ABSTRACT
Productive infection of Trichoplusia ni cells by the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) leads to expression of ∼156 viral genes and results in dramatic cell remodeling. How the cell transcriptome responds to viral infection was unknown due to the lack of a reference genome and transcriptome for T. ni. We used an ∼60-Gb RNA sequencing (RNA-seq) data set from infected and uninfected T. ni cells to generate and annotate a de novo transcriptome assembly of approximately 70,322 T. ni unigenes (assembled transcripts), representing the 48-h infection cycle. Using differential gene expression analysis, we found that the majority of host transcripts were downregulated after 6 h postinfection (p.i.) and throughout the remainder of the infection. In contrast, 5.7% (4,028) of the T. ni unigenes were upregulated during the early period (0 to 6 h p.i.), followed by a decrease through the remainder of the infection cycle. Also, a small subset of genes related to metabolism and stress response showed a significant elevation of transcript levels at 18 and 24 h p.i. but a decrease thereafter. We also examined the responses of genes belonging to a number of specific pathways of interest, including stress responses, apoptosis, immunity, and protein trafficking. We identified specific pathway members that were upregulated during the early phase of the infection. Combined with the parallel analysis of AcMNPV expression, these results provide both a broad and a detailed view of how baculovirus infection impacts the host cell transcriptome to evade cellular defensive responses, to modify cellular biosynthetic pathways, and to remodel cell structure.
IMPORTANCE Baculoviruses are insect-specific DNA viruses that are highly pathogenic to their insect hosts. In addition to their use for biological control of certain insects, baculoviruses also serve as viral vectors for numerous biotechnological applications, such as mammalian cell transduction and protein expression for vaccine production. While there is considerable information regarding viral gene expression in infected cells, little is known regarding responses of the host cell to baculovirus infection. In these studies, we assembled a cell transcriptome from the host Trichoplusia ni and used that transcriptome to analyze changes in host cell gene expression throughout the infection cycle. The study was performed in parallel with a prior study of changes in viral gene expression. Combined, these studies provide an unprecedented new level of detail and an overview of events in the infection cycle, and they will stimulate new experimental approaches to understand, modify, and utilize baculoviruses for a variety of applications.
INTRODUCTION
Baculoviridae is a family of enveloped viruses with circular, double-stranded DNA genomes ranging from approximately 80 to 180 kbp in size. Baculoviruses replicate only in invertebrates, and because many baculoviruses are highly pathogenic to important insect pest species, baculoviruses have been used as biological control agents in agriculture and forestry. Because exceptionally high levels of viral gene expression occur in infected cells, baculoviruses were also developed and are used widely as vectors for protein expression in cultured insect cells (1–3). Applications have included production of proteins for research and medicine, with more specific applications such as therapeutics, subunit vaccines, and virus-like-particle vaccines (4). Baculoviruses have also been developed and used as transduction vectors for protein expression in mammalian cells (5, 6) and for viral display (7, 8). Thus, in addition to their roles in regulating insect populations in natural ecosystems and in agriculture, these large viruses are also used for a wide variety of important biotechnological applications.
Baculoviruses have a complex infection cycle that produces two structurally and functionally distinct types of infectious virions. In nature, the virus is typically found on plant foliage in an environmentally stable form. Infectious baculovirus virions of this type are embedded within a crystalline matrix of protein in occlusion bodies (OBs), which are resistant to heat and desiccation. Virions found within OBs are referred to as occlusion-derived virus (ODV), and this form of the virus is highly infectious to cells of the insect midgut. Once infected, midgut cells produce a second form of the virus that buds from the basal plasma membrane into the insect hemocoel, the open circulatory system. This second form of the virus, referred to as the budded virus (BV), is highly promiscuous and infects most tissues of the animal. Viral entry by both forms of the virus (BV and ODV) delivers nucleocapsids to the nucleus where viral transcription and DNA replication occur. After progeny capsids assemble within the nucleus and package the viral genome, some nucleocapsids are transported out of the nucleus and to the plasma membrane, where they bud to form the BV. This process may occur very rapidly after infection (9). Other nucleocapsids are retained within the nucleus, where they are enveloped to form the ODV. ODVs are later embedded, or occluded, within the crystallized occlusion body protein, Polyhedrin (10). Near the end of the infection cycle, two viral proteins are produced in the cell in enormous quantities. One is the occlusion body protein, Polyhedrin, and the other is P10, a protein associated with maturation of occlusion bodies. The production of enormous quantities of Polyhedrin and P10 and the crystallization of Polyhedrin around ODVs are precisely coordinated events very late in the infection cycle, and little is understood about how viral and cellular gene expression are coordinated to facilitate this process.
The baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has a large genome (∼134 kbp) that encodes ≥156 proteins. In addition to genes that regulate and/or mediate viral transcription, translation, and DNA replication, the virus also encodes a variety of genes that modify cellular and organismal physiology, architecture, and defenses. These viral manipulations include effects on development and behavior of the host insect through hormonal control and perhaps other mechanisms (11–14), a profound modification of cellular physiology and architecture (15–17), and host tissue breakdown, which results in release of the virus into the environment (18–21). The AcMNPV infection is completed in a relatively short time period (approximately 24 to 48 h), producing infectious budded and occluded viruses. Although the timing is variable and depends on experimental conditions, transcription from the viral genome may be detected as early as 1 h postinfection (p.i.), viral DNA replication may begin as early as 4 to 6 h p.i (22, 23), and viral late gene transcription begins simultaneously with or shortly after DNA replication (reviewed in reference 10). A number of cellular responses to baculovirus infection have been documented previously, and those that are observed by light microscopy are collectively referred to as the cytopathic effect (CPE). Examples of specific cellular responses to baculovirus infection include an early rearrangement and induction of actin polymerization (10, 24, 25), the triggering of apoptotic responses (17, 26, 27) and DNA damage responses (28, 29), dramatic modifications of the cell architecture (expansion of the nuclear volume, formation of virogenic stroma within the nucleus, and cell rounding), reduction of host protein synthesis, plasma membrane ruffling (30), modification of the cell cycle (10, 31, 32), and cell lysis (10, 30–32). While many of the cellular responses to AcMNPV infection have been described and documented, it is clear that these responses are complex and that our understanding of them is rudimentary at best.
We recently examined the dynamics of the viral transcriptome throughout the infection cycle in the permissive T. ni cell line Tnms42 (33). In the present study, we examined the dynamics of host T. ni gene expression over the course of AcMNPV infection in the same T. ni cell line. Tnms42 cells are derived from the BTI-Tn-5B1-4 cell line (known as HighFive cells commercially), one of the most widely used cell lines for protein production in research and biotechnology. In addition, T. ni is an important agricultural pest that is highly permissive to AcMNPV and serves as a model for AcMNPV infection at both the organismal and cellular levels. Although T. ni is a species of economic importance and widely used in biotechnology, a reference T. ni genome is not yet available. Therefore, to analyze T. ni gene expression in the absence of a reference genome, we analyzed RNA from both uninfected and AcMNPV-infected Tnms42 cells by strand-specific RNA sequencing (RNA-seq) on the Illumina platform and performed a de novo assembly to generate the transcriptome of T. ni cells. We then measured the expression levels of each of the assembled unigenes (transcripts) throughout a 48-h time course of AcMNPV infection. Our results indicate that 27% of the differentially expressed host transcripts (4,028 of 14,976 unigenes) were upregulated as an initial response to infection, followed by a general global decline in host transcripts as the infection progressed beyond approximately 6 to 12 h. The decline in host transcripts correlated with the replication of the virus and a dramatic increase in viral transcripts. Specific unigenes associated with insect innate immunity and stress responses were upregulated in the early stages of infection (0 to 6 h. p.i.) and then downregulated. We also identified a small group of unigenes that was highly upregulated at either 18 or 24 h. p.i., and this subset was enriched with genes associated with oxidation-reduction, generation of precursor metabolites, and energy. These data provide a detailed view of the initial and continuing host transcriptional responses to baculovirus AcMNPV infection in a model T. ni cell line. In addition, the T. ni transcriptome generated for these studies represents a valuable resource for continuing studies of cell biology and virus-host interactions and for a future T. ni genome annotation.
MATERIALS AND METHODS
Cell line and viruses.
For these studies, uninfected and AcMNPV-infected T. ni cells were used to produce a single large RNA-seq data set that was subsequently used for extensive analysis of AcMNPV transcripts and dynamics of the AcMNPV transcriptome (published separately [33]) and the detailed study of cellular responses to AcMNPV infection (the current study). Methods for propagation of cells, AcMNPV infection, and generation of raw RNA-seq data were reported previously (33) and are briefly summarized below. The cell line Tnms42 is an alphanodavirus-free cell line generated as a subclone of line Tn5B1-4 (HighFive cells). Tnms42 cells were cultured in TNM-FH medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) at 28°C as described previously (33) and infected with wild-type AcMNPV strain E2. The AcMNPV E2 sequence was confirmed by de novo assembly of the genome from the viral transcriptome. Virus titer measurements were performed as previously described (2). For infections, 3 × 106 Tnms42 cells were infected with wild-type (WT) AcMNPV (multiplicity of infection [MOI] = 10) in a T25 flask as described previously (33). After a 1-hour incubation, the inoculum was removed and the cells were rinsed with Grace's medium and cultured in TNM-FH medium supplemented with 10% FBS at 28°C. The time at which the inoculum was removed was designated 0 h postinfection (p.i.). Total RNA was isolated from AcMNPV-infected cells, as well as a set of parallel control cells (uninfected or mock infected), at 0, 6, 12, 18, 24, 36, and 48 h p.i. using a Qiagen RNeasy minikit.
Illumina strand-specific RNA sequencing.
Illumina sequencing libraries were constructed following a modified strand-specific RNA-seq protocol (34) as described previously (33). Briefly, polyadenylated RNA was isolated from 20 μg total RNA using Dynabeads oligo(dT)25 (Invitrogen) and then simultaneously eluted and fragmented in 2× SuperScript III buffer at 94°C for 7 min in the presence of 500 ng hexamer and 100 ng oligo(dT)10VN (5′ p-TTTTTTTTTTVN 3′, IDT). First-strand cDNA synthesis was carried out using SuperScriptIII (Invitrogen) and then purified using AMPure RNA Clean XP (Agencourt). Second-strand cDNA was synthesized using RNase H (NEB) and DNA polymerase I (NEB) with a dUTP mix (final concentration of 1 mM each) at 16°C for 2.5 h. After end repair and dA tailing, the DNA fragments were ligated with the customized TruSeq adapter. The sample was then treated with uracil DNA glycosylase (NEB) and PCR amplified with TruSeq-indexed PCR primers. Sequencing was performed on the Illumina HiSeq2000 platform at Weill Cornell Medical College.
T. ni transcriptome assembly and unigene annotation.
Raw RNA-seq reads were first trimmed with the ShortRead package (35) to remove the low-quality nucleotides and adapter and PCR primer sequences. Reads longer than 40 nucleotides (nt) and with no more than 2 Ns (ambiguous nucleotides) were retained. In addition, reads that mapped to either the rRNA or virus database were discarded. The cleaned reads were assembled using the Trinity package (36). The assembled contigs were further assembled using iAssembler (37) to remove redundancies. All the contigs/unigenes were BLAST searched against the GenBank nonredundant (nr) database with an E value of 1 × 10−5.
Identification of differentially expressed unigenes and cluster analysis.
To identify differentially expressed unigenes, the cleaned reads were aligned to the assembled unigenes using Bowtie (38), allowing 1 nt mismatch. Based on the alignments, the expression of each unigene derived from different samples was estimated and normalized to RPKM (reads per kilobase of exon model per million mapped reads) (39). The edgeR package (40) was used to identify differentially expressed unigenes at different times postinfection, by comparing data from virus-infected cells to either parallel control or 0-h uninfected control cells. Genes with a FDR (false discovery rate) lower than 0.05 were categorized as differentially expressed. Cluster analysis was performed using hierarchical clustering by the complete-linkage clustering method in the Cluster 3.0 package (41).
Analysis of expression patterns of specific functional groups and pathways.
T. ni homologs of several specific pathways and functional groups were identified using a tBLASTn search query against the assembled T. ni transcriptome. Protein homologs used for queries included those from yeasts, human, Drosophila melanogaster, Bombyx mori, Spodoptera exigua, or Spodoptera frugiperda. The T. ni unigenes identified by these searches and the cellular homologs for members of each functional group are listed in Tables S9 to S20 in the supplemental material along with the calculated RPKM values above a background value of 5.
Gene ontology analysis.
To identify gene ontology (GO) terms that were significantly enriched within selected groups of differentially expressed genes, we applied a GO term enrichment analysis tool (http://bioinfo.bti.cornell.edu/tool/GO/GO_enrich.html) that is based on the previously described GO::TermFinder tool (42). The tool identifies overrepresented GO terms from specified lists of genes, and uses three multitest correction methods (simulation, Bonferroni, and false discovery rate [FDR]).
Data accession numbers.
Illumina RNA-seq data were deposited in the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra/?term=SRA057390) under accession number SRA057390. This Transcriptome Shotgun Assembly (TSA) project has been deposited at DDBJ/EMBL/GenBank under accession number GBKU00000000 (http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=GBKU01). The version described in this paper is the first version, GBKU01000000 (http://www.ncbi.nlm.nih.gov/nuccore/GBKU00000000.1).
RESULTS AND DISCUSSION
Assembly and annotation of the T. ni Tnms42 transcriptome.
To analyze cellular responses to AcMNPV infection, a T. ni cell line (Tnms42) was infected with wild-type baculovirus AcMNPV (E2 strain) at a multiplicity of infection (MOI) of 10. RNAs were isolated and analyzed at seven time points through the infection cycle and were also obtained from control (mock-infected) cells. From the collective data set of 586,818,840 Illumina reads (strand-specific RNA-seq reads of 101 bases each), 175,797,789 reads, or approximately 30% of the total reads, were mapped to the AcMNPV genome (see Table S1 in the supplemental material) and were analyzed separately in a prior study (33). After removing viral reads, rRNA, and low-quality reads, we obtained 341,342,181 high-quality reads that were subsequently used to assemble a T. ni transcriptome, which contained 70,322 transcripts (unigenes) with an N50 value of 1,681 nt and a mean length of 850 nt (Table 1). BLAST analysis identified 22,305 T. ni unigenes (∼31%) with similarities to existing GenBank entries (see Table S2 in the supplemental material). Approximately 44% of the unigenes with lengths longer than 300 bp were annotated. Gene ontology (GO) terms were assigned to the T. ni unigenes based on their sequence matches in the UniProt and Pfam domain databases. A total of 19,230 (27.3%) unigenes were assigned at least one GO term, among which 16,527 (23.5%), 15,347 (21.8%), and 15,990 (22.7%) unigenes were assigned GO terms in the categories of cellular components, molecular functions, and biological processes, respectively. In addition, a substantial number of unigenes (14,777, or approximately 21%) were assigned GO terms in two or more categories.
TABLE 1.
Assembly statistics for the Tnms42 transcriptome
| Statistic | Value |
|---|---|
| N25 | 3,128 |
| N50 | 1,681 |
| N75 | 636 |
| Longest | 21,553 |
| Mean | 851.1 |
| Median | 395 |
| Shortest | 201 |
| N_Contigs | 70,810a |
| Annotated | 22,723 |
Includes both T. ni and AcMNPV unigenes. T. ni unigenes represent approximately 70,322 of the total unigenes.
General cellular responses to AcMNPV infection.
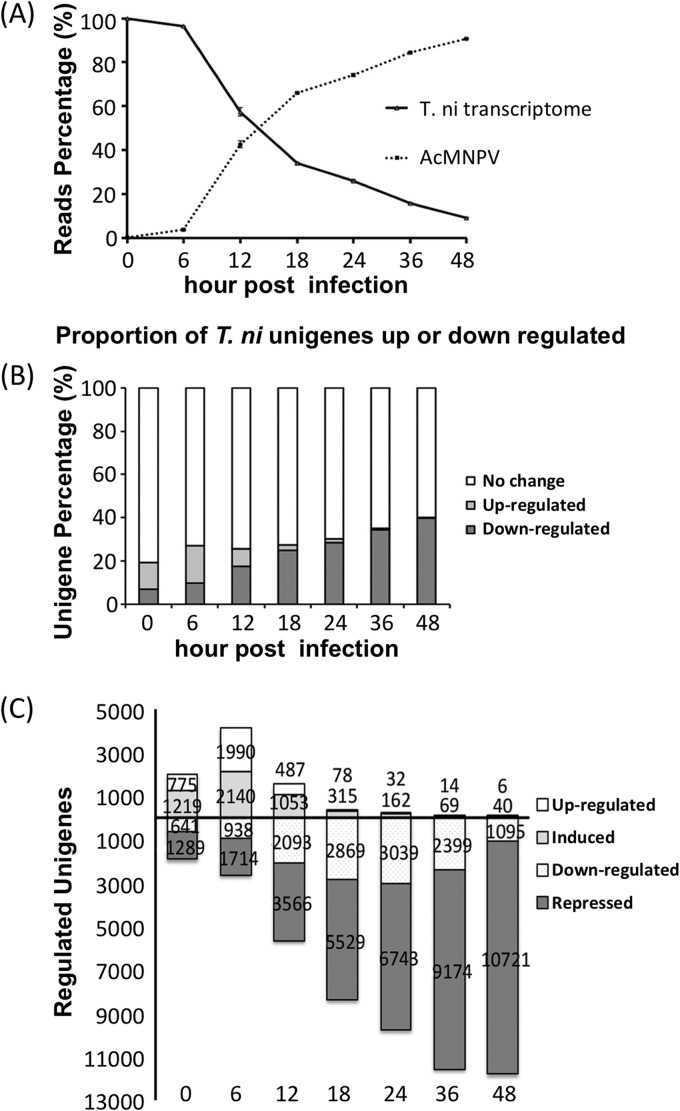
To monitor the expression of T. ni unigenes at various times postinfection, the filtered RNA-seq reads from each time point were mapped to the assembled T. ni transcriptome, and unigene expression levels were calculated as RPKM values (39). Read counts and average RPKM values for each unigene are provided in Tables S3 and S4 in the supplemental material, respectively. In a prior study of viral transcription, we examined overall viral mRNA levels based on the reads mapped to the AcMNPV genome and compared with total reads at various times after infection (33). In the current study, nonviral reads were analyzed by first mapping them to the assembled T. ni transcriptome. As a relative proportion, viral mRNAs began to increase dramatically after 6 h p.i. and exceeded host mRNA levels by 18 h p.i. (Fig. 1A). By 48 h p.i., only approximately 10% of the measured total mRNA levels were derived from the host. To analyze changes in T. ni unigene expression resulting from viral infection, we compared the expression level of each unigene at each time point postinfection, with that of the same unigene from (i) mock-infected control cells at 0 h postinfection or (ii) mock-infected control cells at the same time postinfection (a parallel time point). Both comparisons showed similar trends, with the proportion of downregulated T. ni unigenes increasing throughout infection. Viral infection typically results in arrest of cell division, and effects on the cell cycle have been documented (10, 32, 43). In contrast, uninfected or mock-infected cells continue to grow and divide and may reach relatively high densities compared to infected cells. Thus, stress responses or other responses related to cell density may occur in the control uninfected cells at later times postinfection. Therefore, for comparisons of differential expression resulting from viral infection, we used 0-h mock-infected cells as the control representing the uninfected cell state. At 0 h p.i. (one hour after addition of the virus inoculum), approximately 6 to 9% of the total T. ni unigene population was downregulated (Fig. 1B). By 48 h p.i., 39 to 40% of the T. ni unigenes were downregulated. In contrast, the proportion of T. ni unigenes that were upregulated decreased as infection progressed. After the 1-h adsorption period (0 h p.i.), approximately 12% of the T. ni unigene population was upregulated, but that proportion decreased to approximately 0.3 to 0.6% by 48 h p.i. Thus, the majority of T. ni unigenes that showed detectable changes were downregulated and only a small proportion were upregulated during the infection.
FIG 1.
(A) Transcript profile of AcMNPV mRNA versus T. ni Tnms42 mRNA by comparison of Illumina RNA-seq reads. Each data point represents Illumina reads as a percentage of total reads. (B) Proportions of unigenes up- or downregulated at various times postinfection. For each time point, the proportion of upregulated unigenes, downregulated unigenes, and unigenes with no significant change or for which data are indeterminate are shown as a combined single bar equal to 100%. (C) The numbers of unigenes up- and downregulated are indicated above and below the horizontal line, respectively. Upregulated unigenes are subdivided into those that are induced (unigenes with RPKM values of <5 for uninfected controls) and those that are upregulated (unigenes with RPKM values of ≥5 for uninfected controls). Downregulated unigenes are also subdivided into those that are repressed (unigenes with RPKM values of <5 for the specified time point postinfection) and those that are downregulated (unigenes with RPKM values of ≥5 for the specified time point postinfection).
To identify significantly up- or downregulated unigenes and to avoid artifacts resulting from unigenes that are expressed only at very low levels, we selected only those unigenes for which expression levels were ≥5 RPKM in either the control or infected cells (P ≤ 0.05) at each of the time points postinfection. For reference, an RPKM value of 3 represents a sensitivity of approximately 1 mRNA per cell in a mammalian cell (39). Thus, genes expressed at very low levels both before and after infection are not represented in this analysis, as they are thought to be insignificant and are categorized as indeterminate and grouped with those genes with no detectable change following infection (Fig. 1B).
We further subdivided the regulated unigenes based on the degree of up- or downregulation. For unigenes in which one of the RPKM values was below 5, these were classified as “induced” or “repressed” genes, as appropriate (e.g., a gene with an RPKM value below 5 in the 0-h control and with an RPKM value of >5 at a time point postinfection was classified as “induced”). For unigenes in which both RPKM values were ≥5, these unigenes were classified as up- or downregulated unigenes. The numbers of unigenes corresponding to the four categories (induced, upregulated, repressed, and downregulated) are illustrated in Fig. 1C. In addition, to better understand the magnitude of up- and downregulation, the numbers of unigenes that correspond to increasing fold changes in unigene expression levels in response to infection are also indicated as having 2- to 5-, 5- to 10-, and ≥10-fold changes in expression in Table 2. After filtering, the numbers of unigenes detected as regulated in response to infection are 3,924 (0 h), 6,782 (6 h), 7,199 (12 h), 8,791 (18 h), 9,976 (24 h), 11,656 (36 h), and 11,862 (48 h), respectively (Table 2). In summary, we identified approximately 17% of the total T. ni transcripts that are significantly regulated as a result of infection.
TABLE 2.
Numbers of up- and downregulated unigenes at various times postinfection
| Unigene category | Fold changea | No. of genes at: |
||||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 6 h | 12 h | 18 h | 24 h | 36 h | 48 h | ||
| Up-regulated | Inducedb | 1,219 | 2,140 | 1,053 | 315 | 162 | 69 | 40 |
| ≥10 | 1 | 2 | 3 | |||||
| 5–10 | 19 | 61 | 11 | 9 | 5 | 5 | 3 | |
| 2–5 | 756 | 1,928 | 474 | 66 | 27 | 9 | 3 | |
| Down-regulated | 2–5 | 600 | 817 | 1,794 | 2,400 | 2,382 | 1,267 | 164 |
| 5–10 | 21 | 94 | 245 | 376 | 518 | 831 | 318 | |
| ≥10 | 20 | 27 | 54 | 93 | 139 | 301 | 613 | |
| Repressedb | 1,289 | 1,714 | 3,566 | 5,529 | 6,743 | 9,174 | 10,721 | |
| Total regulated | 3,924 | 6,782 | 7,199 | 8,791 | 9,976 | 11,656 | 11,862 | |
To eliminate artifacts from unigenes with very low levels of expression, these data include only unigenes with RPKM values of >5 in both the control, and from infected cells at each time point (P ≤ 0.05).
Includes only unigenes with RPKM values below 5 in the control (induced) and after infection (repressed).
As a group, unigenes that were either upregulated or induced at 0 h p.i. represented 2.8% of the total transcriptome (1,994 unigenes). The number of upregulated or induced unigenes increased at 6 h p.i. (4,130 unigenes, representing 5.9% of the transcriptome) but decreased thereafter, such that by 18 h, they represented less than 1% of the transcriptome, and by 48 h, they represented less than 0.1%. At 48 h p.i., only 6 unigenes were detected as upregulated and 40 unigenes as induced. Thus, the upregulated and induced unigenes show a trend, with numbers peaking at 6 h p.i. and decreasing rather dramatically thereafter. In contrast to the trend observed with upregulated and induced unigenes, we observed increasing numbers of downregulated and repressed unigenes as the infection cycle progressed. At 0 h p.i., approximately 2.7% (1,930 unigenes) of the transcriptome was downregulated/repressed, and this was followed by increasing numbers of downregulated/repressed unigenes throughout the infection cycle: 3.7% (2,652 unigenes) at 6 h, 8.0% (5,659 unigenes) at 12 h, and 16.8% (11,816 unigenes) by 48 h (Fig. 1C; Table 2).
The most highly upregulated, induced, downregulated, and repressed unigenes (in response to virus infection) are identified and detailed in Tables 3, 4, 5, and 6, respectively. The most highly upregulated unigenes were identified as those encoding the NADH dehydrogenase subunit (UN031997; 20.7-fold), cytochrome b (UN005222, 16.2-fold), and cytochrome oxidase subunit III (UN068092; 11.7-fold) (Table 3). Unigenes induced to the highest levels upon viral infection were identified as those encoding 4-hydroxyphenylpyruvate dioxygenase (UN59730, UN59731, and UN59732), a predicted peroxidase (UN008509 and UN008507), NADH dehydrogenase subunit 2 (UN067842), an unknown protein (UN027117), and a serine protease (UN038864 and UN038865), a putative sprouty gene (UN043663) (an antagonist of FGF signaling that inhibits branching of the trachea in Drosophila [44]), and a 3-dehydroecdysone 3α-reductase gene (UN035226) (Table 4). In the highly upregulated or induced unigenes, we also noted that several genes encode proteins associated with mitochondria and energy/metabolism, such as cytochrome oxidase subunit III (UN068092), NADH dehydrogenase subunit 2 (UN067842), and ATP synthase F0 (UN067090). Xue et al. (45) also identified host genes related to the mitochondrial respiration pathway as highly upregulated over the course of Bombyx mori NPV (BmNPV) infection of B. mori Bm5 cells (45). In addition, prior studies showed that AcMNPV-infected Sf9 cells have an increased metabolic flux through the tricarboxylic acid (TCA) cycle (46). Thus, results from transcriptome studies of AcMNPV-infected T. ni cells confirm that cellular energy/metabolism pathways are modified during the baculovirus AcMNPV infection (45–49), and here we document the effects on the regulation of transcripts from these pathways. Prior studies have also shown that supplementing the culture medium of Sf9 cells with glutamine, pyruvate, or alpha-ketoglutarate can increase the yield of baculovirus-expressed proteins and budded virus (47, 49). The expression of most of the highly induced T. ni unigenes peaked in the late stage of infection (Table 4), suggesting either a potential role in viral processes (such as viral DNA replication, late viral gene expression, or virion assembly), a role in managing cellular metabolism late in infection, or both.
TABLE 3.
Most highly upregulated unigenes, with basal expression levels of >5 RPKM
| h p.i. | Unigene ID | RPKM |
Fold change | Accession no. | Annotation | E value | |
|---|---|---|---|---|---|---|---|
| Control | Infected | ||||||
| 18 | UN031997 | 10.49 | 217.83 | 20.7730 | YP_003208283 | NADH dehydrogenase subunit 4 (Diatraea saccharalis) | 9.00E-111 |
| 12 | UN005222 | 79.53 | 1,291.41 | 16.2382 | YP_003587419 | Cytochrome b (Parnassius bremeri) | 4.00E-133 |
| 18 | UN068092 | 185.71 | 2,178.42 | 11.7305 | ABN04114 | Cytochrome oxidase subunit III (Spodoptera exigua) | 8.00E-113 |
| 6 | UN029233 | 7.34 | 83.12 | 11.3279 | |||
| 12 | UN018930 | 5.57 | 53.43 | 9.5994 | |||
| 18 | UN034582 | 20.54 | 194.73 | 9.4787 | BAD27419 | NADH dehydrogenase subunit 5 (Plusia festucae) | 5.00E-96 |
| 6 | UN013935 | 5.11 | 47.09 | 9.2092 | XP_001651935 | Epoxide hydrolase (Aedes aegypti) | 3.00E-81 |
| 18 | UN024533 | 321.85 | 2,858.45 | 8.8814 | YP_003433778 | Cytochrome c oxidase subunit III (Zonosagitta nagae) | 1.00E-139 |
| 6 | UN016530 | 9.98 | 85.25 | 8.5456 | NP_001166775 | Cuticular protein hypothetical 2 (Bombyx mori) | 7.00E-36 |
| 6 | UN068416 | 22.51 | 189.26 | 8.4083 | |||
| 6 | UN065933 | 18.89 | 149.68 | 7.9227 | NP_001040107 | Chaperonin subunit 4 delta (Bombyx mori) | 3.00E-40 |
| 6 | UN069751 | 10.74 | 84.81 | 7.8939 | |||
| 36 | UN067090 | 183.61 | 1,408.77 | 7.6727 | YP_004021055 | ATP synthase F0 subunit 6 (Helicoverpa armigera) | 5.00E-71 |
| 6 | UN029232 | 35.61 | 272.20 | 7.6440 | |||
| 6 | UN001119 | 13.82 | 103.69 | 7.5001 | |||
| 6 | UN004359 | 12.59 | 93.49 | 7.4280 | NP_989049 | Proteasomal ubiquitin receptor ADRM1 [Xenopus (Silurana) tropicalis] | 3.00E-09 |
| 18 | UN025814 | 6.12 | 45.16 | 7.3834 | XP_001648636 | Hypothetical protein AaeL_AAEL014389 (Aedes aegypti) | 1.00E-23 |
| 6 | UN004358 | 11.28 | 82.40 | 7.3056 | NP_989049 | Proteasomal ubiquitin receptor ADRM1 [Xenopus (Silurana) tropicalis] | 9.00E-10 |
| 6 | UN014990 | 89.12 | 651.01 | 7.3051 | CAI38848 | Adipokinetic hormone precursor (Spodoptera frugiperda) | 5.00E-13 |
| 12 | UN013935 | 5.11 | 37.29 | 7.2915 | XP_001651935 | Epoxide hydrolase (Aedes aegypti) | 3.00E-81 |
TABLE 4.
Most highly induced unigenes, with basal expression levels of <5 RPKM
| h p.i. | Unigene ID | RPKM |
Fold changea | Accession no. | Annotation | E value | |
|---|---|---|---|---|---|---|---|
| Control | Infected | ||||||
| 24 | UN059730 | 3.89 | 430.40 | 110.67 | XP_973835 | Predicted: similar to 4-hydroxyphenylpyruvate dioxygenase (Tribolium castaneum) | 3.00E-39 |
| 24 | UN059732 | 1.89 | 406.12 | 214.65 | XP_973835 | Predicted: similar to 4-hydroxyphenylpyruvate dioxygenase (Tribolium castaneum) | 9.00E-28 |
| 24 | UN059731 | 1.53 | 311.85 | 203.40 | XP_973835 | Predicted: similar to 4-hydroxyphenylpyruvate dioxygenase (Tribolium castaneum) | 2.00E-28 |
| 36 | UN008509 | 0.52 | 212.13 | 404.15 | XP_623940 | Predicted: peroxidase (Apis mellifera) | 5.00E-59 |
| 36 | UN008507 | 0.52 | 211.80 | 403.75 | XP_623940 | Predicted: peroxidase (Apis mellifera) | 5.00E-59 |
| 36 | UN008506 | 0.42 | 204.95 | 482.35 | XP_002072893 | GK13447 (Drosophila willistoni) | 1.00E-21 |
| 36 | UN008508 | 0.42 | 204.87 | 488.60 | XP_002072893 | GK13447 (Drosophila willistoni) | 1.00E-21 |
| 18 | UN068862 | 2.22 | 202.65 | 91.40 | XP_001490517 | Predicted: hypothetical protein LOC100056916 (Equus caballus) | 5.00E-11 |
| 18 | UN067842 | 0.35 | 111.11 | 320.27 | ADO13501 | NADH dehydrogenase subunit 2 (Maruca vitrata) | 5.00E-37 |
| 48 | UN018195 | 1.82 | 103.75 | 56.91 | |||
| 18 | UN027117 | 1.78 | 83.77 | 47.10 | ACY69027 | Unknown (Helicoverpa armigera) | 2.00E-32 |
| 36 | UN068863 | 0.00 | 60.38 | Undefined | |||
| 18 | UN038864 | 0.75 | 58.84 | 78.27 | XP_001868268 | Thymus-specific serine protease (Culex quinquefasciatus) | 5.00E-81 |
| 18 | UN038865 | 0.87 | 56.05 | 64.69 | XP_001868268 | Thymus-specific serine protease (Culex quinquefasciatus) | 4.00E-81 |
| 0 | UN043663 | 3.86 | 44.59 | 11.55 | XP_002430973 | Sprouty, putative (Pediculus humanus corporis) | 2.00E-23 |
| 18 | UN068864 | 0.00 | 42.97 | Undefined | |||
| 6 | UN006074 | 3.69 | 40.51 | 10.97 | |||
| 12 | UN018933 | 3.60 | 40.49 | 11.24 | |||
| 6 | UN035226 | 3.17 | 40.12 | 12.67 | AAF70499 | 3-Dehydroecdysone 3α-reductase (Spodoptera littoralis) | 1.00E-54 |
| 6 | UN027771 | 2.86 | 39.33 | 13.74 | |||
Undefined, division by zero.
TABLE 5.
Most highly downregulated unigenes, with basal expression levels of >5 RPKM
| h p.i. | Unigene ID | RPKM |
Fold change | Accession no. | Annotation | E value | |
|---|---|---|---|---|---|---|---|
| Control | Infected | ||||||
| 36 | UN017433 | 768.51 | 5.73 | −134.12 | NP_001040294 | Protease inhibitor 1 (Bombyx mori) | 4.00E-17 |
| 48 | UN060276 | 1,083.05 | 10.01 | −108.20 | |||
| 48 | UN002341 | 738.75 | 8.76 | −84.33 | |||
| 12 | UN070049 | 954.6317267 | 11.65854117 | −81.88 | EFZ21973 | Hypothetical protein SINV_08163 (Solenopsis invicta) | 8.00E-08 |
| 48 | UN059424 | 401.24 | 5.11 | −78.52 | |||
| 48 | UN026539 | 461.50 | 5.94 | −77.69 | XP_001606841 | Predicted: asparaginyl-tRNA synthetase, cytoplasmic-like (Nasonia vitripennis) | 3.00E-196 |
| 48 | UN038604 | 376.10 | 5.07 | −74.18 | AAD09281 | Immune-related Hdd13 (Hyphantria cunea) | 1.00E-91 |
| 48 | UN039460 | 376.28 | 5.27 | −71.40 | ADH16761 | Microsomal glutathione transferase (Heliothis virescens) | 3.00E-56 |
| 12 | UN065168 | 463.95 | 6.93 | −66.95 | ADJ96631 | Carboxylesterase (Helicoverpa armigera) | 9.00E-64 |
| 48 | UN028887 | 638.85 | 10.11 | −63.19 | XP_973680 | Predicted: similar to X box binding protein-1 CG9415-PA (Tribolium castaneum) | 2.00E-23 |
| 48 | UN040932 | 31,667.95 | 504.81 | −62.73 | BAB33421 | Putative senescence-associated protein (Pisum sativum) | 6.00E-39 |
| 36 | UN006916 | 465.37 | 7.44 | −62.55 | |||
| 48 | UN003297 | 1,034.69 | 16.81 | −61.55 | |||
| 48 | UN049959 | 308.75 | 5.16 | −59.84 | ACM78945 | Heat shock protein 70 (Spodoptera exigua) | 4.00E-282 |
| 12 | UN043911 | 363.54 | 6.31 | −57.61 | |||
| 48 | UN068918 | 650.29 | 11.36 | −57.24 | |||
| 36 | UN062607 | 450.56 | 8.18 | −55.08 | EFA01172 | Hypothetical protein TcasGA2_TC010495 (Tribolium castaneum) | 9.00E-155 |
| 48 | UN003134 | 460.94 | 8.65 | −53.29 | |||
| 48 | UN026538 | 438.65 | 8.24 | −53.23 | XP_001606841 | Predicted: asparaginyl-tRNA synthetase, cytoplasmic-like (Nasonia vitripennis) | 3.00E-231 |
| 36 | UN042088 | 355.60 | 6.79 | −52.37 | XP_001956217 | GF24709 (Drosophila ananassae) | 3.00E-54 |
| 24 | UN031769 | 273.37 | 5.36 | −51.00 | |||
TABLE 6.
Most highly repressed unigenes, with basal expression levels of <5 RPKM
| h p.i. | Unigene ID | RPKM |
Fold change | Accession no. | Annotation | E value | |
|---|---|---|---|---|---|---|---|
| Control | infected | ||||||
| 12 | UN004928 | 595.66 | 0.81 | −735.38 | AAS79891 | Gst1 (Spodoptera litura) | 3.00E-59 |
| 48 | UN006916 | 465.37 | 3.46 | −134.50 | |||
| 48 | UN065168 | 463.95 | 1.01 | −459.36 | ADJ96631 | Carboxylesterase (Helicoverpa armigera) | 9.00E-64 |
| 18 | UN001380 | 463.20 | 2.50 | −185.28 | ADJ96632 | Carboxylesterase (Helicoverpa armigera) | 4.00E-232 |
| 18 | UN043911 | 363.54 | 4.35 | −83.57 | |||
| 48 | UN042088 | 355.60 | 3.90 | −91.18 | XP_001956217 | GF24709 (Drosophila ananassae) | 3.00E-54 |
| 48 | UN053524 | 275.91 | 4.83 | −57.12 | ABK23217 | Unknown (Picea sitchensis) | 2.00E-87 |
| 12 | UN019562 | 266.84 | 3.84 | −69.49 | ADE05550 | Carboxylesterase (Helicoverpa armigera) | 6.00E-35 |
| 48 | UN036559 | 250.15 | 4.78 | −52.33 | XP_003394720 | Predicted: hypothetical protein LOC100647415, isoform 2 (Bombus terrestris) | 1.00E-131 |
| 48 | UN036558 | 236.95 | 4.51 | −52.54 | EFN89744 | Hypothetical protein EAI_01992 (Harpegnathos saltato) | 2.00E-137 |
| 48 | UN053959 | 232.42 | 3.52 | −66.03 | XP_974195 | Predicted: similar to sterol regulatory element-binding protein 1 (Tribolium castaneum) | 4.00E-64 |
| 6 | UN030103 | 229.51 | 0.95 | −241.59 | ACX53802 | Hydroxybutyrate dehydrogenase (Heliothis virescens) | 1.00E-79 |
| 48 | UN024251 | 217.19 | 0.79 | −274.92 | ACZ04356 | Hemolin (Trichoplusia ni) | 9.00E-245 |
| 48 | UN070693 | 209.45 | 2.70 | −77.57 | XP_973504 | Predicted: similar to LOC398543 protein (Tribolium castaneum) | 5.00E-19 |
| 48 | UN067023 | 204.59 | 3.92 | −52.19 | AAF61949 | Insulin-related peptide binding protein (Spodoptera frugiperda) | 6.00E-90 |
| 48 | UN052628 | 200.75 | 3.29 | −61.02 | XP_002042993 | GM16365 (Drosophila sechellia) | 6.00E-263 |
| 48 | UN063467 | 200.43 | 4.17 | −48.06 | |||
| 48 | UN058541 | 191.24 | 4.49 | −42.59 | NP_001091784 | C-type lectin 10 (Bombyx mori) | 4.00E-18 |
| 48 | UN070694 | 184.26 | 2.54 | −72.54 | XP_973504 | Predicted: similar to LOC398543 protein (Tribolium castaneum) | 4.00E-19 |
| 36 | UN061625 | 175.83 | 4.90 | −35.88 | AAF90148 | Tetraspanin D76 (Manduca sexta) | 3.00E-125 |
In the late stage of infection, the host cell mRNA decreased to less than 10% of the total mRNA content (Fig. 1A), and approximately 40% of the host transcripts (unigenes) were downregulated (Fig. 1B). The most dramatically downregulated unigenes were identified as those encoding protease inhibitor 1, a serine protease inhibitor (UN017433; −135-fold downregulated), asparaginyl-tRNA synthetase (UN026539 and UN026538; −78 and −53-fold downregulated), and immune-related Hdd13 (UN038604; −74-fold downregulated) (Table 5). Interestingly, heat shock protein 70 gene (hsp70) mRNA was also greatly downregulated (UN049959; −60-fold downregulated). In a prior study, hsp70 was found to be slightly induced in AcMNPV-infected Sf9 cells and possibly important for viral DNA replication (50, 51). In addition, in a prior microarray study, three hsp70 genes were reported to be differentially regulated in Sf9 cells infected with AcMNPV (52). While one hsp70 transcript was gradually downregulated throughout the infection, two other hsp70 genes were dramatically upregulated at 6 h p.i., and the degree of upregulation declined as infection progressed (52).
In the present study, we also found that hemolin mRNA was repressed during AcMNPV infection of T. ni cells (Table 6). Hemolin is a hemolymph protein that is induced by bacterial infection of a number of lepidopterans (53–56). Functionally, it appears that hemolin may act as an opsonin and/or may function in pattern recognition of pathogens (57). In one prior study, it was found that the hemolin gene was induced in the Chinese oak moth (Antheraea pernyi) when it was infected with a baculovirus (AnpeNPV) (58). In addition, knockdown of the hemolin gene by RNA interference (RNAi) resulted in an apparent enhancement of AnpeNPV infection, suggesting that hemolin played an antiviral role. The downregulation of the hemolin gene in AcMNPV-infected T. ni cells may therefore represent viral repression of an antiviral response by the cell. Additional studies will be necessary to determine whether suppression of the hemolin gene plays any role in the response of T. ni cells or larvae to AcMNPV. In summary, we observed that a large number of host unigenes were downregulated or repressed as the infection progressed. Tables 5 and 6 list the most dramatic examples of transcripts that were present at relatively high levels in uninfected controls (RPKM values ranging from 175 to 595) but were severely downregulated (Table 5) or repressed (Table 6) to levels below an RPKM value of 5 in infected cells.
GO analysis of differentially expressed unigenes during viral infection.
To identify differentially expressed unigenes and to more accurately determine the magnitude of changes, we examined only unigenes with expression levels of ≥5 RPKM in either the control or infected cells. We then selected unigenes that were >10-fold upregulated for GO term enrichment analysis (FDR < 0.05) (see Materials and Methods). All the significantly upregulated unigenes identified from all time points were assigned to 215 processes (see Table S5 in the supplemental material). At 0 h p.i. (representing the point 1 h after addition of the virus), the ≥10-fold-upregulated host unigenes were enriched in the following categories: processes relative to pattern specification process (6 out of 21 unigenes), regionalization (6 unigenes), regulation of cell communication (5 unigenes), and positive regulation of gene expression (4 unigenes). At 6 h p.i., the ≥10-fold-upregulated gene set was enriched in those involved in lipid localization/transport (6 out of 91 unigenes) and the steroid catabolic process (4 unigenes) (see Table S5 in the supplemental material). During the late phase of infection (after 12 h p.i.), the most prominent GO biological processes observed for ≥10-fold-upregulated genes were establishment and maintenance of cellular component localization, molecule transport, oxidation-reduction, and vesicle-mediated transport/endocytosis (see Table S5 in the supplemental material). The downregulated and suppressed unigenes identified from all time points were assigned to 6,691 processes (see Table S6 in the supplemental material). The total number of downregulated or suppressed unigenes increased dramatically after viral infection (Table 2), and we did not observe specific patterns from the population.
Dramatic changes in transcripts at 6 h p.i.
In a prior study (33), we observed that viral DNA and RNA levels increased dramatically after 6 h p.i. and viral late genes were highly active, indicating initiation of the late phase of the infection cycle around 6 h p.i. or between 6 and 12 h p.i. To examine significant changes or regulation in host cell gene expression during the initial stages of the infection, we analyzed T. ni unigenes that are highly expressed at 6 h p.i. By examining unigenes with RPKM values of ≥100 before or immediately after infection (control, 0 or 6 h p.i.), we identified 24 T. ni unigene mRNAs that were induced 5- to 9-fold at 6 h p.i., compared to uninfected cells (see Table S7 in the supplemental material), including a NADH dehydrogenase subunit 4L mRNA (UN014990) and a 3-dehydroecdysone 3b-reductase mRNA (UN029241). The functions of other unigenes were unknown. In addition, using the same approach, we found an additional 112 of these highly expressed unigenes that are induced >3-fold at 6 h p.i. (see Table S7 in the supplemental material). Based on the GO term analysis, these genes are assigned functions relative to transport activity, protein binding, hydrolase activity, nucleotide binding, enzyme regulator activity, and transferase activity (Fig. 2). Interestingly, a 3-dehydroecdysone 3b-reductase gene was previously reported as induced in T. ni larvae challenged with bacteria (59). In healthy insects, 3-dehydroecdysone 3b-reductase mediates production of ecdysone from 3-dehydroecdysone, which is secreted from prothoracic glands. The gene is expressed in the fat body during development, and the protein is found in the hemolymph and the integument (59). Of particular note is the fact that AcMNPV encodes a secreted enzyme, EGT (ecdysteroid UDP-glucosyl transferase), that catalyzes the conjugation of galactose and ecdysteroids, resulting in reduced levels of active ecdysteroids in the hemolymph and preventing molting of infected host larvae (16, 60). The viral egt gene is transcribed early after infection in cultured cells, with peak transcript levels occurring within the first 3 to 6 h p.i. and substantial transcript levels remaining throughout infection (33, 61). The secreted extracellular EGT protein accumulates throughout infection. The upregulation of the cellular 3-dehydroecdysone 3b-reductase gene may be a defensive response to viral infection, but it is unclear how this response is activated in cultured cells and whether this activation is related to the presence of the viral EGT protein. However, these results indicate that even in cultured cells, a mechanism for regulating ecdysone metabolism is activated in response to baculovirus infection. It would be of interest to determine whether transcription of the cellular 3-dehydroecdysone 3b-reductase gene is upregulated by infection with an EGT-null virus and thus whether this cellular upregulation is dependent on viral expression of EGT or some other factor. The upregulation of the 3-dehydroecdysone 3b-reductase gene as a response to both bacterial challenge in the larva and viral challenge in a clonal cell line could suggest that 3-dehydroecdysone 3b-reductase plays a general defensive role in response to infection by microorganisms.
FIG 2.
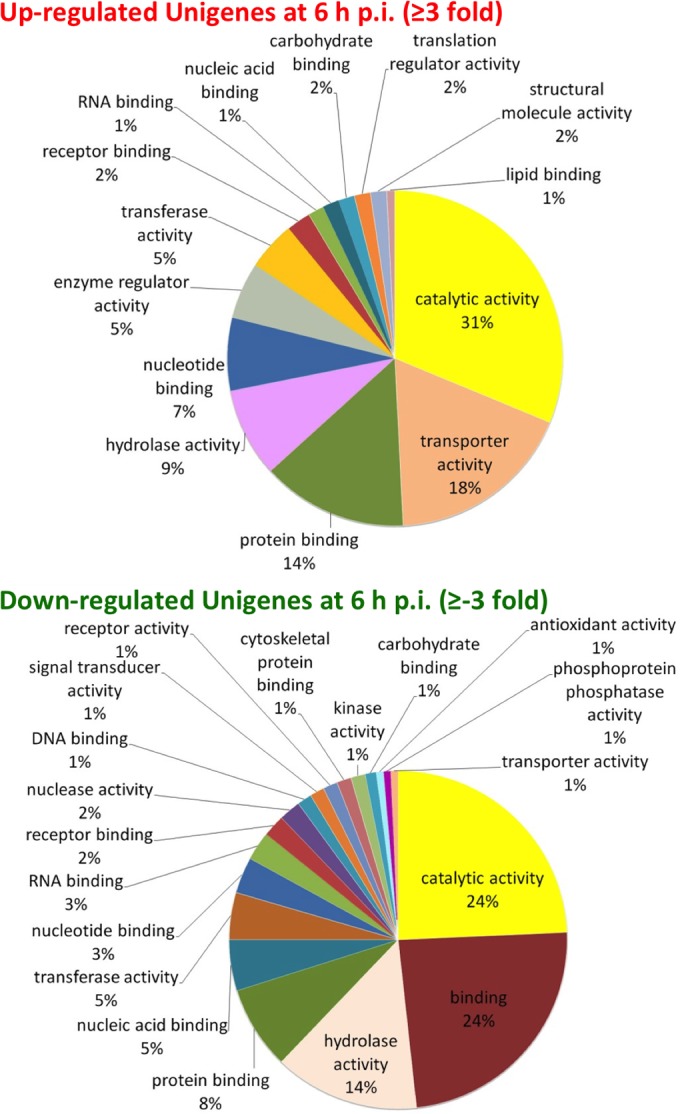
Differentially expressed transcripts among different biological processes. (Top) Distributions of upregulated unigenes at 6 h p.i.; (B) distributions of downregulated unigenes at 6 h p.i. Gene ontology (GO) analysis of differentially expressed T. ni transcripts was performed as described in Materials and Methods.
In contrast to mRNAs that are induced or upregulated in response to AcMNPV infection, we identified a number of T. ni mRNAs that were highly expressed in cells prior to infection (RPKM values of >100, ranging from 100 to 32,000) but for which mRNAs were rapidly and dramatically reduced within the first 6 h postinfection. We identified 25 T. ni unigenes for which mRNA levels were decreased ≥−10-fold compared with the levels observed prior to infection (see Table S7 in the supplemental material). These highly downregulated or repressed unigenes include those encoding hydroxybutyrate dehydrogenase (UN030103), hemolin (UN024251), the facilitated trehalose transporter Tret1 (UN003843), and ATP-citrate synthase (UN002915 and UN002916). In addition, we identified 78 T. ni unigenes for which mRNA levels decreased ≥−3-fold at 6 h p.i., compared with that from the uninfected control.
Cluster analysis of T. ni unigene expression patterns.
To categorize the expression patterns of T. ni unigenes after viral infection, we performed a cluster analysis of unigenes with RPKM values of ≥5, a level estimated to represent ≥1 mRNA molecule per cell (39). A total of 14,824 unigenes that fit this category were selected. The cluster analysis revealed 10 distinct groups (Fig. 3, G0 to G9; also, see Table S8 in the supplemental material) based on the temporal transcript abundance as well as patterns of transcript abundance through the infection cycle. Expression of approximately 23% of the T. ni unigenes began to decrease immediately after viral infection (G1, G8, and G9), while around 76% of the unigenes increased after infection but peaked at either 0 h p.i. (G6 and G7) or 6 h p.i. (G0, G2, G3, and G4) and then decreased through the remainder of the infection time course. Interestingly, one small group of unigenes (G5) represents transcripts that are absent or present in very low abundance in the uninfected control but increased in abundance after viral infection and then peaked at times ranging from 18 to 36 h p.i. (Fig. 3, G5; also, see Table S8 in the supplemental material). Some of these unigenes encode homologs of NADH dehydrogenase subunits, cytochrome oxidase subunits, ATP synthase subunits, and thymus-specific serine protease. Thus, within this small subgroup are included a number of genes with functions related to energy metabolism and regulation. We further examined the unigenes that were ≥2-fold upregulated at either 18 h or 24 h in the G5 group, using GO analysis. We found that those unigenes are enriched for genes related to oxidation-reduction processes, generation of precursor metabolites and energy (Table 7), oxidoreductase activity, and metal ion binding (Table 8).
FIG 3.
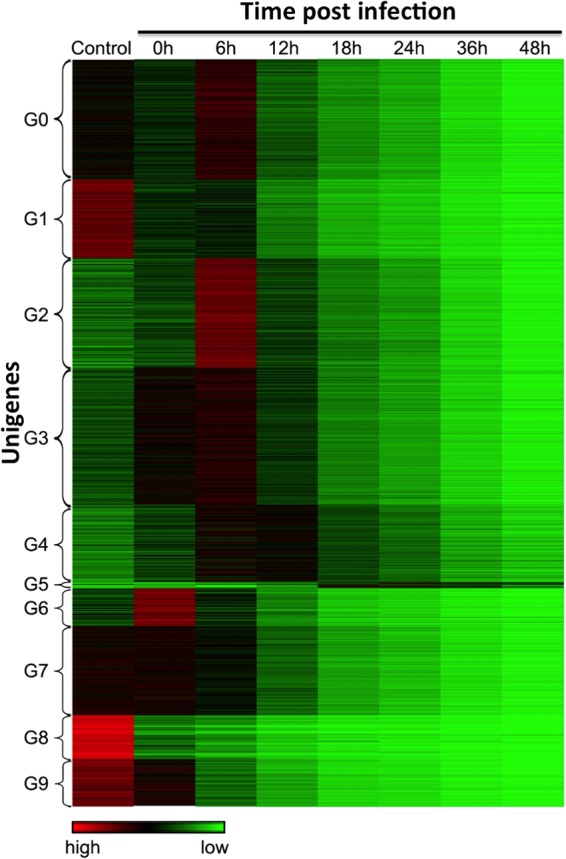
Cluster analysis of T. ni Tnms42 gene expression patterns throughout the 48-h AcMNPV infection cycle. A heat map of normalized gene expression levels from 0 to 48 h p.i. shows each gene as a single horizontal line. Relative expression levels are represented as high (red), intermediate (black), and low (green). Based on gene expression levels and patterns of expression, unigenes were clustered into groups G1 to G9 (indicated on the left).
TABLE 7.
Unigenes >2-fold upregulated in cluster group 5 based on biological process (P ≤ 0.01)
| Gene ontology term | Cluster frequency [no. out of 124 unigenes (%)] | Genome frequency of use [no. out of 70,810 unigenes (%)] | Raw P value | Corrected P value | Unigenes annotated to the term |
|---|---|---|---|---|---|
| Oxidative phosphorylation | 10 (8.1) | 98 (0.1) | 2.30E-15 | 0 | UN034582, UN059811, UN024533, UN002246, UN000652, UN068092, UN031997, UN067842, UN007978, UN066222 |
| Oxidation-reduction process | 23 (18.5) | 1,761 (2.5) | 6.12E-14 | 0 | UN059732, UN059001, UN008506, UN059811, UN024533, UN002823, UN008508, UN002246, UN000652, UN059002, UN008509, UN066222, UN007978, UN063198, UN034582, UN008507, UN064750, UN063199, UN059731, UN068092, UN031997, UN059730, UN067842 |
| Electron transport chain | 11 (8.9) | 233 (0.3) | 4.71E-13 | 0 | UN034582, UN059811, UN024533, UN002823, UN002246, UN000652, UN068092, UN031997, UN067842, UN007978, UN066222 |
| Generation of precursor metabolites and energy | 16 (12.9) | 864 (1.2) | 3.36E-12 | 0 | UN063198, UN059001, UN034582, UN059811, UN024533, UN002823, UN063199, UN002246, UN000652, UN068092, UN067134, UN031997, UN059002, UN067842, UN066222, UN007978 |
| Cellular respiration | 11 (8.9) | 289 (0.4) | 4.86E-12 | 0 | UN034582, UN059811, UN024533, UN002823, UN002246, UN000652, UN068092, UN031997, UN067842, UN007978, UN066222 |
| Energy derivation by oxidation of organic compounds | 13 (10.5) | 575 (0.8) | 3.52E-11 | 0 | UN063198, UN034582, UN059811, UN024533, UN002823, UN063199, UN002246, UN000652, UN068092, UN031997, UN067842, UN066222, UN007978 |
| ATP synthesis coupled electron transport | 7 (5.6) | 78 (0.1) | 1.01E-10 | 0 | UN031997, UN034582, UN059811, UN067842, UN068092, UN066222, UN007978 |
| Respiratory electron transport chain | 8 (6.5) | 145 (0.2) | 2.29E-10 | 0 | UN034582, UN059811, UN002823, UN068092, UN031997, UN067842, UN007978, UN066222 |
| Reactive oxygen species metabolic process | 7 (5.6) | 130 (0.2) | 3.75E-09 | 0 | UN059002, UN059001, UN008506, UN008509, UN008507, UN064750, UN008508 |
| Cuticle development involved in collagen and cuticulin-based cuticle molting cycle | 4 (3.2) | 21 (0.0) | 5.23E-08 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Superoxide metabolic process | 5 (4.0) | 61 (0.1) | 8.34E-08 | 0 | UN059002, UN059001, UN008509, UN008507, UN064750 |
| Post-embryonic body morphogenesis | 4 (3.2) | 24 (0.0) | 9.26E-08 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Mitochondrial ATP synthesis coupled electron transport | 5 (4.0) | 73 (0.1) | 2.07E-07 | 0 | UN031997, UN067842, UN068092, UN066222, UN007978 |
| Response to reactive oxygen species | 8 (6.5) | 366 (0.5) | 3.08E-07 | 0 | UN059001, UN008506, UN008507, UN008508, UN039179, UN059002, UN062469, UN008509 |
| Removal of superoxide radicals | 4 (3.2) | 32 (0.0) | 3.10E-07 | 0 | UN059002, UN059001, UN008509, UN008507 |
| Cellular response to oxygen radical | 4 (3.2) | 32 (0.0) | 3.10E-07 | 0 | UN059002, UN059001, UN008509, UN008507 |
| Cellular response to superoxide | 4 (3.2) | 32 (0.0) | 3.10E-07 | 0 | UN059002, UN059001, UN008509, UN008507 |
| Mitochondrial electron transport, NADH to ubiquinone | 4 (3.2) | 42 (0.1) | 9.52E-07 | 0 | UN031997, UN067842, UN066222, UN007978 |
| Cellular response to reactive oxygen species | 6 (4.8) | 186 (0.3) | 1.04E-06 | 0 | UN059002, UN059001, UN008506, UN008509, UN008507, UN008508 |
| Phosphorylation | 15 (12.1) | 1,913 (2.7) | 1.42E-06 | 0 | UN034582, UN059811, UN024533, UN002246, UN000652, UN068092, UN067134, UN031997, UN062469, UN039179, UN018195, UN006433, UN067842, UN066222, UN007978 |
| Response to superoxide | 4 (3.2) | 47 (0.1) | 1.50E-06 | 0 | UN059002, UN059001, UN008509, UN008507 |
| Response to oxygen radical | 4 (3.2) | 47 (0.1) | 1.50E-06 | 0 | UN059002, UN059001, UN008509, UN008507 |
| Peptide cross-linking | 4 (3.2) | 52 (0.1) | 2.27E-06 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Collagen and cuticulin-based cuticle development | 4 (3.2) | 54 (0.1) | 2.64E-06 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Cellular response to disaccharide stimulus | 2 (1.6) | 2 (0.0) | 3.04E-06 | 0 | UN059002, UN059001 |
| Cellular response to sucrose stimulus | 2 (1.6) | 2 (0.0) | 3.04E-06 | 0 | UN059002, UN059001 |
| Cellular response to ozone | 2 (1.6) | 2 (0.0) | 3.04E-06 | 0 | UN059002, UN059001 |
| Cellular response to light intensity | 2 (1.6) | 2 (0.0) | 3.04E-06 | 0 | UN059002, UN059001 |
| Hydrogen peroxide catabolic process | 4 (3.2) | 60 (0.1) | 4.04E-06 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Response to hydrogen peroxide | 6 (4.8) | 277 (0.4) | 1.02E-05 | 0 | UN062469, UN039179, UN008506, UN008509, UN008507, UN008508 |
| Tyrosine catabolic process | 3 (2.4) | 24 (0.0) | 1.03E-05 | 0 | UN059732, UN059730, UN059731 |
| Cuticle development involved in protein-based cuticle molting cycle | 4 (3.2) | 76 (0.1) | 1.04E-05 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Response to methotrexate | 4 (3.2) | 78 (0.1) | 1.15E-05 | 0 | UN001769, UN001910, UN002605, UN002604 |
| Phosphorus metabolic process | 15 (12.1) | 2,292 (3.2) | 1.24E-05 | 0 | UN034582, UN059811, UN024533, UN002246, UN000652, UN068092, UN067134, UN031997, UN062469, UN039179, UN018195, UN006433, UN067842, UN066222, UN007978 |
| Phosphate-containing compound metabolic process | 15 (12.1) | 2,292 (3.2) | 1.24E-05 | 0 | UN034582, UN059811, UN024533, UN002246, UN000652, UN068092, UN067134, UN031997, UN062469, UN039179, UN018195, UN006433, UN067842, UN066222, UN007978 |
| Metabolic process | 47 (37.9) | 14,933 (21.1) | 1.38E-05 | 0 | UN059001, UN024533, UN002823, UN008508, UN002246, UN000652, UN054195, UN038867, UN007978, UN063198, UN025811, UN034582, UN025815, UN025812, UN063199, UN059731, UN062469, UN038865, UN006433, UN067842, UN054804, UN059732, UN008506, UN059811, UN002605, UN025813, UN025814, UN067134, UN000260, UN059002, UN001910, UN039179, UN008509, UN038866, UN067090, UN002604, UN066222, UN056001, UN001769, UN008507, UN064750, UN068092, UN031997, UN018195, UN059730, UN038864, UN056002 |
| Hydrogen peroxide metabolic process | 4 (3.2) | 83 (0.1) | 1.47E-05 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Cellular response to oxidative stress | 6 (4.8) | 303 (0.4) | 1.70E-05 | 0 | UN059002, UN059001, UN008506, UN008509, UN008507, UN008508 |
| Response to unfolded protein | 6 (4.8) | 320 (0.5) | 2.31E-05 | 0.00051 | UN001769, UN062469, UN039179, UN001910, UN002605, UN002604 |
TABLE 8.
Unigenes >2-fold upregulated in cluster group 5 based on molecular function (P ≤ 0.01)
| Gene ontology term | Cluster frequency [no. out of 124 unigenes (%)] | Genome frequency of use [no. out of 70,810 unigenes (%)] | Raw P value | Corrected P value | Genes annotated to the term |
|---|---|---|---|---|---|
| Oxidoreductase activity | 25 (20.2) | 1,666 (2.4) | 1.82E-16 | 0 | UN059732, UN059001, UN008506, UN059811, UN024533, UN002823, UN008508, UN002246, UN000652, UN059002, UN008509, UN066222, UN007978, UN063198, UN034582, UN008507, UN064750, UN063199, UN059731, UN068092, UN031997, UN059730, UN038865, UN067842, UN038864 |
| NADH dehydrogenase (ubiquinone) activity | 6 (4.8) | 58 (0.1) | 9.58E-10 | 0 | UN031997, UN034582, UN059811, UN067842, UN066222, UN007978 |
| NADH dehydrogenase (quinone) activity | 6 (4.8) | 59 (0.1) | 1.06E-09 | 0 | UN031997, UN034582, UN059811, UN067842, UN066222, UN007978 |
| NADH dehydrogenase activity | 6 (4.8) | 64 (0.1) | 1.76E-09 | 0 | UN031997, UN034582, UN059811, UN067842, UN066222, UN007978 |
| Hydrogen ion transmembrane transporter activity | 7 (5.6) | 149 (0.2) | 9.70E-09 | 0 | UN037449, UN024533, UN000267, UN067090, UN002246, UN000652, UN068092 |
| Prostaglandin-endoperoxide synthase activity | 4 (3.2) | 16 (0.0) | 1.60E-08 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Antioxidant activity | 7 (5.6) | 166 (0.2) | 2.04E-08 | 0 | UN059002, UN059001, UN008506, UN008509, UN008507, UN064750, UN008508 |
| Monovalent inorganic cation transmembrane transporter activity | 9 (7.3) | 381 (0.5) | 2.83E-08 | 0 | UN063198, UN037449, UN024533, UN000267, UN067090, UN063199, UN002246, UN000652, UN068092 |
| Oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor | 6 (4.8) | 104 (0.1) | 3.36E-08 | 0 | UN031997, UN034582, UN059811, UN067842, UN066222, UN007978 |
| Ligand-dependent nuclear receptor binding | 5 (4.0) | 67 (0.1) | 1.34E-07 | 0 | UN025811, UN025813, UN025815, UN025812, UN025814 |
| 4-Hydroxyphenylpyruvate dioxygenase activity | 3 (2.4) | 7 (0.0) | 1.82E-07 | 0 | UN059732, UN059730, UN059731 |
| Lactoylglutathione lyase activity | 3 (2.4) | 8 (0.0) | 2.91E-07 | 0 | UN059732, UN059730, UN059731 |
| Oxidoreductase activity, acting on NADH or NADPH | 6 (4.8) | 160 (0.2) | 4.33E-07 | 0 | UN031997, UN034582, UN059811, UN067842, UN066222, UN007978 |
| Cytochrome c oxidase activity | 4 (3.2) | 35 (0.0) | 4.49E-07 | 0 | UN024533, UN002246, UN000652, UN068092 |
| Heme-copper terminal oxidase activity | 4 (3.2) | 35 (0.0) | 4.49E-07 | 0 | UN024533, UN002246, UN000652, UN068092 |
| Oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor | 4 (3.2) | 35 (0.0) | 4.49E-07 | 0 | UN024533, UN002246, UN000652, UN068092 |
| Unfolded protein binding | 6 (4.8) | 183 (0.3) | 9.51E-07 | 0 | UN001769, UN062469, UN039179, UN001910, UN002605, UN002604 |
| Oxidoreductase activity, acting on a heme group of donors | 4 (3.2) | 44 (0.1) | 1.15E-06 | 0 | UN024533, UN002246, UN000652, UN068092 |
| Inorganic cation transmembrane transporter activity | 9 (7.3) | 596 (0.8) | 1.20E-06 | 0 | UN063198, UN037449, UN024533, UN000267, UN067090, UN063199, UN002246, UN000652, UN068092 |
| Heme binding | 7 (5.6) | 333 (0.5) | 2.24E-06 | 0 | UN008506, UN008509, UN024533, UN008507, UN008508, UN002246, UN000652 |
| Superoxide dismutase activity | 3 (2.4) | 16 (0.0) | 2.88E-06 | 0 | UN059002, UN059001, UN064750 |
| Oxidoreductase activity, acting on superoxide radicals as acceptor | 3 (2.4) | 16 (0.0) | 2.88E-06 | 0 | UN059002, UN059001, UN064750 |
| Tetrapyrrole binding | 7 (5.6) | 346 (0.5) | 2.88E-06 | 0 | UN008506, UN008509, UN024533, UN008507, UN008508, UN002246, UN000652 |
| Cation transmembrane transporter activity | 9 (7.3) | 763 (1.1) | 8.81E-06 | 0 | UN063198, UN037449, UN024533, UN000267, UN067090, UN063199, UN002246, UN000652, UN068092 |
| Channel conductance-controlling ATPase activity | 2 (1.6) | 4 (0.0) | 1.82E-05 | 0 | UN063198, UN063199 |
| Iron ion binding | 7 (5.6) | 488 (0.7) | 2.65E-05 | 0 | UN008506, UN008509, UN024533, UN008507, UN008508, UN002246, UN000652 |
| Catalytic activity | 38 (30.6) | 11,243 (15.9) | 2.92E-05 | 0 | UN059001, UN024533, UN002823, UN008508, UN002246, UN000652, UN054195, UN038867, UN007978, UN063198, UN034582, UN063199, UN059731, UN038865, UN006433, UN067842, UN054804, UN008506, UN059732, UN059811, UN002605, UN067134, UN001910, UN059002, UN008509, UN051939, UN038866, UN067090, UN002604, UN066222, UN001769, UN008507, UN064750, UN068092, UN031997, UN018195, UN059730, UN038864 |
| Carbon-sulfur lyase activity | 3 (2.4) | 37 (0.1) | 3.89E-05 | 0 | UN059732, UN059730, UN059731 |
| Peroxidase activity | 4 (3.2) | 129 (0.2) | 8.32E-05 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Oxidoreductase activity, acting on peroxide as an acceptor | 4 (3.2) | 129 (0.2) | 8.32E-05 | 0 | UN008506, UN008509, UN008507, UN008508 |
| Sulfonylurea receptor activity | 2 (1.6) | 8 (0.0) | 8.45E-05 | 0 | UN063198, UN063199 |
| Ion transmembrane transporter activity | 9 (7.3) | 1,031 (1.5) | 9.06E-05 | 0 | UN063198, UN037449, UN024533, UN000267, UN067090, UN063199 |
Pathways associated with viral processes. (i) Entry and egress.
The entry of budded baculovirus into host cells generally occurs through a receptor-mediated endocytosis pathway (62–64). After virion fusion within the endosome, nucleocapsids are released into the cytoplasm and transported to the nuclear membrane via actin filaments (25, 65, 66). Nucleocapsids then enter the nucleus through a modified or distended nuclear pore (25, 67, 68). Following viral gene expression, DNA replication, and nucleocapsid assembly in the nucleus, the progeny nucleocapsids exit the nucleus by a poorly understood mechanism and are transported to the plasma membrane, where they interact with the plasma membrane and assemble with envelope proteins into budded virions that are released by budding from the cell surface. These complex processes involve numerous membrane systems and their associated protein complexes, including the plasma membrane, nuclear membranes, ER, Golgi, vesicular transport systems, etc. Previously, we found that proteins of the endosomal sorting complex required for transport (ESCRT) pathway were involved in both viral entry and egress (69). To determine whether viral infection impacts gene expression for members of those membrane and transport systems, we examined the effects of infection on mRNAs encoding members of the clathrin-mediated endocytosis (CME) pathway, the ESCRT pathway, the nuclear pore complex (NPC), the transcription/export complex (TREX), and several other relevant complexes (see Fig. S1 to S5 in the supplemental material). For the majority of unigenes involved in these pathways, the general trend was slight to moderate upregulation immediately upon infection (0 and/or 6 h p.i.) followed by gradual reduction in RPKM values as the infection progressed (see Fig. S1 to S5 in the supplemental material). However, in most of these pathways, we identified specific unigenes with a more dramatic upregulation or higher levels of transcripts during the period from 0 to 12 h following infection. These may represent host genes that either are regulated by the virus or respond to viral infection as a defensive or stress response.
(ii) Endocytosis.
HSC70 (UN059101), a heat shock protein, is constitutively expressed in most cells. Normally, HSC70 plays a role in the protein folding and also in the disassembly of clathrin-coated vesicles. Interestingly, upon infection by AcMNPV, HSC70 mRNA spiked dramatically at 0 h p.i. (one hour after addition of the viral inoculum) and declined thereafter (see Fig. S1 [HSC70] in the supplemental material). While most of the endocytosis-related mRNAs declined gradually through infection, several mRNAs were either highly expressed or significantly upregulated at 6 h p.i. These mRNAs included homologs of the mRNAs for clathrin light chain (UN033438 and UN033439), Arp3 (UN064270), AP2M1 (UN007037), and AP2S1 (UN063500). After 6 h p.i., all these mRNAs declined in abundance. Arp3 transcripts were most dramatically upregulated, with a >2-fold increase in transcripts at 0 h p.i. and a >3-fold increase at 6 h p.i. Afterwards, levels of Arp3 transcripts decreased dramatically between 6 and 18 h p.i.
(iii) ESCRT pathway and associated proteins.
Many viruses utilize components of the cellular ESCRT pathway to initiate and execute virion budding and scission from the cellular plasma membrane (70, 71). In a prior study of AcMNPV-infected cells (69), it was found that inhibition of the cellular ESCRT pathway negatively impacted both viral entry and egress. In this study, we examined the effects of AcMNPV infection on transcripts of the ESCRT pathway and associated proteins (see Fig. S2 to S4 in the supplemental material). Several of the ESCRT pathway homologs (encoding Vps28 and CHMP2B) were substantially upregulated within 1 h after virus addition (0 h p.i.), and another partially overlapping subset of these genes was upregulated (some dramatically) at 6 h p.i. (those encoding Vps28, EAP20, CHMP5, CHMP2B, and LIP5) (see Fig. S2A in the supplemental material). For all of these, however, transcript levels decreased between 6 and 12 h p.i. and beyond. Indeed, all the ESCRT homolog transcripts decreased after 12 h p.i., as was typical for host cell transcripts in general. We also examined transcripts of ESCRT-associated and other VPS proteins. We identified a number of those T. ni homologs that were substantially upregulated at 0 and/or 6 h p.i., including genes for VPS8, VPS33A, VPS41 (see Fig. S2B in the supplemental material), Ang2, Are1, SNX5, VPS35, VPS29 (see Fig. S3A in the supplemental material), dynamin-like protein, VPS45, LEPR (see Fig. S3B in the supplemental material), and Akt/PKB (see Fig. S4A in the supplemental material). Although the ESCRT pathway is important for viral entry and exit, the specific roles of components within these pathways in relation to viral infection are not known. The observation that transcripts representing homologs of specific members of this pathway are upregulated soon after viral infection while others are not may indicate regulation of these pathways by the virus to benefit successful viral entry, replication, and/or egress. Detailed mechanistic studies will be required to understand the significance of the current genome-level observations.
(iv) Signaling molecules and stress.
Transcripts encoding representative signaling molecules were also examined in our analysis of host cell responses (see Fig. S4B in the supplemental material). Among the genes examined, one (encoding Ras85D [UN031780, UN050418, and UN067243]) was upregulated at 0 h p.i., and two others (encoding mitogen-activated protein kinase kinase [MAPKK] 3/4/6 [UN023339] and MAPKK 4 [UN028639, UN028641, UN049071, UN066579, UN066580, and UN069980]) were primarily upregulated at 6 h postinfection.
(v) Nuclear entry and exit.
During viral entry, nucleocapsids of AcMNPV are transported to and appear to interact with the nuclear pore complex before moving through the nuclear pore and subsequently uncoating in the nucleus (67, 68). Egress of progeny nucleocapsids from the nucleus is not well studied but does not appear to involve movement through nuclear pores. To examine effects of infection on proteins associated with nucleocapsid entry into the nucleus, and subsequent events related to mRNA export, we examined nuclear pore complex (NPC) proteins and the transcription/export complex (TREX) (see Fig. S5 in the supplemental material). Of the 15 NPC transcripts examined, three were substantially upregulated at by 6 h p.i. These included homologs of genes encoding Rae1, Nup43-like protein, and Nup50-like protein (see Fig. S5A in the supplemental material). We found that of the 13 TREX complex component genes examined, only those for DDX39A/B and ALYREF were only slightly upregulated at 0 h and that for DDX39B was upregulated at 6 h p.i. (see Fig. S5B in the supplemental material).
Antiviral, immune, and stress response genes.
Antiviral responses in insects include small interfering RNAs and Piwi RNAs (siRNAs and piRNAs), apoptosis, and perhaps protein kinase RNA-activated (PKR) and innate immune and stress responses (72–75). How permissive and nonpermissive cells respond defensively to baculovirus infection has not been well characterized. To examine this question, we analyzed the transcriptional responses of genes associated with several pathways associated with known or potential antiviral defenses.
Small RNAs.
While the RNAi response is an antiviral response more typically associated with double-stranded RNAs (dsRNAs) produced during infection by RNA viruses, DNA viruses are also known to elicit an RNAi response (76, 77). Several lines of circumstantial evidence also suggest that baculoviruses might modify or suppress the RNAi response. We previously found that relatively large numbers of overlapping baculovirus transcripts occur in the AcMNPV infection cycle (33), potentially resulting in dsRNA and a RNAi response. In addition, it was also previously observed that an alphanodavirus (TNCL virus) that persistently infects Tn5B1-4 cells appears to be released from suppression upon infection of the cell by AcMNPV (78). Previous studies have also documented viral siRNAs directed against specific hotspots in the Helicoverpa armigera SNPV (HaSNPV) genome (77). Together, these observations suggest that a cellular RNAi response may occur in AcMNPV-infected cells, a response that may be inhibited by AcMNPV. To examine transcripts of RNAi pathway components during AcMNPV infection, we identified and examined levels of transcripts (unigenes) that represent T. ni homologs of the genes for Argonaute (Ago-1, Ago-2, and Ago-3), Dicer (Dcr-1 and Dcr-2), Drosha, Pasha, Piwi, Aubergine, R2D2, and R3D1. At 6 h p.i., transcripts of R3D1 were >2-fold upregulated and those of Ago-3 were slightly upregulated (see Fig. S6 in the supplemental material). All others remained at low levels or were downregulated as the infection progressed. In the case of two genes (encoding Dcr-2 and Ago-2), transcript levels were relatively high in uninfected cells and were then decreased dramatically immediately following infection (see Fig. S6 in the supplemental material). Interestingly, Dcr-2 and Ago-2 are key components of insect cell siRNA responses to long dsRNAs. Dcr-2 is an RNase III enzyme that processes dsRNAs into the 21-nt guide RNAs that direct cleavage of the homologous target RNA by Ago-2, another RNase III found in the RISC complex. Since dsRNAs are present in substantial quantities in AcMNPV-infected cells (33), a dramatic downregulation of these specific components of the RNAi pathway could be important for successful viral replication. While other mechanisms may directly inhibit the RNAi pathway at the protein level, downregulation of the transcripts encoding these key proteins may provide an important or redundant mechanism for inhibiting the cell response to baculovirus dsRNAs. Future experiments to understand the functional effects of these transcriptional changes will be required to determine whether these changes have biological significance, and such experiments should provide important insight into the potential viral regulation of the host RNAi pathway.
Apoptosis.
Apoptosis is a major antiviral defense mechanism at the cellular level. A number of viruses directly inhibit apoptosis by expressing inhibitors of the initiator and/or effector caspases that regulate and mediate apoptosis. Baculoviruses encode several inhibitors of apoptosis, including P35 or P35-like proteins, and a variety of inhibitor of apoptosis (IAP) proteins that are similar to cellular IAP proteins (26). To determine whether the host cell apoptotic cascade is modified at the transcriptional level by AcMNPV infection, we measured the effects of AcMNPV infection on the transcript levels of a variety of apoptosis-related genes (see Fig. S7A in the supplemental material). For the purpose of this analysis, caspase genes are named according to the homologs identified in B. mori, D. melanogaster, S. exigua, and S. frugiperda (see Table S16 and Fig. S7A in the supplemental material). We observed relatively dramatic increases in transcript levels of genes for cytochrome c, caspase-5, and VIAF1 after addition of the virus and through 6 h p.i. These increases in transcript levels were followed by a reduction of their levels through the remainder of the infection cycle. In contrast, transcript levels of most other apoptosis-associated genes examined were very minimally increased at 6 h p.i. and then reduced through the remainder of the infection. In particular, transcript levels of a caspase-1 gene were relatively high in uninfected cells and dropped precipitously between 1 and 12 h following virus addition (see Fig. S7 in the supplemental material). Caspase-1 of Sf9 cells is an effector caspase that was previously identified as a target of AcMNPV P35 (79, 80). The viral p35 gene is expressed immediately after infection, and transcript levels remain high through the infection cycle (33), indicating that the P35 inhibitor protein is continually present. In addition to the inhibition of caspase proteins by the P35 protein, the observed decreases in the levels of caspase transcripts (such as that of caspase-1) should also result in a reduced likelihood of triggering apoptosis. It will be of interest in future studies to examine both the causes and effects of changes in transcript levels of these host genes associated with the apoptotic cascade.
Heat shock proteins.
Heat shock proteins (HSPs) represent a class of stress response genes, and some of the best studied HSPs are associated with protein folding or unfolding. Of the approximately 20 HSP family members examined in this study, we found that viral infection resulted in substantial upregulation of only 2 members, a homolog of Hsp25.4 and a 10-kDa mitochondrial heat shock protein homolog (see Fig. S7B in the supplemental material). After 6 h p.i., almost all of the heat shock protein RNAs declined, some precipitously.
Reactive oxygen species (ROS).
Levels of reactive oxygen species may rise in cells under stress, and in some cases the production of ROS is thought to serve as a defensive response to infection or invasion. The response may be direct, resulting in damage to viral and cellular structures, or indirect, with ROS serving in a signaling role (81). Prior evidence of oxidative stress induced by AcMNPV infection includes increased oxidation of proteins and membrane lipids (82, 83). Baculoviruses such as AcMNPV encode a Cu/Zn superoxide dismutase (SOD) which is expressed at moderate levels beginning between 6 and 12 h p.i. and extending through the infection cycle (33). The viral SOD is thought to scavenge reactive oxygen species and may therefore defend the virus against ROS-mediated defensive measures of the cell (84). Of the T. ni ROS-related genes examined, only the gene encoding superoxide dismutase 2 (Mn) showed a dramatic increase in transcript levels at 6 h p.i. (see Fig. S8A in the supplemental material), while several other ROS-related genes (encoding Gpx, superoxide dismutase SOD, and oxidative stress response 1) increased modestly through 6 h p.i. and then decreased.
Innate immunity.
Innate immunity genes represent an extremely important set of genes that modulate very specific and complex responses by insects to microbes such as bacteria and fungi. In addition, innate immunity may play an important role in viral infections, although little is known regarding antiviral responses and innate immune mechanisms. Genes associated with innate immunity encode proteins responsible for three general activities: microbe recognition, signaling, and execution of antimicrobial responses. Responses may include activation of other pathways, leading to the expression of proteins such as antimicrobial peptides. Over 200 innate immunity-related genes have been identified in the genomes of B. mori, D. melanogaster, and Anopheles gambiae (85–87). In the current study, we examined selected representatives from gene groups associated with several signaling pathways (encoding NF-κB-IkB, Toll, IMD, JNK, and JAK/STAT) (see Fig. S8B to S10 in the supplemental material). In contrast to other groups, genes of the NF-κB-IkB group were all downregulated from the beginning of the infection (see Fig. S8B in the supplemental material). The most abundant member (the NF-κB1 gene) was present at higher levels initially, but transcript levels dropped precipitously during the first 18 h following infection. Indeed, NF-κB1 levels were reduced approximately −2-fold within the first hour after exposure to the virus and were reduced approximately −7-fold by 18 h p.i. The other groups of innate-immunity-associated genes generally declined through the infection cycle. However, transcript levels of several specific genes increased dramatically by 1 h following virus addition (0 h p.i.) and these included genes for UEV1A, Bendless/UBC13 (see Fig. S9A in the supplemental material), Domeless, P38B (see Fig. S9B in the supplemental material), JUN, and Puckered (see Fig. S10A in the supplemental material). Another group of these genes had somewhat distinctive transcript level spikes at 6 h p.i., including those for Bendless/UBC13, UEV1A, SKPA, MPK2/P38A/P38B, and Hemipterous isoform C (see Fig. S9 and S10 in the supplemental material). These specific early spikes and/or precipitous declines in transcripts associated with innate immunity may have implications with regard to how AcMNPV successfully overcomes organismal immune functions, and future studies should build on these detailed initial observations at the genome level.
Histone deacetylases (HDACs) and sirtuins.
In the case of some large DNA viruses, such as the herpesviruses, the state of histone acetylation is known to modulate the activity of various early promoters (88). Indeed, histone deacetylase (HDAC) enzymes and the state of histone acetylation play significant roles in the infection cycles of a number of viruses (89). This is not surprising, as in healthy cells, chromatin remodeling plays important roles in regulating gene expression. This remodeling is accomplished through the action of histone deacetylase enzymes that are responsible for condensing chromatin when histones are deacetylated. Thus, gene expression of both cellular and viral genes may be regulated by the cellular HDACs (89, 90). Prior studies of AcMNPV-infected Sf9 cells showed that addition of an inhibitor of HDACs (sodium butyrate) led to a reduction in gene expression from a baculovirus late promoter (91), although the precise mechanism of this inhibition remains unknown. We examined the effects of AcMNPV infection on the transcript levels of several HDAC and sirtuin gene homologs from the T. ni transcriptome (see Fig. S10B in the supplemental material). Of those genes with transcript levels above RPKM values of 5, only two T. ni homologs (encoding HDAC1 and Sirt6) had slightly increased transcript levels at 6 h p.i., and in all cases transcript levels decreased afterwards. Thus, AcMNPV infection appears to generate no exceptional dramatic effect on the transcript levels of HDAC and sirtuins following infection, and all transcripts decrease following infection, as is generally true of host cell transcripts.
Summary.
AcMNPV infection of a permissive cell such as the T. ni cell line Tnms42 leads to a dramatic remodeling of the cell and the production of large quantities of viral mRNA, viral DNA, and viral structural and nonstructural proteins. At the level of the transcriptome, viral transcripts accumulate gradually over the first 6 h p.i. but increase immensely between 6 and 18 h p.i. and continue to increase substantially through 48 h p.i. (Fig. 1A). This overall decrease in host cell transcripts, as a percentage of all transcripts, was also reflected in the patterns observed for each specific gene group analyzed (see Fig. S1 to S10 in the supplemental material). Among the most highly upregulated unigenes, we identified homologs of genes encoding NADH dehydrogenase subunit, cytochrome b, and cytochrome oxidase subunit III (11- to 20-fold upregulated). Unigenes induced to the highest levels after viral infection include homologs of genes for 4-hydroxyphenylpyruvate dioxygenase, peroxidase, NADH dehydrogenase subunit 2, a serine protease, Sprouty, and 3-dehydroecdysone 3α-reductase (Tables 3 and 4). Within the group of the highly upregulated or induced host genes, we noted that several genes encode proteins associated with mitochondria and energy/metabolism, suggesting a changing cellular metabolism as infection progresses. We also cataloged a number of cellular transcripts that were dramatically downregulated or repressed. These include homologs of genes for protease inhibitor 1, asparaginyl-tRNA synthetase, immune-related Hdd13, and heat shock protein 70 (hsp70) genes (Table 5). Whether these changes are directed by the virus or represent cellular defensive responses or simply side effects of viral replication by this virulent pathogen remains to be determined by more detailed studies of these newly identified candidate genes and pathways.
Because the 6-h p.i. time point represents a time prior to the dramatic increases in viral DNA and viral transcripts (Fig. 1A) (33), the initial 6 to 7 h p.i. represents a sufficient period of time to observe early cellular reactions to infection. Our analysis of differentially expressed host transcripts at 0 and 6 h p.i. showed that host genes that were upregulated and induced >10-fold were enriched in the following biological process categories: pattern specification, regionalization, regulation of cell communication, positive regulation of gene expression, lipid localization and transport, and steroid catabolism (see Table S5 in the supplemental material). These categories suggest a variety of cellular responses that include signaling and defensive responses as well as changes related to the overall structural and physiological remodeling that accompanies viral transcription, translation, DNA replication, membrane modifications, and assembly of viral components. Although a dramatic enrichment was not clearly observed from the downregulated and repressed genes, specific examples of sudden and dramatic downregulation were observed for certain transcripts that were highly abundant in uninfected control cells (see Table S7 in the supplemental material).
Of the approximately 436 T. ni gene homologs that we examined as part of functional pathways (see Tables S9 to S20 and Fig. S1 to S10), we identified approximately 15 genes that were substantially upregulated (2- to 3-fold) within the first 6 to 7 h following infection. These included genes for Arp3, Rae1, Bendless/UBC13, cytochrome c, Hsp25.4, superoxide dismutase 2, 10-kDa mitochondrial heat shock protein, R3D1, Nup43-like, Akt/PKB, MAP kinase kinase 3/4/6, MAP kinase kinase 4, LEPR, VPS28, and CHMP2B. While we can only speculate on how the changes in regulation of these genes may contribute to successful viral infection or how they relate to host defensive responses, the identification of these changes will permit many new targeted studies that should lead to a detailed understanding of how the baculovirus and host cell genomes interact in permissive cells. This study and future studies should permit the enhancement of viral infection for applications in biological insect control and for many current and future biomedical and biotechnological applications of baculoviruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jenny Z. Xiang and Ying Shao for help and assistance with Illumina sequencing, and Maria Huang, and Zen Yi for assistance and advice on bioinformatic analysis and approaches. We thank Abigail McSweeney for technical assistance and Lisa Strassheim for comments on the manuscript.
This work was supported by grants from DARPA and the NSF (IOS-1354421) to G.W.B., a fellowship from the Taiwan National Science Council and CUHK direct grant (4053052) to Y.-R.C., and Boyce Thompson Institute project 1255.
Footnotes
Published ahead of print 17 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02243-14.
REFERENCES
- 1. Hitchman RB, Possee RD, Siaterli E, Richards KS, Clayton AJ, Bird LE, Owens RJ, Carpentier DE, King FL, Danquah JO, Spink KG, King LA. 2010. Improved expression of secreted and membrane-targeted proteins in insect cells. Biotechnol. Appl. Biochem. 56:85–93. 10.1042/BA20090130. [DOI] [PubMed] [Google Scholar]
- 2. O'Reilly DR, Miller LK, Luckow VA. 1992. Baculovirus expression vectors, a laboratory manual. W. H. Freeman and Co., New York, NY. [Google Scholar]
- 3. Summers MD, Smith GE. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin no. 1555. [Google Scholar]
- 4. Mena JA, Kamen AA. 2011. Insect cell technology is a versatile and robust vaccine manufacturing platform. Expert Rev. Vaccines 10:1063–1081. 10.1586/erv.11.24. [DOI] [PubMed] [Google Scholar]
- 5. Airenne KJ, Hu YC, Kost TA, Smith RH, Kotin RM, Ono C, Matsuura Y, Wang S, Yla-Herttuala S. 2013. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol. Ther. 21:739–749. 10.1038/mt.2012.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barsoum J, Brown R, McKee M, Boyce FM. 1997. Efficient transduction of mammalian cells by a recombinant baculovirus having the vesicular stomatitis virus G glycoprotein. Hum. Gene Ther. 8:2011–2018. 10.1089/hum.1997.8.17-2011. [DOI] [PubMed] [Google Scholar]
- 7. Xu XG, Wang ZS, Zhang Q, Li ZC, Zhao HN, Li W, Tong DW, Liu HJ. 2011. Baculovirus surface display of E envelope glycoprotein of Japanese encephalitis virus and its immunogenicity of the displayed proteins in mouse and swine models. Vaccine 29:636–643. 10.1016/j.vaccine.2010.11.045. [DOI] [PubMed] [Google Scholar]
- 8. Zhou J, Blissard GW. 2008. Display of heterologous proteins on gp64null baculovirus virions and enhanced budding mediated by a VSV G-stem construct. J. Virol. 82:1368–1377. 10.1128/JVI.02007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Granados RR, Lawler KA. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297–308. 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- 10. Rohrmann GF. 2011. Baculovirus molecular biology, 2nd ed. National Library of Medicine, Bethesda, MD: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=bacvir. [Google Scholar]
- 11. Hoover K, Grove M, Gardner M, Hughes DP, McNeil J, Slavicek J. 2011. A gene for an extended phenotype. Science 333:1401. 10.1126/science.1209199. [DOI] [PubMed] [Google Scholar]
- 12. Katsuma S, Koyano Y, Kang W, Kokusho R, Kamita SG, Shimada T. 2012. The baculovirus uses a captured host phosphatase to induce enhanced locomotory activity in host caterpillars. PLoS Pathog. 8:e1002644. 10.1371/journal.ppat.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Means JC, Passarelli AL. 2010. Viral fibroblast growth factor, matrix metalloproteases, and caspases are associated with enhancing systemic infection by baculoviruses. Proc. Natl. Acad. Sci. U. S. A. 107:9825–9830. 10.1073/pnas.0913582107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Houte S, Ros VI, Mastenbroek TG, Vendrig NJ, Hoover K, Spitzen J, van Oers MM. 2012. Protein tyrosine phosphatase-induced hyperactivity is a conserved strategy of a subset of baculoviruses to manipulate lepidopteran host behavior. PLoS One 7:e46933. 10.1371/journal.pone.0046933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dever TE, Sripriya R, McLachlin JR, Lu J, Fabian JR, Kimball SR, Miller LK. 1998. Disruption of cellular translational control by a viral truncated eukaryotic translation initiation factor 2alpha kinase homolog. Proc. Natl. Acad. Sci. U. S. A. 95:4164–4169. 10.1073/pnas.95.8.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Reilly DR, Miller LK. 1989. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245:1110–1112. 10.1126/science.2505387. [DOI] [PubMed] [Google Scholar]
- 17. Clem RJ, Fechheimer M, Miller LK. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388–1390. 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 18. Ohkawa T, Majima K, Maeda S. 1994. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J. Virol. 68:6619–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Slack JM, Kuzio J, Faulkner P. 1995. Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple necular polyhedrosis virus. J. Gen. Virol. 76:1091–1098. 10.1099/0022-1317-76-5-1091. [DOI] [PubMed] [Google Scholar]
- 20. Hawtin RE, Arnold K, Ayres MD, Zanotto PMDA, Howard SC, Gooday GW, Chappell LH, Kitts PA, King LA, Possee RD. 1995. Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology 212:673–685. 10.1006/viro.1995.1525. [DOI] [PubMed] [Google Scholar]
- 21. Hodgson JJ, Arif BM, Krell PJ. 2011. Interaction of Autographa californica multiple nucleopolyhedrovirus proV-CATH with CHIA as a mechanism for proV-CATH cellular retention. J. Virol. 85:3918–3929. 10.1128/JVI.02165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tjia S, Carstens EB, Doerfler W. 1979. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus. II. The viral DNA and kinetics of its replication. Virology 99:399–409. [DOI] [PubMed] [Google Scholar]
- 23. Theilmann DA, Stewart S. 1991. Identification and characterization of the IE-1 gene of Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 180:492–508. 10.1016/0042-6822(91)90063-H. [DOI] [PubMed] [Google Scholar]
- 24. Charlton CA, Volkman LE. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J. Virol. 65:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190:187–195. 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clem RJ. 2007. Baculoviruses and apoptosis: a diversity of genes and responses. Curr. Drug Targets 8:1069–1074. 10.2174/138945007782151405. [DOI] [PubMed] [Google Scholar]
- 27. Fisher AJ, Cruz W, Zoog SJ, Schneider CL, Friesen PD. 1999. Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J. 18:2031–2039. 10.1093/emboj/18.8.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitchell JK, Friesen PD. 2012. Baculoviruses modulate a proapoptotic DNA damage response to promote virus multiplication. J. Virol. 86:13542–13553. 10.1128/JVI.02246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang N, Wu W, Yang K, Passarelli AL, Rohrmann GF, Clem RJ. 2011. Baculovirus infection induces a DNA damage response that is required for efficient viral replication. J. Virol. 85:12547–12556. 10.1128/JVI.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manji GA, Friesen PD. 2001. Apoptosis in motion: an apical P35-insensitive caspase mediates programmed cell death in insect cells. J. Biol. Chem. 23:23. 10.1074/jbc.M010179200. [DOI] [PubMed] [Google Scholar]
- 31. Ikeda M, Kobayashi M. 1999. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 258:176–188. 10.1006/viro.1999.9706. [DOI] [PubMed] [Google Scholar]
- 32. Braunagel SC, Parr R, Belyavskyi M, Summers MD. 1998. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology 244:195–211. 10.1006/viro.1998.9097. [DOI] [PubMed] [Google Scholar]
- 33. Chen YR, Zhong S, Fei Z, Hashimoto Y, Xiang JZ, Zhang S, Blissard GW. 2013. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) in Trichoplusia ni cells. J. Virol. 87:6391–6405. 10.1128/JVI.00194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ. 2011. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc. 2011:940–949. 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 35. Morgan M, Anders S, Lawrence M, Aboyoun P, Pages H, Gentleman R. 2009. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25:2607–2608. 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29:644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng Y, Zhao L, Gao J, Fei Z. 2011. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12:453. 10.1186/1471-2105-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628. 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Hoon MJ, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20:1453–1454. 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 42. Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prikhod'ko EA, Miller LK. 1998. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 72:684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. 1998. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92:253–263. 10.1016/S0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- 45. Xue J, Qiao N, Zhang W, Cheng RL, Zhang XQ, Bao YY, Xu YP, Gu LZ, Han JD, Zhang CX. 2012. Dynamic interactions between Bombyx mori nucleopolyhedrovirus and its host cells revealed by transcriptome analysis. J. Virol. 86:7345–7359. 10.1128/JVI.07217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernal V, Carinhas N, Yokomizo AY, Carrondo MJ, Alves PM. 2009. Cell density effect in the baculovirus-insect cells system: a quantitative analysis of energetic metabolism. Biotechnol. Bioeng. 104:162–180. 10.1002/bit.22364. [DOI] [PubMed] [Google Scholar]
- 47. Carinhas N, Bernal V, Monteiro F, Carrondo MJ, Oliveira R, Alves PM. 2010. Improving baculovirus production at high cell density through manipulation of energy metabolism. Metab. Eng. 12:39–52. 10.1016/j.ymben.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 48. Nguyen Q, Chan LC, Nielsen LK, Reid S. 2013. Genome scale analysis of differential mRNA expression of Helicoverpa zea insect cells infected with a H. armigera baculovirus. Virology 444:158–170. 10.1016/j.virol.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 49. Palomares LA, López S, Ramírez OT. 2004. Utilization of oxygen uptake rate to assess the role of glucose and glutamine in the metabolism of infected insect cell cultures. Biochem. Eng. J. 19:87–93. 10.1016/j.bej.2003.12.002. [DOI] [Google Scholar]
- 50. Lyupina YV, Zatsepina OG, Timokhova AV, Orlova OV, Kostyuchenko MV, Beljelarskaya SN, Evgen'ev MB, Mikhailov VS. 2011. New insights into the induction of the heat shock proteins in baculovirus infected insect cells. Virology 421:34–41. 10.1016/j.virol.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 51. Lyupina YV, Dmitrieva SB, Timokhova AV, Beljelarskaya SN, Zatsepina OG, Evgen'ev MB, Mikhailov VS. 2010. An important role of the heat shock response in infected cells for replication of baculoviruses. Virology 406:336–341. 10.1016/j.virol.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 52. Salem TZ, Zhang F, Xie Y, Thiem SM. 2011. Comprehensive analysis of host gene expression in Autographa californica nucleopolyhedrovirus-infected Spodoptera frugiperda cells. Virology 412:167–178. 10.1016/j.virol.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Faye I, Pye A, Rasmuson T, Boman HG, Boman IA. 1975. Insect immunity. 11. Simultaneous induction of antibacterial activity and selection synthesis of some hemolymph proteins in diapausing pupae of Hyalophora cecropia and Samia cynthia. Infect. Immun. 12:1426–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sun SC, Lindstrom I, Boman HG, Faye I, Schmidt O. 1990. Hemolin: an insect-immune protein belonging to the immunoglobulin superfamily. Science 250:1729–1732. 10.1126/science.2270488. [DOI] [PubMed] [Google Scholar]
- 55. Ladendorff NE, Kanost MR. 1990. Isolation and characterization of bacteria-induced protein P4 from hemolymph of Manduca sexta. Arch. Insect Biochem. Physiol. 15:33–41. 10.1002/arch.940150104. [DOI] [PubMed] [Google Scholar]
- 56. Lee KY, Horodyski FM, Valaitis AP, Denlinger DL. 2002. Molecular characterization of the insect immune protein hemolin and its high induction during embryonic diapause in the gypsy moth, Lymantria dispar. Insect Biochem. Mol. Biol. 32:1457–1467. 10.1016/S0965-1748(02)00066-8. [DOI] [PubMed] [Google Scholar]
- 57. Faye I, Kanost M. 1998. Function and regulation of hemolin, p 173–188 In Brey PT, Hultmark D. (ed), Molecular mechanisms of immune responses in insects. Chapman & Hall, London, United Kingdom. [Google Scholar]
- 58. Hirai M, Terenius O, Li W, Faye I. 2004. Baculovirus and dsRNA induce hemolin, but no antibacterial activity, in Antheraea pernyi. Insect Mol. Biol. 13:399–405. 10.1111/j.0962-1075.2004.00497.x. [DOI] [PubMed] [Google Scholar]
- 59. Lundström A, Kang D, Liu G, Fernandez C, Warren JT, Gilbert LI, Steiner H. 2002. A protein from the cabbage looper, Trichoplusia ni, regulated by a bacterial infection is homologous to 3-dehydroecdysone 3β-reductase. Insect Biochem. Mol. Biol. 32:829–837. 10.1016/S0965-1748(01)00145-X. [DOI] [PubMed] [Google Scholar]
- 60. O'Reilly DR, Brown MR, Miller LK. 1992. Alteration of ecdysteroid metabolism due to baculovirus infection of the fall armyworm Spodoptera frugiperda host ecdysteroids are conjugated with galactose. Insect Biochem. Mol. Biol. 22:313–320. 10.1016/0965-1748(92)90069-Q. [DOI] [Google Scholar]
- 61. O'Reilly DR, Miller LK. 1990. Regulation of expression of a baculovirus ecdysteroid UDPglucosyltransferase gene J. Virol. 64:1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Long G, Pan X, Kormelink R, Vlak JM. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830–8833. 10.1128/JVI.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hefferon K, Oomens A, Monsma S, Finnerty C, Blissard GW. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455–468. 10.1006/viro.1999.9758. [DOI] [PubMed] [Google Scholar]
- 64. Volkman LE, Goldsmith PA. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143:185–195. 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- 65. Mueller J, Pfanzelter J, Winkler C, Narita A, Le Clainche C, Nemethova M, Carlier MF, Maeda Y, Welch MD, Ohkawa T, Schmeiser C, Resch GP, Small JV. 2014. Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 12:e1001765. 10.1371/journal.pbio.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D'Alessio JA, Cande WZ, Volkman LE, Welch MD. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467. 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- 67. Au S, Panté N. 2012. Nuclear transport of baculovirus: revealing the nuclear pore complex passage. J. Struct. Biol. 177:90–98. 10.1016/j.jsb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 68. Au S, Wu W, Pante N. 2013. Baculovirus nuclear import: open, nuclear pore complex (NPC) sesame. Viruses 5:1885–1900. 10.3390/v5071885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Z, Blissard GW. 2012. Cellular VPS4 is required for efficient entry and egress of budded virions of Autographa californica multiple nucleopolyhedrovirus. J. Virol. 86:459–472. 10.1128/JVI.06049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McCullough J, Colf LA, Sundquist WI. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82:663–692. 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. 2011. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci. 36:199–210. 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 72. Clem RJ, Popham HJR, Shelby KS. 2010. Antiviral responses in insects: apoptosis and humoral responses, p 453 In Asgari S, Johnson KN. (ed), Insect virology. Caister Academic Press, Poole, United Kingdom. [Google Scholar]
- 73. Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060. 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ding S-W. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644. 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 75. Fulda S, Gorman AM, Hori O, Samali A. 2010. Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010:214074. 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bronkhorst AW, van Cleef KWR, Vodovar N, İnce İA, Blanc H, Vlak JM, Saleh M-C, van Rij RP. 2012. The DNA virus invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. U. S. A. 109:E3604–E3613. 10.1073/pnas.1207213109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jayachandran B, Hussain M, Asgari S. 2012. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86:13729–13734. 10.1128/JVI.02041-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li T-C, Scotti PD, Miyamura T, Takeda N. 2007. Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 81:10890–10896. 10.1128/JVI.00807-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ahmad M, Srinivasula SM, Wang L, Litwack G, Fernandes-Alnemri T, Alnemri ES. 1997. Spodoptera frugiperda caspase-1, a novel insect death protease that cleaves the nuclear immunophilin FKBP46, is the target of the baculovirus antiapoptotic protein p35. J. Biol. Chem. 272:1421–1424. 10.1074/jbc.272.3.1421. [DOI] [PubMed] [Google Scholar]
- 80. Means JC, Clem RJ. 2008. Evolution and function of the p35 family of apoptosis inhibitors. Future Virol. 3:383–391. 10.2217/17460794.3.4.383. [DOI] [Google Scholar]
- 81. West AP, Shadel GS, Ghosh S. 2011. Mitochondria in innate immune responses. Nat. Rev. Immunol. 11:389–402. 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Y, Oberley LW, Murhammer DW. 2001. Evidence of oxidative stress following the viral infection of two lepidopteran insect cell lines. Free Radic. Biol. Med. 31:1448–1455. 10.1016/S0891-5849(01)00728-6. [DOI] [PubMed] [Google Scholar]
- 83. Wang Y, Oberley LW, Howe D, Jarvis DL, Chauhan G, Murhammer DW. 2004. Effect of expression of manganese superoxide dismutase in baculovirus-infected insect cells. Appl. Biochem. Biotechnol. 119:181–193. 10.1385/ABAB:119:2:181. [DOI] [PubMed] [Google Scholar]
- 84. Tomalski MD, Eldridge R, Miller LK. 1991. A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology 184:149–161. 10.1016/0042-6822(91)90831-U. [DOI] [PubMed] [Google Scholar]
- 85. Kounatidis I, Ligoxygakis P. 2012. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2:120075. 10.1098/rsob.120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kanost MR, Nardi JB. 2010. Innate immune responses of Manduca sexta. In Goldsmith MR, Marec F. (ed), Molecular biology and genetics of the Lepidoptera. CRC Press, Boca Raton, FL. [Google Scholar]
- 87. Tanaka H, Ishibashi J, Fujita K, Nakajima Y, Sagisaka A, Tomimoto K, Suzuki N, Yoshiyama M, Kaneko Y, Iwasaki T, Sunagawa T, Yamaji K, Asaoka A, Mita K, Yamakawa M. 2008. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 38:1087–1110. 10.1016/j.ibmb.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 88. Cuevas-Bennett C, Shenk T. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525–9536. 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Herbein G, Wendling D. 2010. Histone deacetylases in viral infections. Clin. Epigenet. 1:13–24. 10.1007/s13148-010-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Guise AJ, Budayeva HG, Diner BA, Cristea IM. 2013. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses 5:1607–1632. 10.3390/v5071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Peng Y, Song J, Lu J, Chen X. 2007. The histone deacetylase inhibitor sodium butyrate inhibits baculovirus-mediated transgene expression in Sf9 cells. J. Biotechnol. 131:180–187. 10.1016/j.jbiotec.2007.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.