Abstract
Many Gram-negative bacteria use N-acyl-l-homoserine lactones (AHLs) as quorum-sensing signal molecules. We have reported that Acinetobacter strains isolated from activated sludge have AHL-degrading activity. In this study, we cloned the amiE gene as an AHL-degradative gene from the genomic library of Acinetobacter sp. strain Ooi24. High-performance liquid chromatography analysis revealed that AmiE functions as an AHL acylase, which hydrolyzes the amide bond of AHL. AmiE showed a high level of degrading activity against AHLs with long acyl chains but no activity against AHLs with acyl chains shorter than eight carbons. AmiE showed homology with a member of the amidases (EC 3.5.1.4) but not with any known AHL acylase enzymes. An amino acid sequence of AmiE from Ooi24 showed greater than 99% identities with uncharacterized proteins from Acinetobacter ursingii CIP 107286 and Acinetobacter sp. strain CIP 102129, but it was not found in the draft or complete genome sequences of other Acinetobacter strains. The presence of transposase-like genes around the amiE genes of these three Acinetobacter strains suggests that amiE is transferred by a putative transposon. Furthermore, the expression of AmiE in Pseudomonas aeruginosa PAO1 reduced AHL accumulation and elastase activity, which were regulated by AHL-mediated quorum sensing.
INTRODUCTION
Quorum sensing is a cell-to-cell communication system that is used by many species of bacteria and that depends on their population densities (1). Their communications are regulated by an autoinducer (AI), a self-produced signal molecule specific for a certain bacterium (1). Once the AI reaches a threshold concentration, gene transcription is activated, resulting in the expression of phenotypes such as motility, adhesion, biofilm formation, toxicity, and pathogenicity (2). N-Acyl-l-homoserine lactone (AHL) is one of the most common AIs produced by Gram-negative bacteria (3). It consists of an alkyl chain of various lengths appended to a lactone ring via an amide bond. The variation in length, saturation, and side chain substitution in the alkyl chain gives specificity to the signal molecule, which enables intraspecies communication (3). During the last decade, many AHL-degradative genes have been cloned from various microorganisms (4). AHL lactonase catalyzes AHL ring opening by hydrolyzing lactones, and AHL acylase hydrolyzes the amide bond of AHL (4).
The activated sludge process is a wastewater treatment widely used for sewage and human waste (5). Recent studies of the activated sludge process have focused on its microbiological aspects to improve the biodegradation capability and process management techniques (6, 7). The internal environment of these activated sludge flocs is similar to that of biofilms; high cell densities are present, allowing bacterial activation via bacterial cell-to-cell communication (6). It has been reported that the AHL-mediated quorum-sensing system affects the wastewater treatment process that uses activated sludge (8). It has been observed that quorum sensing is associated with the formation of a biofouling layer on the membrane surface in a membrane bioreactor (MBR) wastewater treatment system (7). The use of AHL-degrading enzymes has been investigated as an approach to controlling membrane biofouling in wastewater treatment systems (9).
AHL-producing and AHL-degrading bacteria coexist in various ecosystems, including the fish intestine and plant surfaces, and have various strategies for gaining a competitive advantage (10, 11). In a previous study, we reported that AHL-producing and AHL-degrading strains coexist in activated sludge obtained from sewage treatment plants in Japan, and the most dominant AHL-degrading isolates were assigned to the genus Acinetobacter (12). One of the AHL-degrading Acinetobacter strains, Acinetobacter sp. strain Ooi24, had the strongest AHL-degrading activity and showed putative AHL acylase activity (12). Although AHL lactonase activity has been detected in Acinetobacter sp. strain GG2, isolated from the rhizosphere of ginger (13), no reports of the cloning of AHL acylase genes from the genus Acinetobacter have been published. In this study, we report the cloning and characterization of a gene that encodes a novel AHL acylase from Acinetobacter sp. Ooi24.
MATERIALS AND METHODS
Bacterial strains, plasmids, compounds, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α and Pseudomonas aeruginosa PAO1 (14) were grown at 37°C in Luria-Bertani (LB) medium (15). AHL biosensor strains Chromobacterium violaceum CV026 and VIR07, which respond to short-chain and long-chain AHLs, respectively, were grown at 30°C in LB medium (16, 17). Solid bacterial media were prepared by adding agar to the liquid medium to a final concentration of 1.5%. Antibiotics were added as required at final concentrations of 100 μg/ml for ampicillin, 10 μg/ml for chloramphenicol, and 50 μg/ml for gentamicin. The AHLs used in this study, N-hexanoyl-l-homoserine lactone (C4-HSL), N-hexanoyl-l-homoserine lactone (C6-HSL), N-octanoyl-l-homoserine lactone (C8-HSL), N-decanoyl-l-homoserine lactone (C10-HSL), N-dodecanoyl-l-homoserine lactone (C12-HSL), N-(3-oxohexanoyl)-l-homoserine lactone, N-(3-oxooctanoyl)-l-homoserine lactone, N-(3-oxodecanoyl)-l-homoserine lactone (3OC10-HSL), and N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL), were synthesized using a previously described method (18). The identities and purities of all compounds were verified by 1H nuclear magnetic resonance. AHLs were dissolved in dimethyl sulfoxide (DMSO) to prepare 10 mM stock solutions.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− supE44 Δ(lacZYA-argF)U169 ϕ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Nippon Gene |
| Chromobacterium violaceum | ||
| CV026 | ATCC 31532 derivative, cviI::Tn5 xylE Kmr Smr | 16 |
| VIR07 | ATCC 12472 derivative, cviI::Kmr Apr | 17 |
| Acinetobacter sp. Ooi24 | AHL-degrading strain isolated from activated sludge | 12 |
| Pseudomonas aeruginosa PAO1 | Wild-type strain | 14 |
| Plasmids | ||
| pUC118 | Cloning vector, Apr | TaKaRa Bio |
| pAO24-1 | 5,356-bp Sau3AI fragment from Ooi24 genomic DNA in pUC118 | This study |
| pGEM-T Easy | Cloning vector, Apr | Promega |
| pGEM-amiE | pGEM-T Easy containing amiE from Ooi24 | This study |
| pLas28 | pSTV28 vector containing lasI from PAO1, Cmr | 26 |
| pBBR1MCS5 | Broad-host-range cloning vector, Gmr | 22 |
| pBBR1-ahlS | pBBR1MCS5 containing ahlS from Solibacillus silvestris StLB046 | 23 |
| pBBR1-amiE | pBBR1MCS5 containing amiE from Ooi24 | This study |
Identification of the AHL-degradative gene from Acinetobacter sp. Ooi24.
For the cloning of the AHL-degradative gene, a pUC118-based genomic library of Ooi24 was constructed according to a previously described method (19). Briefly, chromosomal DNA of Ooi24 was partially digested with Sau3AI, and the fragments were inserted into the BamHI site of cloning vector pUC118 that had been dephosphorylated by bacterial alkaline phosphatase (TaKaRa Bio, Shiga, Japan). The genomic library of Ooi24 and the pLas28 plasmid were transformed into E. coli. Then, E. coli harboring the genomic library and pLas28 was inoculated into LB agar medium prepared in 96-well plates. After incubation at 30°C for 24 h, VIR07 was also inoculated into the lower position of the well. After incubation at 30°C for 24 h, the colonies that did not induce the production of purple pigments by VIR07 were selected as AHL-degradative clones. Positive clones were sequenced using a BigDye Terminator (version 3.1) sequencing kit and an ABI 3500 genetic analyzer (Applied Biosystems, Tokyo, Japan).
To amplify the upstream regions of the genomic fragment in the positive clone, we carried out inverse PCR. The chromosomal DNA of Ooi24 was digested with EcoRV and then self-ligated. The inverse primers 5′-TAA AAG AGG TCC TGG AGT GAG CAG CAC TGC TGC G-3′ and 5′-AAC TAA GTC GAT TGT CCA TGT CGG CAT CTA CGC C-3′ were designed in accordance with the nucleotide sequences of the genomic fragment in the positive clone. PCR was performed with KOD FX Neo DNA polymerase (Toyobo, Osaka, Japan) using the following cycling parameters: 98°C for 10 s, 60°C for 30 s, and 68°C for 5 min for 30 cycles.
Detection of the AHL-degrading activity of amiE.
The amiE-coding region on the Ooi24 genome was amplified with Blend Taq DNA polymerase (Toyobo) and the following primers: 5′-TAC TTG TTC GC AAT GTG TGA TGG CAC GC-3′ and 5′-CCA CGT TTA TTG AGC AAT GTC CAA ACA ATG GG-3′. PCR was performed with the following cycling parameters: 94°C for 30 s, 60°C for 30 s, and 74°C for 3 min for 30 cycles. The PCR products were cloned into the pGEM-T Easy cloning vector (Promega, Tokyo, Japan) for construction of pGEM-amiE. The full-grown culture of E. coli harboring pGEM-amiE was inoculated into fresh LB medium (a 1% inoculum) with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and cultivated for 16 h. Cells were collected via centrifugation, washed with 50 mM phosphate buffer (pH 7.4), and resuspended in 1 ml phosphate buffer. Then, 2 μl of AHL stock solution was mixed with the cell suspension at a final concentration of 20 μM. For the control, 2 μl of DMSO was mixed with the cell suspension. After incubation at 30°C with shaking, the cells were removed via centrifugation for preparation of the supernatant. The residual AHLs were detected using the AHL biosensors CV026 and VIR07. An overnight culture of CV026 or VIR07 was mixed with 25 ml LB agar medium, and the mixture was poured onto the plates. AHL samples were applied to paper discs with 8-mm diameters (Advantec, Tokyo, Japan), and the discs were placed onto LB agar plates containing CV026 or VIR07. The assay plates were incubated overnight at 30°C, and the appearance of pigment was assessed. The residual amounts of AHL were calculated using relationship equations based on the sizes of the purple zones and known amounts of AHLs (20).
Identification of AHL acylase activity.
Detection of AHL acylase activity was performed according to a previously described method with modification (20, 21). The full-grown culture of E. coli harboring pGEM-amiE was inoculated into fresh LB medium (a 1% inoculum) with 1 mM IPTG and cultivated for 16 h. Cells were collected via centrifugation, washed with 50 mM phosphate buffer (pH 7.4), and resuspended in 1 ml of phosphate buffer containing 500 μM 3OC10-HSL. The reaction mixture was incubated at 30°C for 6 h and centrifuged to remove the cells. Then, 150 μl of supernatant was mixed with 150 μl of saturated borax solution and 300 μl of dansyl chloride (10 mg/ml in acetone) at 40°C for 1 h. As a control, phosphate buffer containing 500 μM l-homoserine lactone (HSL) was dansylated in parallel with the same reagents.
Twenty microliters of sample was chromatographed on a high-performance liquid chromatography (HPLC) system (Jasco, Tokyo, Japan) with a UV-visible detector set at 270 nm and a Mightysil RP-18GP column (250 mm by 4.6 mm, 5-μm particle diameter; Kanto Kagaku, Tokyo, Japan). The initial mobile phase was water-acetonitrile-acetic acid (75:25:0.05 [vol/vol/vol]), and the elution was created with 100% acetonitrile running at 2 ml/min according to the following profile: 0 to 2.5 min, 0%; 2.5 to 7.5 min, 0 to 100%; and 7.5 to 10 min, 100%.
Effect of AmiE on production of AHLs and virulence factors in P. aeruginosa.
The amiE-coding region on the Ooi24 genome was amplified with Blend Taq DNA polymerase and the primers 5′-TCT AAG CTT CCG ATC ATG AGC TTC AAT ATT GCA CC-3′ and 5′-TCT GGA TCC TCG TCA ATC AAT TGA TTT CTA GTC GG-3′ containing the HindIII and BamHI restriction sites (underlined), respectively. PCR was performed with the following cycling parameters: 94°C for 30 s, 60°C for 30 s, and 74°C for 1.5 min for 30 cycles. The PCR products were cut with HindIII and BamHI digestion and inserted into the HindIII and BamHI sites of the broad-host-range vector pBBR1MCS5 (22) for construction of pBBR1-amiE. The pBBR1-ahlS plasmid, which contains the AHL lactonase gene, ahlS, from Solibacillus silvestris StLB046, was used as a positive control (23). The plasmids pBBR1MCS5, pBBR1-amiE, and pBBR1-ahlS were transformed into P. aeruginosa PAO1 via electroporation (24). The full-grown culture of PAO1 harboring plasmids was inoculated into fresh LB medium (a 1% inoculum) with 1 mM IPTG, cultivated for 15 h, and centrifuged to remove the cells.
For the extraction of AHLs, 600 μl of culture supernatant was mixed with an equal volume of ethyl acetate and vortexed for 10 min. The ethyl acetate layer was evaporated to dryness and dissolved in 100 μl of dimethyl sulfoxide. C4-HSL and 3OC12-HSL were detected using the AHL biosensors, as described above. The elastase activity in the culture supernatant of PAO1 harboring plasmids was quantified with an elastin-Congo red assay using a previously described method (25).
Nucleotide sequence accession numbers.
The nucleotide sequence of 16S rRNA from Acinetobacter sp. Ooi24 was deposited in the DNA Data Bank of Japan (DDBJ)/EMBL/GenBank databases under accession no. AB933637. The nucleotide sequences of amiE and its flanking open reading frames (ORFs) from Acinetobacter sp. Ooi24 were deposited under accession no. AB933638.
RESULTS
An AHL-degradative gene is present in the genome of Acinetobacter sp. Ooi24.
Acinetobacter sp. Ooi24 showed degrading activity against C10-HSL but poor activity against C6-HSL (12). Since most AHL acylases degrade AHLs that have an acyl chain longer than eight carbons (4), we used the E. coli/pLas28 reporter system to clone the AHL-degradative gene. The pLas28 reporter plasmid carried the las quorum-sensing system genes of P. aeruginosa PAO1, which contains the AHL synthase gene (lasI) (26). Ooi24 is also able to degrade 3OC12-HSL as well as C10-HSL (data not shown). E. coli harboring pLas28 produced 3OC12-HSL and induced violacein production by the VIR07 reporter strain. The prepared pUC118-based genomic library of Ooi24 was transformed into E. coli harboring pLas28. The formed colonies were transferred to LB agar medium prepared in 96-well plates, and the released 3OC12-HSL was visualized through violacein production by VIR07.
When approximately 10,000 transformants were screened, one positive clone, designated pAO24-1, did not stimulate violacein production by VIR07. pAO24-1 contained the 5,356-bp genomic DNA fragment in the BamHI site of pUC118. After amplification of the upstream region of the genomic DNA fragment by inverse PCR, the sequence of the extended genomic DNA fragment (6,445 bp) contained two incomplete ORFs and five complete ORFs (Fig. 1A). The incomplete ORFs, orf1 and orf7, showed similarities to endoribonuclease and single-stranded DNA-specific exonuclease, respectively. The complete ORFs orf2, orf3, and orf4 showed similarities to FMN reductase, an AraC-family transcriptional regulator, and amidase, respectively. The two complete ORFs orf5 and orf6 were predicted to encode an IS4-family transposase.
FIG 1.
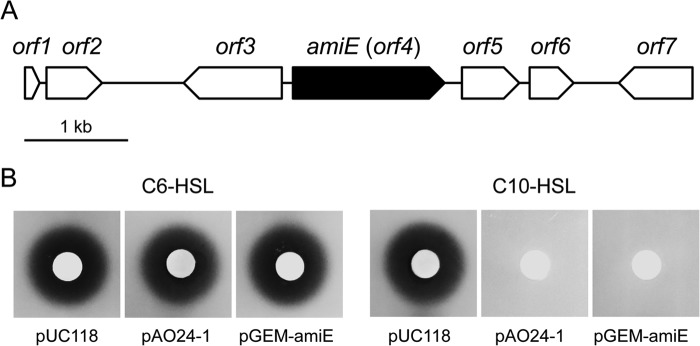
(A) Arrangement of the predicted ORFs on the original genomic clone pAO24-1. The scale represents a 1-kb length of nucleotides. Filled arrow, the amiE gene; open arrows, the other ORFs. (B) AHL-degrading activity of Escherichia coli harboring pUC118, pAO24-1, and pGEM-amiE. Subcultures of E. coli harboring these plasmids were mixed with 10 μM C6-HSL or C10-HSL and incubated at 37°C for 8 h. The residual AHL was detected using Chromobacterium violaceum CV026 (for C6-HSL) or VIR07 (for C10-HSL).
Because sequencing analysis revealed that only orf4, designated amiE, encoded a member of the hydrolase family, we next determined whether amiE works as an AHL-degradative gene. We amplified amiE by PCR and subcloned it into the pGEM-T Easy vector for construction of pGEM-amiE. E. coli harboring pAO24-1 or pGEM-amiE was inoculated into LB medium containing 10 μM C6-HSL or C10-HSL. After incubation for 9 h, residual C6-HSL and C10-HSL in the culture supernatant were detected using CV026 and VIR07, respectively. Compared with the activity of the control, E. coli harboring pAO24-1 and pGEM-amiE showed obvious C10-HSL-degrading activity because of the vanishing of violacein production (Fig. 1B). These results suggested that the amiE gene product has AHL-degrading activity. In contrast, E. coli harboring each plasmid showed no degrading activity with C6-HSL (Fig. 1B). In contrast to amiE, the other four complete ORFs did not show any degrading activity against C6-HSL and C10-HSL (data not shown).
AmiE encodes a long-chain AHL acylase.
To determine whether AmiE functions as an AHL acylase, we used HPLC to analyze the structure of 3OC10-HSL digested by AmiE. The phosphate buffer containing 500 μM 3OC10-HSL was incubated for 6 h at 30°C with E. coli harboring pGEM-T Easy or pGEM-amiE. The detection of HSL liberated from 3OC10-HSL by AHL acylase activity was performed via dansylation of the free amine of HSL. The results of HPLC analysis are shown in Fig. 2. Fractionation of 3OC10-HSL treated with E. coli harboring pGEM-amiE revealed one specific peak at a retention time of 6.56 min. The specific peak of the HSL standard was also detected at a retention time of 6.56 min. In contrast, 3OC10-HSL treated with E. coli harboring pGEM-T Easy displayed no distinct peak at this retention time. The reaction mixture with E. coli harboring each plasmid which was not mixed with 3OC10-HSL did not display the specific peak of the HSL standard. These results demonstrate that AmiE works as an AHL acylase that hydrolyzes the amide bond of 3OC10-HSL. To analyze the substrate specificity of AmiE, we mixed a cell suspension of E. coli harboring pGEM-amiE with a wide range of AHLs. AmiE degraded C8-HSL, C10-HSL, and C12-HSL and showed the highest degrading activity against C10-HSL, but it did not degrade C6-HSL (Fig. 3A). In addition, E. coli harboring pGEM-amiE did not degrade C4-HSL, which has the shortest acyl chain (data not shown). AmiE showed higher degradation activity toward the 3-oxo-unsubstituted AHLs than toward the 3-oxo-substituted AHLs (Fig. 3B).
FIG 2.
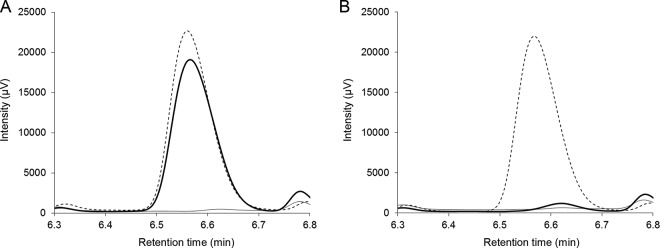
HPLC analysis of the products of 3OC10-HSL treated with E. coli harboring pGEM-amiE (A) and pGEM-T Easy (B). 3OC10-HSL (500 μM) was incubated for 6 h at 30°C with E. coli harboring each plasmid. HSL liberated from 3OC10-HSL was dansylated and fractionated with HPLC (bold line). Dotted and gray lines, the specific peak of 500 μM HSL and the control without 3OC10-HSL, respectively.
FIG 3.
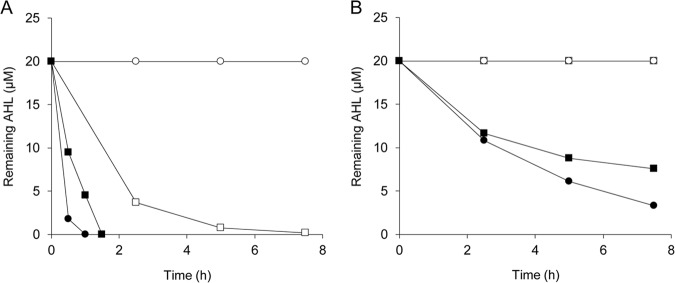
AHL-degrading activity of E. coli harboring pGEM-amiE. The cell suspension of E. coli harboring pGEM-amiE was mixed with 20 μM 3-oxo-unsubstituted (A) or 3-oxo-substituted (B) AHLs with acyl chain lengths of 6 (open circles), 8 (open squares), 10 (filled circles), and 12 (filled squares) carbons and incubated at 30°C with shaking. The remaining AHLs in the culture supernatant were visualized by spotting the supernatant onto plates mixed with C. violaceum CV026 or VIR07. Residual amounts of AHLs were calculated using relationship equations based on the size of the purple zones.
AmiE is a new AHL acylase.
To evaluate the novelty of AmiE, we determined the phylogenetic relationship between AmiE and known AHL acylases from various bacteria using the neighbor-joining method with the ClustalW program (Fig. 4) (27). AiiD from Ralstonia sp. strain XJ12B (21), AhlM from Streptomyces sp. strain M664 (28), PvdQ from P. aeruginosa PAO1 (29), and Aac from Shewanella sp. strain MIB015 (20) belong to the aculeacin A acylase protein family (EC 3.5.1.70). PA0305 and QuiP from P. aeruginosa PAO1 (25, 30) and AiiC from Anabaena sp. strain PCC 7120 (31) belong to the penicillin G acylase protein family (EC 3.5.1.11). However, AmiE showed no significant similarity with these protein families. A Basic Local Alignment Search Tool (BLAST) search revealed that AmiE showed homology to known amidases (EC 3.5.1.4), which are enzymes that catalyze the hydrolysis of monocarboxylic acid amide to produce ammonia and monocarboxylate (32). The putative gene product of amiE from Ooi24 was a 490-amino-acid protein and showed 36.9% and 37.1% sequence identities with AmiE from Azospirillum sp. strain B510 (UniProt accession no. D3NY90) and Amycolatopsis orientalis HCCB10007 (UniProt accession no. R4SWV0), respectively (Fig. 4). The BLAST search revealed that AmiE from Ooi24 showed 100% sequence identity with an uncharacterized protein from Acinetobacter ursingii CIP 107286 (UniProt accession no. N9BV73) and 99.8% sequence identity with a protein of Acinetobacter sp. strain CIP 102129 (UniProt accession no. N8UB59).
FIG 4.
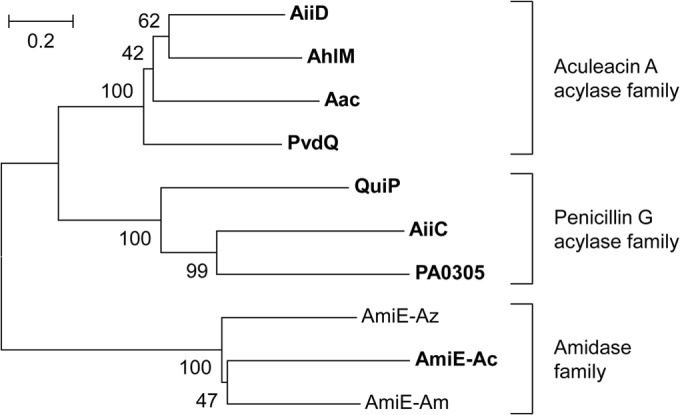
Phylogenetic tree based on the amino acid sequences of AiiD, AhlM, PvdQ, Aac, PA0305, AiiC, QuiP, and AmiE from Acinetobacter sp. Ooi24 (AmiE-Ac), Azospirillum sp. B510 (AmiE-Az), and A. orientalis HCCB10007 (AmiE-Am). The phylogenetic tree was constructed using the neighbor-joining method with the ClustalW program of the MEGA (version 6) package (36). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The scale bar represents 0.2 substitution per amino acid position. The known AHL acylases and Acinetobacter sp. Ooi24 AmiE are shown in bold.
AmiE reduces the production of AHLs and virulence factors in P. aeruginosa.
To evaluate the anti-quorum-sensing activity of AmiE, we evaluated the potential use of the heterologous expression of amiE to interfere with quorum sensing in P. aeruginosa PAO1. In P. aeruginosa PAO1, LasI and RhlI are responsible for the synthesis of the las and rhl signals 3OC12-HSL and C4-HSL, respectively (33). The production of C4-HSL and 3OC12-HSL was detected in PAO1 harboring pBBR1MCS5 but not in PAO1 harboring pBBR1-ahlS (data not shown). Although AmiE does not have C4-HSL-degrading activity, neither C4-HSL nor 3OC12-HSL was detected in the supernatant of PAO1 harboring pBBR1-amiE (data not shown). Because the las and rhl quorum-sensing systems cross-regulate each other and C4-HSL is not detected in the lasI mutant of PAO1 (34), it was assumed that the degradation of 3OC12-HSL by AmiE affected the production of C4-HSL in PAO1. One of them, 3OC12-HSL, regulates the production of the elastase enzyme as a virulence-enhancing factor in P. aeruginosa (33). The elastase activities of PAO1 harboring pBBR1-ahlS and pBBR1-amiE were drastically decreased (more than 50%) compared with the elastase activity of PAO1 harboring pBBR1MCS5 (Fig. 5). In a previous study, the overexpression of another AHL acylase gene, pa0305, resulted in an approximately 50% reduction in elastase activity in P. aeruginosa PAO1 (25). Neither PA0305 nor AmiE had C4-HSL-degrading activity (25). These results indicate that the expression of amiE in PAO1 contributes to the self-degradation of 3OC12-HSL and reduces the AHL-mediated expression of elastase.
FIG 5.
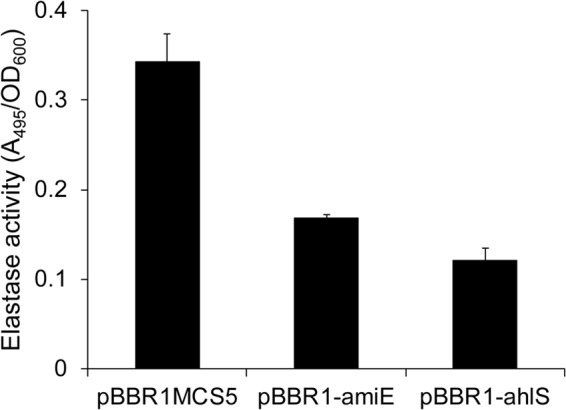
The elastase activities in the culture supernatants of PAO1 harboring pBBR1MCS5, pBBR1-ahlS, and pBBR1-amiE were measured using the elastin-Congo red assay. The absorbance of the supernatants measured at 495 nm was divided by the optical density at 600 nm (OD600) of the culture. The results were reproduced in triplicate, and the error bars indicate the standard deviations.
DISCUSSION
In this study, we found the novel AHL acylase gene amiE from the genome of Acinetobacter sp. Ooi24. In our previous study, Acinetobacter sp. Ooi24 showed high degrading activity against C10-HSL but very weak degrading activity against C6-HSL (12). Similarly, E. coli harboring the AmiE-expressing plasmid degraded AHLs with acyl chains longer than six carbons and showed the highest degrading activity against C10-HSL, but it did not degrade C6-HSL (Fig. 3A). These results demonstrate that the AHL-degrading activity of E. coli harboring the AmiE-expressing plasmid corresponds to that of Ooi24. Previous studies have reported that most AHL acylases degrade long-chain AHLs more efficiently than short-chain forms (4). Especially, some AHL acylases, Aac, PvdQ, AhlM, and QuiP, are unable to degrade AHLs with acyl chains shorter than eight carbons (4). These results suggest that AmiE has activity similar to the activities of known AHL acylases. In general, enzymes belonging to this protein family (EC 3.5.1) catalyze the hydrolysis of carbon-nitrogen bonds other than peptide bonds. On the other hand, some EC 3.5.1-family enzymes, aculeacin A acylase and penicillin G acylase, are able to catalyze the hydrolysis of the amide bond of AHLs. To the best of our knowledge, the present study is the first to report that AmiE, which belongs to the amidase protein family, has AHL acylase activity as well as activity against other EC 3.5.1-family enzymes.
The amiE gene homolog was also found in the genome sequences of two other Acinetobacter strains, A. ursingii CIP 107286 and Acinetobacter sp. CIP 102129. Although these two Acinetobacter strains are phylogenetically different from Ooi24 (Fig. 6), the nucleotide sequences of amiE from these three strains show almost complete identity, as divergence is limited to one nucleotide position. The amiE gene homolog was not found in the draft or complete genome sequences of other Acinetobacter strains deposited in the DDBJ/EMBL/GenBank databases (data not shown). These results demonstrate that the amiE gene homolog is not conserved in a wide range of Acinetobacter strains but is encoded by the genomes of specific Acinetobacter strains.
FIG 6.
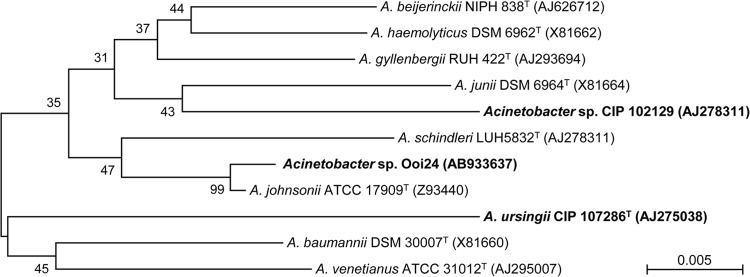
Neighbor-joining trees of the 16S rRNA gene sequences obtained for Acinetobacter strains. For phylogenetic analysis with the 16S rRNA gene, nine type strains of the genus Acinetobacter and two non-type Acinetobacter strains were used. DDBJ/EMBL/GenBank accession numbers appear in parentheses. Acinetobacter strains that contain amiE gene homologs in their genomes are shown in bold. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The scale bar represents 0.005 substitution per nucleotide position. Phylogenetic analyses were conducted with the MEGA (version 6) program (36).
To identify the chromosomal locus of amiE, we compared the upstream and downstream regions of the amiE genes in Acinetobacter strains. Those in A. ursingii CIP 107286 and Acinetobacter sp. CIP 102129 were obtained from the whole-genome shotgun sequences deposited in the DDBJ/EMBL/GenBank databases under accession no. APQA01000004 and APPA01000039, respectively. Interestingly, the nucleotide sequences of the upstream regions of amiE from these strains, which contain genes encoding phytanoyl coenzyme A dioxygenase, endoribonuclease, FMN reductase, and the AraC family transcriptional regulator, showed almost complete identity, as divergence is limited to less than two nucleotide positions (Fig. 7). In addition, many transposase-like genes were found around these highly conserved sequences (Fig. 7). The amino acid sequence of AmiE showed no similarity with any amidase-family proteins from Acinetobacter strains but showed 46.0% and 39.9% identities with amidases from the gammaproteobacteria Thalassolituus oleivorans MIL-1 (UniProt accession no. M5E2I7) and Marinobacter algicola DG893 (UniProt accession no. A6F504), respectively. These results strongly suggest that amiE is transferred by a putative transposon carrying acylhomoserine lactone acylase.
FIG 7.
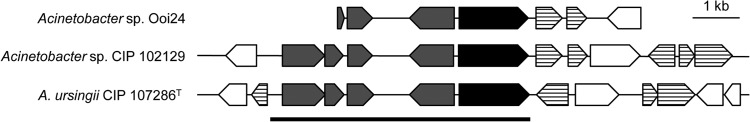
Arrangement of the predicted ORFs in the upstream and downstream regions of the amiE genes in the genomes of Acinetobacter sp. Ooi24 and CIP 102129 and A. ursingii CIP 107286. The scale bar represents a 1-kb length of nucleotides. Filled, gray, and lined arrows, the amiE gene, conserved genes, and putative transposon genes, respectively.
In summary, our study reports that AmiE from Acinetobacter sp. Ooi24, which belongs to the amidase family of proteins, has AHL acylase activity. The putative transposon, including amiE, was found in the chromosome of three Acinetobacter strains. It is possible that the horizontal transfer of amiE among Acinetobacter strains is observed and influences the quorum-sensing system in activated sludge. Quorum sensing is reportedly associated with the formation of a biofouling layer on the membrane surface of membrane bioreactor (MBR) wastewater treatment systems (7). Lee et al. (35) demonstrated that cross-linked AHL acylase molecules in magnetically separable mesoporous silica work together as an antifouling material. The AHL acylase activity of Acinetobacter strains might also perform as an antifouling agent in MBR systems.
ACKNOWLEDGMENT
This work was supported by Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (JST).
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346. 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.de Kievit TR, Iglewski BH. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839–4849. 10.1128/IAI.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsek MR, Greenberg EP. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. U. S. A. 97:8789–8793. 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uroz S, Dessaux Y, Oger P. 2009. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chembiochem 10:205–216. 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- 5.Sanin FD, Clarkson WW, Vesilind PA. 2011. Sludge engineering: the treatment and disposal of wastewater sludges. DEStech Publications, Inc, Lancaster, PA. [Google Scholar]
- 6.Valle A, Bailey MJ, Whiteley AS, Manefield M. 2004. N-acyl-l-homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ. Microbiol. 6:424–433. 10.1111/j.1462-2920.2004.00581.x. [DOI] [PubMed] [Google Scholar]
- 7.Yeon KM, Cheong WS, Oh HS, Lee WN, Hwang BK, Lee CH, Beyenal H, Lewandowski Z. 2009. Quorum sensing: a new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol. 43:380–385. 10.1021/es8019275. [DOI] [PubMed] [Google Scholar]
- 8.Chong G, Kimyon O, Rice SA, Kjelleberg S, Manefield M. 2012. The presence and role of bacterial quorum sensing in activated sludge. Microb. Biotechnol. 5:621–633. 10.1111/j.1751-7915.2012.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lade H, Paul D, Kweon JH. 2014. Quorum quenching mediated approaches for control of membrane biofouling. Int. J. Biol. Sci. 10:547–562. 10.7150/ijbs.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morohoshi T, Someya N, Ikeda T. 2009. Novel N-acylhomoserine lactone-degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 73:2124–2127. 10.1271/bbb.90283. [DOI] [PubMed] [Google Scholar]
- 11.Morohoshi T, Ebata A, Nakazawa S, Kato N, Ikeda T. 2005. N-Acyl homoserine lactone-producing or -degrading bacteria isolated from the intestinal microbial flora of ayu fish (Plecoglossus altivelis). Microbes Environ. 20:264–268. 10.1264/jsme2.20.264. [DOI] [Google Scholar]
- 12.Ochiai S, Morohoshi T, Kurabeishi A, Shinozaki M, Fujita H, Sawada I, Ikeda T. 2013. Production and degradation of N-acylhomoserine lactone quorum sensing signal molecules in bacteria isolated from activated sludge. Biosci. Biotechnol. Biochem. 77:2436–2440. 10.1271/bbb.130553. [DOI] [PubMed] [Google Scholar]
- 13.Chan K-G, Atkinson S, Mathee K, Sam C-K, Chhabra SR, Cámara M, Koh C-L, Williams P. 2011. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 11:51. 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holloway BW, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 16.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711. 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 17.Morohoshi T, Kato M, Fukamachi K, Kato N, Ikeda T. 2008. N-Acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 279:124–130. 10.1111/j.1574-6968.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- 18.Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. 2003. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J. Med. Chem. 46:97–104. 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 19.Wang WZ, Morohoshi T, Ikenoya M, Someya N, Ikeda T. 2010. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 76:2524–2530. 10.1128/AEM.02738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morohoshi T, Nakazawa S, Ebata A, Kato N, Ikeda T. 2008. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci. Biotechnol. Biochem. 72:1887–1893. 10.1271/bbb.80139. [DOI] [PubMed] [Google Scholar]
- 21.Lin YH, Xu JL, Hu J, Wang LH, Ong SL, Leadbetter JR, Zhang LH. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849–860. 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 22.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Morohoshi T, Tominaga Y, Someya N, Ikeda T. 2012. Complete genome sequence and characterization of the N-acylhomoserine lactone-degrading gene of the potato leaf-associated Solibacillus silvestris. J. Biosci. Bioeng. 113:20–25. 10.1016/j.jbiosc.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Smith AW, Iglewski BH. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. 10.1093/nar/17.24.10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wahjudi M, Papaioannou E, Hendrawati O, van Assen AH, van Merkerk R, Cool RH, Poelarends GJ, Quax WJ. 2011. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology 157:2042–2055. 10.1099/mic.0.043935-0. [DOI] [PubMed] [Google Scholar]
- 26.Imura J, Kashima K, Kusano M, Ikeda T, Morohoshi T. 2010. Piecewise affine systems approach to control of biological networks. Philos. Trans. A Math. Phys. Eng. Sci. 368:4977–4993. 10.1098/rsta.2010.0176. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SY, Kang HO, Jang HS, Lee JK, Koo BT, Yum DY. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632–2641. 10.1128/AEM.71.5.2632-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang JJ, Han JI, Zhang LH, Leadbetter JR. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941–5949. 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang JJ, Petersen A, Whiteley M, Leadbetter JR. 2006. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72:1190–1197. 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero M, Diggle SP, Heeb S, Camara M, Otero A. 2008. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 280:73–80. 10.1111/j.1574-6968.2007.01046.x. [DOI] [PubMed] [Google Scholar]
- 32.Skouloubris S, Labigne A, De Reuse H. 2001. The AmiE aliphatic amidase and AmiF formamidase of Helicobacter pylori: natural evolution of two enzyme paralogues. Mol. Microbiol. 40:596–609. 10.1046/j.1365-2958.2001.02400.x. [DOI] [PubMed] [Google Scholar]
- 33.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67:5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steindler L, Bertani I, De Sordi L, Schwager S, Eberl L, Venturi V. 2009. LasI/R and RhlI/R quorum sensing in a strain of Pseudomonas aeruginosa beneficial to plants. Appl. Environ. Microbiol. 75:5131–5140. 10.1128/AEM.02914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee B, Yeon K-M, Shim J, Kim S-R, Lee C-H, Lee J, Kim J. 2014. Effective antifouling using quorum-quenching acylase stabilized in magnetically-separable mesoporous silica. Biomacromolecules 15:1153–1159. 10.1021/bm401595q. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


