Abstract
Bacteriophages are present in every environment that supports bacterial growth, including manmade ecological niches. Virulent phages may even slow or, in more severe cases, interrupt bioprocesses driven by bacteria. Escherichia coli is one of the most widely used bacteria for large-scale bioprocesses; however, literature describing phage-host interactions in this industrial context is sparse. Here, we describe phage MED1 isolated from a failed industrial process. Phage MED1 (Microviridae family, with a single-stranded DNA [ssDNA] genome) is highly similar to the archetypal phage phiX174, sharing >95% identity between their genomic sequences. Whole-genome phylogenetic analysis of 52 microvirus genomes from public databases revealed three genotypes (alpha3, G4, and phiX174). Phage MED1 belongs to the phiX174 group. We analyzed the distribution of single nucleotide variants in MED1 and 18 other phiX174-like genomes and found that there are more missense mutations in genes G, B, and E than in the other genes of these genomes. Gene G encodes the spike protein, involved in host attachment. The evolution of this protein likely results from the selective pressure on phages to rapidly adapt to the molecular diversity found at the surface of their hosts.
INTRODUCTION
Bacteriophages play a critical role in controlling bacterial ecosystems (1). They are thought to maintain genetic diversity by targeting the most abundant bacterial strains, as described by the “killing the winner” hypothesis (2, 3), and contribute to bacterial evolution through horizontal gene transfer (4). Bacterial viruses may play an important role in balancing natural bacterial ecosystems, but their bactericidal nature can create a significant imbalance in biotechnological processes. Humans have used bacteria for millennia to transform foods, improving flavors and increasing preservation time (5), but it is only since the late 20th century that we have used microorganisms to produce molecules with commercial value. Biotechnological processes driven by microorganisms, usually in monoculture or using very few strains, are susceptible to interference by virulent phages (6, 7). Despite the industry's acknowledgment that phages pose a significant risk for large-volume fermentation processes, and with the exception of the dairy industry, literature on the impact of phages on bioindustry processing is sparse. The milk fermentation industry has contributed most to the advancement of knowledge on the interaction of industrial bacterial strains and their phages. Many efficient phage control strategies have been implemented in the dairy industry, for example, training of employees, improved plant design, rotation of starter strains, and use of phage resistant starter strains (reviewed in references 6–8).
Escherichia coli is widely used in the biotechnological industry because it is easily genetically modified, grows fast, and is cost efficient for producing high biomass yields. E. coli variants resistant to virulent phages T1 and T5 (Siphoviridae family, double-stranded genome, and noncontractile tail) are commercially available and are commonly used in research and by the industry to produce biomolecules of interest. In these variants, the fhuA gene, which codes for an outer membrane transporter and receptor of phages T1 and T5, has been inactivated (9). These bacterial cells are impervious to phages T1 and T5 because the phages are unable to adsorb to bacterial surface components to initiate the infection process (10). However, the ongoing arms race evolution between bacteria and phages led to the emergence of T-odd phages that can infect fhuA mutants (11).
We report here a case study where a bioprocess driven by E. coli was inhibited by a virulent phage. Interestingly, this phage belongs to the Microviridae family (single-stranded DNA [ssDNA] viruses and tail-less) and is highly similar to the archetypal phage phiX174. Phages of the Microviridae family are small icosahedral viruses with a 5.3- to 6.1-kb genome generally coding for 11 proteins. Three proteins have structural roles: gpF, gpG, and gpH. Gene F codes for the major capsid protein that assembles into a procapsid, guided by gpB and gpD, the internal and external scaffolding proteins, respectively (12). The mature capsid harbors 12 spikes, each composed of five subunits of gpG. The spikes are involved in recognizing the bacterial host. The exact function of the gpH protein, also part of the spike structure, was recently elucidated. Virus attachment to the bacterial surface triggers a change in gpH conformation, which then forms a tail-like structure used to translocate the viral DNA into the bacterial cytoplasm (13). The gpA, gpC, and gpJ proteins are involved in DNA replication and packaging, while the roles of the nonessential gpA* and gpK proteins are still undefined (12). The gpE protein is involved in cell lysis (12). According to the International Committee on Taxonomy of Viruses (ICTV) (14), the Microviridae family comprises the subfamily Gokushovirinae, which includes three genera (Chlamydiamicrovirus, Bdellomicrovirus, and Spiromicrovirus). The genus Microvirus is not included in the Gokushovirinae, and it includes five species (G4, phiX174, St-1, phiK, and alpha3). The classification of these Microvirus phages is based on host range and temperature sensitivity (14).
The small genome and the short latent period of the Microviridae also make them an attractive evolutionary model. Many studies have been conducted to better understand how ssDNA viruses evolve, and at least two different approaches have been used to investigate Microviridae evolution. The first approach involves replication of viruses in a chemostat with and without different selective pressures for many generations (15–23). The second approach is based on isolating and sequencing novel members of the virus family (24) and surveying environmental sequences (25). A recent review on evolutionary studies showed that Microviridae phages undergo parallel evolution, meaning that a common ancestor evolved the same molecular substitutions even when different selective pressures were used (26). Moreover, some degree of convergent evolution occurred, since the substitutions arising from laboratory experiments were also observed in the natural virus population. Some studies focused on the adaptation of phiX174 to different hosts and revealed that host-specific evolution occurred mainly in the major capsid protein (gpF) (18, 27). However, others found that mutations in gpH appeared to be necessary for specific interactions with host receptors, while mutations in the major capsid protein appeared to play a role in capsid stability (15). Those authors suggested that these structural alterations could play an important role in adaptation to a novel host.
We report here the characterization of a Microviridae phage isolated from a failed E. coli-driven biotechnological process. The genome sequence of this phage reveals evolutionary patterns not previously observed for this phage species (phiX174), shedding light on host adaptation of Microviridae.
MATERIALS AND METHODS
Bacterial strain, phage, and growth conditions.
The host strain E. coli SMQ-1277 was grown in tryptic soy broth (TSB) medium at 37°C with aeration. The phage was isolated as previously described (28, 29). Briefly, a sample containing phages was filtered through a 0.45-μm syringe filter, the filtrate was serially diluted, and the dilutions were added to E. coli SMQ-1277. The mixture was then quickly added to TSB soft agar (containing 0.75% agar) and poured onto TSB agar plates. To improve lysis, TSB medium was supplemented with 25 μM MgSO4. Phage MED1 was purified three times from isolated plaques before genome sequencing (30). Phage MED1 and E. coli SMQ-1277 were deposited at the Félix d'Hérelle Reference Center for Bacterial Viruses (http://www.phage.ulaval.ca/) under accession numbers HER504 and HER1504, respectively. For host range analyses, E. coli strains HER1024 (also named B or ATCC 11303; host of phages T1, T2, T3, T4, T5, T6, and T7), HER1036 (C or ATCC 13706; host of phage phiX174), HER1144 (K12S; host of phage lambda), HER1382 (host of phage HK97), and HER1462 (C-3000; host of phage MS2) were obtained from the Félix d'Hérelle Center.
Electron microscopy.
Phage MED1 was observed by electron microscopy as previously described (31). Briefly, 1.5 ml of phage lysate was centrifuged at 23,500 × g for 1 h at 4°C, and the pellet was washed twice with ammonium acetate (0.1 M; pH 7.0). The resulting solution containing phage was then used to prepare observation grids. The grids were stained with phosphotungstic acid (2%; pH 7.0) and observed with a JEOL 1230 electron microscope.
DNA extraction, genome sequencing, and analysis.
Viral nucleic acid was extracted using a QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions, except that we used 60 μl of Tris-EDTA (TE) buffer (pH 8.0) for the final elution. Second-strand synthesis was conducted with the Clone Miner cDNA library construction kit (Invitrogen) according to the manufacturer's instructions, with the following modifications: isoamyl alcohol was not added to the phenol-chloroform solution used to extract the DNA after second-strand synthesis, and the double-stranded DNA (dsDNA) was dissolved in 25 μl of TE buffer (pH 8.0). The sequencing library was prepared using a Nextera DNA Sample Prep kit (Illumina) according to the manufacturer's instructions. The library was sequenced on a MiSeq system (Illumina) using a MiSeq reagent kit (2 × 151 nucleotides [nt]; Illumina). De novo assembly was performed with Ray assembler version 2.2.0-devel, using a kmer size of 31 (32).
Whole-genome phylogeny.
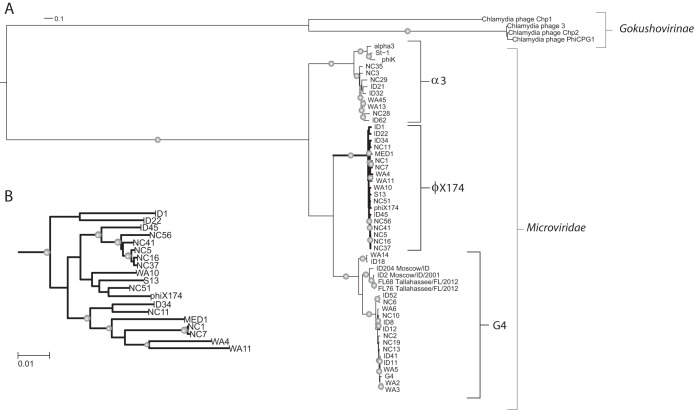
The genomes (n = 55) were selected by searching the NCBI nucleotide database for the term “microvirus” and excluding uncultured sequences to avoid working with chimeric sequences and phiX174 variants from evolutionary studies. Phylogenetic analysis of the complete genomes was conducted by defining the origin of all genomes as the start codon of the gpA gene. The genomes were aligned using MAFFT (v7.130b) (33) with the L-INS-i parameters, and the alignment was converted to the PHYLIP format using BioPython (34) in-house scripts. The most probable nucleic acid substitution model was selected using jModelTest (v2.1.4) (35), and we used this output in PhyML to generate the tree (36). Branch support values were calculated using the Shimodaira-Hasegawa-like procedure (37). The leaves of the tree were renamed using the Newick utilities package to render them compliant with the PHYLIP format and to convert them back to human-readable names after calculating the maximum likelihood (38). Finally, the tree was visualized using the Web interface iTOL (http://itol.embl.de/) (39). The phylogenetic tree was pruned using Python ETE 2.2rev1026 package (40) to focus only on the phiX174 group (see Fig. 3B).
FIG 3.
Maximum likelihood phylogenetic tree of the microvirus genome sequences retrieved from the NCBI database and MED1. (A) Phylogenetic tree showing all phage genomes. (B) Pruned tree showing only phiX174 species. The trees indicate that this group of phages is divided into three genotypes, strongly supported by branch support values of >95% (indicated by gray spheres on the branches). Phages within the same species are very similar, while interspecies diversity is significant.
Comparative genomics.
The type phage of each group was used for the intergenotype comparison. The schematic representation of the genome was manually constructed in Adobe Illustrator. The percent identity was calculated from a Protein-Protein BLAST analysis (package 2.2.28+) (41), using the number of identical amino acids divided by the length of the longest protein from the pairwise comparison.
Protein phylogenies and evolution of the phiX174 species.
The gene sequences extracted from GenBank files were aligned using MAFFT with the parameter G-INS-i (33) to improve global alignment. Single nucleotide variants (SNVs) were extracted from the pairwise comparison of all unique sequence combinations. The impact of each SNV on the protein sequence (nonsense versus missense) was reported in a table, which was used to generate Fig. 4. The graphs were generated using the ggplot2 R package (version 0.9.3.1) (42). The protein phylogenies were analyzed in a way similar to the analysis of genome phylogeny. First, the protein sequences were aligned using MAFFT with the parameter G-INS-i. The protein sequences from phage ID18 (G4 species) were included in the phylogenetic analysis as an outlier, which was later removed from the tree using the ETE 2.2rev1026 package, preserving the branch length for the phiX174 group (40). The most probable amino acid substitution model was determined using ProtTest (3.2) (43), which was then implemented in PhyML to estimate the best phylogenetic trees. The branch support values were calculated using the Shimodaira-Hasegawa-like approach (37). The trees were rendered using ETE 2.2rev1026 (40). Finally, the variable sites identified in gpG were mapped in the structure of phiX174 (MMDB accession number 52636; PDB accession number 2BPA) (44) using Cn3D software (45). The blue-red color gradient is based on the conservation score calculated by Jalview (46) for each residue.
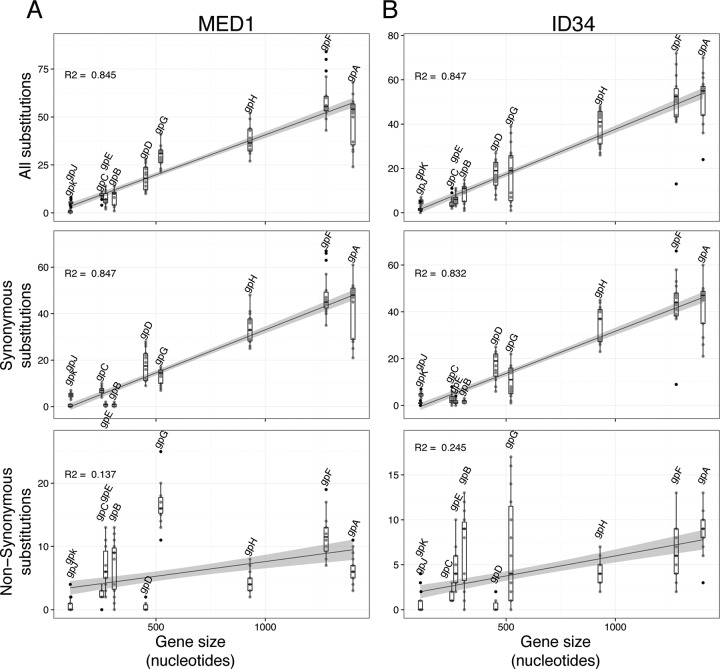
FIG 4.
Frequency of SNVs for MED1 (A) and ID34 (B) as a function of the size of the genes. (Top) Frequency of all SNVs; (middle) frequency of SNVs corresponding to synonymous mutations; (bottom) frequency of SNVs corresponding to nonsynonymous mutations. The results indicate that the number of synonymous mutations correlates with the size of the gene (R2 = 0.92), unlike the number of nonsynonymous mutations, which does not (R2 = −0.0185). This shows that the selective pressure is not the same for all genes, as the number of mutations found in gene G is greater than in the other genes. The gray shading shows the 95% confidence interval for the linear regression.
Nucleotide sequence accession number.
The complete annotated genomic sequence of phage MED1 was deposited in GenBank under accession number KJ997912.
RESULTS AND DISCUSSION
Isolation of the phage.
A phage-contaminated sample was obtained from a biotechnological company using E. coli as one of its production platforms. The pH-controlled organic acid fermentation process suffered from a ∼10-fold decrease in rate of production after about 5 h. The rate of production was monitored as the rate of base addition. Bacterial growth also arrested, but complete lysis was not observed in the industrial fermentor. A sample from the industrial fermentation broth was filtered (0.45 mm), and phage plaques were observed in a plaque assay against the E. coli production organism. The virulent phage MED1 was isolated from the resulting plaques (28, 29).
Optimizing the concentration of MgSO4 in the medium significantly increased phage MED1 titers. The most efficient infection was obtained with a concentration of 25 μM MgSO4 (data not shown). Although tailed phages are more frequently reported to be contaminants in cases like this, we observed, by electron microscopy, that MED1 was similar in morphology to the archetypal phage phiX174 (Fig. 1). This is surprising, since to our knowledge, Microviridae phages have not been previously reported to interfere with industrial bioprocesses. We tested the host range of MED1 against the host strains of coliphages T1, T2, T3, T4, T5, T6, T7, phiX174, λ, HK97, and MS2, and it infects only the host strain of phiX174 (HER1036) and E. coli SMQ-1277. To further characterize MED1, we sequenced its genome and compared it to other phages belonging to the Microviridae family.
FIG 1.
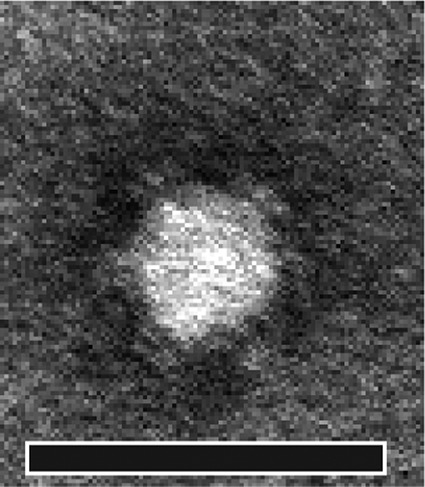
Electron micrograph of MED1. Bar, 50 nm.
Genome sequencing.
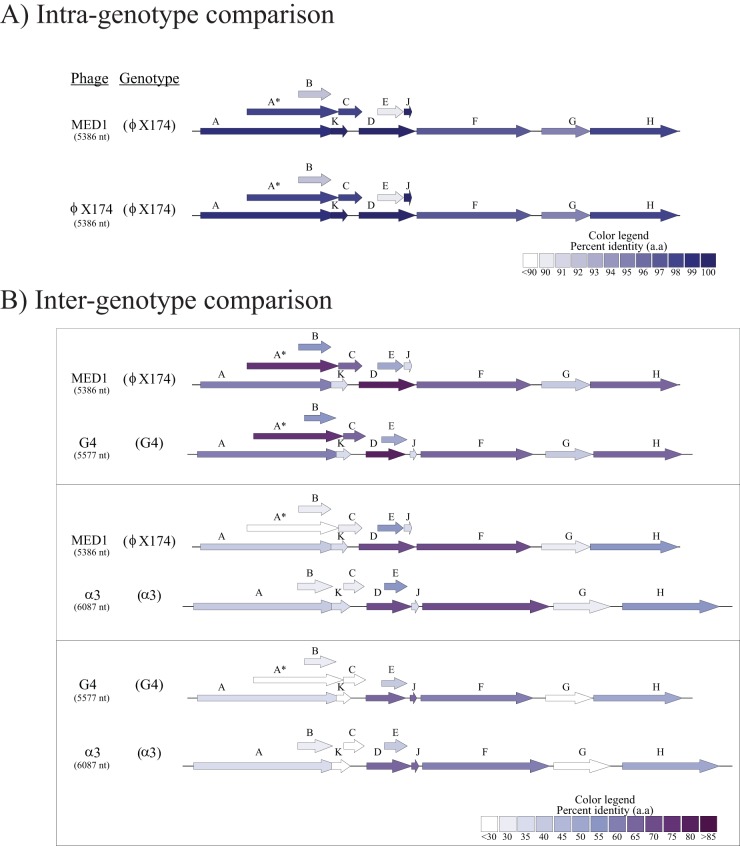
We used a nucleic acid extraction column to extract the MED1 genome, convert the ssDNA to dsDNA, and sequence the resulting dsDNA using a MiSeq apparatus. We assembled the genome into a single contig of 5,386 nt with an average coverage of 842-fold. A nucleotide BLAST analysis against the NCBI nonredundant database confirmed that MED1 belongs to the Microviridae family, sharing >95% identity with phiX174. The genome of MED1 codes for the same 11 proteins encoded by the phage phiX174 genome, and genome synteny is highly conserved across all 52 genomes belonging to the Microvirus genus (Fig. 2), with the exception of the alpha3 species, reported to code for five additional putative proteins not found in phiX174 and G4 species. When members of the genus Microvirus are compared, there is considerable diversity between species and often <40% identity between their amino acid sequences. Conversely, intergenotype comparison reveals an astonishingly high level of conservation (Fig. 2).
FIG 2.
Genetic alignment of one representative of each phage group and MED1. Arrows represent the proteins encoded by the genomes. The colors represent the percent identity shared by the proteins. White arrows represent proteins sharing less identity than the lower limit of the scales shown in the legend. Note that the scales are different for panels A and B. (A) Intragenotype comparison of MED1 and phiX174. All genes share >90% identity between protein sequences. (B) Intergenotype comparison of MED1 with the type phage of the other phage species belonging to the genus Microvirus. Although highly conserved genome synteny was observed between the microviruses, their protein sequences remain highly diverse across the different species, with some sharing <30% identity. a.a, amino acids.
Whole-genome phylogeny.
We conducted a phylogenetic analysis to classify phage MED1. As indicated previously, the ICTV recognizes five species of the Microvirus genus, phiX174, phiK, G4, alpha3, and St-1, based on host range and temperature sensitivity (14). However, a phylogenetic analysis by Rokyta and colleagues (24) suggested that the genus Microvirus should be subdivided into three species: phiX174, alpha3, and G4. Those researchers suggested that the species phiK should belong to the alpha3 species (24), but they did not analyze the St-1 species.
Our analysis of 56 genomes included all genomes belonging to the Microviridae available in the NCBI GenBank database (55 genomes) and our MED1. Of these 56 genomes, 52 belong to the Microvirus genus, while the remaining four phages belong to the Gokushovirinae subfamily (Fig. 3). Our phylogenetic analysis shows that all of these phages group within three species: phiX174, alpha3, and G4. The phiK and St-1 phages cluster with the alpha3 species. On the basis of our observations and those of Rokyta et al. (24), we propose to adapt the classification of the Microviridae and divide the Microvirus genus into three species, namely, phiX174, alpha3, and G4. Phylogenetic analysis based on genome alignment reveals that MED1 is most closely related to phages NC1, NC7, WA4, and WA11. This grouping is supported by a branch support value of 97.9% (Fig. 3B).
Comparison of MED1 with the phiX174 species.
First, we investigated the distribution of SNVs across the coding sequences of the phages belonging to the phiX174 species. The box plots in Fig. 4 show the number of SNVs per gene, where each data point within the box represents a pairwise comparison between a gene from a reference phage and the orthologous gene from the other 18 phiX174-like phages. Thus, 19 versions of this figure were generated for the phiX174 species (one for each phage) (data not shown). We provide two typical examples in Fig. 4. Pairwise comparison of all phage genes revealed that the total number of SNVs per gene correlates with the size of that gene (average R2 = 0.719; standard deviation [SD] = 0.133) (Fig. 4, top). We reached the same conclusion for the number of SNVs leading to a synonymous substitution (average R2 = 0.697; SD = 0.135) (Fig. 4, middle). The linear regression model did not apply when we considered only SNVs resulting in a nonsynonymous substitution (average R2 = 0.231; SD = 0.081) (Fig. 4, bottom). Even though genes B, E, and G are relatively small, they accumulate more missense mutations than the other genes harbored by the phiX174-like phages.
The products of genes B and E are encoded within genes A and D (Fig. 2), respectively, using an alternate reading frame. Gene B and E sequences must be more permissive to maintain the functionality of genes A and D, which code for the phage DNA replication protein and the external scaffolding protein, respectively. The gpA and gpD proteins require stringent structural constraints to conserve their function. Gene B was exchangeable between phages phiX174, G4, and alpha3, even when gpB orthologs shared <70% amino acid sequence identity. Thus, gpB appears inherently tolerant to mutations (47).
The gpG protein is not encoded within another gene but still shows evidence of diversification (Fig. 4 and 5). This gene codes for the major spike proteins and plays a role in host recognition (48). The capsid spikes of phage phiX174 are composed of a gpG homopentamer and one copy of gpH (49). The gpH pilot protein guides phage DNA across the cell wall and bacterial membranes (48). It was recently demonstrated that adsorption of the virus to the host triggers a conformational change of gpH, which forms a tail-like structure responsible for translocation of the phage DNA into the host cell (13). While gpH is highly conserved within the phiX174 species, our phylogenetic analysis of gpG reveals that this protein is prone to mutation (Fig. 5). This suggests that the phiX174 phages rapidly adapt to the genetic landscape of their host, especially to the genetic diversity found in genes coding for cell surface components.
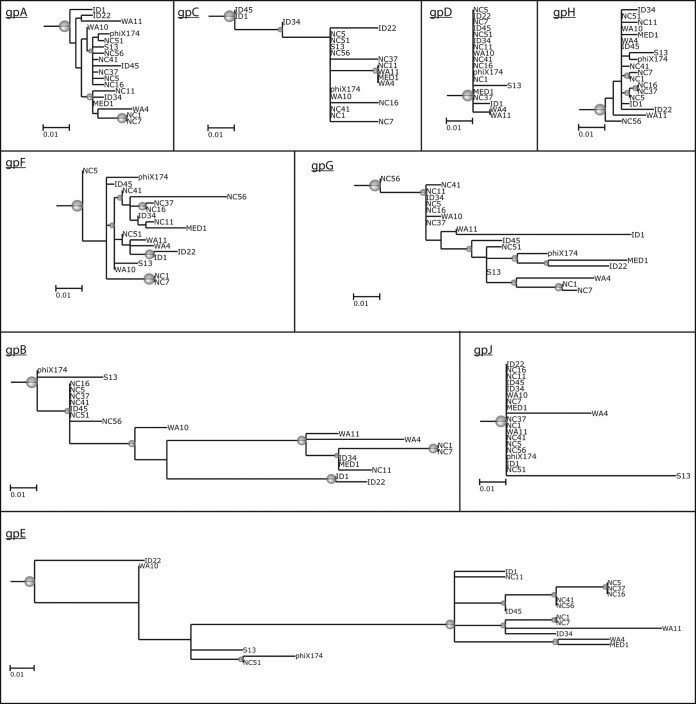
FIG 5.
Phylogenies of the protein alignments of phiX174-like phages. Phage ID18 (G4 species) was used as the outgroup, which was then pruned from the final tree. Gray spheres indicate branches with support values of >95%. The size of the sphere is proportional to the branch support values. This figure shows that gpG is among the most diverse proteins, along with gpB and gpE, encoded within the gpA and gpD genes, respectively.
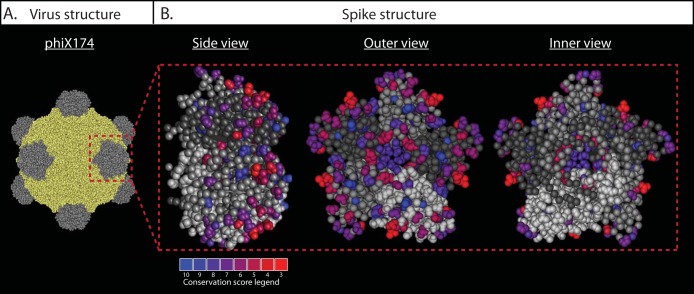
In an evolutionary study of eight Microviridae phages, Rokyta and colleagues observed that the majority of mutations detected at the end of an evolutionary walk were missense changes that occurred in genes coding for structural proteins (gpF, gpG, gpH, and gpJ) (23). Five missense mutations were found in the G gene. These mutations probably did not affect attachment to the bacterial host, since the predicted location of the amino acid change was at the junction of the spike structure and the capsid protein gpF (23). We also analyzed the positions of gpG mutations in the spike structure for phiX174-like phages (Fig. 6). Unlike the previous study (23), we found that the less conserved residues were located at the surface of the spike structure (Fig. 6). This suggests that the observed variability in gpG protein sequences is involved in host recognition and attachment (Fig. 6). Interestingly, others reported that the gpF (major capsid) protein was the principal component mutated when coliphage phiX174 evolved to infect Salmonella as an alternative host. Those researchers postulated that since the mutations were in the vicinity of the gpF/gpG interface, the affected residues could be involved in host attachment. Previous studies also mapped mutations resulting in host range alterations to genes G, H, and F (50–52).
FIG 6.
Distribution of gpG variable sites in the phiX174-like phages. (A) Structure of the complete mature phiX174 virion, where the capsid protein is yellow and the spike protein is gray. (B) Structure of the spike. The blue-red color gradient indicates the conservation score of the residues. The less-conserved residues are represented in red and the most conserved ones in blue. The sites conserved in all phages are represented in gray. Different shades of gray are used to differentiate gpG monomers in the spike structure. Most of the less-conserved residues are at the surface of the spike structure, probably involved in attachment to the host.
We also analyzed the phylogeny of the proteins encoded by all phages belonging to the phiX174 species. With the exception of gpB and gpE, gpG has the longest branches (Fig. 5). The phylogeny of gpG also indicates that the observed diversity in gpG sequences could result from horizontal gene transfers, since the tree topology is different from most proteins and from the whole-genome phylogeny (Fig. 2 and 5). It must be stressed that most phiX174 evolutionary studies examine the evolution of the virus under a given condition or selective pressure, while in this study, we analyzed viral evolution by comparing the genomes of phages isolated from different sources and using different E. coli host strains. Thus, the variability observed for gpG is likely a snapshot of the viral diversity occurring in these different samples. The variability in gpG was also analyzed in the G4 and alpha3 species, and the same trend was observed: gene G accumulates more missense mutations than all other genes. These observations contrast with previous results, where very little variation was detected in gpG (26).
Bacteriophages are an indissociable component of bacterial ecosystems, and they constantly evolve with their hosts. Phages also represent a nonnegligible risk for bioindustry, from foods to biopharmaceuticals, since they may cause delays in bioprocesses and variability in the quality of the final product, ultimately impacting cost and production. We described a phage belonging to the Microviridae family (ssDNA phage) isolated from a bioprocess driven by E. coli, which provided a unique opportunity to study its evolution outside a laboratory environment. The analysis of its genome revealed that this phage is highly similar to the archetypal phage phiX174. The phylogeny and distribution of SNVs in Microviridae coding sequences indicated that gene G accumulates missense mutations to a greater extent than most of the other genes. This gene plays a role in host recognition and adsorption, requiring it to rapidly respond to changes in the host surface to remain competitive.
Microviridae have been well studied because their small genome encodes only a few proteins. Phage phiX174 was the very first whole genome to be sequenced and the first virus for which the complete structure was determined. While phages similar to phiX174 have been reported to be abundant in the environment, they are challenging to isolate (25). We believe that isolation and characterization of additional Microviridae phages are key to a better understanding of the evolution of members of this phage family in their natural environment.
ACKNOWLEDGMENTS
We thank Barbara-Ann Conway for editorial assistance.
J.C. holds a Tier 1 Canada Research Chair in Medical Genomics. S.M. holds a Tier 1 Canada Research Chair in Bacteriophages.
Footnotes
Published ahead of print 5 September 2014
REFERENCES
- 1.Brüssow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16. 10.1016/S0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- 2.Waterbury JB, Valois FW. 1993. Resistance to cooccurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinbauer MG, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1–11. 10.1046/j.1462-2920.2003.00539.x. [DOI] [PubMed] [Google Scholar]
- 4.Zinder ND, Lederberg J. 1952. Genetic exchange in Salmonella. J. Bacteriol. 64:679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutkins RW. 2006. Microbiology and technology of fermented foods. Blackwell Publishing, Chicago, IL. [Google Scholar]
- 6.Samson JE, Moineau S. 2013. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 4:347–368. 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- 7.Labrie S, Moineau S. 2010. Bacteriophages in industrial food processing: incidence and control in industrial fermentation, p 199–216 In Sabour PM, Griffiths MW. (ed), Bacteriophages in the control of food- and waterborne pathogens. ASM Press, Washington, DC. [Google Scholar]
- 8.Garneau JE, Moineau S. 2011. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10(Suppl 1):S20. 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidel W. 1958. Bacterial viruses; with particular reference to adsorption/penetration. Annu. Rev. Microbiol. 12:27–48. 10.1146/annurev.mi.12.100158.000331. [DOI] [PubMed] [Google Scholar]
- 10.Braun V, Schaller K, Wolff H. 1973. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323:87–97. 10.1016/0005-2736(73)90433-1. [DOI] [PubMed] [Google Scholar]
- 11.Langenscheid J, Killmann H, Braun V. 2004. A FhuA mutant of Escherichia coli is infected by phage T1—independent of TonB. FEMS Microbiol. Lett. 234:133–137. 10.1111/j.1574-6968.2004.tb09524.x. [DOI] [PubMed] [Google Scholar]
- 12.Fane BA, Brentlinger KL, Burch AD, Chen M, Hafenstein S, Moore E, Novak CR, Uchiyama A. 2006. øX174 et al. The Microviridae, p 129–145 In Calendar R, Abedon ST. (ed), The bacteriophages. Oxford University Press, New York, NY. [Google Scholar]
- 13.Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA. 2014. Icosahedral bacteriophage ΦX174 forms a tail for DNA transport during infection. Nature 505:432–435. 10.1038/nature12816. [DOI] [PubMed] [Google Scholar]
- 14.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed). 2012. Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Academic Press, London, United Kingdom. [Google Scholar]
- 15.Pepin KM, Domsic J, McKenna R. 2008. Genomic evolution in a virus under specific selection for host recognition. Infect. Genet. Evol. 8:825–834. 10.1016/j.meegid.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Wichman HA, Millstein J, Bull JJ. 2005. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics 170:19–31. 10.1534/genetics.104.034488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vale PF, Choisy M, Froissart R, Sanjuán R, Gandon S. 2012. The distribution of mutational fitness effects of phage φX174 on different hosts. Evolution 66:3495–3507. 10.1111/j.1558-5646.2012.01691.x. [DOI] [PubMed] [Google Scholar]
- 18.Crill WD, Wichman HA, Bull JJ. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull JJ, Millstein J, Orcutt J, Wichman HA. 2006. Evolutionary feedback mediated through population density, illustrated with viruses in chemostats. Am. Nat. 167:E39–E51. 10.1086/499374. [DOI] [PubMed] [Google Scholar]
- 20.Bull JJ, Badgett MR, Wichman HA. 2000. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 17:942–950. 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- 21.Brown CJ, Millstein J, Williams CJ, Wichman HA. 2013. Selection affects genes involved in replication during long-term evolution in experimental populations of the bacteriophage φX174. PLoS One 8:e60401. 10.1371/journal.pone.0060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CJ, Stancik AD, Roychoudhury P, Krone SM. 2013. Adaptive regulatory substitutions affect multiple stages in the life cycle of the bacteriophage φX174. BMC Evol. Biol. 13:66. 10.1186/1471-2148-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rokyta DR, Abdo Z, Wichman HA. 2009. The genetics of adaptation for eight microvirid bacteriophages. J. Mol. Evol. 69:229–239. 10.1007/s00239-009-9267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rokyta DR, Burch CL, Caudle SB, Wichman HA. 2006. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 188:1134–1142. 10.1128/JB.188.3.1134-1142.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labonté JM, Suttle CA. 2013. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 7:2169–2177. 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichman HA, Brown CJ. 2010. Experimental evolution of viruses: Microviridae as a model system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:2495–2501. 10.1098/rstb.2010.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wichman HA, Scott LA, Yarber CD, Bull JJ. 2000. Experimental evolution recapitulates natural evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1677–1684. 10.1098/rstb.2000.0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bissonnette F, Labrie S, Deveau H, Lamoureux M, Moineau S. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged cheddar cheese. J. Dairy Sci. 83:620–627. 10.3168/jds.S0022-0302(00)74921-6. [DOI] [PubMed] [Google Scholar]
- 29.Moineau S, Fortier J, Ackermann H-W, Pandian S. 1992. Characterization of lactococcal bacteriophages from Québec cheese plants. Can. J. Microbiol. 38:875–882. 10.1139/m92-143. [DOI] [Google Scholar]
- 30.Moineau S, Pandian S, Klaenhammer TR. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortier L-C, Moineau S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl. Environ. Microbiol. 73:7358–7366. 10.1128/AEM.00582-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 17:1519–1533. 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyml 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Shimodaira H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492–508. 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 38.Junier T, Zdobnov EM. 2010. The newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26:1669–1670. 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478. 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huerta-Cepas J, Dopazo J, Gabaldón T. 2010. ETE: a python environment for tree exploration. BMC Bioinformatics 11:24. 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham H. 2009. Ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 43.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenna R, Ilag LL, Rossmann MG. 1994. Analysis of the single-stranded DNA bacteriophage phi X174, refined at a resolution of 3.0 Å. J. Mol. Biol. 237:517–543. 10.1006/jmbi.1994.1253. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. 2000. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 25:300–302. 10.1016/S0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 46.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burch AD, Ta J, Fane BA. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J. Mol. Biol. 286:95–104. 10.1006/jmbi.1998.2450. [DOI] [PubMed] [Google Scholar]
- 48.Jazwinski SM, Lindberg AA, Kornberg A. 1975. The gene H spike protein of bacteriophages phiX174 and S13. I. Functions in phage-receptor recognition and in transfection. Virology 66:283–293. [DOI] [PubMed] [Google Scholar]
- 49.Olson NH, Baker TS, Willingmann P, Incardona NL. 1992. The three-dimensional structure of frozen-hydrated bacteriophage φX174. J. Struct. Biol. 108:168–175. 10.1016/1047-8477(92)90016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newbold JE, Sinsheimer RL. 1970. The process of infection with bacteriophage phiX174. XXXII. Early steps in the infection process: attachment, eclipse and DNA penetration. J. Mol. Biol. 49:49–66. [DOI] [PubMed] [Google Scholar]
- 51.Weisbeek PJ, van de Pol JH, van Arkel GA. 1973. Mapping of host range mutants of bacteriophage φX174. Virology 52:408–416. 10.1016/0042-6822(73)90335-8. [DOI] [PubMed] [Google Scholar]
- 52.Dowell CE, Jansz HS, Zandberg J. 1981. Infection of Escherichia coli K-12 by bacteriophage φX-174. Virology 114:252–255. 10.1016/0042-6822(81)90271-3. [DOI] [PubMed] [Google Scholar]