ABSTRACT
In healthy individuals, the functional immune system effectively confines human cytomegalovirus (CMV) replication, while viral immune evasion and persistence preclude sterile immunity. Mouse CMV (MCMV) is a well-established model to study the delicate CMV-host balance. Effective control of MCMV infection depends on the induction of protective type I interferon (IFN-I) responses. Nevertheless, it is unclear whether in professional antigen-presenting cell subsets MCMV-encoded evasins inhibit the induction of IFN-I responses. Upon MCMV treatment, enhanced expression of MCMV immediate-early and early proteins was detected in bone marrow cultures of macrophages and myeloid dendritic cells compared with plasmacytoid dendritic cell cultures, whereas plasmacytoid dendritic cells mounted more vigorous IFN-I responses. Experiments with Toll-like receptor (TLR)- and/or RIG-I like helicase (RLH)-deficient cell subsets revealed that upon MCMV treatment of myeloid cells, IFN-I responses were triggered independently of TLR and RLH signaling, whereas in plasmacytoid dendritic cells, IFN-I induction was strictly TLR dependent. Macrophages and myeloid dendritic cells treated with either UV-inactivated MCMV or live MCMV that lacked the STAT2 antagonist M27 mounted significantly higher IFN-I responses than cells treated with live wild-type MCMV. In contrast, plasmacytoid dendritic cells responded similarly to UV-inactivated and live MCMV. These experiments illustrated that M27 not only inhibited IFN-I-mediated receptor signaling, but also evaded the induction of IFN responses in myeloid dendritic cells. Furthermore, we found that additional MCMV-encoded evasins were needed to efficiently shut off IFN-I responses of macrophages, but not of myeloid dendritic cells, thus further elucidating the subtle adjustment of the host-pathogen balance.
IMPORTANCE MCMV may induce IFN-I responses in fibroblasts and epithelial cells, as well as in antigen-presenting cell subsets. We focused on the analysis of IFN-I responses of antigen-presenting cell subsets, including plasmacytoid dendritic cells, myeloid dendritic cells, and macrophages, which are all triggered by MCMV to mount IFN-I responses. Interestingly, myeloid dendritic cells and macrophages, but not plasmacytoid dendritic cells, are readily MCMV infected and support viral gene expression. As expected from previous studies, plasmacytoid dendritic cells sense MCMV Toll-like receptor 9 (TLR9) dependently, whereas in myeloid cells, IFN-I induction is entirely TLR and RLH independent. MCMV-encoded M27 does not impair the IFN-I induction of plasmacytoid dendritic cells, while in myeloid dendritic cells, it reduces IFN-I responses. In macrophages, M27 plus other, not yet identified evasins profoundly inhibit the induction of IFN-I responses. Collectively, these results illustrate that MCMV has evolved diverse mechanisms to differentially modulate IFN-I responses in single immune cell subsets.
INTRODUCTION
Mouse cytomegalovirus (MCMV) and human cytomegalovirus (CMV) are obligatory species-specific viruses. On the amino acid level, they share approximately 60% identical sequences within the central region of the genome (1). Although both viruses have developed a plethora of divergent species-specific evasins, the overall pathobiology of mouse and human CMV show certain similarities. In the mouse as well as in the human system, cellular immunity and interferons (IFN) play prominent roles in protection against CMV infection (2). IFN are essential to confine CMV replication and to promote the effector function of CD8+ T cells in vivo (3, 4). Upon MCMV infection, dendritic cells (DC) are one major source of type I IFN (IFN-I) (5–7).
Among other proinflammatory cytokines, IFN-I expression is induced upon the engagement of pattern recognition receptors (PRR) expressed by cells of the innate immune system (8). PRR comprise Toll-like receptors (TLR); cytosolic RNA detection systems, such as RIG-I (retinoic acid-inducible gene I)-like helicases (RLH); DNA sensors, including DAI, IFI16, AIM2, and other AIM-like receptors (ALR); and C-type lectin receptors (CLR) (9, 10). Upon triggering by their cognate ligands, TLR dimerize, undergo conformational changes, and recruit their adaptor proteins (11). All TLR, except TLR3, use the adaptor MyD88 (myeloid differentiation primary response gene 88), whereas TLR3 recruits TRIF (TIR domain-containing adapter-inducing IFN-β) for downstream signaling. Endosomal TLR3, TLR7/8, and TLR9 recognize nucleic acids, while TLR1, TLR2, TLR4, and TLR5 are expressed on the cell surface and recognize external pathogen determinants. CARDIF (CARD adaptor-inducing IFN-β) is localized to the outer mitochondrial membrane and recruits activated RLH, as well as their downstream signaling molecules (12). Recent studies revealed the existence of a new family of cytosolic nucleic acid sensors. This family includes the well-known double-stranded RNA (dsRNA)-sensing 2′-5′-oligoadenylate synthase (OAS) proteins and the DNA sensor cyclic GMP-AMP (cGAMP) synthase (cGAS) (reviewed in reference 13). cGAS functions in a classical PRR pathway that monitors the cytosol for the presence of DNA and triggers IFN-I production and antiviral gene expression through activation of stimulator of IFN genes (STING). In contrast, OAS proteins function as nucleic acid sensors in a more immediate antiviral restriction pathway by impeding translation (14).
While upon MCMV infection, the first wave of IFN-I is contributed by lymphotoxin-triggered splenic stroma, the second wave is primarily conferred by splenic plasmacytoid DC (pDC), which are triggered in a MyD88-dependent manner (15). Evasion of immune cell-derived cytokines, cytotoxic T lymphocytes (CTL), and natural killer (NK) cells has been studied extensively in MCMV-infected fibroblasts and epithelial cells (16–18). Nevertheless, little is known about how MCMV triggers different immune cell subsets to mount IFN-I responses and how MCMV evades the induction of IFN-I responses in antigen-presenting cell subsets. In a previous study, the MCMV gene M27, encoding a 79-kDa protein, was identified. It selectively binds and downregulates signal transducer and activator of transcription 2 (STAT2), and induces its polyubiquitination and subsequent proteasomal degradation by recruiting STAT2 to damage-specific DNA binding protein 1 (DDB1)-containing cullin ubiquitin ligase complexes. Thus, MCMV-encoded M27 interferes with signal transduction of antiviral IFN-I responses (19, 20). Deletion of M27 rendered MCMV dramatically more vulnerable to the antiviral effects elicited by IFN-I/II in vitro and in vivo. In their study, Zimmermann et al. investigated M27-mediated effects in infected murine fibroblasts. Despite the existence of a well-known feed-forward loop for the induction of IFN-I responses, M27 did not affect the induction of IFN-I responses in MCMV-permissive fibroblasts (21). The effect of M27 in MCMV infection of DC and macrophages (Mϕ) remains to be elucidated. DC and Mϕ are very heterogeneous groups of immune cells and play multiple roles at the interface between innate and adaptive immunity (22). Conventional myeloid DC (mDC) and Mϕ derive from a common myeloid progenitor, while pDC are also of myeloid origin but derive from a different progenitor than mDC and Mϕ (23). Whereas mDC and Mϕ show enhanced phagocytosis, antigen presentation, and costimulation activities, pDC are the main source of IFN-I in response to various viruses, including MCMV (24). In light of this information, we sought to investigate MCMV evasion of selected antigen-presenting cell subsets. Specifically, we studied whether M27 affected the MCMV-mediated induction of the IFN-I responses of pDC, mDC, and Mϕ.
MATERIALS AND METHODS
Mice and viruses.
C57BL/6 mice (aged 8 to 12 weeks) were purchased from Harlan Winkelmann. IFNAR−/− mice (25), which have been backcrossed 20 times on the C57BL/6 background, and MyD88−/− TRIF−/− mice (26), CARDIF−/− (27) mice, as well as MyD88−/− TRIF−/− CARDIF−/− (51) mice, were described previously. IFN-β reporter mice (messenger of IFN-β [MOB] [28]) expressing yellow fluorescent protein (YFP) from a bicistronic mRNA linked via an internal ribosome entry site (IRES) to IFN-β from within the endogenous ifnb gene locus, and IFN-α6 reporter mice expressing green fluorescent protein (GFP) instead of IFN-α6 (messenger of IFN-α6 [MOA] [29]), as well as double-reporter mice (messenger of IFN-β and IFN-α6 [MOBA] [30]), were used. IFN-β reporter mice carrying the luciferase gene in one allele of the IFN-β gene (IFN-βΔβ-luc/wt) were used (31). All mice were bred under specific-pathogen-free conditions at the mouse facility of the Helmholtz Centre for Infection Research, Braunschweig, Germany, or at the Twincore, Centre for Experimental and Clinical Infection Research, Hannover, Germany. All animal experiments were performed in compliance with the German animal protection law (TierSchG BGBI S. 1105; 25.05.1998). The mice were handled in accordance with good animal practice as defined by the Federation for Laboratory Animal Science Associations (FELASA). All animal experiments were approved by the responsible state office (Lower Saxony State Office of Consumer Protection and Food Safety). In this study, the MCMV strain Smith (1), the recombinant bacterial artificial chromosome (BAC)-derived wild-type (WT) MCMV MW97.01 (32), M27-deficient MCMV (MCMV-ΔM27) (19), as well as MCMV-Δm157/eGFP, expressing enhanced GFP (eGFP) under the control of the minimal CMV promoter within the m157 genomic region (MCMV-GFP), were used. UV irradiation of the virus with 0.25 J/cm2 was performed on ice using a UV cross-linker (MCMV-UV).
Generation of bone marrow-derived antigen-presenting cell subsets.
Femurs and tibias of mice were flushed with mouse medium (RPMI medium supplemented with 10% [vol/vol] fetal calf serum [FCS], 10 mM HEPES, 1 mM sodium pyruvate, 2 mM Glutamax [Gibco], 100 U/ml penicillin [Gibco], 100 μg/ml streptomycin [Gibco], and 0.1 mM 2-mercaptoethanol) to isolate bone marrow (BM) cells. To obtain pDC cultures, after red blood cell (RBC) lysis (using RBC lysing buffer [Sigma]), cells were washed and seeded at a density of 2 × 106 cells/ml in mouse medium supplemented with Flt-3L (100 ng/ml; R&D Systems) and incubated for 8 days. The medium of Flt-3L cultures was changed once at day 4 by replacing two-thirds of the cell culture volume with fresh medium supplemented with Flt-3L. To obtain mDC cultures, BM cells were seeded at a density of 1 × 106 cells/ml in mouse medium supplemented with granulocyte-macrophage colony-stimulating factor (GM-CSF) (100 ng/ml; R&D Systems), and the medium was changed at days 4, 6, and 7. To generate Mϕ cultures, 5 × 105 cells/ml were seeded in mouse medium supplemented with 10% L929 cell-conditioned medium (LCCM) as a source of macrophage colony-stimulating factor (M-CSF) (33). After 3 days of incubation, half of the medium was replaced with mouse medium containing 10% LCCM. At day 6, Mϕ were harvested from culture plates by incubating the cells for 15 min in 1 mM EDTA in phosphate-buffered saline (PBS). The Flt-3L cultures contained approximately 30 to 45% Siglec-H+ CD11c intermediate (CD11cint) pDC, the GM-CSF cultures contained approximately 70 to 80% CD11c+ CD11b+ mDC, and the M-CSF cultures typically contained more than 80% F4/80+ CD11b+ macrophages.
In vitro stimulation of bone marrow-derived immune cell subsets.
For in vitro stimulation experiments, pDC, mDC, and Mϕ were seeded at a density of 1 × 106 cells/ml in 96-well plates. The cells were treated for 18 h with the indicated MCMV preparations at a multiplicity of infection (MOI) of 0.3. As controls, pDC were stimulated with 0.5 μM of the TLR9 agonist CpG-oligodinucleotide (ODN) 1585 (Invivogen), whereas mDC and Mϕ were stimulated with 1 μg/ml poly(I·C) (Invivogen).
Flow cytometry.
For flow cytometric analysis of single-cell suspensions, cells were stained with combinations of antibodies specifically binding CD11b (M1/70.15; Caltag), CD11c (HL3; BD), F4/80 (Cl:A3-1; AbD Serotec), Siglec-H (eBio440c; eBioscience), CD69 (H1.2F3; BD), or CD86 (GL1; BD). For blocking of nonspecific Fcγ-receptor interactions, either murine poly-IgG produced in cell culture or CD16/CD32-specific antibody (2.4G2; BD) was used. Cells (5 × 105 to 5 × 106) were stained in 50 μl fluorescence-activated cell sorter (FACS) buffer (2% [wt/vol] bovine serum albumin [BSA], 20 mM EDTA, 0.2% sodium azide in PBS). Staining was performed for 15 to 20 min at 4°C. Dead cells were excluded from FACS analysis by using an Aqua LIVE/DEAD fixable stain kit (Invitrogen) that was applied after surface marker staining according to the manufacturer's instructions. The cells were subsequently washed with 1 ml FACS buffer and resuspended in 200 to 250 μl of FACS buffer supplemented with 0.25% paraformaldehyde (PFA). Samples were measured using a FACS LSR II (BD), and data were analyzed with FlowJo 7.6.5 software (TreeStar). pDC were purified from bone marrow cultures by magnetic activated cell sorter (MACS) using the AutoMACS Pro and anti-mPDCA-1 MicroBeads (Miltenyi) according to the manufacturer's instructions.
Cytokine measurement by ELISA.
For the determination of IFN-I levels in cell-free supernatants, IFN-α and IFN-β enzyme-linked immunosorbent assay (ELISA) methods were applied (eBioscience, PBL Biomedical Laboratories) following the manufacturer's instructions. Supernatants were harvested and stored at −20°C until they were tested for IFN-I. For enhancement of the sensitivity of the IFN-α ELISA method, samples were incubated for 1 h at room temperature (RT) on coated plates, followed by overnight incubation at 4°C.
Gene expression analysis.
For quantification of mRNA expression, RNA was extracted from 1 × 106 pDC, mDC, or Mϕ using a NucleoSpin RNA Kit (Qiagen), following the manufacturer's instructions. Total RNA (200 ng) was used for cDNA synthesis using a PrimeScript FirstStrand cDNA synthesis kit (TaKaRa) according to the manufacturer's instructions. Primers and SYBR green (Bioline) were added to 10 ng of the original RNA, and 1/10 of the reaction mixture was used for quantitative PCR (qPCR). PCRs were run in a LightCycler 480 (Roche). The fold changes of target genes were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as housekeeping gene (34). The following primers were used in this study: Isg15 (fwd, GAGCTAGAGCCTGCAGCAAT, and rev, TTCTGGGCAATCTGCTTCTT), 2′,5′-OAS (fwd, GGATGCCTGGGAGAGAATCG, and rev, TCGCCTGCTCTTCGAAACTG), and GAPDH (fwd, TGCACCACCAACTGCTTAGC, and rev, GGCATGGACTGTGGTCATGAG). IFN-α4 and IFN-β mRNAs were quantified with a QuantiFast Probe Assay by Qiagen (catalog no. QF00372162 and QF00237027). The fold changes of IFN-I subtypes were normalized to peptidylprolyl isomerase A (Ppia) (catalog no. QF00531286). The fold change was calculated as follows, according to reference 35: R (ratio) = 2−(ΔCT infected group − ΔCT control group), where ΔCT = CT target gene − CT housekeeping gene.
Protein analysis.
Cell lysates were analyzed on 8% acrylamide gels in SDS-PAGE and transferred to Hybond-ECL membranes (GE Healthcare) using a wet transfer cell (Bio-Rad). The membranes were blocked for 1 h at room temperature in 5% milk, followed by incubation with Croma101-, Croma103-, and M57.01-specific antibodies. Upon overnight incubation at 4°C, an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Dako) was used for 1 h at RT. Signals were visualized by chemiluminescence using the ECL Select (Amersham) LAS 3000 Imaging System (Fujifilm).
Determination of CMV replication kinetics.
Multistep growth in vitro was analyzed by infecting pDC, mDC, and macrophages in 24-well plates with MCMV at a multiplicity of infection of 0.1. After infection, the cells were not washed and were directly incubated in RPMI supplemented as described above. At specific time points postinfection (p.i.), the supernatants of the infected cells were harvested, cleared of cellular debris, and frozen at −80°C. Infectious virus was determined by a standard plaque assay on primary C57BL/6 murine embryonic fibroblasts (MEFs).
Statistics.
Statistical analyses were performed using GraphPad Prism 6.02 software (GraphPad). With this software, one-tailed, nonparametric Mann-Whitney U (unpaired) or Wilcoxon (paired) tests, as well as nonparametric Kruskal-Wallis (unpaired) tests or 2-way analyses of variance (ANOVA), were performed. ANOVA always included either Dunn's or Bonferroni correction for multiple comparisons.
RESULTS
mDC and Mϕ are more susceptible to MCMV infection than pDC and show enhanced viral gene expression, while pDC mount higher IFN-I responses.
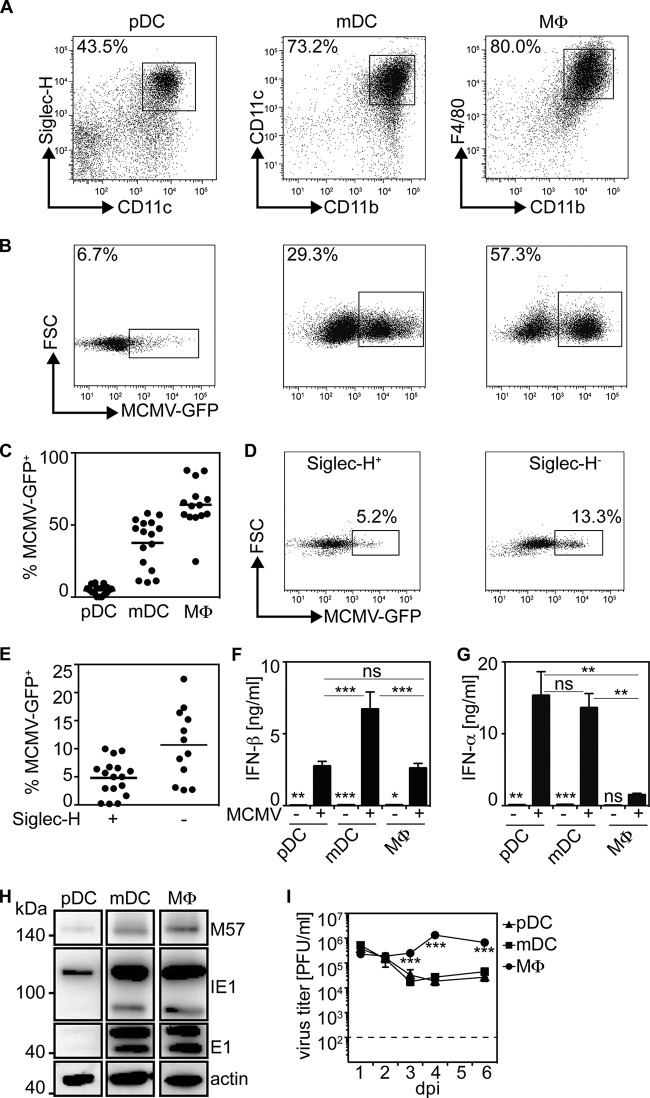
Previous studies showed that MCMV readily infects and replicates in macrophages (36). Here, we studied MCMV-mediated induction of IFN-I responses in bone marrow-derived in vitro-differentiated pDC, mDC, and Mϕ (Fig. 1A). To examine their susceptibility to MCMV infection, cultures of antigen-presenting cells were infected with MCMV-GFP at an MOI of 0.3, and after 18 h of incubation, the surface markers Siglec-H, CD11b, CD11c, and F4/80, which are generally used to distinguish pDC, mDC, and Mϕ (30, 37), were stained and the GFP expression of Siglec-H+ CD11cint pDC, CD11b+ CD11c+ mDC, and CD11b+ F4/80+ Mϕ was analyzed cytofluometrically. Under such conditions, pDC cultures showed 6.7% GFP+ Siglec-H+ CD11cint pDC, while mDC cultures contained 29.3% GFP+ CD11b+ CD11c+ mDC and Mϕ cultures contained 57.3% GFP+ F4/80+ CD11b+ macrophages (Fig. 1B). In multiple repetitions of this experiment, we confirmed that upon MCMV-GFP treatment, a very low percentage of pDC was GFP+, whereas mDC showed a moderately enhanced and Mϕ a high percentage of GFP+ cells (Fig. 1C). pDC cultures that typically comprised approximately 40% Siglec-H+ pDC and 60% Siglec-H− conventional DC (cDC) contained 13.3% GFP+ cDC (Fig. 1D and E), indicating that cDC were more readily infected than pDC. Analysis of the IFN-β and IFN-α contents of cell-free supernatants revealed that mDC cultures contained significantly enhanced IFN-β concentrations compared with pDC and Mϕ cultures (Fig. 1F). On the other hand, significantly increased IFN-α concentrations were detected in pDC and mDC cultures (Fig. 1G). To determine viral gene expression in MCMV-treated antigen-presenting cell subsets, the expression levels of the immediate-early gene 1 (IE1), early gene 1 (E1), and M57 expressed with early gene kinetics were studied by Western blotting. IE1 is expressed directly upon infection and does not depend on cis-activation by other viral gene products, whereas E1 and M57 expression depends on the presence of viral proteins and is required for viral DNA replication (38). While in pDC cultures IE1 and M57 expression was weak and E1 expression was even less abundant, mDC and Mϕ cultures showed enhanced IE1, E1, and M57 expression (Fig. 1H). Furthermore, MCMV replication was supported only in Mϕ, as indicated by higher virus titers found in Mϕ cultures than in pDC and mDC cultures (Fig. 1I). These data supported the hypothesis that pDC were not susceptible to MCMV infection, that in mDC MCMV infection was abortive, and that only Mϕ were susceptible and permissive to MCMV infection and replication.
FIG 1.
mDC and Mϕ are more susceptible to MCMV infection and viral gene expression than pDC. pDC, mDC, and Mϕ bone marrow culture cells (2 × 105) derived from C57BL/6 mice were infected with MCMV-GFP at an MOI of 0.3. After 18 h of incubation, supernatants and cells were analyzed by ELISA and flow cytometry, respectively. (A) Gating of pDC, mDC, and Mϕ among the corresponding bone marrow cultures. (B and C) Flow cytometric analysis of Siglec-H+ CD11cint pDC, CD11c+ CD11b+ mDC, and F4/80+ CD11b+ Mϕ in MCMV-GFP-treated pDC, mDC, and Mϕ cultures, respectively (the dots represent independent experiments, and the bars indicate the means). (D and E) GFP expression of Siglec-H+ pDC and Siglec-H− conventional DC in MCMV-GFP-treated pDC cultures 18 h p.i. (the dots represent independent experiments, and the bars indicate the means). (F and G) Analysis of the IFN-β and IFN-α contents of cell-free supernatants of pDC, mDC, and Mϕ cultures 18 h p.i. (n = 4 to 8; means and standard errors of the mean [SEM]). (H) Western blot analysis of IE1, E1, and M57 in pDC, mDC, and Mϕ cultures. (I) MCMV replication in pDC, mDC, and Mϕ cultures (***, P ≤ 0.0001; **, P ≤ 0.005; *, P ≤ 0.05; Mann-Whitney U test and 2-way ANOVA for the analysis of MCMV titers; means ± SEM). ns, not significant.
To test whether upon MCMV-GFP treatment viral gene expression (as indicated by GFP expression) was a prerequisite for the induction of IFN-I responses, experiments were performed with cells derived from IFN-β reporter mice (MOB). Upon MCMV-GFP treatment of MOB pDC, approximately 7.9% of Siglec-H+ CD11cint pDC were GFP+, whereas 13% of the cells were YFP+, i.e., the cells expressed IFN-β, while only a minor fraction of the cells were GFP+ YFP+ double positive (1.7%) (Fig. 2A, top row). In MCMV-GFP-treated MOB mDC cultures, 46.8% of CD11b+ CD11c+ mDC were GFP+ and 2% were YFP+, whereas only 0.8% were GFP+ YFP+ (Fig. 2A, middle row). Finally, infected MOB Mϕ cultures contained 49.7% GFP+, 0.8% YFP+, and 0.1% GFP+ YFP+ F4/80+CD11b+ macrophages (Fig. 2A, bottom row). Collectively, these data confirmed that upon MCMV treatment, reduced percentages of pDC exhibited viral gene expression compared with mDC and Mϕ. Conversely, pDC showed enhanced percentages of cells expressing IFN-β compared with mDC and Mϕ. Furthermore, mDC and Mϕ contained reduced percentages of infected cells expressing IFN-β compared with pDC (Fig. 2B). These data implied that in mDC and Mϕ, MCMV infection inhibited the induction of IFN-β, whereas in pDC, IFN-β expression was less efficiently repressed.
FIG 2.
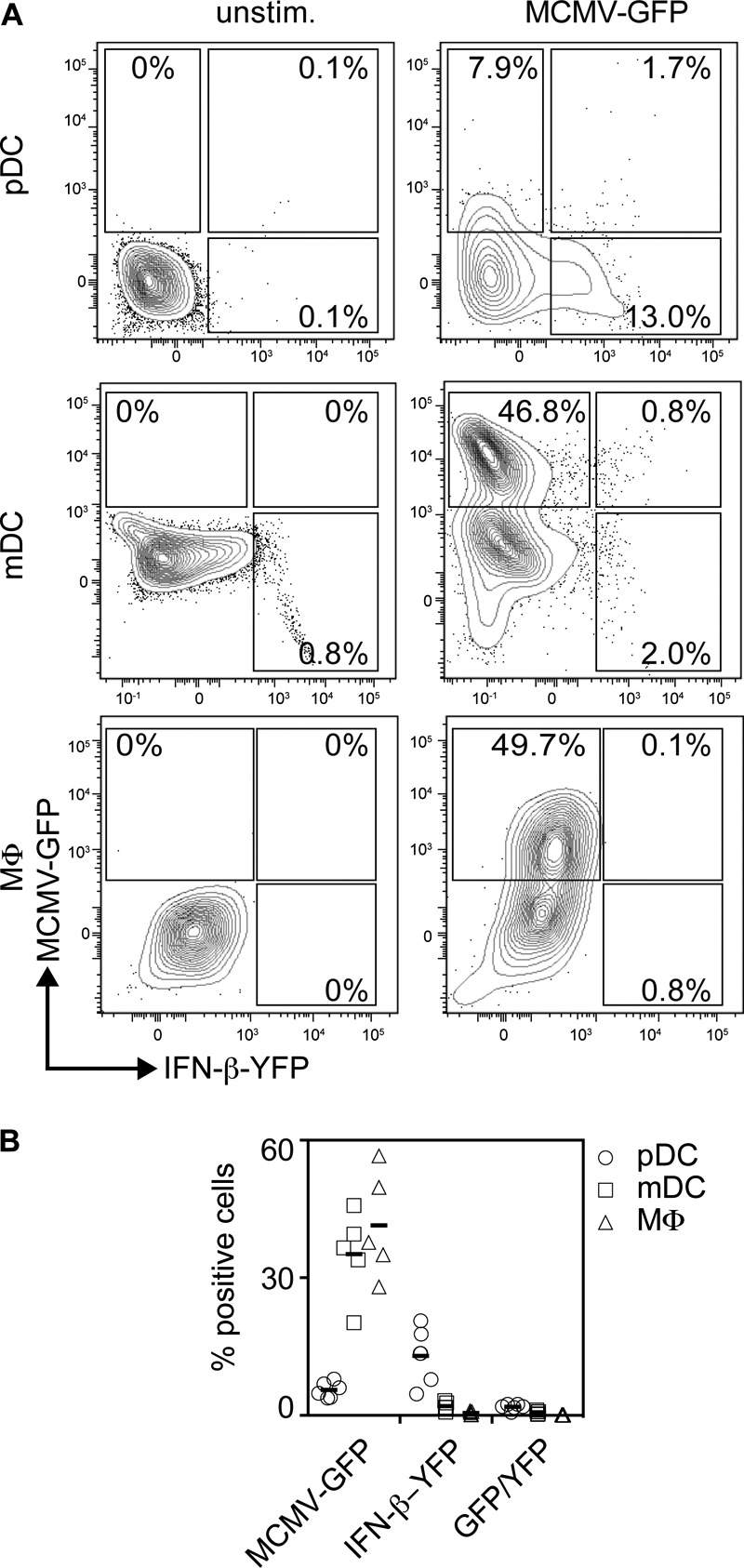
Upon MCMV-GFP incubation, pDC show higher percentages of IFN-β-expressing cells than mDC and Mϕ. pDC, mDC, and Mϕ (2 × 105) derived from MOB mice were treated with MCMV-GFP at an MOI of 0.3. The cells were gated as described in the legend to Fig. 1. (A) Dot plots showing induction of IFN-β (YFP) and infection with MCMV-GFP (GFP) of MOB-derived pDC, mDC, and Mϕ. Unstimulated control cells (unstim.) are shown on the left, and cells treated with MCMV-GFP 18 h p.i. are shown on the right. One representative experiment out of five similar ones is shown. (B) Data from all five experiments.
MCMV triggers IFN-I responses that in pDC are MyD88 dependent, whereas in mDC and Mϕ, the IFN-I induction is MyD88, TRIF, and CARDIF independent.
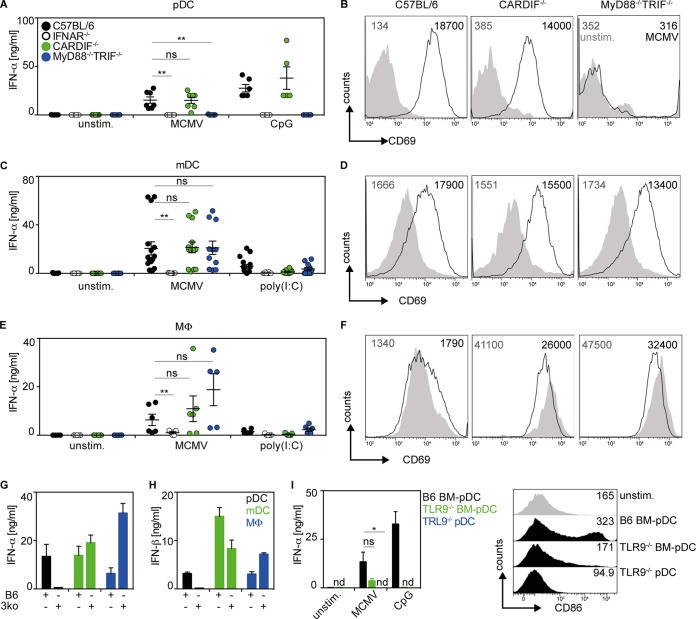
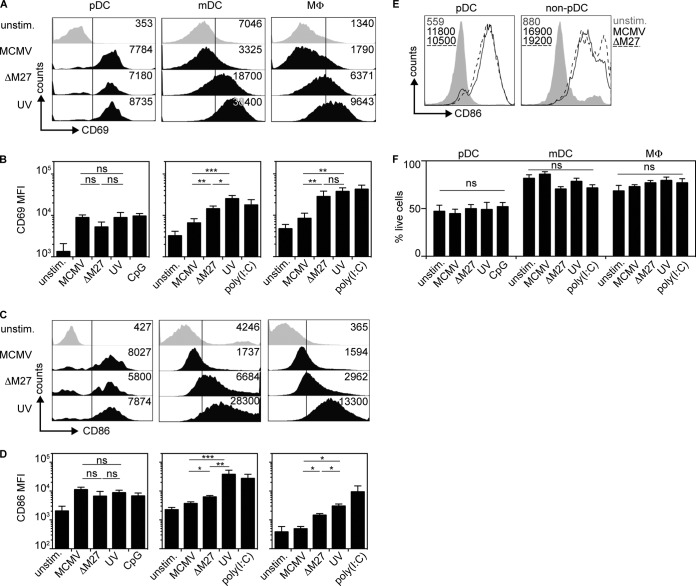
To characterize the mechanism of MCMV sensing, pDC, mDC, and Mϕ derived from C57BL/6 (WT), IFNAR−/− (deficient for the IFN-I receptor), CARDIF−/− (deficient for RLH signaling), and MyD88−/− TRIF−/− (devoid of TLR signaling) mice were studied. Upon MCMV treatment of WT and CARDIF−/− pDC cultures, cell-free supernatants of both setups contained similar quantities of IFN-α (Fig. 3A). In contrast, no IFN-α was detected in MCMV-treated IFNAR−/− or MyD88−/− TRIF−/− pDC cultures (Fig. 3A). Control experiments with pDC purified by MACS from pDC cultures verified that upon MCMV treatment, IFN responses were entirely TLR9 dependent (Fig. 3I). In line with their capacity to mount IFN responses, MCMV-treated WT and CARDIF−/− pDC showed enhanced expression of the activation marker CD69, while MyD88−/− TRIF−/− pDC did not (Fig. 3B). A similar upregulation was observed for the costimulatory molecule CD86 (data not shown). These data confirmed that pDC sensed MCMV in a TLR-dependent manner.
FIG 3.
Upon MCMV incubation, IFN-I production by pDC is MyD88/TRIF dependent, whereas in mDC and Mϕ, IFN-I production is MyD88, TRIF, and CARDIF independent. Bone marrow culture cells (2 × 105) generated from the indicated mouse strains were infected with MCMV at an MOI of 0.3 or treated with artificial nucleic acids, such as CpG-ODN1585 (CpG), a TLR9 agonist, or poly(I·C), a TLR3 and RIG-I agonist. (A, C, and E) IFN-α concentrations of cell-free supernatants were analyzed by ELISA 18 h p.i. (A) pDC. (C) mDC. (E) Mϕ. **, P ≤ 0.01; Mann-Whitney U test. (B, D, and F) CD69 expression was analyzed by flow cytometry 18 h p.i. (B) pDC. (D) mDC. (F) Mϕ. The numbers represent mean fluorescence intensities (MFI). Gray: unstimulated; black: MCMV stimulated. Shown is one representative out of four independently conducted experiments. (G and H) IFN-α and IFN-β ELISA of cell-free supernatants of WT (B6) and MyD88−/− TRIF−/− CARDIF−/− (3ko) pDC, mDC, and Mϕ cultures 18 h after stimulation with MCMV (n = 3 to 5). (I) IFN-α and CD86 expression of WT pDC and TLR9−/− pDC cultures (BM-pDC) and MACS-sorted TLR9−/− pDC (pDC) 18 h after stimulation with MCMV (n = 3). The symbols in panels A, C, and E represent single experiments (means ± SEM; *, P ≤ 0.05; Mann-Whitney U test). unstim., unstimulated control; ns, not significant; nd, not detected.
MCMV-infected mDC cultures derived from C57BL/6, CARDIF−/−, or MyD88−/− TRIF−/− mice produced overall similar amounts of IFN-α (Fig. 3C). To determine the interplay between TLR and RLH signaling, mDC derived from MyD88−/− TRIF−/− CARDIF−/− mice were studied. Of note, MCMV-stimulated MyD88−/− TRIF−/− CARDIF−/− mDC cultures mounted IFN-α and IFN-β responses similar to those of WT mDC cultures (Fig. 3G and H). On the other hand, no IFN-α production was detected in MCMV-treated IFNAR−/− mDC cultures. In line with the induction of IFN-α responses, upregulation of the activation markers CD69 (Fig. 3D) and CD86 (data not shown) was also detected in WT, CARDIF−/−, and MyD88−/− TRIF−/− mDC cultures (Fig. 3D). These results confirmed that mDC sensed MCMV in a TLR- and RLH-independent manner, whereas IFNAR feedback was needed to mount IFN-α responses. WT, CARDIF−/−, and MyD88−/− TRIF−/− Mϕ cultures treated with MCMV also produced comparable amounts of IFN-α (Fig. 3E). CD69 expression was only marginally enhanced on MCMV-infected WT Mϕ and was even downregulated on CARDIF−/− or MyD88−/− TRIF−/− Mϕ (Fig. 3F). Similar results were obtained for the activation marker CD86 (data not shown). Reminiscent of the experiments with mDC, MCMV-treated MyD88−/− TRIF−/− CARDIF−/− Mϕ cultures mounted enhanced IFN-α and IFN-β responses compared with WT Mϕ cultures (Fig. 3G and H). These results indicated that, similar to mDC, Mϕ also sensed MCMV in an entirely TLR- and RLH-independent manner and that MCMV seemed to evade upregulation of the activation markers CD69 and CD86 in Mϕ.
In pDC and mDC, MCMV evasion does not affect the distribution of cells producing either IFN-α or IFN-β or both cytokines.
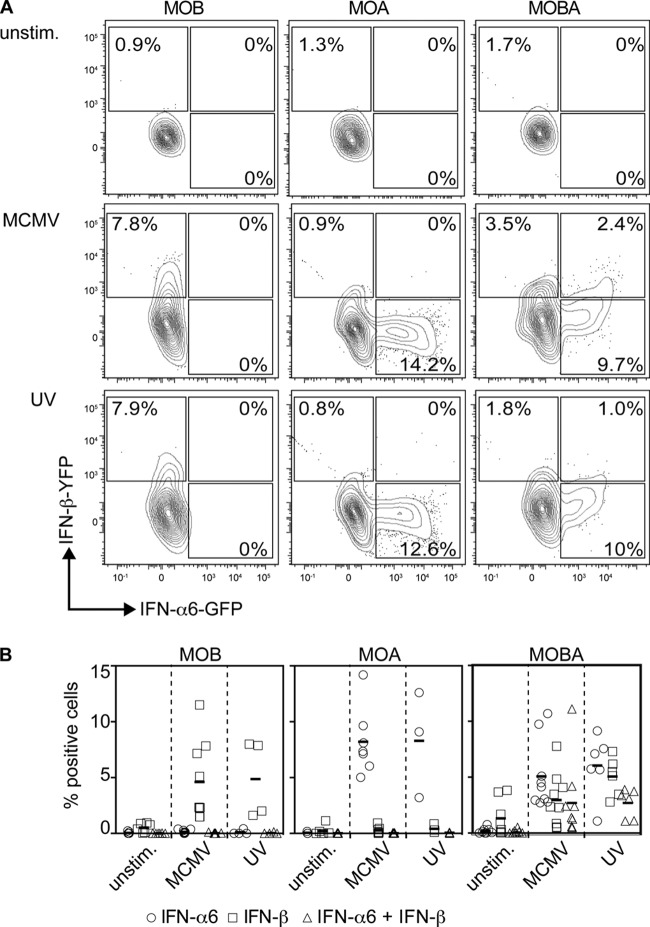
To study IFN-α and IFN-β responses on the single-cell level, the above-mentioned MOB mice were intercrossed with mice expressing GFP under the control of the IFN-α6 promoter (IFN-α6-GFP; MOA) to obtain MOBA mice as reporters for IFN-β and IFN-α6. After MCMV treatment of MOB pDC cultures, 7.8% of Siglec-H+ CD11cint pDC were YFP+ (indicating IFN-β expression), and in accordance with the absence of the IFN-α6-GFP allele, none of the cells were GFP+ (Fig. 4A, middle row, left plot). MCMV stimulation of MOA pDC resulted in 14.2% GFP+ Siglec-H+ CD11cint pDC (indicating IFN-α6 expression), and due to the absence of the MOB allele, only background signals were detected in the YFP channel (Fig. 4A, middle row, middle plot). Upon MCMV treatment of MOBA pDC cultures, 3.5% YFP+, 9.7% GFP+, and 2.4% GFP+ YFP+ Siglec-H+ CD11cint pDC were found (Fig. 4A, middle row, right plot). Thus, upon MCMV stimulation of pDC, a higher percentage of cells expressed only IFN-α6, whereas smaller percentages expressed IFN-β or a combination of IFN-α6 and IFN-β. Interestingly, upon MCMV-UV treatment of MOBA pDC cultures, a very similar distribution of single- and double-positive cells was detected, i.e., 1.8% YFP+, 10% GFP+, and 1% GFP+ YFP+ pDC (Fig. 4A, bottom row, right plot). These results indicated that in MCMV-stimulated pDC, MCMV-conferred evasion did not affect the distribution of single IFN-β- or IFN-α- or double IFN-β- and IFN-α-expressing pDC.
FIG 4.
Live and UV-inactivated MCMV trigger similar percentages of IFN-α+, IFN-β+, and IFN-α+ IFN-β+ pDC. pDC (2 × 105) derived from MOB, MOA, and MOBA mice were treated with live or UV-inactivated MCMV at an MOI of 0.3. The cells were gated as described in the legend to Fig. 1A, and IFN-α6-producing (indicated by GFP) versus IFN-β-producing (indicated by YFP) cells were plotted. One experiment out of five similar ones is shown. (B) Percentages from all experiments. Consistent results were obtained in four or five independent experiments.
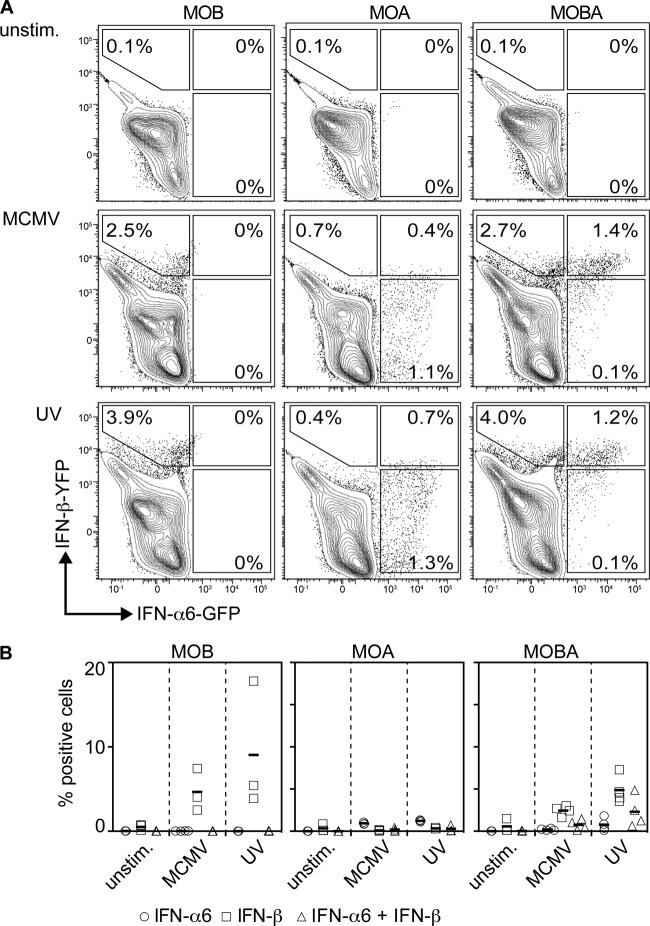
Upon MCMV treatment of MOBA mDC cultures, 2.7% YFP+, 0.1% GFP+, and 1.4% GFP+ YFP+ CD11b+ CD11c+ mDC were detected (Fig. 5A, middle row, right plot). On the other hand, treatment of MOBA mDC cultures with MCMV-UV resulted in 4% YFP+, 0.1% GFP+, and 1.2% GFP+ YFP+ CD11b+ CD11c+ mDC (Fig. 5A, bottom row, right plot). Thus, upon MCMV or MCMV-UV treatment, the distribution of IFN-β- or IFN-α6-expressing mDC or IFN-α6- and IFN-β-expressing mDC was not considerably changed. Furthermore, upon MCMV treatment, pDC primarily expressed IFN-α6, whereas mDC showed stronger IFN-β expression. These observations further supported the notion that upon MCMV infection, pDC are important IFN-α producers.
FIG 5.
MCMV evasion does not affect the distribution of IFN-α+, IFN-β+, and IFN-α+ IFN-β+ mDC. mDC (2 × 105) derived from MOB, MOA, and MOBA mice were treated with live or UV-inactivated MCMV at an MOI of 0.3. The cells were gated as shown in Fig. 1A, and IFN-α6-expressing (indicated by GFP) versus IFN-β-expressing (indicated by YFP) cells were plotted. One experiment out of three or four independent experiments is shown. (B) Percentages from all experiments. Consistent results were obtained in three or four independent experiments.
M27 affects the induction of IFN-I responses in mDC and Mϕ, but not in pDC.
To study how MCMV evasion affects the IFN responses of different immune cell subsets, pDC, mDC, and Mϕ cultures were treated with MCMV, MCMV-ΔM27, and MCMV-UV. Interestingly, in pDC, all three virus preparations induced similar CD69 upregulation (Fig. 6A and B, left). In mDC and Mϕ, only moderate CD69 induction was detected after MCMV treatment, whereas MCMV-ΔM27 and MCMV-UV triggered enhanced CD69 upregulation (Fig. 6A and B, middle and right). Similar results were observed for the induction of CD86 (Fig. 6C and D). As expected, upon MACS sorting of pDC and non-pDC from pDC cultures, MCMV-ΔM27 induced enhanced CD86 expression only in non-pDC and not in pDC (Fig. 6E). Of note, the overall cell viability was not affected by treatment with different MCMV preparations (Fig. 6F).
FIG 6.
M27-mediated evasion and induction of activation markers of MCMV-treated mDC and Mϕ, but not pDC. pDC, mDC, and Mϕ (2 × 105) derived from WT mice were infected with MCMV, MCMV-ΔM27 (ΔM27), or UV-inactivated MCMV (UV) at an MOI of 0.3. (A) CD69 expression of pDC, mDC, and Mϕ. (B) Statistical analysis of CD69 expression (MFI) corresponding to panel A. (C) CD86 expression of pDC, mDC, and Mϕ. (D) Statistical analysis of CD86 expression (MFI) corresponding to panel C. (E) CD86 expression of MACS-sorted pDC 18 h after stimulation with MCMV or MCMV-ΔM27. MFI values are coded to match the curves. (F) Percentages of live cells as measured by Aqua LIVE/DEAD staining of all three cell subsets in all experimental parameters (*, P ≤ 0.05; **, P ≤ 0.003; ***, P ≤ 0.0001; Mann-Whitney U test; means and SEM; n = 5 to 7).
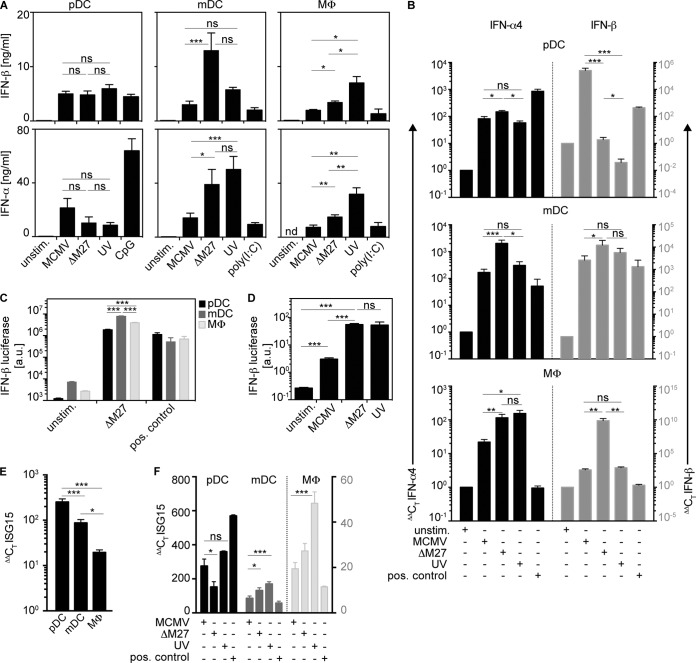
The analysis of cell-free supernatants revealed that MCMV-, MCMV-ΔM27-, and MCMV-UV-treated pDC cultures mounted IFN-β and IFN-α responses of similar magnitudes (Fig. 7A, left). In contrast, mDC and Mϕ cultures stimulated with MCMV-ΔM27 or MCMV-UV mounted significantly enhanced IFN-β and IFN-α responses compared with MCMV stimulation (Fig. 7A, middle and right). These results indicated that MCMV infection primarily evaded IFN-I induction in mDC and Mϕ, but not in pDC. Of note, Mϕ treated with MCMV-UV produced more IFN-I than after treatment with MCMV-ΔM27 (Fig. 7A, right), whereas mDC produced similarly high quantities of IFN-I upon treatment with MCMV-ΔM27 or MCMV-UV (Fig. 7A, middle). To further study IFN induction on the mRNA level, quantitative reverse transcription (qRT)-PCR analysis of IFN-β and IFN-α4 mRNA levels was performed. Upon treatment with MCMV, MCMV-ΔM27, or MCMV-UV, pDC cultures showed overall comparable IFN-β mRNA levels and even enhanced IFN-α4 mRNA levels upon MCMV treatment (Fig. 7B). In contrast, in mDC and Mϕ cultures, IFN-β and IFN-α4 mRNA levels were enhanced upon infection with MCMV-ΔM27 compared with MCMV (Fig. 7B). Upon infection with MCMV-UV, the mRNA levels for IFN-β and IFN-α4 were only moderately, if at all, increased (Fig. 7B). Thus, the qRT-PCR analysis further supported the data obtained by the IFN-I ELISA. To study this effect in a second model, pDC, mDC, and Mϕ were generated from IFN-βΔβ-luc/wt IFN-β luciferase reporter mice, and the luciferase expression, as a measure of IFN-β induction, was determined upon incubation with MCMV. Incubation with MCMV-ΔM27 induced enhanced luciferase expression in mDC and Mϕ cultures compared with pDC cultures (Fig. 7C). Furthermore, splenocytes isolated from IFN-βΔβ-luc/wt mice incubated with MCMV-ΔM27 or MCMV-UV also showed higher luciferase induction than upon incubation with MCMV (Fig. 7D). Next, to study whether such IFN-I responses also triggered pDC, mDC, and Mϕ, ISG15 mRNA induction was analyzed. Upon MCMV incubation, ISG15 induction in pDC cultures was more abundant than in mDC and Mϕ (Fig. 7E). ISG15 mRNA levels were not further increased when pDC were incubated with MCMV-ΔM27 or MCMV-UV instead of WT MCMV, while this effect was observed in mDC and Mϕ cultures (Fig. 7F).
FIG 7.
M27-mediated evasion affects IFN-I production of MCMV-treated mDC and Mϕ, but not pDC. pDC, mDC, and Mϕ (2 × 105) derived from WT mice were infected with MCMV, MCMV-ΔM27 (ΔM27), or UV-inactivated MCMV (UV) at an MOI of 0.3. (A) IFN-β and IFN-α concentrations of the cell-free supernatants were analyzed by ELISA 18 h p.i. (n = 7). (B) qRT-PCR analysis of mRNA levels of IFN-α4 (black bars) and IFN-β (gray bars) (n = 4 to 6). The corresponding cells from panel A were used. (C) Luciferase expression of IFN-βΔβ-luc/wt-derived pDC, mDC, and Mϕ cultures incubated with MCMV-ΔM27 (n = 3). (D) Luciferase expression of IFN-βΔβ-luc/wt-derived splenocytes incubated with MCMV, MCMV-UV, and MCMV-ΔM27 (n = 6). (E) ISG15 mRNA expression levels in pDC, mDC, and Mϕ upon incubation with MCMV (n = 3). (F) ISG15 mRNA expression levels in pDC, mDC (left axis, black numbers), and Mϕ (right axis, gray numbers) upon incubation with MCMV, MCMV-ΔM27, or UV-inactivated MCMV (n = 3) (*, P ≤ 0.05; **, P ≤ 0.003; ***, P ≤ 0.0001; Mann-Whitney U test and Kruskal-Wallis test with Dunn's correction for multiple comparison; means and SEM).
Collectively, these data indicated that MCMV, MCMV-ΔM27, and MCMV-UV triggered pDC to mount IFN-I responses of similar magnitudes, while IFN-I responses of mDC and Mϕ triggered by MCMV-ΔM27 or MCMV-UV were significantly enhanced compared with MCMV-stimulated cells. Furthermore, these results implied that in mDC, M27 was critically involved in the inhibition of MCMV-induced IFN-I responses, whereas in Mϕ, in addition to M27, other MCMV-encoded IFN evasion mechanisms were effective.
DISCUSSION
During the long period of coevolution of MCMV with its host, it is very likely that viral evasion drove the diversification of host immune defense mechanisms. Thus, MCMV provides a unique opportunity to study mechanisms that affect the host-pathogen balance. Previous studies showed that MCMV suppressed IFNAR signaling of MCMV-permissive fibroblasts (19, 39). Furthermore, other MCMV genes may reduce the induction of IFN-I responses in fibroblasts (21). Here, we addressed MCMV-mediated induction of IFN-I responses in different immune cell subsets. As reported previously by others (6), we confirmed that MCMV triggered pDC to mount IFN responses in a TLR9-dependent manner. Interestingly, under such conditions, MCMV-encoded evasins did not affect the magnitude of IFN responses. This notion was further supported by the observation that pDC were not infected by MCMV, indicating that MCMV infection was not a prerequisite for pDC activation. On the other hand, MCMV also triggered mDC and Mϕ to mount IFN-I responses. MCMV-exposed Mϕ produced less IFN-I than pDC and mDC. In MCMV-stimulated Mϕ, primarily IFN-β was expressed, whereas pDC mostly expressed IFN-α. In mDC and Mϕ, IFN-I was induced independently of TLR and RLH signaling. Interestingly, in pDC, M27-mediated evasion was not effective, whereas in mDC, M27 impeded IFN induction nonredundantly, while in Mϕ, in addition to M27, other, not yet identified evasins were effective.
Experiments with MCMV-GFP revealed that MCMV infection and gene expression were less efficient in pDC than in mDC and Mϕ, whereas pDC mounted higher IFN responses upon MCMV exposure. In many viral infections, pDC are important IFN-I producers (24, 40). Constitutive expression of the transcription factor IRF7 enables pDC to rapidly respond to TLR9/MyD88-dependent triggering, resulting in the immediate production of large amounts of IFN-I (41). Because pDC cultures contain approximately 40% Siglec-H+ CD11cint pDC while the remaining cells primarily comprise conventional CD11c+ CD11b+ DC (cDC), we also analyzed viral gene expression in cDC isolated from pDC cultures. Interestingly, cDC showed enhanced percentages of GFP+ cells compared with pDC. Therefore, infected cDC presumably triggered uninfected pDC to mount IFN responses. Indeed, in MCMV-GFP-treated MOB pDC cultures, primarily uninfected pDC expressed IFN-β, which did not show detectable signs of viral gene expression. Thus, it is conceivable that pDC were triggered by apoptotic bodies or exosomes derived from infected cells and therefore showed some minor GFP expression. This notion is in line with previous studies reporting that pDC were overall resistant to productive infection with MCMV (42). The kind of danger signal released by infected cDC still remains to be elucidated. The danger signals described so far are compounds normally confined to the intracellular space of cells, such as cGAMP, ATP, cellular DNA, and heat shock proteins (43). Usually, IFN-I-mediated IFNAR triggering induces an antiviral state in cells. However, when IFNAR−/− pDC were used in MCMV stimulation experiments, only slightly enhanced percentages of infected cells were detected (data not shown). This observation indicated that IFNAR triggering of pDC only partially enhanced protection of pDC.
The observation that an enhanced percentage of mDC and Mϕ were infected and showed MCMV gene expression, including synthesis of viral proteins, implied that MCMV-encoded evasins that might affect the antiviral responses of mDC and Mϕ were also expressed. This notion was supported by experiments with MOB-derived mDC and Mϕ, which showed that basically none of the infected cells (and only a minor percentage of uninfected cells) were able to express IFN-β. Of note, detection of a minor IFN-β-producing mDC subset could also be explained by low-level constitutive IFN-β expression, even though such basal IFN-β expression has so far been described only for different tissues and stromal cells, but not for mDC (31). Upon MCMV treatment, lower percentages of YFP-positive mDC produced more IFN-β than higher percentages of YFP-positive pDC (compare Fig. 1F and 2A/B, as well as 4 and 5). This observation could be explained by mDC producing IFN-α and IFN-β simultaneously, whereas after a short phase of IFN-β expression, pDC primarily produce IFN-α.
We found that in pDC MCMV-mediated triggering of IFN-I responses was TLR9/MyD88 dependent, which was in accordance with previous studies (6, 44). This TLR9/MyD88-dependent sensing of MCMV was also reflected by CD86 and CD69 upregulation only in WT and CARDIF−/− pDC, but not in MyD88−/− pDC. Interestingly, mDC and Mϕ sensed MCMV in an entirely TLR- and RLH-independent manner. CD86 and CD69, as well as IFN-I, expression was increased in WT, CARDIF−/−, MyD88−/− TRIF−/−, and MyD88−/− TRIF−/− CARDIF−/− mDC. These results are in accordance with a previous publication, in which mDC derived from TLR9−/− and MyD88−/− mice still produced IFN-I upon MCMV infection, while TLR9−/− and MyD88−/− pDC did not (45). Our experiments additionally ruled out a role of the adaptor protein CARDIF (the main adaptor protein of RLH signaling) in mDC and Mϕ, arguing for the triggering of a completely different sensing pathway than the classical TLR and RLH routes. It is very likely that the newly described cytosolic DNA receptor cGAS plays a central role in the recognition of MCMV by mDC and Mϕ (46). Of note, MCMV-ΔM27- and MCMV-UV-treated mDC and Mϕ mounted higher IFN-I responses than after WT MCMV treatment. This was also reflected in the observation that ISG15 mRNA was less abundantly induced in MCMV-infected mDC and Mϕ than in MCMV-ΔM27- or MCMV-UV-infected mDC and Mϕ. The observation that attenuated or inactivated virus triggered stronger IFN-I responses than WT MCMV indicated that countermeasures, including M27 expression, substantially inhibited IFN-I induction. Interestingly, upon MCMV stimulation of the BM-derived immune cell subsets tested here, the induction of IFN-α responses was dependent on IFNAR feedback. The role of positive feedback in the induction of IFN-α has been controversial, and particularly for pDC, a feedback-independent IFN-I-triggering pathway has been claimed (47). Nevertheless, our results clearly indicate that pDC, mDC, and Mϕ require IFNAR triggering to mount IFN-I responses upon stimulation with MCMV. The distribution of IFN-β- and IFN-α6-producing pDC and mDC did not change upon stimulation with MCMV or MCMV-UV. Thus, it is more likely that instead of an overall decrease in percentages of IFN-I-producing cell subsets, the IFN-I production of single cells was diminished.
Our finding that pDC are the main source of IFN-α in response to MCMV is consistent with previous studies that came to similar conclusions in the context of in vivo studies (5, 7, 28). Nevertheless, in our experiments, mDC also produced ample amounts of IFN-α, putting them on the list of important IFN-I producers in the context of MCMV infection, as well. Upon MCMV treatment, Mϕ derived from WT mice produced significantly less IFN-I than pDC and mDC. This was also reflected by the lack of upregulation of CD69 and CD86 on Mϕ treated with MCMV, while pDC and mDC exhibited strong upregulation of both markers. MCMV is known to limit CD86 surface expression by the expression of m147.5 (48), which might exhibit stronger effects in permissive Mϕ than in pDC and mDC. The additional increase in CD86 on mDC and Mϕ upon incubation with UV-inactivated MCMV in comparison with MCMV-ΔM27 stimulation can be attributed to loss of m147.5.
Experiments with MCMV-ΔM27 and MCMV-UV revealed that in Mϕ, in addition to M27, other MCMV-encoded evasion mechanisms must be active, while in mDC, MCMV-mediated IFN-I evasion was primarily conferred by M27. Previously, it was reported that Mϕ played an important role in conferring resistance to MCMV infection and that MCMV-infected Mϕ showed reduced major histocompatibility complex (MHC) class II expression levels (49). Thus, MCMV infection affected the biological function of Mϕ more than that of many other cell subsets (36, 50). This notion was further highlighted by our observation that MCMV specifically inhibited effective IFN-I induction and activation marker upregulation of Mϕ.
ACKNOWLEDGMENTS
We thank Shizuo Akira for providing the IFN-α6-GFP mouse; Roland Lang, who supplied L929 cells as a source of MCS-F for Mϕ differentiation; and Stipan Jonjic, who provided Croma101-, Croma103-, and M57.01-specific antibodies.
This study was supported by funding from the Helmholtz Virtual Institute (VH-VI-424 Viral Strategies of Immune Evasion) to M.M., L.C.-S., H.H., and U.K. and by funding from the Hannover Biomedical Research School (HBRS) and the Center for Infection Biology (ZIB) to M.D. The StrucMed Program of the Hannover Medical School granted funding to I.L.
We declare no competing financial interests.
Footnotes
Published ahead of print 17 September 2014
REFERENCES
- 1. Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Luccaronin P, Jonjic S, Koszinowski UH. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J. Exp. Med. 188:1047–1054. 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dag F, Dolken L, Holzki J, Drabig A, Weingartner A, Schwerk J, Lienenklaus S, Conte I, Geffers R, Davenport C, Rand U, Koster M, Weiss S, Adler B, Wirth D, Messerle M, Hauser H, Cicin-Sain L. 2014. Reversible silencing of cytomegalovirus genomes by type I interferon governs virus latency. PLoS Pathog. 10:e1003962. 10.1371/journal.ppat.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. 2007. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol. Cell Biol. 85:46–54. 10.1038/sj.icb.7100013. [DOI] [PubMed] [Google Scholar]
- 5. Dalod M, Salazar-Mather TP, Malmgaard L, Lewis C, Asselin-Paturel C, Briere F, Trinchieri G, Biron CA. 2002. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 195:517–528. 10.1084/jem.20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. 2004. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21:107–119. 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 7. Zucchini N, Bessou G, Robbins SH, Chasson L, Raper A, Crocker PR, Dalod M. 2008. Individual plasmacytoid dendritic cells are major contributors to the production of multiple innate cytokines in an organ-specific manner during viral infection. Int. Immunol. 20:45–56. 10.1093/intimm/dxm119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iwasaki A, Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987–995. 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 9. Goubau D, Deddouche S, Reis ESC. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paludan SR, Bowie AG. 2013. Immune sensing of DNA. Immunity 38:870–880. 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240–273. 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reikine S, Nguyen JB, Modis Y. 2014. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 5:342. 10.3389/fimmu.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hornung V, Hartmann R, Ablasser A, Hopfner KP. 2014. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 14:521–528. 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kristiansen H, Scherer CA, McVean M, Iadonato SP, Vends S, Thavachelvam K, Steffensen TB, Horan KA, Kuri T, Weber F, Paludan SR, Hartmann R. 2010. Extracellular 2′-5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J. Virol. 84:11898–11904. 10.1128/JVI.01003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider K, Loewendorf A, De Trez C, Fulton J, Rhode A, Shumway H, Ha S, Patterson G, Pfeffer K, Nedospasov SA, Ware CF, Benedict CA. 2008. Lymphotoxin-mediated crosstalk between B cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe 3:67–76. 10.1016/j.chom.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krmpoti A, Busch DH, Bubi I, Gebhardt F, Hengel H, Hasan M, Scalzo AA, Koszinowski UH, Jonji S. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529–535. 10.1038/ni799. [DOI] [PubMed] [Google Scholar]
- 17. Lenac T, Arapovic J, Traven L, Krmpotic A, Jonjic S. 2008. Murine cytomegalovirus regulation of NKG2D ligands. Med. Microbiol. Immunol. 197:159–166. 10.1007/s00430-008-0080-7. [DOI] [PubMed] [Google Scholar]
- 18. Marshall EE, Geballe AP. 2009. Multifaceted evasion of the interferon response by cytomegalovirus. J. Interferon Cytokine Res. 29:609–619. 10.1089/jir.2009.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zimmermann A, Trilling M, Wagner M, Wilborn M, Bubic I, Jonjic S, Koszinowski U, Hengel H. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-gamma signaling and antiviral responses. J. Exp. Med. 201:1543–1553. 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trilling M, Le VT, Fiedler M, Zimmermann A, Bleifuss E, Hengel H. 2011. Identification of DNA-damage DNA-binding protein 1 as a conditional essential factor for cytomegalovirus replication in interferon-gamma-stimulated cells. PLoS Pathog. 7:e1002069. 10.1371/journal.ppat.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le VT, Trilling M, Zimmermann A, Hengel H. 2008. Mouse cytomegalovirus inhibits beta interferon (IFN-beta) gene expression and controls activation pathways of the IFN-beta enhanceosome. J. Gen. Virol. 89:1131–1141. 10.1099/vir.0.83538-0. [DOI] [PubMed] [Google Scholar]
- 22. Murray PJ, Wynn TA. 2011. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11:723–737. 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Merad M, Sathe P, Helft J, Miller J, Mortha A. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604. 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baranek T, Zucchini N, Dalod M. 2009. Plasmacytoid dendritic cells and the control of herpesvirus infections. Viruses 1:383–419. 10.3390/v1030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muller U, Steinhoff U, Reis LFL, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type-I and type-II interferons in antiviral defense. Science 264:1918–1921. 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 26. Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143–150. 10.1016/S1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 27. Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803. 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheu S, Dresing P, Locksley RM. 2008. Visualization of IFNbeta production by plasmacytoid versus conventional dendritic cells under specific stimulation conditions in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:20416–20421. 10.1073/pnas.0808537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. 2007. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity 27:240–252. 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 30. Frenz T, Graalmann L, Detje CN, Doring M, Grabski E, Scheu S, Kalinke U. 2014. Independent of plasmacytoid dendritic cell (pDC) infection, pDC triggered by virus-infected cells mount enhanced type I IFN responses of different composition as opposed to pDC stimulated with free virus. J. Immunol. 193:2496–2503. 10.4049/jimmunol.1400215. [DOI] [PubMed] [Google Scholar]
- 31. Lienenklaus S, Cornitescu M, Zietara N, Lyszkiewicz M, Gekara N, Jablonska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S. 2009. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J. Immunol. 183:3229–3236. 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 32. Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mergenthaler H, Dörmer P, Staber F, Hültner L. 1982. The effect of two different types of colony-stimulating factor on the expression of aminopeptidase on marrow-derived murine macrophages. Exp. Hematol. 10:789–797. [PubMed] [Google Scholar]
- 34. Stockinger S, Reutterer B, Schaljo B, Schellack C, Brunner S, Materna T, Yamamoto M, Akira S, Taniguchi T, Murray PJ, Muller M, Decker T. 2004. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 173:7416–7425. 10.4049/jimmunol.173.12.7416. [DOI] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36. Hengel H, Reusch U, Geginat G, Holtappels R, Ruppert T, Hellebrand E, Koszinowski UH. 2000. Macrophages escape inhibition of major histocompatibility complex class I-dependent antigen presentation by cytomegalovirus. J. Virol. 74:7861–7868. 10.1128/JVI.74.17.7861-7868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang CQ, Yu X, Cao Q, Wang Y, Zheng GP, Tan TK, Zhao H, Zhao Y, Wang YP, Harris DCH. 2013. Characterization of murine macrophages from bone marrow, spleen and peritoneum. BMC Immunol. 14:6. 10.1186/1471-2172-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reddehase MJ, Koszinowski UH. 1984. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature 312:369–371. 10.1038/312369a0. [DOI] [PubMed] [Google Scholar]
- 39. Trilling M, Le VT, Rashidi-Alavijeh J, Katschinski B, Scheller J, Rose-John S, Androsiac GE, Jonjic S, Poli V, Pfeffer K, Hengel H. 2014. “Activated” STAT proteins: a paradoxical consequence of inhibited JAK-STAT signaling in cytomegalovirus-infected cells. J. Immunol. 192:447–458. 10.4049/jimmunol.1203516. [DOI] [PubMed] [Google Scholar]
- 40. Colonna M, Trinchieri G, Liu YJ. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5:1219–1226. 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 41. Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. 2005. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434:1035–1040. 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 42. Dalod M, Hamilton T, Salomon R, Salazar-Mather TP, Henry SC, Hamilton JD, Biron CA. 2003. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 197:885–898. 10.1084/jem.20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matzinger P. 2002. The danger model: a renewed sense of self. Science 296:301–305. 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 44. Tabeta K, Georgel P, Janssen E, Du X, Hoebe K, Crozat K, Mudd S, Shamel L, Sovath S, Goode J, Alexopoulou L, Flavell RA, Beutler B. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 101:3516–3521. 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andoniou CE, van Dommelen SL, Voigt V, Andrews DM, Brizard G, Asselin-Paturel C, Delale T, Stacey KJ, Trinchieri G, Degli-Esposti MA. 2005. Interaction between conventional dendritic cells and natural killer cells is integral to the activation of effective antiviral immunity. Nat. Immunol. 6:1011–1019. 10.1038/ni1244. [DOI] [PubMed] [Google Scholar]
- 46. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barchet W, Cella M, Odermatt B, Asselin-Paturel C, Colonna M, Kalinke U. 2002. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 195:507–516. 10.1084/jem.20011666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loewendorf A, Kruger C, Borst EM, Wagner M, Just U, Messerle M. 2004. Identification of a mouse cytomegalovirus gene selectively targeting CD86 expression on antigen-presenting cells. J. Virol. 78:13062–13071. 10.1128/JVI.78.23.13062-13071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Redpath S, Angulo A, Gascoigne NR, Ghazal P. 1999. Murine cytomegalovirus infection down-regulates MHC class II expression on macrophages by induction of IL-10. J. Immunol. 162:6701–6707. [PubMed] [Google Scholar]
- 50. Heise MT, Pollock JL, O'Guin A, Barkon ML, Bromley S, Virgin HW. 1998. Murine cytomegalovirus infection inhibits IFNγ-induced MHC class II expression on macrophages: the role of type I interferon. Virology 241:331–344. 10.1006/viro.1997.8969. [DOI] [PubMed] [Google Scholar]
- 51. Spanier J, Lienenklaus S, Paijo J, Kessler A, Borst K, Heindorf S, Baker DP, Kröger A, Weiss S, Detje CN, Staeheli P, Kalinke U. 2014. Concomitant TLR/RLH signaling of radioresistant and radiosensitive cells is essential for protection against vesicular stomatitis virus infection. J. Immunol. 193:3045–3054. 10.4049/jimmunol.1400959. [DOI] [PubMed] [Google Scholar]