Abstract
Biofilm formation is a complex process in which many factors are involved. Bacterial swarming motility and exopolysaccharides both contribute to biofilm formation, yet it is unclear how bacteria coordinate swarming motility and exopolysaccharide production. Psl and Pel are two key biofilm matrix exopolysaccharides in Pseudomonas aeruginosa. This opportunistic pathogen has three types of motility, swimming, twitching, and swarming. In this study, we found that elevated Psl and/or Pel production reduced the swarming motility of P. aeruginosa but had little effect on swimming and twitching. The reduction was due to decreased rhamnolipid production with no relation to the transcription of rhlAB, two key genes involved in the biosynthesis of rhamnolipids. Rhamnolipid-negative rhlR and rhlAB mutants synthesized more Psl, whereas exopolysaccharide-deficient strains exhibited a hyperswarming phenotype. These results suggest that competition for common sugar precursors catalyzed by AlgC could be a tactic for P. aeruginosa to balance the synthesis of exopolysaccharides and rhamnolipids and to control bacterial motility and biofilm formation inversely because the biosynthesis of rhamnolipids, Psl, and Pel requires AlgC to provide the sugar precursors and an additional algC gene enhances the biosynthesis of Psl and rhamnolipids. In addition, our data indicate that the increase in RhlI/RhlR expression attenuated Psl production. This implied that the quorum-sensing signals could regulate exopolysaccharide biosynthesis indirectly in bacterial communities. In summary, this study represents a mechanism that bacteria utilize to coordinate swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production, which is critical for biofilm formation and bacterial survival in the environment.
INTRODUCTION
Pseudomonas aeruginosa is a ubiquitous environmental microorganism. This Gram-negative bacterium is also an opportunistic pathogen that can infect immunocompromised individuals suffering from cystic fibrosis (CF), cancer, severe burn wounds, and AIDS. P. aeruginosa causes severe nosocomial infections because of its capacity to form biofilms on medical devices and in the lungs of immunocompromised patients (1, 2). Thus, it is an important model organism for the study of biofilm. Bacterial biofilms are surface-associated and multicellular-structure communities of microorganisms. Biofilm formation is a complex process that involves many factors such as the production of extracellular polymeric substances, quorum-sensing (QS) signals, and bacterial motility. However, how bacteria coordinate these multiple factors remains elusive.
Exopolysaccharide is an important biofilm matrix component. Studies have shown that exopolysaccharide production impacts biofilm formation (3–5). In P. aeruginosa, Psl and Pel are two major exopolysaccharides that serve as biofilm matrix components (4, 6–8). Psl is a repeating pentasaccharide containing d-mannose, d-glucose, and l-rhamnose that is an essential matrix component used by P. aeruginosa to initiate biofilm formation and maintain biofilm structure (3, 4, 8, 9). Loss of Psl production greatly attenuates P. aeruginosa biofilm formation, yet enhanced Psl production results in a hyperbiofilm with many mushroom-like microcolonies (3). In addition, Psl can also function as a signal to stimulate P. aeruginosa biofilm formation (10). Pel is a glucose-rich exopolysaccharide that promotes bacterial cell-cell aggregation and is required for the formation of pellicles at the air-liquid interface of standing cultures (5–7). Ma et al. (11) have reported that the biosynthesis of both Psl and Pel requires AlgC, a bifunctional enzyme with both phosphomannomutase and phosphoglucomutase activities, which provides the common sugar precursors mannose-1-phosphate (Man-1-P) and glucose-1-phosphate (Glc-1-P). Glc-1-P is shunted into distinct metabolic pathways to synthesize more immediate precursors, UDP-d-Glc and TDP-l-Rha for the biosynthesis of Psl and UDP-d-Glc for the biosynthesis of Pel (9, 12, 13). It was suggested that there was competition for common sugar precursors between the biosynthesis pathways of Psl and Pel according to the phenomenon that induction of Pel results in a reduction of Psl levels at the posttranscriptional level (11).
Bacterial motility also contributes to biofilm formation and architecture. P. aeruginosa has at least three types of motility, including flagellum-driven swimming, type IV pilus (T4P)-mediated twitching along surfaces, and swarming on semisolid surfaces. Flagella and T4P both contribute to the initial attachment of P. aeruginosa, the first step in biofilm formation (14). Shrout et al. (15) showed that swarming motility affects biofilm architecture. Hyperswarming motility results in a flat and uniform biofilm, while lower swarming motility leads to bacterial aggregates and the formation of microcolonies. Swarming motility is a result of complex multicellular activity that requires flagella and T4P, as well as the presence of biosurfactants (16–18). The biosurfactant produced by P. aeruginosa is mainly rhamnolipids (RL), which contains mono-RL, di-RL, and 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAA) (19, 20). The biosynthesis of RL is a sequential process. The acyltransferase RhlA is involved in the synthesis of HAA (19, 21). The rhamnosyl transferase RhlB catalyzes the transfer of TDP-l-Rha to a molecule of HAA to form a mono-RL. The second rhamnosyltransferase, RhlC, sequentially transfers the rhamnosyl group to mono-RL to form di-RL (22). The transcription of both the rhlAB operon and rhlC is regulated by the RhlI/RhlR QS system (23, 24). Deletion of either rhlI or rhlR results in loss of RL production. The biosynthesis of RL also requires AlgC because TDP-l-Rha is converted from the sugar precursor Glc-1-P, which is catalyzed by AlgC (25).
It has been known that a concentration of the intracellular second messenger cyclic-di-GMP (c-di-GMP) can regulate exopolysaccharide production and bacterial motility. A high level of intracellular c-di-GMP reduces motility and elevates the biosynthesis of exopolysaccharides, including Psl and Pel. In contrast, a low level of c-di-GMP increases motility and reduces exopolysaccharide production. The bacterial motility regulated by c-di-GMP usually affects the activities of flagellum and T4P (10, 26–28). However, it is unclear whether there are any c-di-GMP-independent mechanisms to balance motility and exopolysaccharide production. Recently, Wang et al. (29) reported that bacterial migration mediated by T4P led to the formation of a Psl polysaccharide fiber matrix, which plays a significant role in ensuring efficient biofilm formation, especially under high-shear flow conditions. In the present study, we showed that exopolysaccharide production affected the swarming motility of P. aeruginosa by controlling RL production. At the same time, our data also imply a plausible indirect way for P. aeruginosa to coordinate exopolysaccharide production via the QS system.
MATERIALS AND METHODS
Bacterial strains and media.
The P. aeruginosa strains used in the study are shown in Table 1. Unless otherwise indicated, P. aeruginosa was typically cultured in Luria-Bertani medium without sodium chloride (LBNS). For RL extraction, rhlAB-lacZ transcriptional analysis, and Psl polysaccharide preparation, corresponding strains were grown in nutrient broth (8 g/liter of Oxoid Nutrient Broth and 5 g/liter of d-glucose) at 37°C.
TABLE 1.
P. aeruginosa strains used in this study
| Strain | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| PAO1 | Wild type; Psl+ Pel+ | 45 |
| WFPA801 | Psl-inducible strain; PBAD-psl PAO1 | 3 |
| WFPA831 | Pel-inducible strain; PBAD-pel PAO1 | 11 |
| WFPA833 | Psl and Pel double-inducible strain; PBAD-psl and PBAD-pel PAO1 | This study |
| IMPA114 | PrhlAB-lacZ integrated into attB site of PAO1 | This study |
| IMPA115 | PrhlAB-lacZ integrated into attB site of WFPA801 | This study |
| IMPA116 | PrhlAB-lacZ integrated into attB site of WFPA831 | This study |
| IMPA117 | PrhlAB-lacZ integrated into attB site of WFPA833 | This study |
| PAO1/pUCP18 | PAO1 containing pUCP18 vector; Apr | This study |
| PAO1/pLPS188 | PAO1 containing pLPS188 constitutively expressing algC; Apr | 11 |
| PAO1/rhlRI | PAO1 containing plasmid with PBAD-rhlRI in vector pHerd20T; Apr | This study |
| ΔrhlR/rhlRI mutant | PAO1ΔrhlR containing plasmid with PBAD-rhlRI in vector pHerd20T; Apr | This study |
| PAO1ΔrhlAB | PAO1, rhlAB in-frame deletion | This study |
| PAO1ΔfliC | PAO1, fliC (encodes flagellin) in-frame deletion | 29 |
| PAO1ΔpilA | PAO1, pilA (encodes pilin) in-frame deletion | 29 |
| PAO1ΔalgC | PAO1, algC::Tc Tcr | 46 |
| WFPA803 | PAO1, ΔalgC PBAD-psl | 11 |
| CIM1 | PAO1, ΔalgC PBAD-pel | 11 |
| PAO1ΔrhlR | PAO1, rhlR::Gm Gmr | 47 |
| PAO1ΔpslA | PAO1, pslA in-frame deletion | 9 |
| MJK8 | RSCV isolated from aged PAO1 biofilm | 41 |
| MJK8Δpsl | MJK8, pslBCDE in-frame deletion | 41 |
| MJK8Δpel | MJK8, pelA in-frame deletion | 41 |
| MJK8ΔpslΔpel | MJK8, pslBCDE and pelA in-frame deletions | 41 |
Abbreviations: Tcr, tetracycline resistant; Gmr, gentamicin resistant; Apr, ampicillin resistant.
Motility assay.
Twitching (1.2% agar) and swimming (0.3% agar) motility assays were performed as previously described by using LBNS or Jensen's medium (30, 31). Swarming motility assays were performed as previously described, by using nutrient broth with 0.5% Difco Bacto agar and different concentrations of arabinose. After inoculation, swarming plates were incubated for another 12 to 24 h at 37°C until swarming zones formed. Images were captured by Chemidoc XRS+, MAGELAB (Bio-Rad, USA). The swarming zone diameter was calculated as the average of the largest and smallest diameters of a corresponding swarming colony. At least three replicates were measured for each sample.
Construction of transcriptional fusion strains, rhlAB deletion mutant, and rhlRI-inducible strains.
For transcriptional fusion strain construction, a 432-bp DNA fragment containing the promoter of the rhlAB operon was amplified from the genomic DNA of strain PAO1 by PCR with primers PrhlAF1 (5′-GGGGTACCAAAGCCTGACGCCAGAGC-3′) and PrhlAR1 (5′-CGGGATCCTTCACACCTCCCAAAAATTTTC-3′) (italics denote restriction enzyme sites). The PCR product was digested with KpnI and BamHI and then ligated to a KpnI/BamHI-cut mini-CTX-lacZ vector to generate plasmid pSW4 (a fusion of rhlAB with lacZ). pSW4 was conjugated into the chromosomal attB sites of the P. aeruginosa strains as described previously (32). For in-frame rhlAB deletion, the following PCR primers were used (italics denote restriction enzyme sites): primer 1, F1-del-rhlAB (5′-CCCAAGCTTGCACGCTGAGCAAATTGTTC-3′), primer 2, R1-del-rhlAB (5′-GAGTATCCATATGAACGGTGCTGGCATAACAG-3′), primer 3, F2-del-rhlAB (5′-GAGTATCCATATGGAACGGCAGACAAGTAAC-3′); primer 4, R2-del-rhlAB (5′-CCGGAATTCTCCAATACCACCAACCTG-3′). Deletion alleles were cloned into the pEX18Gm vector, and the resulting plasmid was used to knock out rhlAB by allelic exchange (33). For rhlRI-inducible strain construction, primer 5 (5′-CTAGTCTAGAATGAGGAATGACGGAGGC-3′) and primer 6 (5′-CCCAAGCTTTCACACCGCCATCGACAG-3′) (italics denote restriction enzyme sites) were used to amplify the 1.5-kb rhlRI fragment. rhlRI were cloned into the pHerd20T vector to get plasmid PBAD-rhlRI. This plasmid was transformed into strain PAO1 and its isogenic ΔrhlR mutant, respectively, to obtain the rhlRI-inducible PAO/rhlRI and ΔrhlR/rhlRI mutant strains.
β-Galactosidase assays.
β-Galactosidase activity was quantitatively assayed as previously described, with modifications (34). P. aeruginosa strains were grown in nutrient broth with or without arabinose at 37°C to mid-log phase (optical density at 600 nm [OD600], ∼0.5). One-milliliter culture aliquots were resuspended in 200 μl of Z buffer and subjected to three freeze-thaw cycles. Cell lysates were assayed for β-galactosidase activity and also for total protein concentration with a BCA assay kit (Pierce, USA). All β-galactosidase activities (Miller units) were normalized by the total protein per milliliter of aliquot. All samples were assayed in triplicate.
RL extraction, quantitation, and TLC detection.
After 2 days of growth in nutrient broth with or without arabinose, RL were extracted as previously described (36). Briefly, cells were removed from the culture by centrifugation (10 min at 6,000 × g) and the supernatant was acidified with concentrated HCl until it reached pH 2.0. An equal volume of a chloroform-methanol mixture was added to the acidified supernatant, and then the sample was vortexed for 1 min. The lower organic phase was collected, evaporated to dryness, and then resuspended in 3 ml of methanol. The RL in the extract were measured by using the anthrone colorimetric assay, and the concentration was obtained by using a rhamnose standard curve (21). The RL levels produced by different strains were normalized to the level of RL obtained from PAO1. The samples were also analyzed by thin-layer chromatography (TLC) on silica gel G plates with a mobile phase consisting of 80% chloroform, 18% methanol, and 2% acetic acid, and RL were examined with a 25-mg/ml solution of α-naphthol. RL standards were obtained from the Huzhou Zijin Co. (China) (the main peaks of mono-RL and di-RL are at m/z 503 and 649, respectively).
LC-MS analysis of RL.
Quantification comparison of mono-RL (Rha-C10-C10) and di-RL (Rha-Rha-C10-C10) was performed by using liquid chromatography-mass spectrometry (LC-MS) as previously described, with modifications (19, 37). The analyses were performed with an Agilent 1260/6460 LC/MSD triple-quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA) by electrospray ionization in the negative-ion mode. Samples (5 μl) diluted 3,000-fold were introduced by high-performance LC (HPLC) with an Agilent ZORBAX SB-Aq reverse-phase column (100 by 2.1 mm; particle size, 5 μm). Runs were carried out in 5 mM HCOONH4 with a multistep acetonitrile gradient of 20% from 0 to 3 min, a linear gradient of 20 to 90% from 3 to 20 min, 90% from 20 to 26 min, a linear gradient of 90 to 20% from 26 to 28 min, and 20% from 28 to 40 min. The HPLC flow rate was 0.3 ml/min. The MS conditions were nebulizer gas (N2) at 35 lb/in2, dry gas (N2) at 12 liters/min, a dry temperature of 350°C, and a capillary voltage of 4,000 V. The scanning mass range was 200 to 900 Da. The relative abundances of mono-RL (m/z 503) and di-RL (m/z 649) in different samples were, respectively, normalized to the level in PAO1.
Drop collapse assays.
Drop collapse assays were performed as previously described, with minor modifications (19, 38). Briefly, after 2 days of incubation at 37°C in nutrient broth with or without arabinose, culture samples were centrifuged at 6,000 × g for 10 min to remove the bacteria. The supernatant was filtered through a 0.22-μm membrane and serially diluted with Milli-Q distilled H2O. Aliquots (10 μl) of the diluted supernatant were spotted onto the lid surface of a Nunc 96-well plate to detect bead formation. Samples that did not form a bead were defined as having drop collapse activity.
Immunoblot analysis to detect Psl polysaccharide and PelC protein.
Psl polysaccharide was extracted from an overnight culture with an OD of 5. Psl extracts were detected by immunoblotting with anti-Psl serum as described previously (9). The induction of Pel was assayed by immunoblotting with anti-PelC serum, with modifications (5). Briefly, 1 ml of culture (OD600 of 0.5) was harvested and resuspended in 200 μl PBS. After mixing with an equal volume of 2× Laemmli buffer and boiling for 5 min, the protein concentration was measured with the BCA assay (Pierce, USA). Equal total protein amounts were loaded onto polyvinylidene difluoride membrane for immunoblotting. Psl and Pel polysaccharides were relatively quantified according to gray value calculations after dot blotting experiments and normalized to those of corresponding reference strains.
RESULTS
Increased Psl and Pel production reduces P. aeruginosa swarming motility.
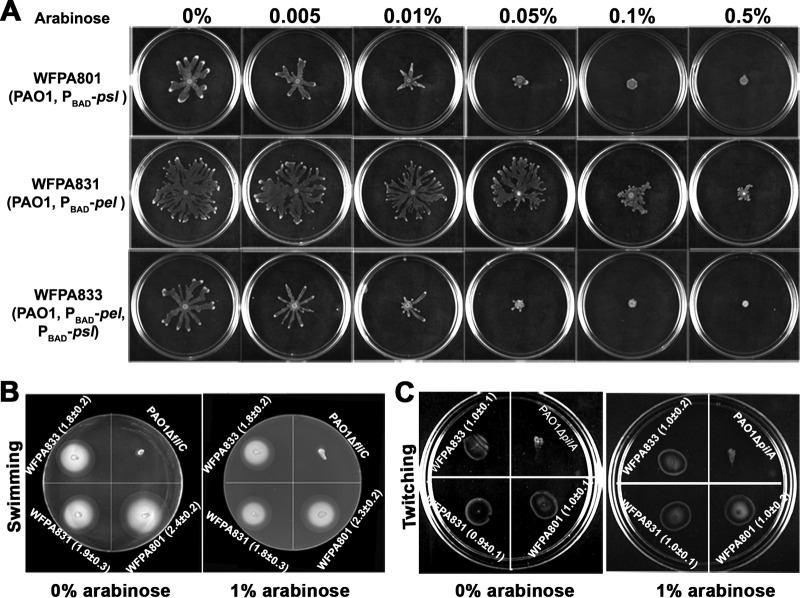
To investigate whether the production of Psl and/or Pel polysaccharides affects P. aeruginosa motility, we used several arabinose-inducible strains derived from P. aeruginosa PAO1, in which the promoter of Psl and/or the Pel gene operon was replaced with the PBAD promoter on the chromosome (Table 1). Thus, Psl and/or Pel production can be induced by arabinose (the inducer). Induction of Psl/Pel was tested by immunoblotting, and 1% arabinose displayed maximum induction (polysaccharides Psl and PelC were increased approximately 3- to 4-fold by the addition of 1% arabinose) (11; data not shown). The swarming assay showed that an increase in Psl and/or Pel production significantly reduced swarming ability (Fig. 1). Addition of 0.005 to 0.5% (wt/vol) arabinose caused a gradual reduction of swarming zones in the three polysaccharide-inducible strains, Psl-inducible strain WFPA801, Pel-inducible strain WFPA831, and Psl and Pel double-inducible strain WFPA833. WFPA801 exhibited a more severe effect than WFPA831, as the swarming zone of WFPA801 was totally abolished by 0.5% arabinose while WFPA831 still had a visible swarming zone at the same concentration (Fig. 1A). In contrast, arabinose had little effect on the swarming zone of wild-type strain PAO1, even at a concentration of 1% (Fig. 2B). Furthermore, 1% arabinose had no impact on the motility of the ΔfliC (flagellum-deficient), ΔpilA (T4P-deficient), and ΔalgC (RL synthesis-deficient) negative-control mutant strains (data not shown). These results indicated that increased Psl/Pel production reduced bacterial swarming motility. Since swarming motility requires the presence of flagella and T4P (16–18), we examined the effect of Psl and/or Pel production on flagellum-mediated swimming and T4P-driven twitching motility. The results showed that the swimming (Fig. 1B) and twitching (Fig. 1C) abilities of these three strains were similar at 1 and 0% arabinose, suggesting that Psl/Pel production did not affect the functions of flagella and T4P. These results indicated that reduction of swarming ability by Psl and/or Pel production may occur through a mechanism other than control of the activities of flagella and T4P.
FIG 1.
Motility of Psl/Pel-inducible strains WFPA801, WFPA831, and WFPA833 at various concentrations of arabinose (inducer). (A) Swarming patterns at 0 to 0.5% arabinose. (B) Swimming zones at 0 or 1% arabinose. (C) Twitching zones at 0 or 1% arabinose. The values in the images are the diameters of the corresponding swimming or twitching zones. PAO1 ΔfliC and PAO ΔpilA were used as negative controls for swimming and twitching motility, respectively.
FIG 2.
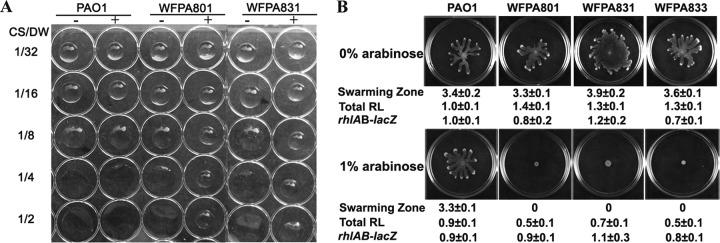
Overproduction of Psl and/or Pel polysaccharide decreases RL production. (A) The drop collapse assay shows a reduction of RL in Psl/Pel-induced strains. The image shows the bead formation of the culture supernatants of the Psl/Pel-inducible strains grown with and without arabinose. CS, culture supernatants; DW, distilled water; +, 1% arabinose; −, no arabinose. (B) Swarming patterns, swarming zones, total RL production, and corresponding rhlAB transcription of PAO1, WFPA801, WFPA831, and WFPA833 with 0 or 1% arabinose. Shown under each image are the corresponding quantitative values of swarming (average zone diameters in centimeters), total RL content, and rhlAB-lacZ transcription. The RL extracts were normalized to the level of PAO1 in the absence of arabinose (1 = 4.9 g/liter total RL by dry-weight calculation or 284.3 mg/liter RL by the anthrone colorimetric assay). Transcription of rhlAB was determined by the β-galactosidase activity of a PrhlAB-lacZ fusion integrated into the chromosomes of the corresponding strains. The values were also normalized to those of PAO1 without arabinose (1 = 116 Miller units). 0, no swarming zone.
Overproduction of Psl and/or Pel decreases RL production.
Swarming motility requires the presence of biosurfactants in addition to a flagellum and T4P (16–18). Thus, we hypothesized that the effect of Psl/Pel production on swarming may be through the control of biosurfactant synthesis. The surface tension of liquid drops correlates with the biosurfactant concentration (38). Thus, drop collapse on a hydrophobic surface provides a quick way to estimate the biosurfactant concentration of the fermentation broth. In the assay, culture supernatants were serially diluted with water and drops containing larger amounts of biosurfactants would require more dilution to reach the point where collapsing is prevented. We utilized this assay to examine the culture supernatant of wild-type strain PAO1 and Psl/Pel-inducible strains grown with and without arabinose. The culture supernatant of PAO1 showed similar drop collapse patterns in the presence of 0 and 1% arabinose (Fig. 2A), suggesting that addition of arabinose did not affect drop collapse and that the concentrations of biosurfactant were similar in the PAO1 cultures with and without arabinose. In contrast, the drop collapse of WFPA801 and WFPA831 appeared different when cultures grown with and without arabinose were compared (Fig. 2A). Without arabinose, all of the cultures exhibited similar drop collapse patterns and their drop collapse was inhibited at an 8-fold dilution. With 1% arabinose, a 4-fold dilution did not lead to drop collapse of WFPA831 and even a 2-fold dilution did not collapse the WFPA801 culture drop (Fig. 2A). This result suggested that there were less biosurfactants when Psl/Pel was overproduced (1% arabinose) than when Psl/Pel was not induced (0% arabinose).
To quantify the amount of biosurfactants, we extracted and compared the total RL from cultures of three Psl/Pel-inducible strains grown with 0 or 1% arabinose. The swarming zones of these strains are also shown under the corresponding images in Fig. 2B. Wild-type strain PAO1 showed similar swarming zones and RL production levels in the presence of 0 and 1% arabinose (indicated by the values under the corresponding images in Fig. 2B). In contrast, the swarming of the three inducible strains was totally inhibited by 1% arabinose (Fig. 2B, compare the upper and the lower panels). Consistently, the total amount of RL extracted from three inducible strains was also significantly decreased by 1% arabinose (lower panel of Fig. 2B). The total RL levels in strains WFPA801, WFPA831, and WFPA833 were reduced to 48, 65, and 46% of the PAO1 level, respectively. These results were consistent with the drop collapse assay and indicate that elevated Psl/Pel production reduced the biosynthesis of RL.
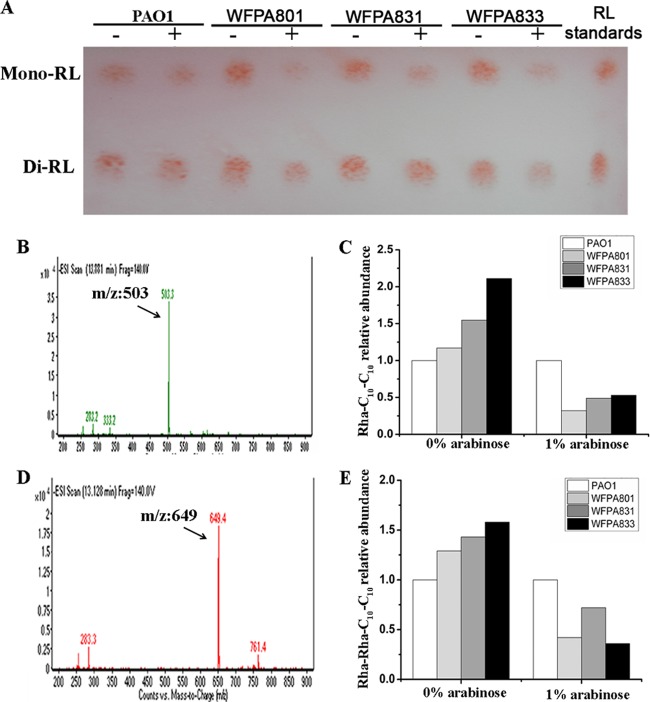
RL produced by P. aeruginosa usually contains a mixture of mono-RL and di-RL (39). To further investigate the effect of Psl/Pel on RL production, we first used TLC to examine the total RL for the separation of mono-RL and di-RL on the basis of their different retention factors (Fig. 3A). The TLC results showed remarkably reduced mono-RL and di-RL levels under the Psl-induced condition (WFPA801 plus 1% arabinose), as well as the Psl and Pel double-induced condition (WFPA833 plus 1% arabinose) (Fig. 3A). As for Pel-inducible strain WFPA831, only mono-RL showed a visible reduction with 1% arabinose (Fig. 3A). The corresponding RL extracts were also quantified by LC-MS (Fig. 3B to E), and the results further confirmed our conclusion from TLC analysis. Rha-C10-C10 and Rha-Rha-C10-C10 were the main peaks of mono-RL and di-RL (15, 19, 37). The corresponding quantification of the two peaks is shown in Fig. 3B to E. The LC-MS results show that there was a remarkable reduction in Rha-C10-C10 from the three inducible strains with 1% arabinose compared to that from PAO1 (>50% reduction) (Fig. 3C). Rha-Rha-C10-C10 from WFPA801 and WFPA833 also showed a 50% reduction compared to that from PAO1 with 1% arabinose, but WFPA831 showed an only 25% reduction under the same conditions (Fig. 3E). Interestingly, Rha-C10-C10 and Rha-Rha-C10-C10 from the three inducible strains grown without arabinose were increased to different degrees compared to those from PAO1 (Fig. 3C and E). This was consistent with the results of the total RL extract shown in the upper panel of Fig. 2B, in which RL of three Pel/Pel-inducible strains without inducer exhibited an approximately 30% increase compared to the PAO1 level. Taken together, our data indicate that enhanced Psl or Pel production reduced the synthesis of the biosurfactant RL, which could be the reason why overproduced Psl/Pel decreased the swarming motility of P. aeruginosa.
FIG 3.
TLC and LC-MS analyses of total RL extracts from the PAO1 and Psl/Pel-inducible strains. (A) TLC analysis of RL extracted from cultures of the corresponding strains grown in the absence (−) or presence (+) of 1% arabinose. (B) LC-MS peak of Rha-C10-C10 (m/z 503) (mono-RL). (C) Relative abundances of Rha-C10-C10 in corresponding RL extracts analyzed by LC-MS. (D) LC-MS peak of Rha-Rha-C10-C10 (m/z 649) (di-RL). (E) Relative abundances of Rha-Rha-C10-C10 in corresponding RL extracts analyzed by LC-MS. The relative abundances of Rha-C10-C10 and Rha-Rha-C10-C10 were normalized to the levels in PAO1 (respectively, 3,646,111 and 1,653,883 without arabinose and 2,722,080 and 2,223,691 with 1% arabinose). The corresponding peaks of mono-RL and di-RL are indicated in panels B and C. ESI, electrospray ionization.
To determine whether the reduction of RL occurred at the gene transcriptional level, we constructed a PrhlAB-lacZ transcriptional fusion in the wild-type and Psl- and/or Pel-inducible strains to detect rhlAB operon transcription. β-Galactosidase analysis showed that the transcription of rhlAB was similar in all of the strains tested, regardless of the concentration of arabinose with which the cultures were supplemented (Fig. 2B), indicating that the production of Psl and Pel does not affect the transcription of rhlAB and suggesting that RL reduction by polysaccharides occurs posttranscriptionally.
RL-deficient strains produced more Psl.
To investigate whether RL production impacts Psl synthesis, we examined two RL-deficient strains, rhlR and rhlAB mutants. Both of these mutants have lost the ability to synthesize RL and thus cannot swarm (Fig. 4A). The detection of Psl by anti-Psl serum showed that the two mutants both produced more Psl than PAO1 did (Fig. 4A). These data indicated that abolishing RL synthesis in P. aeruginosa can increase Psl production.
FIG 4.
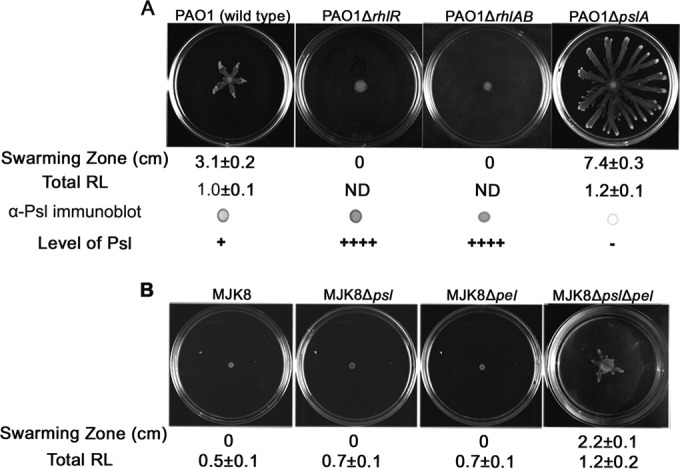
Swarming ability and total RL and/or Psl production of RL-deficient mutants and psl and pel mutant strains. (A) Swarming pattern, corresponding swarming zones, Psl levels, and total RL production of PAO1 and the PAO1 isogenic rhlR, rhlAB, pslA, and galU mutants. (B) Swarming and RL production of MJK8, the isogenic psl and/or pel mutant, and native Psl-negative strain PA14. The mean swarming zone diameter and total RL production are presented under each corresponding image. The RL extracts were normalized to the level of PAO1 (214 mg/liter). Psl was detected by immunoblotting with Psl antiserum. ++++, Psl level 4-fold that of PAO1; −, Psl negative; 0, no swarming zone; ND, not determined.
Psl-negative strains had a hyperswarming phenotype.
To test whether loss of Psl also affects swarming, we used several Psl-negative strains. Deletion of pslA, the first gene of the psl operon, resulted in loss of Psl production (9). Thus, we first tested the swarming of the ΔpslA mutant, which exhibited a hyperswarming phenotype. Its swarming zone was 2.4-fold larger than that of PAO1 (Fig. 4A). Consistently, the total RL level of the ΔpslA mutant was 20% greater than that of PAO1 (Fig. 4A).
Growth of P. aeruginosa in biofilms and chronic CF airway infections generate rugose small-colony variants (RSCVs) (40). Both clinic- and in vitro-derived biofilm RSCVs display increased production of the Pel and Psl polysaccharides and loss of motility due to elevated cellular c-di-GMP levels (40). To investigate whether the effect of polysaccharide production on swarming holds true in RSCVs, we examined MJK8, an RSCV derived from a laboratory-grown biofilm of PAO1, and its isogenic pel and psl mutants (Table 1) (41). MJK8 and its pel or psl single deletion mutant could not swarm; however, the swarming of a pel psl double mutant did recover to 70% of the PAO1 level (Fig. 4B). This is a remarkable change because flagellum biosynthesis and activity are inhibited by the high level of c-di-GMP in MJK8 (41). Strikingly, the total RL production of MJK8ΔpelΔpsl was higher than that of PAO1 and similar to that of the PAO1ΔpslA strain (Fig. 4A). Moreover, the total RL production of MJK8 was also similar to that of WFPA833 (PBAD-psl and PBAD-pel) at 1% arabinose (Fig. 2B, lower panel). These data indicate that the impact of Psl/Pel production on swarming and RL biosynthesis not only was found in the artificial PBAD-induced system but also occurred in a physiologically relevant polysaccharide-overproducing strain.
Coordinated synthesis of exopolysaccharides and RL is likely mediated by competition for common sugar precursors.
Overproduced Psl/Pel decreased the synthesis of RL, yet a lack of RL synthesis increased the production of Psl. This phenomenon was similar to the previous report about the biosynthesis of Psl, alginate, and lipopolysaccharide (LPS), which competed with each other for the common sugar precursor Man-1-P converted by AlgC (11). It has been suggested that the Psl and Pel synthesis pathways compete for the sugar precursor Glc-1-P catalyzed by AlgC (11). As RL synthesis also requires AlgC to provide the common sugar precursor Glc-1-P (25) and the above-described data indicate coordinated biosynthesis of Psl/Pel and RL, this suggested that there was very likely competition for common sugar precursors among the biosynthesis pathways of RL, Psl, and Pel (Fig. 5A). Since AlgC is the only enzyme in P. aeruginosa to catalyze Glc-1-P for Psl, RL, and Pel, we assumed that an additional copy of algC expressed by a plasmid should increase the production of Psl and RL. To test this hypothesis, we compared the Psl and RL production and swarming of PAO1 containing plasmid pLPS188 with constitutively expressed algC (PAO1/algC) and those of PAO1 with the vector (PAO1/vector). In support of the hypothesis, PAO1/algC showed a hyperswarming pattern and produced more RL and Psl than PAO1/vector (Fig. 5B to D). In summary, our data suggest that the biosynthesis of Psl, Pel, and RL involves the same sugar precursors and leads to the coordination of swarming motility and the biosynthesis of exopolysaccharides Psl and Pel.
FIG 5.
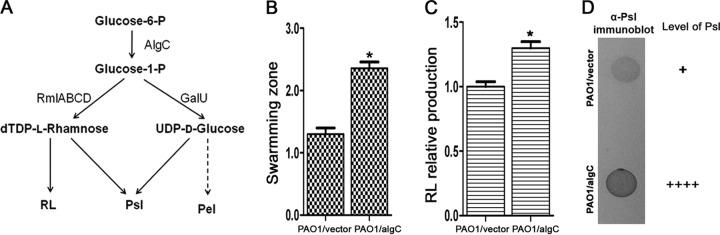
The biosynthesis pathways of RL, Psl, and Pel have the sugar precursor Glc-1-P (catalyzed by AlgC) in common. (A) Schematic representation of the P. aeruginosa PAO1 metabolic routes for the biosynthesis of Psl, Pel, and RL. The solid lines represent known metabolic routes, and the dashed line represents a hypothetical route. (B) Swarming zone of PAO1/algC (algC was constitutively expressed by plasmid pLPS188.) and control strain PAO1/vector. (C) Comparison of RL production in PAO1/algC and PAO1/vector. The amount of RL was normalized to the PAO1/vector level (261 mg/liter). (D) Comparison of PAO1/algC and PAO1/vector Psl levels. *, P < 0.05.
Enhanced expression of RhlR/RhlI can decrease Psl production.
RL production is strictly regulated by the RhlR/RhlI QS system. Once RhlR binds the QS signal molecule synthesized by RhlI, it activates the transcription of rhlAB to synthesize RL. On the basis of the precursor competition theory proposed above (Fig. 5A), increasing RL would reduce Psl production if more common sugar precursors were used for RL biosynthesis. Then enhanced expression of RhlR/RhlI would decrease the synthesis of Psl. To test this hypothesis, we constructed PBAD-rhlRI by using pHerd20T as the vector. The PBAD-rhlRI plasmid was transformed into PAO1 and the rhlR mutant, respectively, to obtain two rhlRI-inducible strains, PAO1/rhlRI and the ΔrhlR/rhlRI mutant. The Psl synthesis of both strains reduced gradually while the expression of rhlRI was induced by various concentrations of arabinose (0.2 to 1%) (Fig. 6B). Their swarming zone size (Fig. 6A) and RL production (Fig. 6B) were also increased accordingly. These data suggest that the RhlR/RhlI QS system can influence Psl production through regulation of RL expression. These results also confirm that there was competition for the common sugar precursors between the biosynthesis pathways of Psl and RL, given that increased RL biosynthesis reduced Psl production (Fig. 6B) and vice versa (Fig. 2B).
FIG 6.
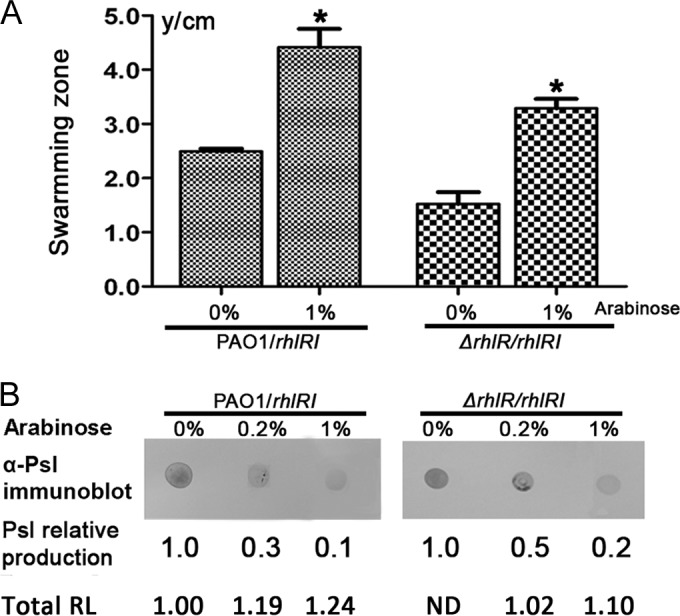
Effect of RhlR/RhlI QS system on Psl polysaccharide biosynthesis. (A) Swarming zones of rhlRI-inducible PAO/rhlRI and ΔrhlR/rhlRI mutant strains grown with or without arabinose. (B) Psl and RL production of PAO/rhlRI and the ΔrhlR/rhlRI mutant at various concentrations of arabinose. The amount of RL was normalized to the PAO1/vector level (214 ± 3 mg/liter). *, P < 0.05; ND, not determined.
DISCUSSION
Exopolysaccharides and motility are significant contributing factors in the formation of bacterial biofilms. Understanding how exopolysaccharides and motility cooperate to shape the architecture of biofilm will shed light on the development of strategies against biofilm-related concerns, such as chronic and persistent infections. In this study, we demonstrated that the production of exopolysaccharides Psl and Pel affected bacterial swarming but not swimming and twitching. This was accomplished by control of RL production.
Recently, it was discovered that Psl can also function as a signaling molecule to stimulate diguanylate cyclases to produce more intracellular second messenger c-di-GMP (10), which is known to regulate the production of polysaccharide and bacterial motility. Elevated intracellular concentrations of c-di-GMP reduced bacterial motility by affecting flagellum and T4P activities (10, 26–28). Our data showed that production of Psl and Pel had little impact on bacterial swimming and twitching motility (Fig. 1B and C), indicating that flagellum and T4P activities were not inhibited. This also suggested that c-di-GMP contributed little to the effect of Psl and/or Pel on swarming motility and RL production. More evidence came from the results obtained with MJK8. This RSCV strain has a high level of cellular c-di-GMP (41); even so, deletion of psl and pel from MJK8 recovers its RL production and some swarming motility (Fig. 4B).
We proposed that biosynthesis of Psl, Pel, and RL competes for common sugar precursors. Psl is a repeating pentasaccharide containing d-Man, d-Glc, and l-rhamnose (9). The rhamnose and glucose precursors are both converted from Glc-1-P synthesized by AlgC (Fig. 6A). d-Man is converted from Man-1-P (which is also catalyzed by AlgC) and is shared by the Psl, alginate, and LPS biosynthesis pathways. We showed in this study that Psl production led to more severe inhibition of swarming motility and RL synthesis than did that of the glucose-rich exopolysaccharide Pel (Fig. 1 to 3). One explanation for this phenomenon is that the synthesis of Psl may use more sugar precursors catalyzed by AlgC than that of Pel does. P. aeruginosa RL usually contains a mixture of mono-RL and di-RL. Various RL play different roles in swarming motility. Mono-RL acts as a wetting agent, di-RL promotes tendril formation and migration, and they work together to form the swarming pattern of P. aeruginosa (39). Our TLC and LC-MS results indicate that Pel production decreased mono-RL similar to that of Psl production yet had little effect on di-RL. This can be a way to fine tune the ratio of mono-RL and di-RL. RL not only play critical roles in P. aeruginosa biofilm formation (42, 43) but also serve as important biosurfactants because of their broad applications in oil recovery, cosmetics, environmental protection, etc. (20). Thus, our results could also provide potential strategies for industry to upgrade RL production.
QS circuits allow bacteria to coordinate their gene expression in a cell density-dependent manner. P. aeruginosa employs three QS signaling systems (LasR/LasI, RhlR/RhlI, and PQS). So far, little is known about the involvement of QS in the regulation of exopolysaccharides in P. aeruginosa. Our data suggest that the RhlR/RhlI signaling system may indirectly control Psl and/or Pel production by coordinating RL production. The synthesis of RL is regulated by the RhlR/RhlI system, which is activated during the maturation stage of P. aeruginosa biofilm development (44). It has been reported that little Psl is found in the center of the microcolony at the maturation stage, which is associated with the preparation of future seeding dispersal (4). The center of the microcolony is the microenvironment with the densest bacterial population and the upregulated RhlR/RhlI, which activates the synthesis of RL. Increasing the synthesis of RL enhances swarming and may further reduce exopolysaccharide production. This may be a strategy by which P. aeruginosa balances polysaccharide production and bacterial motility during biofilm development to allow seeding dispersal to occur. Consistent with this scenario, the biofilm of an RL hyperproducer yields earlier seeding dispersal than that of wild-type strain PAO1 (43).
Swarming is the fastest known bacterial mode of surface translocation and enables the rapid colonization of a nutrient-rich environment and host tissues. In this study, we revealed how P. aeruginosa coordinates the production of biofilm exopolysaccharides and swarming motility without affecting the flagella and T4P. This was accomplished by competition for sugar precursors common to the biosynthesis pathways of exopolysaccharides and the biosurfactant RL. Since RL synthesis in P. aeruginosa is regulated by the QS signaling system and a reduction or increase in RL and RhlRI can impact exopolysaccharide production, our data suggest a plausible strategy by which QS signals may regulate biofilm matrix exopolysaccharides and swarming motility in bacterial communities in a coordinating manner.
ACKNOWLEDGMENTS
We thank Alan K. Chang at Liaoning University and Di Wang at the Institute of Microbiology, Chinese Academy of Sciences, for their contribution to the revision of the manuscript and Matthew Parsek at the University of Washington for providing the anti-PelC serum and MJK8 strains.
This work was supported by Chinese Academy of Science grant KSCXZ-YW-BR-5 (L.Z.M.), National Natural Science Foundation of China grants 31270177 (L.Z.M.) and 31400094 (S.W.), and the National Basic Research Program of China (973 Program, grant 2014CB846002) (L.Z.M.).
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Wei Q, Ma L. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 14:20983–21005. 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L-Y, Jackson K, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213–8221. 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5(3):e1000354. 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14:1913–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457–4465. 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690. 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Wang S, Wang D, Parsek MR, Wozniak DJ. 2012. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 65:377–380. 10.1111/j.1574-695X.2012.00934.x. [DOI] [PubMed] [Google Scholar]
- 9.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638. 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 109:20632–20636. 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. 2012. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14:1995–2005. 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocchetta HL, Burrows LL, Lam JS. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63:523–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghafoor A, Jordens Z, Rehm BHA. 2013. Role of PelF in Pel polysaccharide biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 79:2968–2978. 10.1128/AEM.03666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304. 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 15.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277. 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 16.Köhler T, Curty LK, Barja F, van Delden C, Pechère J-C. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990–5996. 10.1128/JB.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66:493–520. 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 18.Leech AJ, Mattick JS. 2006. Effect of site-specific mutations in different phosphotransfer domains of the chemosensory protein ChpA on Pseudomonas aeruginosa motility. J. Bacteriol. 188:8479–8486. 10.1128/JB.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Déziel E, Lépine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 20.Soberón-Chávez G, Lépine F, Déziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68:718–725. 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhu K, Rock CO. 2008. RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 190:3147–3154. 10.1128/JB.00080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochsner UA, Fiechter A, Reiser J. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269:19787–19795. [PubMed] [Google Scholar]
- 23.Ochsner UA, Reiser J. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 92:6424–6428. 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberón-Chávez G. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708–718. 10.1046/j.1365-2958.2001.02420.x. [DOI] [PubMed] [Google Scholar]
- 25.Olvera C, Goldberg JB, Sánchez R, Soberón-Chávez G. 1999. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179:85–90. 10.1111/j.1574-6968.1999.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuchma SL, Ballok AE, Merritt JH, Hammond JH, Lu W, Rabinowitz JD, O'Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 192:2950–2964. 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Römling U. 2013. Microbiology: bacterial communities as capitalist economies. Nature 497:321–322. 10.1038/nature12103. [DOI] [PubMed] [Google Scholar]
- 28.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc. Natl. Acad. Sci. U. S. A. 110:18478–18483. 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Parsek MR, Wozniak DJ, Ma LZ. 2013. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 15:2238–2253. 10.1111/1462-2920.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:4885–4890. 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen SE, Fecycz IT, Campbell JN. 1980. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J. Bacteriol. 144:844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoang T, Kutchma A, Becher A, Schweizer H. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–71. 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- 33.Hoang T, Karkhoff-Schweizer R, Kutchma A, Schweizer H. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 28:77–86. [DOI] [PubMed] [Google Scholar]
- 34.Hassett DJ, Woodruff W, Wozniak DJ, Vasil ML, Cohen MS, Ohman DE. 1993. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding iron- and manganese-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J. Bacteriol. 175:7658–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Hori K, Marsudi S, Unno H. 2002. Simultaneous production of polyhydroxyalkanoates and rhamnolipids by Pseudomonas aeruginosa. Biotechnol. Bioeng. 78:699–707. 10.1002/bit.10248. [DOI] [PubMed] [Google Scholar]
- 37.Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R. 1999. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1440:244–252. 10.1016/S1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 38.Jain DK, Collins-Thompson DL, Lee H, Trevors JT. 1991. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods 13:271–279. 10.1016/0167-7012(91)90064-W. [DOI] [Google Scholar]
- 39.Tremblay J, Richardson A-P, Lépine F, Déziel E. 2007. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9:2622–2630. 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 40.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191:3492–3503. 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirisits MJ, Prost L, Starkey M, Parsek MR. 2005. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 71:4809–4821. 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davey ME, Caiazza NC, O'Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027–1036. 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boles BR, Thoendel M, Singh PK. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57:1210–1223. 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 44.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154. 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GKS, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 46.Coyne MJ, Russell KS, Coyle CL, Goldberg JB. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuster M, Greenberg EP. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8:287. 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]