Abstract
Members of the Enterobacteriaceae with extended-spectrum beta-lactamases (ESBLs) of the CTX-M type have disseminated rapidly in recent years and have become a threat to public health. In parallel with the CTX-M type expansion, the consumption and widespread use of silver-containing products has increased. To determine the carriage rates of silver resistance genes in different Escherichia coli populations, the presence of three silver resistance genes (silE, silP, and silS) and genes encoding CTX-M-, TEM-, and SHV-type enzymes were explored in E. coli isolates of human (n = 105) and avian (n = 111) origin. The antibiotic profiles were also determined. Isolates harboring CTX-M genes were further characterized, and phenotypic silver resistance was examined. The silE gene was present in 13 of the isolates. All of them were of human origin. Eleven of these isolates harbored ESBLs of the CTX-M type (P = 0.007), and eight of them were typed as CTX-M-15 and three as CTX-M-14. None of the silE-positive isolates was related to the O25b-ST131 clone, but 10 out of 13 belonged to the ST10 or ST58 complexes. Phenotypic silver resistance (silver nitrate MIC > 512 mg/liter) was observed after silver exposure in 12 of them, and a concomitant reduced susceptibility to piperacillin-tazobactam developed in three. In conclusion, 12% of the human E. coli isolates but none of the avian isolates harbored silver resistance genes. This indicates another route for or level of silver exposure for humans than that caused by common environmental contamination. Since silE-positive isolates were significantly more often found in CTX-M-positive isolates, it is possible that silver may exert a selective pressure on CTX-M-producing E. coli isolates.
INTRODUCTION
The prevalence of pathogenic bacteria with antibiotic multiresistance has increased rapidly in recent years, and they have become a serious threat to public health. During 2007, approximately 25,000 patients died due to therapeutic failure in Europe alone, and Gram-negative bacteria were the cause in about two-thirds of the cases (1).
Beta-lactamases of the extended-spectrum type (ESBLs) are major players in this development, and they emerged in the 1980s as TEM and SHV derivates (2, 3). These enzymes are usually harbored by members of the family Enterobacteriaceae and hydrolyze penicillins, cephalosporins, and monobactams. Today, the CTX-M enzymes are the most prevalent group of ESBLs. They have spread in a pandemic fashion, causing outbreaks in hospitals and communities worldwide (4–6). Furthermore, they have disseminated into distant environmental niches where there are no or few human activities (7–9).
Escherichia coli clone O25b-ST131 has been the dominant ESBL lineage in about 25% of the human E. coli cases (10), and it usually produces CTX-M-14 or CTX-M-15. During the years 2005 to 2007, there was a major outbreak at Uppsala University Hospital caused by a CTX-M-15-producing Klebsiella pneumoniae strain (11). A 41-kbp resistance region, highly similar to the resistance regions of two plasmids isolated previously from E. coli strains belonging to the outbreak lineage ST131, was found on one of its plasmids. This plasmid carried, in addition, a complex of sil genes (12).
Bacteria can develop resistance to silver (13) through inducible efflux mechanisms encoded by sil or pco (cop) genes (14, 15). sil genes can be harbored on plasmids carrying antibiotic resistance genes (16, 17), and silver can thereby exert an indirect selective pressure. Silver may also affect antibiotic resistance more directly by selecting for porin deficiency and thereby mediate a cross-resistance to beta-lactams (18).
Since the turn of the century, the consumption of biocides has increased (1). One of the more popular biocides is silver, which is used as a disinfectant and preservative in a wide and mounting range of health care and consumer products, e.g., silver-containing wound dressings, silver-treated catheters and textiles, and washing machines releasing silver ions (19, 20). Although humans and animals are naturally exposed to silver at low concentrations through food and water (21, 22), the exposure level in both hospitals and in the society is probably higher today than when the silver consumption was mainly confined to the photographic, battery, and car industries.
The aim of the present study was to investigate the distribution of silver resistance genes in seven E. coli populations derived from humans and wild birds. The four avian populations had various levels of exposure to human activity.
MATERIALS AND METHODS
Bacterial isolates.
A total of 216 E. coli isolates were included in the study. The isolates were fecal samples from humans (n = 105) and wild birds (n = 111) collected during the years 2006 to 2008. The human samples were all unidentified before culturing and belonged to one of the following groups: (i) randomly chosen routine samples from patients with diarrhea in Uppsala County (n = 52), (ii) randomly chosen samples from patients admitted to Uppsala University Hospital during the major K. pneumoniae outbreak and thereby automatically taking part in a surveillance program of ESBL producers (n = 34) (11), and (iii) samples from healthy travelers participating in a study in which the risk of acquiring ESBL-producing Enterobacteriaceae during travels outside Scandinavia was investigated (n = 19) (23). The avian samples were collected from four wild bird populations from the following areas and with an increasing exposure to human activities: Commander Islands, Russia (n = 26) (24); Kalmar (n = 27) (25) and Uppsala (n = 17), Sweden; and Marseille, France (n = 41) (26) (Table 1).
TABLE 1.
Origin and genetic properties of the isolates included in the study
| Beta-lactamase | No. of isolatesa |
||||||
|---|---|---|---|---|---|---|---|
| Human |
Avian |
||||||
| Patients with diarrhea (Uppsala) | Patients in surveillance programs |
Herring gulls (Larus argentatus) (Commander Islands) | Black-headed gulls (Larus ridibundus) (Kalmar) | Mallards (Anas platyrhynchos) (Uppsala) | Yellow-legged gulls (Larus michahellis) (Marseille) | ||
| Uppsala | Outside Scandinavia | ||||||
| Total | 52 | 34 | 19 | 26 | 27 | 17 | 41 |
| CTX-M-15 | 0 | 21 | 11 | 1 | 1 | 0 | 1 |
| CTX-M-14 | 0 | 11 | 2 | 0 | 1 | 0 | 0 |
| CTX-M-9 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| CTX-M-1 | 0 | 0 | 1 | 0 | 0 | 0 | 8 |
| CTX-M-27 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| silE | 2 | 8 | 3 | 0 | 0 | 0 | 0 |
| merA | 5 | 8 | 4 | 0 | 0 | 0 | 5 |
| TEM | NA | 27 | 9 | NA | NA | NA | 9 |
| SHV | NA | 2 | 1 | NA | NA | NA | 2 |
NA, not applicable.
The avian E. coli samples isolated at the Department of Clinical Microbiology, Uppsala University Hospital, were collected with the M40 Transystem (Copan Diagnostics, USA). After the swabs had been rotated in fresh feces from mallard ducks living at a pond in central Uppsala, they were put into the transport medium and immediately brought to the laboratory. These samples and those collected from the patient group with diarrhea were plated on cysteine lactose electrolyte-deficient agar or blood agar (Becton, Dickinson Company, USA). After overnight incubation at 35°C, E. coli was identified using standard laboratory procedures or the Vitek 2AutoMicrobic system (bioMérieux, USA). Collection and culture of the remaining isolates were performed as described earlier (9, 11, 23, 25, 26). All isolates were frozen at −70°C before further analysis.
Antibiotic susceptibility.
Susceptibility testing was carried out on Iso-Sensitest agar with the disc diffusion method (Oxoid Ltd., England) according to the recommendations of the Swedish Reference Group for Antibiotics (www.srga.org) at that time point. The following antibiotics were included: cefpodoxime, ceftibuten, piperacillin-tazobactam, cefotaxime, meropenem, ciprofloxacin, gentamicin, and cotrimoxazole. The plates were incubated for 18 to 20 h at 35°C. Species-related breakpoints defined by SRGA were used for the categorization of isolates as susceptible, indeterminate, or resistant.
The first criterion for ESBL production was resistance to cefpodoxime (inhibition zone ≤ 24 mm). Phenotypic confirmation of ESBL production was performed by disc diffusion synergy test according to the method described by Jarlier et al. (27).
DNA preparation and amplification.
DNA was prepared by heating the bacterial suspension to 95°C for 10 min. Two μl of the prepared template was amplified in Taq or HotStar MasterMix (Qiagen, Germany). All PCRs were carried out in a GeneAmp PCR system 9700 cycler (PE Applied Biosystems, USA). For each PCR, denaturation lasted 2 to 15 min at 94°C, followed by 30 to 35 cycles as follows: denaturation at 94°C for 30 to 60s, annealing at a primer-specific temperature for 30 to 60s, elongation at 72°C for 1 to 3 min, and a final extension step at 72°C for 12 min. (For details about primer sequences and annealing temperatures, see the supplemental material.) The PCR products were separated by electrophoresis on 1.0% agarose gels (Shelton Scientific Inc., USA) containing ethidium bromide. After completed electrophoresis, the gels were photographed and the bands analyzed. The sizes of the different PCR products were compared with a DNA molecular weight marker (Fermentas, USA). The positive controls were provided by the Department of Clinical Microbiology, Uppsala University Hospital, unless otherwise indicated. Before use, all products were sequenced to verify their specificity.
Detection of genes for resistance to silver and mercury.
For detection of silver resistance genes, a PCR method described by Percival et al. (28) was used. All strains were screened for the silE gene. Additional PCR amplifications were carried out for the silP and silS genes whenever the silE gene was present in an isolate. CTX-M-15-producing K. pneumoniae strain U-0503875 (the outbreak strain) with the complete sil gene cassette was included as a positive control.
Since resistance to mercury is widely dispersed in the environment, resistance to this toxic heavy metal was investigated for comparison by screening for the merA gene, as described earlier (29). As a positive control, strain ESBL0610747 was used.
Molecular detection and characterization of beta-lactamases.
All isolates with phenotypic ESBL production were screened for genes encoding blaCTX-M phylogenetic lineage groups I, II, III, and IV and for blaTEM and blaSHV genes with PCR, as described by Pitout et al. or elsewhere (11, 30). Isolates positive for CTX-M genes were further typed by sequencing (11). Amplified products were purified by using the ExoSAP-IT purification kit (Amersham Biosciences, United Kingdom). Sequencing was performed using the BigDye Terminator sequencing kit (version 3.1) and an automatic DNA sequencer (Applied Biosystems, USA). The sequences obtained were edited with Vector NTI Advance 10 software (Invitrogen, USA) and compared with published sequences, employing the NCBI Basic Local Alignment Search Tool (BLAST).
PCR detection of the O25b-ST131 clone.
All ESBL-positive isolates were screened for the pabB gene using an O25b-ST131 clone allele-specific PCR described by Clermont et al. (31). It was slightly modified before use. Briefly, DNA templates for PCR were generated by boiling bacteria in distilled water (dH2O) for 5 min, followed by centrifugation at 13,000 rpm for 10 min. Two microliters of the supernatant was used as the DNA template in each PCR, which had a total volume of 25 μl. HotStarTaq master mix kit (Qiagen, Germany) and a 400 nM concentration of each primer (Eurogentec S.A., Belgium) were used. Amplification was performed on a Perkin-Elmer Cetus DNA thermal cycler. E. coli strain 65917 was the positive control (24).
Susceptibility to silver nitrate.
The MIC of silver nitrate was determined for all isolates positive for the silE gene. Bacteria were suspended in Iso-Sensitest broth (Oxoid Ltd., USA) containing silver nitrate at concentrations ranging from 4 to 512 mg/liter. The final bacterial concentration was 105 CFU/ml. After 24 h of incubation, the MIC was recorded as the lowest concentration yielding no visible growth. A silver nitrate MIC of >512 mg/liter classified the bacterium as silver resistant. E. coli strain ATCC 25922 was used as a control.
To investigate whether phenotypic silver resistance could be induced, one CFU of each isolate was inoculated into Iso-Sensitest broth. After an overnight incubation at 37°C, 10 μl of the bacterial suspension was inoculated into a series of tubes with increasing concentrations of silver nitrate (16 to 512 mg/liter) in the same broth. The tubes were incubated at 37°C overnight, and the new inocula were transferred from the tube with the highest silver nitrate concentration and still-visible growth to a new series of tubes. The experiment was repeated until a MIC of silver nitrate of >512 mg/liter was reached or 10 passages had been carried out.
To control how stable the silver resistance was, all silver-resistant isolates were subcultured five times on blood agar (Acumedia Manufacturers, Lansing, MI, USA). After each passage, at least five colonies from each isolate were tested to determine if they were still able to grow in Iso-Sensitest broth (Oxoid Ltd.) containing 512 mg/liter of silver nitrate broth.
Susceptibility to cefpodoxime, ceftibuten, piperacillin-tazobactam, cefotaxime, meropenem, ciprofloxacin, gentamicin, and cotrimoxazole was tested with the disc diffusion method after silver exposure. A change of the inhibition zone diameter of more than 5 mm was defined as significant.
Outer membrane protein profiles.
Outer membrane protein profiles were examined (32) before and after silver exposure in six randomly chosen isolates (R09, S10, P03, S08, P18, and F23). In brief, bacteria were grown to late logarithmic phase at 37°C in Mueller-Hinton broth. An amount of culture equivalent to two units of optical density at 600 nm (OD600) was centrifuged for 10 min at 2,335 × g, washed with 100 mM Tris-HCl (pH 8.0) with 20% sucrose, and incubated on ice for 10 min. Cells were centrifuged again and suspended in 1 ml 100 mM Tris-HCl (pH 8.0) with 20% sucrose and 10 mM EDTA. Lysozyme was added to a final concentration of 100 μg/ml and incubated on ice for 10 min. MgSO4, RNase A, and DNase I were added (final concentrations of 20 mM, 5 μg/ml, and 5 μg/ml, respectively), and cells were disrupted with 5 freeze-thaw cycles in dry ice–ethanol and a room temperature water bath. After the 6th freezing, samples were left to thaw on ice for 2 to 3 h. Membranes were pelleted for 25 min at 16,100 × g and washed and pelleted three times in 1 ml 20 mM NaPO4 (pH 7.0) and 0.5% N-lauroylsarcosine. The protein extracts were suspended in 60 μl Laemmli sample buffer (80 mM Tris-HCl [pH 6.8], 3% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue), boiled for 5 min, and subjected to SDS-PAGE (11% polyacrylamide, 6 M urea).
Multilocus sequence typing (MLST).
All silE-positive isolates (n = 13) were typed with multilocus sequencing, together with 10 randomly chosen ESBL-producing avian isolates and 8 ESBL-producing silE-negative human isolates. Amplification of adk, fumC, gyrB, icd, mdh, purA, and recA was carried out according to an established protocol (33) after modifications: the primer pairs were linked with universal tags, and the PCR step was performed in a Lightcycler (Roche, Switzerland). Sequence data were edited using SeqTrace (34), ClustalW (35), Jalview (36), and the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). With the eBURST program (version 3; http://eburst.mlst.net) (37), a population snapshot of the E. coli populations from the E. coli MLST database (downloaded July 2013) was performed with related and unrelated STs. The stringent group definition of clonal complexes (CCs) was used, i.e., only STs that shared identical alleles at ≥6 of 7 loci were grouped (37).
Statistical analyses.
Differences in the distributions of silE and merA genes in the various E. coli populations of human and avian origin were analyzed with Fisher's exact test. A difference was considered statistically significant if P was ≤0.05.
RESULTS
Genes encoding resistance to silver and mercury.
The silE gene was detected in 13 (6%) of the isolates examined. They were all isolated from human samples (P < 0.0001), and three of them were acquired during travels to India (n = 2) and Tajikistan (n = 1). Ten of the 13 isolates positive for the silE gene harbored, in addition, silS and silP genes. Of the remaining three isolates, two lacked the silP gene and one isolate carried the silE gene alone. See Tables 1 and 2 for distribution.
TABLE 2.
Characteristics of the silE positive strains in the study
| Isolatea | Presence of silP/presence of silS | ESBL type | TEM/SHV type | No. of passages needed to induce silver resistance (affected antibiotic susceptibility) | Sequence type and clonal complex |
|---|---|---|---|---|---|
| P03c | +/+ | CTX-M-15 | TEM/− | 8 (ceftibuten, ciprofloxacin) | ST1598; CC ST10 |
| P09 | +/+ | CTX-M-15 | TEM/− | 3 | ST388 |
| P14c | +/+ | CTX-M-15 | TEM/− | 9 (piperacillin-tazobactam, cotrimoxazole) | ST205; CC ST58 |
| P18 | +/+ | CTX-M-15 | TEM/− | 5 | ST127 |
| P21 | +/+ | CTX-M-15 | TEM/− | 7 (ceftibuten) | ST1312; CC ST10 |
| S08 | +/+ | CTX-M-14 | −/− | 7 | ST58; CC ST58 |
| S10 | −/+ | CTX-M-14 | TEM/− | 3 | ST940; CC ST58 |
| S11b,c | +/+ | CTX-M-14 | TEM/− | 2 (ciprofloxacin) | ST10; CC ST10 |
| R07c | +/+ | CTX-M-15 | TEM/− | 2 (piperacillin-tazobactam) | ST424; CC ST58 |
| R09 | +/+ | CTX-M-15 | −/− | 6 | ST940; CC ST58 |
| R20 | +/+ | CTX-M-15 | TEM/− | 3 (ceftibuten) | ST155; CC ST58 |
| F23 | −/+ | NAd | NA | 6 (piperacillin-tazobactam, gentamicin, cotrimoxazole) | 409 |
| F32 | −/− | NA | NA | NA | ST10; CC ST10 |
Designations with P and S indicate isolates from patients in surveillance programs at Uppsala University Hospital (group 2); R indicates isolates from healthy travelers (group 3); F indicates isolates from patients with diarrhea (group 1).
merA-positive strain.
Multiresistant strain.
NA, not applicable.
The merA gene was detected in 22 (10%) of the 216 isolates. Seventeen isolates were of human origin, whereas 5 were of avian origin (Table 1). Although the mercury resistance gene was present in avian samples (5%), the gene was significantly more frequent (16%) in human isolates (P = 0.004). Furthermore, the merA gene was detected only in birds from the Marseille area. A single isolate carried merA and silE genes simultaneously, and it was isolated from a stool sample from a patient taking part in the surveillance program during the K. pneumoniae outbreak.
ESBL-positive isolates.
A total of 72 isolates (34%) exhibited a phenotypic ESBL profile. Of these isolates, 63 (88%) carried ESBL genes of the CTX-M type. Beta-lactamases of the TEM and SHV types were found in the remaining 9 isolates (Table 1).
Fifty-three (76%) of the ESBL producers were isolated from human samples. The predominant ESBL type was CTX-M-15 (60%), followed by CTX-M-14 (25%). Among the CTX-M producers, 11 (85%) of the silE-positive isolates were observed (P = 0.007). The most common ESBL type among the carriers of silE gene was CTX-M-15 (n = 8) followed by CTX-M-14 (n = 3). Two of the three isolates with only one or two sil genes did not produce ESBL (Table 2).
Four CTX-M-positive isolates belonged to the pandemic O25b-ST131 clone, of which one was of avian origin. They carried CTX-M-15 (n = 3) or CTX-M-14 (n = 1). The most frequent ESBL type in avian samples was CTX-M-1 (42%). None of the O25b-ST131 positive isolates harbored silE or merA, and none of the merA-carrying birds from the Marseille area were colonized with CTX-M-producing E. coli.
Resistance to antibiotics and silver nitrate.
In general, resistance to any of the tested antibiotics was rare in ESBL-negative isolates, with the exception of resistance to cotrimoxazole. In addition, avian isolates were more susceptible to the tested antibiotics than human isolates independent of ESBL status. Multiresistance, defined as resistance to cephalosporins and three other classes of antibiotics, was present in 13 of the human ESBL-positive isolates. The corresponding figure for avian ESBL-positive isolates was 1. All isolates were susceptible to carbapenems. More detailed information is provided in the supplemental material.
silE-positive isolates were more likely to be resistant to cotrimoxazole (92% versus 40%; P = 0.0005) and gentamicin (46% versus 17%; P = 0.022) than silE-negative isolates. In merA-positive isolates, decreased susceptibility to cotrimoxazole was significantly more frequent (73% versus 23%; P < 0.0001). This was also true for gentamicin (27% versus 9%; P = 0.014). The isolate positive for both silE and merA was resistant to cotrimoxazole, gentamicin, and ciprofloxacin.
Silver nitrate MICs for silE-positive and silE-negative isolates were in the range of 8 to 32 mg/liter before silver exposure. After exposure, 12 of 13 isolates developed silver resistance. Two to nine passages (mean, 5.1 ± 2.4) were necessary to reach a MIC of >512 mg/liter. The silver-resistant phenotype remained stable after five passages in the absence of silver. Of the isolates with both genetic and phenotypic silver resistance, 11 (92%) produced CTX-M enzymes, and 10 (83%) of these had all three sil genes. For details, see Table 2.
Silver exposure decreased the inhibition zones of ceftibuten (n = 6), piperacillin-tazobactam (n = 5), cefotaxime (n = 2) and gentamicin (n = 1) with more than 5 mm. For ciprofloxacin and cotrimoxazole, both increases and decreases of the inhibition zones were observed. The changes in inhibition zones were clinically relevant for seven isolates, i.e., they changed their classification from susceptible or indeterminate to indeterminate or resistant or the inverse (for ceftibuten R20, P21 [I to R], and P03 [S to R]; for piperacillin-tazobactam, R07 [S to I], P14, and F23 [S to R]; for gentamicin, F23 [S to R]; for ciprofloxacin, S11 and P03 [R to S]; for cotrimoxazole, P14 [R to I] and F23 [S to R]).
Four of the six randomly selected isolates had deficient expression of OmpF before silver exposure. After exposure, three isolates had lost the expression of at least one more porin. Most common was loss of the OmpC porin (Fig. 1). None of the losses yielded an obvious pattern in terms of changed susceptibility to the tested antibiotics.
FIG 1.
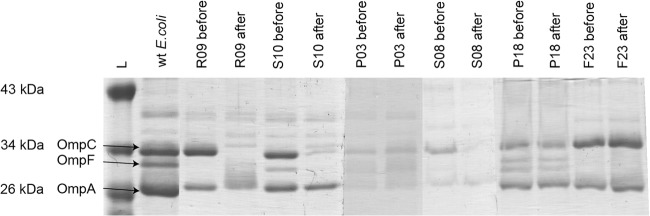
OMP profiles. Lanes (left to right): markers (L), wild-type (wt) E. coli, and isolates R09, S10, P03, S08, P18, and F23 before and after silver exposure. OmpC, OmpF, and OmpA are indicated with arrows for wt E. coli.
MLST and eBURST analyses.
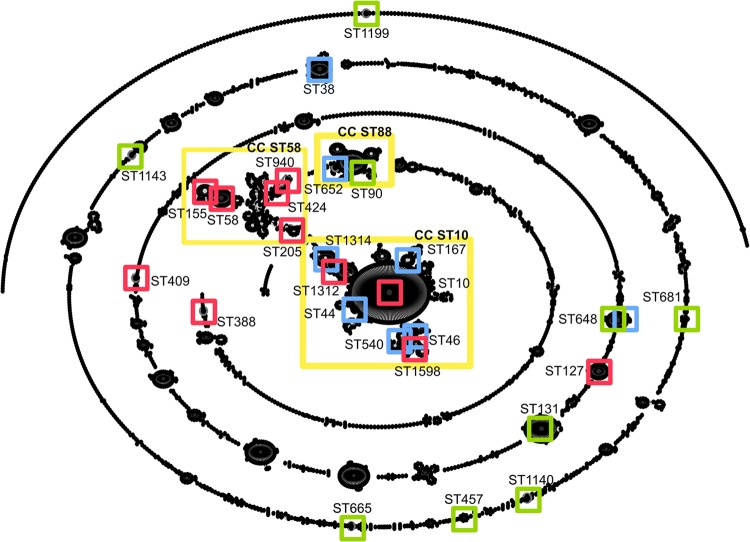
The MLST analysis of the 31 isolates identified 27 STs, and the eBURST analysis identified 11 CCs. Two complexes predominated, CC ST10 and CC ST58. Together they accounted for 48% of the isolates, which were all derived from human samples. Both humans and birds shared three CCs: CC ST131, CC ST648, and CC ST665. Furthermore, the majority of the human ESBL-producing strains clustered in the same two complexes: 77% of the silE-positive and 63% of the human silE-negative isolates clustered in the ST10 complex and ST58 complex (Fig. 2). The other silE-positive isolates were assigned to ST127 (P18), ST388 (P9), and ST409 (F23, a non-ESBL producer).
FIG 2.
Population snapshot of clusters of E. coli based on the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) with related and unrelated STs. STs identified in this study are indicated as follows: red squares, silE-positive isolates; light blue squares, human silE-negative isolates; green squares, avian isolates; and yellow boxes, clonal complexes (CC).
DISCUSSION
In the present study, the distribution of sil genes was investigated in seven different E. coli populations originating from humans and wild birds. The avian stool samples made it possible to investigate indirectly the natural exposure to silver and how increased exposure to human activities, including the production and consumption of silver-containing products, might affect the presence of genes encoding resistance to silver. The results of the study showed that sil genes were not detected in any of the avian samples, but they were found in 12% of the human stool samples. A clear majority of these isolates harbored ESBL of CTX-M types 15 and 14. Both enzymes are part of relatively recent evolutionary events. According to GenBank, the CTX-M-14 sequence was submitted from China in 2000 (accession no. AF252622) and the CTX-M-15 sequence came from India in 2001 (accession no. AY044436).
It is impossible for humans and animals, including birds, to avoid natural low-level exposure to silver. The finding that only human isolates harbored silver resistance genes, therefore, suggests a route for or level of exposure other than that caused by common environmental contamination. Although both the silver and mercury resistance operons are frequently found on plasmids harboring antibiotic resistance genes, sil and merA genes were found together only in a single multiresistant ESBL-producing isolate. A common source for the selection of these genes therefore does not seem very likely.
Resistance to mercury is one of the most widely observed phenotypes in bacteria (38). Not surprisingly, the merA gene was found in both human and avian isolates, but the frequency of this gene in human isolates was only slightly higher than that found for the sil genes. Bacterial flora from wild birds is sometimes used as an indicator of environmental contamination secondary to human activity. None of the merA-positive isolates were obtained from Swedish or Arctic birds. The only source was birds feeding from the Marseille city dump or in its surroundings (26). Interestingly, the avian E. coli isolates with the merA gene carried only older types of ESBLs, i.e., TEM and SHV, although several birds from this area were colonized with E. coli producing CTX-M-1, the dominant ESBL type in the human population in southern France (39). Furthermore, silE-positive isolates were more frequently resistant to antibiotics than merA-positive isolates. By being associated with relatively recent types of CTX-M with more multiresistant profiles, it is possible that the sil genes are coselected by antibiotics to a larger extent than the merA gene in the human population.
Although sil genes were found, none of the sil-positive isolates were phenotypically silver resistant prior to silver exposure. Silver resistance was, however, easily induced in vitro, and in some cases this was associated with loss of porins and/or altered susceptibility to antibiotics. Loss of porins is known to cause decreased susceptibility to beta-lactams (18, 32, 40). A problematic decrease in susceptibility to piperacillin-tazobactam, one of the few treatment alternatives left in more severe infections caused by ESBL-producing E. coli, was noticed. How this decrease was related to porin deficiency was, however, not obvious. For the other classes of antibiotics, the outcome of silver exposure was quite unpredictable.
Some ESBL-producing E. coli strains seem to be more prone to spread than others. Despite the successful dissemination of the pandemic clone ST131 to secluded areas, such as the Arctic, and its high virulence (6, 9), none of the silE-positive isolates belonged to this clone. Instead, nearly half of them belonged to CCs ST10 and ST58. ST10 and ST58 have both a global distribution, and they have been isolated from humans as well as animals (41–44). The K. pneumoniae outbreak in Uppsala was also caused by a global clone, ST16 (12), which in recent years has been associated with several clinically important carbapenemases (45). Interestingly, it has been suggested that resistance to silver ions represents a potent fitness factor in members of the Enterobacter cloacae complex (46).
In conclusion, 12% of the human E. coli isolates but none of the avian isolates harbored silver resistance genes. The sil genes were found among E. coli isolates carrying CTX-M-14 or CTX-M-15, two globally very successful ESBLs that emerged at the turn of the century. The potential for silver products to directly and/or indirectly select for ESBL producers and other multiresistant strains in humans and animals needs to be further investigated.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by grants from the Collaborative Foundation Uppsala-Västerås.
Footnotes
Published ahead of print 15 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01803-14.
REFERENCES
- 1.ECDC/EMEA Joint Working Group. 2009. The bacterial challenge: time to react. EMEA/576176/2009. ECDC/EMEA, Stockholm, Sweden. [Google Scholar]
- 2.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Duval J. 1987. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet ii:302–306. [DOI] [PubMed] [Google Scholar]
- 3.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315–317. 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 4.Cantón R, Coque TM. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466–475. 10.1016/j.mib.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Coque TM, Novais Â, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195–200. 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273–281. 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 7.Bartoloni A, Bartalesi F, Mantella A, Dell'Amico E, Roselli M, Strohmeyer M, Barahona HG, Barrón VP, Paradisi F, Rossolini GM. 2004. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J. Infect. Dis. 189:1291–1294. 10.1086/382191. [DOI] [PubMed] [Google Scholar]
- 8.Österblad M, Norrdahl K, Korpimäki E, Huovinen P. 2001. Antibiotic resistance. How wild are wild mammals? Nature 409:37–38. 10.1038/35051173. [DOI] [PubMed] [Google Scholar]
- 9.Sjölund M, Bonnedahl J, Hernandez J, Bengtsson S, Cederbrant G, Pinhassi J, Kahlmeter G, Olsen B. 2008. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 14:70–72. 10.3201/eid1401.070704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66:1–14. 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 11.Lytsy B, Sandegren L, Tano E, Torell E, Andersson DI, Melhus Å. 2008. The first major extended-spectrum beta-lactamase outbreak in Scandinavia was caused by clonal spread of a multiresistant Klebsiella pneumoniae producing CTX-M-15. APMIS 116:302–308. 10.1111/j.1600-0463.2008.00922.x. [DOI] [PubMed] [Google Scholar]
- 12.Sandegren L, Linkevicius M, Lytsy B, Melhus Å, Andersson DI. 2012. Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67:74–83. 10.1093/jac/dkr405. [DOI] [PubMed] [Google Scholar]
- 13.McHugh GL, Moellering RC, Hopkins CC, Swartz MN. 1975. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet i:235–240. [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, Matsui K, Lo JF, Silver S. 1999. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 5:183–188. 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 15.Odermatt A, Krapf R, Solioz M. 1994. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem. Biophys. Res. Commun. 202:44–48. 10.1006/bbrc.1994.1891. [DOI] [PubMed] [Google Scholar]
- 16.Gupta A, Phung LT, Taylor DE, Silver S. 2001. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 147:3393–3402. [DOI] [PubMed] [Google Scholar]
- 17.Johnson TJ, Siek KE, Johnson SJ, Nolan LK. 2005. DNA Sequence and comparative genomics of pAPEC-O2-R, an avian pathogenic Escherichia coli transmissible R plasmid. Antimicrob. Agents Chemother. 49:4681–4688. 10.1128/AAC.49.11.4681-4688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XZ, Nikaido H, Williams KE. 1997. Silver-resistant mutants of Escherichia coli display active efflux of Ag+ and are deficient in porins. J. Bacteriol. 179:6127–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SCENIHR. 2009. Assessment of the antibiotic resistance effects of biocides. European Commission, Brussels, Belgium. [Google Scholar]
- 20.Silver S, Phung LT, Silver G. 2006. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J. Ind. Microbiol. Biotechnol. 33:627–634. 10.1007/s10295-006-0139-7. [DOI] [PubMed] [Google Scholar]
- 21.Silver S. 2003. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27:341–353. 10.1016/S0168-6445(03)00047-0. [DOI] [PubMed] [Google Scholar]
- 22.WHO. 2002. Silver and silver compounds: environmental aspects. WHO, Geneva, Switzerland. [Google Scholar]
- 23.Tängdén T, Cars O, Melhus Å, Löwdin E. 2010. Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum β-lactamases: a prospective study with Swedish volunteers. Antimicrob. Agents Chemother. 54:3564–3568. 10.1128/AAC.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez J, Bonnedahl J, Eliasson I, Wallensten A, Comstedt P, Johansson A, Granholm S, Melhus Å, Olsen B, Drobni M. 2010. Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in glaucous-winged gull at remote Commander Islands, Russia. Environ. Microbiol. Rep. 2:329–332. 10.1111/j.1758-2229.2010.00142.x. [DOI] [PubMed] [Google Scholar]
- 25.Bonnedahl J, Drobni P, Johansson A, Hernandez J, Melhus Å, Stedt J, Olsen B, Drobni M. 2010. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65:1939–1944. 10.1093/jac/dkq222. [DOI] [PubMed] [Google Scholar]
- 26.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus Å, Kahlmeter G, Waldenström J, Johansson A, Olsen B. 2009. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 4:e5958. 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarlier V, Nicolas MH, Fournier G, Philippon A. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867–878. 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 28.Percival SL, Woods E, Nutekpor M, Bowler P, Radford A, Cochrane C. 2008. Prevalence of silver resistance in bacteria isolated from diabetic foot ulcers and efficacy of silver-containing wound dressings. Ostomy Wound Manage. 54:30–40. [PubMed] [Google Scholar]
- 29.Novais Â, Cantón R, Valverde A, Machado E, Galán JC, Peixe L, Carattoli A, Baquero F, Coque TM. 2006. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob. Agents Chemother. 50:2741–2750. 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitout JD, Hossain A, Hanson ND. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 42:5715–5721. 10.1128/JCM.42.12.5715-5721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, Mulvey MR, Nordmann P, Ruppé E, Sarthou JL, Frank T, Vimont S, Arlet G, Branger C, Woodford N, Denamur E. 2009. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother. 64:274–277. 10.1093/jac/dkp194. [DOI] [PubMed] [Google Scholar]
- 32.Adler M, Anjum M, Andersson DI, Sandegren L. 2013. Influence of acquired beta-lactamases on the evolution of spontaneous carbapenem resistance in Escherichia coli. J. Antimicrob. Chemother. 68:51–59. 10.1093/jac/dks368. [DOI] [PubMed] [Google Scholar]
- 33.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stucky BJ. 2012. SeqTrace: a graphical tool for rapidly processing DNA sequencing chromatograms. J. Biomol. Tech. 23:90–93. 10.7171/jbt.12-2303-004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 36.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530. 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osborn AM, Bruce KD, Strike P, Ritchie DA. 1997. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19:239–262. 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 39.Galas M, Decousser JW, Breton N, Godard T, Allouch PY, Pina P. 2008. Nationwide study of the prevalence, characteristics, and molecular epidemiology of extended-spectrum-β-lactamase-producing Enterobacteriaceae in France. Antimicrob. Agents Chemother. 52:786–789. 10.1128/AAC.00906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 55:1485–1493. 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben Sallem R, Ben Slama K, Estepa V, Jouini A, Gharsa H, Klibi N, Sáenz Y, Ruiz-Larrea F, Boudabous A, Torres C. 2012. Prevalence and characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates in healthy volunteers in Tunisia. Eur. J. Clin. Microbiol. Infect. Dis. 31:1511–1516. 10.1007/s10096-011-1471-z. [DOI] [PubMed] [Google Scholar]
- 42.Blaak H, Hamidjaja RA, van Hoek AH, de Heer L, de Roda Husman AM, Schets FM. 2014. Detection of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli on flies at poultry farms. Appl. Environ. Microbiol. 80:239–246. 10.1128/AEM.02616-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahmen S, Haenni M, Châtre P, Madec JY. 2013. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J. Antimicrob. Chemother. 68:2797–2801. 10.1093/jac/dkt291. [DOI] [PubMed] [Google Scholar]
- 44.Hansen F, Olsen SS, Heltberg O, Justesen US, Fuglsang-Damgaard D, Knudsen JD, Hammerum AM. 2014. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb. Drug Resist. 20:316–324. 10.1089/mdr.2013.0157. [DOI] [PubMed] [Google Scholar]
- 45.Peirano G, Ahmed-Bentley J, Fuller J, Rubin JE, Pitout JD. 2014. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J. Clin. Microbiol. 52:1575–1581. 10.1128/JCM.00162-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kremer AN, Hoffmann H. 2012. Subtractive hybridization yields a silver resistance determinant unique to nosocomial pathogens in the Enterobacter cloacae complex. J. Clin. Microbiol. 50:3249–3257. 10.1128/JCM.00885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.