Abstract
Despite recent advances in metagenomic and single-cell genomic sequencing to investigate uncultivated microbial diversity and metabolic potential, fundamental questions related to population structure, interactions, and biogeochemical roles of candidate divisions remain. Numerous molecular surveys suggest that stratified ecosystems manifesting anoxic, sulfidic, and/or methane-rich conditions are enriched in these enigmatic microbes. Here we describe diversity, abundance, and cooccurrence patterns of uncultivated microbial communities inhabiting the permanently stratified waters of meromictic Sakinaw Lake, British Columbia, Canada, using 454 sequencing of the small-subunit rRNA gene with three-domain resolution. Operational taxonomic units (OTUs) were affiliated with 64 phyla, including more than 25 candidate divisions. Pronounced trends in community structure were observed for all three domains with eukaryotic sequences vanishing almost completely below the mixolimnion, followed by a rapid and sustained increase in methanogen-affiliated (∼10%) and unassigned (∼60%) archaeal sequences as well as bacterial OTUs affiliated with Chloroflexi (∼22%) and candidate divisions (∼28%). Network analysis revealed highly correlated, depth-dependent cooccurrence patterns between Chloroflexi, candidate divisions WWE1, OP9/JS1, OP8, and OD1, methanogens, and unassigned archaeal OTUs indicating niche partitioning and putative syntrophic growth modes. Indeed, pathway reconstruction using recently published Sakinaw Lake single-cell genomes affiliated with OP9/JS1 and OP8 revealed complete coverage of the Wood-Ljungdahl pathway with potential to drive syntrophic acetate oxidation to hydrogen and carbon dioxide under methanogenic conditions. Taken together, these observations point to previously unrecognized syntrophic networks in meromictic lake ecosystems with the potential to inform design and operation of anaerobic methanogenic bioreactors.
INTRODUCTION
Over the past 2 decades, cultivation-independent approaches have identified at least 60 major branch points (phyla or divisions) in the bacterial and archaeal domains of life (1). Approximately half of these branch points represent candidate divisions with no known cultivated representatives, so-called microbial dark matter (MDM) (2). Emerging lines of evidence suggest that bacteria affiliated with BD1-5, OP11, OP9/JS1, OP1, and WWE1 candidate divisions harbor genes encoding components of fermentation, hydrogen or sulfur metabolic pathways supporting cometabolic or syntrophic growth modes under anaerobic conditions (3–6). Consistent with this, a recent single-cell genomic study of OP9 bacteria from hot spring sediments suggested a dependence on exogenous vitamins sourced from surrounding microbial community members (3). Such public good dynamics could represent a common organizing principle in structuring microbial community interaction networks (7, 8) and help explain the resistance of most environmental microorganisms, including candidate divisions to clonal isolation (7, 9).
While advances in metagenomic and single-cell genomic sequencing have begun to open a window on the metabolic potential of many candidate divisions, fundamental questions relating to their population structure, interactions, and biogeochemical roles remain. Recent molecular surveys indicate that natural and engineered ecosystems with anoxic, sulfidic, and/or methane-rich conditions tend to harbor a diversity of candidate divisions (6, 10–12). Among ecosystems manifesting these biogeochemical conditions, permanently stratified meromictic lakes provide tractable models in which to study MDM population structure, function, and dynamics. In these aquatic ecosystems, the water column partitions into oxic fresh surface waters referred to as the mixolimnion, a redox transition zone, and anoxic, often hydrogen sulfide (H2S)- and methane (CH4)-rich bottom waters, referred to as the monimolimnion (13, 14).
Previous studies have identified MDM in geographically isolated meromictic lakes with different degrees of spatial and taxonomic resolution. For example, small-subunit (SSU) rRNA gene clone library sequencing in Lake Pavin in France identified at least five bacterial candidate divisions, including OP1, OP3, OP10, OP11, and WS5 that partitioned between the redox transition zone and monimolimnion (15). A more recent study using a combination of SSU rRNA gene clone library sequencing and PhyloChip hybridization in Mahoney Lake in Canada identified at least eight bacterial candidate divisions, including OP1, OP8, OP9/JS1, OP11, TM6, WS1, WS3, and ZB2 (16). A higher-throughput study in Arctic Lake A identified SSU rRNA gene pyrotag sequences affiliated with at least four candidate divisions, including OP3, OP8, OP9, and OP11 that increased in abundance within the monimolimnion (17).
Here we describe the microbial community inhabiting Sakinaw Lake, a meromictic lake, on the Sunshine Coast of British Columbia, Canada, using a pyrosequencing approach targeting the SSU rRNA gene, with specific emphasis on describing the population structure of MDM across defined redox transition zones. We apply hierarchical clustering and indicator species analyses to identify operational taxonomic units (OTUs) differentially associated with the mixolimnion, upper and lower part of the transition zone, and monimolimnion in relation to environmental parameter data. Network analysis is then used to chart potential interactions between prevalent OTUs, including many candidate divisions. Metabolic reconstruction based on recently published near-complete Sakinaw Lake single-cell amplified genome sequences reinforces network connectivity patterns, revealing putative syntrophic relationships among candidate divisions and between candidate divisions and other microbial community members (2).
MATERIALS AND METHODS
Sampling.
Water samples from Sakinaw Lake, British Columbia, Canada, were taken at deep basin station S1 (49°40.968′N 124°00.119′W) using a combination of 12-liter Niskin and 8-liter GO-FLO bottles on 6 June 2007, 23 October 2007, 21 May 2008, 5 August 2009, 5 January 2010, 27 January 2011, and 24 May 2011 (see Table S1 in the supplemental material). A total of 66 samples were collected from different depths covering the mixolimnion (5 m to 30 m), transition zone (33 m to 55 m), and monimolimnion (60 m to 120 m). Conductivity temperature and depth were measured with a Sea-Bird SBE19 CTD device measuring conductivity, temperature, and depth (Sea-Bird Electronics Inc., Bellevue, WA, USA). Samples were kept at 4°C in the dark and subsequently processed for environmental DNA extraction. Additionally, transition metal, sulfide (H2S), and sulfate (SO42−) concentrations were determined for samples collected on 24 May 2011.
Environmental DNA.
Approximately 6 h after sample collection, 2 liters of water was filtered through a 0.22-μm Sterivex GV filter (Millipore) without a prefilter using a Masterflex L/S 7553–70 peristaltic pump (Cole-Parmer) for DNA extraction. DNA was extracted from Sterivex filters by the method of Zaikova and colleagues (18) and DeLong and colleagues (19). The DNA extraction protocol can be viewed as a visualized experiment at http://www.jove.com/video/1352/ (20).
Enumeration of cells by flow cytometry.
Enumeration of total cells was performed by the method of Zaikova and colleagues (18). Briefly, water samples were fixed in 4% (wt/vol) formaldehyde and stored at 4°C in the dark for approximately 18 h prior to processing for flow cytometry. Nucleic acids were stained with SYBR green (Invitrogen), and cells were counted with a fluorescence-activated cell sorting (FACS) LSRII flow cytometer equipped with an air-cooled argon laser (Becton Dickinson). Cell counts were estimated using a known concentration of 6-μm fluorescent beads (Invitrogen).
Chemical profiling.
All salinities are on the TEOS-10 reference composition salinity scale, with the salinity anomaly assumed to be zero (21). Dissolved oxygen concentrations were determined by Winkler titrations (22). Hydrogen sulfide concentration was measured from water samples fixed with 2% final concentration of zinc acetate and analyzed in the lab using the methylene blue method of Cline et al. (23). The concentrations of transition metals, including iron (Fe), manganese (Mn), and arsenic (As), and SO42− were determined at Maxxam Analytics (Burnaby, British Columbia, Canada) using Standard Methods for the Examination of Water and Wastewater published by the American Public Health Association (24). Sulfate was determined with automated colorimetry according to the standard protocol SM 4500 SO42− within 24 h after sample collection (24). Samples for dissolved fractions of Fe, Mn, and As were preserved with nitric acid (HNO3) and filtered through a 0.45-μm membrane filter prior to analysis with inductively coupled plasma mass spectrometry based on standard protocol EPA 200.8 (25) within 14 days of sampling. Dissolved methane (CH4) was measured from 13 samples (water depths of 5 m, 10 m, 25 m, 30 m, 33 m, 36 m, 40 m, 45 m, 50 m, 55 m, 60 m, 80 m, and 120 m) collected in June 2007 by gas chromatography coupled to mass spectrometry (GC-MS) using the static headspace equilibrium technique of Zaikova et al. (18). As CH4 concentrations in the monimolimnion of Sakinaw Lake exceed detection limits of the applied method, our data are estimates and indicate that CH4 concentrations in the deep Sakinaw Lake waters exceed atmospheric saturation values.
PCR amplification of SSU rRNA gene for pyrotag sequencing.
Environmental DNA extracts described above were amplified using previously published three-domain primers targeting the V6-V8 region of the SSU rRNA gene (26): 926F (5′-cct atc ccc tgt gtg cct tgg cag tct cag AAA CTY AAA KGA ATT GRC GG-3′) and 1392R (5′-cca tct cat ccc tgc gtg tct ccg act cag-XXXXX-ACG GGC GGT GTG TRC-3′). Primer sequences were modified by the addition of 454 A or B adapter sequences (shown in lowercase type). In addition, the reverse primer included a 5-bp bar code designated XXXXX for multiplexing of samples during sequencing.
Twenty-five microliter PCRs were performed in triplicate and pooled to minimize PCR bias. Each reaction mixture contained between 1 and 10 ng of target DNA, 0.5 μl Taq DNA polymerase (Bioshop Inc., Canada), 2.5 μl Bioshop 10× buffer provided in the Bioshop Taq-polymerase kit, 1.5 μl of 25 mM Bioshop MgCl2, 2.5 μl of 10 mM deoxynucleoside triphosphates (dNTPs) (Agilent Technologies), and 0.5 μl of 10 mM (each) primer. The thermal cycler protocol started with an initial denaturation at 95°C for 3 min and then 25 cycles, with 1 cycle consisting of 30 s at 95°C, 45 s at 55°C, 90 s at 72°C, and 45 s at 55°C. The final step, an extension step, was 10 min at 72°C. PCR products were purified using the QIAquick PCR purification kit (Qiagen), eluted in 20 mM Tris (pH 8), quantified using the Quant-it Picogreen dsDNA reagent. SSU rRNA amplicons were pooled at 30 ng DNA for each sample. Emulsion PCR and sequencing of the PCR amplicons were performed at the McGill University and Génome Québec Innovation Center on a Roche 454 GS FLX titanium platform according to the manufacturer's instructions.
Pyrotag sequence analysis.
Pyrotag sequences were analyzed using the Quantitative Insights Into Microbial Ecology (QIIME) software package (27). To minimize the removal of false-positive singleton OTUs, 901,664 pyrotag sequences generated from 66 samples collected in Sakinaw Lake between 2007 and 2011 were clustered together (see Table S1 in the supplemental material). Reads with length shorter than 200 bases, ambiguous bases, and homopolymer sequences were removed prior to chimera detection. Chimeras were detected and removed using the chimera slayer provided in the QIIME software package. Sequences were then clustered at 97% identity using uclust with furthest linkage algorithm. Prior to taxonomic assignment, singleton OTUs (OTUs represented by one read) were omitted, leaving 23,231 OTUs (Table S2). To generate an OTU table specific for the May 2011 data set, the filter_otus_by_sample.py script was used, leaving 181,464 sequences and 12,908 OTUs for downstream analysis. The average number of reads per sample in the Sakinaw Lake May 2011 data set was 16,323, with the exception of the 33-m-deep sample, which had ∼50% fewer reads. Representative sequences from each nonsingleton OTU were queried against the SILVA database release 111 (28) and the Greengenes database (29) using BLAST (30).
Overall, the comparison between SILVA and Greengenes databases revealed similar taxonomic assignments for bacterial OTUs. However, significant differences for archaeal OTU assignments were observed between the two databases. Approximately twice as many reads were assigned to Methanomicrobia using the Greengenes database than when the SILVA database was used. This difference could be mapped back to a single abundant OTU, approaching 10% of total sequences at a depth of 45 m that was assigned to Methanomicrobiales using the Greengenes database and to Halobacteriales using the SILVA database. BLAST-based comparisons indicated that sequences comprising this OTU shared ∼90% identity with Halobacteriales reference sequences in SILVA and 90% identity with Methanomicrobiales reference sequences in Greengenes. Moreover, a number of putative archaeal OTUs associated with the redox transition zone and monimolimnion (depths of 33 m to 120 m) were identified with “no blast hits” using Greengenes but assigned to Halobacteriales using SILVA. Because both databases are not well annotated for Halobacteria (31), the Sakinaw Lake data set was mapped onto the full-length Halobacteria SSU rRNA gene database generated by Youssef and colleagues (31) using the program CD-HIT (cd-hit-est-2d) (32). The data set could not be clustered with the Halobacteria reference sequences at 99%, 97%, or 95% identity. Even for 90% sequence identity, only 981 reads (0.5%) clustered with the Halobacteria reference sequences. Given these uncertainties in putative archaeal OTU affiliation, we designated them “unassigned Archaea” until full-length sequences or reference genomes become available to support more-in-depth phylogenetic analysis.
Statistical analyses.
Microbial community richness was determined using scripts implemented in the QIIME package. OTU tables were rarefied starting with 10 sequences to a maximum of 6,000 sequences. Ten iterations per sample were calculated with 100 sequences between each step. Hierarchical cluster analysis of microbial community compositional profiles and environmental parameters was conducted in the R statistical environment (Development Core Team, 2011; http://www.R-project.org/) using Manhattan distance measures for clustering microbial community compositional profiles (33) and Euclidean distance for environmental parameters (34). Prior to analysis, pyrotag data sets were normalized to the total number of reads per sample, and environmental parameter data were transformed to the same order of magnitude so that each variable had equal weight.
Multilevel indicator species analysis (ISA) was performed to identify OTUs specifically associated with different water column compartments (mixolimnion [5 m, 20 m, and 30 m], upper part of the transition zone [33 m, 36 m, 40 m, and 45 m], lower part of the transition zone [50 m and 55 m], and monimolimnion [60 m, 80 m, and 120 m]). Dufrene and Legendre established a method to determine an indicator species by its relatedness to a user-defined environment, e.g., group of samples, where each species is treated individually, and its indicator value is determined according to its abundance value (35). De Caceres and colleagues recently extended the algorithm for ISA to include a multilevel pattern analysis (36). Arguing that species with higher adaptability for different environmental factors are indicative for specific environment combinations, this analysis additionally determines indicator species for combinations of environments. The ISA/multilevel pattern analysis calculates P values with Monte Carlo simulations and returns indicator values (IVs) and values with α ≤ 0.05. The IVs fall between zero and one, where one is considered a true indicator.
Cooccurrence network.
To generate a robust network emphasizing cooccurrences between prevalent OTUs in water column compartments rather than individual depth intervals, Spearman's rank correlation was used. Spearman's rank correlation coefficients were calculated using a custom perl script, “correlation_network.pl” (https://github.com/hallamlab/utilities/tree/master/correlation_network). The initial data set consisted of 12,900 OTUs. To simplify the network, we retained OTUs with at least 10 reads appearing in at least three samples leaving 1,528 OTUs with Spearman's rank correlations equal to or greater than 0.99. The resulting cooccurrence network contained 130,101 edges, each with a positive correlation. The network was visualized with a force-directed layout, using Cytoscape 2.8.3 (37). Network properties were calculated with the “Network Analysis” plug-in. Nodes in the cooccurrence network corresponded to individual OTUs, and edges were defined by computed correlations between corresponding OTU pairs. The layout revealed four distinct modules, which persisted after lowering the correlation coefficient cutoff for edge creation to 0.90, reinforcing the robustness of the network.
Pathway reconstruction.
To evaluate the metabolic potential for syntrophic acetate oxidation (SAO) among highly connected candidate divisions, publicly available single-cell genomes from Sakinaw Lake affiliated with OP9/JS1, OP8, and WWE1 were searched for coverage of the Wood-Ljungdahl pathway using the Joint Genome Institute integrated microbial genomes expert review portal (IMG-ER) (http://img.jgi.doe.gov/).
Nucleotide sequence accession number.
The sequences reported in this study have been deposited in the NCBI BioProject database (www.ncbi.nlm.nih.gov/bioproject) under BioProject accession no. PRJNA257655 (identification no. [ID] 257655).
RESULTS
Site location and physicochemical properties.
Sakinaw Lake is a permanently stratified lake located on the Sunshine Coast of British Columbia, Canada (49°40.8′N, 124°00.39′W) at an elevation of ∼5 m above sea level. Originally a fjord opening to the Strait of Georgia, Sakinaw Lake was almost completely isolated from the ocean due to coastal uplift ∼11,000 years ago following the last ice age (38). For thousands of years, a small stream named Sakinaw Creek was the only conduit between Sakinaw Lake and the Strait of Georgia. In 1952, Sakinaw Creek was dammed to better manage water levels for development projects in the surrounding watershed preventing seawater ingress. The lake consists of two basins: a relatively shallow freshwater basin 49 m deep and a more extensive salt stratified basin 140 m deep (39). A gradual salinity increase in the transition zone is followed by a series of three <0.5-g/kg salinity steps punctuating the monimolimnion.
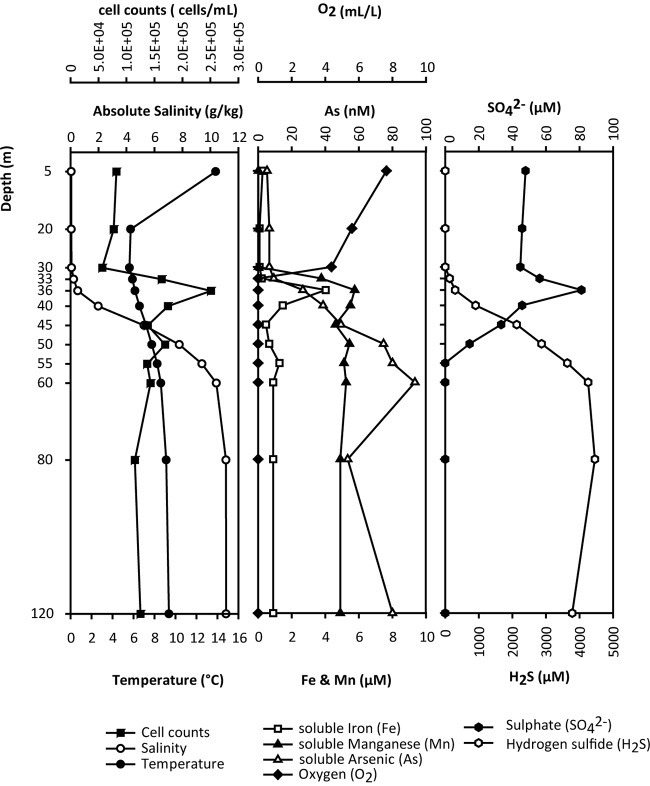
Measurements of absolute salinity (SA), temperature, and oxygen between 2007 and 2011 revealed a remarkable stability in the salinity gradient throughout the water column as well as consistent temperatures below a depth of 36 m (see Table S3 in the supplemental material). Temperature and oxygen (O2) profiles in the mixolimnion changed between different sampling time points, which was expected due to seasonal influences. To determine the availability of terminal electron acceptors (TEAs) for microbial energy metabolism in different water column compartments, O2, iron (Fe), manganese (Mn), arsenic (As), and SO42− measurements were plotted as a function of depth and compared to measurements of SA, sulfide (H2S), and total bacterial cell counts (Fig. 1). The resulting profiles were then compared to geographically distinct meromictic lakes to determine common baseline conditions (Table S4). The mixolimnion in Sakinaw Lake (5 m to 30 m) was constituted of entirely fresh, oxygen-rich water. In the transition zone (30 m to 55 m), salinity and the concentration of alternative TEAs, including Fe, Mn, and SO42− gradually increased, whereas O2 concentrations decreased. The oxic-anoxic interface was located at a depth of 33 m. Methane concentrations, which were determined for samples collected in June 2007 increased below 33 m and reached saturation at 45 m (data not shown), forming a sulfate methane transition zone (SMTZ). Peak concentrations of Fe, Mn, and SO42− were observed in the micromolar range (8.1 μM, 4.2 μM, and 57.5 μM, respectively) at a depth of 36 m and corresponded with increased microbial cell abundance (∼2.8 × 105 cells/ml). Sulfide concentrations in the lower part of the redox transition zone and the monimolimnion (50 m to 120 m) were high (4.5 mM) and resulted in the removal of soluble Fe from the water column by precipitation into iron sulfide (FeS) and pyrite (FeS2) (40). The peak concentration of As was in the nanomolar range (93.4 nM) at a depth of 60 m.
FIG 1.
Environmental parameters in Sakinaw Lake on 24 May 2011. The salinity gradient revealed water column compartmentalization into the mixolimnion (5 m to 30 m), redox transition zone (33 m to 55 m) and monimolimnion (60 m to 120 m). Seasonal temperature influences resulted in a temperature gradient within the mixolimnion, whereas temperatures in the transition zone and monimolimnion remained unchanged. The peak in microbial cell counts appeared at a depth of 36 m where oxygen was depleted and maximum concentrations of alternative terminal electron acceptors iron (Fe), manganese (Mn), and sulfate (SO42−) were measured. Maximum concentrations of arsenic (As) were located at a depth of 60 m. Sulfide was absent in the mixolimnion and increased to millimolar concentrations in the transition zone and monimolimnion.
Relationship between community structure and physicochemical characteristics.
To better understand how the physicochemical properties of the Sakinaw Lake water column influence microbial community diversity, we performed 454 pyrosequencing of the SSU rRNA gene with three-domain resolution. Although OTUs were generated from a time series, we focused on data sets obtained from the 24 May 2011 sampling campaign given the availability of extensive physicochemical information spanning defined water column redox gradients (see Materials and Methods). With this approach, the number of nonsingleton OTUs available for downstream analysis increased compared to clustering of the May 2011 data set alone. Indeed, a number of singletons in May 2011 were found in clusters containing >200 reads in the time series consistent with the presence of conditionally rare taxa in the Sakinaw Lake water column (see Table S2 in the supplemental material) (41).
Approximately 75% of the organisms in the May 2011 data set were assigned to the domain Bacteria, and 25% of these were assigned to candidate divisions. Collectively, bacterial OTUs contained the majority of pyrotag reads in all but the 45-m-deep sample where almost 60% of the reads were assigned to the domain Archaea (Fig. 2A). Eukaryotes, although prevalent in oxygenated surface waters, vanished almost completely below the mixolimnion. Richness estimates based on count data indicated that the mixolimnion had fewer OTUs than samples from the transition zone and monimolimnion (Fig. 2B). This pattern was consistent for all samples collected between 2007 and 2011 (see Fig. S1 in the supplemental material).
FIG 2.
Relationship between community structure and physicochemical characteristics of the Sakinaw Lake water column. (A) Bacterial operational taxonomic units (OTUs) contained the majority of pyrotag reads in all samples but the sample taken at a depth of 45 m, where almost 60% of the reads were assigned to Archaea. Eukaryotes vanished almost completely below the mixolimnion. (B) Richness estimates based on count data indicated that the mixolimnion had fewer OTUs than samples from the transition zone and monimolimnion. (C) Hierarchical clustering of the OTU abundance and environmental parameter data (temperature, SA, TEAs, and H2S).
Hierarchical clustering of the microbial community composition profiles and selected environmental parameter data (temperature, SA, TEAs, and H2S) reflected the stratified nature of the lake ecosystem (Fig. 2C). The resulting dendrograms revealed identical clustering patterns between OTU abundance and environmental parameters for almost all depth intervals. Both dendrograms suggest that water column partitioning in Sakinaw Lake did not easily conform to a three-compartment meromixis model, i.e., mixolimnion, redox transition zone, and monimolimnion. This was not unexpected, as the division of the water column into three compartments is based solely on hydrodynamic conditions and does not reflect partitioning of the water column into distinct environmental niches. On the basis of these observations, we operationally divided the transition zone into an upper and lower part to more accurately reflect potential niche partitioning.
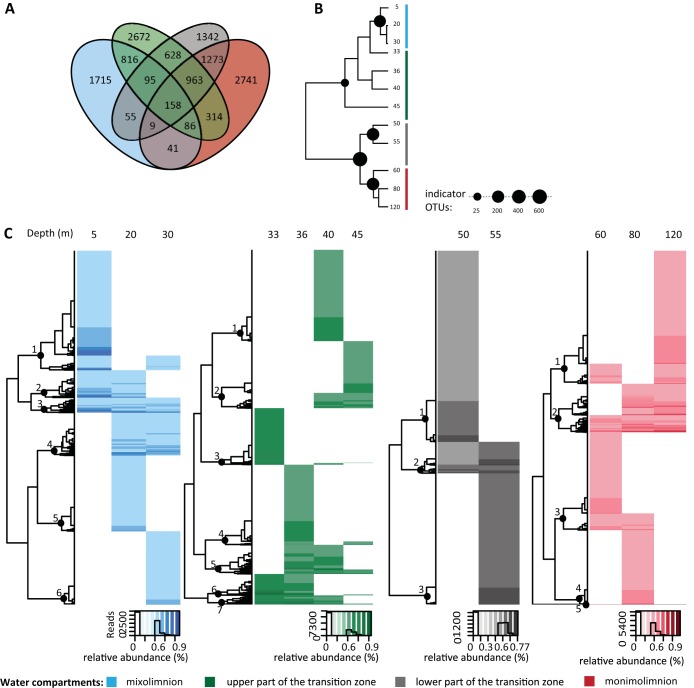
To identify how OTUs partitioned between the mixolimnion, upper part of the transition zone, lower part of the transition zone, and monimolimnion, we conducted a four-way set difference analysis of the presence of OTUs (Fig. 3A). Approximately 1% of the OTUs were shared between all four water column compartments, with the majority of OTUs exhibiting mutually exclusive distribution patterns (13% unique to the mixolimnion, 21% unique to the upper transition zone, 10% unique to the lower transition zone, and 21% unique to the monimolimnion) (Fig. 3A). Operational taxonomic units associated with a given water column compartment that are absent or rare in other compartments are potential ecological indicators. To identify potential indicator OTUs, we performed a multilevel indicator species analysis. With this analysis we were able to determine indicator species for individual water column compartments and compartment combinations. Only 5 to 10% of the compartment-specific OTUs were identified as indicator species (Fig. 3B; see Fig. S2 in the supplemental material). Indeed, the majority of OTUs that were identified as compartment specific in the four-way set difference analysis exhibited fine-scale depth partitioning (Fig. 3C and Table S5). Indicators shared between compartments were also uncommon with the exception of the lower part of the transition zone and monimolimnion. In these two compartments, ∼44% of the OTUs were identified as indicators. Overall, the majority of OTU indicators were affiliated with unassigned Archaea (25.63%) and candidate phyla (21.3%), followed by Chloroflexi (16.2%). For a detailed taxonomic breakdown of indicator OTUs, see Table S6.
FIG 3.
Partitioning of operational taxonomic units (OTUs) within and between the mixolimnion, upper part of the transition zone, lower part of the transition zone, and monimolimnion. (A) The Venn diagram revealed mutually exclusive distribution patterns for the majority of OTUs. (B) Number of indicator OTUs in different water column compartments. (C) OTUs that were identified as water compartment specific using the four-way set difference analysis in panel A exhibited a fine-scale depth partitioning as revealed by a cluster analysis of the associated OTUs. For identification of OTUs within individual clusters of the water column compartments, the clusters were labeled with numbers and associated OTUs were listed in Table S5 in the supplemental material.
Taxonomic composition.
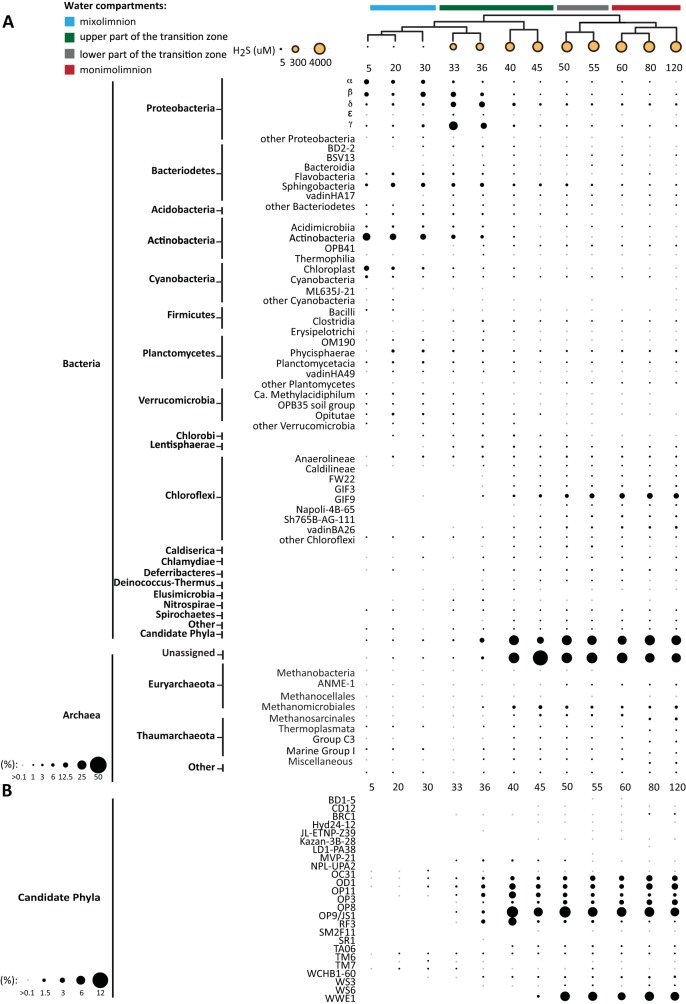
To determine how the taxonomic composition of the microbial community was distributed within and between water column compartments, we plotted bacterial and archaeal OTUs based on relative abundance (Fig. 4). The presence of eukaryotes in Sakinaw Lake is described in the supplemental material (see Fig. S3 in the supplemental material). All taxa exhibiting intermediate abundance (>0.1%) or above are represented in Fig. 4 and also Fig. S3 in the supplemental material. However, for visualization purposes, only taxa >1% are shown to scale.
FIG 4.
(A) Relative abundances of bacterial and archaeal phyla/classes based on relative OTU abundance. All taxa exhibiting intermediate abundance (>0.1%) are represented in the figure; however, for visualization purposes, only taxa with abundance of >1% are shown to scale. Sulfide concentrations at the individual depths are represented as dots where the radius correlates to the sulfide concentration. (A) Bacterial and archaeal taxa exhibited water compartment-specific distribution patterns. Microbial dark matter comprised of candidate phyla and unassigned Archaea was predominately recovered from the sulfide-rich part of the water column. (B) Distribution and abundance of all bacterial candidate divisions in Sakinaw Lake.
The most abundant taxa in the mixolimnion were Alphaproteobacteria (11.8%) the majority of which were affiliated with SAR11 (9.7%), Betaproteobacteria (10%) mostly affiliated with Burkholderiales (8.1%), Bacteriodetes (13.2%), Actinobacteria (22.3%), Cyanobacteria (16.9%), Planctomycetes (8.3%), and Verrucomicrobia (1.1%) (Fig. 4A; see Table S2 in the supplemental material). Archaea were almost absent from the mixolimnion, although Methanomicrobia were identified among members of the rare biosphere (<0.1%) (Fig. 4B and Table S2) (42, 43). Depth-specific trends within the mixolimnion were observed for several taxa, including Cyanobacteria, which decreased rapidly below 5 m, and Planctomycetes and Verrucomicrobia, which increased below 5 m. Overall, the bacterial composition in the mixolimnion of Sakinaw Lake was similar to other freshwater and meromictic lake ecosystems (17, 44, 45).
Proteobacteria dominated the upper part of the transition zone, including Betaproteobacteria (12.5%), mostly affiliated with Burkholderiales (7.7%), Deltaproteobacteria (12.9%) mostly affiliated with Desulfurobacterales (4%) and Syntrophobacterales (9.6%), and Gammaproteobacteria (23.8%) mostly affiliated with the Methylococcales (22.4%). In addition to proteobacterial groups, Bacteriodetes (13.2%), Actinobacteria (22.3%), Cyanobacteria (16.9%), Plantomycetes (8.3%), Verrucomicrobia (1.1%), Chlorobiales (1.8%), and Chloroflexi (8.5%) were also abundant in the upper part of the transition zone (Fig. 4A; see Table S2 in the supplemental material). Interestingly, between depths of 33 m and 40 m, a 10-fold increase in candidate divisions, including OP3 (4%), OP8 (4%), OP11 (4%), OP9/JS1 (8%), RF3 (5.6%), and WWE1 (7%) was observed (Fig. 4A and B). The upper part of the transition zone was further marked by a distinct increase in archaeal OTU abundance. Several of these OTUs were affiliated with the methanogenic Methanomicrobiales (7%) and Methanosarcinales (3%), ammonia-oxidizing Thaumarchaeota (1.4%), and anaerobic methane-oxidizing (ANME) archaea belonged to the rare biosphere (<1%). The remaining archaeal OTUs (44.5%) shared similar identity to Halobacteriales and Methanomicrobiales and could not be assigned to a specific phylum. Overall, the microbial community structure in the upper part of the transition zone was similar to other methane-rich meromictic lake ecosystems (17, 46). However, the diversity and abundance of bacterial candidate divisions and the relative proportion of unassigned Archaea are unprecedented. Indeed, 25 of the currently estimated 30 bacterial candidate divisions were recovered from the upper part of the transition zone. Their recently established affiliation to superphyla and the availability of single amplified genomes (SAGs) is summarized in Table S7 (2).
The taxonomic compositions of the lower part of the transition zone and the monimolimnion were similar for the samples from depths of 40 and 45 m of the upper part of the transition zone. The most abundant Bacteria were affiliated with candidate divisions OP3 (1.5%), OP8 (3.6%), OP9/JS1 (6.1%), OP11 (4.1%), WWE1 (6.1%), and Chloroflexi (22.3%). Depth-specific trends were observed for the Chloroflexi, which increased in abundance between 45 m and 120 m. This trend has also been reported for Chloroflexi in meromictic Arctic Lake A and Lake Pavin in France (15, 17). Similar to the upper part of the transition zone, OTUs affiliated with methanogenic Methanomicrobiales (5.2%) and Methanosarcinales (4.5%) and unassigned Archaea (30.5%) were also recovered. Moreover, ANME-1 abundance approached 1% at a depth of 50 m and remained present throughout the monimolimnion.
Cooccurrence analysis.
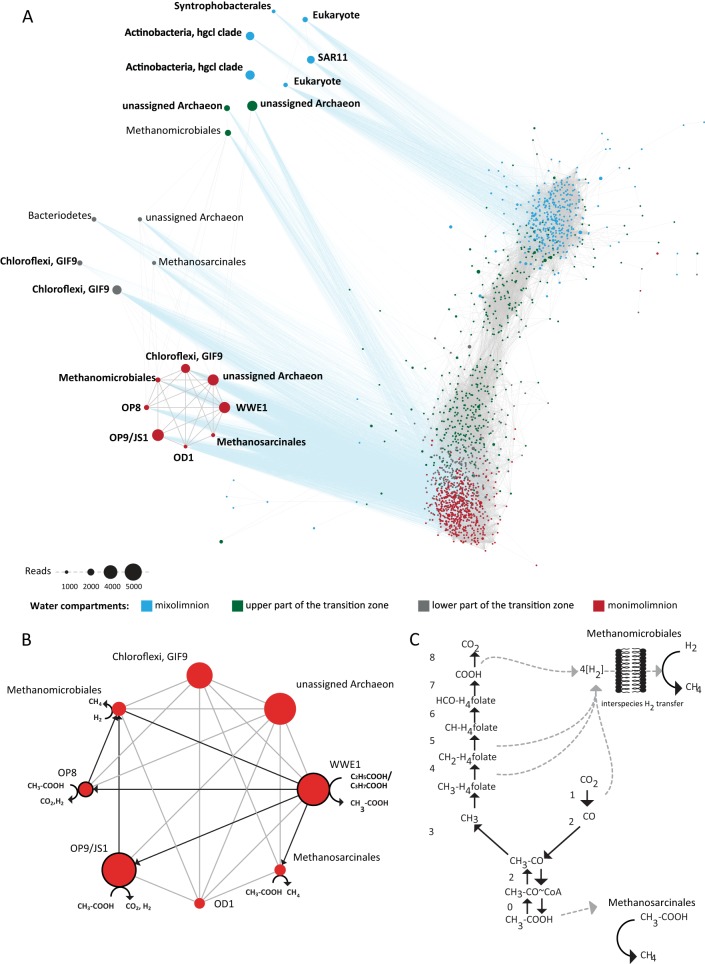
To identify putative interactions between microbial community members in the Sakinaw Lake water column, we constructed a cooccurrence network. The resulting network contained 130,101 positively correlated cooccurrences (edges) between 1,528 OTUs (nodes) and was composed of four modules corresponding to previously defined water column compartments. Twenty-two OTUs in the network contained >1,000 reads, and collectively, these OTUs represented 40% of total reads in the network (Fig. 5A). Three of these OTUs were indicators for the mixolimnion, one was a multilevel indicator for the mixolimnion and upper part of the transition zone, eight were multilevel indicators for the lower part of the transition zone and monimolimnion, and four were multilevel indicators for the upper part of the transition zone, lower part of the transition zone, and monimolimnion (see Fig. S2 and Table S8 in the supplemental material). Eight of the multilevel indicators revealed highest abundance in the monimolimnion and displayed a unique correlation pattern that was not observed for other abundant OTUs in the network. Closer inspection of these OTUs revealed linkages between OP8, OP9/JS1, OD1, WWE1, Chloroflexi, Methanomicrobiales, Methanosarcinales, and one unassigned archaeal OTU (Fig. 5B). Previous studies have posited a role for OP8, OP9/JS1, OD1, WWE1, and Chloroflexi in providing methanogenic substrates, specifically acetate and hydrogen (3, 5, 6, 11, 47).
FIG 5.
(A) Cooccurrence network depicting OTUs present in at least three samples with a minimum of 10 reads using Spearman's rank correlations at a correlation coefficient of 0.99. To better resolve potential interdependencies between abundant indicator OTUs (indicator OTUs were marked with boldface letters) and other abundant OTUs (≥1,000 reads), associated nodes were presented outside the main network, without altering edge sets. Edges that connected these 22 nodes to nodes in the main network are colored blue. (B) Eight of the most prevalent multilevel indicators in the monimolimnion displayed a unique correlation pattern that was not observed for other abundant OTUs in the network. (C) Single amplified genomes affiliated with OP9/JS1 and OP8 encode all components of the Wood-Ljungdahl pathway. The components of the Wood-Ljungdahl pathway are as follows: compound 1/2, CO dehydrogenase/acetyl-CoA synthase; compound 3, trimethylamine:corrinoid methyltransferase; compound 4, ATP:corrinoid adenosyltransferase; compound 5, methylenetetrahydrofolate reductase; compound 6, 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase; compound 7, formyltetrahydrofolate synthetase; compound 8, formate dehydrogenase. In contrast, WWE1 genomes harbored genes for the carbonyl branch of the Wood-Ljungdahl pathway. The components of the carbonyl branch of the Wood-Ljungdahl pathway are as follows: compound 1/2, CO dehydrogenase/acetyl-CoA synthase; compound 3, 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase. Additionally, WWE1 encodes compound 0 (ADP-forming acetyl-CoA synthase).
Linkages between OP8, OP9/JS1, and hydrogenotrophic methanogenic Methanomicrobiales in the monimolimnion are consistent with interspecies hydrogen transfer and competition for acetate during syntrophic acetate oxidation (SAO). In support of this observation, no linkages were observed between OP8 and OP9/JS1 or between OP8, OP9/JS1, and aceticlastic methanogens affiliated with Methanosarcinales. Conversely, linkages between WWE1, Methanomicrobiales, and Methanosarcinales suggest that this candidate division is unlikely to perform SAO. To validate network results, we evaluated whether publicly available near-complete (see also Table S9 in the supplemental material) Sakinaw Lake OP8, OP9/JS1, and WWE1 SAGs encode components of the Wood-Ljungdahl pathway, which is proposed to run in reverse during SAO (2). Candidate divisions OP8 and OP9/JS1 harbored a complete set of genes encoding the carbonyl and methyl branches of the Wood-Ljungdahl pathway, while WWE1 harbored only a subset, including genes encoding 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase and CO-dehydrogenase/acetyl coenzyme A (acetyl-CoA) synthase (Fig. 5C) (48).
DISCUSSION
The water column in Sakinaw Lake (British Columbia, Canada) is a highly stratified ecosystem, in which microbial community members partition into distinct subpopulations on the basis of water column redox gradients. This stratification is stabilized by a steep salinity gradient that persists below the oxygen interface. Richness estimates revealed higher diversity in the redox transition zone and monimolimnion than in the mixolimnion, which is likely supported by a continuous supply of nutrients and substrates as previously described for other stratified lakes (49, 50). Microbial community cluster analysis mirrored patterns observed for physicochemical parameters. This distribution pattern promotes hypotheses related to redox-driven niche partitioning and metabolic coupling in the Sakinaw Lake water column.
The microbial community structure in the oxygen-rich, sunlit, and entirely fresh mixolimnion (5 m to 30 m) is dominated by Actinobacteria, Cyanobacteria, and Alphaproteobacteria affiliated with SAR11 consistent with other freshwater ecosystems recently described by Newton and colleagues (51). Moreover, eukaryotic OTUs ware also abundant in surface waters and strong correlations (Spearmann's rank correlations of ≥0.99) between SAR11 and eukaryotic opisthokonta as well as SAR supergroup OTUs were identified, indicating potential grazing relationships.
The microaerophilic upper part of the redox transition zone (between 33 and 45 m deep) provides a habitat for abundant aerobic methane oxidizers affiliated with the Methylococcales (22.4% at a depth of 33 m). As O2 concentrations decrease below 33 m, Proteobacteria abundance increases, consistent with patterns observed in other meromictic lake ecosystems (17, 45, 52–54). At depths between 33 and 36 m, where concentrations of SO42−, Fe, and Mn are at their highest, OTUs affiliated with putative sulfate-reducing Deltaproteobacteria are abundant. Interestingly, candidate phylum RF3 OTUs increased between 36 and 40 m within the SMTZ. In marine sediments, the anaerobic oxidation of methane (AOM) is associated with similar gradients of H2S and CH4 (55). AOM is driven by syntrophic interactions between sulfate-reducing bacteria (SRB) and ANME (56, 57). While no genomic sequence information is currently available for RF3, its niche in the Sakinaw Lake water column suggests a potential role in sulfur or methane cycling. Moreover, OTUs affiliated with ANME-1 were recovered from the rare biosphere in the upper part of the redox transition zone and increased in abundance within the monimolimnion, consistent with water column AOM potential.
As the water becomes more sulfidic in the lower part of the redox transition zone between 50 and 55 m and the monimolimnion between 60 and 120 m, the diversity and abundance of MDM, with the potential to mediate cometabolic or syntrophic interactions, increases (3–6). With more than 25 candidate divisions accounting for 40% of SSU rRNA gene sequences and high numbers of unassigned Archaea, MDM enrichment in Sakinaw Lake is unprecedented. Abundant bacterial candidate divisions were affiliated with OD1, OP3, OP8, OP9/JS1, OP11, and WWE1. With the exception of WWE1, all of these candidate divisions have been previously recovered from other meromictic lake ecosystems (10, 16, 17). Furthermore, genomic potential for OD1, OP3, OP9/JS1, and OP11 has been recently inferred from metagenomic or single-cell genomic approaches (3, 6, 58, 59).
Metagenomic reconstruction of OD1 and OP11 genomes revealed the potential for fermentative metabolism as well as polysulfide reduction to H2S (6). Perry inspected sulfur chemistry in Sakinaw Lake in the early 1990s and reported a remarkably high concentration of polysulfide below the oxygen-sulfide interface (40). Based on this evidence, OD1 and OP11 could contribute to the high H2S concentrations in Sakinaw Lake through polysulfide reduction. Candidate division OP3 has been proposed to be magnetotactic with the ability for anaerobic respiration (58, 59). The availability of alternative TEAs in Sakinaw Lake is restricted to the redox transition zone, suggesting that OP3 in the monimolimnion could encode an alternative energy metabolism to previously studied OP3 genomes. The genomic potential for OP9/JS1 recovered from hot spring sediments revealed a saccharolytic, fermentative lifestyle with the potential for cellulose degradation and hydrogen production (3). In contrast, enrichment cultures from acetate-amended sulfate-rich, anoxic marine sediments revealed [13C]acetate uptake by OP9/JS1 (60). On the basis of these observations, Webster and colleagues (60) suggested that OP9/JS1 could be acetate oxidizers using SO42− as a terminal electron acceptor. In Sakinaw Lake, this would be possible only in the upper part of the redox transition zone where SO42− is available suggesting a fermentative lifestyle below the transition zone. In support of this conclusion, sulfate-reducing genes such as dsr could not be recovered from recently published OP9/JS1 SAGs from Sakinaw Lake (2).
In addition to bacterial candidate divisions, Chloroflexi and methanogenic Archaea affiliated with Methanomicrobiales and Methanosarcinales are abundant in the lower part of the redox transition zone and monimolimnion. In many aquatic ecosystems, sediments are considered the main source of CH4 production. However, the potential role of water column methanogenesis in Sakinaw Lake should not be underestimated and has been previously reported for other meromictic lakes. Under methanogenic conditions, microbial communities commonly consist of primary and secondary fermenting bacteria, which degrade polymeric substrates into hydrogen (H2), carbon dioxide (CO2), and organic acids, including acetate, formate, propionate, and butyrate. These substrates in turn are used by hydrogenotrophic and aceticlastic methanogens to convert CO2 and acetate, respectively, into methane (61). Energy yields during the degradation of organic matter into CH4 are low, and syntrophic interactions between community members are needed to make the process energetically more favorable (62). A common strategy used by well-studied syntrophs affiliated with the Syntrophobacterales and hydrogenotrophic methanogens is to overcome energy constraints by interspecies hydrogen transfer (63, 64). A similar interaction is established during syntrophic acetate oxidation (SAO), where acetogenic bacteria are proposed to run the Wood-Ljungdahl cycle in reverse while transferring four hydrogen molecules to methanogens (48).
Consistent with syntrophic growth modes associated with water column methanogenesis, Syntrophobacterales and Methanomicrobiales were abundant in the lower part of the transition zone and monimolimnion of Sakinaw Lake. Moreover, several MDM, including WWE1, OP9/JS1, OP8, and OD1 manifested statistically significant cooccurrence (Spearman's rank correlations of ≥0.99) patterns among themselves and between known methanogens and putative fermentative Chloroflexi (5, 6, 47). These cooccurrence patterns are consistent with syntrophic interactions driving interspecies hydrogen transfer between OP8, OP9/JS1, and Methanomicrobiales as well as competition for acetate between OP8, OP9/JS1, and Methanosarcinales. Metabolic reconstruction focused on the Wood-Ljungdahl pathway using recently published Sakinaw Lake OP8 and OP9/JS1 SAG sequences revealed complete pathway coverage in both candidate divisions with the potential to mediate SAO. In contrast, 5,10-methylene-tetrahydrofolate dehydrogenase/methenyl tetrahydrofolate cyclohydrolase and CO dehydrogenase/acetyl-CoA synthase were the only Wood-Ljungdahl pathway components identified in WWE1. Given that WWE1, but not OP8 and OP9/JS1, were linked to aceticlastic methanogens in the network, we suggest that this candidate division has the potential to produce acetate as a metabolic end product from butyrate oxidation, as recently proposed for WWE1 in a terephthalate-degrading methanogenic bioreactor (11). Syntrophic acetate oxidation has been speculated to be an important syntrophic pathway in methanogenic bioreactors (48, 65). The Sakinaw Lake water column thereby provides an interesting convergence of natural and engineered ecosystems with potential applications for design and operation of anaerobic bioreactors built for bioremediation and energy generation.
In conclusion, Sakinaw Lake is a model ecosystem in which to study population structure and metabolic potential of MDM. By combining cooccurrence networks with SAG-enabled metabolic reconstruction, we posit that SAO in the monimolimnion is linked to active methane cycling. Network analysis has previously been combined with biogeochemical measurements to predict physiology and function of candidate divisions (66); however, to our knowledge, this study is the first to reconstruct syntrophic relationships based on convergent cooccurrence patterns and single-cell genomic evidence. Process-oriented studies combined with phylogenetic staining and high-spatial-resolution secondary ion mass spectrometry (NanoSIMs) are now needed to validate predicted syntrophic interactions and provide quantitative insights into methane cycling within the Sakinaw Lake water column.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris Payne, Elena Zaikova, and Craig Mewis for technical support in the field, Eugene Kuatsjah and Sam Kheirandish for help in sample processing, and all members of the Hallam, Crowe, and Pawlowicz laboratories for helpful comments along the way. We also thank Christian Rinke at the Joint Genome Institute for help with the taxonomic assignments of MDM groups.
This work was performed under the auspices of the Natural Sciences and Engineering Research Council (NSERC) of Canada, Canada Foundation for Innovation (CFI), and the Canadian Institute for Advanced Research (CIFAR) through grants awarded to S.J.H. and J.T.B. K.M.K was supported by the Tula Foundation-funded Centre for Microbial Diversity and Evolution, and E.A.G. was supported by a 4-year fellowship (4YF) from the University of British Columbia.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 29 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01774-14.
REFERENCES
- 1.Hugenholtz P, Kyrpides NC. 2009. A changing of the guard. Environ. Microbiol. 11:551–553. 10.1111/j.1462-2920.2009.01888.x. [DOI] [PubMed] [Google Scholar]
- 2.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 3.Dodsworth JA, Blainey PC, Murugapiran SK, Swingley WD, Ross CA, Tringe SG, Chain PS, Scholz MB, Lo CC, Raymond J, Quake SR, Hedlund BP. 2013. Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage. Nat. Commun. 4:1854. 10.1038/ncomms2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R, Riviere D, Ganesan A, Daegelen P, Sghir A, Cohen GN, Medigue C, Weissenbach J, Le Paslier D. 2008. “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 190:2572–2579. 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, Lipton MS, Long PE, Williams KH, Banfield JF. 2014. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 8:1452–1463. 10.1038/ismej.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 7.Cordero OX, Ventouras LA, DeLong EF, Polz MF. 2012. Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc. Natl. Acad. Sci. U. S. A. 109:20059–20064. 10.1073/pnas.1213344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3(2):e00036-12. 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haruta S, Kato S, Yamamoto K, Igarashi Y. 2009. Intertwined interspecies relationships: approaches to untangle the microbial network. Environ. Microbiol. 11:2963–2969. 10.1111/j.1462-2920.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- 10.Borrel G, Lehours AC, Bardot C, Bailly X, Fonty G. 2010. Members of candidate divisions OP11, OD1 and SR1 are widespread along the water column of the meromictic Lake Pavin (France). Arch. Microbiol. 192:559–567. 10.1007/s00203-010-0578-4. [DOI] [PubMed] [Google Scholar]
- 11.Lykidis A, Chen CL, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O, Liu WT. 2011. Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J. 5:122–130. 10.1038/ismej.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybak M, Dickman M. 1988. Paleoecological reconstruction of changes in the productivity of a small, meromictic lake in southern Ontario, Canada. Hydrobiologia 169:293–306. 10.1007/BF00007552. [DOI] [Google Scholar]
- 13.Boehrer B, Schultze M. 2008. Stratification of lakes. Rev. Geophys. 46:RG2005. 10.1029/2006RG000210. [DOI] [Google Scholar]
- 14.Hakala A. 2004. Meromixis as a part of lake evolution - observations and a revised classification of true meromictic lakes in Finland. Boreal Environ. Res. 9:37–53. [Google Scholar]
- 15.Lehours AC, Evans P, Bardot C, Joblin K, Gerard F. 2007. Phylogenetic diversity of archaea and bacteria in the anoxic zone of a meromictic lake (Lake Pavin, France). Appl. Environ. Microbiol. 73:2016–2019. 10.1128/AEM.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klepac-Ceraj V, Hayes CA, Gilhooly WP, Lyons TW, Kolter R, Pearson A. 2012. Microbial diversity under extreme euxinia: Mahoney Lake, Canada. Geobiology 10:223–235. 10.1111/j.1472-4669.2012.00317.x. [DOI] [PubMed] [Google Scholar]
- 17.Comeau AM, Harding T, Galand PE, Vincent WF, Lovejoy C. 2012. Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters. Sci. Rep. 2:604. 10.1038/srep00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaikova E, Walsh DA, Stilwell CP, Mohn WW, Tortell PD, Hallam SJ. 2010. Microbial community dynamics in a seasonally anoxic fjord: Saanich Inlet, British Columbia. Environ. Microbiol. 12:172–191. 10.1111/j.1462-2920.2009.02058.x. [DOI] [PubMed] [Google Scholar]
- 19.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503. 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 20.Wright JJ, Lee S, Zaikova E, Walsh DA, Hallam SJ. 2009. DNA extraction from 0.22 microM Sterivex filters and cesium chloride density gradient centrifugation. J. Vis. Exp. 2009(31):pii=1352. 10.3791/1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Intergovernmental Oceanographic Commission. 2010. The international thermodynamic equation of seawater – 2010: calculation and use of thermodynamic properties. Manuals and guides 56. Intergovernmental Oceanographic Commission (IOC), United Nations Educational, Scientific and Cultural Organization (UNESCO), Paris, France. [Google Scholar]
- 22.Winkler LW. 1888. Die Bestimmung des in Wasser gelösten Sauerstoffen. Ber. Dtsch. Chem. Ges. 21:2843–2855. [Google Scholar]
- 23.Cline JD. 1968. Spectrophotometric determination of hydrogensulfide in natural waters. Limnol. Oceanogr. 14:454–458. [Google Scholar]
- 24.Clesceri LS, Greenberg AE, Eaton AD. (ed). 1999. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC. [Google Scholar]
- 25.US Environmental Protection Agency. 1994. Method 200.8. Determination of trace elements in waters and wastes by inductively coupled plasma - mass spectrometry. Environmental Monitoring Systems Laboratory, Office of Research and Development, US Environmental Protection Agency, Cincinnati, OH. [Google Scholar]
- 26.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647. 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 27.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- 31.Youssef NH, Ashlock-Savage KN, Elshahed MS. 2012. Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl. Environ. Microbiol. 78:1332–1344. 10.1128/AEM.07420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 33.McCune B, Grace JB. 2002. Analysis of ecological communities. MjM Software Design, Gleneden Beach, OR. [Google Scholar]
- 34.Guler C, Thyne GD, McCray JE, Turner AK. 2002. Evaluation of graphical and multivariate statistical methods for classification of water chemistry data. Hydrogeol. J. 10:455–474. 10.1007/s10040-002-0196-6. [DOI] [Google Scholar]
- 35.Dufrene M, Legendre P. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67:345–366. [Google Scholar]
- 36.De Caceres M, Legendre P, Moretti M. 2010. Improving indicator species analysis by combining groups of sites. Oikos 119:1674–1684. 10.1111/j.1600-0706.2010.18334.x. [DOI] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clague JJ, James TS. 2002. History and isostatic effects of the last ice sheet in southern British Columbia. Quaternary Sci. Rev. 21:71–87. 10.1016/S0277-3791(01)00070-1. [DOI] [Google Scholar]
- 39.Vagle S, Hume J, McLaughlin F, MacIsaac E, Shortreed K. 2010. A methane bubble curtain in meromictic Sakinaw Lake, British Columbia. Limnol. Oceanogr. 55:1313–1326. 10.4319/lo.2010.55.3.1313. [DOI] [Google Scholar]
- 40.Perry KA. 1990. The chemical limnology of two meromictic lakes with emphasis on pyrite formation. Ph.D. thesis University of British Columbia, Vancouver, Canada. [Google Scholar]
- 41.Shade A, Jones SE, Caporaso JG, Handelsman J, Knight R, Fierer N, Gilbert JA. 2014. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5(4):e01371-14. 10.1128/mBio.01371-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bell EM, Laybourn-Parry J. 1999. Annual plankton dynamics in an Antarctic saline lake. Freshwater Biol. 41:507–519. 10.1046/j.1365-2427.1999.00396.x. [DOI] [Google Scholar]
- 43.Garcia-Gil LJ, Vicente E, Camacho A, Borrego CM, Vila X, Cristina XP, Rodriguez-Gonzalez J. 1999. Vertical distribution of photosynthetic sulphur bacteria linked to saline gradients in Lake ‘El Tobar' (Cuenca, Spain). Aquatic Microb. Ecol. 20:299–303. 10.3354/ame020299. [DOI] [Google Scholar]
- 44.Gregersen LH, Habicht KS, Peduzzi S, Tonolla M, Canfield DE, Miller M, Cox RP, Frigaard NU. 2009. Dominance of a clonal green sulfur bacterial population in a stratified lake. FEMS Microbiol. Ecol. 70(1):30–41. 10.1111/j.1574-6941.2009.00737.x. [DOI] [PubMed] [Google Scholar]
- 45.Pasche N, Schmid M, Vazquez F, Schubert CJ, Wuest A, Kessler JD, Pack MA, Reeburgh WS, Burgmann H. 2011. Methane sources and sinks in Lake Kivu. J. Geophys. Res. 116:G03006. 10.1029/2011JG001690. [DOI] [Google Scholar]
- 46.Lehours AC, Bardot C, Thenot A, Debroas D, Fonty G. 2005. Anaerobic microbial communities in Lake Pavin, a unique meromictic lake in France. Appl. Environ. Microbiol. 71:7389–7400. 10.1128/AEM.71.11.7389-7400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, Williams KH, Tringe SG, Banfield JF. 2013. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. 10.1186/2049-2618-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller B, Sun L, Schnurer A. 2013. First insights into the syntrophic acetate-oxidizing bacteria–a genetic study. MicrobiologyOpen 2:35–53. 10.1002/mbo3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shade A, Read JS, Welkie DG, Kratz TK, Wu CH, McMahon KD. 2011. Resistance, resilience and recovery: aquatic bacterial dynamics after water column disturbance. Environ. Microbiol. 13:2752–2767. 10.1111/j.1462-2920.2011.02546.x. [DOI] [PubMed] [Google Scholar]
- 50.Shade A, Read JS, Youngblut ND, Fierer N, Knight R, Kratz TK, Lottig NR, Roden EE, Stanley EH, Stombaugh J, Whitaker RJ, Wu CH, McMahon KD. 2012. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 6:2153–2167. 10.1038/ismej.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75:14–49. 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biderre-Petit C, Jezequel D, Dugat-Bony E, Lopes F, Kuever J, Borrel G, Viollier E, Fonty G, Peyret P. 2011. Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol. Ecol. 77:533–545. 10.1111/j.1574-6941.2011.01134.x. [DOI] [PubMed] [Google Scholar]
- 53.Peduzzi S, Tonolla M, Hahn D. 2003. Vertical distribution of sulfate-reducing bacteria in the chemocline of Lake Cadagno, Switzerland, over an annual cycle. Aquat. Microb. Ecol. 30:295–302. 10.3354/ame030295. [DOI] [Google Scholar]
- 54.Tonolla M, Peduzzi S, Demarta A, Peduzzi R, Hahn D. 2004. Phototropic sulfur and sulfate-reducing bacteria in the chemocline of meromictic Lake Cadagno, Switzerland. J. Limnol. 63:161–170. 10.4081/jlimnol.2004.161. [DOI] [Google Scholar]
- 55.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334. 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 56.Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483–5491. 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457–1462. 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 58.Glockner J, Kube M, Shrestha PM, Weber M, Glockner FO, Reinhardt R, Liesack W. 2010. Phylogenetic diversity and metagenomics of candidate division OP3. Environ. Microbiol. 12:1218–1229. 10.1111/j.1462-2920.2010.02164.x. [DOI] [PubMed] [Google Scholar]
- 59.Kolinko S, Jogler C, Katzmann E, Wanner G, Peplies J, Schuler D. 2012. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 14:1709–1721. 10.1111/j.1462-2920.2011.02609.x. [DOI] [PubMed] [Google Scholar]
- 60.Webster G, Watt LC, Rinna J, Fry JC, Evershed RP, Parkes RJ, Weightman AJ. 2006. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ. Microbiol. 8:1575–1589. 10.1111/j.1462-2920.2006.01048.x. [DOI] [PubMed] [Google Scholar]
- 61.Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu. Rev. Microbiol. 66:429–452. 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 62.Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plugge CM, Zhang W, Scholten JC, Stams AJ. 2011. Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2:81. 10.3389/fmicb.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7:568–577. 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 65.Karakashev D, Batstone DJ, Trably E, Angelidaki I. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 72:5138–5141. 10.1128/AEM.00489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peura S, Eiler A, Bertilsson S, Nykanen H, Tiirola M, Jones RI. 2012. Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J. 6:1640–1652. 10.1038/ismej.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.