabilis, human, and D. discoideum Skp1 proteins. There was no interaction with human and D. discoideum Skp1 proteins although PBCV-1 protein A607R interacted with C. variabilis Skp1 (GenBank accession number EFN54040). These results validate the specificity that chlorovirus-encoded ANK family proteins interact with their partnered vSkp1 exclusively and one of two Skp1 proteins from the host alga.
vSkp1PBCV-1 interacts with endogenous cellular F-box proteins and PBCV-1 ANK repeat proteins.
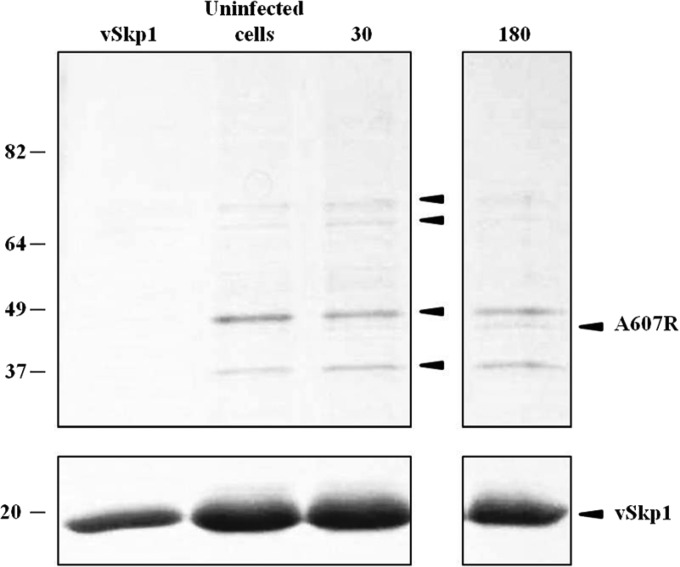
Although the results from the Y2H screen were convincing, it cannot be assumed that an interaction observed with the yeast system also occurs in a functional assay. To address this concern, we used absorption chromatography to test endogenous protein interactions. We constructed a plasmid to overexpress the His-tagged vSkp1PBCV-1 protein and immobilized it on a Ni2+ column. Three cell lysates were applied separately to either the viral Skp1-charged resin or an uncharged resin: (i) uninfected C. variabilis cells, (ii) C. variabilis cells at 30 min p.i., and (iii) C. variabilis cells at 180 min p.i. The His-tagged vSkp1PBCV-1 and the binding partners contained in the lysates were subsequently eluted from the Ni2+ column. Eluted proteins were resolved by SDS-PAGE with Coomassie blue staining. A subtle band was detected in the viral lysate precipitate that was absent in the uninfected control precipitate, which contained four bands native to the host C. variabilis (Fig. 10). No proteins were detected in the lysates applied to the uncharged resin. The viral lysate-specific band, as well as the four bands from the uninfected host cells, was excised, digested, and analyzed by nano-LC-MS/MS. All detected proteins corresponded to either cellular F-box proteins from C. variabilis or PBCV-1-encoded ANK proteins. A protein of 17 kDa in size was identified as the recombinant vSkp1PBCV-1 protein in each lysate. In the uninfected lysate, proteins of 35 kDa, 44 kDa, 67 kDa, and 76 kDa were identified as C. variabilis F-box proteins consisting of 331 (GenBank EFN51283), 393 (GenBank EFN54870), 621 (UniProt E1ZSC7), and 739 (GenBank EFN59680) amino acid residues, respectively. In the viral infected cell lysates, one protein below the 49-kDa marker appeared at 30 min p.i. and increased in intensity at 180 min p.i. This band corresponded to PBCV-1 ANK protein A607R. Interestingly, the proteins identified as C. variabilis F-box proteins of 67 kDa and 76 kDa decreased in the viral infected cell lysates as the viral ANK protein increased. There are two plausible explanations for this observation: (i) the chlorovirus ANK proteins are competing with cellular F-box proteins for vSkp1 binding, or (ii) the host transcriptional machinery and protein synthesis are downregulated later in infection.
FIG 10.
Ni2+ pulldown assay confirms vSkp1PBCV-1 interaction with F-box and ANK family proteins. vSkp1PBCV-1-charged resin was subjected to three separate lysates from uninfected and PBCV-1-infected C. variabilis cells at 30 and 180 min p.i. Proteins were eluted from the column and resolved by SDS/PAGE. Binding proteins were excised and analyzed by MS. Pulldown results confirm that vSkp1PBCV-1 interacts with four endogenous cellular F-box proteins from C. variabilis (designated by black arrows in third column) and PBCV-1 ANK family protein, A607R (fourth column). Cellular F-box proteins bound to vSkp1 are identified according to their accession number in descending order by their molecular sizes: GenBank accession number EFN59680, UniProt E1ZSC7, GenBank EFN54870, and GenBank EFN51283, respectively.
DISCUSSION
The widespread prevalence of F-box proteins emphasizes their importance in intracellular regulation, particularly in substrate recruitment to the SCF complex (35, 48–50). They represent almost 2.3% of the protein-coding genes in the model plant Arabidopsis thaliana (50, 51), and increasing evidence suggests that plants use the ubiquitin-proteasome system (UPS) to recognize and combat pathogen invasion (7). Most of the characterized F-box proteins are associated with the cellular SCF complex and function as the substrate recognition domain for the SCF complex. Using protein sequence alignment and bioinformatics, we identified eight ANK proteins coded by chlorovirus PBCV-1 which contain N-terminal ankyrin repeats together with possible C-terminal Skp1-binding domains. Previous studies have shown that poxviruses, which have the same domain organization as the chloroviruses, encode ANK proteins. These poxvirus ANK proteins interact with the cellular Skp1 using a C-terminal domain similar to an F-box (9, 19–23). Considering that this domain arrangement does not exist in cellular F-box proteins (40), the conservation of these ANK genes in chloroviruses encouraged us to speculate that PBCV-1 ANK proteins interact with Skp1 and thus play an intimate role in modulating host cell SCF-mediated ubiquitination. Y2H screening and pulldown methods identified protein-protein interactions between Skp1 members and their interacting F-box-like-containing proteins. This approach established that vSkp1PBCV-1 interacts with PBCV-1 ANK proteins A682L and A607R. The failure to interact with the other six PBCV-1 ANK proteins may be related to several factors, which were beyond the scope of this study. One possibility is that posttranslational modifications might be required for either the Skp1 or the remaining six ANK proteins to interact. Examples of such posttranslational regulation are seen in Dictyostelium (52). Moreover, vSkp1ATCV-1 and vSkp1FR483 interacted with their own respective PBCV-1 A607R homologs, Z568R and N710R, respectively. Collectively, our data indicate that chloroviruses encode a vSkp1 capable of interaction with viral ANK proteins via a C-terminal domain with functional similarity to F-box domains.
As noted above, the chlorovirus Skp1-binding domains, like those of the poxviruses, are located at the C terminus of ANK proteins, which is the opposite of the location in cellular F-box proteins (19). Despite the lack of obvious homology of the C-terminal region of PBCV-1 ANK proteins to F-box domains, they share some key residues with human F-box protein Skp2 that contacts cellular Skp1 (Fig. 2). Here, we show that deleting the putative Skp1-binding domain in PBCV-1 A682L resulted in a protein that no longer interacted with its counterpart vSkp1. Serial domain deletions of A682L from the N terminus further support an essential Skp1-binding region required for protein interaction while in tandem with at least two ankyrin repeats. These results suggest that this domain combination is the minimum size required for binding to Skp1 family members. Additionally, a signature dipeptide of cellular F-box proteins, Leu-Pro, located at the beginning of the putative viral Skp1-binding domain, was present in three PBCV-1 ANK proteins. Replacement of this dipeptide with Ala-Ala in A682L prevented protein interaction with vSkp1PBCV-1. However, it is important to note that one of the viral ANK proteins (A607R) lacks the signature Leu-Pro residues, and it binds to vSkp1PBCV-1, even though its C terminus has fewer common residues with a canonical F-box domain.
A comprehensive phylogenetic analysis of PBCV-1 A607R identified a 51-member family of ANK proteins coded by all sequenced chloroviruses. This conserved class of ANK proteins constitutes a newly discovered protein family, with one member present in all 41 annotated chloroviruses. Using multiple-sequence alignment methods, we showed high sequence identity among these ANK proteins according to the respective virus families. Further phylogenetic analysis of the ANK and putative Skp1-binding domains from this ANK protein family revealed three monophyletic clades separated according to the three host algae, suggesting partnered function tailored to the host alga (Fig. 4). From this result, we speculate that this unique class of ANK genes serves as a repertoire of partnering proteins that function with chlorovirus-encoded vSkp1s. Our results indicate that a degenerative F-box-like motif resides in these ANK proteins that is capable of recognizing the host and vSkp1 proteins exclusively.
While the protein sequences of the C-terminal domains of the viral ANK proteins have weak homology to a canonical F-box domain, they do have bona fide F-box-like function based on protein-protein interactions with Skp1, predicted structural similarities to Skp1-canonical F-box interactions, the presence of core conserved Leu-Pro residues in some of the proteins, and a domain organization similar to that of the chordopox NCLDV member that has ANK proteins containing a C-terminal PRANC domain. The weak sequence homology to the canonical F-box consensus is a significant finding because it reveals a much wider range of primary amino acid sequences that can fold into domains that mediate Skp1 binding than was previously appreciated. Rather than weakening the evidence for the F-box-like function of the chlorovirus ANK proteins, the low sequence homology suggests that Skp1-interacting motifs bearing F-box-like function exist in a wider range of repeat-containing proteins, such as ANK-containing proteins, than previously envisioned. This observation is likely to enhance the discovery of new F-box-like proteins.
In addition to encoding F-box-like ANK proteins, chloroviruses encode a candidate virulence factor, a Skp1 homolog, for intercepting the ubiquitin-proteasome system. While multiple Skp1 homologs exist in cellular organisms (28) including C. variabilis, virus-encoded Skp1 genes have only been reported in algal viruses, with the exception of the recently sequenced pandoraviruses (53). Sharing the domain architecture conserved among Skp1 family members (Fig. 5), chlorovirus vSkp1 homologs contain two separate protein interaction motifs: a POZ (pox virus and zinc finger) domain located in the N-terminal region (1 to 61 amino acids) and an Skp1 domain that resides in the C-terminal region (68 to 145 amino acids). In cellular Skp1 proteins, the POZ domain mediates contact with SCF subunit Cul1 while the Skp1 domain is responsible for binding the F-box-containing proteins. Phylogenetic analyses clearly demonstrate host-based clusters of the conserved cellular homolog, further suggesting specialized functionality that caters to their host-virus interactions. Together, the presence of bipartite domains and monophyly clusters according to virus host suggests that chlorovirus vSkp1 proteins are uniquely expressed to complement a counterpart ANK family protein, thereby mimicking cellular Skp1–F-box binding properties.
As the only chlorovirus that does not encode a vSkp1 protein, NC64A virus KS-1B represents an outlier of the 41 sequenced and annotated chloroviruses. The absence of the vSkp1 in this single example questions the gene's importance in pathogenesis. Furthering speculation, four spontaneously derived, large deletion mutants of PBCV-1 suggest that the vSkp1 gene is not an essential gene for virus replication in a laboratory environment, according to the study of Landstein et al. (54). In this later study, one PBCV-1 mutant contained a 37-kb deletion at the left end of its 331-kb genome, which encoded 26 single-copy open reading frames including vSkp1. Interestingly, replication of the deletion mutants is attenuated as their burst sizes were half that of the parent virus burst size, and their plaque sizes were consequently smaller. This reduced virulence and plaque phenotype suggest a potential importance for vSkp1 in chlorovirus replication within the limits of the mutant analysis that included several other genes in the deletion.
Adding to the complexity of pathogenesis, some viruses, including the chloroviruses, encode their own ubiquitin homologs to enable replication and evade cellular immune defenses (55, 56). Depending on their needs, viruses have developed means to enhance or inhibit ubiquitination of specific substrates. In fact, 11 SAG viruses, including ATCV-1, and one NC64A virus, NY-2A, encode their own viral ubiquitin (Z203L and B699L, respectively), sharing up to 97% sequence identity with human ubiquitin. Besides encoding ubiquitin, many viruses encode putative E3 ligases and deubiquitylating enzymes, as well as adaptor proteins. For example, in addition to PBCV-1-encoded protein vSkp1 (A39L), PBCV-1 encodes putative enzymes involved in protein degradation, including a ubiquitin C-terminal hydrolase (A105L) and RING finger E3 ubiquitin ligase (A481L) (43). Together, this ensemble of virus-encoded ubiquitin-related genes creates an arsenal of proteins that threaten the host's UPS machinery. These viral proteins may engage the ubiquitin system through ubiquitin-binding motifs, another important specificity-providing component of the ubiquitin system. Determining the true physiological substrates of virally encoded enzymes will be challenging, as is determining their effect on viral replication in vivo.
ACKNOWLEDGMENTS
We thank James Gurnon and Irina Agarkova for their technical help and maintaining the culture conditions and David Dunigan and Chris West for helpful discussions.
This research was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 1 T32 AIO60547 (E.A.N.) from the National Institute of Allergy and Infectious Diseases, NSF-EPSCoR grant EPS-1004094 (J.L.V.E.), Stanley Medical Research Institute 11R-0003 (J.L.V.E.), NIH grant P20 RR15635 from the COBRE Program of the National Center for Research Resources (J.L.V.E.), and the Department of Energy (DE-EE00003373) Consortium for Algae Biofuels Commercialization (G.A.O.).
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1. Gao G, Luo H. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84:5–14. 10.1139/y05-144. [DOI] [PubMed] [Google Scholar]
- 2. Ulrich HD, Walden H. 2010. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11:479–489. 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 3. Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703–709. 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 4. Surjit M, Varshney B, Lal SK. 2012. The ORF2 glycoprotein of hepatitis E virus inhibits cellular NF-κB activity by blocking ubiquitination mediated proteasomal degradation of IκBαin human hepatoma cells. BMC Biochem. 13:7. 10.1186/1471-2091-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thiel H, Hleibieh K, Gilmer D, Varrelmann M. 2012. The p25 pathogenicity factor of beet necrotic yellow vein virus targets the sugar beet 26S proteasome involved in the induction of a hypersensitive resistance response via interaction with an F-box protein. Mol. Plant Microbe Interact. 25:1058–1072. 10.1094/MPMI-03-12-0057-R. [DOI] [PubMed] [Google Scholar]
- 6. Correa RL, Bruckner FP, de Souza Cascardo R, Alfenas-Zerbini P. 2013. The role of F-box proteins during viral infection. Int. J. Mol. Sci. 14:4030–4049. 10.3390/ijms14024030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Magori S, Citovsky V. 2011. Hijacking of the host SCF ubiquitin ligase machinery by plant pathogens. Front. Plant Sci. 2:87. 10.3389/fpls.2011.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuink TJ, Crosby WL, Hooykaas PJ. 2001. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11:258–262. 10.1016/S0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 9. van Buuren N, Couturier B, Xiong Y, Barry M. 2008. Ectromelia virus encodes a novel family of F-box proteins that interact with the SCF complex. J. Virol. 82:9917–9927. 10.1128/JVI.00953-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. 2004. Inventory and functional analysis of the large HRP regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type iii secretion system. Mol. Microbiol. 53:115–128. 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- 11. Both GW. 2002. Identification of a unique family of F-box proteins in atadenoviruses. Virology 304:425–433. 10.1006/viro.2002.1734. [DOI] [PubMed] [Google Scholar]
- 12. Aronson MN, Meyer AD, Gyorgyey J, Katul L, Vetten HJ, Gronenborn B, Timchenko T. 2000. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 74:2967–2972. 10.1128/JVI.74.7.2967-2972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. 2006. F-box-like domain in the polerovirus protein p0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U. S. A. 103:1994–1999. 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 18:132–139. 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Werden SJ, Lanchbury J, Shattuck D, Neff C, Dufford M, McFadden G. 2009. The myxoma virus M-T5 ankyrin repeat host range protein is a novel adaptor that coordinately links the cellular signaling pathways mediated by Akt and Skp1 in virus-infected cells. J. Virol. 83:12068–12083. 10.1128/JVI.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Werden SJ, McFadden G. 2008. The role of cell signaling in poxvirus tropism: the case of the M-T5 host range protein of myxoma virus. Biochim. Biophys. Acta 1784:228–237. 10.1016/j.bbapap.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 17. Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435–1448. 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siozios S, Ioannidis P, Klasson L, Andersson SG, Braig HR, Bourtzis K. 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8:e55390. 10.1371/journal.pone.0055390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular Scf1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. U. S. A. 105:10955–10960. 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanie S, Gelfi J, Bertagnoli S, Camus-Bouclainville C. 2010. MNF, an ankyrin repeat protein of myxoma virus, is part of a native cellular SCF complex during viral infection. Virol. J. 7:56. 10.1186/1743-422X-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buttigieg K, Laidlaw SM, Ross C, Davies M, Goodbourn S, Skinner MA. 2013. Genetic screen of a library of chimeric poxviruses identifies an ankyrin repeat protein involved in resistance to the avian type I interferon response. J. Virol. 87:5028–5040. 10.1128/JVI.02738-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-κB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 83:4140–4152. 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sperling KM, Schwantes A, Schnierle BS, Sutter G. 2008. The highly conserved orthopoxvirus 68k ankyrin-like protein is part of a cellular SCF ubiquitin ligase complex. Virology 374:234–239. 10.1016/j.virol.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 24. Muniz JR, Guo K, Kershaw NJ, Ayinampudi V, von Delft F, Babon JJ, Bullock AN. 2013. Molecular architecture of the ankyrin SOCS box family of Cul5-dependent E3 ubiquitin ligases. J. Mol. Biol. 425:3166–3177. 10.1016/j.jmb.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yutin N, Koonin EV. 2012. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol. J. 9:161. 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Proschold T, Darienko T, Silva PC, Reisser W, Krienitz L. 2011. The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 13:350–364. 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 27. Van Etten JL, Dunigan DD. 2012. Chloroviruses: not your everyday plant virus. Trends Plant Sci. 17:1–8. 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW. 2007. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50:873–885. 10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 29. Inoue H, Nojima H, Okayama H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28. 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 30. Gietz D, St Jean A, Woods RA, Schiestl RH. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Etten JL, Burbank DE, Xia Y, Meints RH. 1983. Growth cycle of a virus, PBCV-1, that infects Chlorella-like algae. Virology 126:117–125. 10.1016/0042-6822(83)90466-X. [DOI] [PubMed] [Google Scholar]
- 32. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350–4354. 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4:363–371. 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34. Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. Patchdock and Symmdock: servers for rigid and symmetric docking. Nucleic Acids Res. 33:W363–W367. 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381–386. 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 36. Kipreos ET, Pagano M. 2000. The F-box protein family. Genome Biol. 1:REVIEWS3002. 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool Web services from the EMBL-EBI. Nucleic Acids Res. 41:W597–W600. 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dereeper A, Audic S, Claverie JM, Blanc G. 2010. Blast-explorer helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mercer AA, Fleming SB, Ueda N. 2005. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes 31:127–133. 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- 41. Barry M, van Buuren N, Burles K, Mottet K, Wang Q, Teale A. 2010. Poxvirus exploitation of the ubiquitin-proteasome system. Viruses 2:2356–2380. 10.3390/v2102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hellmann H, Estelle M. 2002. Plant development: regulation by protein degradation. Science 297:793–797. 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- 43. Dunigan DD, Cerny RL, Bauman AT, Roach JC, Lane LC, Agarkova IV, Wulser K, Yanai-Balser GM, Gurnon JR, Vitek JC, Kronschnabel BJ, Jeanniard A, Blanc G, Upton C, Duncan GA, McClung OW, Ma F, Van Etten JL. 2012. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J. Virol. 86:8821–8834. 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kawasaki T, Tanaka M, Fujie M, Usami S, Yamada T. 2004. Immediate early genes expressed in chlorovirus infections. Virology 318:214–223. 10.1016/j.virol.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 45. Blanc G, Mozar M, Agarkova IV, Gurnon JR, Yanai-Balser G, Rowe JM, Xia Y, Riethoven JJ, Dunigan DD, Van Etten JL. 2014. Deep RNA sequencing reveals hidden features and dynamics of early gene transcription in Paramecium bursaria chlorella virus 1. PLoS One 9:e90989. 10.1371/journal.pone.0090989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41:D344–D347. 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Russell ID, Grancell AS, Sorger PK. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933–950. 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Willems AR, Schwab M, Tyers M. 2004. A hitchhiker's guide to the Cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695:133–170. 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 49. Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. 1996. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263–274. 10.1016/S0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 50. Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 99:11519–11524. 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hua Z, Zou C, Shiu SH, Vierstra RD. 2011. Phylogenetic comparison of F-box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS One 6:e16219. 10.1371/journal.pone.0016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. West CM, Wang ZA, van der Wel H. 2010. A cytoplasmic prolyl hydroxylation and glycosylation pathway modifies Skp1 and regulates O2-dependent development in Dictyostelium. Biochim. Biophys. Acta 1800:160–171. 10.1016/j.bbagen.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 54. Landstein D, Burbank DE, Nietfeldt JW, Van Etten JL. 1995. Large deletions in antigenic variants of the chlorella virus PBCV-1. Virology 214:413–420. 10.1006/viro.1995.0051. [DOI] [PubMed] [Google Scholar]
- 55. Albrecht M, Kuhne Y, Ballmer-Weber BK, Becker WM, Holzhauser T, Lauer I, Reuter A, Randow S, Falk S, Wangorsch A, Lidholm J, Reese G, Vieths S. 2009. Relevance of IgE binding to short peptides for the allergenic activity of food allergens. J. Allergy Clin. Immunol. 124:328–336.e6. 10.1016/j.jaci.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 56. Kahloul S, HajSalah El Beji I, Boulaflous A, Ferchichi A, Kong H, Mouzeyar S, Bouzidi MF. 2013. Structural, expression and interaction analysis of rice SKP1-like genes. DNA Res. 20:67–78. 10.1093/dnares/dss034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, Van Etten JL, Blanc G. 2013. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14:158. 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]