Abstract
Persister cells, which are tolerant to antimicrobials, contribute to biofilm recalcitrance to therapeutic agents. In turn, the ability to kill persister cells is believed to significantly improve efforts in eradicating biofilm-related, chronic infections. While much research has focused on elucidating the mechanism(s) by which persister cells form, little is known about the mechanism or factors that enable persister cells to revert to an active and susceptible state. Here, we demonstrate that cis-2-decenoic acid (cis-DA), a fatty acid signaling molecule, is able to change the status of Pseudomonas aeruginosa and Escherichia coli persister cells from a dormant to a metabolically active state without an increase in cell number. This cell awakening is supported by an increase of the persister cells' respiratory activity together with changes in protein abundance and increases of the transcript expression levels of several metabolic markers, including acpP, 16S rRNA, atpH, and ppx. Given that most antimicrobials target actively growing cells, we also explored the effect of cis-DA on enhancing antibiotic efficacy in killing persister cells due to their inability to keep a persister cell state. Compared to antimicrobial treatment alone, combinational treatments of persister cell subpopulations with antimicrobials and cis-DA resulted in a significantly greater decrease in cell viability. In addition, the presence of cis-DA led to a decrease in the number of persister cells isolated. We thus demonstrate the ability of a fatty acid signaling molecule to revert bacterial cells from a tolerant phenotype to a metabolically active, antimicrobial-sensitive state.
INTRODUCTION
Persister cells are considered to be a subpopulation of stochastically produced, nongrowing (dormant) cells present in biofilm and planktonic bacterial cultures. Persister cells account for 10−6 to 10−4 of the total cell population of mid-exponential-phase cells and up to 1% of the total cell population of stationary-phase cells and biofilms, a pattern resembling that of a quorum-sensing mechanism (1–3). Tolerance to antimicrobials is one of the key characteristics of this subpopulation, as persisters escape killing by antimicrobials such as fluoroquinolones, which can kill slow-growing bacteria but not dormant cells (4). It is therefore not surprising that persister cells are considered to play a major role in the resilience of bacterial populations and have recently been isolated from patients with candidiasis, from cystic fibrosis patients with chronic lung infections, and from Mycobacterium tuberculosis biofilms responsible for chronic tuberculosis (5–8).
Several mechanisms have been described to contribute to persister cell formation. For instance, several genes involved in energy generation and cell maintenance have been shown to be downregulated in persister cells, further indicating that persisters are nongrowing, dormant cells (1). Among these genes were members of several operons involved in oxidative phosphorylation, including NADH dehydrogenase, ATP synthase, and cytochrome O-ubiquinol oxidase (9). The stringent response has also been linked with persister cells, where increased concentrations of polyphosphate (polyP) compounds, such as ppGpp, were present compared to concentrations in nonpersister cells. Moreover, arrest of protein synthesis accomplished by the use of tetracycline has been shown to correlate with a 103- to 104-fold increase in the number of persister cells in planktonic cultures (10). Persister cell formation has also been shown to be related to increased expression levels of chromosomal and plasmid-encoded toxin-antitoxin (TA) modules capable of inducing stasis, resulting in increased tolerance to lethal shock, DNA-damaging conditions, and antimicrobials (2, 11, 12).
In addition to understanding how persister cells are formed, research has also focused on eliminating persister cells, either by eradicating persister cells or by inducing persister cells to revert from a dormant to an active state. Eradication of persister cells has been accomplished by exposure of the cells to reactive oxygen species, weak electrical currents, and 3-[4-(4-methoxyphenyl)piperazin-1-yl]piperidin-4-yl biphenyl-4-carboxylate (C10) (13–15). Likewise, activation of the Clp protease by acyldepsipeptide-4 (ADEP4) has been demonstrated to enhance the efficacy of rifampin in killing persister cells formed by Staphylococcus aureus (16). Other approaches to reanimate persister cells include the use of metabolic stimuli. For instance, Pascoe et al. demonstrated that spent medium has a resuscitating effect on S. aureus persister cells, as indicated by the finding of a >600-fold increase in bacterial growth (17). Similarly, the addition of mannitol, glucose, fructose, and pyruvate to persister cells isolated from Escherichia coli and S. aureus has been demonstrated to increase the central metabolism, increase the respiration of persister cells, and increase the ability of aminoglycosides to permeate membranes (18). Those authors furthermore demonstrated that exposure to mannitol resulted in E. coli persister cells being significantly more susceptible to gentamicin, resulting in a reduction of their viability to the point of eradication (18). Likewise, the addition of the quorum sensing inhibitor (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one (BF8) to Pseudomonas aeruginosa persister cells has been shown to sensitize them to ciprofloxacin and tobramycin, with the effect hypothesized to be the result of changes in the cells' metabolism (19).
Recently, a family of fatty acid signaling molecules has been identified in several Gram-negative bacteria, including Xanthomonas campestris, Burkholderia cenocepacia, and P. aeruginosa (20–22). cis-2-Decenoic acid (cis-DA), originally isolated from P. aeruginosa, induces P. aeruginosa biofilms to disperse by inducing cells to transition from a biofilm to a planktonic (free-swimming) phenotype, with only a small percentage of cells remaining surface attached (22). A similar dispersion response was noted for various other Gram-negative and Gram-positive biofilms as well as for Candida albicans biofilms (22). In addition to inducing dispersion, cis-DA was found to increase the recovery of cells of several bacterial species on agar plates and to increase the killing efficacy of antimicrobials against P. aeruginosa biofilms (23, 24). The presence of cis-DA together with antimicrobials has also been found to eradicate E. coli and Klebsiella pneumoniae mixed-species biofilms grown on catheters and to remove preformed biofilms of Bacillus subtilis, Salmonella enterica, S. aureus, and E. coli (25, 26). cis-DA also improves methicillin-resistant S. aureus (MRSA) biofilm reduction when used adjunctively with daptomycin, vancomycin, and linezolid (27). Together, these findings indicated that cis-DA has cross-species and cross-kingdom dispersion activity and can lead to increased cell recovery and increased efficacy of antibiotics. These observations led us to ask whether cis-DA is able to revert persister cells to a metabolically active and susceptible state. In this work, we demonstrate that while cis-DA cannot be used as a carbon source for growth, exposure of P. aeruginosa and E. coli persister cells derived from biofilm and planktonic populations to nanomolar concentrations of cis-DA results in an increase of respiratory activity, concomitant with elevated 16S rRNA, ATP synthase, and exopolyphosphatase levels. This signaling molecule also acts synergistically with antimicrobials, enhancing killing and enabling eradication of persister cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas aeruginosa PA14 and Escherichia coli BW25113 were used throughout this study. All cultures were grown overnight in Difco LB Lennox broth (BD) in flasks at 220 rpm at 37°C, unless indicated otherwise.
Persister cell isolation.
Biofilm and planktonic persister cell populations of P. aeruginosa and E. coli were isolated by relying on activation of the SOS response, as previously described, using ciprofloxacin (4, 28–30). For biofilm persister subpopulations, P. aeruginosa or E. coli biofilm cultures were grown in a tube reactor system at 22°C, using L/S 14 Masterflex peroxide-cured silicone tubing with 5% LB pumped through at a rate of 10.8 ml/h (22, 31, 32). Each tube reactor was inoculated with 2 ml of a standardized culture grown overnight (optical density at 600 nm [OD600] of 0.8) and incubated, under static conditions, for a period of 1 h to facilitate cell attachment. Following 1 h, the flow was initiated, and biofilms were allowed to develop for a period of 6 days. Following 6 days of growth, mature biofilms were exposed to saline (0.85% NaCl in water) or ciprofloxacin (150 μg/ml) in saline, and viability was monitored at 0, 1, 3, 5, and 24 h. At each time point, biofilms were harvested (using the rolling pin method) into centrifuge tubes containing 1 ml of saline with 1% MgCl2 · 7H2O, homogenized, serially diluted, and drop plated onto plate count agar (PCA) plates with 1% MgCl2 · 7H2O. Viability was determined following 24 h of incubation at 37°C. Bacterial viability was also visualized by using confocal microscopy and the Live/Dead BacLight bacterial viability kit, where SYTO9 labels all bacteria while propidium iodide labels only dead bacteria (Life Technologies). For the planktonic persister subpopulation, planktonic cultures grown overnight were diluted to 1% in fresh medium and grown at 37°C with agitation (220 rpm) for a period of 24 h. Cells were then collected (16,000 × g for 5 min at 4°C), washed twice with saline (16,000 × g for 5 min at 4°C), and subsequently resuspended in either saline or ciprofloxacin (20 μg/ml) in saline to a final OD600 of 0.8. Cultures were subsequently incubated at 37°C with agitation (220 rpm) for a period of 24 h. Viability was determined after 0, 1, 3, 5, and 24 h, as described above.
Persister isolation in the presence of cis-DA.
Planktonic persister cell populations of P. aeruginosa and E. coli were isolated in the presence of cis-2-decenoic acid (cis-DA) (Carbosynth Ltd., Compton, United Kingdom). P. aeruginosa was exposed to 100 nM cis-DA, while E. coli was exposed to 310 nM cis-DA. The concentrations of cis-DA used were based on previous studies (22). Planktonic cultures grown overnight were diluted to 1% in fresh medium and grown at 37°C with agitation (220 rpm) for a period of 24 h. Cells were collected (16,000 × g for 5 min at 4°C), washed twice with saline (16,000 × g for 5 min at 4°C), and subsequently resuspended, to a final OD600 of 0.8, in either saline or cis-DA in saline. Cultures were subsequently incubated at 37°C with agitation (220 rpm) for a period of 24 h. Following 24 h, cultured cells were processed as described above for persister cell isolation.
Confirmation and maintenance of the persister cell state.
Persister cells were isolated from planktonic cultures of P. aeruginosa and E. coli as described above. To confirm that their persister cell state was maintained, persister cells were exposed to saline or ciprofloxacin in saline for a period of 24 h at 37°C. Cell viability was assessed as described above. The persister state was indicated by the maintenance of stable cell viability. For determination of the effect of cis-DA on persister cell maintenance, persister cells were exposed to cis-DA or saline for a period of 24 h at 37°C. P. aeruginosa was exposed to 100 nM cis-DA, while E. coli was exposed to 310 nM cis-DA. Following 24 h of exposure, the resulting persister cell population was washed twice with saline and resuspended in either saline or ciprofloxacin (20 μg/ml) in saline. Cell viability was determined at 0, 2, 5, and 24 h, as described above. Loss of the persister state was indicated by a loss of cell viability.
Killing efficacy assays.
Persister cells were exposed to antimicrobials alone and antimicrobials in combination with the fatty acid signaling molecule cis-DA. For assays with planktonic persister cells, P. aeruginosa or E. coli planktonic persister cells were pelleted and resuspended in 50 ml of saline, and aliquots of 6 ml were subjected to one of the following treatments: saline, cis-DA (100 nM or 310 nM) in saline, antimicrobials in saline, or antimicrobials with cis-DA (100 nM or 310 nM) in saline. Cultures were incubated at 37°C with shaking at 220 rpm for 24 h. Cell viability was determined at 0, 1, 3, 5, and 24 h, as described above. The antimicrobials used were ciprofloxacin (20 μg/ml), tobramycin (20 μg/ml), and tetracycline (100 μg/ml). For assays with biofilm persister cells, P. aeruginosa or E. coli biofilm persister cells were exposed to either saline, cis-DA in saline, ciprofloxacin (150 μg/ml) in saline, or ciprofloxacin (150 μg/ml) and cis-DA in saline for 24 h, after which time the remaining biofilms were collected into centrifuge tubes and processed for viability, as described above.
Quantification of respiratory activity in biofilms.
Respiratory activity was assessed by using CTC (5-cyano-2,3-ditolyl tetrazolium chloride), a monotetrazolium redox dye which produces a CTC-formazan (CTF) fluorescent complex (indicated by cells stained in red) when it is biologically reduced, indicating respiration (metabolic activity). P. aeruginosa and E. coli biofilms were grown in flow cell reactors (BioSurface Technologies), as described previously (22, 31), in 5% LB for a period of 6 days and subsequently exposed to ciprofloxacin (150 μg/ml) for a period of 18 h. The remaining biofilm population, consisting of persister cells only, was exposed to saline for a period of 30 min and subsequently to either saline or cis-DA (100 nM or 310 nM) in saline for a further 30 min. Treatments were performed together with the stains SYTO40 (5 μM) (Invitrogen) and CTC (5 mM) (Life Technologies, NY). CTC has previously been used to determine the respiratory activity of bacteria within biofilms (33). Biofilm architecture and metabolic activity were assessed by confocal scanning laser microscopy (CSLM) using a Leica Confocal TCS SP5 imaging system with a DMI 6000 inverted microscope (Leica Microsystems, Wetzlar, Germany) and Leica LAS AF software (Leica). Quantitative analysis of images was performed using COMSTAT (34). Relative fluorescence was quantified using the Intensity Luminance V1 software program (C. N. H. Marques and S. A. Craver, http://bingweb.binghamton.edu/~scraver/IL.html). The percentage of metabolic activity was determined by comparing the relative fluorescence of CTC to that of SYTO40 prior to and following treatments.
Use of cis-DA as a source of carbon to support growth.
Planktonic growth curves were performed for E. coli and P. aeruginosa with 2 different carbon sources, glucose and cis-DA. Planktonic cultures grown overnight were washed twice with saline (16,000 × g for 5 min at 4°C) and subsequently diluted to 1% in minimal EPRI (Electric Power Research Institute) medium (22, 35) supplemented with 100 nM, 300 nM, or 1,000 nM of either glucose or cis-DA. Cultures were grown at 37°C in 96-well microtiter plates, with OD595 measurements being performed every 30 min (DTX880 multimode detector; Beckman Coulter, CA).
Biofilm dispersion assays.
Biofilm dispersion in P. aeruginosa persister and total cell populations was determined using standard continuous-culture tube reactor methodology (22, 32, 36). Briefly, biofilms were cultured as described above, using a continuous-flow tube reactor system. Persister cells were isolated as described above. Subsequently, the treatment was switched from ciprofloxacin to either saline or cis-DA (100 nM) in saline for a period of 1 h. Controls consisted of biofilms exposed to saline for a period of 18 h, instead of ciprofloxacin. Controls were subsequently exposed to either saline or cis-DA (100 nM) in saline. Dispersion was evaluated by assessing the turbidity of the effluent at 600 nm. An increase of the effluent absorbance is indicative of dispersion.
Identification of cytoplasmic and membrane proteins.
Persister cells derived from planktonic and biofilm populations, exposed to saline or cis-DA in saline, were collected by centrifugation (16,000 × g for 5 min at 4°C). The resulting pellet was resuspended in 500 μl of TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA with 0.3 μg/ml phenylmethylsulfonyl fluoride [PMSF]) and lysed by sonication (six times for 10 s at 5 W). Samples were subsequently centrifuged (21,200 × g for 2 min at 4°C) to pellet unbroken cells. The resulting supernatant was further centrifuged at 30,000 × g to remove any remaining cell debris. The supernatant was subsequently spun at 100,000 × g for 90 min at 4°C. The supernatant, containing the cytoplasmic protein fraction, was retained. The pellet was resuspended in 1 ml of ice-cold TE buffer and centrifuged at 100,000 × g for 90 min at 4°C. The final pellet, containing the membrane protein fraction, was resuspended in 200 μl of TE buffer with 1% CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate}. Determination of adequate protein fractionation was achieved by the absence of catalase activity in the membrane fraction but its presence in the cytoplasmic fraction, determined with 1 mM hydrogen peroxide as the substrate (37, 38). This ensured the absence of contaminants from extracellular materials. The catalase control consisted of commercially available catalase (purified from Aspergillus) (39). Protein concentrations of each sample were determined via a modified Lowry assay (kit number 23240; Thermo Scientific, Rockford, IL) (40). Samples were subsequently mixed with SDS sample buffer and heat denatured at 100°C for 10 min. Proteins present within each fraction were visualized by SDS-PAGE analysis using 8% and 15% SDS-polyacrylamide gels and silver staining, performed as described previously (31). Image analysis and differences in protein abundance were analyzed using Image J. De novo or differentially abundant proteins were excised from the SDS-polyacrylamide gels, digested with trypsin, and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a QStarXL mass spectrometer (Applied Biosystems), as previously described (36).
Quantitative reverse transcriptase PCR.
Relative transcription levels in biofilms of P. aeruginosa and E. coli upon exposure to saline or cis-DA were evaluated. Exposure to saline or cis-DA in saline was performed for a period of 1 h for P. aeruginosa and for a period of 6 h for E. coli. We also compared the persister cell population transcription profile to that of the total cell population. The total cell population, composed of persister and nonpersister cells, consisted of the biofilm population exposed solely to saline for a period of 18 h. On the other hand, the persister cell population consisted of the cell population exposed to ciprofloxacin in saline for a period of 18 h. RNA was extracted from RNA Protect (Qiagen)-treated total biofilm and planktonic samples, as well as from persister cells, using the RNeasy minikit (Qiagen), with residual DNA being degraded using the DNase I amplification-grade kit (Invitrogen). A total of 1 μg of RNA was used for cDNA synthesis (36, 41, 42), and cDNA was generated by using a RETROscript kit (Ambion). Quantitative reverse transcriptase PCR (qRT-PCR) was performed with an Eppendorf Mastercycler Ep Realplex instrument (Eppendorf AG, Hamburg, Germany) and the Kapa SYBR Fast qPCR kit (Kapa Biosystems, Woburn, MA) (41) with the oligonucleotides (obtained from Integrated DNA Technologies, Coralville, IA) listed in Table 1. Relative transcript quantitation was accomplished by using Ep Realplex software (Eppendorf AG), with the transcript abundance (based on the threshold cycle [CT] value) being normalized to mreB (control) before the determination of transcript abundance ratios. Verification of single-product amplification was carried out by analysis of the melting curves.
TABLE 1.
Nucleotide primers used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| P. aeruginosa PA14 | |
| HDA_FW | GACTCCTACGGGAGGCAGCAGT |
| HDA_RV | GTATTACCGCGGCTGCTGGCAC |
| acpP_FW | GAACGCGTTAAGAAGATCG |
| acpP_RV | GGATTTCGGTCTCGAATTC |
| mreB_FW | CTGTCGATCGACCTGGG |
| mreB_RV | CAGCCATCGGCTCTTCG |
| PA14_21030_FW | CAGGAAGGTGTTCGTCAC |
| PA14_21030_RV | CCGGAACCGATCATGATG |
| atpH_FW | CAAAGAGCCTCAGCTGAC |
| atpH_RV | CTTTGCTCAAGGTGAAGG |
| PA14_16710_FW | TCAGTCACTACGAGGTGATC |
| PA14_16710_RV | ATCGCCAGGTCGGAGAC |
| uvrB_FW | GAGAAGGACTCCTCGATC |
| uvrB_RV | TGTCGATCACATCGCCAC |
| PA14_19410_FW | TCGAATCCGCGGAAGTTG |
| PA14_19410_RV | GTCCATCCACTCGTTGAG |
| PA14_33240_FW | GGACTCCTATGGCGATAC |
| PA14_33240_RV | TTGCAGGCGGAACAGTTC |
| PA14_01730_FW | CGAACTATCCTTATCACACG |
| PA14_01730_RV | CATGAAGCGCTTGATGGTAC |
| ppx_FW | GCATGCCGCAAAAACCTG |
| ppx_RV | CCAGTTGAACCTTCTCGC |
| ppk1_FW | GAAACCGTAGTGGCGAAC |
| ppk1_RV | GAGAGCTCGCGATGAATG |
| E. coli BW25113 | |
| 16S rRNA_FW | CAGCCACACTGGAACTGAGAC |
| 16S rRNA_RV | GCTTCTTCTGCGGGTAACGTC |
| acpP_FW | GAACAGCTGGGCGTTAAGAG |
| acpP_RV | CCAGCTCAACGGTGTCAAGAG |
| mreB_FW | GTCCATTGACCTGGGTACTGC |
| mreB_RV | CATCTGCTTCGCGTCATGACC |
| atpA_FW | GGTTAACACTCTGGGTGCACC |
| atpA_RV | GTCTGTACCGGCTGATCTACG |
| gadA_FW | GAAGCTGCAGGCAAACCAACG |
| gadA_RV | CGTTCAGAGAGGTCGTACAGG |
| oppA_FW | CAGCGATCTTGACGGTCATCC |
| oppA_RV | CGTTGTAGTCAGCACACCAGC |
| groL_FW | CGAACTGGACGTGGTTGAAGG |
| groL_RV | AACACCACCACCAGCAACCAC |
| tnaA_FW | ACACCATTCCGACTCACCAGG |
| tnaA_RV | GATCTGCTCGATGGTCCAGTC |
| tufA_FW | CGTCACTACGCACACGTAGAC |
| tufA_RV | GGTAACAACGGTACCACGACC |
| fbaB_FW | GTGGAGCAGGCGTTCAACATG |
| fbaB_RV | CCACAGCACTGTCACCATACC |
| ppk_FW | GTGCATGAGATGGAAGCCAGC |
| ppk_RV | CCTGTAACTCAACCACCACGG |
| frdA_FW | GGTTGTGTGAGCAGGATGTCG |
| frdA_RV | CCTTTACGCCATTCGTGCCAG |
MICs.
MICs of ciprofloxacin for P. aeruginosa and E. coli were determined in 5% and 100% LB by standard methods (43).
Statistical analysis.
One-way analysis of variance (ANOVA) was performed for multivariant analysis followed by Tukey's or Dunnett's multiple-comparison tests by using GraphPad Prism V 6.0a.
RESULTS
Induction of the SOS response enables the isolation of P. aeruginosa persister cells from biofilms.
While the presence of persister cell populations has been well documented for E. coli under both biofilm and planktonic conditions, little is known concerning the persister population present in biofilms of P. aeruginosa. To determine whether persister cells can be isolated from P. aeruginosa biofilms, we utilized the continuous-culture tube reactor system to grow biofilms (22, 31, 32). Persister cells were isolated through SOS response selection upon exposure to ciprofloxacin. The use of ciprofloxacin or ampicillin is the main standardized procedure for persister cell isolation and has been widely used in previous studies (2, 4, 14, 19, 29, 30, 44). To ensure adequate isolation of persister cell populations from P. aeruginosa biofilms, we used E. coli cultures as a control, where persister cell isolation from both planktonic and biofilm populations is well documented (2, 4, 14, 19, 29, 30, 44).
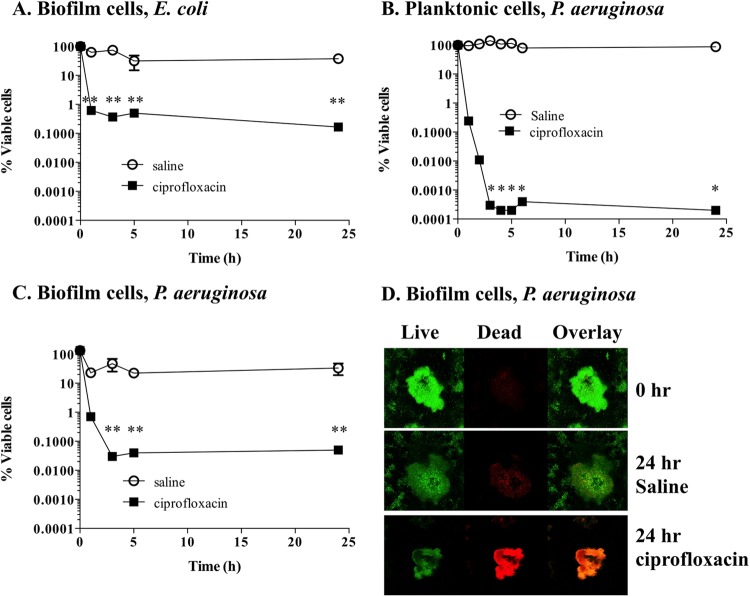
Persister cell populations were isolated from E. coli cells grown planktonically and as biofilms. Exposure of planktonic stationary-phase and biofilm cultures of E. coli to ciprofloxacin resulted in a typical biphasic killing curve, where a significant decrease (P < 0.01) of cell viability within the initial 3 h of treatment was observed (Fig. 1A). The decrease in cell viability was followed by a viability plateau, which remained constant from the 3-h point until the end of the experiment. Based on the definition of persister cells provided by Keren et al. (2), all cells that were recovered during the plateau phase were assumed to be persister cells. By doing so, the final cell recovery of persister cells in planktonic populations (not shown) and biofilm populations (Fig. 1A) was <0.1%, an observation consistent with previous findings (45).
FIG 1.
Isolation of persister cells from planktonic and biofilm populations. Stationary-phase planktonic cultures and 6-day biofilms were exposed to saline or ciprofloxacin in saline for a period of 24 h. Cell viability was determined throughout the experiment. (A to C) E. coli biofilm populations (A), P. aeruginosa planktonic populations (B), and P. aeruginosa biofilm populations (C). (D) Cell viability of P. aeruginosa biofilms was also assessed by using SYTO9 (live cells) and propidium iodide (dead cells) at 0 h and at 24 h for saline- and ciprofloxacin-exposed cells. The averages of data from 3 experiments with 2 replicates per experiment are shown. Error bars indicate standard deviations (*, P < 0.001; **, P < 0.01 [significantly different from persister cells treated with saline, as indicated by one-way ANOVA]).
Following consistent isolation of persisters from E. coli cultures, we used identical procedures to isolate persister cells from P. aeruginosa biofilms and planktonic cells. To optimize the isolation of persister cell subpopulations from P. aeruginosa, we initially performed ciprofloxacin dose-dependent survival curves (see Fig. S1 in the supplemental material). By doing so, we observed that when concentrations of ciprofloxacin of ≥10 μg/ml were used, planktonic P. aeruginosa cells had a biphasic survival typically observed when persister cells are present in a population (see Fig. S1 in the supplemental material). Similar survival curves were performed for biofilms with ciprofloxacin concentrations between 50 and 300 μg/ml (not shown). Based on the observations that the ciprofloxacin MIC for P. aeruginosa and E. coli was 1 μg/ml together with the results from the ciprofloxacin dose-dependent survival curves, we used concentrations of ciprofloxacin in the order of 20× MIC (20 μg/ml) for isolation of persister populations derived from planktonic cultures and 150× MIC (150 μg/ml) for isolation of persister populations derived from biofilms.
Exposure of planktonic stationary-phase cultures of P. aeruginosa to ciprofloxacin resulted in significantly decreased cell viability (P < 0.001) within the initial 3 h of treatment. No further decrease in viability was observed after 3 h of treatment (Fig. 1B). The final cell recovery of persister cells in planktonic populations was <0.001% for P. aeruginosa (Fig. 1B). The low percentage of persister cells is consistent with previous findings (45) and suggests that the persister population present in P. aeruginosa planktonic cells is significantly smaller than the one observed for E. coli. Persister cells present in biofilms were isolated from 6-day-old P. aeruginosa biofilms grown under continuous-flow conditions through exposure to ciprofloxacin (150 μg/ml) for a period of 24 h (Fig. 1C). Similarly to the planktonic cultures, cell viability significantly decreased (P < 0.001) with the initial 3-h exposure to ciprofloxacin and remained constant from that point onwards, with a final cell recovery of <0.1% compared to the control (saline), an observation consistent with E. coli persister cell isolation.
Confirmation of the persister cell state.
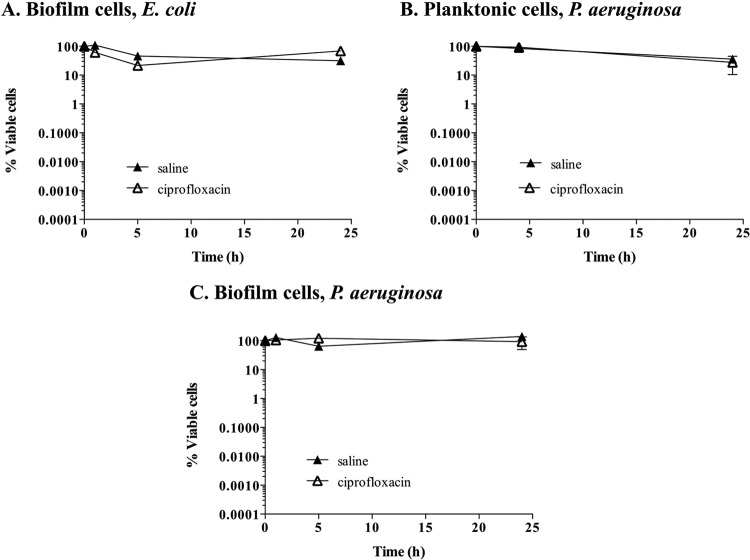
As the isolation of persister cells from biofilm cultures grown in tube reactors has not been previously described, we decided to confirm the persister cell state in E. coli and P. aeruginosa populations following 18 h of exposure to ciprofloxacin. We made use of the knowledge that persister cells are tolerant to antimicrobials and assessed the viability of persister cells upon exposure to antimicrobials for several hours (28). E. coli was once again used as a control. Following persister cell isolation, persister subpopulations of E. coli biofilms were exposed to either saline or ciprofloxacin for an additional 24 h. No significant difference in viability (P > 0.1) between persister subpopulations treated with saline and those treated with ciprofloxacin was noted (Fig. 2A). The finding that ciprofloxacin treatment did not result in further killing confirmed the persister state of the isolated subpopulation.
FIG 2.
Confirmation of the persister cell state. Persister cells isolated from planktonic and biofilm populations were exposed to saline or ciprofloxacin in saline for 24 h. Cell viability was determined throughout the experiment. Shown are E. coli biofilm populations (A), P. aeruginosa planktonic populations (B), and P. aeruginosa biofilm populations (C). The averages of data from 3 experiments with 2 replicates per experiment are shown. Error bars indicate standard deviations.
We likewise confirmed the persister cell state of P. aeruginosa persister cells obtained from both planktonic and biofilm cultures by exposing persister cells to either saline or ciprofloxacin for an additional 24 h. The absence of a significant decrease (P > 0.1) in cell viability (no further killing by ciprofloxacin) confirmed their persister state (Fig. 2B and C). Similar to E. coli persister cells, treatment with ciprofloxacin did not result in further killing of P. aeruginosa persister cells isolated from both planktonic and biofilm cultures.
To further confirm that cells within P. aeruginosa biofilms were in a persister cell state, the resulting population after 18 h of ciprofloxacin treatment was analyzed by microscopy with a Live/Dead BacLight bacterial viability kit. We hypothesized that following extended ciprofloxacin exposure, the only remaining viable population consists of persister cells, as ciprofloxacin kills and can lyse nonpersister cells (46–48). Prior to ciprofloxacin treatment or following exposure to saline for 24 h, the vast majority of P. aeruginosa biofilm cells appeared to be viable, as indicated by the retention of SYTO9, indicative of the cells being alive, with only a small percentage of cells being stained red (Fig. 1D). However, following treatment with ciprofloxacin, biofilm cells retained both SYTO9 and propidium iodide. This finding indicated that the membrane was compromised and, thus, that the cells were either lysed or dead (Fig. 1D). Our findings are in strong support of our methodology resulting in the isolation of persister cells from planktonic and biofilm populations of P. aeruginosa.
The presence of cis-DA influences the number of persister cells isolated.
Previous findings indicated that exposure to cis-DA resulted in increased cell recovery of several bacterial species on agar plates (23), suggesting that cis-DA somehow affects viability or stimulates bacterial growth. This observation led us to ask whether cis-DA can affect the size of persister populations isolated (Fig. 3). Therefore, isolation of persister cells of planktonic populations of E. coli and P. aeruginosa was performed by using ciprofloxacin in the presence and absence of cis-DA. The addition of cis-DA resulted in a significant 1- to 2-log decrease (P < 0.01) in the number of persister cells compared to the number of cells with ciprofloxacin alone for both P. aeruginosa (Fig. 3A) and E. coli (Fig. 3B). Our findings suggested that cis-DA contributes to a reduction of the persister cell population regardless of growth conditions or bacterial species.
FIG 3.
Effect of cis-DA on the percent persister cell subpopulation. P. aeruginosa (A) and E. coli (B) persister cells were isolated from stationary-phase planktonic cultures in the presence and absence of cis-DA. Cultures were exposed to ciprofloxacin in saline (20 μg/ml), ciprofloxacin with cis-DA in saline, and cis-DA in saline for a period of 24 h. Cell viability was determined throughout the experiment. The averages of data from 3 experiments with 2 replicates per experiment are shown. Error bars indicate standard deviations (*, P < 0.01 [significantly different from persister cells treated with ciprofloxacin, as indicated by one-way ANOVA]).
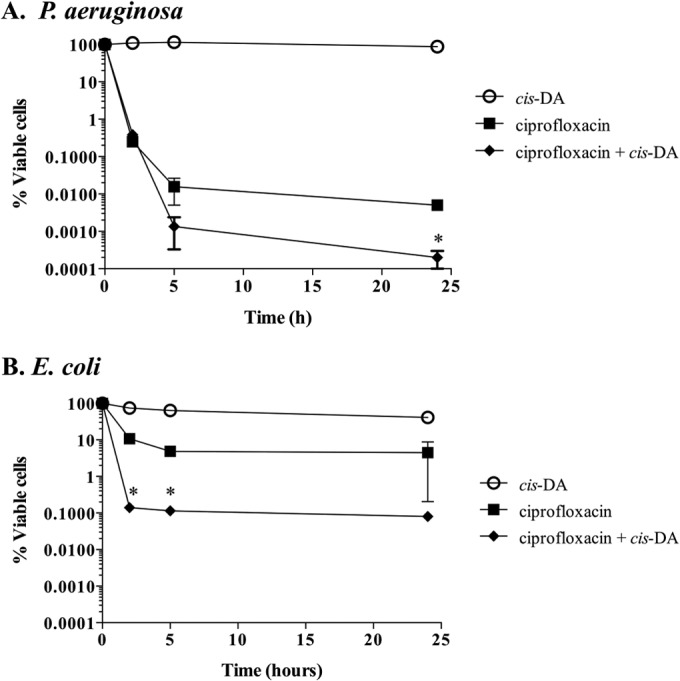
Exposure of persister cells to cis-DA in combination with ciprofloxacin results in cell number reduction or cell eradication.
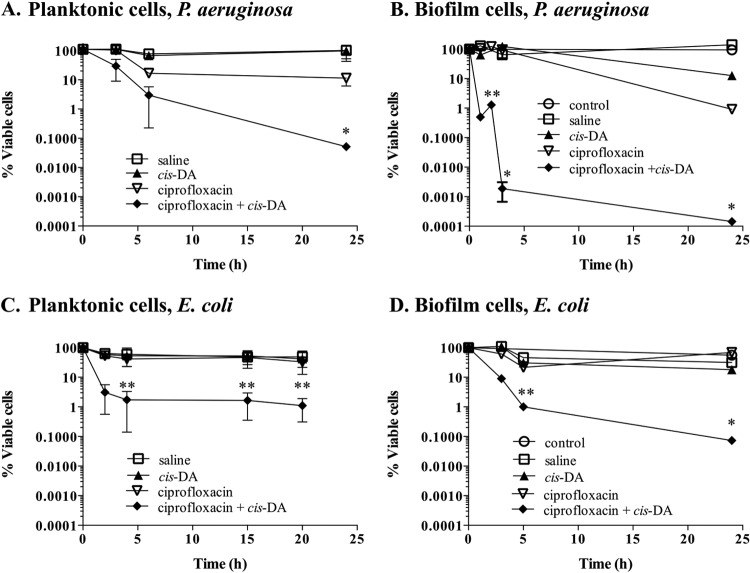
To further confirm a role of cis-DA in altering the persister cell state, we next asked whether cis-DA is likewise capable of altering the persister cell state of isolated persister cells. To do so, we made use of the finding that cis-DA enhances the efficacy of several classes of antimicrobials. For instance, cotreatment of P. aeruginosa PAO1 biofilm cells with cis-DA and tobramycin or ciprofloxacin resulted in 1- and 2-log decreases in viability, respectively, compared to treatment with tobramycin and ciprofloxacin alone (24). The presence of cis-DA together with antimicrobials has also been found to reduce the viability of S. aureus biofilm cells, eradicate mixed-species biofilms, and remove preformed biofilms of B. subtilis, S. enterica, S. aureus, and E. coli from surfaces (25–27). These observations, together with the fact that cis-DA leads to a reduction in the number of persister cells isolated (Fig. 3), led us to ask whether the presence of cis-DA was capable of breaking persistence, leading to a reversion of the persister cells' tolerant state. Thus, we investigated whether exposure of isolated persister cells to antimicrobials in the presence of cis-DA led to a decrease in cell viability and, eventually, cell eradication. P. aeruginosa and E. coli persister cells isolated from biofilm and planktonic populations were exposed to ciprofloxacin at the concentrations indicated, in the presence or absence of cis-DA. In the absence of cis-DA, ciprofloxacin treatment alone had no effect on the viability of persister cells, as indicated by the finding that no decrease in the number of persister cells was noted (Fig. 4). Furthermore, exposure of persister cells to cis-DA alone had no effect on the overall viability of the persister cell population. In contrast, however, exposure of persister cells to ciprofloxacin in conjunction with cis-DA resulted in a significant reduction of cell viability. Overall, a 3.5-log reduction in viability was noted for planktonic culture-derived persister subpopulations of P. aeruginosa (Fig. 4A), while a 2-log reduction was observed for planktonic culture-derived persister subpopulations of E. coli (Fig. 4C). Exposure of persister cells from P. aeruginosa and E. coli biofilm populations to ciprofloxacin in conjunction with cis-DA correlated with 6-log and 4-log reductions, respectively (Fig. 4B and D).
FIG 4.
Tolerance of persister cells to ciprofloxacin is reduced in the presence of cis-DA. Persister cell subpopulations of P. aeruginosa planktonic populations (A), P. aeruginosa biofilms (B), E. coli planktonic populations (C), and E. coli biofilms (D) were exposed to saline, cis-DA in saline, ciprofloxacin in saline, and ciprofloxacin with cis-DA in saline. Biofilm and planktonic populations were exposed to 150 μg/ml and 20 μg/ml of ciprofloxacin, respectively. Experiments were performed for a period of 24 h, and cell viability was determined throughout the experiment. Experiments with persister cells derived from biofilms had one additional control, a total biofilm population treated with cis-DA in saline (control). P. aeruginosa cells were exposed to 100 nM cis-DA, and E. coli cells were exposed to 310 nM cis-DA. The averages of data from 3 experiments are shown. Error bars indicate standard deviations (*, P < 0.001; **, P < 0.01 [significantly different from persister cells treated with ciprofloxacin, saline, and cis-DA, as indicated by one-way ANOVA]).
Our findings strongly suggested that cis-DA renders persister populations susceptible to ciprofloxacin. However, whether cis-DA acts by increasing the efficacy of ciprofloxacin or instead alters the persister cell state is not clear. We hypothesized that if cis-DA acts by increasing the efficacy of ciprofloxacin, pretreatment of persister cells with cis-DA prior to ciprofloxacin treatment would have no effect. However, if cis-DA alters the persister cell state, by returning persister cells to an active metabolic state (18), exposure of isolated persister cells to cis-DA prior to ciprofloxacin treatment would correlated with a decrease in viability. To test this, we exposed planktonic P. aeruginosa and E. coli persister cells to cis-DA or saline for 20 h preceding exposure to ciprofloxacin (20 μg/ml) alone. While preexposure of persister cells to saline resulted in no change in the viability of the persister population upon exposure to ciprofloxacin, preexposure to cis-DA led to a significant 2- to 3-log decrease (P < 0.001) in the number of viable cells of P. aeruginosa (Fig. 5A) and E. coli (Fig. 5B).
FIG 5.
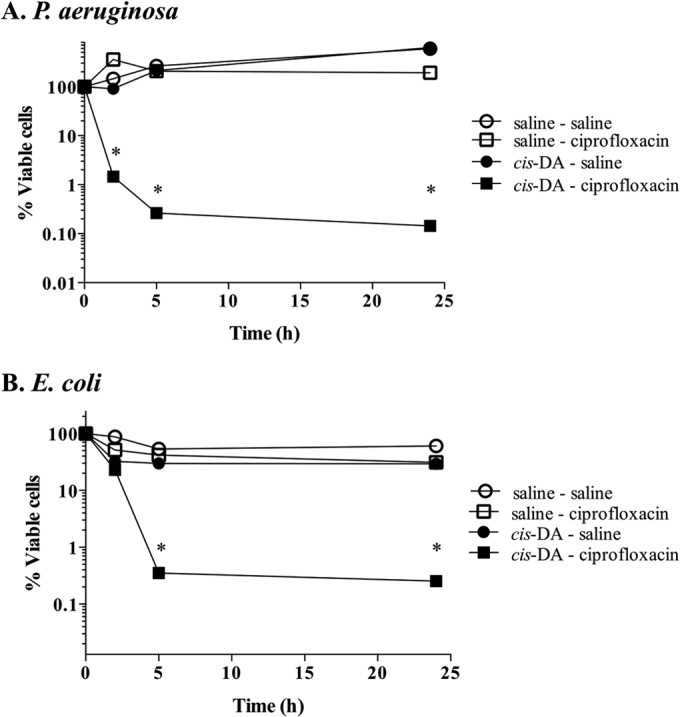
Effect of cis-DA on maintenance of the persister state. P. aeruginosa (A) and E. coli (B) persister cells were exposed to cis-DA in saline or saline for a period of 20 h prior to exposure to saline or ciprofloxacin in saline for a further 24 h. Viability was determined throughout the experiment. The averages of data from 3 experiments are shown. Error bars indicate standard deviations (*, P < 0.001 [significantly different from persister cells treated with saline, as indicated by one-way ANOVA]).
These findings indicated that cis-DA contributes to a reduction of the persister cell population and affects the persister cell state by altering the physiological status of the persister population of P. aeruginosa and E. coli rather than by enhancing the efficacy of the antibiotic itself.
The presence of cis-DA reduces persister cell viability when used in conjunction with tobramycin and tetracycline.
We postulated that if cis-DA altered the physiological status of persister cells, cis-DA should likewise cause a reduction in viability of persister cell populations when persister cells are exposed to antimicrobial agents other than ciprofloxacin (Fig. 4), a quinolone antibiotic (49). We therefore exposed planktonic P. aeruginosa persister cells to the polyketide antibiotic tetracycline (100 μg/ml) and the aminoglycoside tobramycin (20 μg/ml) (50, 51). Exposure of planktonic P. aeruginosa persister cells to 20 μg/ml tobramycin (Fig. 6A) resulted in a slight decrease of cell viability at 3 h, with no further reduction at 24 h. Exposure of planktonic persister cells to 100 μg/ml tetracycline (Fig. 6B) alone did not result in a reduction of cell viability. However, exposure of persister cells to tobramycin and cis-DA (100 nM) resulted in a >1-log decrease of cell viability (Fig. 6A). Likewise, exposure of persister cells to tetracycline and cis-DA resulted in a >1-log decrease of cell viability (Fig. 6B). Together, these findings of increased killing of persister cells in the presence of cis-DA, regardless of the antimicrobials used, support the notion that cis-DA awakens persister cells and reverts them to a susceptible state.
FIG 6.
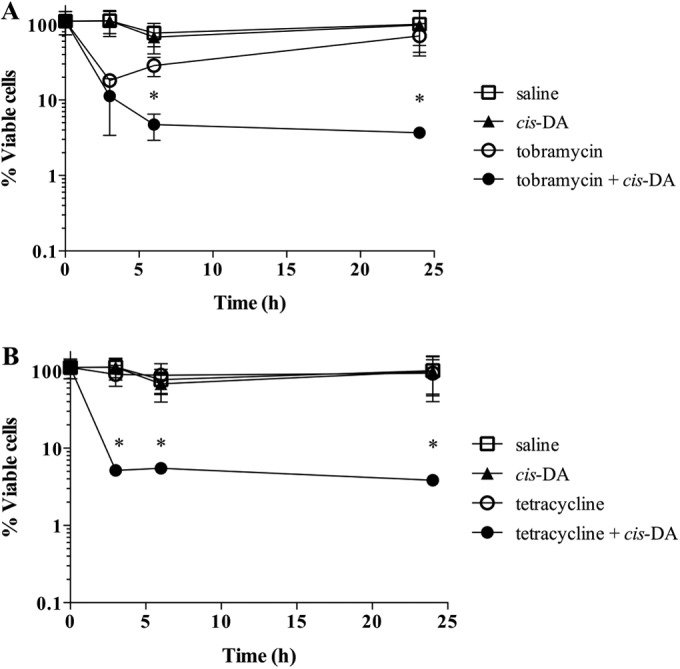
Tolerance of persister cells to tobramycin and tetracycline in the presence of cis-DA. Persister cell subpopulations of P. aeruginosa planktonic populations were exposed to tobramycin (A) and tetracycline (B) in the presence and absence of cis-DA (100 nM) for a period of 24 h. Shown are data for treatments with saline, cis-DA in saline, antimicrobial in saline, and antimicrobial and cis-DA in saline. Tetracycline was used at a concentration of 100 μg/ml, and tobramycin was used at a concentration of 20 μg/ml. Viability was determined throughout the experiment. The averages of data from 3 experiments are shown. Error bars indicate standard deviations (*, P < 0.01 [significantly different from persister cells treated with saline, cis-DA, and antimicrobials, as indicated by one-way ANOVA]).
cis-DA is not utilized as a carbon source for bacterial growth.
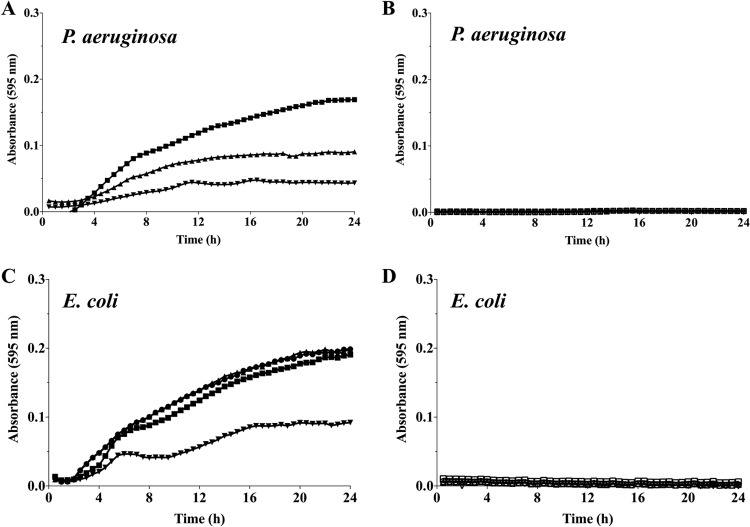
We next asked whether persister cell awakening was a result of cis-DA being utilized as a carbon source. If cis-DA was utilized as a carbon source, it could influence the metabolic rates of the persister population and thus stimulate active cell growth. In order to assess this, growth of E. coli and P. aeruginosa planktonic cultures in EPRI liquid medium using either cis-DA or glucose as the sole carbon source was assessed. Both glucose and cis-DA were provided at concentrations ranging from 100 nM to 1,000 nM. With glucose as a sole carbon source, growth of P. aeruginosa was observed at all concentrations tested, with logarithmic growth being initiated within the initial 4 h of growth (Fig. 7A). No growth, however, was detected in the presence of cis-DA regardless of the concentrations used (Fig. 7B). Likewise, no growth of E. coli was observed when cis-DA was used as the sole carbon source, independent of the concentration used (Fig. 7D). This is in contrast to the logarithmic growth observed in the presence of glucose (Fig. 7C). These findings indicated that cis-DA does not serve as a carbon source.
FIG 7.
Use of cis-DA as a carbon source. Planktonic cultures of P. aeruginosa and E. coli were grown in EPRI medium containing either glucose or cis-DA as a carbon source. The concentrations of glucose (closed symbols) (A and C) and cis-DA (open symbols) (B and D) used were 100 nM (▼/▽), 300 nM (▲/△), and 1,000 nM (◼/◻). Results correspond to representative data from 3 experiments with a total of at least 10 biological replicates.
Exposure to cis-DA does not induce dispersion of biofilm-derived persister cells.
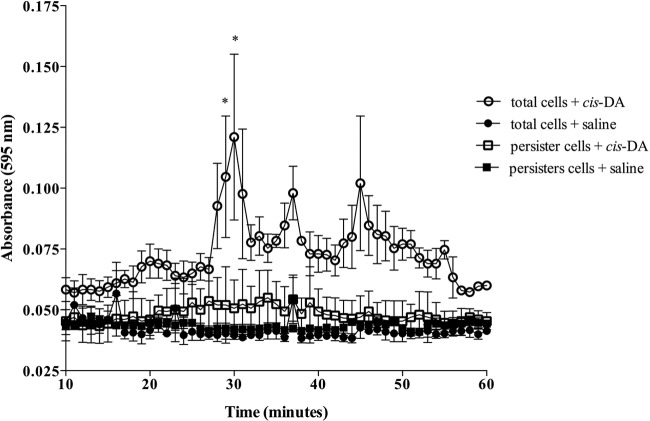
To exclude the possibility that the reversion of the persister cell state and the reduction in persister cell isolation upon exposure to cis-DA were due to dispersion, we determined the dispersion response of persister cell subpopulations using standard continuous-culture tube reactor methodology (22, 31, 32, 36, 52). To achieve this, P. aeruginosa biofilm persister cells were exposed to cis-DA (100 nM) or saline for a period of 1 h under continuous-flow conditions (Fig. 8). Controls consisted of P. aeruginosa biofilms, composed of total cell populations, which were exposed to saline for 18 h instead of ciprofloxacin prior to exposure to saline or cis-DA for a period of 1 h (Fig. 8). A dispersion response, revealed by an increase of the effluent turbidity, was observed after 25 min of exposure to cis-DA in total cell populations and continued until approximately 55 min (Fig. 8). No dispersion response, however, was observed for persister cell populations exposed to cis-DA.
FIG 8.
Persister cells do not disperse in the presence of cis-DA. Biofilms were composed of total cell populations (exposed to saline for a period of 18 h) or persister cell populations (exposed to ciprofloxacin at 150 μg/ml for 18 h). Biofilms were exposed to cis-DA (100 nM) in saline or saline for a period of 60 min, and dispersion was evaluated by measuring the absorbance at a wavelength of 595 nm. The averages of data from 3 experiments are shown. Error bars indicate standard deviations (*, P < 0.01 [significantly different from persister cells treated with saline, as indicated by one-way ANOVA]).
To further confirm that cis-DA does not induce dispersion of biofilm-derived P. aeruginosa persister cells, flow-cell-grown biofilm-derived P. aeruginosa persister cells were exposed to saline or cis-DA (100 nM) for a period of 1 h and stained with SYTO40. No significant difference in biomass accumulation over a period of 1 h was noted. Our observations were confirmed by COMSTAT analysis, which indicated no significant difference in biofilm thickness, the portion of the slice occupied by the bacteria, and total biomass prior to and following exposure to cis-DA (Table 2). These results indicated that while cis-DA induces dispersion of biofilms composed of total cells, cis-DA does not induce dispersion of biofilm-derived persister cells.
TABLE 2.
COMSTAT analysis of P. aeruginosa biofilms composed solely of persister cells and exposed to saline or cis-DA (100 nM) in saline for a period of 30 mina
| Parameter | Avg value for treatment ± SD |
|||
|---|---|---|---|---|
| Saline |
cis-DA |
|||
| SYTO40 | CTC | SYTO40 | CTC | |
| Total biomass (μm3/μm2) | 22.56 ± 22.30 | 0.0003 ± 0.0006 | 15.82 ± 17.75 | 1.01 ± 1.00 |
| Portion of slice occupied by bacteria (%) | 56.53 ± 32.49 | 0.05 ± 0.025 | 67.89 ± 24.66 | 19.06 ± 15.06 |
| Thickness (μm) | 26.84 ± 25.47 | 0 ± 0 | 20.14 ± 19.26 | 1.36 ± 1.29 |
| Roughness coefficient (dimensionless; range, zero to infinity) | 0.76 ± 0.59 | 0 ± 0 | 0.45 ± 0.43 | 1.25 ± 0.62 |
| Surface area of biomass in image stack (μm2) | 3.9E+6 ± 7.2E+5 | 1.3E+3 ± 4.7E+2 | 3.9E+6 ± 4.7E+5 | 1.3E+5 ± 9.2E+4 |
| Surface-to-biovolume ratio (μm2/μm3) | 1.44 ± 11 | 9.07 ± 6.39 | 1.85 ± 1.26 | 6.85 ± 3.44 |
| Maximum thickness (μm) | 41.70 ± 33.27 | 0 ± 0 | 41.13 ± 30.26 | 10.28 ± 6.86 |
Total cells were stained with SYTO40, and respiratory activity was monitored with CTC stain. Data are averages ± standard deviations of data for 15 replicates from triplicate experiments.
Respiratory activity of persister cells is increased upon exposure to cis-DA.
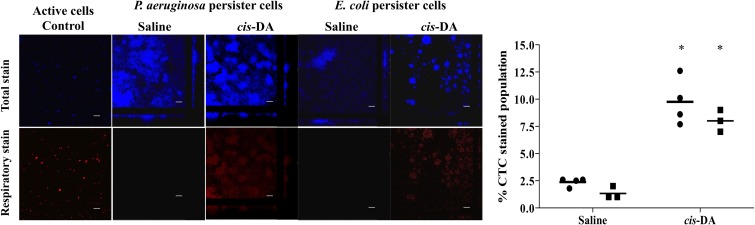
Considering that cis-DA contributes to persister cells returning to a susceptible state without being used as a carbon source or inducing dispersion, we next asked whether cis-DA awakens persister cells by returning them to an active metabolic state. We therefore made use of CTC (5-cyano-2,3-ditolyl tetrazolium chloride) stain, which produces an insoluble, red fluorescent compound when it is biologically reduced, indicating respiration (metabolic activity). As the number of persister cells is higher in biofilms than in planktonic cultures, we made use of biofilm-derived persister cell populations that were exposed to saline or cis-DA for a period of 1 h. Cells were counterstained with SYTO40, which presented a blue color to visualize cells regardless of their metabolic state. No CTC staining (no visible red staining) was detected when P. aeruginosa and E. coli biofilm persister cells were exposed to saline alone. Overall, exposure to saline resulted in a 2.5% increase in CTC staining in P. aeruginosa cells and a 1% increase in CTC staining in E. coli cells, suggesting that persister cells display very low respiratory activity (Fig. 9). This finding furthermore suggests that persister cell populations are not dormant or metabolically inactive, as previously indicated (53). In contrast, exposure to cis-DA resulted in cells staining red, which is indicative of respiratory/metabolic activity. Quantitative analysis revealed a 10% increase of CTC staining (P < 0.001) upon exposure to cis-DA in P. aeruginosa cells. Similar results were obtained for persister cells formed by E. coli biofilms, where an 8% increase in CTC staining was observed upon exposure to cis-DA (Fig. 9). Moreover, while no significant difference in the total populations prior to and after cis-DA exposure was noted, CTC-stained cells were found to be distributed throughout the biofilm microcolonies, as indicated by increases in CTC-stained total biomass, the portion of the slice occupied by the bacteria, average thickness, and maximum thickness. While our analysis indicated that 8 to 10% of the persister cell population was rendered metabolically active upon exposure to cis-DA, respiratory activity measurements using CTC commonly underestimate the metabolically active population (54). It is thus likely that cis-DA affects a higher percentage of persister cells. It is of interest to note that 85.8% ± 26.1% of active mid-exponential-phase planktonic populations were stained with CTC and SYTO40.
FIG 9.
Respiratory activity of persister cells upon exposure to cis-DA. Respiratory activity of P. aeruginosa and E. coli biofilm persister cells was evaluated by the ability of the cells to metabolize CTC into fluorescent formazan following 1 h of incubation with saline or cis-DA in saline. SYTO40 was used to stain the overall population. The percentage of the CTC-stained population of persister cells compared to the SYTO40-stained population was calculated. Error bars indicate standard deviations. Symbols in the graph correspond to P. aeruginosa (●) and E. coli (■). Bar = 25 μm (*, P < 0.001 [significantly different from persister cells treated with saline alone, as indicated by one-way ANOVA]).
These observations indicated that cis-DA contributes to an increase in respiratory activity and that, as a result, cells reverted from a low metabolic state to a susceptible, active metabolic state.
Exposure to cis-DA leads to increased abundance or de novo synthesis of proteins involved in cell repair and metabolism.
Exiting from dormancy requires a sudden burst of transcription and translation (55). Thus, to further confirm the awakening of persister cells due to a transition from a low metabolic state in the presence of cis-DA, SDS-PAGE was used to evaluate the effect of cis-DA on the abundance of proteins in planktonic and biofilm persister cells of P. aeruginosa and E. coli. Membrane and cytoplasmic fractions were isolated, and adequate fractionation was confirmed by the presence of catalase activity in the cytoplasmic fractions and by the absence of catalase activity in the membrane fractions, as previously described (37, 38). Protein bands selected for identification (Tables 3 and 4; see also Fig. S2 and S3 in the supplemental material) were excised from SDS gels, tryptically digested, and identified by LC-MS/MS. Although individual protein bands were selected, certain samples contained more than one protein due to the fact that SDS-PAGE gels separate proteins based on their size only, and thus, ideal fractionation is not always achieved.
TABLE 3.
Membrane and cytoplasmic proteins isolated from P. aeruginosa persister cells upon exposure to saline or cis-DA (100 nM) in saline and identified by LC-MS/MS
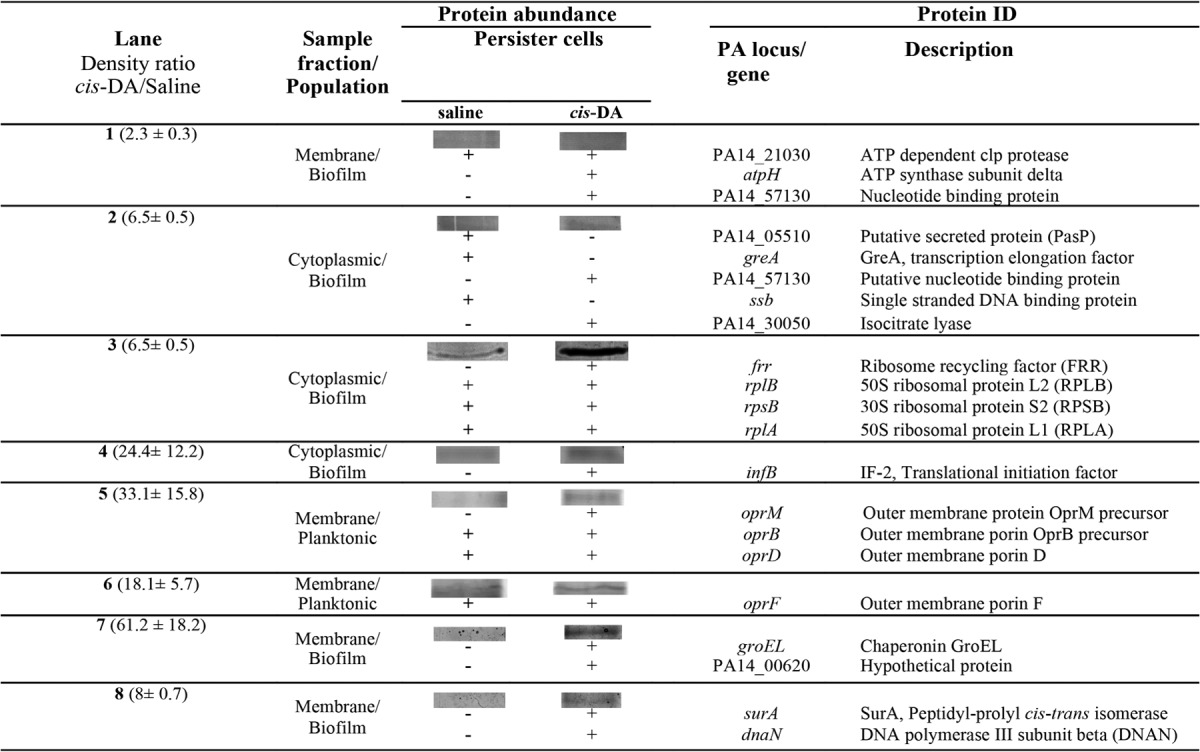
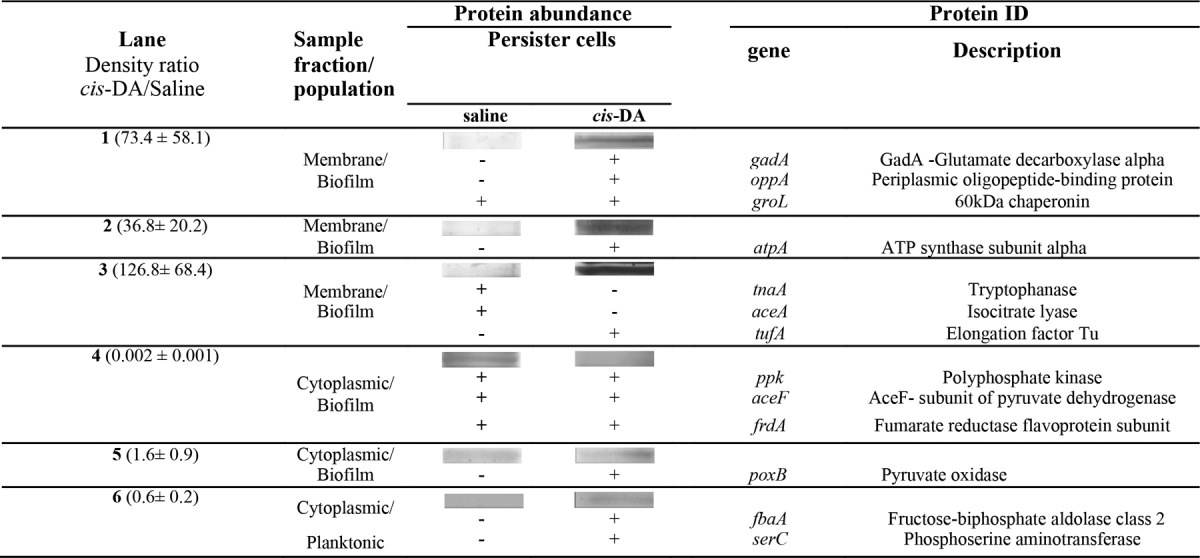
TABLE 4.
Membrane and cytoplasmic proteins isolated from E. coli persister cells upon exposure to saline or cis-DA (310 nM) in saline and identified by LC-MS/MS
In P. aeruginosa, the presence of cis-DA led to de novo production of proteins involved in metabolism and virulence (isocitrate lyase), protein repair and chaperoning (GroEL and SurA), protein translation (translational initiation factor 2 [IF-2] and ribosomal recycling factor [FRR]), antibiotic resistance and transport (OprM), ATP synthesis (AtpH), and DNA/RNA binding (PA14_57130, a nucleotide binding protein, and DNA polymerase III subunit beta) (56–66). Proteins that presented higher abundances in P. aeruginosa following exposure to cis-DA were involved in antibiotic resistance and transport (OprD and OprF), protein degradation (ATP-dependent Clp protease), and DNA/RNA binding (50S ribosomal protein L1 [RPLA] and 50S ribosomal protein L2 [RPLB]) (67–73).
In E. coli, exposure to cis-DA led to de novo production of proteins involved in transport (OppA), protein translation (TufA), metabolism (PoxB, FbaA, GadA, and SerC), and ATP synthesis and ATP hydrolysis (AtpA) (74–80). Proteins identified in E. coli under both cis-DA and saline conditions were involved in several functions, including metabolism (pyruvate dehydrogenase, AceF, and FrdA), ATP hydrolysis (polyphosphate kinase [PPK]), and protein repair (GroL) (81–85).
Overall, the presence of cis-DA led to an increase in the abundance of proteins involved in protein initiation, translation, cleavage, and repair; ATP synthesis and hydrolysis; transport; metabolism; and DNA and RNA binding (Tables 3 and 4). This finding provides evidence that the cells are using energy through AtpH and AtpA to initiate protein degradation of misfolded proteins (GroEL and GroL), degrade misfolded or unwanted proteins (ATP-dependent Clp protease, a ClpP2 homolog), initiate protein synthesis (IF-2 and TufA), bind to ribosomes (RPLB and RPLA), and increase transport (OprM, OprD, OprF, and OppA). In addition, exposure to cis-DA resulted in a decrease in the abundance of PPK, indicating that polyphosphate (polyP) degradation might be occurring instead (86–88).
Relative transcript abundances of cell metabolic markers are increased in the presence of cis-DA.
Our findings support the hypothesis that cis-DA increases the metabolic status of cells, likely by not only initiating protein repair but also enhancing protein synthesis and ATP synthesis and hydrolysis. To determine whether the noted increase in protein abundance was due to cis-DA enhancing transcription or just enhancing the translation of previously produced mRNA, qRT-PCR was carried out, focusing on the transcript abundances of certain genes encoding proteins found to be differentially produced upon exposure to cis-DA (Tables 3 and 4).
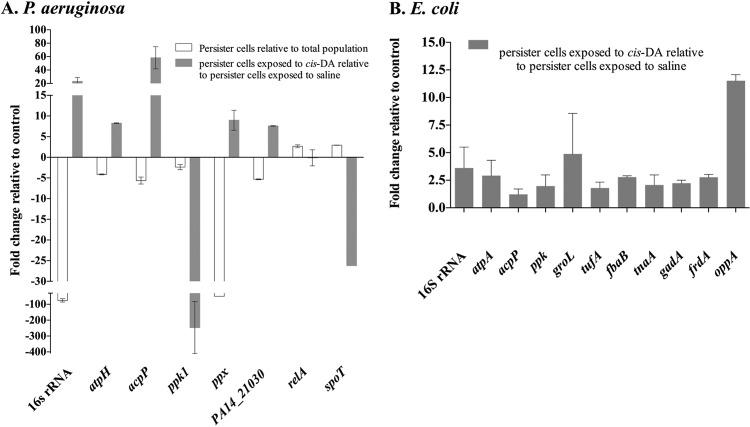
Exposure of P. aeruginosa biofilm persister populations to cis-DA for a period of 1 h resulted in a 245-fold decrease in ppk transcript abundance and an 8-fold increase in PA14_21030 transcript abundance compared to exposure to saline alone (Fig. 10A). Isolated persister cells of P. aeruginosa exhibited 2.4- and 5.3-fold decreases of the relative abundances of ppk and PA14_21030, respectively, compared to the total population (Fig. 10A). When the effect of exposure of E. coli persister cells to cis-DA was evaluated, there were increases of the relative abundances of genes related to the proteins identified. The greatest increase in transcript abundance observed was 12-fold for oppA, while a 5-fold increase in transcript abundance was observed for groL, a 3-fold increase was observed for fbaB, and 2-fold increases were observed for ppk, tnaA, gadA, and tufA (Fig. 10B).
FIG 10.
Transcription abundance of metabolic activity markers. Persister cells were selected from biofilm cultures of P. aeruginosa (A) and E. coli (B). The relative expression levels of the metabolic activity markers 16S rRNA, acpP, atpH, atpA, ppk, and ppx were evaluated upon exposure of persister cells to cis-DA in saline and compared to the levels upon exposure to saline alone. P. aeruginosa was exposed to cis-DA for a period of 1 h, while E. coli was exposed for a period of 6 h. The relative expression level for P. aeruginosa persister cells derived from biofilms was also compared to that for the overall nonpersister population. In addition, the relative expression levels of relA and spoT, known to be involved in persister cell formation, were also evaluated for P. aeruginosa. Genes related to some proteins present in higher abundance upon exposure of persister cells were also evaluated. The CT value of the housekeeping gene mreB remained constant throughout the different treatments (P > 0.5 by ANOVA and no difference between treatments by Tukey's multiple-comparison test). Experiments were carried out at least in triplicate. Error bars denote standard deviations. A significant change was considered to occur when a 2-fold change in the relative expression level occurred.
To further establish whether the increase of the cells' metabolic status was concurrent with an increase in respiratory activity (Fig. 9), we determined the transcript abundances of 16S rRNA, acpP, atpH, and ppx. Both 16S rRNA and acpP have previously been related to the cell metabolic status and cell growth status (81, 82). The ATP synthase genes atpH (δ subunit) and atpA (α subunit) have been demonstrated to be involved in proton motive force and ATP production in bacteria (89, 90). Polyphosphate kinase (ppk) and exopolyphosphatase (ppx) have been demonstrated to be involved in the production and hydrolysis, respectively, of inorganic polyPs, which can then be available for ATP production or gene regulation (in the case of polyP synthesis) (86–88).
Exposure of P. aeruginosa biofilm persister populations to cis-DA for a period of 1 h resulted in 58-, 23-, 4.1-, and 10-fold increases in relative abundances of acpP, 16S rRNA, atpH, and ppx, respectively, compared to saline treatment (Fig. 10A). Similarly to P. aeruginosa, exposure of E. coli biofilm persister populations to cis-DA (310 nM) resulted in increased relative abundances of 16S rRNA (4-fold) and atpA (3-fold) compared to treatment with saline alone (Fig. 10B).
When persister cell populations of P. aeruginosa were compared to total cell populations, decreased relative abundances of major metabolism-associated genes were observed (5.6-, 75-, 4-, and 50-fold decreases for acpP, 16S rRNA, atpH, and ppx, respectively), while no change was observed for ppk (Fig. 10A). These results are in contrast to those obtained for persister cells exposed to cis-DA, which exhibited relative increases in the abundances of the above-mentioned genes compared to persister cells exposed to saline (Fig. 10A). Overall, these data suggest that cis-DA is capable of returning persister cells to a metabolically active state that is comparable to the metabolic state observed for the total population.
These results give further evidence that persister cells are awakening in the presence of cis-DA, as increased transcript abundances of metabolic activity markers (16S rRNA, acpP, and atpH-atpA) were observed upon exposure of persister cells to cis-DA compared to saline-exposed samples. Likewise, increased transcript abundances were also observed for genes related to the proteins that presented higher abundances upon exposure of persister cells to cis-DA, further supporting the change in the persister cells' metabolic status.
Relative transcript abundance of SOS response genes is reduced or unchanged in the presence of cis-DA.
Elevated levels of persister cell formation were previously associated with high ppGpp levels (11). In this study, we induced the SOS response by means of ciprofloxacin to select for persister cell populations. Increased levels of ppGpp, an alarmone expressed in response to harsh conditions, are known to inhibit rRNA synthesis, a classical characteristic of the stringent/SOS response that leads to a decrease in cell metabolism (91). This phenotypic loss indicates that the synthesis of ppGpp is responsible for the rapid transition from an active to a quiescent state under stress conditions (11). Those previous findings, together with our results demonstrating a significant decrease in the relative abundance of ppk (involved in the pathway of ppGpp formation [92]) upon exposure of P. aeruginosa persister cells to cis-DA, led us to investigate the relative abundances of relA and spoT. Both relA and spoT have been demonstrated to be involved in the triggering of the SOS response (93). P. aeruginosa persister cells exposed to cis-DA demonstrated either no change in or reduced relative abundances of relA and spoT. However, the abundances of relA and spoT increased by 2.7- and 3-fold, respectively, during the formation of persister cells (Fig. 10A), a finding that is in agreement with persister cell formation being achieved by the triggering of the SOS response, which involves global regulators such as relA and spoT (11). These results further support that, upon exposure to cis-DA, the cell metabolic status is increased and that persister cells are reverting from a tolerant to an antimicrobial-susceptible state.
DISCUSSION
Persister cells are thought to be an infinitesimal fraction of the overall bacterial population. These cells are commonly considered to be dormant and are known to play an important role in the antimicrobial resilience of bacteria in infections (28). Currently, a wide range of literature has been published on research involving persister cell populations, focusing on the determination of genes and pathways that lead to persister cell formation (reviewed in reference 94). However, little research has focused on the awakening of persister cells. Our study focuses on the “breaking” of cells from the persistent state into a metabolically active (awake) state, which is sensitive to antimicrobials as a result of exposure to the signaling molecule cis-2-decenoic acid (cis-DA).
Exposure of persister cells of P. aeruginosa and E. coli to cis-DA seems to jump-start the cells' metabolism by mobilizing energy sources to repair misfolded proteins (GroEL), degrade misfolded or unwanted proteins (ClpP2 homolog), initiate transcription and translation (initiation of protein synthesis, IF-2, and 16S rRNA), initiate membrane repair (increased expression of acyl carrier protein [AcpP]), and produce ATP (ATP synthase). We also observed increased levels of transcription of 16S rRNA, atpH, atpA, ppx, and PA14_21030 (Fig. 10), previously demonstrated to be related to an increase in growth and the cell metabolic rate (81, 95). The notion of cis-DA awakening persister cells is further supported by the finding that the transcript abundance of ppx is increased, while the transcript abundances of ppk, relA, and spoT are decreased (Fig. 10). These findings suggest the possibility of polyP degradation with the gained energy being used for the cells' awakening in the presence of cis-DA. PolyP is a product of polyphosphate kinase (PPK) activity and is believed to be the source of ATP for many reactions, including DNA transcription, RNA synthesis, and protein structure (96).
While neither P. aeruginosa nor E. coli utilizes cis-DA as a carbon source (Fig. 7), the presence of cis-DA nevertheless correlates with increased respiratory activity of persister cells (Fig. 9) without inducing a biofilm dispersion response (Fig. 8). These findings support the idea of cis-DA being used as a signaling molecule to enhance metabolic activity. The finding of a signaling molecule and not a carbon source enhancing the metabolic activity of persister cells is novel. It is known, however, that persister cells revert to a susceptible state, or to an awakened state, upon reinoculation into fresh medium (2, 28), in the presence of sugar metabolites (18), in the presence of environmental signals (97, 98), by stochastic exit from dormancy (99, 100), or upon the addition of spent medium to preformed persister cells (17). Under these conditions, reversion to a susceptible state correlates with a sudden burst of transcription and translation (55) and the production of proteins necessary to perform cell repair (101). However, no reports have demonstrated that persister cells awake due to the presence of a specific signaling molecule.
Our findings of increased transcript abundances of genes relevant to metabolism together with increased CTC staining (indicating respiratory activity) and higher abundances of proteins involved in reversion to a state of active metabolism strongly demonstrate that exposure of persister cells to the signaling molecule cis-DA results in a reversion from a dormant-tolerant to an active-susceptible state. Thus, in addition to the previously described properties of cis-DA and other members of the DSF (diffuse signal factor) family in dispersion, cell aggregation, and virulence (22, 102–105), our findings suggest a novel role of cis-DA and fatty acid signaling systems in awakening persister cell populations, thus rendering them susceptible to antimicrobials.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Karin Sauer for providing guidance, proofreading the manuscript, and providing the P. aeruginosa PA14 strain. We also acknowledge Tom Wood for providing the E. coli strain.
This work was funded by Binghamton University structural funds.
Footnotes
Published ahead of print 5 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01576-14.
REFERENCES
- 1.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 2.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13–18. 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 3.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren I, Shah D, Spoering A. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172–8180. 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaFleur MD, Kumamoto CA, Lewis K. 2006. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 50:3839–3846. 10.1128/AAC.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Mulcahy LR, Burns JL, Lory S, Lewis K. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192:6191–6199. 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69:164–174. 10.1111/j.1365-2958.2008.06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis K. 2005. Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.) 70:267–274 http://protein.bio.msu.ru/biokhimiya/contents/v70/full/70020327.html. [DOI] [PubMed] [Google Scholar]
- 10.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57:1468–1473. 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korch SB, Henderson TA, Hill TM. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199–1213. 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 12.Scherrer R, Moyed HS. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. 2012. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc. Natl. Acad. Sci. U. S. A. 109:12147–12152. 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niepa THR, Gilbert JL, Ren D. 2012. Controlling Pseudomonas aeruginosa persister cells by weak electrochemical currents and synergistic effects with tobramycin. Biomaterials 33:7356–7365. 10.1016/j.biomaterials.2012.06.092. [DOI] [PubMed] [Google Scholar]
- 15.Kim JS, Heo P, Yang TJ, Lee KS, Cho DH, Kim BT, Suh JH, Lim HJ, Shin D, Kim SK, Kweon DH. 2011. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob. Agents Chemother. 55:5380–5383. 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pascoe B, Dams L, Wilkinson TS, Harris LG, Bodger O, Mack D, Davies AP. 2014. Dormant cells of Staphylococcus aureus are resuscitated by spent culture supernatant. PLoS One 9:e85998. 10.1371/journal.pone.0085998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J, Bahar AA, Syed H, Ren D. 2012. Reverting antibiotic tolerance of Pseudomonas aeruginosa PAO1 persister cells by (Z)-4-bromo-5-(bromomethylene)-3-methylfuran-2(5H)-one. PLoS One 7:e45778. 10.1371/journal.pone.0045778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. U. S. A. 100:10995–11000. 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. 2009. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J. Bacteriol. 191:5013–5019. 10.1128/JB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies DG, Marques CNH. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191:1393–1403. 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amari DT, Marques CNH, Ferreira A, Silverman I, Davies DG. 2010. Resuscitation of bacteria by exposure to the intercellular communication molecule, cis-2-decenoic acid, abstr D-2096 Abstr. 110th Gen. Meet. Am. Soc. Microbiol. [Google Scholar]
- 24.Marques CNH, Dolacky SD, Guttenplan SB, Payabyab EC, Davies DG. 2007. Pseudomonas aeruginosa antimicrobial challenge in combination with a biofilm dispersal agent, abstr B-308:166 Abstr. 4th ASM Conf. Biofilms. ASM Press, Washington, DC. [Google Scholar]
- 25.Sepehr S, Rahmani-Badi A, Babaie-Naiej H, Soudi MR. 2014. Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS One 9:e101677. 10.1371/journal.pone.0101677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahmani-Badi A, Sepehr S, Mohammadi P, Soudi MR, Babaie-Naiej H, Fallahi H. 2014. A combination of cis-2-decenoic acid and antibiotics eradicates pre-established catheter-associated biofilms. J. Med. Microbiol. 10.1099/jmm.0.075374-0. [DOI] [PubMed] [Google Scholar]
- 27.Jennings J, Courtney H, Haggard W. 2012. cis-2-Decenoic acid inhibits S. aureus growth and biofilm in vitro: a pilot study. Clin. Orthop. Relat. Res. 470:2663–2670. 10.1007/s11999-012-2388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sufya N, Allison DG, Gilbert P. 2003. Clonal variation in maximum specific growth rate and susceptibility towards antimicrobials. J. Appl. Microbiol. 95:1261–1267. 10.1046/j.1365-2672.2003.02079.x. [DOI] [PubMed] [Google Scholar]
- 29.Dörr T, Lewis K, Vulić M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Möker N, Dean CR, Tao J. 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192:1946–1955. 10.1128/JB.01231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauer K, Camper A, Ehrlich G, Costerton J, Davies D. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154. 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312–7326. 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaule G, Flemming HC, Ridgway HF. 1993. Use of 5-cyano-2,3-ditolyl tetrazolium chloride for quantifying planktonic and sessile respiring bacteria in drinking water. Appl. Environ. Microbiol. 59:3850–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 http://mic.sgmjournals.org/content/146/10/2395.full. [DOI] [PubMed] [Google Scholar]
- 35.Amari DT, Marques CNH, Davies DG. 2013. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J. Bacteriol. 195:4600–4610. 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 5:e1000668. 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen. J. Bacteriol. 177:6536–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsuwon J, Anderson AJ. 1990. Catalase and superoxide dismutase of root-colonizing saprophytic fluorescent pseudomonads. Appl. Environ. Microbiol. 56:3576–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194:2904–2915. 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- 41.Petrova OE, Sauer K. 2010. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J. Bacteriol. 192:5275–5288. 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114–8126. 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48:5–16. 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 44.Hong SH, Wang X, O'Connor HF, Benedik MJ, Wood TK. 2012. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 5:509–522. 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoering A, Lewis K. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746–6751. 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brazas MD, Hancock REW. 2005. Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:3222–3227. 10.1128/AAC.49.8.3222-3227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elliott TSJ, Shelton A, Grennwood D. 1987. The response of Escherichia coli to ciprofloxacin and norfloxacin. J. Med. Microbiol. 23:83–88. 10.1099/00222615-23-1-83. [DOI] [PubMed] [Google Scholar]
- 48.Crosby HA, Bion JF, Penn CW, Elliott TS. 1994. Antibiotic-induced release of endotoxin from bacteria in vitro. J. Med. Microbiol. 40:23–30. 10.1099/00222615-40-1-23. [DOI] [PubMed] [Google Scholar]
- 49.Smith JT. 1986. The mode of action of 4-quinolones and possible mechanisms of resistance. J. Antimicrob. Chemother. 18:21–29. [DOI] [PubMed] [Google Scholar]
- 50.Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232–260. 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goffic F, Capmau M-L, Tangy F, Baillarge M. 1979. Mechanism of action of aminoglycoside antibiotics. Binding studies of tobramycin and its 6′-N-acetyl derivative to the bacterial ribosome and its subunits. Eur. J. Biochem. 102:73–81. [DOI] [PubMed] [Google Scholar]
- 52.Allegrucci M, Hu FZ, Shen K, Hayes J, Ehrlich GD, Post JC, Sauer K. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325–2335. 10.1128/JB.188.7.2325-2335.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56. 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 54.Ullrich S, Karrasch B, Hoppe H, Jeskulke K, Mehrens M. 1996. Toxic effects on bacterial metabolism of the redox dye 5-cyano-2,3-ditolyl tetrazolium chloride. Appl. Environ. Microbiol. 62:4587–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zengler K. 2009. Central role of the cell in microbial ecology. Microbiol. Mol. Biol. Rev. 73:712–729. 10.1128/MMBR.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castanié-Cornet MP, Bruel N, Genevaux P. 2014. Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim. Biophys. Acta 1843:1442–1456. 10.1016/j.bbamcr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 57.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473–2484. 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rouviere PE, Gross CA. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170–3182. 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 59.Steiner-Mosonyi M, Creuzenet C, Keates RAB, Strub BR, Mangroo D. 2004. The Pseudomonas aeruginosa initiation factor IF-2 is responsible for formylation-independent protein initiation in P. aeruginosa. J. Biol. Chem. 279:52262–52269. 10.1074/jbc.M408086200. [DOI] [PubMed] [Google Scholar]
- 60.Inokuchi Y, Hirashima A, Sekine Y, Janosi L, Kaji A. 2000. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 19:3788–3798. 10.1093/emboj/19.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs DE, Bianco N. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322–3327. 10.1128/AAC.44.12.3322-3327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Massoni SC, Sandler SJ. 2013. Specificity in suppression of SOS expression by recA4162 and uvrD303. DNA Repair (Amst.) 12:1072–1080. 10.1016/j.dnarep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Díaz-Pérez AL, Román-Doval C, Díaz-Pérez C, Cervantes C, Sosa-Aguirre CR, López-Meza JE, Campos-García J. 2007. Identification of the aceA gene encoding isocitrate lyase required for the growth of Pseudomonas aeruginosa on acetate, acyclic terpenes and leucine. FEMS Microbiol. Lett. 269:309–316. 10.1111/j.1574-6968.2007.00654.x. [DOI] [PubMed] [Google Scholar]
- 65.Yahr TL, Wolfgang MC. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631–640. 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 66.Lindsey TL, Hagins JM, Sokol PA, Silo-Suh LA. 2008. Virulence determinants from a cystic fibrosis isolate of Pseudomonas aeruginosa include isocitrate lyase. Microbiology 154:1616–1627. 10.1099/mic.0.2007/014506-0. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. 2012. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int. J. Med. Microbiol. 302:63–68. 10.1016/j.ijmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nestorovich EM, Sugawara E, Nikaido H, Bezrukov SM. 2006. Pseudomonas aeruginosa porin OprF: properties of the channel. J. Biol. Chem. 281:16230–16237. 10.1074/jbc.M600650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fito-Boncompte L, Chapalain A, Bouffartigues E, Chaker H, Lesouhaitier O, Gicquel G, Bazire A, Madi A, Connil N, Véron W, Taupin L, Toussaint B, Cornelis P, Wei Q, Shioya K, Déziel E, Feuilloley MGJ, Orange N, Dufour A, Chevalier S. 2011. Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immun. 79:1176–1186. 10.1128/IAI.00850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wylie JL, Worobec EA. 1995. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. J. Bacteriol. 177:3021–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wylie JL, Bernegger-Egli C, O'Neil JD, Worobec EA. 1993. Biophysical characterization of OprB, a glucose-inducible porin of Pseudomonas aeruginosa. J. Bioenerg. Biomembr. 25:547–556. 10.1007/BF01108411. [DOI] [PubMed] [Google Scholar]
- 72.Ochs MM, McCusker MP, Bains M, Hancock REW. 1999. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob. Agents Chemother. 43:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith CK, Baker TA, Sauer RT. 1999. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. U. S. A. 96:6678–6682. 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Acosta MBR, Ferreira RCC, Padilla G, Ferreira LCS, Costa SOP. 2000. Altered expression of oligopeptide-binding protein (OppA) and aminoglycoside resistance in laboratory and clinical Escherichia coli strains. J. Med. Microbiol. 49:409–413 http://jmm.sgmjournals.org/content/49/5/409.short. [DOI] [PubMed] [Google Scholar]
- 75.Andrews JC, Blevins TC, Short SA. 1986. Regulation of peptide transport in Escherichia coli: induction of the trp-linked operon encoding the oligopeptide permease. J. Bacteriol. 165:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weijland A, Harmark K, Cool RH, Anborgh PH, Parmeggiani A. 1992. Elongation factor Tu: a molecular switch in protein biosynthesis. Mol. Microbiol. 6:683–688. 10.1111/j.1365-2958.1992.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 77.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309–1320. 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 78.De Biase D, Tramonti A, Bossa F, Visca P. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198–1211. 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 79.Kasimoglu E, Park SJ, Malek J, Tseng CP, Gunsalus RP. 1996. Transcriptional regulation of the proton-translocating ATPase (atpIBEFHAGDC) operon of Escherichia coli: control by cell growth rate. J. Bacteriol. 178:5563–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klein SG, Serchi T, Hoffmann L, Blömeke B, Gutleb AC. 2013. An improved 3D tetraculture system mimicking the cellular organisation at the alveolar barrier to study the potential toxic effects of particles on the lung. Part. Fibre Toxicol. 10:31. 10.1186/1743-8977-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pérez-Osorio AC, Williamson KS, Franklin MJ. 2010. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J. Bacteriol. 192:2991–3000. 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williamson KS, Richards LA, Perez-Osorio AC, Pitts B, McInnerney K, Stewart PS, Franklin MJ. 2012. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J. Bacteriol. 194:2062–2073. 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stephens PE, Darlison MG, Lewis HM, Guest JR. 1983. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur. J. Biochem. 133:481–489. [DOI] [PubMed] [Google Scholar]
- 84.Tseng CP, Hansen AK, Cotter P, Gunsalus RP. 1994. Effect of cell growth rate on expression of the anaerobic respiratory pathway operons frdABCD, dmsABC, and narGHJI of Escherichia coli. J. Bacteriol. 176:6599–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tseng CP, Albrecht J, Gunsalus RP. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Akiyama M, Crooke E, Kornberg A. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633–639. [PubMed] [Google Scholar]
- 87.Akiyama M, Crooke E, Kornberg A. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 267:22556–22561. [PubMed] [Google Scholar]
- 88.Kornberg A. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89–125. 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 89.Dimroth P, von Ballmoos C, Meier T. 2006. Catalytic and mechanical cycles in F-ATP synthases. Fourth in the Cycles Review Series. EMBO Rep. 7:276–282. 10.1038/sj.embor.7400646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Von Ballmoos C, Cook GM, Dimroth P. 2008. Unique rotary ATP synthase and its biological diversity. Annu. Rev. Biophys. 37:43–64. 10.1146/annurev.biophys.37.032807.130018. [DOI] [PubMed] [Google Scholar]
- 91.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51. 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 92.Kim HY, Schlictman D, Shankar S, Xie Z, Chakrabarty AM, Kornberg A. 1998. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol. Microbiol. 27:717–725. 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 93.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. 2011. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect. Immun. 79:4094–4104. 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lewis K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372. 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 95.Lenz AP, Williamson KS, Pitts B, Stewart PS, Franklin MJ. 2008. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 74:4463–4471. 10.1128/AEM.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brown MRW, Kornberg A. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284–290. 10.1016/j.tibs.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 97.Dworkin J, Shah IM. 2010. Exit from dormancy in microbial organisms. Nat. Rev. Microbiol. 8:890–896. 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 98.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. 2006. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 14:271–276. 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 99.Buerger S, Spoering A, Gavrish E, Leslin C, Ling L, Epstein SS. 2012. Microbial scout hypothesis, stochastic exit from dormancy, and the nature of slow growers. Appl. Environ. Microbiol. 78:3221–3228. 10.1128/AEM.07307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Epstein SS. 2009. Microbial awakenings. Nature 457:1083. 10.1038/4571083a. [DOI] [PubMed] [Google Scholar]
- 101.Halvorson HO, Vary JC, Steinberg W. 1966. Developmental changes during the formation and breaking of the dormant state in bacteria. Annu. Rev. Microbiol. 20:169–188. 10.1146/annurev.mi.20.100166.001125. [DOI] [PubMed] [Google Scholar]
- 102.Newman KL, Chatterjee S, Ho KA, Lindow SE. 2008. Virulence of plant pathogenic bacteria attenuated by degradation of fatty acid cell-to-cell signaling factors. Mol. Plant Microbe Interact. 21:326–334. 10.1094/MPMI-21-3-0326. [DOI] [PubMed] [Google Scholar]
- 103.Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, Avison MB, Ryan RP, Dow JM. 2007. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 189:4964–4968. 10.1128/JB.00310-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Huang T-P, Wong ACL. 2007. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 73:5034–5040. 10.1128/AEM.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chatterjee S, Wistrom C, Lindow SE. 2008. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc. Natl. Acad. Sci. U. S. A. 105:2670–2675. 10.1073/pnas.0712236105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.