Abstract
Circular bacteriocins are a group of N-to-C-terminally linked antimicrobial peptides, produced by Gram-positive bacteria of the phylum Firmicutes. Circular bacteriocins generally exhibit broad-spectrum antimicrobial activity, including against common food-borne pathogens, such as Clostridium and Listeria spp. These peptides are further known for their high pH and thermal stability, as well as for resistance to many proteolytic enzymes, properties which make this group of bacteriocins highly promising for potential industrial applications and their biosynthesis of particular interest as a possible model system for the synthesis of highly stable bioactive peptides. In this review, we summarize the current knowledge on this group of bacteriocins, with emphasis on the recent progress in understanding circular bacteriocin genetics, biosynthesis, and mode of action; in addition, we highlight the current challenges and future perspectives for the application of these peptides.
CHARACTERISTICS AND CLASSIFICATION
The circular bacteriocins constitute a group of ribosomally synthesized antimicrobial peptides characterized by their N-to-C-terminal covalent linkage, forming a structurally conserved circular peptide backbone. In this review, we will consistently use the term circular instead of cyclic, to clearly distinguish the ribosomal head-to-tail-ligated bacteriocins from other, nonribosomally or intramolecularly cyclized peptides. Circular bacteriocins are synthesized as linear precursors, containing a leader sequence of variable size which is cleaved off during maturation (1). The mature circular peptides themselves range from 58 to 70 amino acid residues, corresponding to masses of approximately 5.6 to 7.2 kDa. So far, the circular bacteriocins that have been identified are produced by Gram-positive bacteria of the phylum Firmicutes, mainly lactic acid bacteria but a few also from Bacillus and Clostridium genera. The isolation sources range from fermented foods to dairy products, meat, and mammalian feces and wound exudate (Table 1). The circular bacteriocins are generally broad-spectrum antimicrobials and are known for their pH and thermal stability, as well as for their resistance to many proteolytic enzymes, properties which make this group of bacteriocins especially interesting for potential industrial applications. In the classification scheme of Gram-positive bacteriocins (2–4), circular bacteriocins have been considered to belong to the class II nonmodified peptides and are often ascribed to subclass IIc. However, this issue has been debated, and some have made a case for this group being assigned to a new class, i.e., class IV or V bacteriocins (5–7). In any case, the circular bacteriocins are clearly distinct from other Gram-positive bacteriocins and are homogenous enough to be considered a separate group.
TABLE 1.
General properties of the circular bacteriocinsa
| Subgroup | Bacteriocinb | GenBank accession no. | Producer organism | Isolation source | Length (aa) of: |
Molecular mass (Da) of circular peptide | pI | Net charge | GRAVYc | |
|---|---|---|---|---|---|---|---|---|---|---|
| Leader sequence | Mature peptide | |||||||||
| i | Garvicin ML | EKF52513 | Lactococcus garvieae | Duck intestines | 3 | 60 | 6,007.2 | 10.13 | +5 | 0.89 |
| i | AS-48 | CAA72917 | Enterococcus faecalis | Human wound exudate | 35 | 70 | 7,149.5 | 10.09 | +6 | 0.54 |
| i | Uberolysin A | ABG48503 | Streptococcus uberis | Cow mammary secretion | 6 | 70 | 7,048.3 | 9.60 | +3 | 0.94 |
| i | Carnocyclin A | ACC93994 | Carnobacterium maltaromaticum | Fresh pork | 4 | 60 | 5,862.0 | 10.00 | +4 | 1.06 |
| i | Amylocyclin A | NAd | Bacillus amyloliquefaciens | Soil | 48 | 64 | 6,381.6 | 9.82 | +5 | 0.85 |
| i | Leucocyclicin Q | BAL14584 | Leuconostoc mesenteroides | Japanese pickles | 2 | 61 | 6,115.2 | 9.53 | +2 | 0.74 |
| i | Circularin A | CAD97580 | Clostridium beijerinckii | Soil | 3 | 69 | 6,771.0 | 10.46 | +4 | 1.01 |
| i | Lactocyclicin Q | BAH29711 | Lactococcus sp. | Cheese | 2 | 61 | 6,060.1 | 9.70 | +2 | 0.83 |
| ii | GassericinA/reutericin A | BAH08712 | Lactobacillus gasseri/Lactobacillus reuteri | Human infant feces | 33 | 58 | 5,653.6 | 6.75 | 0 | 0.10 |
| ii | Butyrivibriocin A | AAC69560 | Butyrivibrio fibrisolvens | Rumen isolate | 22 | 58 | 5,981.9 | 4.03 | −2 | 1.00 |
Physicochemical properties were calculated using ProtParam (56), where molecular mass was calculated for the circularized peptides, whereas the remaining parameters were calculated for the linear forms.
Garvicin ML (GarML), enterocin AS-48 (AS-48), uberolysin A (UblA), carnocyclin A (CclA), amylocyclicin (Acn), leucocyclicin Q (LcyQ), circularin A (CirA), lactocyclicin Q (LycQ), gassericin A (GaaA), and butyrivibriocin AR10 (BviA).
GRAVY, grand average of hydropathicity, calculated as the sum of hydropathy values of all amino acids divided by the total number of residues, i.e., increasing positive score indicates greater hydrophobicity.
NA, not available.
To date, 10 circular bacteriocins have been characterized, and these are again subdivided into two subgroups based on physicochemical characteristics and level of sequence identity (2, 8). Subgroup i includes the most highly characterized circular bacteriocin, enterocin AS-48, as well as carnocyclin A (9), circularin A (10), uberolysin (11), lactocyclicin Q (12), leucocyclicin (13), garvicin ML (14), and amylocyclin (15) (Table 1). A circular bacteriocin recently identified from Enterococcus faecalis, named enterocin RM6, was shown to be identical to enterocin AS-48 (16). The subgroup i peptides are characterized by having several positively charged amino acid residues (overall cationic charge) and high isoelectric points (pIs of ∼10) (Table 1). An alignment of these peptides, as shown in Fig. 1, clearly illustrates that the sequence similarity is limited and there are few conserved residues within the peptides of subgroup i. The exceptions are the two peptides lactocyclicin Q and leucocyclicin Q, which share 71.4% sequence identity and thus appear to be evolutionarily more closely related than the rest of the circular bacteriocins.
FIG 1.

Protein sequence alignments of the leaders and mature peptides of the subgroup i (top) and ii (bottom) circular bacteriocins, where the level of sequence identity/conservation is indicated by color scale (the light-to-dark scale indicating low to high identity/conservation) and the consensus sequence for each subgroup is displayed below the subgroup alignment. Alignments were produced using T-Coffee (57) and visualized using Jalview (58).
The subgroup ii circular bacteriocins currently comprise only two members, gassericin A (10) and butyrivibriocin AR10 (17). These peptides display a high level of sequence identity (44.8%) and also differ from the subgroup i bacteriocins by their low isoelectric points (pIs of ∼4 and ∼7) (Table 1). The circular peptide reutericin 6 was initially identified as another subgroup ii circular bacteriocin, with high similarity to gassericin A but containing different levels of d-Ala. However, later analysis revealed that it is in fact identical to gassericin A (with neither peptide actually containing d-Ala) (18).
The circular antimicrobial peptide subtilosin A, produced by Bacillus subtilis, was also initially considered to belong to the circular bacteriocins. However, this and a few other circular peptides, including the sporulation killing factor (SKF) of Bacillus thuringiensis (19), contain thioether linkages and are structurally and genetically distinct from the circular bacteriocins. These peptides have therefore been reclassified as members of a group known as the sactibiotics (20).
GENETICS AND BIOSYNTHESIS
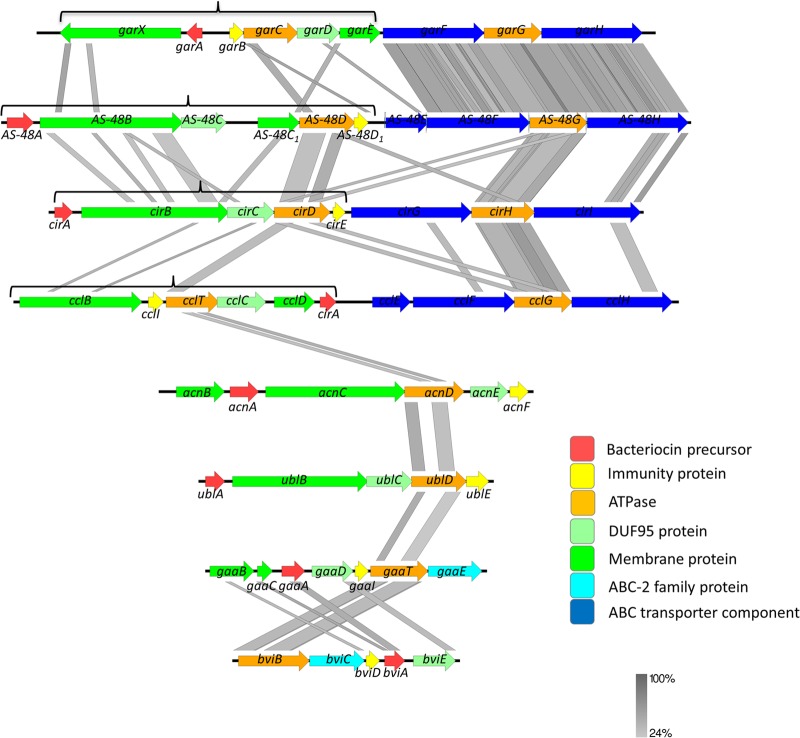
The genetic background has been described for a majority of the circular bacteriocins, and functional analysis of the gene clusters of enterocin AS-48 and a few other circular bacteriocins (circularin A, gassericin A, carnocyclin A, and most recently, garvicin ML) has also provided some insight into the roles of the encoded proteins in the biosynthesis of these peptides. Figure 2 displays the gene clusters of all circular bacteriocins which have been genetically characterized to date, either chromosomally located or carried on plasmids. Despite limited sequence similarity (average 20 to 30% pairwise reciprocal ortholog identity) between the encoded proteins, these gene clusters share a number of traits and similar architecture, which could indicate that there are also commonalities in the biosynthetic machinery and processes of circular bacteriocins.
FIG 2.
Genetic organization of the characterized circular bacteriocin gene clusters: garvicin ML (garAXBCDEFGH) (24), enterocin AS-48 (AS-48ABCC1DD1EFGH) (1), circularin A (cirABCDEGHI) (21), carnocyclin A (cclBITCDAEFGH) (22), amylocyclicin (acnBACDEF) (15), uberolysin A (ublABCDE) (11), gassericin A (gaaBCADITE) (26), and butyrivibriocin AR10 (bviBCDAE) (17). The genes are colored according to function/characteristics, as indicated by the key, and homology at the protein sequence level is indicated by greyscale blocks. The minimal subsets of genes required for biosynthesis and immunity are indicated by black braces above the respective gene clusters. The figure was produced using EasyFig (59).
The majority of proteins encoded in circular bacteriocin gene clusters are predicted to be largely hydrophobic and, thus, are assumed to be membrane associated (Fig. 2). A possible inference is that circular bacteriocin biosynthesis takes place at the membrane interface, but this has yet to be experimentally verified. The minimal set of genes required for bacteriocin production and immunity in general comprises 5 to 7 genes (Fig. 2). These include, first and foremost, the bacteriocin structural gene (e.g., AS-48A), which encodes a precursor peptide containing a leader sequence ranging from 2 to 48 amino acid residues in length. Second, all gene clusters contain a gene encoding a very short, cationic (high-pI), and hydrophobic protein (e.g., AS-48D1) that has been shown to provide a basal level of immunity toward the cognate bacteriocin (21, 22). Furthermore, the minimal gene set includes an ATPase (e.g., AS-48D), one or more putative membrane protein(s) with unknown function (e.g., AS-48C1), and an integral membrane protein belonging to the DUF 95 protein family (e.g., AS-48C). Distinctly, most subgroup i gene clusters encode a very large (477 to 581 residues) protein (e.g., AS-48B) with no known functional domains or homology but with a predicted transmembrane topology (23). Subgroup ii gene clusters, however, encode a membrane protein (160 to 174 residues) that is similar to the ABC-2 subfamily of membrane transporters. Most of the subgroup i gene clusters also have an accessory operon (3 to 4 genes) (e.g., AS-48EFGH) encoding an ABC transporter complex, consisting of a permease, an ATPase, and a predicted extracellular protein. This putative ABC transporter complex has been shown not to be essential or, indeed, to have any significant effect on either bacteriocin production or immunity (1, 21, 22, 24). Based on the apparent conservation of these genes between subgroup i circular bacteriocin gene clusters (Fig. 2), however, it is difficult to rule out the possibility that an independent function or accessory role of these ABC transporters could be elucidated in future studies. In the case of enterocin AS-48, it has indeed been shown to provide a small increase in the immunity level and enhancement of the production of bacteriocin (25).
As mentioned, all circular bacteriocin gene clusters contain a gene coding for a short (49 to 88 amino acid residues), high-pI, hydrophobic protein termed the immunity protein (Fig. 2). These proteins have a predicted structure of two α-helical segments and appear to be membrane associated (26). These immunity proteins have been demonstrated to provide a basal level of immunity to their respective bacteriocins; however, it has become clear that full immunity requires the combined activity of several of the encoded proteins (15, 21, 22, 24). In this respect, the circular bacteriocins seem similar to the lantibiotics (class I), which require a specialized ABC transporter in addition to the immunity peptide to achieve full immunity (27).
Biosynthesis of circular bacteriocins requires three steps: cleavage of the leader sequence, circularization, and export out of the cell (Fig. 3). Despite several functional studies on circular bacteriocins in recent years, there is much that remains unknown about this process. Specifically, the mechanisms involved, the enzymes responsible, and the potential coupling of these steps are still largely unanswered questions (5, 28).
FIG 3.
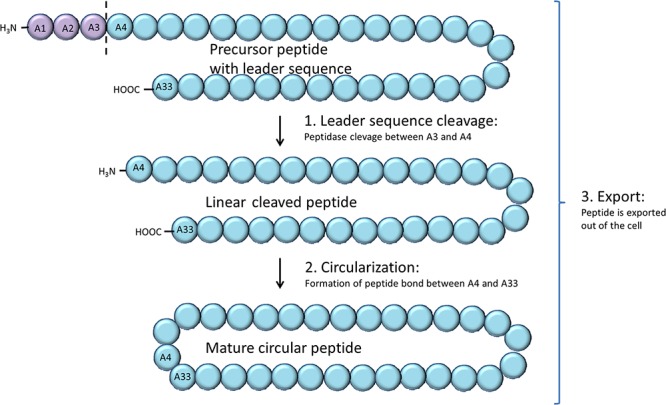
Illustration of the three steps of circular bacteriocin biosynthesis. The 3-amino-acid leader peptide (mauve) of the bacteriocin precursor is removed by cleavage (dashed line) by an unidentified peptidase. The cleaved linear peptide is then circularized by condensation of the N- and C-terminal amino and carboxyl groups, forming a new peptide bond and resulting in the mature circular peptide. The peptide is exported out of the producer cell by a dedicated transporter complex, possibly concomitant with circularization.
The removal of the leader sequence has been assumed to be the first step in biosynthesis and a requirement for further processing into the mature peptide, but this was not experimentally verified until quite recently. For the circular bacteriocin garvicin ML, leader sequence cleavage was demonstrated to precede circularization in time and occur independently from the circularization reaction (24). Contrary to the previous model, this finding implies that the leader sequence is not required as a recognition signal for the circularization reaction and/or for export out of the cell. It is, however, conceivable that the leader functions as a critical control checkpoint for the biosynthetic machinery, i.e., that export and circularization are only possible when the processed N-terminal end of the mature bacteriocin is accessible.
The leader sequences of the circular bacteriocins range from 2 to 48 residues (Fig. 1), containing mainly hydrophobic residues but often with a charged amino acid in the −1 position, i.e., the residue that has been shown to affect the specificity of the cleavage reaction of enterocin AS-48 (29). As no peptidase/protease functional domains have been characterized in any of the proteins encoded by circular gene clusters, the entity responsible for leader sequence cleavage remains enigmatic. In the case of garvicin ML, evidence suggests that leader sequence cleavage is not performed by any of the proteins encoded by the respective gene cluster, and it is proposed that an as-yet-unidentified host-encoded peptidase is required for leader peptide cleavage (24). These results may thus explain the lack of peptidases/proteases in the gene clusters and, furthermore, may reflect a situation which is more similar to the mechanisms of other bacterial head-to-tail-linked circular peptides, such as TrbC from Escherichia coli (30) and VirB2 from Agrobacterium tumefaciens (29), which are N-terminally cleaved by chromosomally encoded signal peptidases (SPases). The requirement for accessory determinants, i.e., factors not encoded within the respective gene cluster, has been suggested previously for enterocin AS-48 (31, 32) and may also explain the apparent problems in expressing circular bacteriocins outside the genus or species of the original producer (26, 31) and in obtaining stable heterologous production (21). However, the leader sequences of the circular bacteriocins, being highly variable both in size and sequence (Fig. 1), differ markedly from the known recognition site of the bacterial SPase I (33). Thus, it is likely that one or more yet-uncharacterized peptidases with different specificities are required for leader sequence cleavage in circular bacteriocin biosynthesis.
The circularization reaction is perhaps the most intriguing aspect of circular bacteriocin biosynthesis and the area where knowledge is most scarce. It is, however, clear that specific features in the mature peptide sequence are essential for circularization to occur. The N- and C-terminal ends of the circular bacteriocins, which are likely to contribute to this process, consist mainly of stretches of hydrophobic residues (Fig. 1). The circularization point in each case appears to be located internal to an alpha helix in the structure of the peptides (Fig. 4), requiring the circularization process to occur in a largely hydrophobic and sterically hindered region. Mutational analysis of enterocin AS-48 has shown that substitutions of the first and last residues of the mature peptide affect the circularization reaction: a change of Met to Ala at position 1 (Met1Ala) lowered the circularization efficiency significantly, whereas replacement of the C-terminal Trp with Ala resulted in the detection of both the circular and a small amount of the linear form of the peptide, thus establishing that the nature of both the N- and the C-terminal residues is critical to the efficiency of the circularization process (29). Notably, most of the circular bacteriocins contain several aromatic and/or small hydrophobic residues in the N terminus, which may indicate that the hydrophobic nature of these residues is important for biosynthesis (including circularization) and/or antimicrobial activity.
FIG 4.
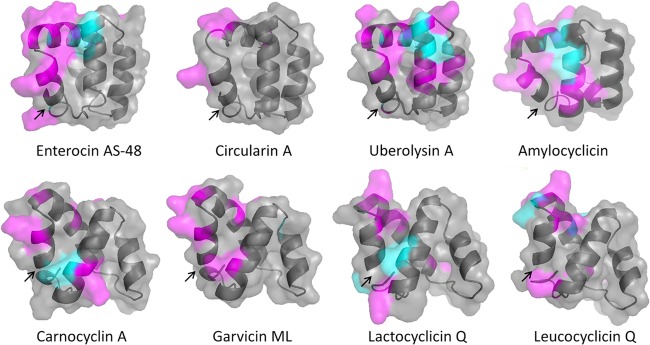
3-D structures of subgroup i circular bacteriocins shown as cartoon rendering with a transparent surface, where positively charged residues are colored in magenta and negatively charged residues in cyan and with the circularization points indicated by arrows. Structures were predicted by homology modeling (SWISS-MODEL) based on the alignment shown in Fig. 1, and images were rendered using the PyMOL Molecular Graphics system (version 1.5.0.4, Schrödinger, LLC.). The solution structure of enterocin AS-48 (PDB ID 1E68) was used as the template for homology modeling of circularin A, uberolysin A, and amylocyclicin, whereas the solution structure of carnocyclin A (PDB ID 2KJF) was used as the template for homology modeling of garvicin ML, lactocyclicin Q, and leucocyclicin Q. While the longer peptides (circularin A, uberolysin A, and amylocyclicin) are predicted to form 5 α-helices, like enterocin AS-48, the shorter peptides (garvicin ML, lactocyclicin Q, and leucocyclicin Q) are predicted to form 4 α-helices, like carnocyclin A. Structures of subgroup ii peptides gassericin A and butyrivibriocin AR10 are not included in this figure, as these peptides do not share sufficient sequence identity to any of the determined structures for reliable prediction.
Looking to other types of head-to-tail-linked circular peptides, it becomes clear that circular bacteriocins stand out in that they contain no C-terminal extension and that the N-terminal leader appears to have no role in the circularization reaction. This is in contrast to the aforementioned TrbC peptide from E. coli, as well as the plant cyclotide kalata B1, which require both N- and C-terminal regions with more or less conserved motifs for circularization to occur. The VirB2 peptide is more similar to circular bacteriocins in that it also lacks a C-terminal extension. The VirB2 peptide is processed by the removal of a 47-residue N-terminal extension by a general signal peptidase and further circularized in rapid succession either by an unknown enzyme or by the same peptidase. Interestingly, circularization occurs in the absence of the plasmid-encoded virB operon, indicating that processing of VirB2 is entirely performed by chromosomally encoded factors (34). When it comes to the enzyme(s) responsible for performing the circularization reaction of circular bacteriocins, however, it has become evident that these are encoded within the bacteriocin gene cluster. Functional analysis of the garvicin ML gene cluster revealed that circularization was aborted in the absence of either of two operons, garX and garBCDE (24). Similar analyses of the enterocin AS-48, circularin A, and carnocyclin A gene clusters have also shown that minimal sets of four and five genes are required for the production and/or secretion of these peptides, respectively (Fig. 2) (1, 21, 22). Taken together, it appears that no single protein but, rather, several proteins are involved in the mechanism of circularization of these bacteriocins. Interestingly, this minimal subset invariably includes three putative transmembrane proteins and an ATPase, which could presumably form a membrane-located protein complex. It is thus tempting to speculate that the processes of circularization and export out of the cell could be jointly performed by such a complex; however, this remains to be investigated.
STRUCTURE AND PHYSICOCHEMICAL PROPERTIES
The solution structures of enterocin AS-48 and carnocyclin A have been solved, revealing a common structural motif for the subgroup i circular bacteriocins (Fig. 4). Both peptides have a compact globular structure consisting of four (carnocyclin A) or five (enterocin AS-48) α-helices enclosing a hydrophobic core (8, 35). The point of circularization is located inside an α-helical segment (Fig. 4), which for enterocin AS-48 has been demonstrated to have a pronounced effect on the stability of the structure (35). The presence of several basic amino acid residues imparts a localized positive charge on the surface of the structure. The overall structure is similar to the saposin fold (excluding the intrachain disulfide bonds) found in the mammalian antimicrobial and cytotoxic peptide NK lysin (35), and a similar structure was recently determined also for the linear bacteriocins enterocin 7a and 7b (36). Protein structure homology modeling of the remaining circular bacteriocins suggests that this structure is shared among the subgroup i bacteriocins (Fig. 4) (8). The 3-dimensional (3-D) structures of the subgroup ii peptides butyrivibriocin AR10 and gassericin A have not yet been solved, but structure predictions suggest that these also consist of predominantly α-helical regions and could possibly form similar structures (8). These two peptides do not display the localized charges of subgroup i peptides, but the significance of this is uncertain.
Their compact and circular structures are believed to be the reason why circular bacteriocins in general exhibit very high thermal and pH stability and may even render the peptides resistant to degradation by many proteolytic enzymes by reducing the availability of possible cleavage sites (23). Furthermore, the shared 3-D structures could imply a common mode of action of these antimicrobial peptides. In particular, the positively charged patches on the surface of the structures are thought to be the driving force behind the initial attraction to and subsequent insertion into the negatively charged phospholipid layer of the target cell membrane (8). In the case of garvicin ML, knockout strains producing linear but not circular forms of this bacteriocin displayed no antimicrobial activity, suggesting that the circular structure of garvicin ML is essential for its antimicrobial activity (24). It is conceivable that the head-to-tail peptide bond is important for retention of the 3-D structure, and consequently, that loss of structural integrity could affect the interaction of the bacteriocin with the target cell. In contrast, characterization of enterocin AS-48 has indicated that linear forms of this peptide retain antimicrobial activity to some extent (37), indicating that the circular form is not essential for antimicrobial activity but could be more important for stabilizing the structure (37, 38).
ANTIMICROBIAL SPECTRUM AND MODE OF ACTION
Circular bacteriocins have broad antimicrobial activity spectra. Reflecting their producer organisms, the antimicrobial activities are generally directed against Gram-positive bacteria of the phylum Firmicutes, i.e., mainly lactic acid bacteria (order Lactobacillales), bacilli, and clostridia (Table 2). Antimicrobial activity against other Gram-positive phyla has also been reported. Notably, the antibacterial spectra of circular bacteriocins include common food-borne pathogens within the Listeria and Clostridium genera and, in a few cases, encompass nosocomial pathogens, such as enterococci and staphylococci. A few circular bacteriocins (enterocin AS-48, leucocyclicin Q, and lactocyclicin Q) are shown under certain conditions to display antimicrobial activity against E. coli, although at significantly higher concentrations than are required for activity against the Gram-positive bacteria (12, 13, 39). Antimicrobial activity of carnocyclin A against Gram-negative species has been shown to occur when the integrity of the cellular outer membrane is perturbed by the metal-chelating agent EDTA, which could indicate that it is restricted access to the membrane that inhibits activity toward Gram-negative bacteria (40). Such a combined antimicrobial affect with EDTA has also been observed in other bacteriocins, including nisin (40, 41).
TABLE 2.
Antimicrobial activity spectra of the circular bacteriocins
| Bacteriocina | Subgroup | Producer organism | Activity against members ofb: |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Firmicutes |
Proteobacteria |
Actinobacteria: Actinomycetales |
||||||||||||||||||||||||
|
Lactobacillales |
Bacillales |
Clostridiales |
Enterobacteriales |
Other |
||||||||||||||||||||||
| Lactobacillus | Leuconostoc | Pediococcus | Lactococcus | Streptococcus | Enterococcus | Carnobacterium | Weissella | Staphylococcus | Bacillus | Paenibacillus | Listeria | Clostridium | Ruminococcus | Eubacterium | Lachnospira | Escherichia | Salmonella | Shigella | Klebsiella | Pseudomonas | Agrobacterium | Clavibacter | Micrococcus | |||
| GarML | i | L. garvieae | + | + | + | + | + | + | + | − | ||||||||||||||||
| CclA | i | Cb. maltaromaticum | + | + | + | + | + | + | + | − | − | − | ||||||||||||||
| CirA | i | C. beijerinckii | + | + | + | + | ||||||||||||||||||||
| AS-48 | i | Ec. faecalis | + | + | + | + | + | + | + | + | + | |||||||||||||||
| UblA | i | S. uberis | + | + | + | + | − | |||||||||||||||||||
| LycQ | i | Lactococcus sp. | + | + | + | + | + | W | ||||||||||||||||||
| LcyQ | i | Ln. mesenteroides | + | + | + | + | + | + | W | |||||||||||||||||
| Acn | i | B. amyloliquefaciens | − | + | + | − | − | − | + | + | ||||||||||||||||
| GaaA | ii | Lb. gasseri | + | + | + | + | ||||||||||||||||||||
| BviA | ii | Bv. fibrisolvens | + | + | + | + | + | + | ||||||||||||||||||
Garvicin ML (14), carnocyclin A (29), circularin A (6), enterocin AS-48 (39), uberolysin A (11), lactocyclicin Q (12), leucocyclicin Q (13), gassericin A (60), amylocyclicin (15), and butyrivibriocin AR10 (61).
Antimicrobial activity is denoted as +, weak activity as W, and no activity as −, while blank cells show that activity was not determined. The values given here reflect the antimicrobial spectra of these bacteriocins as reported in the original papers, but in some cases only the highly/moderately sensitive strains are included, and the values may not reflect the complete activity spectrum due to low sampling and/or bias.
Like other class II bacteriocins, the circular bacteriocins are generally believed to act by disruption of the integrity of the membrane of target cells (23). The shared structural and physicochemical features of subgroup i circular bacteriocins suggest that these antimicrobials have a common functional mechanism. However, the studies that have been carried out have so far highlighted distinct differences in the modes of action within this subgroup. Studies have shown that enterocin AS-48 forms nonselective pores in liposomes, leading to leakage of ions and low-molecular-weight compounds (42). Furthermore, enterocin AS-48 has been shown to oligomerize at physiological pH, and the suggested mechanism for molecular function of AS-48 involves a structural transition of this complex from a water-soluble form to a membrane-bound state (43). Several studies have also shown that other class II bacteriocins become structured in hydrophobic or membranelike solvents (44, 45). Carnocyclin A has been shown to form pores in lipid membranes, but unlike the pores formed by enterocin AS-48, those of carnocyclin A are anion selective and voltage dependent (46).
For circular bacteriocins, it has been a controversial issue whether or not these antimicrobial peptides require a receptor molecule for target recognition of the sensitive cell. For many groups of class II bacteriocins, however, it has become evident that the antimicrobial activity is receptor mediated, i.e., by recognition of specific proteins on the membrane of target cells. This is probably the reason why bacteriocins in general are much more potent (about 1,000-fold) than eukaryotic antimicrobial peptides (AMPs), the latter group being known to act nonspecifically, i.e., without need of a receptor on target cells (47). Types of receptors appear to be targeted by several related and sometimes also nonrelated bacteriocins. For instance, the permease mannose-phosphotransferase (PTS) is targeted by many pediocin-like bacteriocins, in addition to the unrelated lactococcins A and B (48), whereas a zinc-dependent membrane-bound protease belonging to the M50 family is targeted by several related leaderless bacteriocins (49; unpublished results), and the undecaprenyl pyrophosphate phosphatase UppP (a membrane protein involved in peptidoglycan synthesis) is targeted by the two-peptide bacteriocins lactococcin G and enterocin 1071 (50).
Although the modes of action of most circular bacteriocins have not yet been determined, two of the most studied peptides, enterocin AS-48 and carnocyclin A, have been shown to permeabilize liposomes and/or lipid bilayers (42, 46), supporting the notion that direct interaction with the cell membrane is sufficient for antimicrobial activity and that a target receptor is not required for this group of bacteriocins. Also, studies of the circular bacteriocin gassericin A have indicated that this bacteriocin does not require a target receptor (51). However, the conclusions of these studies are undermined by the fact that the experiments were performed at bacteriocin concentrations significantly higher than that required for antimicrobial activity in vivo. Notably, studies of garvicin ML have identified a maltose ABC transporter complex which facilitates high sensitivity to this bacteriocin, while its absence causes resistance, and thus, it is likely to constitute a target receptor for this bacteriocin (52). Interestingly, at very high concentrations of bacteriocin, receptor-independent killing was observed, consistent with the studies of enterocin AS-48 and carnocyclin A. Together, these studies may thus suggest that, like that of nisin (53), the mode of action of circular bacteriocins is concentration dependent, with a nonspecific activity at higher bacteriocin concentrations and a specific activity at lower bacteriocin concentrations, the latter probably involving a receptor on target cells. As several other circular bacteriocins share significant structural similarly to garvicin ML, it is possible that other circular bacteriocins could also utilize the maltose ABC transporter or related permeases as receptors.
CHALLENGES AND FUTURE PERSPECTIVES
With emerging development of resistance to classical antibiotics in recent years, there is an ever increasing need for novel antimicrobial agents for medical use. Additionally, there is a demand for antimicrobials and antimicrobial-producing bacteria for use as natural preservatives, starter cultures, and probiotics in the food and feed industry. Bacteriocins have many favorable characteristics in this context, and circular bacteriocins particularly so, due to their broad-spectrum activity and exceptional stability.
There are still only 10 circular bacteriocins that have been characterized, and the low sequence identity between them makes predicting novel circular bacteriocins from available genomic sequences difficult. The presence of the gene for the conserved DUF95 protein in the gene clusters encoding these peptides could, however, be an indicator of the distribution of circular bacteriocins in other species; this protein domain appears to be widespread in Bacteria (977 species, mostly Firmicutes) and is also found in Archaea (120 species) (source, Pfam version 27.0, European Bioinformatics Institute). Although all of these proteins may not necessarily be encoded as part of circular bacteriocin gene clusters, this could indicate that there are still many circular bacteriocins to be discovered, which could have activity against other food-spoilage bacteria or pathogens and thus prove to be valuable antimicrobials.
However, the use of circular bacteriocins for such applications requires first and foremost a detailed understanding of the molecular mechanisms which govern the target recognition and mode of action of these antimicrobials. Not only is this knowledge essential for the rational use and type of application, but it can also contribute to minimizing the potential development of resistance resulting from use of these antimicrobials. In this aspect of circular bacteriocin biology, however, there is much that remains to be elucidated, and future studies should thus be aimed at clarifying both the mode of action and resistance development mechanisms of these peptides. Accordingly, elucidating immunity mechanism(s) could in turn allow for the bioengineering of bacteriocins which are not targeted by specific resistance mechanisms.
Several studies have indicated problems in expressing circular bacteriocins outside the genus or species of the original producer (26, 31) and in obtaining stable heterologous production (21). As recent findings have suggested, one possible reason for this could be the requirement for a host-encoded signal peptidase that is responsible for cleaving off the leader sequence, which could be essential for further processing into the mature peptide (24). Certainly, characterizing the full complement of factors required for circular bacteriocin biosynthesis is essential for establishing stable heterologous expression systems for these peptides, which could also allow bacteriocin production in industrially relevant bacterial species and scale.
Concerning the biosynthesis of circular bacteriocins, there is still much that remains unknown, although there is increasing evidence that protein complexes are involved in both the circularization and export of these peptides and in producer self-immunity mechanism(s). Future investigations should thus be aimed at further characterizing the interaction between the components of these complexes, to elucidate the functional domains involved and to gain a detailed understanding of these mechanisms. A better understanding of these processes could in turn allow for rational bioengineering of not only bacteriocins but also other types of bioactive peptides with enhanced properties such as increased thermal and pH stability, resistance to proteases, solubility, potency, and modified target specificity. Indeed, several of these traits have already been engineered for derivatives of the lantibiotic nisin (54), most notably enhanced activity against both Gram-positive and Gram-negative pathogens (55), which demonstrates the real potential of bioengineering in creating tailor-made peptides for specific applications.
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Martinez-Bueno M, Valdivia E, Galvez A, Coyette J, Maqueda M. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347–358. 10.1046/j.1365-2958.1998.00682.x. [DOI] [PubMed] [Google Scholar]
- 2.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788. 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 3.Nissen-Meyer J, Rogne P, Oppegard C, Haugen HS, Kristiansen PE. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by Gram-positive bacteria. Curr. Pharm. Biotechnol. 10:19–37. 10.2174/138920109787048661. [DOI] [PubMed] [Google Scholar]
- 4.Rea M, Ross RP, Cotter P, Hill C. 2011. Classification of bacteriocins from Gram-positive bacteria, p 29–53 In Drider D, Rebuffat S. (ed), Prokaryotic antimicrobial peptides. Springer, New York, NY. [Google Scholar]
- 5.Maqueda M, Sanchez-Hidalgo M, Fernandez M, Montalban-Lopez M, Valdivia E, Martinez-Bueno M. 2008. Genetic features of circular bacteriocins produced by Gram-positive bacteria. FEMS Microbiol. Rev. 32:2–22. 10.1111/j.1574-6976.2007.00087.x. [DOI] [PubMed] [Google Scholar]
- 6.Kemperman R, Kuipers A, Karsens H, Nauta A, Kuipers O, Kok J. 2003. Identification and characterization of two novel clostridial bacteriocins, circularin A and closticin 574. Appl. Environ. Microbiol. 69:1589–1597. 10.1128/AEM.69.3.1589-1597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng NCK, Wescombe PA, Burton JP, Jack RW, Tagg JR. 2007. The diversity of bacteriocins in gram-positive bacteria, p 45–92 Riley MA, Chavan MA. (ed), Bacteriocins: ecology and evolution. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 8.Martin-Visscher LA, Gong X, Duszyk M, Vederas JC. 2009. The three-dimensional structure of carnocyclin A reveals that many circular bacteriocins share a common structural motif. J. Biol. Chem. 284:28674–28681. 10.1074/jbc.M109.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Visscher LA, van Belkum MJ, Garneau-Tsodikova S, Whittal RM, Zheng J, McMullen LM, Vederas JC. 2008. Isolation and characterization of carnocyclin a, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756–4763. 10.1128/AEM.00817-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai Y, Kemperman R, Kok J, Saito T. 2004. The circular bacteriocins gassericin A and circularin A. Curr. Protein Pept. Sci. 5:393–398. 10.2174/1389203043379549. [DOI] [PubMed] [Google Scholar]
- 11.Wirawan RE, Swanson KM, Kleffmann T, Jack RW, Tagg JR. 2007. Uberolysin: a novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 153:1619–1630. 10.1099/mic.0.2006/005967-0. [DOI] [PubMed] [Google Scholar]
- 12.Sawa N, Zendo T, Kiyofuji J, Fujita K, Himeno K, Nakayama J, Sonomoto K. 2009. Identification and characterization of lactocyclicin Q, a novel cyclic bacteriocin produced by Lactococcus sp. strain QU 12. Appl. Environ. Microbiol. 75:1552–1558. 10.1128/AEM.02299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda Y, Ono H, Kitagawa H, Ito H, Mu F, Sawa N, Zendo T, Sonomoto K. 2011. Identification and characterization of leucocyclicin Q, a novel cyclic bacteriocin produced by Leuconostoc mesenteroides TK41401. Appl. Environ. Microbiol. 77:8164–8170. 10.1128/AEM.06348-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrero J, Brede DA, Skaugen M, Diep DB, Herranz C, Nes IF, Cintas LM, Hernandez PE. 2011. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos). Appl. Environ. Microbiol. 77:369–373. 10.1128/AEM.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz R, Vater J, Budiharjo A, Wang Z, He Y, Dietel K, Schwecke T, Herfort S, Lasch P, Borriss R. 2014. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 196:1842–1852. 10.1128/JB.01474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang E, Zhang L, Chung YK, Zheng Z, Yousef AE. 2013. Characterization and application of enterocin RM6, a bacteriocin from Enterococcus faecalis. Biomed Res. Int. 2013:206917. 10.1155/2013/206917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalmokoff ML, Cyr TD, Hefford MA, Whitford MF, Teather RM. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can. J. Microbiol. 49:763–773. 10.1139/w03-101. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa K, Kawai Y, Ito Y, Nakamura K, Chujo T, Nishimura J, Kitazawa H, Saito T. 2010. HPLC purification and re-evaluation of chemical identity of two circular bacteriocins, gassericin A and reutericin 6. Lett. Appl. Microbiol. 50:406–411. 10.1111/j.1472-765X.2010.02810.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu WT, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. 2010. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 107:16286–16290. 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy K, O'Sullivan O, Rea MC, Cotter PD, Ross RP, Hill C. 2011. Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters. PLoS One 6:e20852. 10.1371/journal.pone.0020852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemperman R, Jonker M, Nauta A, Kuipers OP, Kok J. 2003. Functional analysis of the gene cluster involved in production of the bacteriocin circularin A by Clostridium beijerinckii ATCC 25752. Appl. Environ. Microbiol. 69:5839–5848. 10.1128/AEM.69.10.5839-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2010. Cloning and characterization of the gene cluster involved in the production of the circular bacteriocin carnocyclin A. Probiotics Antimicrob. Proteins 2:218–225. 10.1007/s12602-010-9056-1. [DOI] [PubMed] [Google Scholar]
- 23.van Belkum MJ, Martin-Visscher LA, Vederas JC. 2011. Structure and genetics of circular bacteriocins. Trends Microbiol. 19:411–418. 10.1016/j.tim.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Gabrielsen C, Brede DA, Salehian Z, Nes IF, Diep DB. 2014. Functional genetic analysis of the GarML gene cluster in Lactococcus garvieae DCC43 gives new insights into circular bacteriocin biosynthesis. J. Bacteriol. 196:911–919. 10.1128/JB.01115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz M, Valdivia E, Martinez-Bueno M, Fernandez M, Soler-Gonzalez AS, Ramirez-Rodrigo H, Maqueda M. 2003. Characterization of a new operon, as-48EFGH, from the as-48 gene cluster involved in immunity to enterocin AS-48. Appl. Environ. Microbiol. 69:1229–1236. 10.1128/AEM.69.2.1229-1236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai Y, Kusnadi J, Kemperman R, Kok J, Ito Y, Endo M, Arakawa K, Uchida H, Nishimura J, Kitazawa H, Saito T. 2009. DNA sequencing and homologous expression of a small peptide conferring immunity to gassericin A, a circular bacteriocin produced by Lactobacillus gasseri LA39. Appl. Environ. Microbiol. 75:1324–1330. 10.1128/AEM.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Draper LA, Ross RP, Hill C, Cotter PD. 2008. Lantibiotic immunity. Curr. Protein Pept. Sci. 9:39–49. 10.2174/138920308783565750. [DOI] [PubMed] [Google Scholar]
- 28.Conlan BF, Gillon AD, Craik DJ, Anderson MA. 2010. Circular proteins and mechanisms of cyclization. Biopolymers 94:573–583. 10.1002/bip.21422. [DOI] [PubMed] [Google Scholar]
- 29.Cebrian R, Maqueda M, Neira JL, Valdivia E, Martinez-Bueno M, Montalban-Lopez M. 2010. Insights into the functionality of the putative residues involved in enterocin AS-48 maturation. Appl. Environ. Microbiol. 76:7268–7276. 10.1128/AEM.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eisenbrandt R, Kalkum M, Lurz R, Lanka E. 2000. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182:6751–6761. 10.1128/JB.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez M, Martinez-Bueno M, Martin MC, Valdivia E, Maqueda M. 2007. Heterologous expression of enterocin AS-48 in several strains of lactic acid bacteria. J. Appl. Microbiol. 102:1350–1361. 10.1111/j.1365-2672.2006.03194.x. [DOI] [PubMed] [Google Scholar]
- 32.Montalban-Lopez M, Sanchez-Hidalgo M, Cebrian R, Maqueda M. 2012. Discovering the bacterial circular proteins: bacteriocins, cyanobactins, and pilins. J. Biol. Chem. 287:27007–27013. 10.1074/jbc.R112.354688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paetzel M, Karla A, Strynadka NC, Dalbey RE. 2002. Signal peptidases. Chem. Rev. 102:4549–4580. 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 34.Lai EM, Eisenbrandt R, Kalkum M, Lanka E, Kado CI. 2002. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 184:327–330. 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez C, Langdon GM, Bruix M, Galvez A, Valdivia E, Maqueda M, Rico M. 2000. Bacteriocin AS-48, a microbial cyclic polypeptide structurally and functionally related to mammalian NK-lysin. Proc. Natl. Acad. Sci. U. S. A. 97:11221–11226. 10.1073/pnas.210301097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohans CT, Towle KM, Miskolzie M, McKay RT, van Belkum MJ, McMullen LM, Vederas JC. 2013. Solution structures of the linear leaderless bacteriocins enterocin 7A and 7B resemble carnocyclin A, a circular antimicrobial peptide. Biochemistry 52:3987–3994. 10.1021/bi400359z. [DOI] [PubMed] [Google Scholar]
- 37.Montalban-Lopez M, Spolaore B, Pinato O, Martinez-Bueno M, Valdivia E, Maqueda M, Fontana A. 2008. Characterization of linear forms of the circular enterocin AS-48 obtained by limited proteolysis. FEBS Lett. 582:3237–3242. 10.1016/j.febslet.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 38.Montalban-Lopez M, Martinez-Bueno M, Valdivia E, Maqueda M. 2011. Expression of linear permutated variants from circular enterocin AS-48. Biochimie 93:549–555. 10.1016/j.biochi.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. 1989. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against Gram-positive and Gram-negative bacteria and other organisms. Res. Microbiol. 140:57–68. 10.1016/0923-2508(89)90060-0. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Visscher LA, Yoganathan S, Sit CS, Lohans CT, Vederas JC. 2011. The activity of bacteriocins from Carnobacterium maltaromaticum UAL307 against Gram-negative bacteria in combination with EDTA treatment. FEMS Microbiol. Lett. 317:152–159. 10.1111/j.1574-6968.2011.02223.x. [DOI] [PubMed] [Google Scholar]
- 41.Stevens KA, Sheldon BW, Klapes NA, Klaenhammer TR. 1991. Nisin treatment for inactivation of Salmonella species and other Gram-negative bacteria. Appl. Environ. Microbiol. 57:3613–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galvez A, Maqueda M, Martinez-Bueno M, Valdivia E. 1991. Permeation of bacterial cells, permeation of cytoplasmic and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J. Bacteriol. 173:886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Barrena MJ, Martinez-Ripoll M, Galvez A, Valdivia E, Maqueda M, Cruz V, Albert A. 2003. Structure of bacteriocin AS-48: from soluble state to membrane bound state. J. Mol. Biol. 334:541–549. 10.1016/j.jmb.2003.09.060. [DOI] [PubMed] [Google Scholar]
- 44.Rogne P, Fimland G, Nissen-Meyer J, Kristiansen PE. 2008. Three-dimensional structure of the two peptides that constitute the two-peptide bacteriocin lactococcin G. Biochim. Biophys. Acta 1784:543–554. 10.1016/j.bbapap.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Hauge HH, Mantzilas D, Eijsink VG, Nissen-Meyer J. 1999. Membrane-mimicking entities induce structuring of the two-peptide bacteriocins plantaricin E/F and plantaricin J/K. J. Bacteriol. 181:740–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong X, Martin-Visscher LA, Nahirney D, Vederas JC, Duszyk M. 2009. The circular bacteriocin, carnocyclin A, forms anion-selective channels in lipid bilayers. Biochim. Biophys. Acta 1788:1797–1803. 10.1016/j.bbamem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 47.Steinstraesser L, Kraneburg U, Jacobsen F, Al-Benna S. 2011. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 216:322–333. 10.1016/j.imbio.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389. 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uzelac G, Kojic M, Lozo J, Aleksandrzak-Piekarczyk T, Gabrielsen C, Kristensen T, Nes IF, Diep DB, Topisirovic L. 2013. A Zn-dependent metallopeptidase is responsible for sensitivity to LsbB, a class II leaderless bacteriocin of Lactococcus lactis subsp. lactis BGMN1-5. J. Bacteriol. 195:5614–5621. 10.1128/JB.00859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kjos M, Oppegard C, Diep DB, Nes IF, Veening JW, Nissen-Meyer J, Kristensen T. 2014. Sensitivity to the two-peptide bacteriocin lactococcin G is dependent on UppP, an enzyme involved in cell-wall synthesis. Mol. Microbiol. 92:1177–1187. 10.1111/mmi.12632. [DOI] [PubMed] [Google Scholar]
- 51.Kawai Y, Ishii Y, Arakawa K, Uemura K, Saitoh B, Nishimura J, Kitazawa H, Yamazaki Y, Tateno Y, Itoh T, Saito T. 2004. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl. Environ. Microbiol. 70:2906–2911. 10.1128/AEM.70.5.2906-2911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrielsen C, Brede DA, Hernandez PE, Nes IF, Diep DB. 2012. The maltose ABC transporter in Lactococcus lactis facilitates high-level sensitivity to the circular bacteriocin garvicin ML. Antimicrob. Agents Chemother. 56:2908–2915. 10.1128/AAC.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772–1779. 10.1074/jbc.M006770200. [DOI] [PubMed] [Google Scholar]
- 54.Molloy EM, Ross RP, Hill C. 2012. ‘Bac’ to the future: bioengineering lantibiotics for designer purposes. Biochem. Soc. Trans. 40:1492–1497. 10.1042/BST20120193. [DOI] [PubMed] [Google Scholar]
- 55.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. 2012. Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLoS One 7:e46884. 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112:531–552. [DOI] [PubMed] [Google Scholar]
- 57.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217. 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 58.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Itoh T, Fujimoto Y, Kawai Y, Toba T, Saito T. 1995. Inhibition of food-borne pathogenic bacteria by bacteriocins from Lactobacillus gasseri. Lett. Appl. Microbiol. 21:137–141. 10.1111/j.1472-765X.1995.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 61.Kalmokoff ML, Teather RM. 1997. Isolation and characterization of a bacteriocin (butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B. fibrisolvens. Appl. Environ. Microbiol. 63:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]