ABSTRACT
Tick-borne encephalitis (TBE) virus is an important human-pathogenic flavivirus endemic in large parts of Europe and Central and Eastern Asia. Neutralizing antibodies specific for the viral envelope protein E are believed to mediate long-lasting protection after natural infection and vaccination. To study the specificity and individual variation of human antibody responses, we developed immunoassays with recombinant antigens representing viral surface protein domains and domain combinations. These allowed us to dissect and quantify antibody populations of different fine specificities in sera of TBE patients and vaccinees. Postinfection and postvaccination sera both displayed strong individual variation of antibody titers as well as the relative proportions of antibodies to different domains of E, indicating that the immunodominance patterns observed were strongly influenced by individual-specific factors. The contributions of these antibody populations to virus neutralization were quantified by serum depletion analyses and revealed a significantly biased pattern. Antibodies to domain III, in contrast to what was found in mouse immunization studies with TBE and other flaviviruses, did not play any role in the human neutralizing antibody response, which was dominated by antibodies to domains I and II. Importantly, most of the neutralizing activity could be depleted from sera by a dimeric soluble form of the E protein, which is the building block of the icosahedral herringbone-like shell of flaviviruses, suggesting that antibodies to more complex quaternary epitopes involving residues from adjacent dimers play only a minor role in the total response to natural infection and vaccination in humans.
IMPORTANCE Tick-borne encephalitis (TBE) virus is a close relative of yellow fever, dengue, Japanese encephalitis, and West Nile viruses and distributed in large parts of Europe and Central and Eastern Asia. Antibodies to the viral envelope protein E prevent viral attachment and entry into cells and thus mediate virus neutralization and protection from disease. However, the fine specificity and individual variation of neutralizing antibody responses are currently not known. We have therefore developed new in vitro assays for dissecting the antibody populations present in blood serum and determining their contribution to virus neutralization. In our analysis of human postinfection and postvaccination sera, we found an extensive variation of the antibody populations present in sera, indicating substantial influences of individual-specific factors that control the specificity of the antibody response. Our study provides new insights into the immune response to an important human pathogen that is of relevance for the design of novel vaccines.
INTRODUCTION
The genus Flavivirus of the family Flaviviridae comprises several important mosquito- and tick-transmitted human pathogens, including yellow fever (YF), dengue, West Nile (WN), Japanese encephalitis (JE), and tick-borne encephalitis (TBE) viruses (1). For humans, live-attenuated vaccines are available against YF (2) as well as JE (3), and inactivated vaccines are available against JE (3) and TBE (4), but no dengue vaccine has reached the market so far (5). The induction of neutralizing antibodies is generally believed to be essential for long-lived flavivirus immunity (6, 7). Studies with polyclonal and monoclonal antibodies (MAbs) have provided detailed insights into the mechanism of virus neutralization through the inhibition of viral entry functions (7–9). In contrast, less is known about antibody populations with different specificities in polyclonal sera, their relative concentrations, and their contributions to virus neutralization. Nevertheless, deconstructing the antibody specificities in sera is essential for investigating the antibody repertoire produced by long-lived plasma cells (LLPCs) in the bone marrow, which are the main suppliers of antibodies circulating in the blood (10–12). The as-yet-undefined mechanisms of selecting only a subset of cells to generate the LLPC repertoire in the bone marrow are certainly an important factor contributing to the poorly understood phenomenon of antibody immunodominance (13).
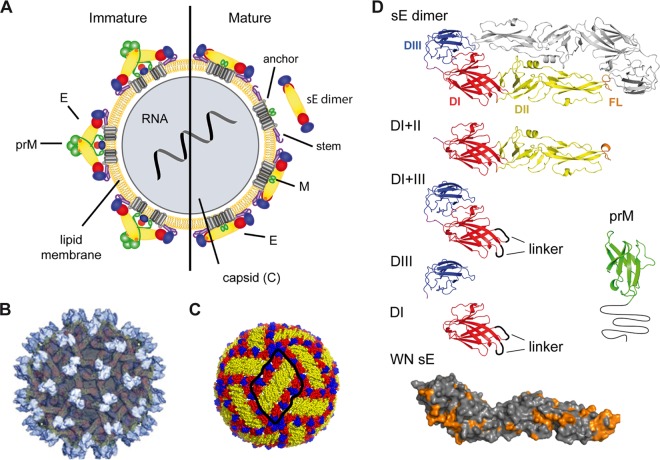
Flaviviruses have a relatively simple molecular organization, with a nucleocapsid (composed of the positive-stranded RNA and the capsid protein C) that is surrounded by a lipid envelope with two membrane-associated proteins, M and E (14). Virus assembly occurs in the endoplasmic reticulum and first leads to the formation of immature, noninfectious forms of the virion containing the precursor of M (prM), which is proteolytically cleaved by furin during exocytosis to generate infectious, M-containing virions (Fig. 1A). Structural details have been elucidated by X-ray crystallography of isolated E proteins from several flaviviruses, a prM-E complex of dengue 2 virus, as well as cryoelectron microscopic studies of both mature and immature dengue and WN virus particles (reviewed in reference 15). Immature virions are studded with 60 spikes of trimers of prM-E heterodimers, whereas mature virions display 90 antiparallel E dimers that form a closed herringbone-like shell (Fig. 1B and C). The externally accessible part of E is composed of three domains (DI, DII, DIII) (Fig. 1D) that are the major targets of neutralizing antibodies, generally believed to mediate long-term protection against flavivirus disease (6, 7).
FIG 1.
Schematic models and structures of immature and mature flavivirus particles and recombinant antigens employed. (A) Schematic representation of a virion with the immature form on the left and the mature form after prM cleavage on the right side. The nucleocapsid, surrounded by a lipid bilayer, is composed of C proteins and contains the viral RNA. In immature virions, prM and E form heterodimers. During virus secretion from host cells, prM is cleaved and E proteins are rearranged into homodimers. sE dimer, soluble form of the E dimer, lacking the stem and anchor regions. Reproduced from reference 27 under the CC BY license. (B) Surface representation of an immature dengue 2 virus as determined by cryoelectron microscopy. A total of 60 trimers of E-prM heterodimers protrude from the surface of immature virions. prM proteins at the tip of the spikes are colored in light gray. E proteins are shown in darker gray. Reprinted from reference 79 with permission of the publisher. (C) E protein arrangement on the surface of mature dengue 2 virus particles. Three E dimers form a raft (encircled in black) of the herringbone-like lattice covering the virion surface. Reproduced from reference 27 under the CC BY license. (D) Ribbon diagrams of the TBE sE dimer and other recombinant antigens used in the study (DI+II, DI+III, DIII, DI, and prM). In the surface representation of WN sE, amino acids conserved between TBE and WN viruses are highlighted in orange. Color code: DI, red; DII, yellow; DIII, blue; the fusion loop (FL) is highlighted in orange. prM (displayed is the structure of the dengue 2 prM [80], assuming that, like E, the overall structure of the TBE prM protein will be similar) is shown in green and black. Gly linkers introduced in constructs of DI and DI+III are shown as black loops.
Studies with both mouse and human MAbs revealed that any site accessible at the virion surface can function as a target for virus neutralization. At least in the mouse system, the most strongly neutralizing MAbs were directed against DIII (reviewed in reference 15), whereas broadly flavivirus cross-reactive antibodies recognizing the highly conserved fusion loop at the tip of DII had no or only low neutralizing activity both in mice and humans (16–19). Consistent with the relatively large size of antibody binding sites in protein antigens (around 1,000 Å2[20, 21]), MAbs were found to bind not only to individual domains and junctions between domains of the same E protein molecule but in some instances also to more complex quaternary antigenic sites, comprising amino acid residues from both molecules of the E dimer (22, 23) or even from adjacent dimers that are found only at the virion surface (24, 25). Evidence for the involvement of such complex antigenic sites has also been obtained from the analysis of human sera, primarily from dengue patients (26) and YF vaccinees (27).
It was the major objective of our work to study the specificities of antibodies in peripheral blood induced by natural TBE virus infection as well as vaccination with the inactivated TBE vaccine, which has proven to be highly effective in the field (28–30). As a specific goal, we wanted to gain insights into the extent of individual variation of antibody reactivities and immunodominance patterns and their possible consequences for virus neutralization through the analysis of relatively large panels of serum samples from both cohorts. For this purpose, we established a platform of immunoassays with recombinant antigens of E and prM, making use of the modular domain organization of E that allows not only the production of the whole protein E but also of its individual domains and combinations thereof. These analyses yielded quantitative information on the proportions of antibody populations directed to different domains of the viral envelope proteins and thus a serological fingerprint for each individual serum sample. By depletion analyses, we were able to assign most of the virus neutralizing activity, both in postinfection and postvaccination sera, to antibodies reacting with DI and DII and/or the junction between the two domains. We also found evidence for neutralization epitopes dependent on the E dimer structure, and the involvement of even more complex quaternary epitopes cannot be excluded. Postinfection sera had a higher frequency of antibodies to prM, whereas a higher degree of flavivirus cross-reactivity was found in postvaccination sera. Independent of these overall characteristics, both groups displayed a substantial degree of individual variability with respect to the antibody specificities found in different sera and their contribution to virus neutralization.
MATERIALS AND METHODS
Study groups/cohorts.
TBE postinfection sera were collected from 38 patients >1.5 years (median of 1.9 years) after admission to the Hospital Ceske Budejovice. A total of 43 healthy persons previously vaccinated against TBE received a booster vaccination (FSME-IMMUN, 0.5 ml; Baxter) at the Department of Virology of the Medical University of Vienna, and blood was withdrawn 2.5 to 5 weeks (median of 24 days) after vaccination (Table 1).
TABLE 1.
Characteristics of the study cohorts of TBE patients and vaccinees
| Characteristic | Value |
|
|---|---|---|
| Patients | Vaccinees | |
| No. | 38 | 43 |
| Gender | 11 female/27 male | 23 female/20 male |
| Median age (range) in yrs at sample collection | 56 (19–77) | 61 (19–78) |
| Median no. of yrs (range) after onset of symptoms | 1.9 (1.6–3.9) | |
| Median no. of days (range) after booster vaccination | 24 (16–43) | |
Ethics statement.
The studies were approved by the ethics committees of the Hospital Ceske Budejovice, Czech Republic (approval number 8/2008), and the Medical University of Vienna, Austria (approval number 590/2007), and all patients gave their written informed consent.
Virus production.
Production and purification of formalin-inactivated TBE virus was performed as described in reference 31. Briefly, TBE virus strain Neudörfl (GenBank accession number U27495) was used to infect primary chicken embryo cells. The supernatant was harvested 48 h postinoculation, and the virus was concentrated by ultracentrifugation. Purification was carried out by rate zonal and equilibrium sucrose density gradient centrifugation. For inactivation, the purified virus was treated with 37% formalin at a final dilution of 1:2,000 at 37°C for 24 h.
Production of recombinant proteins.
All recombinant proteins of TBE virus were derived from strain Neudörfl (GenBank accession number U27495), and the WN virus soluble E (sE) protein was derived from strain NY99 (GenBank accession number AF196835). The recombinant antigens (displayed in Fig. 1D) contained the following amino acids for TBE virus: sE, 1 to 400; DI+II, 1 to 302; DI+III, 1 to 52 + 8 Gly linker + 137 to 192 + 8 Gly linker + 285 to 400; DI, 1 to 52 + 8 Gly linker + 137 to 192 + 8 Gly linker + 285 to 302; DIII, 302 to 398; prM, 1 to 129 (containing a deletion at the furin cleavage site as described in reference 32). For the WN virus, the sE protein contained amino acids 1 to 400.
With the exception of DIII, all antigens were produced with a tandem Strep-tag in the Drosophila expression system (Invitrogen) as described previously (27). The expression vector pT389 (kindly provided by Thomas Krey and Felix Rey, Institut Pasteur, France) encodes the export signal sequence Bip, an enterokinase cleavage site, and the tandem Strep-tag. Drosophila Schneider 2 cells were stably transfected using blasticidin for selection. Protein expression was induced by the addition of CuSO4, and supernatants were harvested 7 to 10 days after induction. Antigens were purified via affinity chromatography with Strep-Tactin columns (IBA) according to the manufacturer's instructions.
DIII was expressed as a fusion protein with thioredoxin and a C-terminal His tag in Escherichia coli strain BL21 with the pET 32a Xa/LIC vector (Novagen) as described previously (33). Cell lysates were clarified, and the DIII-Trx-His protein was purified via Ni2+ affinity chromatography (GE Healthcare Life Sciences) from the soluble fraction.
Blocking ELISA.
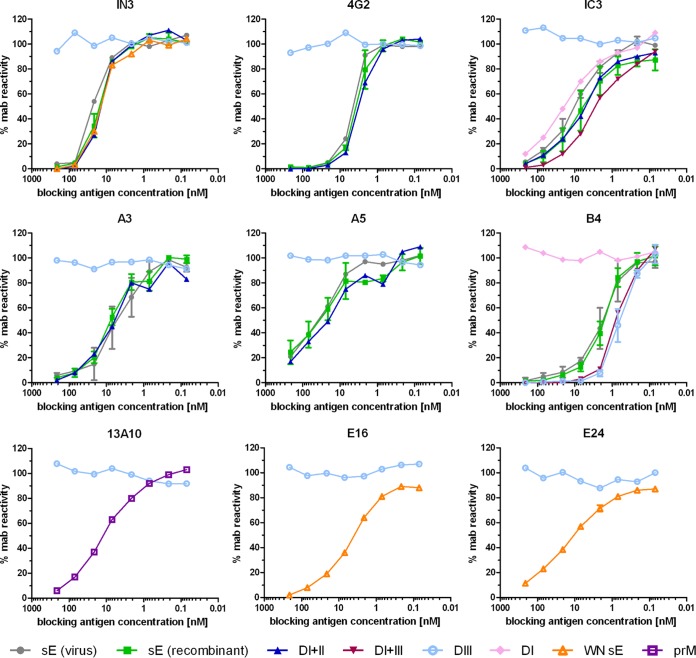
The reactivity of recombinant proteins was analyzed with conformation-sensitive and/or neutralizing MAbs (the specificities are indicated in the legend to Fig. 2) in blocking enzyme-linked immunosorbent assay (ELISA) as described in reference 34. In brief, 3-fold serial dilutions of 212 nM blocking antigen were incubated with a predetermined concentration of MAbs for 1.5 h at 37°C. The mixture was added for 1 h at 37°C to microtiter plates coated with formalin-inactivated TBE or WN virus or TBE virus with a high prM content, depending on the MAb specificity. Antibodies not blocked by their reaction with the antigen in solution were detected using peroxidase-labeled rabbit anti-mouse IgG (Pierce). Results are expressed as percent MAb reactivity (absorbance) in the absence of blocking antigen.
FIG 2.
Blocking ELISA of recombinant antigens with MAbs. The MAbs recognize the following antigenic sites: IN3 and 4G2, flavivirus fusion loop (16); IC3, TBE DI; A3 and A5, TBE DII; B4, TBE DIII (34); 13A10, TBE prM (81); E16 and E24, WN DIII (82). The antigens analyzed are indicated in different colors, as shown at the bottom of the panel.
Chemical cross-linking.
Proteins were cross-linked by dimethylsuberimidate (DMS) treatment as described previously (35). Briefly, purified protein in triethanolamine buffer (pH 8.0) was incubated with 10 mM DMS for 30 min at room temperature (5 μg/ml). Ethanolamine was added to a final concentration of 10 mM and incubated for an additional 15 min to stop the reaction. Proteins were precipitated by treatment with trichloroacetic acid and analyzed before and after cross-linking by SDS-PAGE in a continuous phosphate-buffered system (36).
Sedimentation analysis.
Proteins were analyzed by rate zonal sedimentation in sucrose gradients as described previously (37). Monomer controls were prepared for each protein by incubation with 1% SDS for 30 min at 65°C. A total of 3 μg of SDS-treated and 3 μg of untreated proteins were applied to the gradients and centrifuged for 20 h at 38,000 rpm at 15°C in a Beckman SW40 rotor. The gradients were fractionated, and the amount of antigen in each fraction was quantified by ELISA (38).
IgG ELISA.
ELISAs with human sera were carried out as previously described (16, 27) by using nontreated microtiter plates coated with 50 ng/well of recombinant antigens or 25 ng/well of TBE virions and 3-fold serial dilutions of human sera, starting at 1:100. For detection, biotin-labeled goat anti-human IgG (Pierce) and streptavidin-conjugated peroxidase (Sigma) were used.
ELISA cutoffs were determined as the mean of 8 negative sera plus three standard deviations, as recommended in reference 39. Specific IgG was quantified by the use of a standard serum assigned 1,000 arbitrary units in each assay. Four dilutions of all samples were analyzed, and ELISA units were determined after curve fitting using a 4-parameter logistic regression with GraphPad Prism 5 (GraphPad Software Inc.).
In order to harmonize the results obtained in all ELISAs, we adjusted the units obtained by the factor required to shift the standard curves of the recombinant protein ELISAs in such a way that their linear ranges coincided with that of the virion ELISA. Significances of differences in the ELISA results observed between the two groups were calculated by unpaired two-tailed t tests.
TBE virus neutralization assay.
Virus neutralization tests with human sera were carried out in baby hamster kidney cells (ATCC BHK-21) as described in reference 40. Two-fold serial dilutions of sera, starting at 1:10, were preincubated with 25 50% tissue culture infective doses (TCID50) of TBE virus (strain Neudörfl) for 1 h at 37°C before inoculation of cells and incubation for 3 days. To avoid interference by the depletion antigen, the cell culture supernatant was replaced by fresh medium 1 day after infection.
The presence of virus in the cell culture supernatant was detected by four-layer ELISA as described previously (41). Titers were calculated after curve fitting with a four-parameter logistic regression (Graph Pad Prism 5; GraphPad Software Inc.) using 50% of the absorbance in the absence of antibody as a cutoff (NT50). Titers of ≥10 were considered positive. Significances of differences in the NT50 titers observed between the two groups were calculated by unpaired two-tailed t tests.
Antibody depletion.
Depletion of specific antibodies from human sera was performed essentially as described in reference 33, using Dynabeads (Life Technologies) for His-tagged proteins and Strep-Tactin beads (Qiagen) for Strep-tagged antigens. For this purpose, 10 μg antigen was bound to 33 μl paramagnetic beads for 1 h at room temperature, and after a washing step, 500 μl of 1:5 prediluted serum was added and incubated for 1 h at 37°C. Beads were pelleted by magnetic force, and the supernatant was subjected to two further rounds of depletion. The absence of nonspecific serum binding was confirmed with uncharged beads and the success of depletion by ELISA with the depletion antigen. The depleted serum samples were analyzed in virion ELISA and NT, and the data obtained were expressed as the percentage of the mean values of the nondepleted and mock-depleted sera.
Statistical analyses of depletion results.
NT titers and ELISA units were log transformed and analyzed by a general linear model with normal deviates. Parameters, corresponding to the magnitude of depletion, and their standard errors were estimated by maximum likelihood. Confidence intervals for the percent depletion were calculated based on the standard errors of the parameter estimates by back transformation. Hypotheses about differences in the magnitude of depletion were tested by Wald chi-square tests. P values below 0.05 were considered significant.
RESULTS
Characterization of recombinant antigens for immunoassays.
A panel of recombinant antigens for ELISAs and depletion analysis (Fig. 1D) was produced and purified as described in Materials and Methods. All of these proteins reacted with neutralizing and/or conformation-specific MAbs, and nonspecific binding was not observed using appropriate control antigens (Fig. 2). In the case of the TBE virus E-derived antigens (Fig. 1D), these reactivities corresponded to that of the sE protein isolated from purified virions. In contrast to sE, which is a dimer (42, 43), DI+II was monomeric, as shown by cross-linking and sedimentation analyses (Fig. 3), although it contains sequence elements contributing to dimerization (42). Using these antigens, we established a platform of immunoassays that allowed us to obtain information on relative quantities (expressed as ELISA units) of antibody subsets reacting with the whole dimeric sE protein and different domains or domain combinations of E, as well as prM.
FIG 3.
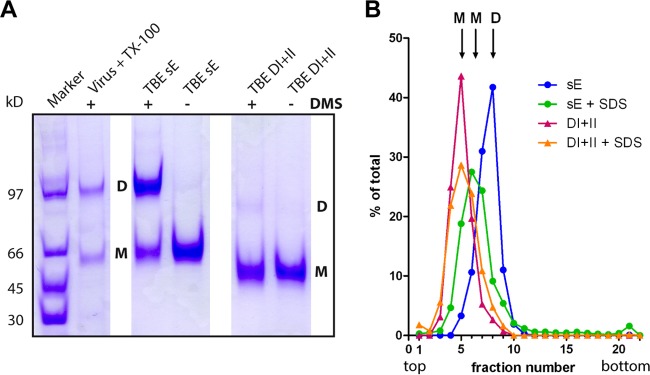
Determination of the oligomeric state of TBE DI+II. (A) SDS-PAGE of recombinant TBE sE and DI+II as well as full-length E obtained from virus particles solubilized with Triton X-100 without (−) and with (+) cross-linking by DMS. Positions of monomers (M) and dimers (D) are indicated. Staining with Coomassie brilliant blue. (B) Sedimentation analysis of TBE antigens. The antigens are color coded as indicated in the panel. Monomeric controls were generated by SDS treatment (+SDS) of the antigens. Sedimentation is from left to right. The antigen content in each fraction was determined by ELISA. Positions of monomers (M) and dimers (D) are indicated.
Reactivities of postinfection and postvaccination sera.
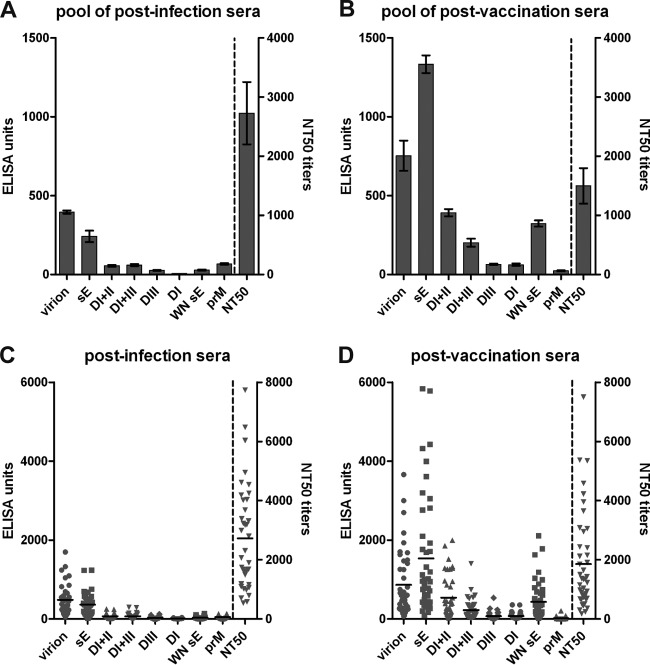
Antibody reactivities were determined with individual sera from 38 TBE patients (collected about 2 years after infection) as well as from 43 TBE vaccinees who had received a booster vaccination about 3 weeks before sample collection. These two cohorts are described in more detail in Materials and Methods, and their key features are listed in Table 1. In addition, serum pools containing identical aliquots of all postinfection and postvaccination sera were analyzed separately. The results of ELISAs and neutralization tests (NTs) are displayed in Fig. 4. In the serum pools, the ELISA reactivities against all antigens were significantly higher after vaccination, with the exception of prM-specific antibodies, which were significantly higher after infection (P < 0.0001) (Fig. 4A and B). The neutralization titer, however, was nonsignificantly lower in the postvaccination pool (NT50 = 1,497) than in the postinfection pool (NT50 = 2,726) (P = 0.0653). The mean values of individual serum analyses (Fig. 4C and D) yielded a pattern similar to that obtained with the serum pools but revealed an extensive degree of individual variation with respect to the quantity of antibodies reacting with the different antigens and also with respect to neutralization titers.
FIG 4.
ELISAs and NTs of TBE postinfection and postvaccination sera. Analysis of serum pools (A, B) and individual sera (C, D) in TBE virion and subunit ELISAs as well as in neutralization tests. Error bars in panels A and B represent standard errors of the means calculated from the results from at least 3 independent experiments. Black horizontal lines in panels C and D represent the means of individual sera.
Calculations of the correlation coefficients of subunit ELISA results as well as NT titers to the virion ELISA results yielded very good positive correlation coefficients (Pearson r = 0.74 to 0.98) (Fig. 5). Only the correlation between prM and virion reactivity in TBE vaccinees was somewhat lower (Pearson r = 0.51).
FIG 5.
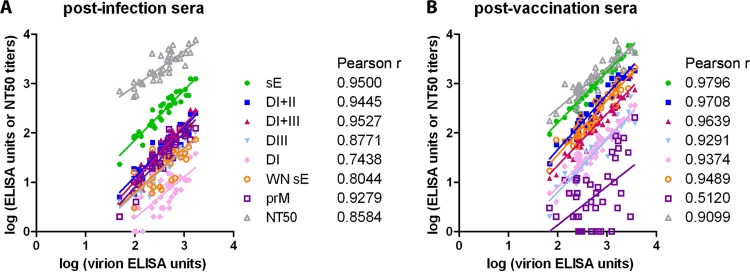
Correlations of individual serum reactivities in different assays. Linear regression between virion ELISA and subunit ELISA units as well as with NT titers. Pearson correlation coefficients (r) are indicated. (A) Postinfection sera; (B) postvaccination sera.
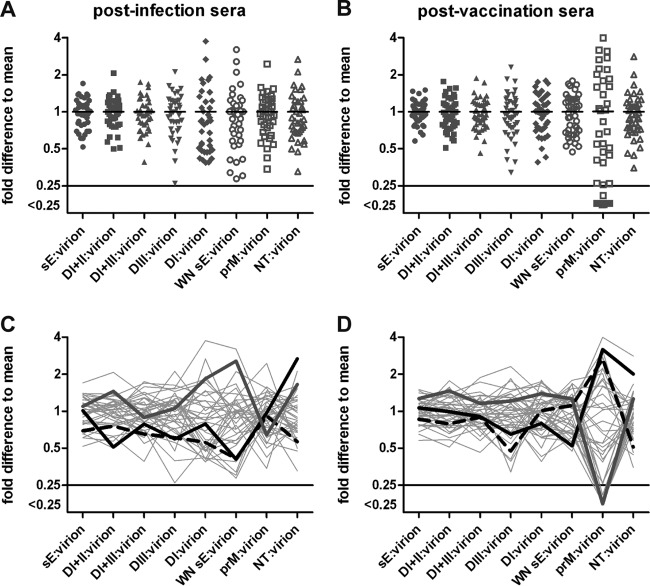
To obtain a standardized measure for the individual variation of the proportions of antibody subsets and to compensate for titer variations, we determined, for each serum, the ratios of subunit ELISA units as well as NT titers relative to virion ELISA units. These individual serum ratios are displayed as deviations from the mean values of all sera in Fig. 6A and B. For all parameters tested, there was substantial variation (including the ratio of NT to virion ELISA) reflecting quantitative differences in the specificities and composition of sera, both from infected individuals and vaccinees. This analysis provided a characteristic reactivity pattern for each serum, representing a fingerprint of TBE virus-specific antibody subsets and their neutralizing activities, shown as gray lines in Fig. 6C and D. The patterns are quite heterogeneous and divergent at the single serum level, consistent with individual-specific factors that shape the specificity of the antibody response. Three sera were selected from both groups and highlighted in Fig. 6C and D to show that sera with similar reactivity patterns can have quite divergent NT-to-virion ELISA ratios (compare black dashed and solid lines) and, vice versa, sera with divergent patterns can have similar NT to virion ELISA ratios (black and gray curves).
FIG 6.
Variation of reactivities of individual sera. (A, B) Ratios of subunit ELISA units and NT titers in relation to virion ELISA units, expressed as fold difference from the mean of all sera. (C, D) Ratios obtained for each individual serum are connected by gray lines. Patterns of three selected sera are highlighted. (A, C) Postinfection sera; (B, D) postvaccination sera.
Depletion analysis.
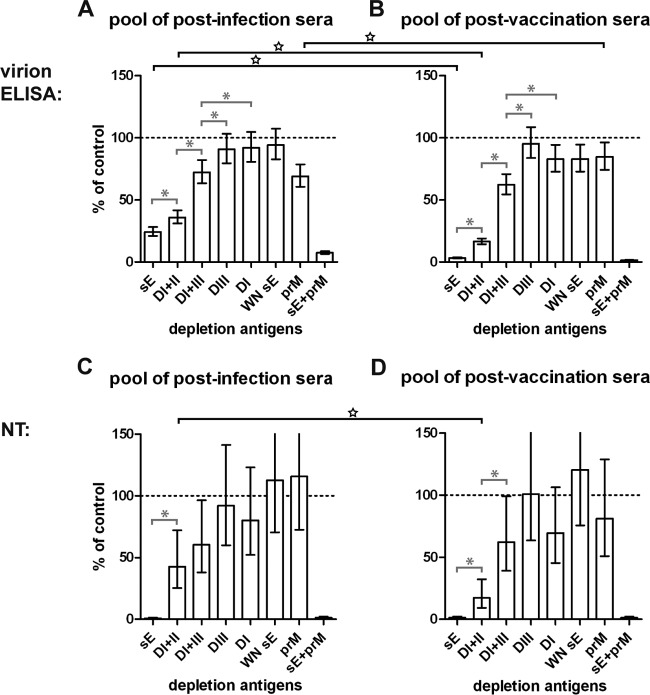
In order to gain quantitative information on the contribution of different antibody subsets to the total TBE virus-specific serum reactivity and to neutralizing activity, we depleted the postinfection and postvaccination serum pools with the recombinant proteins shown in Fig. 1D. As controlled by ELISA with the respective depletion antigens, depletion was complete (below ELISA cutoff and/or less than 5% remaining reactivity) in all instances. The effect of the removal of distinct antibody subsets was then determined in virion ELISA as well as NTs, and the results of these analyses are shown in Fig. 7.
FIG 7.
Depletion of antibody subsets from serum pools with recombinant antigens. Postdepletion analysis of the pools of postinfection sera (A, C) and postvaccination sera (B, D) in virion ELISA (A, B) and NT (C, D). The reactivity of depleted serum pools is given as a percentage of the mean of non- and mock-depleted controls. Depletion is significant when the error bar does not reach the 100% mark (dashed line). Error bars represent confidence intervals of 2 to 4 independent experiments. Gray capped lines with asterisks within each of the panels indicate significant differences between depletions of the same serum pool with different antigens. Black capped lines with stars spanning the panels indicate significant differences between depletions of the postinfection and postvaccination pools with the same antigen. Antigens used for depletion are indicated below the panels.
Depletion with sE resulted in a virtually complete removal of the total virion reactivity in the postvaccination serum pool, compared to about 75% in the postinfection pool. The majority of this residual reactivity, however, could be removed by double depletion with sE and prM. Most importantly, in both serum pools, all of the neutralizing activity was removed by depletion with sE alone, indicating that the epitopes recognized by these antibodies are displayed completely or at least to a large extent on the soluble dimeric E protein. A substantial proportion of virion reactivity and neutralizing activity could even be removed by the monomeric DI+II protein (65% in postinfection and 85% in postvaccination pools; Fig. 7C and D), suggesting that after both vaccination and infection a considerable portion of antibodies recognized antigenic sites that were largely independent of the dimer structure of E.
In none of the cases, depletion with DIII resulted in a significant reduction of virion reactivity and neutralizing activity, indicating that this domain plays a very minor role in the human antibody response to TBE virus, irrespective of its presentation as a replicating or inactivated virus. Similar results were also obtained with the isolated DI, except for a slight reduction of virion reactivity in the postvaccination pool. Significant reduction of virion reactivity and neutralizing activity, however, was observed in both serum pools after depletion with the recombinant protein comprising both DI and DIII, suggesting the involvement of epitopes at the junction between the two domains.
Consistent with the ELISA results shown in Fig. 4, the postinfection sera had a significantly higher proportion of prM-specific antibodies than the postvaccination sera, whereas a slight but significant degree of depletion with the cross-reactive sE from WN virus was found with the postvaccination but not the postinfection pool. In none of these cases a significant depletion effect on neutralizing activity was achieved. The small amount of cross-reactive antibodies was completely depletable with DI+II but not DI+III (data not shown), suggesting that they were directed primarily to conserved sites in DII, although amino acids conserved between TBE and WN sE are found in all domains of E (see Fig. 1D).
Results obtained with individual sera.
To identify possible individual variations in antibody subsets contributing to the depletion patterns found with serum pools (Fig. 7), we selected three postinfection sera (I-16, I-9, I-29) and three postvaccination sera (V-208, V-207, V-229) with relatively high neutralization titers for depletion analyses. Table 2 summarizes the characteristics of the selected sera. The results of the analyses are displayed in Fig. 8 and revealed significant deviations from the average patterns obtained with the serum pools. The most important differences relative to the pool data can be described as follows. (i) Sera I-16 and V-208 contained significantly more DIII-specific antibodies than the pools, which, however, did not contribute significantly to the neutralizing activity of these sera. (ii) In sera I-9 and V-207, the depletion of total virion reactivity with monomeric DI+II was as efficient as that with the whole sE dimer, suggesting a minor role of antibodies to dimer-specific epitopes in these cases. In serum V-207, the DI+II reactive antibodies also accounted for almost all of the neutralizing activity. (iii) In contrast to what was observed with serum V-207, most of the neutralizing activity of serum V-229 could not only be removed by DI+II but also by DI+III, suggesting that DI-specific antibodies dominated the neutralizing response in this case. (iv) Serum I-29 did not contain significant amounts of antibodies depletable with prM, different from the results obtained with the postinfection pool and sera I-9 and I-16.
TABLE 2.
Characteristics of sera selected for depletion analyses
| Serum | Time after onset of symptoms for postinfection sera (yrs) or after booster vaccination for postvaccination sera (days) | Age (yrs) of patient/vaccinee | NT50 titer |
|---|---|---|---|
| Postinfection | |||
| I-16 | 1.7 | 61 | 4,253 |
| I-9 | 1.6 | 72 | 4,540 |
| I-29 | 3.8 | 17 | 7,737 |
| Postvaccination | |||
| V-208 | 30 | 23 | 5,350 |
| V-207 | 22 | 46 | 7,505 |
| V-229 | 28 | 19 | 2,157 |
FIG 8.
Depletion of antibody subsets from individual sera with recombinant antigens. Postdepletion analysis of three postinfection sera (I-16, I-9, I-29) and three postvaccination sera (V-208, V-207, V-229) in virion ELISA (A) and NT (B). The reactivity of depleted sera is given as the percentage of the mean of non- and mock-depleted controls. Depletion is significant when the error bar does not reach the 100% mark (dashed line). Error bars represent confidence intervals of 2 to 4 independent experiments. Gray capped lines with asterisks within each of the panels indicate significant differences between depletions of the same serum with different antigens. Black stars indicate significant differences between depletions of an individual serum and the respective serum pool with the same antigen. Antigens used for depletion are indicated below the panels.
DISCUSSION
In this study, we deconstructed specificities and functional activities of antibody populations induced in the course of natural TBE virus infection and TBE vaccination through the use of immunoassays with recombinant proteins, including single domains and domain combinations of the target for neutralizing antibodies (protein E), as well as prM. The samples were collected late after infection (median of 1.9 years) and 3 to 4 weeks after booster vaccination, and the measured antibodies are therefore believed to be responsible for long-term protection. It was an important finding of our work that the neutralizing activity, both after natural infection and vaccination, could be almost completely removed from serum pools by depletion with the dimeric recombinant sE protein (Fig. 7C and D), suggesting that neutralizing epitopes are largely confined to this building block of the viral envelope. More complex quaternary epitopes (generated only by the herringbone-like arrangement of E at the virion surface) have been structurally characterized through cryoelectron microscopy analyses of certain MAb-derived Fabs complexed to WN virus (24) and dengue 1 virus (25) and were shown to have amino acid contributions from two adjacent dimers present at the virion surface. Also in these cases, however, a large proportion of the antibody contact site was confined to a single E dimer. Depletion with the sE dimer could therefore potentially also lead to the removal of antibodies to such dimer overlapping sites.
Somewhat unexpectedly, our sE depletion data did not conform to those of a related study analyzing dengue postinfection sera, which revealed that the majority of neutralizing antibodies bound only to intact virions but not to soluble E (26). Such discrepancies may be explained by structural differences between TBE and dengue viruses, especially with respect to the stability and/or the curvature of the E protein dimer in solution (44). Whereas the TBE E dimer corresponds in its curvature to that of the virion surface (42) and remains dimeric even at the relatively high dilutions employed in serological assays (Fig. 3) (45), recent observations indicate that the dengue E protein is monomeric in solution (46). This antigen would allow only discrimination between monomer-specific and more complex quaternary epitopes but not between dimer-specific (22, 23) and herringbone-specific (24, 25) epitopes. It also has to be kept in mind that the results of neutralization assays can be influenced by the maturation state of the virus (47, 48) as well as the cell type used in such analyses (49).
Quite remarkably, antibodies to domain III (which forms a prominent protrusion at the surface of flaviviruses [reviewed in reference 15]) played only a marginal role in the responses both to TBE virus infection and vaccination, and depletion of DIII-specific antibodies from the serum pools had no measurable effect on virion binding and neutralization activities (Fig. 7), similar to what was observed in a study with human YF postvaccination sera (27). In a mouse immunization study with inactivated and aluminum hydroxide-adjuvanted purified TBE virions (i.e., an immunogen similar to that in the TBE vaccine), however, a substantial proportion of the total and neutralizing antibody response was directed at DIII (43). These data are consistent with previous observations made with monoclonal and polyclonal antibody responses to West Nile and dengue viruses, which had shown that the dominance of DIII responses typical of mice has no counterpart in humans (reviewed in reference 50). Considering the fact that the antigenically relevant surface of DIII differs strongly among flaviviruses (62% of the surface-exposed residues are not conserved in any pairwise comparisons between TBE, WN, JE, dengue 2, and YF viruses [data not shown]), such disparate findings are difficult to explain by differences in the antibody repertoires of humans and mice. Understanding the mechanisms underlying such a pronounced species-related divergence in antigen recognition could lead to a better understanding of the largely elusive phenomenon of antibody immunodominance in general.
A substantial proportion of virion binding and neutralizing antibodies could be depleted with a monomeric DI+II construct (Fig. 7), although the extent of these depletions was significantly lower than that of the sE depletions in serum pools. Since DIII had no effect in depletion (Fig. 7 and paragraph above), this difference provides evidence for the involvement of E dimer-specific epitopes as addressed before (22, 23) but also indicates that more than 50% of the total virion binding and neutralizing antibody response was directed to epitopes represented by the monomeric E protein. This is reminiscent of data obtained with YF postvaccination sera using a monomeric form of sE as well as YF DI+II (27). The importance of epitopes in DI- and DII-inducing neutralizing antibodies, especially involving residues at the junction between the two domains, has been shown previously in several monoclonal and polyclonal antibody studies with dengue viruses (51–53). Furthermore, the dominance of such antibodies in virus neutralization was recently demonstrated in elegant experiments using recombinant viruses with transplanted amino acid residues at the DI+II hinge region of dengue viruses (54), suggesting that this is a characteristic feature of the human neutralizing antibody response to flaviviruses in general.
Similar to what was found with DIII, antibodies depletable by the isolated DI did not contribute significantly to the neutralizing activity of serum pools after infection and vaccination. However, the construct encompassing DI and DIII removed substantial amounts of virion binding and neutralizing activity from the serum pools, suggesting that epitopes at the junction between the two domains, similar to those at the junction between DI and DII, were involved in these responses. Such an epitope has been identified previously by a dengue-specific monoclonal antibody (5H2) that was shown to recognize DI and the linker between DI and DIII (55). The importance of interdomain epitopes in the antibody response to flavivirus infections in humans has also been deduced from the application of a high-throughput dot blot assay with alanine mutant E proteins for the analysis of dengue postinfection sera (51).
The single-serum analyses of postinfection and postvaccination panels in recombinant protein ELISA platforms demonstrated a substantial individual-specific variation with respect to the composition and specificities of antibody populations in peripheral blood (Fig. 6), similar to those reported in an analysis of the Ig responses to three distinct epitopes of E for dengue fever patients (56) and YF vaccinees (27). Functional consequences of such variations became apparent in depletion analyses of selected sera (Fig. 8) and were observed between individuals (I-16 compared to I-9 and V-208 compared to V-229) of similar age and time since infection and vaccination (Table 2). They thus apparently reflect individual-specific variations in factors controlling the stimulation and selection of specific B cells for antibody production, including germ line Ig repertoire (57, 58), competition between different B cell clones by affinity maturation in germinal centers (59), and poorly understood mechanisms leading to the selection of B cell subsets to become B memory cells or antibody-producing LLPCs in the bone marrow (12, 60).
Although postvaccination sera had significantly higher ELISA titers than postinfection sera against soluble forms of E compared to whole virion (Fig. 4), the patterns obtained in depletion analyses were remarkably similar. Specifically, the complete neutralizing activity could be depleted by the soluble E protein from serum pools of both cohorts, and also other characteristics of the antibody response, such as the immunosilence of DIII, the contribution of DI+II and DI+III junction epitopes (Fig. 7), and the extents of individual variability of antibody fine specificities (Fig. 6), were comparable in the two groups. However, prM antibodies were found more frequently and at higher titers in postinfection sera than after vaccination, and the opposite was true for broadly cross-reactive antibodies. Nevertheless, both of these antibody populations (especially the cross-reactive antibodies induced by vaccination) made up only a minor fraction of the total virion-reactive antibodies and did not contribute to neutralization, as revealed by depletion analyses. The induction of prM-specific antibodies in the course of infection suggests that immature or partially immature virus particles and possibly also soluble forms of prM circulate in the course of TBE virus infections in humans and thus stimulate a prM response, similar to what has also been observed in JE and dengue virus infections (18, 19, 61, 62).
Surprisingly, the broadly flavivirus cross-reactive response was very low after infection, in stark contrast to what has been observed in dengue and WN postinfection sera, in which fusion loop-specific antibodies were found to encompass a significant percentage of the anti-E response (18, 51, 56, 63, 64). The reasons for these discrepancies are not clear yet but could be related to virus-specific differences in the stability of E-E and E-prM complexes, as well as the dynamic motions of E at the virus surfaces (65–70), potentially affecting the transient exposure of antigenic sites that would be cryptic in the closed shell state of mature virus particles (16, 48, 71). Structural features could also contribute to the differences observed between infected and vaccinated individuals, as observed in the reactivity patterns of the recombinant protein ELISA platforms (Fig. 4), and may be a consequence of chemical modification (72) and/or stabilization of E oligomers by formalin (43, 73), adsorption to aluminum hydroxide (74, 75), and different concentrations of soluble antigen forms in the vaccine compared to those produced during infection.
In summary, our study not only highlights similarities and differences in the antibody response to TBE virus infection and vaccination in comparison to other flaviviruses but more generally points to the impact of individual-specific variations of antibody fine specificities and their functional consequences for immune responses to virus infections and vaccines. All of these issues are related to the phenomenon of immunodominance which, in addition to structural features of the antigen, is apparently strongly modulated by mechanisms that are private to each individual. An in-depth understanding of the factors involved in these mechanisms can inform strategies for a more rational design of new vaccines (13, 76), especially against viruses with highly variable target antigens, such as influenza (77) and HIV (78).
ACKNOWLEDGMENTS
We gratefully acknowledge the excellent technical assistance of Jutta Huttecek in conducting the neutralization assays, Andrea Reiter in antigen production, and Walter Holzer in virus production. We thank Michael S. Diamond for providing hybridoma cells secreting E16 and E24 monoclonal antibodies as well as Connie Schmaljohn for MAb 13A10.
This work was supported by the Austrian Science Fund FWF grant P25265-B21 to FXH and intramural funds of the Medical University of Vienna.
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1. Simmonds P, Becher P, Collett MS, Gould EA, Heinz FX, Meyers G, Monath T, Pletnev A, Rice CM, Stiasny K, Thiel HJ, Weiner A, Bukh J. 2011. Family Flaviviridae. In King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB. (ed), Virus taxonomy, vol IX Report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 2. Monath TP, Gershman M, Staples JE, Barrett ADT. 2013. Yellow fever vaccine, p 870–968 In Plotkin SA, Orenstein WA, Offit PA. (ed), Vaccines, 6th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 3. Halstead SB, Jacobson J, Dubischar-Kastner K. 2013. Japanese encephalitis vaccine, p 312–351 In Plotkin SA, Orenstein WA, Offit PA. (ed), Vaccines, 6th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 4. Barrett PN, Portsmouth D, Ehrlich HJ. 2013. Tick-borne encephalitis virus vaccines, p 773–788 In Plotkin SA, Orenstein WA, Offit PA. (ed), Vaccines, 6th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 5. Halstead SB, Thomas SJ. 2013. Dengue vaccines, p 1042–1051 In Plotkin SA, Orenstein WA, Offit PA. (ed), Vaccines, 6th ed. Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 6. Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4:229–238. 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierson TC, Diamond MS. 2008. Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev. Mol. Med. 10:e12. 10.1017/S1462399408000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gollins SW, Porterfield JS. 1986. A new mechanism for the neutralization of enveloped viruses by antiviral antibody. Nature 321:244–246. 10.1038/321244a0. [DOI] [PubMed] [Google Scholar]
- 9. Dowd KA, Pierson TC. 2011. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology 411:306–315. 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slifka MK, Matloubian M, Ahmed R. 1995. Bone marrow is a major site of long-term antibody production after acute viral infection. J. Virol. 69:1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benner R, Hijmans W, Haaijman JJ. 1981. The bone marrow: the major source of serum immunoglobulins, but still a neglected site of antibody formation. Clin. Exp. Immunol. 46:1–8. [PMC free article] [PubMed] [Google Scholar]
- 12. Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, Dekosky BJ, Murrin EM, Wirth MM, Ellington AD, Dorner T, Marcotte EM, Boutz DR, Georgiou G. 2014. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. U. S. A. 111:2259–2264. 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dormitzer PR, Ulmer JB, Rappuoli R. 2008. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 26:659–667. 10.1016/j.tibtech.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindenbach DB, Murray CL, Thiel HJ, Rice CM. 2013. Flaviviridae. In Knipe DM, Howley PM. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 15. Pierson TC, Diamond MS. 2013. Flaviviruses. In Knipe DM, Howley PM. (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 16. Stiasny K, Kiermayr S, Holzmann H, Heinz FX. 2006. Cryptic properties of dominant flavivirus cross-reactive antigenic sites. J. Virol. 80:9557–9568. 10.1128/JVI.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80:12149–12159. 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK. 2008. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J. Virol. 82:6631–6643. 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, Rey FA, Varani L, Whitehead SS, Diamond MS, Harris E, Lanzavecchia A, Sallusto F. 2010. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8:271–283. 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davies DR, Padlan EA, Sheriff S. 1990. Antibody-antigen complexes. Annu. Rev. Biochem. 59:439–473. 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 21. Sundberg EJ, Mariuzza RA. 2002. Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 61:119–160. 10.1016/S0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 22. Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. 2006. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile virus. J. Virol. 80:6982–6992. 10.1128/JVI.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goncalvez AP, Purcell RH, Lai CJ. 2004. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 78:12919–12928. 10.1128/JVI.78.23.12919-12928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. 2010. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. U. S. A. 107:18950–18955. 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 4:139ra83. 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 26. de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U. S. A. 109:7439–7444. 10.3410/f.716398009.791803068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vratskikh O, Stiasny K, Zlatkovic J, Tsouchnikas G, Jarmer J, Karrer U, Roggendorf M, Roggendorf H, Allwinn R, Heinz FX. 2013. Dissection of antibody specificities induced by yellow fever vaccination. PLoS Pathog. 9:e1003458. 10.1371/journal.ppat.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kunz C. 1996. Epidemiology of tick-borne encephalitis and the impact of vaccination on the incidence of disease, p 143–149 In Eibl MM, Huber C, Peter HH, Wahn V. (ed), Symposium in immunology, vol V Springer Verlag, Heidelberg, Germany. [Google Scholar]
- 29. Heinz FX, Holzmann H, Essl A, Kundi M. 2007. Field effectiveness of vaccination against tick-borne encephalitis. Vaccine 25:7559–7567. 10.1016/j.vaccine.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 30. Heinz FX, Stiasny K, Holzmann H, Grgic-Vitek M, Kriz B, Essl A, Kundi M. 2013. Vaccination and tick-borne encephalitis, central Europe. Emerg. Infect. Dis. 19:69–76. 10.3201/eid1901.120458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heinz FX, Kunz C. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57:263–274. 10.1099/0022-1317-57-2-263. [DOI] [PubMed] [Google Scholar]
- 32. Elshuber S, Allison SL, Heinz FX, Mandl CW. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183–191. 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 33. Zlatkovic J, Stiasny K, Heinz FX. 2011. Immunodominance and functional activities of antibody responses to inactivated West Nile virus and recombinant subunit vaccines in mice. J. Virol. 85:1994–2003. 10.1128/JVI.01886-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiermayr S, Stiasny K, Heinz FX. 2009. Impact of quaternary organization on the antigenic structure of the tick-borne encephalitis virus envelope glycoprotein E. J. Virol. 83:8482–8491. 10.1128/JVI.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maizel JV. 1971. Polyacrylamide gel electrophoresis of viral proteins. Methods Virol. 5:179–246. 10.1016/B978-0-12-470205-9.50011-3. [DOI] [Google Scholar]
- 37. Stiasny K, Allison SL, Schalich J, Heinz FX. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784–3790. 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz FX, Stiasny K, Puschner-Auer G, Holzmann H, Allison SL, Mandl CW, Kunz C. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109–117. 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 39. Frey A, Di Canzio J, Zurakowski D. 1998. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221:35–41. 10.1016/S0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
- 40. Stiasny K, Aberle JH, Keller M, Grubeck-Loebenstein B, Heinz FX. 2012. Age affects quantity but not quality of antibody responses after vaccination with an inactivated flavivirus vaccine against tick-borne encephalitis. PLoS One 7:e34145. 10.1371/journal.pone.0034145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heinz FX, Tuma W, Guirakhoo F, Kunz C. 1986. A model study of the use of monoclonal antibodies in capture enzyme immunoassays for antigen quantification exploiting the epitope map of tick-borne encephalitis virus. J. Biol. Stand. 14:133–141. 10.1016/0092-1157(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 42. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291–298. 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 43. Zlatkovic J, Tsouchnikas G, Jarmer J, Koessl C, Stiasny K, Heinz FX. 2013. Aluminum hydroxide influences not only the extent but also the fine specificity and functional activity of antibody responses to tick-borne encephalitis virus in mice. J. Virol. 87:12187–12195. 10.1128/JVI.01690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, Crowe JE, Jr, Lok SM. 2014. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 6:358–371. 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rey FA. 2013. Dengue virus: two hosts, two structures. Nature 497:443–444. 10.1038/497443a. [DOI] [PubMed] [Google Scholar]
- 47. Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. 2014. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J. Virol. 88:11726–11737. 10.1128/JVI.01140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nelson S, Jost CA, Xu Q, Ess J, Martin JE, Oliphant T, Whitehead SS, Durbin AP, Graham BS, Diamond MS, Pierson TC. 2008. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 4:e1000060. 10.1371/journal.ppat.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mukherjee S, Dowd KA, Manhart CJ, Ledgerwood JE, Durbin AP, Whitehead SS, Pierson TC. 2014. Mechanism and significance of cell type-dependent neutralization of flaviviruses. J. Virol. 88:7210–7220. 10.1128/JVI.03690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wahala WM, Silva AM. 2011. The human antibody response to dengue virus infection. Viruses 3:2374–2395. 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin HE, Tsai WY, Liu IJ, Li PC, Liao MY, Tsai JJ, Wu YC, Lai CY, Lu CH, Huang JH, Chang GJ, Wu HC, Wang WK. 2012. Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay. PLoS Negl. Trop. Dis. 6:e1447. 10.1371/journal.pntd.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. VanBlargan LA, Mukherjee S, Dowd KA, Durbin AP, Whitehead SS, Pierson TC. 2013. The type-specific neutralizing antibody response elicited by a dengue vaccine candidate is focused on two amino acids of the envelope protein. PLoS Pathog. 9:e1003761. 10.1371/journal.ppat.1003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith SA, de Alwis R, Kose N, Durbin AP, Whitehead SS, de Silva AM, Crowe JE., Jr 2013. Human monoclonal antibodies derived from memory B cells following live attenuated dengue virus vaccination or natural infection exhibit similar characteristics. J. Infect. Dis. 207:1898–1908. 10.1093/infdis/jit119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Messer WB, de Alwis R, Yount BL, Royal SR, Huynh JP, Smith SA, Crowe JE, Jr, Doranz BJ, Kahle KM, Pfaff JM, White LJ, Sariol CA, de Silva AM, Baric RS. 2014. Dengue virus envelope protein domain I/II hinge determines long-lived serotype-specific dengue immunity. Proc. Natl. Acad. Sci. U. S. A. 111:1939–1944. 10.1073/pnas.1317350111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Lai CJ, Goncalvez AP, Men R, Wernly C, Donau O, Engle RE, Purcell RH. 2007. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J. Virol. 81:12766–12774. 10.1128/JVI.01420-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crill WD, Hughes HR, Delorey MJ, Chang GJ. 2009. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS One 4:e4991. 10.1371/journal.pone.0004991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mathonet P, Ullman CG. 2013. The application of next generation sequencing to the understanding of antibody repertoires. Front. Immunol. 4:265. 10.3389/fimmu.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baumgarth N. 2013. How specific is too specific? B-cell responses to viral infections reveal the importance of breadth over depth. Immunol. Rev. 255:82–94. 10.1111/imr.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chan TD, Brink R. 2012. Affinity-based selection and the germinal center response. Immunol. Rev. 247:11–23. 10.1111/j.1600-065X.2012.01118.x. [DOI] [PubMed] [Google Scholar]
- 60. Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, Marcotte EM, Georgiou G. 2013. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc. Natl. Acad. Sci. U. S. A. 110:2993–2998. 10.1073/pnas.1213737110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cardosa MJ, Wang SM, Sum MS, Tio PH. 2002. Antibodies against prM protein distinguish between previous infection with dengue and Japanese encephalitis viruses. BMC Microbiol. 2:9. 10.1186/1471-2180-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsai WY, Lai CY, Wu YC, Lin HE, Edwards C, Jumnainsong A, Kliks S, Halstead S, Mongkolsapaya J, Screaton GR, Wang WK. 2013. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J. Virol. 87:12562–12575. 10.1128/JVI.00871-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. 2007. Induction of epitope-specific neutralizing antibodies against West Nile virus. J. Virol. 81:11828–11839. 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang J, Lok SM. 2013. Structural changes in dengue virus when exposed to a temperature of 37 degrees C. J. Virol. 87:7585–7592. 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. 2011. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 7:e1002111. 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. 2013. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U. S. A. 110:6795–6799. 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pierson TC, Kuhn RJ. 2012. Capturing a virus while it catches its breath. Structure 20:200–202. 10.1016/j.str.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. 2012. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure 20:303–314. 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 70. Kostyuchenko VA, Chew PL, Ng TS, Lok SM. 2014. Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J. Virol. 88:477–482. 10.1128/JVI.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. 2008. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 15:312–317. 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 72. Metz B, Kersten GF, Baart GJ, de Jong A, Meiring H, ten Hove J, van Steenbergen MJ, Hennink WE, Crommelin DJ, Jiskoot W. 2006. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug. Chem. 17:815–822. 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 73. Fraenkel-Conrat H, Mecham DK. 1949. The reaction of formaldehyde with proteins; demonstration of intermolecular cross-linking by means of osmotic pressure measurements. J. Biol. Chem. 177:477–486. [PubMed] [Google Scholar]
- 74. Jones LS, Peek LJ, Power J, Markham A, Yazzie B, Middaugh CR. 2005. Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. J. Biol. Chem. 280:13406–13414. 10.1074/jbc.M500687200. [DOI] [PubMed] [Google Scholar]
- 75. Morefield GL, Sokolovska A, Jiang D, HogenEsch H, Robinson JP, Hem SL. 2005. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine 23:1588–1595. 10.1016/j.vaccine.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 76. Dormitzer PR, Grandi G, Rappuoli R. 2012. Structural vaccinology starts to deliver. Nat. Rev. Microbiol. 10:807–813. 10.1038/nrmicro2893. [DOI] [PubMed] [Google Scholar]
- 77. Wong SS, Webby RJ. 2013. Traditional and new influenza vaccines. Clin. Microbiol. Rev. 26:476–492. 10.1128/CMR.00097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chiodi F, Weiss RA. 2014. HIV antibodies and the vaccine problem. J. Intern. Med. 275:444–455. 10.1111/joim.12225. [DOI] [PubMed] [Google Scholar]
- 79. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22. 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 80. Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. 2008. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319:1830–1834. 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 81. Iacono-Connors LC, Smith JF, Ksiazek TG, Kelley CL, Schmaljohn CS. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43:125–136. [DOI] [PubMed] [Google Scholar]
- 82. Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522–530. 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]