ABSTRACT
Endogenous retroviruses are the remnants of past retroviral infections that are scattered within mammalian genomes. In humans, most of these elements are old degenerate sequences that have lost their coding properties. The HERV-K(HML2) family is an exception: it recently amplified in the human genome and corresponds to the most active proviruses, with some intact open reading frames and the potential to encode viral particles. Here, using a reconstructed consensus element, we show that HERV-K(HML2) proviruses are able to inhibit Tetherin, a cellular restriction factor that is active against most enveloped viruses and acts by keeping the viral particles attached to the cell surface. More precisely, we identify the Envelope protein (Env) as the viral effector active against Tetherin. Through immunoprecipitation experiments, we show that the recognition of Tetherin is mediated by the surface subunit of Env. Similar to Ebola glycoprotein, HERV-K(HML2) Env does not mediate Tetherin degradation or cell surface removal; therefore, it uses a yet-undescribed mechanism to inactivate Tetherin. We also assessed all natural complete alleles of endogenous HERV-K(HML2) Env described to date for their ability to inhibit Tetherin and found that two of them (out of six) can block Tetherin restriction. However, due to their recent amplification, HERV-K(HML2) elements are extremely polymorphic in the human population, and it is likely that individuals will not all possess the same anti-Tetherin potential. Because of Tetherin's role as a restriction factor capable of inducing innate immune responses, this could have functional consequences for individual responses to infection.
IMPORTANCE Tetherin, a cellular protein initially characterized for its role against HIV-1, has been proven to counteract numerous enveloped viruses. It blocks the release of viral particles from producer cells, keeping them tethered to the cell surface. Several viruses have developed strategies to inhibit Tetherin activity, allowing them to efficiently infect and replicate in their host. Here, we show that human HERV-K(HML2) elements, the remnants of an ancient retroviral infection, possess an anti-Tetherin activity which is mediated by the envelope protein. It is likely that this activity was an important factor that contributed to the recent, human-specific amplification of this family of elements. Also, due to their recent amplification, HERV-K(HML2) elements are highly polymorphic in the human population. Since Tetherin is a mediator of innate immunity, interindividual variations among HERV-K(HML2) Env genes may result in differences in immune responses to infection.
INTRODUCTION
Remnants of past viral infections are scattered within mammalian genomes. Most of those, the endogenous retroviruses (ERVs), were generated after infectious elements inserted into the germ line of their host during the course of a retroviral infection and thereafter were transmitted vertically. In some cases, the newly inserted element undergoes further replication cycles in the germ line, increasing its copy number within the genome. Given time, in the absence of selective pressure, most of the endogenous proviruses degenerate and lose their coding capacity (reviewed in references 1–4). However, the human genome still contains some intact retroviral open reading frames (ORFs), some of which have been conserved and recruited to serve physiological functions (5–7), whereas others are simply too recent to have undergone degeneration. This is the case of HERV-K(HML2), the most active ERV family in the human genome, which is comprised of around 50 proviruses. Although this family initially integrated in the genome of primates a long time ago (approximately 40 million years ago [mya]), most of the proviruses present in the human genome are much younger, with several having inserted at their present location less than 3 mya (i.e., after the divergence between the human and chimpanzee lineages) (8, 9; reviewed in reference 10). Most of them are polymorphic among the human population, and some have been dated to less than 100,000 years (9, 11). There is speculation that the amplification process of this family is still ongoing in humans (12). HERV-K(HML2) elements belong to betaretroviruses, and full-length proviruses encode the canonical retroviral Gag, Pro (protease), Pol (polymerase), and Env (envelope) proteins, as well as Rec, an accessory protein whose functions are similar to those of Rem in mouse mammary tumor virus or Rev in human immunodeficiency virus (HIV) (13, 14). Some of these proviruses are expressed and produce viral particles in tumor-derived cell lines (15–18). Functional assays showed that among the six complete Env ORFs identified in the human databases, one corresponds to a functional protein able to confer infectivity to heterologous retroviral particles (19).
Tetherin (or BST2 or CD317) is an antiviral protein whose activity was initially identified on HIV-1 (20, 21). It has a broad activity, being able to restrict most enveloped viruses against which it has been tested, including HERV-K(HML2) elements (22; reviewed in reference 23). Tetherin is a type II integral protein composed of a short N-terminal cytoplasmic tail containing motifs that mediate clathrin-dependent endocytosis (24), a single-pass transmembrane domain, and an ectodomain attached to the cell membrane at the C-terminal end via a GPI (glycosylphosphatidylinositol) anchor (25). Tetherin is present at the cell membrane as a homodimer (26) and acts against viruses by blocking their egress, leading to the retention of viral particles at the surface of the cells (20, 27). Like other components of innate immunity, Tetherin expression can be induced by type I interferons (28, 29). In addition to its direct antiviral effect, Tetherin is also a sensor of virus assembly and induces NF-κB proinflammatory responses while blocking viral egress (30, 31).
Over time, viruses have developed strategies to evade Tetherin-mediated restriction. Numerous examples of independent viral proteins able to counteract Tetherin have been described in different viral families, suggesting it is important for viruses to escape its action. They include so-called accessory proteins, such as Vpu (from HIV-1 and some simian immunodeficiency virus [SIV] strains [20, 32, 33]) and Nef (in several SIV strains [33–36]), the ubiquitin ligase K5 from human herpesvirus 8 (HHV8) (37–39), and also glycoproteins, including retroviral Env proteins (e.g., in HIV-2 [40], some SIV strains [41], feline immunodeficiency virus [FIV] [42–44], and equine infectious anemia virus [EIAV] [45]) as well as Ebola glycoprotein (46). The mechanisms of action of these proteins are diverse and not always fully understood, but all result in restoring virus release. Since HERV-K(HML2) became endogenous in the primate lineage more than 40 mya, it has had time to evolve in response to its host and acquire beneficial mutations, which could be a reason why it efficiently reamplified very recently in the genome of our ancestors. We reasoned that this recent amplification burst might have been associated with an intrinsic resistance against modern human restriction factors. Indeed, it was previously shown that human TRIM5α has no effect on HERV-K(HML2) infection (47). Thus, we decided to test HERV-K(HML2) for the presence of inhibitory proteins active against Tetherin. Here, we show that this element indeed encodes an inhibitor of human Tetherin able to restore virus release in pseudotyping experiments. This property is embedded in the Env protein, as already observed for other viruses (described above). Through immunoprecipitation experiments, we mapped the interacting domain to the surface (SU) subunit. We also show that, like the Ebola glycoprotein, HERV-K(HML2) Env does not lead to degradation or cell surface removal of Tetherin (48, 49).
MATERIALS AND METHODS
Plasmids.
HIV-1-derived particles were produced using plasmids 8.91 (50), pHR′SIN-cPPT-SEW (51), and phCMV VSV-G (GenBank accession no. AJ318514). The human Tetherin ORF (NM_004335.3) was cloned in a pcDNA3 plasmid. An internally HA-tagged version, cloned in pCR3.1, was kindly provided by S. Neil (52) and was transferred in a lentiviral vector for transduction experiments (BamHI-NotI green fluorescent protein [GFP]-containing fragment from pHR′SIN-cPPT-SEW replaced by the Tetherin-HA ORF). Other primate Tetherin genes all are cloned into pCR3.1 and were obtained from S. Neil.
pCMV-ß (Clontech) is an expression vector for beta-galactosidase. It was used as a control vector (i.e., not encoding a Tetherin inhibitor) and is designated “None” in the figures. It also was used to adjust total DNA content in transfection experiments. Murine leukemia virus amphotropic [MLV ampho] Env (4070A strain) was expressed from a previously described phCMV-based vector (53). HIV2 ROD10 Env ORF was a gift from F. Clavel; point mutations were introduced to match the sequence of HIV2 ROD10 FL Env (described in reference 54), and the modified ORF was cloned into the phCMV vector. pCR3.1 Vpu-HA was a gift from S. Neil (55); in this study, we used a modified version generated by replacing the HA tag with a myc tag.
The expression vector for the cytomegalovirus (CMV)-driven HERV-K(HML2) consensus provirus with the inactivated env gene is described in reference 56. phCMV-HERV-K(HML2) Env-Rec plasmid was constructed by replacing the G protein ORF in phCMV vesicular stomatitis virus glycoprotein (VSV-G) with nucleotides (nt) 6434 to 9214 of the HERV-K consensus provirus (sequence provided in reference 56). Rec cDNA was generated by performing reverse transcription-PCR (RT-PCR) on RNA extracted from 293T cells transfected with phCMV-HERV-K(HML2) Env-Rec using primers surrounding the Rec ORF (FP, TTGCAGAATTCACATTTGAAGTTCTACAATGAAC; RP, TTGCAGAATTCGTGAACAAAGGTCTTTGCATC) and cloned in the phCMV vector. phCMV-HERV-K(HML2) Env is derived from phCMV-HERV-K(HML2) Env-Rec, with point mutations introduced to inactivate splice donor and acceptor sites (SD and SA, respectively) without modifying the Env amino acid sequence (6711, gtggtaagt to gtCgtCTCA; 8395, ttttgtctgttgttagtc to ttCtgCctgCtgCtCgtG; modified nucleotides are shown in uppercase, and positions are according to those of the HERV-K consensus provirus).
Derivatives of the HERV-K(HML2) Env-Rec expression vector were generated from phCMV-HERV-K(HML2) Env-Rec. Env mut1 has a premature stop codon inserted after amino acid (aa) 658 (numbered from the first methionine). Env mut2 has a sequence encoding a glycophospholipid-anchoring motif (GLA, CCAAATAAAGGAAGTGGAACCACTTCAGGTACTACCCGTCTTCTATCTGGGCACACGTGTTTCACGTTGACAGGTTTGCTTGGGACGCTAGTAACCATGGGCTTGCTGACT) (57, 58) inserted after aa 631 [nt 8344 of HERV-K(HML2) consensus provirus] and aa 632, 633, and 634 mutated into stop codons (8344, accattgga to TAATAATga). Env mut3 was derived from Env mut2 by deleting the region corresponding to aa 459 to 630 [nt 7825 to 8340 of HERV-K(HML2) consensus provirus]. Env mut4 was generated by point mutations in phCMV-HERV-K(HML2) Env-Rec, changing aa 462 and 463 into stop codons (7834, agatcc to TgatAA). In all of these plasmids, the Rec ORF is left unchanged.
All of these plasmids were constructed using standard molecular biology techniques, and their sequences were verified by sequencing before use.
Cell culture, transfections, and lentiviral vector production.
293T and HeLa cells were maintained at 37°C, 5% CO2 in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all reagents are from Life Technology). HeLa cells (50% confluence on the day of transfection) were transfected using 1 μg of DNA (500 ng of candidate inhibitor and 500 ng of pCMV-ß control plasmid) and 5 μl Fugene 6 (Promega) per 3.5-cm dish. Lentiviral vectors for the expression of HA-tagged Tetherin were produced by transfecting 293T cells (3 × 106 per 10-cm dish) with 2 μg 8.91, 3 μg pSIN Tetherin-HA, and 0.7 μg phCMV VSV-G using 11.4 μl JetPrime (PolyPlus Transfection). Cell media were replaced 24 h posttransfection. Day 3 supernatants were collected, filtered (0.45 μm), and used for transducing cells either directly or after freezing and storage at −80°C.
Antibodies, Western blot analysis, and cell staining.
Human Tetherin was detected using a rabbit antiserum obtained from the AIDS Research and Reference Reagent Program (diluted 1/20,000 for Western blotting and 1/2,000 for cell staining; generated by Klaus Strebel and Amy Andrew). The HA-tagged version of human Tetherin was detected using anti-HA antibody (clone 3F10) from Roche (diluted 1/5,000). Vpu was detected using HIV-1NL4-3 Vpu rabbit antiserum from the NIH AIDS Research and Reference Reagent Program (diluted 1/20,000; generated by Klaus Strebel). HIV-1 Gag protein was detected using a p24 rabbit antiserum obtained from the NIH AIDS Research and Reference Reagent Program (dilution, 1/5,000). HERV-K(HML2) Gag protein was detected using a rabbit antiserum (1/1,000; described in reference 56). MLV ampho Env protein was detected using a goat antiserum directed against Rauscher leukemia virus gp70 (from the National Cancer Institute, Frederick, MD). HERV-K(HML2) Env protein was detected using a monoclonal antibody (19) (2 μg/ml). GFP was detected with a rabbit polyclonal antibody from Abcam (dilution, 1/20,000). Horseradish peroxidase (HRP)-conjugated secondary antibodies and ECL Plus reagent (GE Healthcare) were used for Western blotting, and an Alexa 488 goat anti-rabbit secondary antibody was used for cell staining (1/100; Molecular Probes).
For Western blot analysis, cells were washed in phosphate-buffered saline (PBS) and lysed for 45 min at 4°C in PBS, 1% NP-40 containing protease inhibitors (cOmplete EDTA-free protease inhibitors cocktail tablets; Roche). Cell debris was removed by a 15-min centrifugation step (4°C, 12,000 rpm). Protein samples (cell lysates or cell supernatants) then were subjected to SDS-PAGE under reducing conditions with a NuPAGE system (using precast Novex 4 to 12% Bis-Tris gels; Life Technology) and transferred onto nitrocellulose membranes with a semidry system. For Tetherin degradation assays, some samples were treated with peptide N-glycosidase F (PNGase F; NEB Biolabs) before SDS-PAGE.
To measure membrane Tetherin levels, HeLa cells were collected 48 h posttransfection in PBS, 5 mM EDTA, blocked for 30 min (PBS, 2% FCS, 0.1% sodium azide), and then incubated for 1 h with anti-Tetherin (or control) serum. Cells were washed thrice before a 45-min incubation with the secondary antibody, washed again 3 times, and finally resuspended in PBS–0.1% sodium azide for fluorescence-activated cell sorter (FACS) analysis. All staining steps were performed at 4°C.
Viral release assays.
In the first assays for Tetherin activity [HIV-1 or HERV-K(HML2)-Gag, measured by Western blotting] (Fig. 1), 293T cells (in 3.5-cm dishes) were transfected with 0.5 μg 8.91 [or 1 μg CMV-driven HERV-K(HML2) provirus with env inactivated], 100 ng phCMV VSV-G, 100 ng candidate inhibitor (or control), 0 to 50 ng pcDNA3-Tetherin, and pCMV-ß control plasmid (to adjust total DNA content up to 1.55 μg) using 8 μl Fugene 6 (Promega) or 3 μl JetPrime (PolyPlus Transfection). Cells were washed and their medium replaced 24 h posttransfection. At day 2, protein samples (cell lysates and supernatants) were collected for Western blot analysis or cells were fixed for electron microscopy (EM) studies.
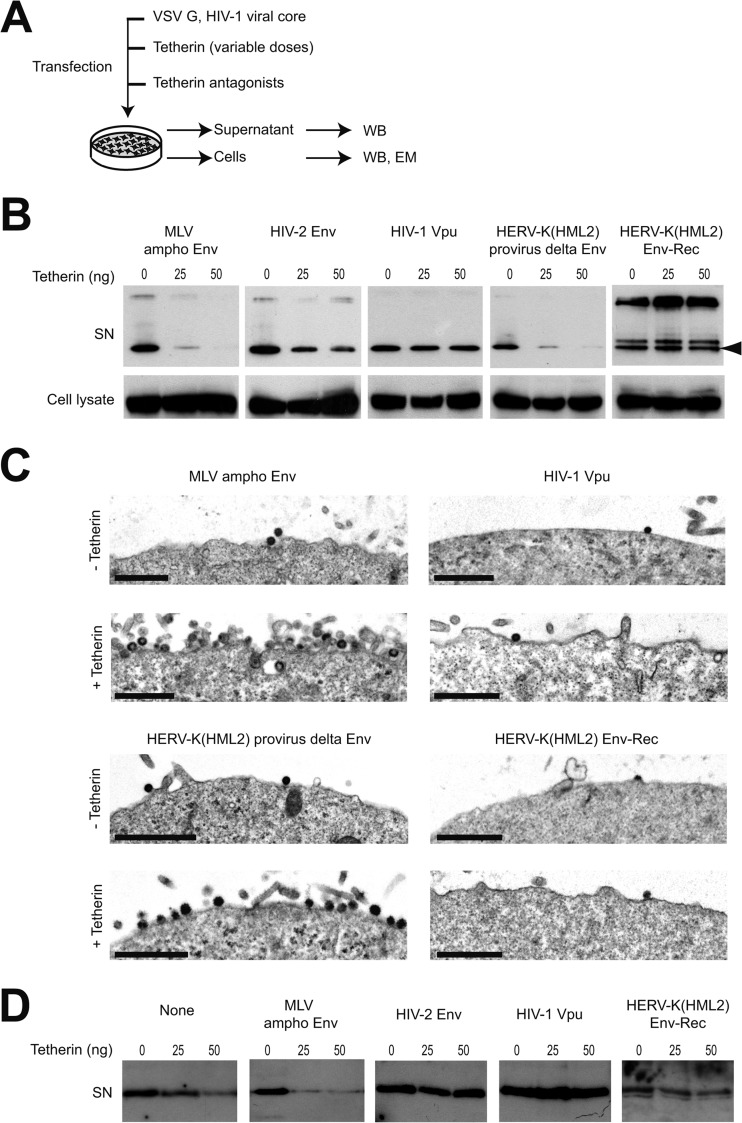
FIG 1.
Tetherin restriction is antagonized by HERV-K(HML2) Env-Rec. (A) Scheme of the experimental procedure. 293T cells were transfected with the indicated plasmids. Two days after transfection, cell supernatants were harvested and cells were either lysed to perform Western blot analysis or prepared for electron microscopy observation. (B) HIV-1 Gag protein content in neat supernatants (SN) was analyzed by Western blotting; p55 levels in corresponding cell lysates are displayed below for comparison. The nature of the potential Tetherin antagonist transfected is indicated above each gel part, and the doses of Tetherin transfected are given above each well (0, 25, or 50 ng). HIV-1 Gag p24 protein is indicated with an arrowhead. The additional bands observed in some supernatant samples (approximately 26 and 35 to 40 kDa) correspond to partially processed Gag protein. (C) Electron microscopy of cells obtained as described for panel A. Shown is a low magnification of cell membranes with viral particles observed by electron microscopy after negative staining. Images obtained with HIV-2 Env (not displayed here) were similar to those observed with HIV-1 Vpu and HERV-K(HML2) Env-Rec. Scale bars correspond to 1 μm. (D) HERV-K Gag protein content in neat supernatant was analyzed by Western blotting. The experimental procedure used was the same as that described for panel A, except the CMV-driven HERV-K(HML2) provirus defective for the envelope gene (instead of HIV-1 viral core) was transfected. Only cell supernatants were analyzed, since the anti-HERV-K(HML2) Gag antibody does not allow its specific detection in cell lysates (the signal/noise ratio is too high).
In the quantitative assay using FACS, 293T cells (in 3.5-cm dishes) were transfected as described above, except 0.75 μg pHR′SIN-cPPT-SEW was added and pcDNA3-Tetherin varied between 0 and 100 ng. Functional assays of the primate Tetherin genes were performed using the same method but with a single Tetherin dose (25 ng). Naive 293T target cells were infected with serial dilutions of day 3 viral supernatants (filtered with 0.45-μm-pore-size filters). The percentage of GFP-positive cells was measured 72 h postinfection by FACS and used to calculate the viral titer of each supernatant. Only samples with 5 to 20% GFP-positive cells were used to calculate titers to ensure that results were in the linear zone of the infection where positive cells have theoretically been infected by only one particle.
Coimmunoprecipitation assay.
293T cells were transduced at a high multiplicity of infection with supernatants containing the Tetherin-HA-expressing lentiviral vector. The day after, the cells were reseeded in 10-cm dishes and transfected with 4 μg of candidate antagonists or control (total amount of DNA was adjusted to 6 μg by adding pCMV-ß control plasmid) using 30 μl Fugene 6 (Roche). Medium was replaced after transfection, and cell lysates were collected at day 2 and used immediately. Immunoprecipitation was performed using a commercial anti-HA immunoprecipitation kit (Sigma). The immunoprecipitates (IP; 20 μl/100 μl per well) and lysates (input; 5 μl/700 μl per well) then were analyzed by Western blotting.
Electron microscopy.
For ultrastructural studies, monolayers of transfected cells were fixed for 1 h at 4°C in glutaraldehyde 2.5% in 0.1 M Sörensen phosphate buffer, pH 7.3. Cells were scraped off following fixation and centrifuged. The fixed pellets were rinsed for 1 h in ice-cold phosphate buffer, postfixed with 1% aqueous osmium tetroxide for 2 h, and dehydrated in increasing concentrations of ethanol prior to Epon embedding. Ultrathin sections were stained with uranyl acetate and lead citrate and examined under a Tecnai 12 Spirit transmission electron microscope (FEI, Hillsboro, OR, USA) at 80 kV.
RESULTS
To assess the presence of anti-Tetherin activity within HERV-K(HML2) proviruses, we used 2 expression vectors that together encode all of the proteins that are expressed by the HERV-K(HML2) consensus genome (56), with one construct being a full-length CMV-driven provirus defective for Env and the other containing only the env region also placed under the control of a CMV promoter. The latter encodes both Env and Rec, since the rec ORF is fully contained within env. These two constructs were tested for Tetherin antagonist activity in an assay in which 293T cells were transfected with expression vectors for (i) HIV-1 structural proteins (Gag, Pol, Tat, and Rev but not Vpu), (ii) VSV-G, (iii) human Tetherin (in increasing amounts), and (iv) candidate antagonists (Fig. 1A). The release of HIV-1-derived particles then was measured 2 days posttransfection by Western blotting experiments. As shown in Fig. 1B, the 2 HERV-K(HML2)-derived plasmids gave different results in this assay. In the presence of the env-defective provirus, the amount of HIV-1 Gag in cell supernatants decreases when the Tetherin amount increases, in agreement with the described inhibitory effect of Tetherin on viral particle release, and also as observed with an unrelated retroviral Env protein (murine leukemia virus amphotropic [MLV ampho] Env protein) used as a negative control. However, in the presence of HERV-K(HML2) Env-Rec, the amount of particles secreted is unaffected by Tetherin expression, indicating Tetherin activity is abrogated by Env-Rec. In the same assay, HIV-1 Vpu and, to a lesser extent, HIV-2 Env also inhibit Tetherin activity, confirming previous results (for example, see references 20 and 40). Since Rec is expressed from the two HERV-K(HML2) constructs we used (the inactivation of env in the provirus does not alter the Rec ORF), these data indicate that HERV-K(HML2) Env possesses anti-Tetherin activity. Unrelated to this, in the HERV-K(HML2) Env-Rec samples, the migration profile of HIV-1 Gag in cell supernatants is modified compared to that of the other proteins we tested, whether Tetherin is expressed or not. This altered maturation profile is not observed with the HERV-K(HML2) provirus defective for Env. This suggests that HERV-K(HML2) Env, in addition to its effect on Tetherin, also modifies in an unknown way the maturation of HIV-1-derived particles.
Some of the cells transfected for the experiments described above were additionally analyzed by electron microscopy (Fig. 1C). Irrespective of the antagonist, in the absence of Tetherin, very few particles are observed at the cell surface, consistent with their being efficiently released in the supernatant. When Tetherin is expressed, however, cells coexpressing the control MLV ampho Env protein or the HERV-K Gag-Pro-Pol proteins, which display no anti-Tetherin activity, are surrounded by numerous particles that are trapped after budding, whereas no such effect is observed in cells coexpressing HERV-K(HML2) Env-Rec or HIV-1 Vpu. Of note, most of the particles we observed display an immature morphology. This might be a side effect of the overexpression of HIV-1 structural proteins due to transient transfection. However, we could detect in all samples a small amount of mature particles displaying the specific shape of HIV-1 particles.
To characterize further the HERV-K(HML2) Env anti-Tetherin activity, we then tested if it could antagonize the tethering of HERV-K(HML2)-encoded particles. To do so, we transfected 293T cells as described above, except that the expression vector for HIV-1 core proteins was replaced by the HERV-K(HML2) provirus defective for Env. As presented in Fig. 1D, the amount of HERV-K p30 in the supernatant of transfected cells decreased with the amount of Tetherin in the presence of the control protein or the ampho MLV envelope, confirming that the budding of HERV-K(HML2) particles is impeded by Tetherin (22). In this assay, all three previously tested inhibitors of Tetherin, namely, HIV-1 Vpu, HIV-2 Env, and HERV-K(HML2) Env-Rec, completely blocked Tetherin activity, indicating that HERV-K(HML2)-encoded anti-Tetherin activity is active on its own particles.
To characterize further the action of HERV-K(HML2) Env-Rec against Tetherin, we then used a quantitative assay based on HIV-1-derived particles in which the amount of infectious particles (containing a GFP reporter gene) released in the supernatant was determined by measuring their titers on target cells using FACS (Fig. 2A). In this assay, Tetherin expression leads to a decrease in viral production in a dose-dependent manner (Fig. 2B). However, the efficiency of viral particle release also depends on the antagonist tested and is lowest when HERV-K(HML2) Env-Rec is coexpressed (data not shown), in agreement with the limited maturation of HIV-1 Gag protein that we observed by Western blotting in Fig. 1B. To allow a direct comparison of the inhibitors and to circumvent the high variability in titers observed in independent experiments, all values were normalized relative to the no-Tetherin condition for each of the potential inhibitors before statistical analysis. As illustrated in Fig. 2B, coexpression of HIV-2 Env or HERV-K(HML2) Env-Rec partially restored the viral particle release at all 3 different Tetherin doses compared to the control proteins (“None” and MLV ampho Env). Under the same conditions, HIV-1 Vpu proved to be much more effective as a Tetherin antagonist, as previously observed. This might be due to its inherently different nature, or higher expression level, compared to those of retroviral envelope proteins. Using the same assay, we tested all of the described natural alleles known for HERV-K(HML2) Env-Rec. The results obtained for the highest Tetherin dose are presented in Fig. 3A. As indicated, 2 out of the 6 tested alleles, namely, K108 and K109, are active against Tetherin in this assay. They correspond to the 2 most functional alleles as tested for other classical virological properties (19), which are recapitulated in Fig. 3B, suggesting that anti-Tetherin activity is linked to the canonical infectious properties of the proteins.
FIG 2.
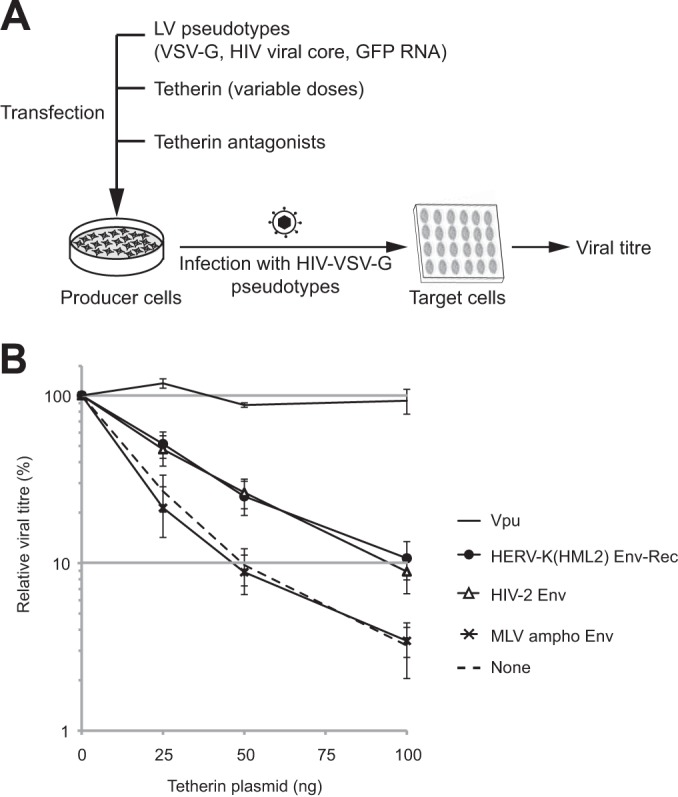
Quantitative assay of the HERV-K(HML2) Env-Rec-mediated inhibition of Tetherin activity. (A) Schematic description of the experimental procedure. 293T cells were transfected with the indicated plasmids. Three days posttransfection, the supernatants of producer cells were harvested to infect target cells. The percentage of infected cells was measured after 72 h by flow cytometry analysis, and viral titers were calculated for each condition. (B) The evolution of viral titers with the amount of transfected Tetherin (0 to 100 ng) is plotted for each potential inhibitor. The 100% value was set for each inhibitor using the titer measured without Tetherin. Error bars represent standard deviations of the means from 10 independent experiments, except for HIV-1 Vpu data (3 experiments).
FIG 3.
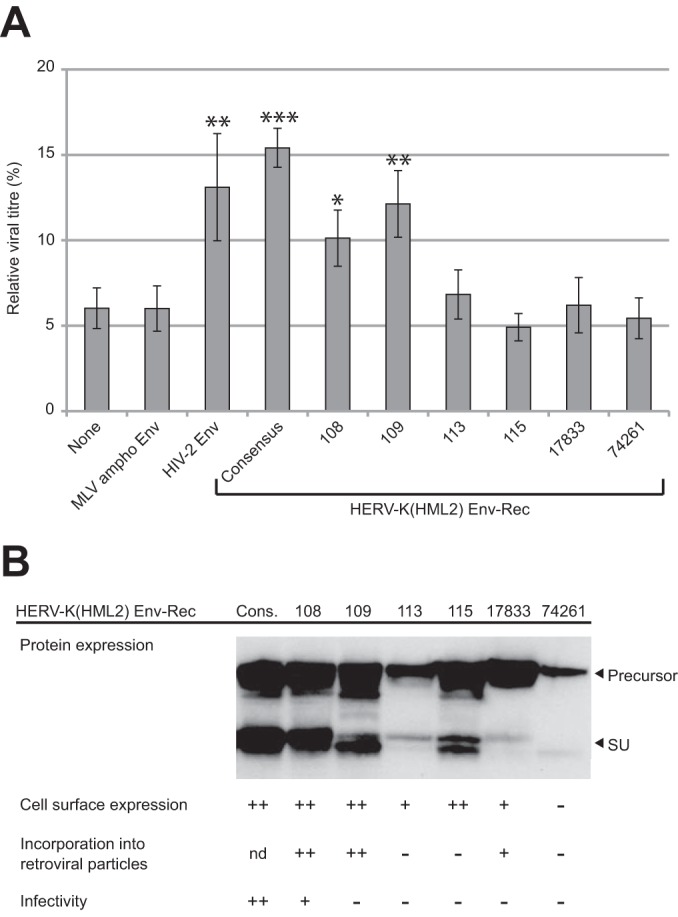
Effect of endogenous alleles of HERV-K(HML2) Env-Rec on Tetherin activity. (A) Previously described endogenous alleles of HERV-K(HML2) Env-Rec were tested for their ability to antagonize Tetherin as described in the legend to Fig. 2A. For each protein tested, the relative viral titer measured with 100 ng of Tetherin is represented (100% corresponds to the value measured in the absence of Tetherin). Under these conditions, HIV-1 Vpu restores the viral titer to 95.3% of the no-Tetherin condition (not shown). For statistical tests, all results were compared to the “None” condition. Asterisks indicate values significantly different from that obtained under the “None” condition (unpaired two-tailed t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Recapitulation of the functional properties of the 6 endogenous alleles of HERV-K(HML2) Env-Rec. Expression levels of the different HERV-K(HML2) Env-Rec alleles were analyzed by Western blotting on 293T cell lysates following transient transfection. Other functional properties of these alleles, which have been previously assessed (19), also are recapitulated. Cons., consensus HERV-K(HML2) element; nd, not determined.
We then used the same assay to ascertain which of the 2 HERV-K(HML2) proteins contained in the Env-Rec expression vector is active against Tetherin. Indeed, the experiment shown in Fig. 1B suggests that Env, and not Rec, is the active effector. However, it is possible that its expression levels obtained from the two constructs were different. To test the effect of each protein individually, we generated expression vectors for each of them. The Rec ORF consists of 2 exons and is completely contained within the env gene (Fig. 4A). cDNA of HERV-K(HML2) Rec was generated using transfected cells and cloned into an expression vector. In parallel, we introduced single point mutations in the Env-Rec expression vector in such a way that the splicing sites normally used to generate the Rec transcript were abolished without altering the amino acid sequence of HERV-K(HML2) Env (see Materials and Methods for more details). We then tested these two proteins for their activity against Tetherin as described above. As shown in Fig. 4B, the expression of the Rec protein has no effect on the antiviral activity of Tetherin, whereas transfection of the cells with the expression vector for Env has an inhibitory effect. This effect observed with Env is not statistically different from that of Env-Rec in an unpaired t test but was consistently slightly lower in all experiments, which might be due to a reduced expression of Env protein alone compared to the Env-Rec expression vector (see the Western blotting profile for each construct under similar conditions in Fig. 4A, right), most probably because Rec is involved in the nuclear export of HERV-K transcripts.
FIG 4.
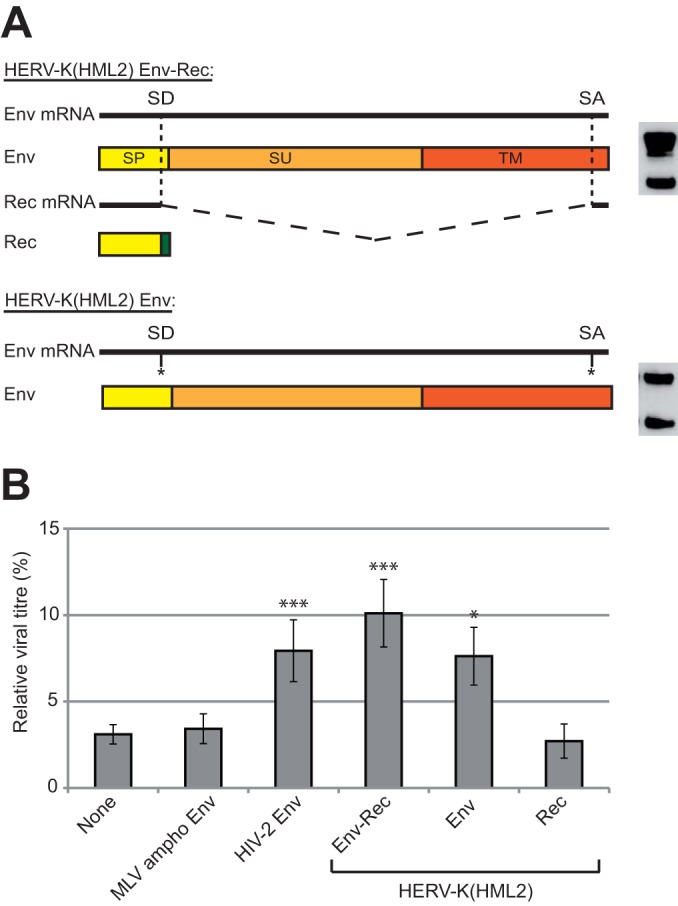
HERV-K(HML2) Env inhibits Tetherin activity. (A) The HERV-K(HML2) Env-Rec plasmid leads to the expression of 2 distinct proteins via alternative splicing: the full-length RNA encodes Env, a glycoprotein that is cleaved into 2 subunits during synthesis (surface subunit, SU, and transmembrane subunit, TM), while internal splicing sites (SD for splice donor and SA for splice acceptor) lead to the production of Rec, an accessory protein whose first exon (in yellow) is contained within the signal peptide (SP) of Env while the second exon (in green) is translated from a different reading frame. An expression vector leading to the production of Env but not Rec was generated by point mutations introduced in the splicing sites. Env expression levels obtained with each of the two constructs are shown on the right. (B) The ability of HERV-K(HML2) Env and Rec to antagonize Tetherin restriction was assayed as described in the legend to Fig. 2A. Results are given as described in the legend to Fig. 3A.
To further characterize the anti-Tetherin activity carried out by HERV-K(HML2) Env protein, we generated a series of mutants presented in Fig. 5A (see Materials and Methods for details). In order to enhance the readout of the experiments, all constructs were derived from the Env-Rec expression vector, which had the strongest anti-Tetherin activity. As shown in Fig. 5B, all mutant proteins are expressed at a similar level in 293T cells, and those designed to have a cell-membrane localization could be detected at the cell surface by FACS. All of these mutants then were tested in the functional assay described above, and the results obtained with the highest Tetherin dose are presented in Fig. 5C. As indicated in the figure, only HERV-K(HML2) Env mut1 still possesses anti-Tetherin activity, with an efficacy similar to that observed with the native form of the protein. This mutant is deleted for most of its cytoplasmic tail, which indicates that this domain is not involved in Tetherin recognition or inhibition. All of the other mutants, which are unable to counteract Tetherin activity, lack a proper transmembrane domain. This demonstrates that the transmembrane domain is required for the anti-Tetherin activity of HERV-K(HML2) envelope protein.
FIG 5.
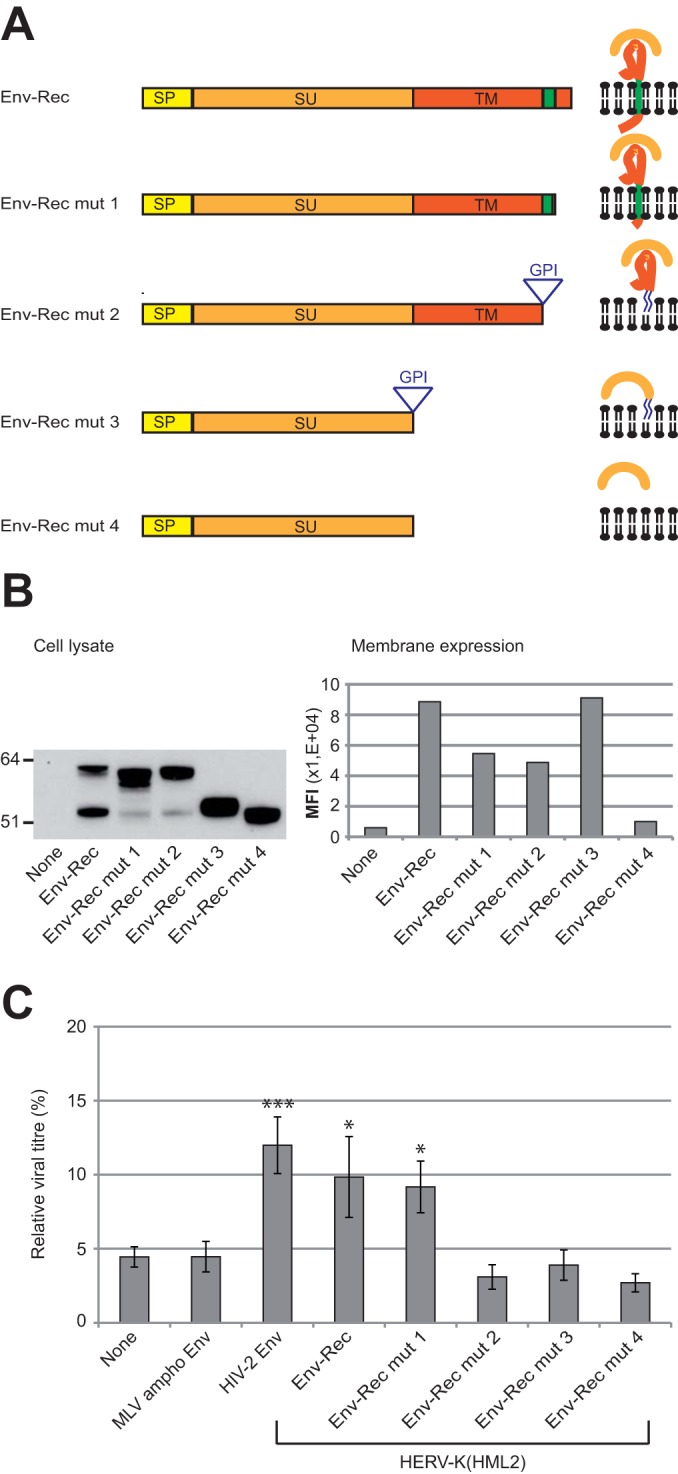
Characterization of HERV-K(HML2) Env domains necessary for inhibiting Tetherin activity. (A) Scheme of the different derivatives of HERV-K(HML2) Env-Rec. All modifications were designed to modify Env without altering the Rec ORF. The different domains of each construct are indicated: SP (signal peptide, yellow), SU (orange), and TM (red). HERV-K(HML2) Env-Rec mut1 corresponds to a C-terminal truncated version of the complete Env (41 aa deleted). HERV-K(HML2) Env-Rec mut2 has its membrane spanning (in green) and cytoplasmic domains replaced by a GPI anchor (the signal for the addition of the GPI anchor is depicted by a triangle in the scheme). HERV-K(HML2) Env-Rec mut3 consists of the SU subunit attached to the cell membrane by a GPI anchor. HERV-K(HML2) Env-Rec mut4 corresponds to the soluble surface subunit. For each construct, the expected protein structure is depicted on the right. (B) Expression levels of Env-Rec and derivatives were measured in transiently transfected 293T cells either by Western blotting on whole-cell lysates (left) or by FACS following cell surface staining (right). The histogram displays the median fluorescence intensities (MFI) of the whole population for each protein tested. (C) The efficiency of the different HERV-K(HML2) Env-Rec derivatives to antagonize Tetherin was assayed as described in the legend to Fig. 2A. The representation of results is the same as that described in the legend to Fig. 3A.
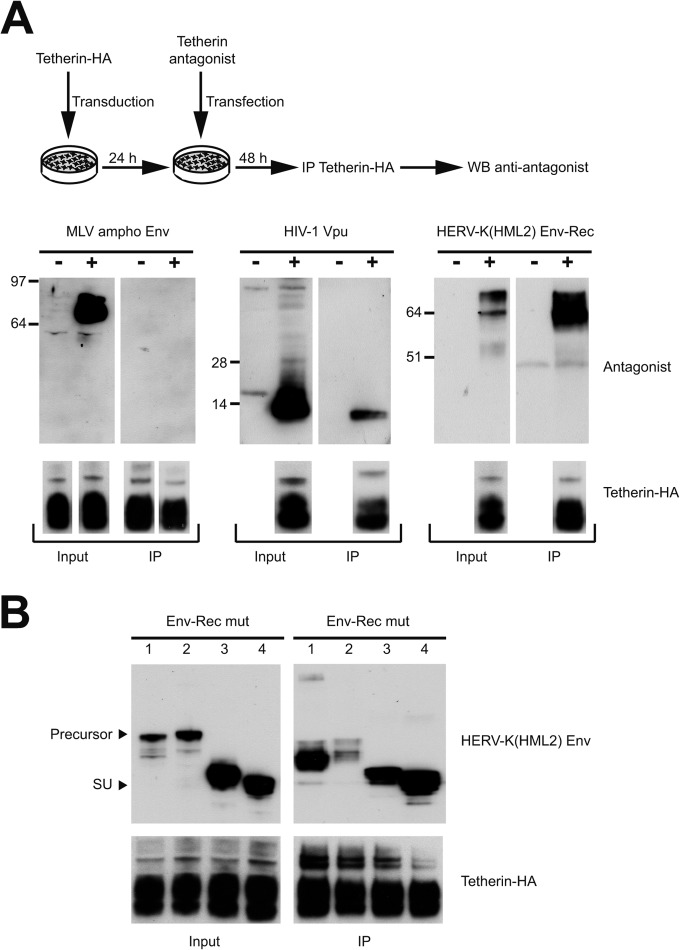
Most of the previously characterized Tetherin antagonists directly interact with it, as demonstrated through coimmunoprecipitation experiments. We tested if this is also the case for HERV-K(HML2) Env. 293T cells were transduced with a lentiviral vector expressing a HA-tagged version of Tetherin and then transfected with antagonists or control proteins. Two days posttransfection, cells were lysed and Tetherin was immunoprecipitated via the HA tag. The presence or absence of each antagonist in the immunoprecipitated fraction then was tested by Western blotting. As shown in Fig. 6A, the control MLV ampho Env protein, which has no anti-Tetherin activity, does not coimmunoprecipitate with it, unlike the well-characterized antagonist HIV-1 Vpu, in agreement with previous studies (55, 59–61). In this assay, HERV-K(HML2) Env also is immunoprecipitated very efficiently by Tetherin, suggesting that the two proteins interact in vivo. In all cases, the HA-tagged Tetherin was recovered in similar amounts in the immunoprecipitated fractions (Fig. 6A, lower), indicating that the nature of the tested antagonist did not bias the analysis. We then mapped the interacting domain within HERV-K(HML2) Env. We tested in coimmunoprecipitation experiments the mutants that we had previously assessed for their activity against Tetherin. As shown in Fig. 6B, all of them interact efficiently with Tetherin, even those unable to block its activity. Mutant 2 seems to be recovered less efficiently than other HERV-K(HML2) derivatives, but it was detected at a level similar to that of HIV-1 Vpu and clearly higher than that of the control ampho Env protein (not detected at all at the immunoprecipitated level despite a higher expression level in the raw input). This may be due to some misfolding in this mutant or even to a slightly different localization at the cell surface. Among this series of mutants, all of which interact with Tetherin, the shortest protein consists of only the SU subunit of the protein, which is responsible for the interaction observed with Tetherin.
FIG 6.
Assay for interaction between HERV-K(HML2) Env and Tetherin. (A) Scheme of the experimental procedure. 293T cells were transduced with an internally HA-tagged Tetherin lentiviral vector and then transfected with different Tetherin antagonists [MLV ampho Env, HIV-1 Vpu, and HERV-K(HML2) Env-Rec]. Cell lysates were harvested 48 h posttransfection and subjected to anti-HA immunoprecipitation. The presence of the Tetherin antagonists or control protein in the immunoprecipitated fraction (IP) was then assessed by Western blotting (top). A fraction of the cell lysates before immunoprecipitation (input) was included to ensure their efficient expression and detection. The presence of similar amounts of Tetherin in all samples (input and IP fractions) was checked by Western blotting in a separate gel run under the same conditions using an anti-HA antibody (lower). The minus antagonist sample was the same for each protein tested and was migrated only once to ensure Tetherin expression; it is presented below the MLV ampho panel. (B) In order to map the interaction domain between Tetherin and HERV-K(HML2) Env-Rec, HERV-K(HML2) Env-Rec derivatives (mut 1, mut 2, mut 3, and mut 4) were assayed for their ability to interact with Tetherin by following the protocol described for panel A. In the top panels, arrows point to the precursor form and SU subunit of HERV-K(HML2) Env-Rec derivatives. As described for panel A, Tetherin expression levels were measured for all samples by Western blotting and are presented below the Env panels.
We then investigated the mechanisms through which HERV-K(HML2) Env inhibits Tetherin activity. First, we tested its effect on the cellular localization of endogenous Tetherin in HeLa cells. As shown in Fig. 7A, the amount of Tetherin expressed at the cell surface was unaffected by HERV-K(HML2) Env-Rec expression. Therefore, the mechanism of action of HERV-K(HML2) Env protein is different from the one occurring with HIV-1 Vpu, which led to a clear downregulation of membrane Tetherin in our assay, in agreement with previous results (21). We then tested another described mechanism of action of known Tetherin antagonists, i.e., Tetherin degradation. For this experiment, 293T cells were transfected as described in the legend to Fig. 2 with all of the plasmids necessary to produce HIV-1-derived particles in the presence or absence of Tetherin inhibitors. The amount of Tetherin was analyzed by Western blotting 2 days posttransfection. As shown in Fig. 7B, confirming previous studies (62–64), the expression of Vpu by the cells results in a nearly complete loss of Tetherin expression, unlike control proteins (“None” and MLV ampho Env). The coexpression of HERV-K(HML2) Env modifies the migration pattern of Tetherin, with the lowest-molecular-weight form seemingly more abundant than when control proteins were expressed. To ensure that the total amount of Tetherin was not modified, we treated the samples with PNGase F, which removes N-linked glycosylations from proteins and renders the migration pattern of Tetherin, a normally heavily glycosylated protein, simpler. This confirmed that HERV-K(HML2) Env, unlike HIV-1 Vpu, does not lead to Tetherin degradation. However, it modifies its glycosylation pattern within the cells.
FIG 7.
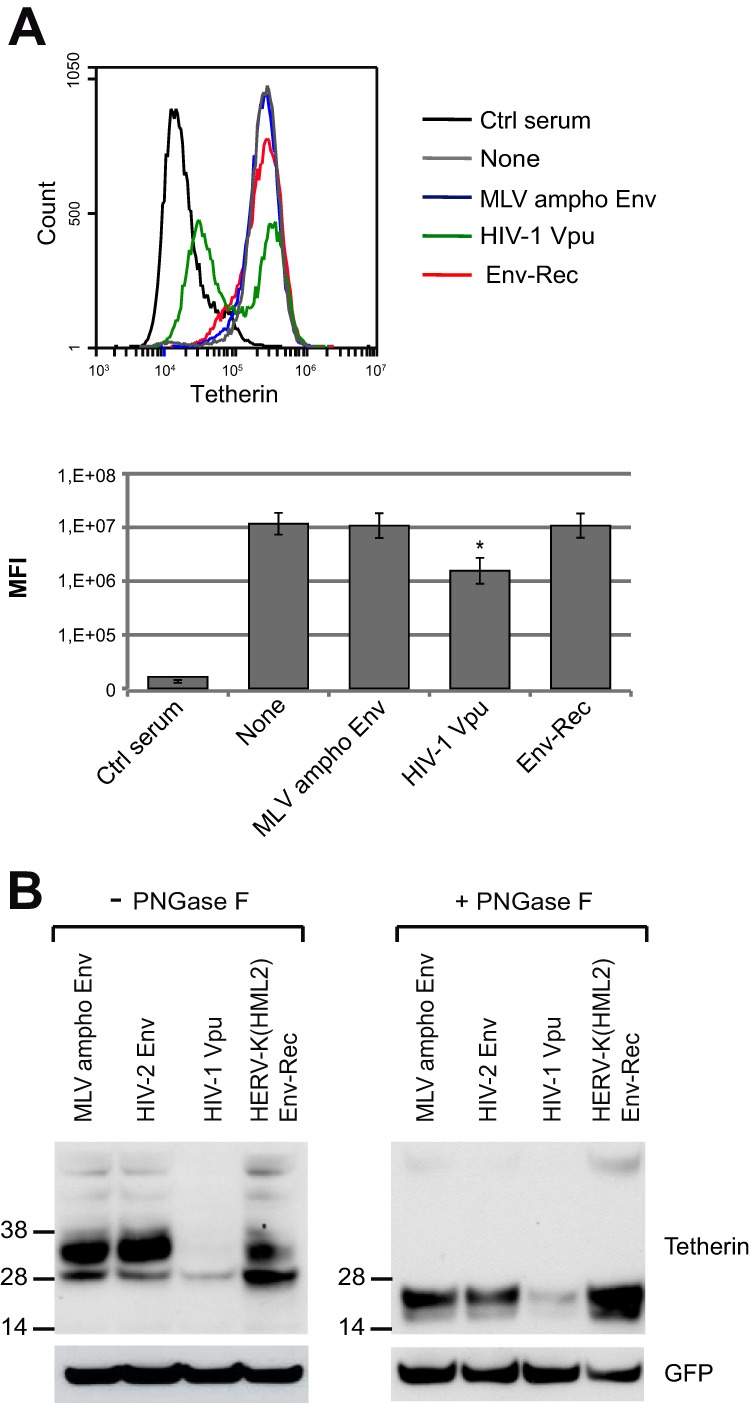
Mechanistic aspects of HERV-K(HML2) Env-Rec inhibition of Tetherin. (A) Assay for cell membrane downregulation of Tetherin by inhibitors. HeLa cells were transfected with expression vectors for Tetherin inhibitors or control proteins. Forty-eight hours posttransfection, cells were stained for cell surface Tetherin. A staining with a control rabbit serum (Ctrl) also was included in the experiment. The histogram on the top corresponds to 1 representative experiment out of 3. Below is displayed the median fluorescence intensities for each antagonist tested. Error bars represent standard deviations of the means from three independent experiments. All results are compared to the “None” condition. Asterisks indicate values significantly different (*, P < 0.05) from that obtained under “None” conditions (unpaired two-tailed t test). (B) Assay for Tetherin degradation by inhibitors. 293T cells were transfected with 8.91, pHR′SIN-cPPT-SEW, VSV-G, Tetherin expression vector, and a Tetherin inhibitor or control protein. Two days after transfection, cell lysates were prepared, treated with PNGase F (right), or left untreated (left). The membrane was stained for Tetherin (top) and then stripped and stained again for GFP (expressed from transfected pHR′SIN-cPPT-SEW) (bottom) to ensure that each well of the gel had been equally loaded.
Finally, we investigated the species specificity of HERV-K(HML2) Env-Rec toward Tetherin. Using the same functional assay as that described above, we tested a series of Tetherin genes from primates. The results obtained are shown in Fig. 8, together with those for control proteins or other Tetherin antagonists, with statistical significance versus the “None” condition indicated. Quite remarkably, HERV-K(HML2) Env-Rec shows a broad tropism, being able to antagonize all primate Tetherin proteins that we tested with nearly the same efficiency (viral titer increased 2.6 to 4.5 times compared to that of the “None” condition). HIV-2 Env shows the same broad efficiency but with slightly lesser effect (1.5- to 2.2-fold increase compared to the “None” condition). In comparison, HIV-1 Vpu protein possesses a very high inhibitory effect against Tetherin from species closely related to humans (chimpanzee and gorilla) but is much less active against the Tetherin from Old World monkey species, confirming previous results (33, 35, 52, 65, 66).
FIG 8.
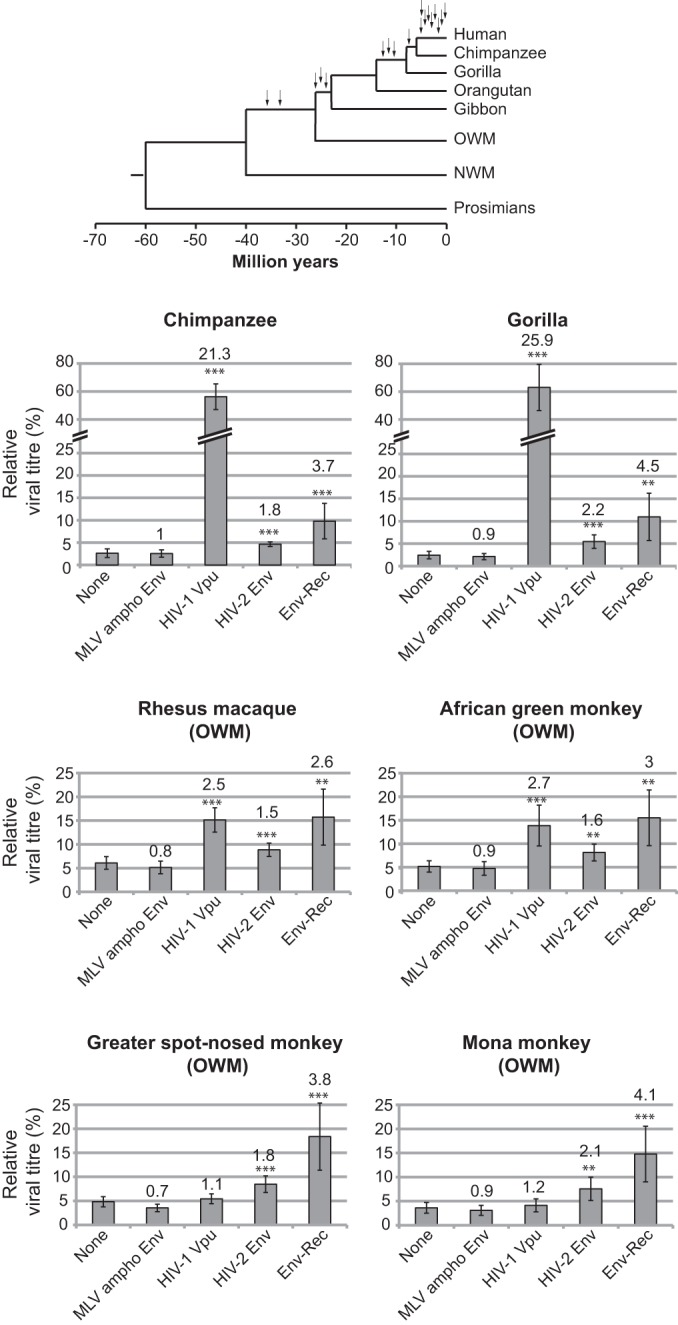
Effect of HERV-K(HML2) Env-Rec on several primate Tetherin proteins. HERV-K(HML2) Env-Rec was tested for its ability to antagonize several primate Tetherin proteins as described in the legend to Fig. 2A. For each Tetherin and antagonist tested, the relative viral titer measured with 25 ng of Tetherin is represented (100% corresponds to the value measured in the absence of Tetherin), with the average fold increase compared to the “None” condition indicated above each antagonist. Statistical tests were done as described in the legend to Fig. 3A using 9 independent experiments. A phylogenetic tree of primates is presented (top). The arrows indicate amplification bursts of HERV-K(HML2) elements during primate evolution (from reference 8) (OGM and NWM, Old and New World monkeys, respectively).
DISCUSSION
In this paper, we have identified Tetherin antagonist activity in human endogenous HERV-K(HML2) elements and mapped it to the Env protein. Through immunoprecipitation experiments, we have shown that the interacting domain is fully contained within the SU moiety. In previous years, numerous viral inhibitors of Tetherin were identified and characterized, showing a huge diversity in the way they interact with Tetherin. In some cases, recognition takes place in the cytoplasm, or within the cell membrane, between the transmembrane domains of the inhibitor and Tetherin. However, binding to Tetherin through its ectodomain is not unheard of: the interacting domain of HIV-2 Env has been localized to the extracellular domain of the protein, even if it has not been determined whether the interacting domain is contained within the SU subunit or the TM ectodomain (40). The functional assays we carried out with mutant proteins also indicate that the presence of the interacting domain is not enough to ensure anti-Tetherin activity, with the envelope transmembrane region being absolutely required. In particular, the GPI-anchored mutants that we generated are completely unable to counteract Tetherin, even though they both contain the SU subunit and are efficiently expressed on the surface of the cells, as shown by flow cytometry. However, it is possible that these GPI-anchored proteins are localized to specific cell surface microdomains where they would be unable to inhibit Tetherin activity. The interactions that we detected through immunoprecipitation experiments could happen during the transport of both proteins within the cells, at a place where the interaction would have no functional consequence for Tetherin activity.
In addition, the functional assays indicated that the cytoplasmic tail of the HERV-K(HML2) Env protein plays no role in its anti-Tetherin activity. This is unlike HIV-2 Env protein, for which the corresponding domain has been shown to be required (40). However, this specific requirement has been more precisely linked to the presence of a GYxxθ motif (where θ is a hydrophobic residue) that plays a role in HIV-2 Env protein retrograde transport from the cell surface to trans-Golgi network (TGN)-associated compartments (40, 67). HERV-K(HML2) Env protein lacks such a motif in the corresponding domain, in good agreement with this region being dispensable for anti-Tetherin activity. These observations also are consistent with the differences observed in the mechanisms of action of these two Tetherin antagonists: HIV-2 Env acts at least partially through depleting Tetherin from the cell membrane, whereas we showed that HERV-K(HML2) Env protein expression has no effect on Tetherin cell surface expression.
Indeed, the mechanism through which HERV-K(HML2) Env protein inhibits Tetherin remains elusive. We tested another mechanism that was shown to play a role in HIV-1 Vpu inhibition, namely, Tetherin degradation (21), and showed that HERV-K(HML2) Env protein expression does not modify the total amount of cellular Tetherin. During these experiments, we repeatedly noticed a slight modification in the glycosylation pattern of Tetherin specifically associated with HERV-K(HML2) Env expression. This was not observed with the “None” control vector or with another retroviral envelope protein devoid of anti-Tetherin activity. However, the Tetherin glycosylation pattern has been shown to be different whether it is stably produced by the cell or expressed through transient transfection (26). Thus, the modification in the glycosylation pattern that we observe may be specific to the overexpression system used and not relevant to normal expression conditions. In addition, in a previous study it was shown that Tetherin remains fully able to block viral egress even in the complete absence of glycosylation (26). Therefore, the modification in the migration pattern that we noted is unlikely to account for the observed Tetherin inhibition. As is the case for other described Tetherin antagonists (e.g., Ebola GP [48, 49, 68]), more experiments will be required to understand the mechanism of action used by HERV-K(HML2) Env protein.
In another set of experiments, we measured the anti-Tetherin activity of HERV-K(HML2) Env-Rec and other antagonists against the Tetherin proteins present in several primate species. HERV-K(HML2) Env-Rec and HIV-2 Env both showed a similar broad spectrum of inhibition, being able to antagonize all 6 genes that we tested with the same efficiency, whereas HIV-1 Vpu is more active, but only on a subset of closely related Tetherin genes. Interestingly, all of the genes tested are from species where the HERV-K(HML2) family is present, suggesting that Tetherin inhibition could have been an ancestral property of this family that helped its amplification in the primate lineage. These data also suggest that the very high efficiency of HIV-1 Vpu comes at the cost of a reduced spectrum of action, unlike what is observed with HIV-2 and HERV-K(HML2) Env proteins. Of note, Ebola GP is another Tetherin antagonist that shows a very broad spectrum of action, being able to antagonize human, primate, and murine Tetherin proteins, as well as artificial constructs with no sequence similarity to Tetherin (48, 49), and also is less efficient than HIV-1 Vpu or HIV-2 Env at inhibiting human Tetherin (48).
Finally, we tested all described alleles of intact HERV-K(HML2) envelope proteins for their anti-Tetherin activity. These genes belong to recent proviruses, all of which integrated after the divergence between the human and chimpanzee lineages, and the corresponding proteins are highly conserved (more than 97% sequence identity, i.e., less than 20 amino acid changes between any 2 sequences). We found that the only Env proteins active against Tetherin were the two that were previously shown to have retained their full or nearly complete set of viral properties. In particular, the results obtained for K115 Env (whose expression level and maturation are similar to those of K108 and K109 Env proteins) indicate that the amount and maturation of Env are not the only criteria required for anti-Tetherin activity. Of note, despite its proper expression, maturation, and localization to the membrane, K115 Env is unable to be incorporated onto HIV-1-derived particles, unlike K108 and K109. Thus, anti-Tetherin activity appears to depend on most of the classical properties of retroviral Env proteins. Interestingly, the HERV-K(HML2) consensus Env, which was reconstructed as the theoretical ancestor of human-specific HERV-K(HML2) provirus Env protein (56), seems to be the best protein for all of the properties we assessed, suggesting that the alleles we tested have been degenerating since their integration in the human genome less than 2 mya. However, because the HERV-K(HML2) family was active within the human genome until recently, it is still characterized by a strong polymorphism, both in the insertion sites and in the exact viral sequences present at specific loci. As a result, it is very likely that some people will possess alleles with a strong anti-Tetherin activity, whereas others may have no functional allele in their genome. This could have physiological consequences, as HERV-K(HML2) elements have been shown to be induced in some HIV-1-positive patients (69, 70; reviewed in reference 71). In this case, HERV-K(HML2)-encoded anti-Tetherin activity could add up to that of Vpu in some, but not all, individuals, provided it is expressed at a high enough level. Moreover, Tetherin has been shown to act as an innate immune activator through type I interferons while blocking viral egress, and at least some of its antagonists, as well as being able to restore viral budding, also inhibit Tetherin signaling activity (30, 31). If the latter property is shared by HERV-K(HML2) Env proteins, which will have to be tested, these Env proteins may modulate innate immune responses, and it should be assessed if their specific induction, observed in several pathologies (reviewed in references 71 and 72), could have a causal role.
ACKNOWLEDGMENTS
We thank F. Clavel (Institut Universitaire d'Hématologie, Hôpital Saint Louis, Paris, France) for the gift of HIV2 ROD10 Env and S. Neil (King's College, London, United Kingdom) for BST2 and Vpu plasmids and his advice concerning available Tetherin antibodies. We also thank G. Cornelis and J. Tsang for helpful discussions and critical readings of the manuscript and S. Souquere for help with the electron microscopy experiments.
This work was supported by grants from the Centre National de la Recherche Scientifique and Ligue Nationale Contre le Cancer (Equipe Labelisée).
Footnotes
Published ahead of print 10 September 2014
REFERENCES
- 1. Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101(Suppl 2):S14572–S14579. 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 10:395–406. 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- 3. Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13:283–296. 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 4. Dewannieux M, Heidmann T. 2013. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr. Opin. Virol. 3:646–656. 10.1016/j.coviro.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5. Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321–3329. 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789. 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 7. Blaise S, de Parseval N, Benit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U. S. A. 100:13013–13018. 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Medstrand P, Mager DL. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian RP, Wildschutte JH, Russo C, Coffin JM. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Magiorkinis G, Belshaw R, Katzourakis A. 2013. “There and back again”: revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120504. 10.1098/rstb.2012.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535. 10.1016/S0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 12. Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79:12507–12514. 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, Cullen BR. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. U. S. A. 96:13404–13408. 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boller K, Konig H, Sauter M, Mueller-Lantzsch N, Lower R, Lower J, Kurth R. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349–353. 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 16. Bieda K, Hoffmann A, Boller K. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591–596. [DOI] [PubMed] [Google Scholar]
- 17. Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Fodinger D, Seppele H, Schanab O, Magin-Lachmann C, Lower R, Jansen B, Pehamberger H, Wolff K. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63:8735–8741. [PubMed] [Google Scholar]
- 18. Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65:4172–4180. 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 19. Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573–15577. 10.1128/JVI.79.24.15573-15577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 21. Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83:1837–1844. 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neil SJ. 2013. The antiviral activities of tetherin. Curr. Top. Microbiol. Immunol. 371:67–104. 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 24. Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. 2007. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci. 120:3850–3858. 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- 25. Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709. 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 26. Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liberatore RA, Bieniasz PD. 2011. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 108:18097–18101. 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Galao RP, Le Tortorec A, Pickering S, Kueck T, Neil SJ. 2012. Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe 12:633–644. 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. 2013. Stimulation of NF-kappaB activity by the HIV restriction factor BST2. J. Virol. 87:2046–2057. 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim ES, Emerman M. 2009. Simian immunodeficiency virus SIVagm from African green monkeys does not antagonize endogenous levels of African green monkey tetherin/BST-2. J. Virol. 83:11673–11681. 10.1128/JVI.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang SJ, Lopez LA, Hauser H, Exline CM, Haworth KG, Cannon PM. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. 10.1186/1742-4690-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartee E, McCormack A, Fruh K. 2006. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2:e107. 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:9672–9681. 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pardieu C, Vigan R, Wilson SJ, Calvi A, Zang T, Bieniasz P, Kellam P, Towers GJ, Neil SJ. 2010. The RING-CH ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog. 6:e1000843. 10.1371/journal.ppat.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83:11966–11978. 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U. S. A. 106:20889–20894. 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Celestino M, Calistri A, Del Vecchio C, Salata C, Chiuppesi F, Pistello M, Borsetti A, Palu G, Parolin C. 2012. Feline tetherin is characterized by a short N-terminal region and is counteracted by the feline immunodeficiency virus envelope glycoprotein. J. Virol. 86:6688–6700. 10.1128/JVI.07037-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dietrich I, McMonagle EL, Petit SJ, Vijayakrishnan S, Logan N, Chan CN, Towers GJ, Hosie MJ, Willett BJ. 2011. Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J. Virol. 85:5840–5852. 10.1128/JVI.00071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morrison JH, Guevara RB, Marcano A, Saenz DT, Fadel HJ, Rogstad DK, Poeschla EM. 2014. FIV envelope glycoproteins antagonize Tetherin through a distinctive mechanism that requires virion incorporation. J. Virol. 88:3255–3272. 10.1128/JVI.03814-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yin X, Hu Z, Gu Q, Wu X, Zheng YH, Wei P, Wang X. 2014. Equine tetherin blocks retrovirus release and its activity is antagonized by equine infectious anemia virus envelope protein. J. Virol. 88:1259–1270. 10.1128/JVI.03148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 106:2886–2891. 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee YN, Bieniasz PD. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3:e10. 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J. Virol. 84:7243–7255. 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuhl A, Banning C, Marzi A, Votteler J, Steffen I, Bertram S, Glowacka I, Konrad A, Sturzl M, Guo JT, Schubert U, Feldmann H, Behrens G, Schindler M, Pohlmann S. 2011. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J. Infect. Dis. 204(Suppl 3):S850–S860. 10.1093/infdis/jir378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875. 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 51. Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, Grez M, Thrasher AJ. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803–813. 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 52. McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5:e1000300. 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sandrin V, Muriaux D, Darlix JL, Cosset FL. 2004. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 78:7153–7164. 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bour SP, Aberham C, Perrin C, Strebel K. 1999. Lack of effect of cytoplasmic tail truncations on human immunodeficiency virus type 2 ROD env particle release activity. J. Virol. 73:778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vigan R, Neil SJ. 2010. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J. Virol. 84:12958–12970. 10.1128/JVI.01699-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556. 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee SW, Kahn ML, Dichek DA. 1992. Expression of an anchored urokinase in the apical endothelial cell membrane. Preservation of enzymatic activity and enhancement of cell surface plasminogen activation. J. Biol. Chem. 267:13020–13027. [PubMed] [Google Scholar]
- 58. Ragheb JA, Anderson WF. 1994. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J. Virol. 68:3207–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dube M, Roy BB, Guiot-Guillain P, Binette J, Mercier J, Chiasson A, Cohen EA. 2010. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 6:e1000856. 10.1371/journal.ppat.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. 2009. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5:e1000574. 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem. 284:35060–35072. 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J. Virol. 83:7931–7947. 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5:e1000450. 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrew AJ, Miyagi E, Strebel K. 2011. Differential effects of human immunodeficiency virus type 1 Vpu on the stability of BST-2/tetherin. J. Virol. 85:2611–2619. 10.1128/JVI.02080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Krausslich HG, Fackler OT, Keppler OT. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285–297. 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 66. Gupta RK, Hue S, Schaller T, Verschoor E, Pillay D, Towers GJ. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, Cannon PM. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lopez LA, Yang SJ, Exline CM, Rengarajan S, Haworth KG, Cannon PM. 2012. Anti-tetherin activities of HIV-1 Vpu and Ebola virus glycoprotein do not involve removal of tetherin from lipid rafts. J. Virol. 86:5467–5480. 10.1128/JVI.06280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bhardwaj N, Maldarelli F, Mellors J, Coffin JM. 2014. HIV-1 infection leads to increased transcription of HERV-K(HML-2) proviruses in vivo but not to increased virion production. J. Virol. 23:01623–01614. 10.1128/JVI.01623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 22:979–984. 10.1089/aid.2006.22.979. [DOI] [PubMed] [Google Scholar]
- 71. Hohn O, Hanke K, Bannert N. 2013. HERV-K(HML-2), the best preserved family of HERVs: endogenization, expression, and implications in health and disease. Front. Oncol. 3:246. 10.3389/fonc.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. 2008. Endogenous retroviruses and cancer. Cell. Mol. Life Sci. 65:3366–3382. 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]