Abstract
Bacteria are rapidly killed on solid copper surfaces, so this material could be useful to limit the spread of multiple-drug-resistant bacteria in hospitals. In Escherichia coli, the DNA-protecting Dps protein and the NADH:ubiquinone oxidoreductase II Ndh were not involved in tolerance to copper ions or survival on solid copper surfaces. Decreased copper tolerance under anaerobic growth conditions in the presence of ascorbate and with melibiose as the carbon source indicated that sodium-dependent symport systems may provide an import route for CuI into the cytoplasm. Glutathione-free ΔcopA ΔgshA double mutants of E. coli were more rapidly inactivated on solid copper surfaces than glutathione-containing wild-type cells. Therefore, while DNA protection by Dps was not required, glutathione was needed to protect the cytoplasm and the DNA against damage mediated by solid copper surfaces, which may explain the differences in the molecular mechanisms of killing between glutathione-containing Gram-negative and glutathione-free Gram-positive bacteria.
INTRODUCTION
Copper surfaces, long known for their beneficial effect on public health, may be useful to limit the spread of antibiotic-resistant bacteria in hospitals, long-term care facilities, and public places (1, 2). In contrast to other surfaces such as stainless steel or plastic material, microorganisms are rapidly killed on the surfaces of copper and its alloys (3). This effect has been demonstrated for bacteria such as pathogenic Escherichia coli strains (4), Pseudomonas aeruginosa (5), methicillin-resistant Staphylococcus aureus (6), Burkholderia cepacia (7), and Salmonella enterica (8), as well as for viruses (9, 10) and yeasts (11). Copper ions need to be released from the copper surfaces to mediate the killing process (12, 13), and there has to be direct contact between the cell and the copper surface (14). Therefore, a variety of parameters influence the success of the inactivation process by interfering with copper release and the availability of the copper ions for the killing action: (i) temperature, (ii) humidity, (iii) ionization/corrosion, (iv) the dry/wet test protocol used, and (v) the content of organic material (6, 15, 16).
If copper surfaces are employed to limit the spread of bacteria in hospitals (17, 18), it is essential to understand the molecular mechanisms behind the killing process, to optimize the handling and cleaning of copper surfaces, and to avoid the evolution of copper-surface-resistant pathogenic bacteria (19). Since copper ions are the mediators of the inactivation process, the presence of efflux pumps that remove copper ions from the cells extends the survival time of bacteria and yeasts on these surfaces, although they are killed later on nevertheless (11, 12, 16, 20). All cells suffered from rapid damage of the membranes (11, 21) by membrane lipid peroxidation (22), oxidative modifications of their proteins (23), and degradation of their DNA (24–26).
On solid copper surfaces, Firmicutes such as S. aureus may be predominantly killed by DNA degradation, but Proteobacteria such as E. coli may be predominantly killed by membrane damage (27). This difference was explained by protection of the nucleic acid by the periplasm of the Gram-negative proteobacteria (27). If this is true, copper import into the cytoplasm should be a slow process in proteobacteria. The zinc importer ZupT is a low-rate transport system for CuII (28) and should supply only minor amounts of copper to the cytoplasm. In contrast, import of the more toxic CuI could be mediated unspecifically and rapidly by sodium or potassium transporters (29). The ionic radius of CuI is 0.96 Å and therefore similar to that of NaI, 0.95 Å (30). NaI ions bind to copper-binding proteins (31) despite the differences in their ligand preferences. Thus, CuI is more likely to use NaI import pathways than those for KI. Time course experiments have revealed that the copA gene for the CuI-exporting P-type ATPase of E. coli is expressed stronger and for a longer time under anaerobic than under aerobic conditions (32) because CuII is anaerobically reduced to CuI and subsequently, more CopA efflux pumps are needed to balance the rapid and unspecific CuI uptake (33). This explains also the necessity of the periplasmic CuI oxidase CueO and the transenvelope CuI/AgI efflux complex CusCBA/CusF to remove CuI from the periplasm by oxidation and by export to the outside, respectively (29, 34–38).
In this study, we tested whether (i) Ndh, (ii) Dps, (iii) sodium-dependent uptake systems, or (iv) glutathione (GST) might be involved in survival of E. coli on solid copper surfaces and its tolerance to copper ions. Copper ions in liquid culture served as a model for copper ions released by solid copper surfaces during the inactivation process. The NADH:ubiquinone oxidoreductase II Ndh might reduce CuII to CuI, thus providing CuI for rapid import of CuI, maybe by NaI-dependent symporters. Ndh is a copper-dependent (39) cupric reductase (40) expressed under aerobic conditions (41, 42). Dps protects the genomes of stationary-phase cells of E. coli by binding nonspecifically to the DNA and sequesters iron in the interior of the dodecameric protein. Although Dps does not store copper, Δdps mutants of E. coli were more sensitive to copper under anaerobic conditions (43). If DNA is a target of copper surface-mediated cell killing in proteobacteria, Δdps mutants should therefore show decreased survival on copper surfaces, especially in cells without copper efflux systems such as CopA. Glutathione is also involved in copper ion tolerance (44). In contrast to most proteobacteria, Gram-positive bacteria contain no glutathione (45–47). The presence or absence of this important cellular thiol compound might explain the fundamental difference in the ways Gram-negative and -positive bacteria are killed on solid copper surfaces.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains (Table 1) were grown in Luria-Bertani (LB) medium or in Tris-buffered mineral salts medium (TMM) (48) containing 2 ml of glycerol and 3 g of Casamino Acids per liter. Anaerobic growth curves were recorded in Hungate tubes in TMM with 2 g/liter of glucose, lactose, or melibiose. Hungate tubes were used for growing and maintaining anaerobic bacteria and culture conditions (49). Solid media contained 20 g of agar/liter. Antibiotics (25 μg/ml of chloramphenicol, 50 μg/ml of kanamycin, and 125 μg/ml of ampicillin) and copper ions as chloride were added when appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| W3110 | Wild type (K-12 derivative) | 72 |
| ECA769 | ΔcopA | 44 |
| ECA464 | ΔcopA ΔcueO ΔcusCFBA | This study |
| ECA657 | Δndh | This study |
| ECA659 | usA ΔcueO Δndh | This study |
| ECA658 | ΔcueO ΔcusCFBA Δndh | This study |
| ECA765 | Δdps | This study |
| ECA766 | ΔcopA Δdps | This study |
| ECA768 | ΔcopA ΔcueO ΔcusCFBA Δdps | This study |
| ECA461 | ΔcopA ΔghsA | 44 |
| ECA462 | ΔcopA ΔghsB | 44 |
| ECA764 | ΔcopA ΔghsA Δdps | This study |
| ECA767 | ΔcopA ΔghsB Δdps | This study |
| Plasmids | ||
| pASK-IBA3 | Vector | IBA GmbH, Göttingen, Germany |
| pECD1256 | pASK-IBA3::gshA | This study |
| pECD1257 | pASK-IBA3::gshB | This study |
Dose-response growth experiments.
LB cultures of E. coli strains were diluted 1:400 into TMM, cultivated overnight, and diluted again 1:400 into fresh TMM. After 2 h of incubation at 37°C, they were diluted 1:400 into fresh TMM with increasing metal cation concentrations and cultivated for 16 h with shaking at 37°C. Optical density was measured at 600 nm using a SmartSpec3000 photometer (Bio-Rad, Munich, Germany). For anaerobic growth, the 2-h cultures were diluted 1:400 into fresh medium in Hungate tubes and cultivated without shaking at 37°C for 16 h. TMM with added resazurin was boiled for reduction, immediately used to fill Hungate tubes, and autoclaved. Optical density was measured using a Spectronic20+ photometer (Milton Roy, Ivyland, PA).
Time-dependent growth experiments.
LB cultures of E. coli strains were diluted 1:400 into TMM, cultivated overnight, and diluted again 1:400 into fresh TMM. After 2 h of incubation at 37°C, they were diluted 1:400 into fresh TMM in 12-well plates (Greiner Bio-One, Frickenhausen, Germany) with 50 μM CuCl2 (final concentration) and cultivated with shaking at 37°C. Optical density was measured every 30 min at 600 nm using a TECAN Infinite 200 PRO reader (TECAN, Männersdorf, Switzerland). Anaerobic growth cultures were measured every 2 h using a Spectronic20+ photometer (Milton Roy).
Gene deletions and other genetic techniques.
Genes were deleted by insertion of resistance cassettes using the λ red recombinase system (50). Initial deletions were performed in E. coli strain BW25113, in which the target genes were exchanged for a chloramphenicol (cat) resistance cassette, and subsequently transferred by general transduction with phage P1 into E. coli strain W3110 or its derivatives. In the resulting mutant strains, the genes were disrupted by insertion of the cat resistance cassette through homologous recombination. Multiple deletions were constructed by FLP recombination target (FRT)-dependent elimination of the respective resistance cassette assisted by flippase from plasmid pCP20 (50) and subsequent general phage P1 transduction. For construction of plasmids pECD1256 and pECD1257, the genes gshA and gshB were amplified by PCR from start to stop codon and cloned with SacII and XhoI in the vector pASK-IBA3 (IBA GmbH, Göttingen, Germany), respectively. Due to the stop codon, no fusion with the C-terminal StrepTagII resulted. Both plasmids were checked by restriction analysis and DNA sequencing. Otherwise, standard molecular genetic techniques were used (51). PCR was performed with Taq or Taq/Pwo DNA polymerase (Roche, Mannheim, Germany). All primer sequences can be obtained upon request.
RNA isolation and RT-PCR.
E. coli wild-type and mutant cells were cultivated as described below for the survival assay. Total RNA was isolated and the reverse transcriptase (RT) reaction performed as previously described (52). To exclude experimental artifacts resulting from DNA contaminations, only RNA preparations that did not generate products in a PCR with chromosomal primers without a previous RT reaction were used. As an endogenous control, rpoZ was used. All cDNAs displayed the same expression level when amplified with primers for the gene rpoZ. A no-template control was performed under identical conditions as for the target genes. Two independent biological samples were used. Details for PCR protocols and primer sequences are available on request.
Glutathione determination.
Overnight cultures were diluted 1:100 into fresh TMM without or with CuCl2 and cultivated with shaking at 37°C until the optical density (600 nm) reached 2.25 ± 0.25 (late exponential phase of growth). Cells representing 5 mg (dry mass) were harvested by centrifugation (15 min at 4,500 × g and 4°C) and washed twice in medium. The pellet was suspended in 3 volumes of 5% sulfosalicylic acid (SSA) and disrupted by two freeze-thawing cycles. Cell debris was removed by centrifugation (15 min at 15,300 × g and 4°C). The supernatant was used to determine the protein concentration with the QuantiPro bicinchoninic acid (BCA) assay kit (Sigma-Aldrich, Taufkirchen, Germany) using bovine serum albumin as a standard and to measure the reduced glutathione (GSH) content with a glutathione assay kit (CS0260; Sigma-Aldrich) according to the manufacturer's instructions. The enzymatic determination of the total amount of glutathione (GSH and oxidized glutathione [GSSG]) after deproteinization with SSA was conducted photometrically using TNB [5,5′dithiobis(2-nitrobenzoic acid)].
Survival on metal surface assay.
As copper alloy coupons, copper C11000 (99.9% Cu), cupronickel C75200 (maximum of 62% Cu, maximum of 18% Ni, and maximum of 21% Zn) and, as a control, stainless steel (AISI 304) were used and treated prior to each experiment (12). As previously described, overnight cultures of E. coli were concentrated 10-fold by centrifugation and cells were suspended in phosphate-buffered saline (PBS) (12). A 40-μl sample (approximately 1.5 × 109 cells) was applied to a sterile cotton swab and spread evenly once across a 2.5- by 2.5-cm metal coupon. All samples dried completely within 5 s after contact with the surfaces. Coupons were transferred in 10 ml of PBS with 20 glass beads (2 mm; Carl Roth, Karlsruhe, Germany) and vortexed vigorously for 1 min to remove the cells. Samples were diluted in PBS and plated on LB agar. Surviving bacteria were counted as CFU.
DNA degradation assay.
For the DNA assay, a 20-fold-higher cell density (cell cultures were concentrated 200-fold in PBS) compared to the survival experiments were spread on the coupons as described above. After cell removal from the coupons into PBS, DNA was isolated with the GeneJet genomic DNA purification kit (Thermo Scientific/Fermentas, St. Leon-Rot, Germany), concentrated in a SpeedVac, separated by agarose gel electrophoresis on a 1% (wt/vol) gel, stained with ethidium bromide, and documented with a UV system (gel imager; INTAS, Göttingen, Germany).
RESULTS
Import of copper ions.
To test the hypothesis that Ndh may reduce CuII released by copper surfaces to CuI, which is subsequently imported into the cytoplasm by sodium symporters, the ndh gene was deleted in the E. coli W3110 wild type, the ΔcueO ΔcusCFBA double mutant, and the ΔcueO single mutant. Tolerance to copper ions of the strains with and without ndh was tested under aerobic and anaerobic conditions, but no difference was found (data not shown). Thus, the Ndh cupric reductase (alone) did not increase copper sensitivity in E. coli.
To test whether sodium-dependent transport systems might use CuI instead of NaI in an unspecific transport process, growth of wild-type E. coli and its ΔcopA ΔcueO ΔcusCFBA triple mutant in the presence of substrates for sodium-dependent import systems was studied. There was no effect on copper tolerance when each strain was cultivated in the presence of various amino acids (Gln, Ser, Pro, Ala, Ile, and Leu) under aerobic conditions, neither in the presence nor in the absence of additional glucose (data not shown).
Copper tolerance of the E. coli W3110 wild type under aerobic conditions with lactose or melibiose as a carbon source was not different from that in glucose; however, the ΔcopA ΔcueO ΔcusCFBA triple mutant was more sensitive to copper on lactose or melibiose than on glucose (Fig. 1). Under anaerobic conditions and in the absence of ascorbate, wild-type cells cultivated in melibiose-containing medium were less tolerant to copper than those on lactose or glucose (Fig. 2A), while no difference was visible in the triple mutant. In the presence of ascorbate, which reduces CuII to CuI, wild-type and triple mutant cells were less tolerant to copper when grown in melibiose than in glucose; cells cultivated on lactose showed an intermediary level of copper tolerance (Fig. 2B and C). Since melibiose is imported by the sodium-dependent MelB system (53, 54) but lactose by the proton-dependent LacY system (55, 56), this may indicate that CuI may be indeed a substrate for this sodium-dependent uptake system in E. coli.
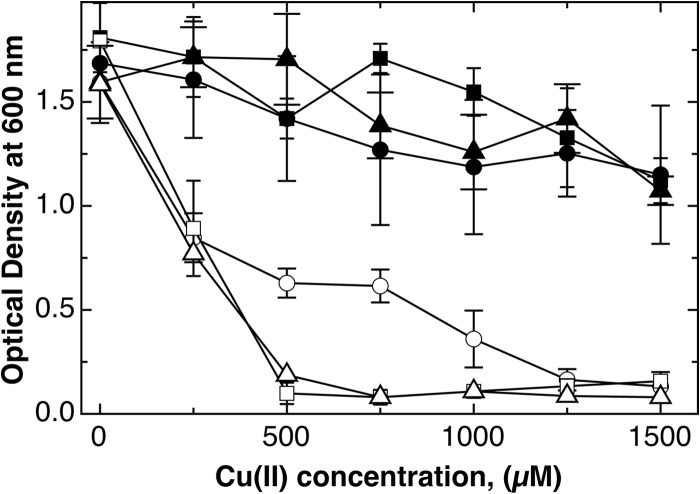
FIG 1.
Aerobic growth of E. coli mutant strains in the presence of different carbon sources. Dose-response curves (16 h at 37°C with shaking in TMM containing 2 g/liter of the respective carbon source) were recorded for E. coli wild-type strain W3110 (closed symbols) and the ΔcopA ΔcueO ΔcusCFBA mutant strain (open symbols) in the presence of glucose (circles), lactose (triangles), or melibiose (squares). The experiment was repeated at least three times; error bars show standard deviations.
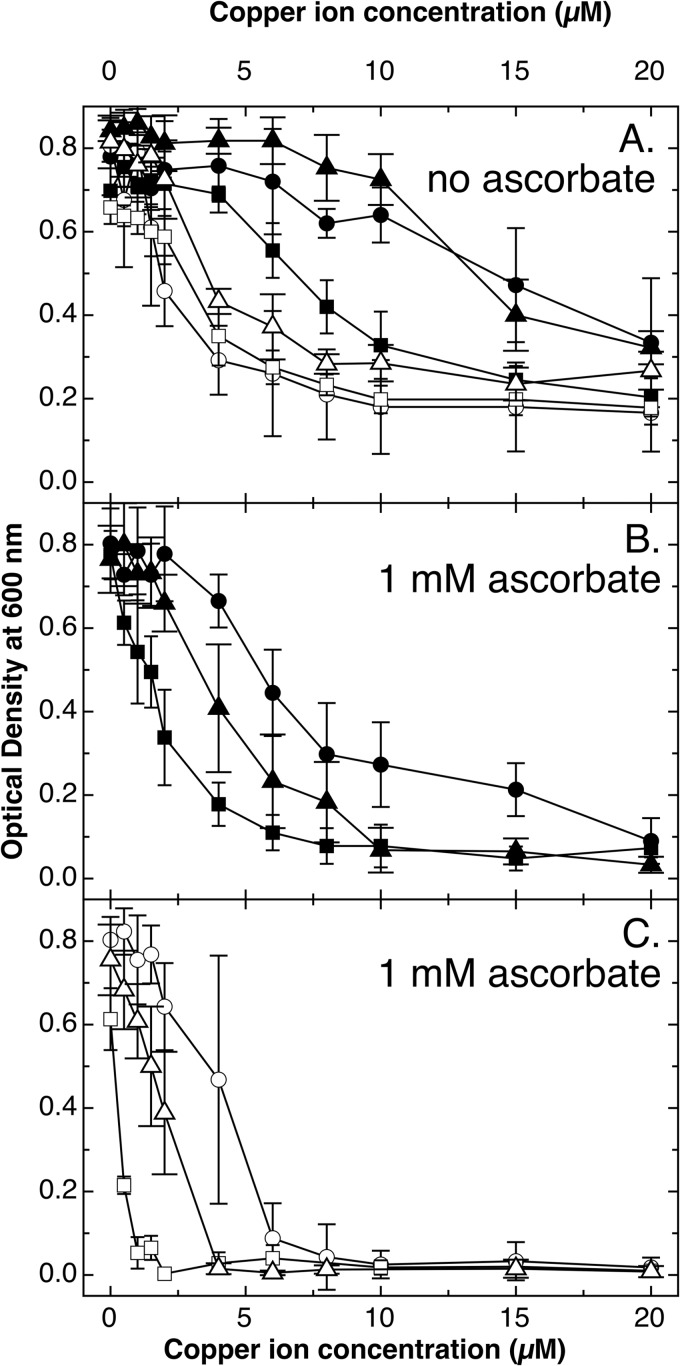
FIG 2.
Anaerobic growth of E. coli mutant strains in the presence of different carbon sources. Dose-response curves (16 h at 37°C in TMM containing carbon source) were recorded for E. coli wild-type strain W3110 (closed symbols) and the ΔcopA ΔcueO ΔcusCFBA mutant strain (open symbols) in the presence of glucose (circles), lactose (triangles), or melibiose (squares). The growth medium contained 1 mM ascorbate to reduce CuII to CuI (B and C) or not (A). The experiment was repeated at least three times; error bars show standard deviations.
Dps was not required for copper tolerance.
To address the question of whether DNA damage is involved in killing of E. coli cells on copper surfaces, the dps gene was deleted in wild-type E. coli strain W3310 and various mutants carrying deletions in genes (copA, cueO, cusCFBA; gshA and gshB for synthesis of glutathione [GSH] via gamma-glutamyl-cysteine) that are involved in copper tolerance (32, 37, 38, 44, 57). RT-PCR controls were done in all experiments to demonstrate the absence or presence of the gene transcripts in the respective mutants (data not shown). The effect of the dps deletion in these strains was subsequently determined in aerobic and anaerobic liquid cultures in the presence of copper ions and on solid copper alloy surfaces.
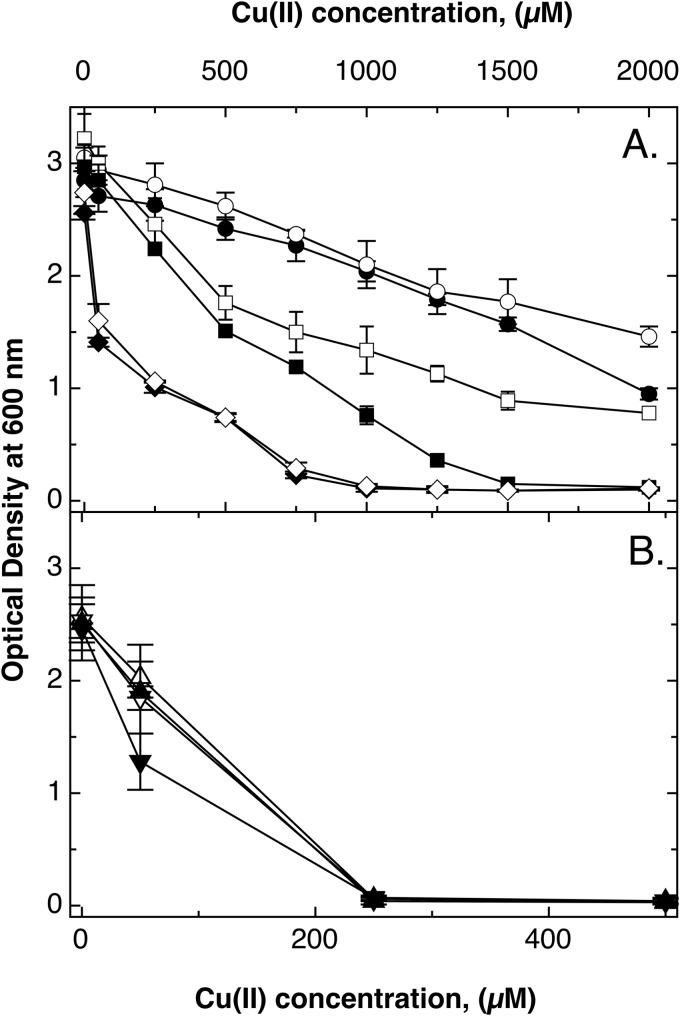
In cultures under aerobic conditions, deletion of dps led to a small effect (Fig. 3A), as published previously (43). To challenge the cells with increased cytoplasmic copper concentrations, the Δdps effect was also tested in a ΔcopA mutant strain. Deletion of copA significantly decreases copper tolerance in E. coli (33, 44), but additional deletion of the dps gene diminished this effect (Fig. 3A) by an unknown mechanism.
FIG 3.
Aerobic growth of E. coli mutant strains with or without dps. Dose-response curves (16 h at 37°C in TMM) were recorded for E. coli wild-type strain W3110 (●) and Δdps (○), ΔcopA (■), ΔcopA Δdps (□), ΔcopA ΔcueO ΔcusCFBA (◆), and ΔcopA ΔcueO ΔcusCFBA Δdps (♢) mutants (A). Panel B shows only the data points up to 500 μM CuCl2 for the ΔcopA ΔghsA (▲), ΔcopA ΔghsB (▼), ΔcopA ΔghsA Δdps (△), and ΔcopA ΔghsB Δdps (▽) mutant strains. The experiment was repeated at least three times; error bars show standard deviations.
The gene dps was deleted in the ΔcopA ΔcueO ΔcusCFBA triple mutant. There was no decrease of tolerance in the resulting Δdps ΔcopA ΔcueO ΔcusCFBA quadruple mutant compared to that of the triple mutant strain (Fig. 3A). Finally, dps was removed from ΔcopA ΔgshA and ΔcopA ΔgshB double mutants that were not able to produce glutathione (ΔgshA) or produced equal amounts of γ-glutamyl-cysteine (γEC) instead (ΔgshB) (44). Again, there was no effect under aerobic growth conditions (Fig. 3B).
The same mutant strains with or without dps were also cultivated under anaerobic conditions in the presence of ascorbate. The dps gene was not important for copper tolerance in E. coli W3110 wild-type cells and increased copper tolerance only a little in the ΔcopA mutant (Fig. 4A). There was also no effect when dps was deleted in the ΔcopA ΔcueO ΔcusCFBA triple mutant (Fig. 4A).
FIG 4.
Anaerobic growth of E. coli mutant strains with or without dps. Dose-response curves (16 h at 37°C in TMM containing 2 g/liter of glucose and 1 mM ascorbate) were recorded for E. coli wild-type strain W3110 (●) and Δdps (○), ΔcopA (■), ΔcopA Δdps (□), ΔcopA ΔcueO ΔcusCFBA (◆), and ΔcopA ΔcueO ΔcusCFBA Δdps (♢) mutants (A) and for the ΔcopA ΔghsA (▲), ΔcopA ΔghsB (▼), ΔcopA ΔghsA Δdps (△), and ΔcopA ΔghsB Δdps (▽) mutants (B). The experiment was repeated at least three times; error bars show standard deviations.
In contrast to findings under aerobic conditions in LB medium (44), the ΔcopA ΔgshB mutant was more tolerant to copper ions than the ΔcopA ΔgshA mutant. This indicated that γEC, which slightly increased copper toxicity under aerobic conditions, was important for copper tolerance under anaerobic conditions. The ΔcopA ΔgshA mutant strain, however, grew poorly even in the absence of copper (Fig. 4B). There was, however, again no effect of Δdps on copper tolerance of either mutant strain with a defect in the glutathione biosynthesis pathway. This indicated that Dps did not contribute to copper tolerance in E. coli.
Survival on surfaces of solid copper alloys.
Survival of the various Δdps and Δgsh mutants on the surfaces of solid copper alloys was determined. For 99.9% “pure” copper (alloy C11000), all strains could not be cultivated after incubation of just a few seconds on this surface (data not shown). To get a better differentiation, the experiments were repeated on cupronickel (alloy C75200; 62% Cu, 18% Ni, and 21% Zn). The number of culturable cells on cupronickel coupons was decreased by a factor of about 10 within a minute in the cases of wild-type strain W3110, the ΔcopA single mutant, the ΔcopA ΔcueO ΔcusCFBA triple mutant, and the ΔcopA ΔgshB double mutant, all with or without dps (see Fig. S1 in the supplemental material for the triple mutant; all other data are not shown).
In contrast, no survivors were found among the ΔcopA ΔgshA double mutants with and without dps after 30 s (see Fig. S1B in the supplemental material), while no difference between these mutants was observed in the case of stainless steel (see Fig. S1A). When DNA was isolated from wild-type and ΔcopA ΔgshA mutant strains, DNA remained intact for 3 h for wild-type cells incubated on stainless steel and for 60 min for mutant cells on steel and wild-type cells on cupronickel, while DNA from mutants on cupronickel showed beginning degradation after only 30 min (see Fig. S2). This clearly indicated that glutathione (or at least presence of γEC) but not the Dps protein may be essential for survival of E. coli on solid copper surfaces and for stability of its nucleic acids.
Glutathione is required for survival of E. coli on solid copper surfaces.
The contribution of GSH was subsequently investigated in more detail with ΔcopA ΔgshA and ΔcopA ΔgshB double mutants that were also complemented in trans with the gshA or the gshB gene cloned into vector pASK-IBA3. To increase the sensitivity of the glutathione determination, the cells were disrupted not by ultrasonication as published previously (44) but by a freeze-thaw cycle. This led to a 15.3-fold-lower release of proteins from the cells (data not shown), enhancing the efficiency of the subsequent deproteinization step, and consequently to a higher quotient of glutathione to released protein. The glutathione content of wild-type E. coli strain W3110 was 97 ± 3 mg of GSH per g of released protein (Table 2), 7-fold higher than the published value of 13.7 ± 1.6 mg of GSH per g of total protein (44). The value did not change when copA was deleted but increased 50% when 50 μM CuII was present in the growth medium. GSH was not detected in ΔgshA and ΔgshB mutant cells even in this assay with increased sensitivity. Expression of the respective gene in trans restored the GSH content of the cells (Table 2) and restored copper tolerance under aerobic (see Fig. S3) and anaerobic (see Fig. S4) conditions.
TABLE 2.
Total glutathione contents in E. coli strains in the absence or presence of 50 μM CuCl2
| Bacterial strain | GSH content (mg/g of released protein) with CuII in growth medium at the indicated concn (μM)a |
|
|---|---|---|
| 0 | 50 | |
| Wild-type W3110 | 97 ± 3 | 146 ± 34 |
| ΔcopA (pASK-IBA3) mutant | 119 ± 19 | 124 ± 28 |
| ΔcopA ΔgshA (pASK-IBA3) mutant | 0 ± 1 | 0 ± 0 |
| ΔcopA ΔgshB (pASK-IBA3) mutant | 3 ± 2 | 0 ± 0 |
| ΔcopA ΔgshA (pASK-IBA3::gshA) mutant | 132 ± 10 | 195 ± 32 |
| ΔcopA ΔgshB (pASK-IBA3::gshB) mutant | 115 ± 14 | 163 ± 27 |
Shown are contents of total glutathione (GSH and GSSG); values are means ± standard deviations.
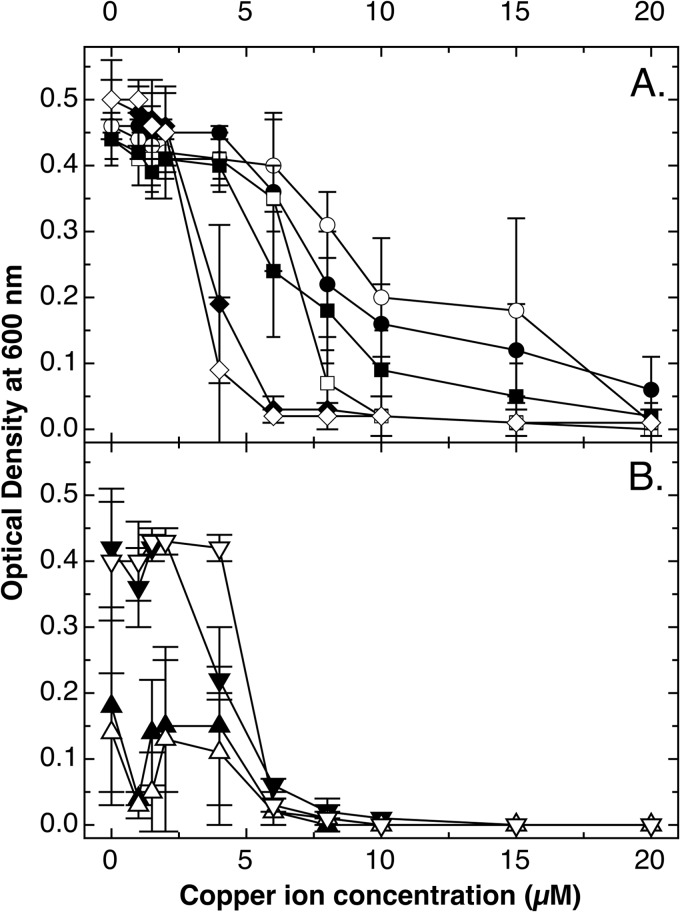
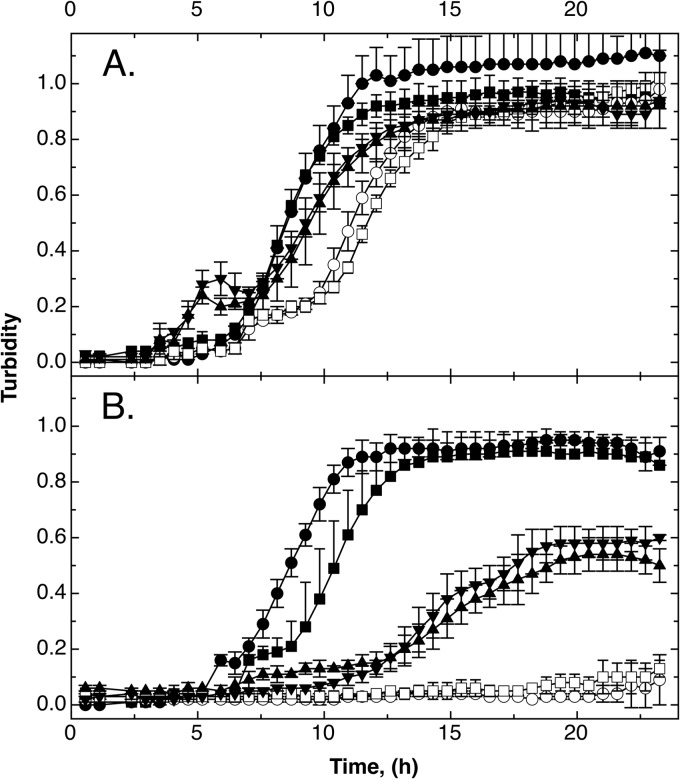
Time-dependent growth experiments under aerobic conditions indicated that at high copper concentrations (500 μM CuCl2), in trans complementation with gshA and ghsB increased the tolerance level of the ΔgshA and ΔgshB mutants, respectively, again but not to the level of the parent strain (Fig. 5B), while tolerance was back to the level of the ΔcopA parent strain at 50 μM CuCl2 (data not shown). Survival rates of the strains tested were not different on stainless steel (Fig. 6A), but the ΔghsA and ΔghsB mutant strains were again rapidly killed on cupronickel alloy (Fig. 6B). Since the ΔgshA mutant was more sensitive than the ΔghsB mutant, the gamma-glutamyl-cysteine present in the ΔghsB strain (44) might offer a small degree of protection. Expression of the gshA and the ghsB gene in trans again restored the tolerance level of the parent strain. Thus, glutathione increased survival of E. coli on metallic copper surfaces.
FIG 5.
Expression of gshA or gshB in trans in a ΔghsA or ΔghsB mutant strain, respectively, restores copper tolerance under aerobic conditions but not completely. Time-dependent growth curves (37°C in TMM containing 2 g/liter of glucose) in the absence (A) or presence (B) of 500 μM CuCl2 were recorded as turbidity at 600 nm in a TECAN reader for E. coli wild-type strain W3110 (●) and ΔcopA (■), ΔcopA ΔghsA (○), and ΔcopA ΔghsB (□) mutants, all containing the vector plasmid pASK3, and ΔcopA ΔghsA(pASK-IBA3::gshA) (▲) and ΔcopA ΔghsB(pASK-IBA3::ghsB) (▼) mutants. The experiment was repeated at least three times; error bars show standard deviations.
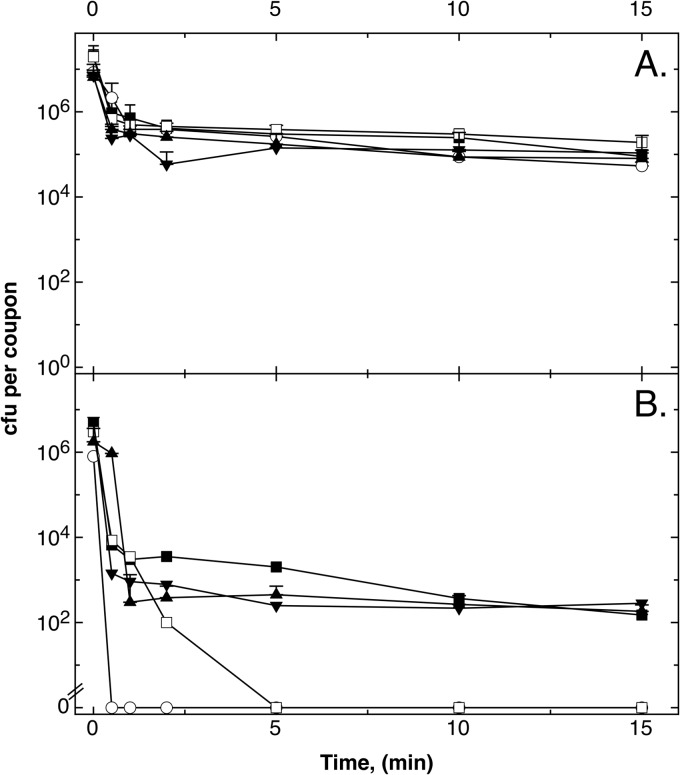
FIG 6.
Expression of gshA or gshB in trans in a ΔghsA or ΔghsB mutant strain, respectively, confers protection from the toxic effects associated with metallic copper surfaces. Cells of E. coli W3110 ΔcopA (■), ΔcopA ΔghsA (○), and ΔcopA ΔghsB (□) mutant strains, all containing the vector plasmid pASK-IBA3, and ΔcopA ΔghsA(pASK-IBA3::gshA) (▲) and ΔcopA ΔghsB(pASK-IBA3::ghsB) (▼) mutant strains were streaked on stainless steel (A) or cupronickel (B). After the indicated periods under ambient conditions (23°C), cells were removed from metal surfaces, diluted, and plated on LB agar. Surviving cells were counted as CFU after 16 h at 37°C. Values are means of results from three repetitions of the experiment; error bars show standard deviations.
DISCUSSION
Bacteria, bacterial biofilms, yeasts, and viruses are inactivated on moist or dry copper surfaces, and this may be useful to control the spread of pathogenic entities in hospitals (5, 7, 9, 17, 58). On the other hand, some bacterial strains are able to survive on solid copper surfaces, such as on coins (19). Evolution of copper surface-resistant pathogenic bacteria, however, has to be prevented if indeed copper surfaces are to be used in hospitals and elsewhere. It is therefore essential to understand the molecular mechanisms underlying the killing process.
Copper ions are released from solid copper surfaces (13) and are bound by the cells (21), which leads to killing. Differences in the cellular morphology or physiology between Gram-negative Proteobacteria and Gram-positive Firmicutes and Actinobacteria may decide how these copper ions kill the respective bacterial cells (27). In Gram-negative bacteria, copper ions need to be imported into the periplasm first. OmpC and ComC (YcfR) play a role here (57, 59). Although a strain deficient in the gene for the NADH:ubiquinone oxidoreductase II (Ndh) suffered a faster inactivation than its parent strain in the presence of copper and tert-butyl hydroperoxide (60), Ndh was not involved in copper tolerance in the absence of this substance (data not shown). So, Ndh seems not to be required for reduction of CuII to CuI. This process can be mediated by respiratory chain components (61).
A comparison of aerobic and anaerobic growth of E. coli (Fig. 3 and 4) shows the higher toxicity of copper ions under anaerobic conditions in the presence of ascorbate. This may indicate that CuI is more toxic than CuII; however, no uptake system was known for copper ions in E. coli except some CuII transport by the ZupT protein as indicated by competition experiments (28). On the other hand, upregulation of copA expression from background expression to about 350 copies per cell took only 2 min after addition of CuII to the cells (32), and this regulatory event is performed by the MerR-type regulator CueR, which is usually constantly bound to its operator on the DNA (62). Thus, a CuI import pathway should exist.
Due to the similarities in charge and ionic diameter, CuI may be transported instead of NaI by sodium-dependent transport systems such as MelB (53, 54) (Fig. 2). In agreement with this, E. coli cells suspended in 0.8% NaCl showed prolonged survival on copper surfaces (16) because the high concentration of NaI may competitively block CuI uptake by these transport systems. NaI binds to copper-binding proteins (31) and vice versa; copper ions also inhibit NaI transport across membranes (63). So, copper ions could be able to enter the cytoplasm of E. coli by mimicking NaI or by ZupT as an import route for CuII (28). As a practical consequence, sodium ions should not be present in solutions used to clean copper surfaces in hospitals.
In the cytoplasm, CuII and GSH yield rapidly CuIGSH2, which reacts with O2 to generate a superoxide radical (64) and CuIIGSSG, which can be reduced back to CuIGSH2 by GSH (65). Nevertheless, the presence of GSH increases copper ion resistance in the absence of the copper-exporting PIB1-type ATPase CopA (44). Copper does not damage DNA by oxidation in vivo in E. coli (66) but destabilizes iron-sulfur clusters in Bacillus subtilis (67) and in E. coli, especially in dehydratases (68), and the copper efflux system CusCBA is needed to protect iron-sulfur clusters (69). This toxic action of copper ions is similar to that of Cd2+, which is also a toxic “soft” transition metal cation (70).
Cells of E. coli and various yeasts suffered extensive membrane damage within minutes after exposure to copper surfaces, but the DNA was not immediately affected (11, 21). Copper leads first to oxidative damage of phospholipids and subsequent loss of membrane integrity and cell death before the DNA is fragmented (22). On the other hand, DNA as a primary target was observed in pathogenic enterococci (24, 26) and methicillin-resistant Staphylococcus aureus cells (25). DNA damage came before an effect on the cytoplasmic membrane in these examples. Indeed, while in Gram-positive bacteria the DNA seems to be the primary target of copper released from solid copper surfaces, Gram-negative bacteria and eukaryotes suffered primarily from damage of the cytoplasmic membrane (27), and damage of the plasma membrane of yeasts by cupric ions has already been described some time ago (71). In addition to differences in cellular morphology, Proteobacteria and Eukaryota contain large intracellular concentrations of glutathione, but Firmicutes such as enterococci or S. aureus contain much smaller concentrations and/or other thiol compounds (45, 46). Moreover, a ΔcopA ΔgshA mutant of E. coli was more rapidly inactivated on cupronickel than its parent strain, and its DNA was also less stable. Taken together, the findings show that glutathione but not Dps was required to protect cytoplasm and DNA of E. coli against damage by copper ions released from solid copper surfaces. This effect may explain some of the differences in the modes of killing of Gram-negative and -positive bacteria on this material.
Supplementary Material
ACKNOWLEDGMENT
We thank Kerstin Helbig for helpful discussions about the GSH assay.
Footnotes
Published ahead of print 5 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02842-14.
REFERENCES
- 1.Grass G, Rensing C, Solioz M. 2011. Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77:1541–1547. 10.1128/AEM.02766-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt MG, Attaway HH, Sharpe PA, John J, Sepkowitz KA, Morgan A, Fairey SE, Singh S, Steed LL, Cantey JR, Freeman KD, Michels HT, Salgado CD. 2012. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J. Clin. Microbiol. 50:2217–2223. 10.1128/JCM.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogers J, Dowsett AB, Dennis PJ, Lee JV, Keevil CW. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noyce JO, Michels H, Keevil CW. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239–4244. 10.1128/AEM.02532-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison JJ, Turner RJ, Joo DA, Stan MA, Chan CS, Allan ND, Vrionis HA, Olson ME, Ceri H. 2008. Copper and quaternary ammonium cations exert synergistic bactericidal and antibiofilm activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:2870–2881. 10.1128/AAC.00203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michels HT, Noyce JO, Keevil CW. 2009. Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett. Appl. Microbiol. 49:191–195. 10.1111/j.1472-765X.2009.02637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim M, Wang F, Lou MM, Xie GL, Li B, Bo Z, Zhang GQ, Liu H, Wareth A. 2011. Copper as an antibacterial agent for human pathogenic multidrug resistant Burkholderia cepacia complex bacteria. J. Biosci. Bioeng. 112:570–576. 10.1016/j.jbiosc.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Zhu LB, Elguindi J, Rensing C, Ravishankar S. 2012. Antimicrobial activity of different copper alloy surfaces against copper resistant and sensitive Salmonella enterica. Food Microbiol. 30:303–310. 10.1016/j.fm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Noyce JO, Michels H, Keevil CW. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl. Environ. Microbiol. 73:2748–2750. 10.1128/AEM.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JY, Dennehy JJ. 2011. Differential bacteriophage mortality on exposure to copper. Appl. Environ. Microbiol. 77:6878–6883. 10.1128/AEM.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quaranta D, Krans T, Santo CE, Elowsky CG, Domaille DW, Chang CJ, Grass G. 2011. Mechanisms of contact-mediated killing of yeast cells on dry metallic copper surfaces. Appl. Environ. Microbiol. 77:416–426. 10.1128/AEM.01704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espírito Santo C, Taudte N, Nies DH, Grass G. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977–986. 10.1128/AEM.01938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molteni C, Abicht HK, Solioz M. 2010. Killing of bacteria by copper surfaces involves dissolved copper. Appl. Environ. Microbiol. 76:4099–4101. 10.1128/AEM.00424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews S, Hans M, Mucklich F, Solioz M. 2013. Contact killing of bacteria on copper is suppressed if bacterial-metal contact is prevented and is induced on iron by copper ions. Appl. Environ. Microbiol. 79:2605–2611. 10.1128/AEM.03608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih HY, Lin YE. 2010. Efficacy of copper-silver ionization in controlling biofilm- and plankton-associated waterborne pathogens. Appl. Environ. Microbiol. 76:2032–2035. 10.1128/AEM.02174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elguindi J, Moffitt S, Hasman H, Andrade C, Raghavan S, Rensing C. 2011. Metallic copper corrosion rates, moisture content, and growth medium influence survival of copper ion-resistant bacteria. Appl. Microbiol. Biotechnol. 89:1963–1970. 10.1007/s00253-010-2980-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikolay A, Huggett S, Tikana L, Grass G, Braun J, Nies DH. 2010. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl. Microbiol. Biotechnol. 87:1875–1879. 10.1007/s00253-010-2640-1. [DOI] [PubMed] [Google Scholar]
- 18.Casey AL, Adams D, Karpanen TJ, Lambert PA, Cookson BD, Nightingale P, Miruszenko L, Shillam R, Christian P, Elliott TS. 2010. Role of copper in reducing hospital environment contamination. J. Hosp. Infect. 74:72–77. 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 19.Santo CE, Morais PV, Grass G. 2010. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76:1341–1348. 10.1128/AEM.01952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elguindi J, Wagner J, Rensing C. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 106:1448–1455. 10.1111/j.1365-2672.2009.04148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espírito Santo C, Lam EW, Elowsky CG, Quaranta D, Domaille DW, Chang CJ, Grass G. 2011. Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol. 77:794–802. 10.1128/AEM.01599-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong R, Kang TY, Michels CA, Gadura N. 2012. Membrane lipid peroxidation in copper alloy-mediated contact killing of Escherichia coli. Appl. Environ. Microbiol. 78:1776–1784. 10.1128/AEM.07068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nandakumar R, Santo CE, Madayiputhiya N, Grass G. 2011. Quantitative proteomic profiling of the Escherichia coli response to metallic copper surfaces. Biometals 24:429–444. 10.1007/s10534-011-9434-5. [DOI] [PubMed] [Google Scholar]
- 24.Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76:5390–5401. 10.1128/AEM.03050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver L, Noyce JO, Michels HT, Keevil CW. 2010. Potential action of copper surfaces on meticillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 109:2200–2205. 10.1111/j.1365-2672.2010.04852.x. [DOI] [PubMed] [Google Scholar]
- 26.Warnes SL, Keevil CW. 2011. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following wet or dry surface contact. Appl. Environ. Microbiol. 77:6049–6059. 10.1128/AEM.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ. Microbiol. 14:1730–1743. 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 28.Taudte N, Grass G. 2010. Point mutations change specificity and kinetics of metal uptake by ZupT from Escherichia coli. Biometals 23:643–656. 10.1007/s10534-010-9319-z. [DOI] [PubMed] [Google Scholar]
- 29.Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper resistance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670–30677. 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 30.Weast RC. 1984. CRC handbook of chemistry and physics, 64th ed. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 31.Bagautdinov B. 2014. The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: characterization of metal-binding sites and metal-induced assembly. Acta Crystallogr. F Struct. Biol. Commun. 70:404–413. 10.1107/S2053230X14003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thieme D, Neubauer P, Nies DH, Grass G. 2008. Sandwich hybridization assay for sensitive detection of dynamic changes in mRNA transcript levels in crude Escherichia coli cell extracts in response to copper ions. Appl. Environ. Microbiol. 74:7463–7470. 10.1128/AEM.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97:652–656. 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grass G, Rensing C. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145–2147. 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, Tollin G, Rensing C, Montfort WR. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:2766–2771. 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh SK, Grass G, Rensing C, Montfort WR. 2004. Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 186:7815–7817. 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke S, Grass G, Nies DH. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965–972. [DOI] [PubMed] [Google Scholar]
- 38.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting CusCFBA efflux system from Escherichia coli. J. Bacteriol. 185:3804–3812. 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapisarda VA, Chehín RN, De Las Rivas J, Rodríguez-Montelongo L, Farías RN, Massa EM. 2002. Evidence for Cu(I)-thiolate ligation and prediction of a putative copper-binding site in the Escherichia coli NADH dehydrogenase-2. Arch. Biochem. Biophys. 405:87–94. 10.1016/S0003-9861(02)00277-1. [DOI] [PubMed] [Google Scholar]
- 40.Rapisarda VA, Montelongo LR, Farias RN, Massa EM. 1999. Characterization of an NADH-linked cupric reductase activity from the Escherichia coli respiratory chain. Arch. Biochem. Biophys. 370:143–150. 10.1006/abbi.1999.1398. [DOI] [PubMed] [Google Scholar]
- 41.Green J, Guest JR. 1994. Regulation of transcription at the ndh promoter of Escherichia coli by FNR and novel factors. Mol. Microbiol. 12:433–434. 10.1111/j.1365-2958.1994.tb01032.x. [DOI] [PubMed] [Google Scholar]
- 42.Spiro S, Roberts RE, Guest JR. 1989. Fnr-dependent repression of the ndh gene of Escherichia coli and metal-ion requirement for FNR-regulated gene expression. Mol. Microbiol. 3:601–608. 10.1111/j.1365-2958.1989.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 43.Thieme D, Grass G. 2010. The Dps protein of Escherichia coli is involved in copper homeostasis. Microbiol. Res. 165:108–115. 10.1016/j.micres.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Helbig K, Bleuel C, Krauss GJ, Nies DH. 2008. Glutathione and transition metal homeostasis in Escherichia coli. J. Bacteriol. 190:5431–5438. 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahey RC. 2001. Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55:333–356. 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 46.Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J. Bacteriol. 133:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newton GL, Fahey RC. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388–394. 10.1007/s00203-002-0469-4. [DOI] [PubMed] [Google Scholar]
- 48.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hungate RE. 1969. A roll tube method for cultivation of strict anaerobes, p 117–132 In Norris JR, Ribbons DW. (ed), Methods in microbiology, vol 3B Academic Press, New York, NY. [Google Scholar]
- 50.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 52.Wiesemann N, Mohr J, Grosse C, Hause G, Reith F, Nies DH. 2013. Influence of copper resistance determinants on gold transformation by Cupriavidus metallidurans strain CH34. J. Bacteriol. 195:2298–2308. 10.1128/JB.01951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson TH, Ding PZ. 2001. Sodium-substrate cotransport in bacteria. Biochim. Biophys. Acta 1505:121–130. 10.1016/S0005-2728(00)00282-6. [DOI] [PubMed] [Google Scholar]
- 54.Jakkula SV, Guan L. 2012. Reduced Na+ affinity increases turnover of Salmonella enterica serovar Typhimurium MelB. J. Bacteriol. 194:5538–5544. 10.1128/JB.01206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaback HR, Smirnova I, Kasho V, Nie YL, Zhou YG. 2011. The alternating access transport mechanism in LacY. J. Membr. Biol. 239:85–93. 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaback HR, Voss J, Wu J. 1997. Helix packing in polytopic membrane proteins: the lactose permease of Escherichia coli. Curr. Opin. Struct. Biol. 7:537–542. 10.1016/S0959-440X(97)80119-4. [DOI] [PubMed] [Google Scholar]
- 57.Egler M, Große C, Grass G, Nies DH. 2005. Role of ECF sigma factor RpoE in heavy metal resistance of Escherichia coli. J. Bacteriol. 187:2297–2307. 10.1128/JB.187.7.2297-2307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casey AL, Adams D, Karpanen TJ, Lambert PA, Cookson BD, Nightingale P, Miruszenko L, Shillam R, Christian P, Elliott TSJ. 2010. Role of copper in reducing hospital environment contamination. J. Hosp. Infect. 74:72–77. 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 59.Mermod M, Magnani D, Solioz M, Stoyanov JV. 2012. The copper-inducible ComR (YcfQ) repressor regulates expression of ComC (YcfR), which affects copper permeability of the outer membrane of Escherichia coli. Biometals 25:33–43. 10.1007/s10534-011-9510-x. [DOI] [PubMed] [Google Scholar]
- 60.Rodríguez-Montelongo L, Volentini SI, Farias RN, Massa EM, Rapisarda VA. 2006. The Cu(II)-reductase NADH dehydrogenase-2 of Escherichia coli improves the bacterial growth in extreme copper concentrations and increases the resistance to the damage caused by copper and hydroperoxide. Arch. Biochem. Biophys. 451:1–7. 10.1016/j.abb.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 61.Volentini SI, Farias RN, Rodriguez-Montelongo L, Rapisarda VA. 2011. Cu(II)-reduction by Escherichia coli cells is dependent on respiratory chain components. Biometals 24:827–835. 10.1007/s10534-011-9436-3. [DOI] [PubMed] [Google Scholar]
- 62.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383–1387. 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 63.Suwalsky M, Ungerer B, Quevedo L, Aguilar F, Sotomayor CP. 1998. Cu2+ ions interact with cell membranes. J. Inorg. Biochem. 70:233–238. 10.1016/S0162-0134(98)10021-1. [DOI] [PubMed] [Google Scholar]
- 64.Speisky H, Gomez M, Burgos-Bravo F, Lopez-Alarcon C, Jullian C, Olea-Azar C, Aliaga ME. 2009. Generation of superoxide radicals by copper-glutathione complexes: redox-consequences associated with their interaction with reduced glutathione. Bioorgan. Med. Chem. 17:1803–1810. 10.1016/j.bmc.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 65.Aliaga ME, Lopez-Alarcon C, Garcia-Rio L, Martin-Pastor M, Speisky H. 2012. Redox-changes associated with the glutathione-dependent ability of the Cu(II)-GSSG complex to generate superoxide. Bioorgan. Med. Chem. 20:2869–2876. 10.1016/j.bmc.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 66.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616–1626. 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192:2512–2524. 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Acad. Sci. U. S. A. 106:8344–8349. 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fung DKC, Lau WY, Chan WT, Yan AX. 2013. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J. Bacteriol. 195:4556–4568. 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Helbig K, Grosse C, Nies DH. 2008. Cadmium toxicity in glutathione mutants of Escherichia coli. J. Bacteriol. 190:5439–5454. 10.1128/JB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohsumi Y, Kitamoto K, Anraku Y. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 170:2676–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bachmann BJ. 1972. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol. Rev. 36:525–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.