Abstract
Leptospirosis is a zoonosis caused by pathogenic Leptospira spp. Most of the outbreaks of leptospirosis occur after floods caused by heavy rain in countries where Leptospira spp. are endemic. It has been believed that the overflow of seawater rarely causes outbreaks of leptospirosis because the leptospires are killed by salt water. On 8 November 2013, a storm surge caused by Super Typhoon Haiyan (Yolanda) inundated the entire coastal areas of Tacloban and Palo in Leyte, Philippines. The present study was carried out in order to determine whether the environmental leptospires in soil were able to survive after the storm surge in the affected areas. We collected 23 wet soil samples along the coastal areas of Tacloban and Palo 2 months after the storm surge. The samples were suspended in HEPES buffer, and the supernatants were cultured in liquid or semisolid Korthof's medium supplemented with five antimicrobial agents to inhibit the growth of contaminants. Leptospires were isolated from primary cultures of 22 out of 23 samples. The DNA of pathogenic Leptospira species was detected in 11 samples (47.8%) by analysis of flaB by nested PCR. Eventually, two pathogenic Leptospira strains were isolated and showed the highest 16S rRNA gene sequence similarity to Leptospira kmetyi. When these isolates were experimentally mixed with soil, they were found to survive in seawater for 4 days. These results show the possibility that leptospires living in soil survived after the storm surge. Our findings may serve as a warning that when seawater inundates the land during a storm surge or a tsunami, an outbreak of leptospirosis could occur in the disaster-stricken area.
INTRODUCTION
Leptospirosis is a zoonosis caused by pathogenic Leptospira spp. The spectrum of symptoms of leptospirosis is extremely broad. The great majority of leptospiral infections are either subclinical or of very mild severity and are accompanied by fever, chills, headache, myalgia, abdominal pain, and conjunctival hyperemia (1). Weil's disease is the most severe form of human leptospirosis, which is characterized by jaundice, hemorrhage, and renal failure (1, 2). More than 50,000 cases of severe leptospirosis are reported each year, with the case fatality rate being higher than 10% (3).
Leptospira species are aerobic, Gram-negative bacteria that are long, thin, flexible rods with a regular right-handed helical coiling pattern (4). The genus Leptospira is classified into 21 species on the basis of 16S rRNA sequence comparisons and DNA-DNA hybridization studies. Phylogenetic tree analysis based on the 16S rRNA sequence shows that Leptospira species are composed of three clusters: pathogenic, intermediate-pathogenic, and nonpathogenic (saprophytic) species.
The pathogenic leptospires infect the renal tubules of reservoir animals (e.g., rats) and are shed through rat urine into the environment, where they can survive in moist soil and surface water for up to several months (5, 6). Humans and other animals become infected mainly through their skin and mucous membranes when they encounter a leptospire-contaminated environment (7). Due to this correlation between organisms in the environment and disease transmission, isolation of pathogenic leptospires from the environment is important for epidemiological studies. However, isolation from environmental samples is usually unsuccessful due to the slow growth of leptospires and the overgrowth of coexisting microorganisms. We previously reported a novel combination of antimicrobial agents (sulfamethoxazole, trimethoprim, amphotericin B, fosfomycin, and 5-fluorouracil [STAFF]) for the selective isolation of leptospires from contaminated samples (8). After this cocktail was incorporated into Leptospira growth medium, it inhibited the growth of contaminants and successfully isolated pathogenic leptospires as well as saprophytic species from environmental samples (9). Using this isolation method and pulsed-field gel electrophoresis (PFGE) analysis, we demonstrated that environmental leptospires survived in the wet soil on dry days and appeared in the surface water on rainy days (9). It is thought that the soil could be a reservoir for leptospires.
The salt resistance of bacteria is usually evaluated by determination of resistance to sodium chloride. Trueba et al. (6) previously reported that Leptospira survived for only short periods of time in 0.85% NaCl solution. It is well-known that the vast majority of seawater has a salinity of approximately 3%. Therefore, it is believed that the penetration of seawater rarely causes the outbreak of leptospirosis, as the leptospires in the environment are killed by salt water.
On 8 November 2013, Typhoon Haiyan, called Typhoon Yolanda in the Philippines, made landfall in Eastern Visayas and caused considerable damage to property and the loss of lives mostly in Leyte Province. According to the National Disaster Risk Reduction and Management Council (NDRRMC), the maximum wind speed and central pressure of this super typhoon at the time of its landfall were 87.5 m/s and 895 hPa, respectively. Tacloban is the largest city in Leyte Province and is low lying, with most parts of the city being at an elevation of less than 10 ft. While strong winds were the predominant cause of physical damage to buildings and infrastructure, the storm surge caused a massive inundation of the entire city of Tacloban and caused severe damage in the affected areas. The height of the storm surge in Tacloban ranged from 4.6 to 7.0 m (10).
The Philippines is a country where leptospirosis is endemic. Leptospirosis outbreaks are usually reported during the rainy season (June to November) and right after the rainy season in flood-prone areas (11). The present study was carried out in order to clarify whether environmental leptospires are able to survive after a storm surge in a country where leptospirosis is endemic. We collected soil samples along the coastal areas in Tacloban and its adjoining municipality, Palo. These were two of the areas worst hit by the storm surge. The samples were analyzed using isolation methods and PCR analysis.
MATERIALS AND METHODS
Procedure for isolation of Leptospira species from environmental samples.
Environmental soil samples were collected in Tacloban and Palo in Leyte Province, Philippines, on 28 and 29 January 2014. These were two of the areas hardest hit by the storm surge during Typhoon Haiyan, or Yolanda (in the vernacular), on 8 November 2013. Soil samples were collected at the coastal areas that were inundated for several minutes or several days by seawater during the super typhoon.
Approximately 10 g of wet soil was collected and placed in sterile 15-ml screw-cap tubes. In order to keep the pH of the samples at about 7, 10 ml of 20 mM HEPES buffer (pH 7.4) was added and mixed into each soil sample. All the tubes were kept in a vertical position for 1 h to allow the sample to settle. After checking the pH of the supernatant using a urine pH test kit (Nippon Chemiphar, Tokyo, Japan), 2 ml was transferred to 2.5 ml 2×-concentrated liquid and semisolid Korthof's media supplemented with 500 μl of 10×-concentrated STAFF (sulfamethoxazole, 400 μg/ml; trimethoprim, 200 μg/ml; amphotericin B, 50 μg/ml; fosfomycin, 4 mg/ml; 5-fluorouracil, 1 mg/ml) (8). These tubes were incubated at 30°C for 28 days and checked daily by dark-field microscopy for the presence of Leptospira, which was confirmed by observing the characteristic thin helical structures with prominent hooked ends and motility. When the presence of Leptospira had been confirmed microscopically, primary cultures were filtered using a 0.2-μm-pore-size membrane filter to remove the contaminants. The filtrate (0.5 ml) was added to 4.5 ml fresh Korthof's medium or semisolid medium without STAFF and cultured at 30°C.
For single-colony isolation, solid medium was prepared by incorporating 1% (wt/vol) agar in the liquid Korthof's medium, and 0.1 ml of diluted leptospiral pure culture was inoculated by evenly spreading the sample over the surface of the medium, as previously described (9). After incubation for 6 to 15 days, five single colonies were picked up with Pasteur pipettes, transferred into fresh liquid Korthof's medium, and incubated at 30°C (9). Leptospires from these single colonies were characterized through the molecular techniques described in the succeeding sections.
Enzyme digestion and PFGE.
PFGE analysis was performed as previously reported (9). Briefly, the bacterial suspension was mixed with melted 1% SeaKem Gold agarose (Lonza, Rockland, ME) to make agarose plugs. The plugs were incubated in 500 mM EDTA (pH 8.0) containing 1 mg of proteinase K per ml and 1% N-lauroylsarcosine at 50°C for 1 h by mixing at 15-min intervals. The plugs were subsequently transferred to phenylmethylsulfonyl fluoride (PMSF) solution (40 μg/ml in Tris-EDTA [TE] buffer, pH 7.6), and the mixture was incubated twice at 50°C for 30 min each time. Then, the plugs were washed once with TE buffer (pH 8.0). The DNA extracted from each agarose block prepared as described above was digested with 40 units of NotI (TaKaRa Bio Inc., Otsu, Shiga, Japan) in a 37°C incubator overnight. PFGE was performed using a contour-clamped homogeneous electric field apparatus (CHEF-DR II apparatus; Bio-Rad Laboratories Inc., Hercules, CA). Electrophoresis was performed at 14°C for 20 h at 6.0 V/cm with a ramped pulse time of 10 to 60 s. After electrophoresis, the gels were stained with 1 mg/liter of ethidium bromide. DNA bands were visualized on a UV transilluminator and were photographed through a red filter.
DNA extraction.
Bacterial DNA was extracted from the supernatant of the soil sample suspension, the primary culture before filtration, and the confluent pure culture of Leptospira isolates using an illustra bacteria genomicPrep Mini Spin kit (GE Healthcare, Buckinghamshire, United Kingdom) following the protocol designated for Gram-negative bacteria.
23S rRNA gene PCR.
The 23S rRNA gene PCR was reported by Woo et al. (12) to be capable of detecting all of Leptospira species. For the 23S rRNA gene PCR, the primers rrl-F (5′-GACCCGAAGCCTGTCGAG-3′) and rrl-R (5′-GCCATGCTTAGTCCCGATTAC-3′) were used. The PCR solution (50 μl) contained 1× Ex Taq buffer (TaKaRa, Otsu, Shiga, Japan), 100 μM each deoxynucleoside triphosphate, 0.25 μM each universal primer, 100 ng of extracted DNA, and 1.25 units of Ex Taq HS DNA polymerase (TaKaRa, Otsu, Shiga, Japan). Leptospiral DNA from the samples were amplified in a thermal cycler (PC-320 program temperature control system; ASTEC Co. Ltd., Fukuoka, Japan) under the following conditions: 30 cycles of 94°C for 20 s, 54°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 6 min. DNA amplification was confirmed by gel (1.5%) electrophoresis.
flaB nested PCR.
Koizumi et al. (13) reported that the flaB nested PCR was capable of detecting pathogenic Leptospira strains. For the first PCR, the primers L-flaB-F1 (5′-CTCACCGTTCTCTAAAGTTCAAC-3′) and L-flaB-R1 (5′-TGAATTCGGTTTCATATTTGCC-3′) were used. Amplification was carried out under the following conditions: an initial cycle of 1 min at 94°C and 25 cycles of 94°C for 10 s, 50°C for 30 s, and 72°C for 1 min. Next, 1 μl of the first-round PCR product was added to 19 μl second-round PCR mixture. For the second PCR, the primers L-flaB-F2 (5′-TGTGCACAAGACGATGAAAGC-3′) and L-flaB-R2 (5′-AACATTGCCGTACCACTCTG-3′) were used. PCR amplification consisted of an initial cycle of 1 min at 94°C, followed by 30 cycles of 10 s at 94°C, 30 s at 55°C, and 50 s at 72°C. A final extension was performed for 7 min at 72°C. PCR amplification was confirmed by electrophoresis on 1.5% agarose gels.
16S rRNA gene sequence determination and phylogenetic analysis.
In order to differentiate the species of the leptospiral isolates, 16S rRNA gene sequences were analyzed as previously described (9). The sequences of the other Leptospira species used for alignment and for calculating levels of homology were obtained from GenBank. Phylogenetic distances were calculated using the neighbor-joining method. The phylogenetic tree was constructed using TREEVIEW software (14).
Test of pathogenicity of isolates in golden Syrian hamsters.
The pathogenicity of the isolates was tested in 4-week-old male golden Syrian hamsters (Japan SLC, Inc., Shizuoka, Japan). The hamsters were inoculated intraperitoneally with 1 × 108 cells of Leptospira isolates in 1 ml of phosphate-buffered saline (PBS), pH 7.4, and observed for 28 days. Blood, urine, liver, kidney, spleen, and lung samples from sacrificed hamsters were cultured in Korthof's medium.
The animal experiments were reviewed and approved by the Ethics Committee on Animal Experiments at the Faculty of Medical Sciences, Kyushu University. The experiments were carried out under the conditions indicated in the Regulations for Animal Experiments of Kyushu University and law 105 and notification 6 of the government of Japan.
Test of isolate survival during exposure to salt solution.
Five milliliters of a confluent culture (1 × 108/ml) of isolates was centrifuged at 4,000 × g for 20 min, and the supernatant was thoroughly removed. The bacteria were washed three times with PBS, resuspended in 5 ml of sterile PBS, 3% sodium chloride solution, artificial seawater (Daigo's artificial seawater SP for marine microalgae medium; Nihon Pharmaceutical Co. Ltd., Tokyo, Japan), or natural seawater collected from Hakata Bay, Fukuoka, Japan, and incubated at room temperature. At 0, 0.5, 3, 12, and 24 h and 2, 3, 4, and 6 days after exposure to salt solution, 100 μl of cell suspension was taken at each time point and centrifuged. The pellet from the suspension was washed with PBS and resuspended in 4 ml of Korthof's medium. The cultures were incubated at 30°C for 30 days. Culturable live cells were detected by checking the cultures daily under a dark-field microscope for the presence of Leptospira spirochetes, which was confirmed by observing their characteristic thin helical structures with prominent hooked ends and motility.
Test of isolate survival in soil during exposure to artificial seawater.
Five milliliters of confluent culture (1 × 108/ml) of isolates was centrifuged, washed with PBS three times, and resuspended in 5 ml of sterile PBS. One milliliter of bacterial suspension was mixed with 2 g of autoclaved soil in quadruplicate, and the mixture was incubated at room temperature overnight. Then, 9 ml of artificial seawater was added to each soil sample containing leptospires and mixed well. The samples were kept in a vertical position and incubated at room temperature. At 0, 2, 3, 4, and 6 days after exposure to seawater, one soil sample at a time was washed with PBS three times, resuspended in 5 ml of Korthof's medium, and incubated at 30°C for 30 days. The cultures were checked daily to determine the presence of Leptospira bacteria. To evaluate the role of soil in the survival of leptospires, 1 ml of bacterial suspension was mixed with 9 ml of artificial seawater or PBS without soil, and the mixture was incubated at room temperature in quadruplicate. Culturable live cells were detected as described above.
Nucleotide sequence accession numbers.
The sequences of isolates MS422 and MS432 obtained in this study have been deposited in GenBank and can be found under accession numbers AB937993 and AB937994, respectively.
RESULTS
Primary culture of coastal soil and detection of pathogenic leptospiral DNA.
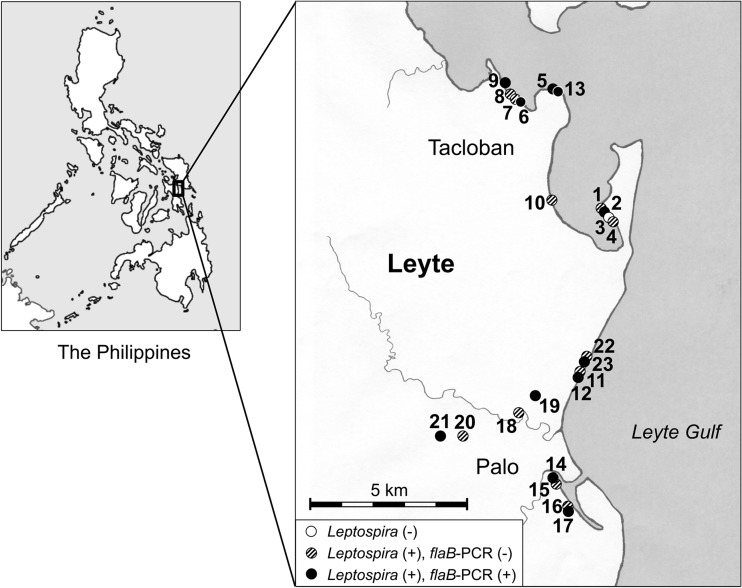
A total of 23 samples (Leyte environmental sample 1 [LES-1] to LES-23) were collected in Tacloban and Palo in Leyte Province, Philippines, 2 months after the storm surge. The locations of the sites where the soil samples were collected were plotted on a map using a geographic information system (GIS) (Fig. 1). Some publications have reported that Leptospira cannot survive for long periods under mildly acidic conditions (15–17). We previously confirmed that the number of motile Leptospira spirochetes gradually decreased after a 10-h exposure to pH 6, while the Leptospira spirochetes survived at pH 7.4 (18). Therefore, we used 20 mM HEPES buffer (pH 7.4) to suspend the collected soil samples. This buffer was able to maintain the pH of all samples at 6.5 to 7.6. In order to analyze the samples, we first performed flaB nested PCR for the detection of pathogenic Leptospira species in the supernatant before starting culture. We found that all the samples were PCR negative (Table 1). During the first 7 days after the initiation of culture in liquid and semisolid Korthof's media, the spirochetes were found by dark-field microscopy in 22 out of 23 samples. These 22 samples were spirochete positive in both liquid and semisolid culture media. For each sample, bacterial DNA was extracted from the mixture of equal parts of the primary liquid culture and the semisolid culture, and rrl PCR was performed. All the spirochete-positive cultures were found to be rrl PCR positive. This result indicated that 22 samples contained organisms which belonged to the genus Leptospira (Table 1; Fig. 1). We then performed a flaB nested PCR to detect the DNA of pathogenic Leptospira strains and found 11 samples to be positive. These data revealed that 11 out of 23 samples (48%) contained culturable pathogenic Leptospira bacteria (Table 1; Fig. 1).
FIG 1.
Locations where the soil samples were collected and results of Leptospira culture and flaB PCR. A total of 23 samples were collected in Tacloban and Palo in Leyte Province, Philippines. The locations of the sites where the soil samples were collected are plotted on the map. Open circle, the primary culture of the collected sample contained no leptospires (1 out of 23 samples; 4%); shaded circles, the primary culture of the collected samples contained leptospires but no DNA of pathogenic Leptospira strains (11 out of 23 samples; 48%); closed circles, the primary culture of the collected samples contained leptospires, and the DNA of pathogenic Leptospira strains was detected (11 out of 23 samples; 48%).
TABLE 1.
Results of culture and PCR of 23 soil samples
| Sample no.a | Resultb |
||||||
|---|---|---|---|---|---|---|---|
| flaB PCR of supernatant of soil suspension | Primary culture |
Subculture of filtrate |
Growth of Leptospira colony by single-colony isolation | ||||
| Growth of Leptospira | rrl PCRc | flaB PCRc | Growth of Leptospira | flaB PCR | |||
| LES-1 | − | L+, S+ | + | − | L+, S− | L− | |
| LES-2 | − | L+, S+ | + | + | L+, S− | L− | |
| LES-3 | − | L+, S+ | + | − | L−, S− | ||
| LES-4 | − | L−, S− | − | − | |||
| LES-5 | − | L+, S+ | + | + | L+, S+ | L−, S+ | S+ |
| LES-6 | − | L+, S+ | + | + | L−, S− | ||
| LES-7 | − | L+, S+ | + | − | L−, S+ | S− | |
| LES-8 | − | L+, S+ | + | − | L+, S− | L− | |
| LES-9 | − | L+, S+ | + | + | L−, S− | ||
| LES-10 | − | L+, S+ | + | − | L−, S− | ||
| LES-11 | − | L+, S+ | + | − | L+, S+ | L−, S− | |
| LES-12 | − | L+, S+ | + | + | L+, S+ | L+, S+ | L+, S+ |
| LES-13 | − | L+, S+ | + | + | L−, S− | ||
| LES-14 | − | L+, S+ | + | + | L−, S+ | S− | |
| LES-15 | − | L+, S+ | + | − | L−, S− | ||
| LES-16 | − | L+, S+ | + | − | L+, S− | L− | |
| LES-17 | − | L+, S+ | + | + | L−, S− | ||
| LES-18 | − | L+, S+ | + | − | L+, S− | L− | |
| LES-19 | − | L+, S+ | + | + | L+, S− | L− | |
| LES-20 | − | L+, S+ | + | − | L+, S− | L− | |
| LES-21 | − | L+, S+ | + | + | L+, S+ | L−, S− | |
| LES-22 | − | L+, S+ | + | − | L−, S− | ||
| LES-23 | − | L+, S+ | + | + | L−, S− | ||
LES, Leyte environmental sample.
L, culture using liquid Korthof's medium; S, culture using semisolid Korthof's medium.
Bacterial DNA was extracted from a mixture of equal volumes of liquid Korthof's medium and semisolid Korthof's medium.
Colony isolation of pathogenic leptospires from primary culture.
Since all the Leptospira-positive cultures contained contaminants, the primary cultures were filtered using a 0.2-μm-pore-size membrane filter. The filtrate was added to fresh liquid or semisolid Korthof's medium and incubated. Leptospires were recovered from 13 out of 22 samples using either liquid or semisolid medium (Table 1). To determine whether these pure cultures of Leptospira contained pathogenic strains or not, a flaB nested PCR was done on the DNA extracted from the pure cultures. One liquid (L) culture (LES-12-L) and two semisolid (S) cultures (LES-5-S, LES-12-S) originating from two samples (LES-5, LES-12) were PCR positive (Table 1). We previously reported that Leptospira-positive environmental samples sometimes contain more than one Leptospira strain (9). Therefore, single-colony isolation was done on the 3 positive bacterial cultures (LES-5-S, LES-12-L, LES-12-S) using solid medium. After subsurface colonies appeared, five single colonies were picked up from each plate and cultured in fresh liquid Korthof's medium. A total of 15 colonies were obtained, and all were found to be flaB PCR positive (Table 1 and Fig. 1).
PFGE analysis of pathogenic Leptospira isolates.
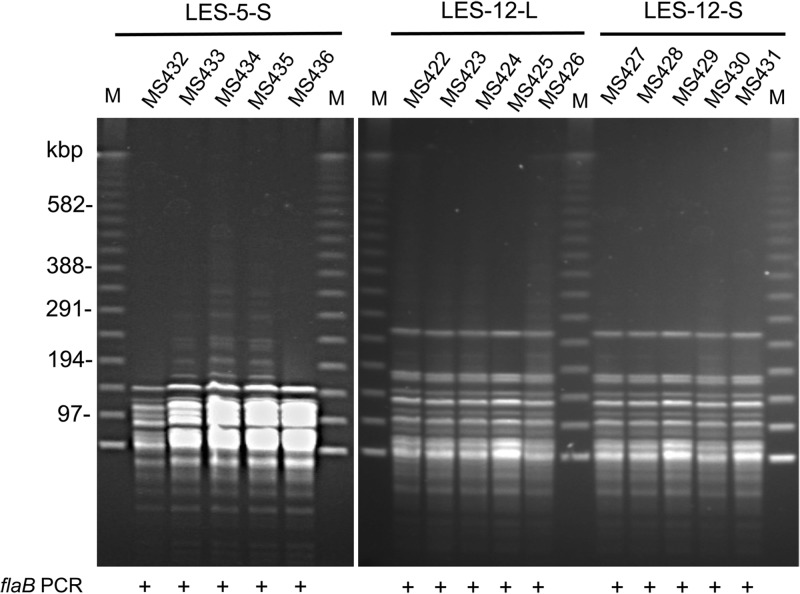
Five strains from three individual pathogenic Leptospira-positive cultures showed identical fingerprint patterns (Fig. 2). The pathogenic leptospires of sample LES-12 were isolated from both liquid medium (LES-12-L) and semisolid medium (LES-12-S). Five isolates from LES-12-L and five isolates from LES-12-S showed identical fingerprint patterns (Fig. 2). A total of two strains of pathogenic Leptospira (here referred to as strains MS432 and MS422) were isolated from two soil samples (LES-5 and LES-12, respectively).
FIG 2.
PFGE fingerprint patterns of pathogenic Leptospira isolates. Single-colony isolation was done on the 3 flaB PCR-positive Leptospira cultures (LES-5-S, LES-12-L, and LES-12-S) using solid medium. Five single colonies were picked up from each plate and cultured in fresh liquid Korthof's medium. A total of 15 strains were obtained, and all the strains were found to be flaB PCR positive. Chromosomal DNA of the strains was digested with the NotI restriction endonuclease. The five isolates from each of the three pathogenic leptospiral cultures showed identical fingerprint patterns. The pathogenic leptospires from sample LES-12 were isolated from both liquid medium (LES-12-L) and semisolid medium (LES-12-S). The five isolates from LES-12-L and the five isolates from LES-12-S showed identical fingerprint patterns. Lanes M, 100-bp size markers.
16S rRNA gene sequence determination and phylogenetic analysis.
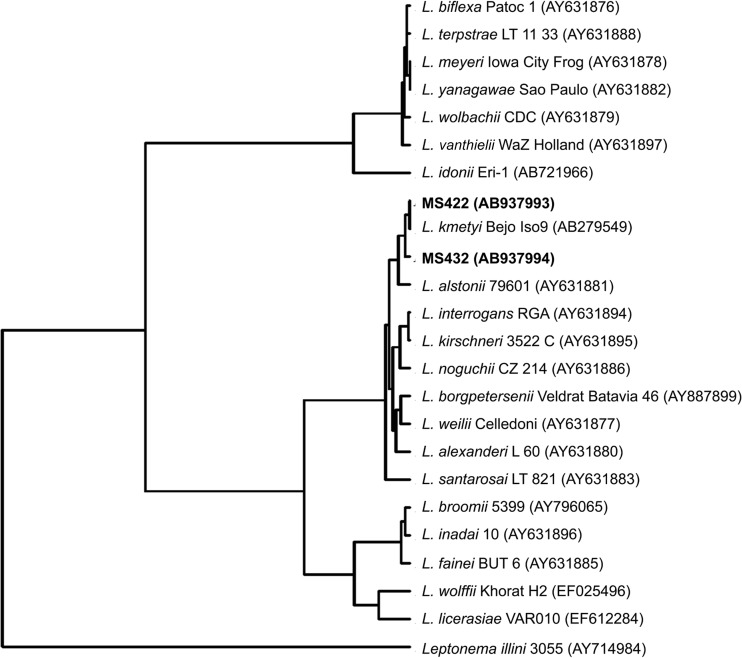
For genomospecies identification, almost full-length 16S rRNA gene sequences of two isolates of pathogenic species (MS432 and MS422) were determined. A phylogenetic tree was constructed on the basis of the 16S rRNA gene sequences of these 2 isolates and type strains of 21 Leptospira genomospecies deposited in GenBank (Fig. 3). The two isolates showed the highest 16S rRNA gene sequence similarity to Leptospira kmetyi.
FIG 3.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequence analysis showing the phylogenetic positions of two isolates from this study (MS422 and MS432). The tree was rooted with the 16S rRNA gene of Leptonema illini strain 3055 (GenBank accession number AY714984). GenBank accession numbers are shown in parentheses.
Pathogenicity of isolates in golden Syrian hamsters.
To examine the pathogenicity of strains MS432 and MS422, hamsters in groups consisting of three hamsters each were inoculated intraperitoneally with 1 × 108 cells of each isolate in 1 ml of PBS and observed for 28 days. No hamsters showed any symptoms of leptospirosis or died after infection. Blood, urine, liver, kidney, spleen, and lung samples from hamsters sacrificed on day 28 after infection were cultured. Leptospires were not recovered from any of the specimens or organs of the infected hamsters.
Survival of isolates belonging to pathogenic species after experimental exposure to salt solution.
Two strains (MS432 and MS422) were tested for their tolerance to salt. After exposure to 3% sodium chloride solution, which possesses the same osmotic pressure as seawater, the isolates were killed within 12 h. In contrast, the isolates survived in artificial seawater for 3 to 4 days (Table 2). We also tested the natural seawater collected from Hakata Bay and obtained similar results (data not shown). Contrary to the common perception, it was revealed that the Leptospira isolates could survive in seawater, but not in sodium chloride solution, for 3 days.
TABLE 2.
Test of isolate survival after exposure to salty solution
| Strain | Salt solution | Survival after the following period of exposure to salt solutiona: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 0.5 h | 3 h | 12 h | 24 h | 2 days | 3 days | 4 days | 6 days | ||
| MS422 | PBS | + | ND | ND | ND | ND | + | ND | + | + |
| 3% NaCl | + | + | + | − | − | − | ND | ND | ND | |
| Seawater | + | + | + | + | + | + | + | + | − | |
| Soil + seawater | + | ND | ND | ND | ND | + | ND | + | − | |
| MS432 | PBS | + | ND | ND | ND | ND | + | ND | + | + |
| 3% NaCl | + | + | + | − | − | − | ND | ND | ND | |
| Seawater | + | + | + | + | + | + | + | − | − | |
| Soil + seawater | + | ND | ND | ND | ND | + | ND | + | − | |
+, culturable live cells were detected; −, culturable live cells were not detected; ND, not done.
In order to replicate the actual situation during the storm surge that occurred in Eastern Visayas in November 2013, each strain (MS432 and MS422) was mixed with autoclaved soil and exposed to artificial seawater. The isolates in soil with seawater survived for 4 days (Table 2). These data demonstrate that the leptospires living in soil became more resistant to seawater.
DISCUSSION
The ecology of environmental leptospires remains to be fully elucidated. In this study, leptospires were isolated from almost all the investigated sites in 2 of the areas hardest hit by Super Typhoon Haiyan. The DNA of pathogenic Leptospira species was detected in about half of the soil samples. It is suggested that leptospires are ubiquitous in the Philippines and are soil bacteria. Based on our results, we hypothesized that soil serves as an important reservoir of pathogenic Leptospira spirochetes, which may cause outbreaks of leptospirosis during floods caused by typhoons and/or heavy rains. Therefore, it is expected that walking through the mud without protective footwear during farm work or water-based recreation or while wading after a flood would place a person at high risk for acquiring infection with Leptospira.
The following are considered to be the reasons for the high rate of detection of pathogenic leptospires in this study. (i) HEPES buffer was used to suspend the soil samples in order to keep the pH at about 7. (ii) A combination of five antimicrobial agents (abbreviated STAFF) was used to inhibit the growth of contaminants and successfully allowed leptospires to grow from contaminated soil samples. (iii) The DNA of Leptospira isolates was extracted from the primary culture at a time when the number of leptospires increased. (iv) To increase the sensitivity and specificity of flaB PCR, nested PCR was performed to detect the DNA of pathogenic Leptospira species. According to the data from our preliminary experiment, the detection limit of the flaB nested PCR was 102 cells of pathogenic Leptospira spp. per ml. It is suggested that the number of pathogenic Leptospira cells in the supernatant of the soil suspension was less than 1 × 102/ml, but their number exceeded the detection limit after use of an enrichment culture method.
Until now, it was believed that leptospires are killed after exposure to seawater because the organisms have a low tolerance for salinity. This hypothesis is probably based on the findings of a previous study concerning the low tolerance of Leptospira to sodium chloride (6). The present study confirmed that the Leptospira isolates were killed within several hours by sodium chloride solution alone. However, the isolates were found to survive in seawater for approximately 3 days. It is known that seawater contains not only Na+ (0.469 mol/kg) and Cl− (0.546 mol/kg) but also Mg2+ (0.0528 mol/kg), SO42− (0.0282 mol/kg), Ca2+ (0.0103 mol/kg), K+ (0.0102 mol/kg), and other microelement ions (19). Salts other than NaCl could support the survival of Leptospira in seawater. Moreover, the isolates mixed with soil were found to survive in seawater for 4 days. If it rains within 4 days after a storm surge, the salt in soil will be diluted by the infiltration of rainwater; therefore, environmental leptospires may be kept alive in soil. The environmental leptospires were actually isolated at a high frequency from soil samples in the coastal areas of 2 of the places that were hardest hit by the storm surge. These results suggest that leptospires living in soil are more resistant to seawater, and there is a possibility that they survived during the storm surge. There is another possibility that the soil became recontaminated from extraneous sources over the 2.5 months after the typhoon struck. However, since we did not perform the survey before or immediately after the typhoon, it is impossible to determine the real situation.
After the storm surge, there were reports of leptospirosis cases (clinical diagnosis) in Leyte and Samar Provinces. In Leyte alone, the numbers of suspected leptospirosis cases reported by WHO in 2013 (20) were 49 on 17 November, 270 on 24 November, 80 on 1 December, 21 on 8 December, and 8 on 15 December. In this study, two isolates (MS432 and MS422) were considered to belong to the pathogenic species L. kmetyi but were found to be avirulent for hamsters. Initially, L. kmetyi was isolated from soil in Malaysia in 2009 (21). Bourhy et al. (22) reported that they first identified L. kmetyi in at least two patients with acute leptospirosis in the French West Indies. Therefore, L. kmetyi is thought to be pathogenic for humans, but our strains did not manifest their virulence in a hamster infection model. On the basis of the isolation of L. kmetyi in this study, it is expected that other virulent leptospires may still exist in soil after a storm surge.
In conclusion, our findings may serve as a warning that when seawater floods the land during a storm surge or tsunami, outbreaks of leptospirosis may occur in the affected areas, similar to the case of a flood caused by heavy rain, because Leptospira species classified into a pathogenic group were isolated from the soil in a coastal area that had been inundated by seawater 2.5 months earlier.
ACKNOWLEDGMENTS
This study was supported by a grant in a scheme of the Science and Technology Research Partnership for Sustainable Development (SATREPS) Program from the Japan Science and Technology Agency (JST) and the Japan International Cooperation Agency (JICA) and by a Grant-in-Aid for Scientific Research (C) (grant no. 25460536) from the Japan Society for the Promotion of Science (JSPS).
We thank Hideko Kameyama for her technical cooperation.
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326. 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771. 10.1016/S1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. 1999. Leptospirosis worldwide, 1999. Wkly. Epidemiol. Rec. 74:237–242. [PubMed] [Google Scholar]
- 4.Carleton O, Charon NW, Allender P, O'Brien S. 1979. Helix handedness of Leptospira interrogans as determined by scanning electron microscopy. J. Bacteriol. 137:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DJ, Self HR. 1955. Observations on the survival of Leptospira australis A in soil and water. J. Hyg. (Lond.) 53:436–444. 10.1017/S0022172400000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trueba G, Zapata S, Madrid K, Cullen P, Haake D. 2004. Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int. Microbiol. 7:35–40. [PubMed] [Google Scholar]
- 7.Adler B, de la Peña Moctezuma A. 2010. Leptospira and leptospirosis. Vet. Microbiol. 140:287–296. 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty A, Miyahara S, Villanueva SY, Saito M, Gloriani NG, Yoshida S. 2011. A novel combination of selective agents for isolation of Leptospira species. Microbiol. Immunol. 55:494–501. 10.1111/j.1348-0421.2011.00347.x. [DOI] [PubMed] [Google Scholar]
- 9.Saito M, Villanueva SY, Chakraborty A, Miyahara S, Segawa T, Asoh T, Ozuru R, Gloriani NG, Yanagihara Y, Yoshida S. 2013. Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl. Environ. Microbiol. 79:601–609. 10.1128/AEM.02728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tajima Y, Yasuda T, Pacheco BM, Cruz EC, Kawasaki K, Nobuoka H, Miyamoto M, Asano Y, Arikawa T, Ortigas NM, Aquino R, Mata W, Valdez J, Briones F. 2013. Initial report of JSCE-PICE joint survey on the storm surge disaster caused by Typhoon Haiyan. Coast. Eng. J. 56:1450006. 10.1142/S0578563414500065. [DOI] [Google Scholar]
- 11.Yanagihara Y, Villanueva SY, Yoshida S, Okamoto Y, Masuzawa T. 2007. Current status of leptospirosis in Japan and Philippines. Comp. Immunol. Microbiol. Infect. Dis. 30:399–413. 10.1016/j.cimid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Woo TH, Patel BK, Smythe LD, Symonds ML, Norris MA, Dohnt MF. 1997. Identification of pathogenic Leptospira genospecies by continuous monitoring of fluorogenic hybridization probes during rapid-cycle PCR. J. Clin. Microbiol. 35:3140–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koizumi N, Muto MM, Akachi S, Okano S, Yamamoto S, Horikawa K, Harada S, Funatsumaru S, Ohnishi M. 2013. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J. Med. Microbiol. 62:630–636. 10.1099/jmm.0.050039-0. [DOI] [PubMed] [Google Scholar]
- 14.Page RD. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358. [DOI] [PubMed] [Google Scholar]
- 15.Chang SL, Buckingham M, Taylor MP. 1948. Studies on Leptospira icterohaemorrhagiae. IV. Survival in water and sewage: destruction in water by halogen compounds, synthetic detergents, and heat. J. Infect. Dis. 82:256–266. 10.1093/infdis/82.3.256. [DOI] [PubMed] [Google Scholar]
- 16.Parker J, Walker M. 2011. Survival of a pathogenic Leptospira serovar in response to combined in vitro pH and temperature stresses. Vet. Microbiol. 152:146–150. 10.1016/j.vetmic.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Smith CE, Turner LH. 1961. The effect of pH on the survival of leptospires in water. Bull. World Health Organ. 24:35–43. [PMC free article] [PubMed] [Google Scholar]
- 18.Asoh T, Saito M, Villanueva SY, Kanemaru T, Gloriani NG, Yoshida S. 2014. Natural defense by saliva and mucosa against oral infection by Leptospira. Can. J. Microbiol. 60:383–389. 10.1139/cjm-2014-0016. [DOI] [PubMed] [Google Scholar]
- 19.Dickson AG, Goyet C. 1994. Handbook of methods for the analysis of the various parameters of the carbon dioxide system in sea water, version 2. US Department of Energy, Washington, DC: http://cdiac.ornl.gov/oceans/DOE_94.pdf Accessed 23 May 2014. [Google Scholar]
- 20.World Health Organization. 2013. Weekly summary report of early warning alert and response network. Post-typhoon week 6. 15th to 21st December 2013. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 21.Slack AT, Khairani-Bejo S, Symonds ML, Dohnt MF, Galloway RL, Steigerwalt AG, Bahaman AR, Craig S, Harrower BJ, Smythe LD. 2009. Leptospira kmetyi sp. nov., isolated from an environmental source in Malaysia. Int. J. Syst. Evol. Microbiol. 59(Pt 4):705–708. 10.1099/ijs.0.002766-0. [DOI] [PubMed] [Google Scholar]
- 22.Bourhy P, Herrmann Storck C, Theodose R, Olive C, Nicolas M, Hochedez P, Lamaury I, Zinini F, Brémont S, Landier A, Cassadou S, Rosine J, Picardeau M. 2013. Serovar diversity of pathogenic Leptospira circulating in the French West Indies. PLoS Negl. Trop. Dis. 7:e2114. 10.1371/journal.pntd.0002114. [DOI] [PMC free article] [PubMed] [Google Scholar]