Abstract
Ring-hydroxylating dioxygenases (RHDs) play a crucial role in the biodegradation of a range of aromatic hydrocarbons found on polluted sites, including polycyclic aromatic hydrocarbons (PAHs). Current knowledge on RHDs comes essentially from studies on culturable bacterial strains, while compelling evidence indicates that pollutant removal is mostly achieved by uncultured species. In this study, a combination of DNA-SIP labeling and metagenomic sequence analysis was implemented to investigate the metabolic potential of main PAH degraders on a polluted site. Following in situ labeling using [13C]phenanthrene, the labeled metagenomic DNA was isolated from soil and subjected to shotgun sequencing. Most annotated sequences were predicted to belong to Betaproteobacteria, especially Rhodocyclaceae and Burkholderiales, which is consistent with previous findings showing that main PAH degraders on this site were affiliated to these taxa. Based on metagenomic data, four RHD gene sets were amplified and cloned from soil DNA. For each set, PCR yielded multiple amplicons with sequences differing by up to 321 nucleotides (17%), reflecting the great genetic diversity prevailing in soil. RHDs were successfully overexpressed in Escherichia coli, but full activity required the coexpression of two electron carrier genes, also cloned from soil DNA. Remarkably, two RHDs exhibited much higher activity when associated with electron carriers from a sphingomonad. The four RHDs showed markedly different preferences for two- and three-ring PAHs but were poorly active on four-ring PAHs. Three RHDs preferentially hydroxylated phenanthrene on the C-1 and C-2 positions rather than on the C-3 and C-4 positions, suggesting that degradation occurred through an alternate pathway.
INTRODUCTION
Bioremediation procedures used to treat polluted sites rely on specialized microorganisms that can transform or utilize organic pollutants as carbon sources. Knowledge on pollutant biodegradation mainly arises from studies on pure strains that have been isolated from contaminated sites. For example, numerous bacterial strains able to degrade polycyclic aromatic hydrocarbons (PAHs) have been used to elucidate relevant degradation pathways and characterize some of the enzymes involved (1, 2). Nevertheless, exploration of the diversity of soil bacteria using culture-independent molecular techniques revealed that soils contain a great taxonomic richness and established that bacterial isolates described thus far represented no more than 5% of the bacterial diversity (3). As a consequence, it could be anticipated that bacteria responsible for PAH removal in situ would be largely unknown and would differ from previously studied isolates. Accordingly, sphingomonads detected on polluted sites by 16S rRNA sequence analysis were found to be different from described species in this taxonomic group (4). In the last decade, the implementation of stable isotope probing (SIP) to track PAH degraders led to the discovery of new bacteria with interesting biodegradation potential (5, 6). Moreover, SIP approaches also revealed that most PAH-degrading bacteria identified in contaminated soils were affiliated with uncultured microorganisms (5, 7–10). Notably, betaproteobacteria were shown to form a dominant subgroup of the phenanthrene-degrading community found in polluted soils, suggesting that they played a major role in PAH degradation in soil (7, 8). Specifically, soil bacteria utilizing phenanthrene included several taxa related to Burkholderiales, as well as unclassified Rhodocyclaceae. Closely related representatives of the latter family have been found in contaminated soils from America (9), Europe (7), and a tropical region of Africa (11). A Rhodocyclaceae member appeared as the main bacterium in a consortium obtained by enrichment from soil after repeated cultivation on a pyrene-containing minimal medium (12). Although the bacterium could not be isolated in pure culture, a metagenomic analysis of the DNA isolated from a simplified consortium consisting of the Rhodocyclaceae member and three other detectable bacterial species gave insights into their metabolic capabilities. Eight sets of genes coding for ring-hydroxylating dioxygenases were identified in separate contigs. Ring-hydroxylating dioxygenases (RHDs) are multicomponent metalloenzymes, which catalyze the first step in the bacterial degradation of various aromatic hydrocarbons. PAH-specific RHDs belonged to two families based on phylogenetic comparison of available sequences in public databases (13–15). Six of the RHD enzymes mentioned above were cloned and shown to catalyze the hydroxylation of several PAHs, including pyrene (12).
The goal of the present study was to examine the metabolic potential of PAH degraders in a polluted soil by combining DNA-SIP with metagenomics. Although this combination has been recognized as a promising new approach in soil bioremediation studies (16), it has not been frequently implemented thus far (17). In the present study, we investigated the soil bacterial community of a facility collecting the road runoffs of a highway. A SIP analysis of phenanthrene-utilizing bacteria in this soil previously demonstrated the preponderance of Betaproteobacteria, especially members of the Acidovorax, Rhodoferax, and Hydrogenophaga genera, as well as unclassified Rhodocyclaceae (7). Moreover, a PCR-based analysis of the diversity of RHDs associated with phenanthrene degradation in the same soil revealed the occurrence of five groups of enzymes, three of which were poorly related to known dioxygenases, with sequence identities in the 60 to 80% range with best matches in databases (18). To get further information on phenanthrene degradation in soil, we have undertaken a metagenomic analysis involving a scaled-up SIP experiment in order to isolate enough labeled DNA for subsequent shotgun sequencing. From the resulting metagenomic data, four sets of RHD genes were cloned and overexpressed in Escherichia coli. The RHDs were found to be distantly related to the enzymes of known bacterial isolates but shared high similarities with the RHDs found in the pyrene-degrading consortium mentioned above. The catalytic properties of the enzymes with respect to the oxidation of two- to four-ring PAHs were determined.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in the present study are listed in Table 1. E. coli strains were grown in rich medium (Luria-Bertani) at 37°C with appropriate antibiotics as previously described (19).
TABLE 1.
Bacterial strains and plasmids used in this study
| Stain or plasmid | Description/genotypea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| NEB 5-α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 ϕ80Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England BioLabs |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Merck, Millipore, Novagen |
| Plasmids | ||
| pJET1.2 | Ampr; cloning plasmid | Thermo Fisher Scientific |
| pET15b | Ampr; expression plasmid | Merck, Millipore |
| pIZ1036 | Kmr; broad-range expression plasmid | 42 |
| pIBA34 | pIZ1036 carrying phnA3 and phnA4 from strain CHY-1 | 18 |
| pJCA1 | pJET1.2 carrying pahAb | This study |
| pJCA3 | pJET1.2 carrying pahAa | This study |
| pJCA5 | pJCA3 carrying pahAb from pJCA1 downstream of pahAa | This study |
| pJCA6/pJCA7 | pJET1.2 carrying two different pahAc5-pahAd5 amplicons | This study |
| pJCA8/pJCA9 | pJET1.2 carrying two different pahAc4-pahAd4 amplicons | This study |
| pJCA10/pJCA11 | pJET1.2 carrying two different pahAc2-pahAd2 amplicons | This study |
| pJCA13/pJCA14 | pJET1.2 carrying two different pahAc8-pahAd8 amplicons | This study |
| pCAE1 | pET15b carrying pahAc2-pahAd2 from pJCA10 | This study |
| pCAE4 | pET15b carrying pahAc8-pahAd8 from pJCA14 | This study |
| pCAE5 | pET15b carrying pahAc4-pahAd4 from pJCA9 | This study |
| pCAE7 | pET15b carrying pahAc5-pahAd5 from pJCA7 | This study |
| pCAZ1 | pIZ1036 carrying the pahAa-pahAb insert from pJCA5 | This study |
Ampr, ampicillin resistance; Kmr, kanamycin resistance.
SIP experiments and isolation of labeled DNA from soil.
Soil sampling was carried out on 29 October 2010, on a previously described study site, which is designed to collect road runoffs from a highway (7). SIP experiments were conducted in 250-ml microcosms containing 20 g of soil. Briefly, the soil of each microcosm was mixed with 2.5 mg of [13C]phenanthrene dissolved in 0.25 ml of dimethyl sulfoxide. Microcosms were closed with rubber stoppers and then incubated in the dark for 6 days at 25°C under static conditions. Microcosms that did not receive phenanthrene were incubated under identical conditions to serve as controls. Mineralization of the labeled substrate was monitored by gas chromatography-mass spectrometry (GC/MS) quantification of 13CO2 in the gas phase (7). DNA was extracted from 10-g lots of wet soil using a PowerMax soil DNA extraction kit (Mo Bio laboratories) and then separated by CsCl isopycnic ultracentrifugation as previously described (7). The heavy fractions of the gradient containing [13C]DNA were subjected to PCR tests as illustrated in Fig. S1 in the supplemental material. The primer pair used (RHD-Beta-Grp1f and RHD-Beta-Grp1r) was previously shown to amplify a 950-bp fragment of the RHD alpha subunit genes from soil Betaproteobacteria (18). Fractions that responded positively to the PCR test were pooled from repeated preparations involving 18 soil DNA extractions, followed by gradient fractionation, yielding a total of 30.6 μg of labeled DNA.
Metagenomic DNA sequencing and analysis.
Sequencing was carried out using 454 pyrosequencing (Roche Biosciences), as well as Illumina technology. An 8-kb paired-end library was constructed according to the protocol for the 454 Titanium apparatus. Two sequencing runs were performed generating 784.5 Mbp of raw data. A library of 362-bp inserts (average size) was constructed according to the Illumina HiSeq 2000 protocol. Six lanes of 100-bp pair-end sequencing were used to generate 130.4 Gbp of raw data.
The Titanium sequences were assembled by Newbler (version MapAsmResearch-04/19/2010-patch-08/17/2010), and the sequences of the scaffolds were corrected using the Illumina sequences (20). Resulting assembly was composed of 69,435 contigs (11,909 of them being larger than 500 bp, summing up to 9,014,532 bp) organized in 288 scaffolds for a cumulative scaffold size of 824,483 bp. Contigs larger than 500 bp were scanned for 16S ribosomal genes by BLASTn similarity search against the Greengenes otu97 database (release 13-5) (21). Contigs with significant matches (>90% nucleic identity over at least 300 bp) were retained for subsequent analysis. Selected sequences were also compared to entrees in the NCBI nucleotide database using BLASTn.
Predictions of coding regions from the obtained set of contigs were performed with MetaGeneAnnotator with default parameters (22), resulting in a total of 85,156 coding sequences. Predicted protein sequences were compared to the UniProtKB database (release 2014/01/24) using the LASSAP implementation of the BLASTp algorithm with a threshold E value of 1e−5 (23).
Cloning of RHD-encoding genes.
PCR amplifications of selected RHD-encoding genes were carried out on a Tpersonal thermocycler (Whatman Biometra), using primer pairs depicted in Table 2 with unfractionated DNA from phenanthrene-spiked soil as the template. Reactions were performed in a 25- or 50-μl total volume containing 1× polymerase buffer, 1.5 mM MgSO4, 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.3 μM concentrations of each primer, 2 ng/μl of metagenomic DNA, 0.02 U/μl of high-fidelity DNA polymerase, and 40 ng/μl of phage T4 gp32 (New England BioLabs). The KOD Hot Start DNA polymerase was most commonly used (Merck Novagen), under the following PCR conditions: DNA denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing for 15 s at a temperature adequate for the chosen primer pair, and extension at 72°C for 30 s/kb. For the amplification of the pahAa and pahAb genes, a touchdown program was implemented whereby the annealing temperature was lowered from 53 to 49°C by 1° increments during the first five cycles of the PCR. Occasionally, the KOD enzyme was replaced by the Q5 high-fidelity DNA polymerase (New England BioLabs) with the following modifications: the extension step was increased to 1 min/kb, and a final 2-min extension was added at the end of the PCR program. PCRs where the template DNA was obtained from soil samples not supplemented with phenanthrene were run as controls.
TABLE 2.
Oligonucleotides used for PCR amplification of RHD genes
| Designation | Sequence (5′–3′)a | Targeted gene(s) |
|---|---|---|
| C095-F | GAAGGAGATATATTATGGATAATAATTTCACCATC | pahAa |
| C095-R | CGTCGACTTAATAAAAACGGTCAAAATGC AACTG | pahAa |
| C113-F | GCTCGAGGAGATATTGTATGAATACCCGTGTCAA | pahAb |
| C113-R | CTTATTCTACTTCTGCCTCAA | pahAb |
| C951-F | GCATATGATAAATATAGATGATCTGATTGA | pahAc2-pahAd2 |
| C341-Rb | CTCGAGTTAAAATAAAGTGTTCATGTTGCTATC | pahAc2-pahAd2 |
| C763-F | GCATATGATGAAGCCAAGCGAGTTGATTGA | pahAc4-pahAd4 |
| C569-F | GCATATGGTCGATGTAAACAGTCTG | pahAc3-pahAd3 |
| C2271-R | GCCTACCCAATGGCTGATGCC | pahAc3-pahAd3 |
| C451-F1 | GAAGGAGATATCATATGAATGAATGGCTGGAGGAG | pahAc5-pahAd5 |
| C451-R | GTCTAGATCAGAAAAACATATTCAGATTTTTATC | pahAc5-pahAd5 |
| C451-F2*c | GAAGGAGATATCATATGAAAAACATTAACTATCAGGAAC | pahAc5-pahAd5 |
| JCA7-R2* | GGCTCGAGATCAGAAAAACATATTCAGA | pahAc5-pahAd5 |
| C5241-F | GCATATGTTCGATATCAAGAATTTAATCAA | pahAc8-pahAd8 |
| C427-R2 | CTCGAGTTACAAGATAAACAACAAGTTTTTCCC | pahAc8-pahAd8 |
Letters in italics indicate restriction sites for one of the following enzymes: NdeI, SalI, XhoI, or XbaI.
This primer was used for the amplification of pahAc4-pahAd4, together with C763-F.
Primers used for subcloning pahAc5-pahAd5 in pET15b are indicated by an asterisk (*).
PCR products were purified by agarose gel electrophoresis, followed by DNA fragment extraction with the NucloSpin Extract II kit (Macherey-Nagel), and then cloned using the CloneJET PCR cloning kit (Thermo Scientific). Plasmid inserts were sequenced on both strands by Eurofins MWG/Operon (Germany). DNA sequences obtained were analyzed using ApE software (http://biologylabs.utah.edu/jorgensen/wayned/ape/) and compared to those in databases using BLASTn available on the EMBL EBI website. Search for proteins similar to our translated sequences was performed in the UniProt knowledgebase using BLASTp. Neighbor-joining analysis of RHD alpha subunit sequences was done on the phylogeny.fr website using the Oneclick option (24).
Construction of plasmids for the overexpression of RHD genes in E. coli.
RHD genes were cloned into plasmid pET15b and overexpressed in strain BL21(DE3). The gene pairs pahAc2-pahAd2 and pahAc8-pahAd8 were subcloned as NdeI-XhoI fragments from plasmids pJCA10 and pJCA14 into pET15b to give plasmids pCAE1 and pCAE4, respectively. Since the pahA4-pahAd4 sequence contains an internal NdeI site, a 2-kb fragment carrying the two genes was recovered after partial digestion of pJCA9 with NdeI and XhoI and subsequently cloned into pET15b to give pCAE5. For subcloning of pahAc5-pahAd5, a 1.8-kb fragment carrying these two genes was amplified with the primers C451F2 and JCA7-R2 (Table 2) using pJCA7 as the template, and then the PCR product was digested by NdeI and XhoI before being cloned into pET15b to give pCAE7.
Plasmid pCAZ1 carrying the pahAa and pahAb genes, which encode an NAD(P)H oxidoreductase and a ferredoxin, respectively, was constructed in two steps. First, the pahAb sequence was isolated from pJCA2 as a XhoI-NcoI fragment and then cloned downstream of pahAa in pJCA3 digested by SalI and NcoI. The resulting plasmid, pJCA5, was cut by XhoI and XbaI to isolate a 1.5-kb fragment encompassing the two genes, which were then cloned into pIZ1036 digested by SalI and XbaI, to give pCAZ1, where the two genes are under the control of the tac promoter (Table 1).
RHD overproduction and assays.
Gene overexpression was performed in strain BL21(DE3), which had been cotransformed with pCAZ1 and one of the pET15b derivatives carrying a pair of RHD genes. In some experiments, plasmid pCAZ1 was replaced by pIBA34, thus allowing the expression of an alternate pair of electron carriers. Recombinant E. coli strains were grown at 37°C in LB medium until the optical density at 600 nm (OD600) reached ∼1.0. The cultures were then induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and further incubated for 21 to 22 h at 25°C under orbital shaking at 180 rpm. The cells were harvested by centrifugation, washed, and resuspended to an OD600 of 2.0 in the minimal M9 medium containing 10 mM glucose. For PAH oxidation assays, the suspension was distributed into 2-ml Eppendorf tubes (1 ml/tube), which had received 1.0 μmol of chosen PAH, applied as a 20 mM stock solution in acetone. Three replicates per PAH were prepared. The cells were incubated for 6 h (naphthalene, biphenyl, and phenanthrene) or 24 h (anthracene, fluorene, pyrene, and fluoranthene) at 25°C with vigorous shaking. The cell suspensions were then centrifuged, and the supernatant fluid was extracted with an equal volume of ethyl acetate in the presence of 10 μM 2,3-dihydroxybiphenyl (Sigma-Aldrich), added as an internal standard. Dried extracts were taken up in 0.2 ml of acetonitrile and analyzed by GC/MS as n-butylboronate derivatives as previously described (25). Dihydrodiols were quantified using calibration curves obtained by analyzing samples of purified naphthalene 1,2-dihydrodiol or phenanthrene 3,4-dihydrodiol in the 5 to 100 μM range (26) and normalized with respect to the internal standard concentration. Pyrene oxidation products were instead acetylated by treating dried extracts with 40 μl of pyridine and 60 μl of acetic anhydride for 30 min at 60°C. Samples were analyzed by GC/MS in single ion monitoring mode (with 260 and 320 as the elected m/z values), using purified pyrene 4,5-dihydrodiol as a standard for calibration. The pyrene dihydrodiol, as well as other diols mentioned above, were prepared as previously described (27). Activities are expressed as the amount of dihydrodiol formed (μM) per hour per ml of culture, normalized to a density of 1.0 (OD600). Fluorene oxidation products were also analyzed after trimethylsilylation as previously described (26).
SDS-PAGE analysis.
Expression of recombinant RHDs in E. coli was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% slab gels using a SE 260 Mighty Small II system (Hoefer). Gels were processed and stained as previously described (26).
Nucleotide sequence accession numbers.
The gene sequences described in the present study have been deposited in the European Nucleotide Archive database under accession numbers HG918050 to HG918067. Metagenomic sequences are available in GenBank under project ID number PRJEB5694.
RESULTS
Overview of sequence predictions from the genomic DNA of soil phenanthrene degraders.
The first goal of the present study was to isolate enough genomic DNA from soil bacteria able to degrade PAHs in order to afford subsequent shotgun sequencing. A large-scale labeling experiment with [13C]phenanthrene as probe was implemented, consisting of several SIP experiments performed according to a procedure that enabled us to identify phenanthrene degraders in the soil of the same study site (7). In soil microcosms, labeled phenanthrene underwent rapid metabolization after a 2-day lag period, as shown by recording the time course of mineralization (see Fig. S2 in the supplemental material). At day 6, DNA was extracted from soil and then subjected to isopycnic separation by repeated CsCl gradient centrifugations. For each run, heavy fractions containing the labeled DNA were selected on the basis of a PCR test as depicted in Fig. S1 in the supplemental material. The primers used in this test targeted phenanthrene-specific RHD genes previously identified by SIP in the same soil (18), The fractions that responded positively to this PCR test most likely contained DNA from soil PAH-degrading bacteria. Pooling the fractions resulted in a total of 30.6 μg of DNA, which was subjected to both Roche 454 and Illumina sequencing.
Analysis of metagenomic data allowed the assembly of 69,435 contigs, including 11,909 contigs longer than 500 bp, the longest comprising 8,050 bp. Sequence annotation revealed that most open reading frames (ORFs) were partial as a consequence of the short length of the contigs. Nevertheless, a total of 85,156 coding sequences were deduced from the metagenomic data, including 63,802 sequences that showed at least one match in the UniprotKB database. About 48% of these protein sequences were predicted to belong to Betaproteobacteria (see Table S1 in the supplemental material). In addition, 11 sequences coding for 16S rRNA subunits were identified, four of which were affiliated with Betaproteobacteria (Table 3). These results are consistent with previous findings showing that dominant PAH degraders in the same soil belonged to this bacterial class (7). Moreover, seven 16S rRNA gene sequences matched the operational taxonomic units (OTU) previously affiliated with members of the Gammaproteobacteria and Betaproteobacteria, which likely contributed greatly to phenanthrene degradation, including two sequences from uncultured Rhodocyclaceae (Table 3).
TABLE 3.
Affiliation of 16S ribosomal sequences identified in the metagenomic data set obtained from labeled DNA
| Proposed affiliation | Contig | Length (bp) | Best-match GenBank accession no. | Query cover (%) | Identity (%) | Relevant OTUa |
|---|---|---|---|---|---|---|
| Acidobacteria | ||||||
| Unclassified | 815 | 1,275 | FQ659841 | 100 | 96 | OTU397 |
| Unclassified | 9991 | 531 | JN178653 | 67 | 97 | NF |
| Alphaproteobacteria | ||||||
| Bradyrhizobiaceae | 6706 | 598 | JN869027 | 86 | 94 | NF |
| Betaproteobacteria | ||||||
| Rhodocyclaceae | 6757 | 600 | FQ660427 | 100 | 99 | OTU17 |
| 8969 | 551 | FQ660504 | 100 | 100 | OTU101 | |
| Burkholderiales | ||||||
| Comamonadaceae | 10394 | 520 | FQ660439 | 100 | 98 | OTU6 |
| 10648 | 519 | FQ659005 | 100 | 99 | OTU154 | |
| Gammaproteobacteria | ||||||
| Unclassified | 1848 | 929 | FQ660401 | 100 | 99 | OTU2 |
| Sinobacteriaceae | 2378 | 848 | FQ660299 | 100 | 99 | OTU153 |
| Xanthomonadaceae | 3372 | 740 | JN868994 | 87 | 93 | NF |
| Xanthomonadales | 8678 | 548 | EF632898 | 67 | 97 | |
| FQ658754 | 38 | 96 | OTU153 |
That is, the OTU previously associated with phenanthrene degraders as deduced from SIP experiments on the same study site (7). NF, no relevant OTU found.
Approximately 510 sequences were annotated as components of dioxygenases possibly involved in the biodegradation of aromatic hydrocarbons (see Fig. S3 in the supplemental material). Half of them would correspond to alpha or beta subunits of RHDs with undefined substrate specificity, whereas the other half included benzoate dioxygenases and enzymes involved in lower steps of the hydrocarbon metabolic pathways, such as phthalate or protocatechuate dioxygenases, and extradiol dioxygenases. Only a few sequences coding for RHD-associated electron carriers were identified, suggesting that the oxygenase components of many of the three-component RHDs might share common electron carriers. We focused here on the RHD sequences found among the 600 longest contigs derived from our metagenomic analysis (see Table S2 in the supplemental material).
Cloning and sequence analysis of four RHD genes and one set of genes coding for related electron carriers.
Contigs 204, 332, 341, 427, 451, and 569 contained ORFs coding potentially for RHD alpha subunits, four of which also showed an ORF for a beta subunit (see Table S2 in the supplemental material). The α and β RHD subunits deduced from the contig 204 sequence showed highest similarities (ca. 60% identity) with the subunits of a RHD annotated as a phenylpropionate dioxygenase in Pseudoxanthomonas spadix, a BTEX degrader affiliated to the Gammaproteobacteria (28). The α subunit also showed a similar relatedness (60% identity) with the subunit of an enzyme characterized as a salicylate hydroxylase in the PAH-degrading Sphingomonas sp. strain CHY-1 (29). In the other five contigs, the identified ORFs were similar to recently described RHD genes, which were obtained from a pyrene-degrading consortium dominated by uncultured Rhodocyclaceae (12). Since the contigs contained partial RHD sequences or lacked one of the subunit genes, we searched in the whole set of contigs for missing sequences using relevant homologous genes described in the above study as queries. Sequences corresponding to the missing 5′ end of the α subunit genes present in contigs 332, 341, and 427 were found in contigs 763, 55951, and 5241, respectively. For the alpha subunit genes present in contigs 451 and 569, candidate beta subunits were found in contigs 3411 and 2271, respectively. Using appropriate DNA primers designed after the extremities of each reconstituted pair of RHD genes (Table 2) and metagenomic DNA from phenanthrene-spiked soil as the template, PCR products of ∼2.0 kb were obtained in all cases except for the reaction with primers C569-F and C2271-R. Sequence analysis of the cloned PCR products revealed that the RHD genes were very similar but not identical to those determined by metagenomic sequencing (Table 4). Moreover, analysis of another amplicon from each PCR yielded a sequence that was different from that of the first clone, and from that in the relevant contig. The number of mismatches between amplicons cloned from the same PCR varied from 12/1955 (0.6% for pJCA13 versus pJCA14) to 321/1906 (17% for pJCA6 versus pJCA7). These results most likely reflect the great diversity of gene sequences within each type of RHD in soil bacteria.
TABLE 4.
Properties of dioxygenase sequences cloned from metagenomic DNA
| Genea | Plasmid | Relevant contig(s) | No. of mismatches/contigb | Product length (aa)c | Identity (%)d |
|---|---|---|---|---|---|
| pahAc2 | pJCA10/pJCA11 | 341 | 178/1863 | 454 | 94.3 |
| pahAd2 | 168 | 80.2 | |||
| pahAc4 | pJCA8/pJCA9 | 763, 332 | 21/1852 | 454 | 87.7 |
| pahAd4 | 168 | 77.5 | |||
| pahAc5 (1) | pJCA6 | 451 | 235/1327 | 459 | 90.4 |
| pahAd5 (1) | 180 | 82 | |||
| pahAc5 (2) | pJCA7 | 451 | 16/1327 | 453 | 89 |
| pahAd5 (2) | 183 | 78.8 | |||
| pahAc8 | pJCA13/pJCA14 | 427 | 9/1653 | 449 | 92.3 |
| pahAd8 | 176 | 86.4 |
Each pair of RHD genes is represented by two different amplicons cloned in the plasmids indicated in column 2. Amplicon sequences are closely similar except for the pahAc5-pahAd5 amplicons [referred to as (1) and (2)], which show 321 mismatches upon sequence alignment.
Number of mismatches between each pahAc-pahAd pair and the gene pair found in the relevant contig.
Amino acid content of gene products is deduced from nucleotide sequences.
That is, the amino acid sequence identities with homologous gene products as previously described by Singleton et al. (12).
Contig 095 contained one gene coding for a NAD(P)H-ferredoxin oxidoreductase, as well as three ORFs very similar in sequence and arrangement to a gene cluster previously described for a pyrene-degrading consortium (12). The reductase gene, designated pahAa in that study, is part of a gene cluster, including pahAb, located about 3 kb upstream and encoding a ferredoxin (Fig. 1). A gene closely similar to pahAb was found in contig 113, which also contained two genes encoding putative transcriptional regulators of the LysR type (best match, Q12EV9; 49% identity) and MarR type (best match, UniProtKB no. D6CQZ4; 51% identity), respectively (Fig. 1). These genes showed an opposite orientation with respect to pahAb and were followed by a gene similar to that coding for a cytochrome B561 in Sideroxydans lithotrophicus (UniProtKB no. D5CTW7; 54% identity).
FIG 1.
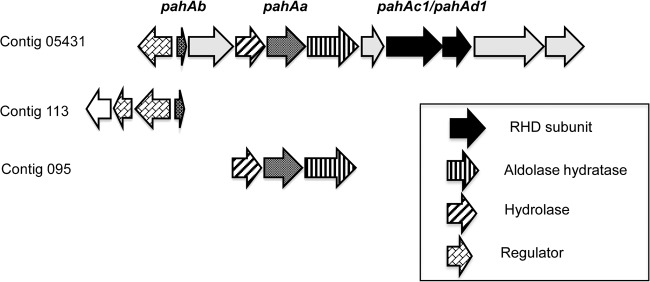
Maps of contigs 095 and 113 containing the RHD-specific electron carrier genes pahAa and pahAb. The genes identified in the two contigs have counterparts in contig 05431 found in a previously described pyrene-degrading bacterial consortium (12). The putative function of genes is depicted by different fill patterns, as indicated.
The two genes homologous to pahAa and pahAb were separately amplified with DNA from phenanthrene-spiked soil as the template. Cloned sequences showed 29 and 5 mismatches compared to those found in contigs 95 and 113, respectively. For the sake of clarity, these genes, as well as the RHD genes described above, were given the same names as their counterparts previously found in a pyrene-degrading consortium (Table 4). Note that control PCRs where template DNA was from soil incubated without phenanthrene yielded no detectable product, except for the pahAc8-pahAd8 gene pair (data not shown). This result provides further evidence that the RHD genes were from main degraders that developed at the expense of added phenanthrene during the 6-day incubation in microcosms.
Heterologous expression of four RHDs and electron carrier preference.
For each set of RHD genes identified here (Table 4), we selected one of the cloned sequences for subcloning in plasmid pET15b and subsequent expression in E. coli BL21(DE3) (see Table 1 and Materials and Methods for details). Hence, the pahAc2-pahAd2, pahAc4-pahAd4, pahAc5-pahAd5, and pahAc8-pahAd8 gene pairs cloned in pCAE1, pCAE5, pCAE7, and pCAE4 were from pJCA10, pJCA9, pJCA7, and pJCA14, respectively. Another plasmid called pCAZ1 (Table 1) was constructed and introduced in BL21(DE3) to coexpress the pahAa and pahAb genes coding for two electron carriers, which were assumed to associate with RHD oxygenase components to form active enzyme complexes. Under appropriate induction conditions, recombinant E. coli strains overproduced two polypeptides of around 50 and 20 kDa, as shown by SDS-PAGE analysis of whole-cell extracts (Fig. 2). The observed polypeptide sizes were consistent with those deduced from gene sequences. In this respect, the PahAd5 product (184 amino acids; expected mass, 21,331 Da) showed a slower migration profile than those of PahAd2 (19,870 Da), PahAd4 (19,821 Da), and PahAd8 (21,186 Da). However, the observed mobility shift of PahAd5 cannot be only explained by its higher molecular mass. In fact, it might be due to its basic character (theoretical isoelectric point around 9.0), whereas the other beta subunits were predicted to be acidic (pI 5.63 and 6.40 for PahAd4 and PahAd8) or near neutral (pI 7.8 for PahAd2). The overexpression of pahAa and pahAb yielded polypeptides with apparent Mr of ∼37,000 and 13,000, in fairly good agreement with the expected masses of 37,385 and 11,635 Da, as deduced from their respective sequences (Fig. 2, lane 6).
FIG 2.
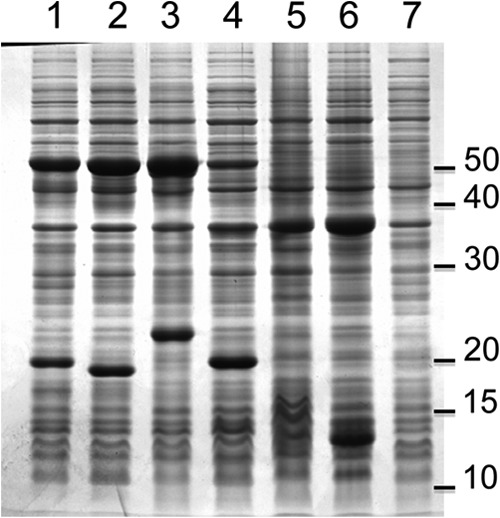
Overexpression of RHD components in recombinant E. coli strains, as illustrated by SDS-PAGE. Whole-cell extracts were prepared from IPTG-induced culture normalized to a bacterial density of 2.0 (OD600). Samples (5 μl) were analyzed by slab gel electrophoresis, followed by Coomassie blue staining. The following RHDs were expressed in the indicated strains: lane 1, PahAc2/PahAd2 in BL21(pCAE1)(pIBA34); lane 2, PahAc4/PahAd4 in BL21(pCAE5)(pIBA34); lane 3, PahAc5/PahAd5 in BL21(pCAE7)(pCAZ1); lane 4, PahAc8/PahAd8 in BL21(pCAE4)(pCAZ1); and lane 5, control strain BL21(pET15b)(pCAZ1). Lane 6 shows a BL21(pJCA5) extract overexpressing PahAa (Mr ≈ 37,000) and PahAa (Mr ≈ 13,000). Lane 7 shows an extract of uninduced BL21(pCAE4)(pCAZ1). The scale on the right indicates the molecular mass markers in kilodaltons.
All four oxygenases converted naphthalene to cis-1,2-dihydroxy 1,2-dihydronaphthalene on condition that the PahAa and PahAb proteins were simultaneously produced in E. coli. No or negligible activity was detected in strains overexpressing the oxygenase components alone (Table 5). The activity of PahAc4/PahAd4 was about 1 order of magnitude lower compared to the other oxygenases, although its expression level in E. coli was similar, as judged from SDS-PAGE (Fig. 2). This suggested that either naphthalene was not a good substrate for this enzyme or the electron carriers were inadequate. The second hypothesis was tested by replacing pCAZ1 by a plasmid (pIBA34) overexpressing PhnA4 and PhnA3, the electron carriers associated to the naphthalene dioxygenase from Sphingomonas CHY-1 (19, 26). The resulting strain BL21(DE3)(pCAE5)(pIBA34) exhibited a dioxygenase activity 20-fold higher than that measured in the strain expressing PahAa and PahAb from pCAZ1 (Table 5). Assuming that the two compared strains showed equivalent expression levels of PahAc4/PahAd4 (data not shown), and similar levels of the electron carriers since pCAZ1 and pIBA34 were derived from the same plasmid, it is concluded that the higher activity found in the strain harboring pIBA34 most likely reflected a better compatibility of the oxygenase with the PhnA4/PhnA3 electron carriers. Likewise, the PahAc2/PahAd2 oxygenase showed a higher activity with PhnA4/PhnA3 than with PahAa/PahAb, although the activity ratio was only 1.5 in that case. These results suggested that the latter two oxygenases might operate in cells related to sphingomonads, consistent with their sequence similarity with PAH dioxygenases from this taxonomic group (see Fig. 3 and discussion below). From the metagenomic data we identified one ferredoxin-encoding gene in contig 9967, whose closest match was a bphA3 gene from Novosphingobium sp. strain PCY (accession no. KF373043). However, at the protein level, sequence similarities with BphA3 from strain PCY (53% identity) or with PhnA3 from strain CHY-1 (51% identity) were relatively low, thus precluding a possible affiliation of that protein to any taxon.
TABLE 5.
Dependence of recombinant dioxygenase activity on the coexpression of appropriate electron carriers
| Oxygenase | Mean activity (μmol/h/ml)a ± SD with: |
||
|---|---|---|---|
| PahAa/PahAb | PhnA4/PhnA3 | No electron carrier | |
| PahAc2/PahAd2 | 2.27 ± 0.25 | 3.39 ± 0.49 | <5 × 10−3 |
| PahAc4/PahAd4 | 0.118 ± 0.035 | 2.63 ± 0.59 | 0.0254 ± 0.0012 |
| PahAc5/PahAd5 | 0.720 ± 0.31 | ND | 0.033 ± 0.0094 |
| PahAc8/PahAd8 | 2.44 ± 0.36 | ND | 0.054 ± 0.0072 |
Activities are expressed as micromoles of dihydrodiol formed per hour per ml of culture normalized to an OD600 of 1.0. Naphthalene was used as the substrate, except for PahAc5/PahAd5 and PahAc8/PahAd8, which were assayed using phenanthrene. ND, not determined.
FIG 3.
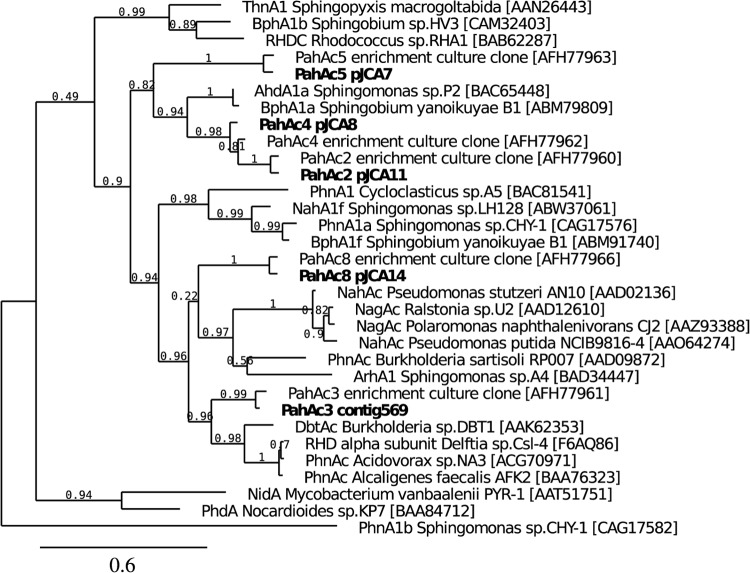
Phylogenetic tree showing the relationships between alpha subunit sequences of selected PAH dioxygenases. The RHD sequences studied here are indicated in boldface letters. The GenBank accession numbers are indicated in brackets. Numbers at the nodes indicate neighbor-joining bootstrap confidence. The sequence of the salicylate hydroxylase from Sphingomonas sp. strain CHY-1 (Phna1b) was used as an outgroup.
Substrate specificity toward two- to four-ring PAHs.
The four recombinant RHDs exhibited a narrow specificity for two- and three-ring PAHs, naphthalene being the preferred substrate (Table 6). PahAc8/PahAd8 showed the broadest substrate range and oxidized biphenyl at about the same rate as naphthalene. The enzyme also oxidized three-ring PAHs at a significant rate, given that the relatively small amount of dihydrodiol detected with fluorene as the substrate reflected only part of the activity. Indeed, analysis of trimethylsilylated fluorene oxidation products generated by PahAc8/PahAd8 allowed the detection of one monohydroxy-fluorene (M+ = 254) and three dihydroxy-fluorene (M+ = 342) by GC/MS (data not shown). PahAc5/PahAd5 appeared as the only RHD able to generate measurable amounts of dihydrodiol from pyrene (Table 6), although the dihydroxylation rate was low compared to that observed with naphthalene (∼0.054%). With phenanthrene as the substrate, the enzyme produced two dihydrodiols with a 1:2 molar ratio, the isomer hydroxylated at the C-3 and C-4 positions being the less abundant. The predominant isomer, showed a retention time (14.63 min versus 14.27 min for the 3,4 isomer) and a mass spectrum distinct from those of the previously identified 9,10-isomer (30). Based on comparisons with previously published data (31), we assumed that the second dihydrodiol formed by PahAc5/PahAd5 was hydroxylated on C-1 and C-2 positions. Likewise, the PahAc2/PahAd2 and PahAc4/PahAd4 oxygenases formed two isomers from phenanthrene with an even higher proportion of the 1,2-isomer (Table 6). Despite small differences in their specific activity, the two enzymes, which share high sequence similarity (81 to 84% identity overall), exhibited the same narrow selectivity for naphthalene and phenanthrene.
TABLE 6.
Activities of recombinant RHDs toward two- to four-ring PAHs
| Substrate | Product formed | Mean dioxygenase activity (μmol/h/ml)a ± SD with: |
|||
|---|---|---|---|---|---|
| PahAc2/PahAd2 | PahAc4/PahAd4 | PahAc5/PahAd5 | PahAc8/PahAd8 | ||
| Naphthalene | cis-1,2-Dihydrodiol | 1.34 ± 0.40 | 2.63 ± 0.59 | 2.65 ± 0.44 | 3.76 ± 0.18 |
| Biphenyl | cis-2,3-Dihydrodiol | – | – | – | 3.75 ± 0.70 |
| Phenanthrene | cis-3,4-Dihydrodiol | 0.173 ± 0.001 | 0.0228 ± 0.0081 | 0.113 ± 0.016 | 0.30 ± 0.075 |
| cis-1,2-Dihydrodiol | 0.806 ± 0.17 | 0.765 ± 0.28 | 0.238 ± 0.066 | ||
| Anthracene | cis-1,2-Dihydrodiol | Traces | Traces | 0.0381 ± 0.0058 | 0.529 ± 0.059 |
| Fluoreneb | Dihydrodiol | – | – | – | (9.1 ± 1.9) × 10−3 |
| Pyrene | cis-4,5-Dihydrodiol | – | – | (1.42 ± 0.21) × 10−3 | – |
Activities are expressed as micromoles of dihydrodiol formed per hour per ml of culture normalized to an OD600 of 1.0. –, no detectable activity.
Fluorene oxidation by PahAc8/PahAd8 also yielded monohydroxyfluorene and dihydroxyfluorene, which were detected as trimethylsilylated derivates (see the text).
DISCUSSION
Combining SIP with shotgun sequencing appears to be a valuable strategy for targeting uncultured microorganisms with desired metabolic functions and for extracting relevant genetic information from soil DNA. As shown in the present study, this approach was successfully used to specifically investigate the potential of PAH-degrading bacteria that predominate in contaminated soil, resulting in a large body of genomic sequences from which interesting new biocatalysts have been cloned and functionally characterized. Ideally, metagenomic data should give upon assembly large contigs representing portions of bacterial chromosomes bearing clusters of genes involved in the same metabolic function, such as pollutant degradation. In the present work, the limited size of the contigs precluded the deciphering of large gene clusters related to PAH degradation. The limited length of the contigs might primarily be a consequence of the complexity of the recovered metagenomic sequences, likely originating from multiple genomes. Despite the sieving effect of SIP, which removed the DNA from bacteria unrelated to phenanthrene metabolism from the analysis, the detection of 10 16S rRNA gene sequences in our metagenomic data indicated that the labeled DNA contained fragments of at least as many genomes. In addition, there seems to be a high degree of polymorphism among some isofunctional genes present in soil bacteria, as exemplified by the fact that cloned amplicons specific for each RHD gene set were different in sequence. This polymorphism might have hindered the assembly of short reads generated by 454 pyrosequencing or led to artifacts due to mosaic assembly. Another possible cause of the contig shortness might be the depth of the sequencing effort, which was limited by the scarcity of labeled DNA recovered from soil. To overcome the problem associated with the low recovery of labeled DNA inherent to SIP, amplification methods such as multiple displacement amplification may be used. Employing this method, Wang et al. were able to assemble a nag gene cluster responsible for the conversion of naphthalene to salicylate (17). Presumably, this gene cluster was part of the genome of an uncultured Acidovorax sp., which was identified as a prevalent naphthalene degrader in situ.
Although partial and fragmented, the information derived from our metagenomic data provides new and valuable insights into the metabolic potential of soil phenanthrene degraders. Based on sequence annotation, a majority of genes would belong to the Betaproteobacteria, one fourth of which appear as Rhodocyclaceae. Even though some of the annotations might be wrong, the high proportion of genome sequences related to Rhodocyclaceae is corroborated by the occurrence of two (of 11) 16S rRNA sequences affiliated to this family in the metagenomic DNA (Table 3). The two 16S rRNA sequences of interest are identical to prevalent ones previously detected by DNA-SIP in the PAH-degrading community of the same soil (OTU17 and OTU101) (7). In addition to Rhodocyclaceae, our metagenomic data also underscored the importance of Gammaproteobacteria, as indicated by the occurrence of four 16S rRNA sequences, which were representative of two unclassified bacterial groups previously referred to as OTU2 and OTU153 (Table 3) (7). Species represented by OTU2 are phylogenetically related to soil bacteria designated Pyr group 2, first identified as pyrene degraders (9) and later shown to also degrade other four-ring PAHs (32). On the other hand, bacteria of the Acidovorax, Rhodoferax, and Hydrogenophaga genera, which were previously identified as prevalent phenanthrene degraders in the studied soil, were poorly represented in the annotations of the sequence data. This observation might reflect some variability of the soil bacterial community, which possibly underwent changes during the 1-year period separating soil sampling for this work and our previous study (7).
As a first step toward a better understanding of the metabolism of soil phenanthrene degraders, we have cloned and characterized four RHDs responsible for the initial attack of PAHs. The enzymes exhibited limited sequence similarity (50 to 70% identity) with well-characterized PAH dioxygenases described thus far, which were isolated from culturable strains (13). A phylogenetic comparison of selected PAH dioxygenases indicated that three enzyme sequences (PahAc2, PahAc4, and PahAc5) clustered with poorly characterized RHDs from sphingomonads (AhdA1a or BphA1a; Fig. 3). On the other hand, the PahAc8 sequence appeared as distant from NahAc/NagAc from Beta- and Gammaproteobacteria as from PhnAc from Betaproteobacteria. PahAc3, which could not be biochemically studied here, was more closely related to PhnAc-type enzymes.
The PahAc sequences also differed from RHDs previously identified by a PCR method used to amplify a region coding for the catalytic domain (314 residues) of the alpha subunit and isolated from the same soil (18). In that study, retrieved RHD sequences were grouped in five main clusters, two of which were related to PAH dioxygenases found in Betaproteobacteria, and one other cluster was related to Alphaproteobacteria (mainly sphingomonads). One sequence was almost identical to the corresponding part of PahAc8. The other three RHDs have sequences that do not match any of the previously detected sequences, perhaps because their bacterial hosts were absent due to changes in the soil bacterial community. Alternatively, the primer pairs we used in our previous study to amplify RHD alpha subunits might have been inadequate to detect pahAc2, pahAc4, and pahAc5. In this respect, sequence analysis predicts that the reverse primers used in our previous work would not correctly hybridize with the coding sequences of the latter three genes. On the other hand, pahAc3 present in contig 569 (see Table S2 in the supplemental material) is closely similar to numerous sequences affiliated to the Betaproteobacteria and referred to as cluster 4 (18). Unfortunately, we have been unable to clone a pahAc3 gene with its associated beta subunit gene, precluding the functional characterization of the corresponding RHD.
Finally, the RHDs that best matched those described here were recently obtained from a bacterial consortium selected by enrichment with pyrene as carbon source (12). Eight RHDs have been cloned from this pyrene-specific consortium, six of which were apparently able to attack two- to four-ring PAHs to various extent. It has been proposed that all of these enzymes belonged to the same bacterial type, related to Rhodocyclaceae, since it appeared as a predominant member of the consortium. Curiously, all eight RHDs except one appear to have counterparts in the phenanthrene-degrading community examined here, since, in addition to the four studied RHDs, sequences similar to pahAc1, pahAc3, and pahAc6 were detected in contigs 38041, 569, and 1214, respectively. Our data also suggest that they might have the same bacterial source. Examples of bacterial isolates with multiple functional PAH dioxygenases are rare, especially among Betaproteobacteria (13). In sphingomonads, up to six sets of RHD-like sequences have been described (33, 34), but only one set appeared to encode a PAH dioxygenase (19), the other enzymes catalyzing the hydroxylation of salicylate and methyl salicylates (29, 35). In mycobacteria, some strains have been shown to synthesize up to three types of PAH dioxygenases with distinct specificities (30, 36).
The substrate range of the four RHDs is relatively narrow and limited to two- and three-ring PAHs. Only one RHD proved capable of utilizing pyrene, which contrasts with a previous report providing evidence that similar enzymes could degrade the four-ring substrate (12). Also, while PahAc8/PahAd8 exhibited the broadest substrate specificity, its counterpart called RHD-8 showed insignificant activity with any PAH except phenanthrene and pyrene. Discrepancies might be due primarily to differences in experimental conditions, although differences in amino acid sequence between homologous RHDs should also be taken into consideration. The percent identity between homologous RHD components in the present and the cited study vary between 77.5 and 94% (Table 4). Marked changes in specificity were observed between PAH dioxygenases from sphingomonads showing equivalent sequence relatedness. Notably, the RHD from Sphingobium yanoikuyae B1 has a preference for biphenyl (37), whereas the RHDs from Sphingomonas strains CHY-1 and LH128 utilize naphthalene as the best substrate (26, 38). Also, biphenyl dioxygenases displaying as high as 99% sequence identity proved markedly different in substrate specificity toward polychlorobiphenyls (39). Hence, compared to the enzymes described here, the RHDs described by Singleton et al. might have a better activity for pyrene because of the bacterial enrichment on this PAH, which preceded RHD gene isolation (12).
When incubated with phenanthrene, three of the studied RHDs generated more 1,2-dihydrodiol than 3,4-dihydrodiol (Table 6), a rather unexpected result since the latter compound is thought to be the most common intermediate in bacterial degradation pathways (2). Although dioxygenation of phenanthrene on the C-1 and C-2 positions has been shown to occur in Sphingomonas strain P2 (40) and in Burkholderia strain C3 (41) and gives rise to effective metabolization, most known RHDs do not generate significant amounts of 1,2-dihydrodiol from phenanthrene. In this respect, a mutant form of the naphthalene dioxygenase from Pseudomonas sp. strain NCIB9816-4 appears to be an exception. Its ability to form an excess of phenanthrene 1,2-dihydrodiol resulted from the replacement of the Phe 352 residue by a valine at the enzyme active site (31). The RHDs described in the present study have a phenylalanine in equivalent position, indicating that their ability to better hydroxylate phenanthrene on the C-1 and C-2 positions is not due to a similar amino acid substitution. Our work provides evidence that this catalytic property might be a common feature of PAH dioxygenases from soil bacteria, suggesting that it could confer a selective advantage to phenanthrene degraders.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the CNRS and the Université Grenoble Alpes for funding. This study was also supported in part by grants from the Rhône-Alpes region and from the ANR Program Labex (ARCANE project ANR-11-LABX-003).
Footnotes
Published ahead of print 15 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01883-14.
REFERENCES
- 1.Doyle E, Muckian L, Hickey AM, Clipson N. 2008. Microbial PAH degradation. Adv. Appl. Microbiol. 65:27–66. 10.1016/S0065-2164(08)00602-3. [DOI] [PubMed] [Google Scholar]
- 2.Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, Tian YS, Yao QH. 2008. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 32:927–955. 10.1111/j.1574-6976.2008.00127.x. [DOI] [PubMed] [Google Scholar]
- 3.Cole JR, Konstantinidis K, Farris RJ, Tiedje JM. 2010. Microbial diversity and phylogeny: extending from rRNAs to genomes, p 1–19 In Liu W-T, Jansson JK. (ed), Environmental molecular microbiology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Leys NMEJ, Ryngaert A, Bastiaens L, Verstraete W, Top EM, Springael D. 2004. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:1944–1955. 10.1128/AEM.70.4.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon CO, Park W, Padmanabhan P, DeRito C, Snape JR, Madsen EL. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. U. S. A. 100:13591–13596. 10.1073/pnas.1735529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton DR, Ramirez LG, Aitken MD. 2009. Characterization of a polycyclic aromatic hydrocarbon degradation gene cluster in a phenanthrene-degrading Acidovorax strain. Appl. Environ. Microbiol. 75:2613–2620. 10.1128/AEM.01955-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, Bru D, Geremia R, Blake G, Jouanneau Y. 2012. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ. Pollut. 162:345–353. 10.1016/j.envpol.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Singleton DR, Powell SN, Sangaiah R, Gold A, Ball LM, Aitken MD. 2005. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl. Environ. Microbiol. 71:1202–1209. 10.1128/AEM.71.3.1202-1209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. 2006. Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol. 8:1736–1745. 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- 10.Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, Vlcek C, Cardenas E, Mackova M, Macek T. 2012. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS One 7:e40653. 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regonne RK, Martin F, Mbawala A, Ngassoum MB, Jouanneau Y. 2013. Identification of soil bacteria able to degrade phenanthrene bound to a hydrophobic sorbent in situ. Environ. Pollut. 180:145–151. 10.1016/j.envpol.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 12.Singleton DR, Hu J, Aitken MD. 2012. Heterologous expression of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes from a novel pyrene-degrading betaproteobacterium. Appl. Environ. Microbiol. 78:3552–3559. 10.1128/AEM.00173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jouanneau Y, Martin F, Krivobok S, Willison JC. 2011. Ring-hydroxylating dioxygenases involved in PAH biodegradation: structure, function, and biodiversity, p 149–175 In Koukkou AI. (ed), Microbial bioremediation of nonmetals: current research. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 14.Kweon O, Kim S-J, Baek S, Chae J-C, Adjei M, Baek D-H, Kim Y-C, Cerniglia C. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem. 9:11. 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam JW, Nojiri H, Yoshida T, Habe H, Yamane H, Omori T. 2001. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci. Biotechnol. Biochem. 65:254–263. 10.1271/bbb.65.254. [DOI] [PubMed] [Google Scholar]
- 16.Uhlik O, Leewis MC, Strejcek M, Musilova L, Mackova M, Leigh MB, Macek T. 2013. Stable isotope probing in the metagenomics era: a bridge toward improved bioremediation. Biotechnol. Adv. 31:154–165. 10.1016/j.biotechadv.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Chen Y, Zhou Q, Huang S, Ning K, Xu J, Kalin RM, Rolfe S, Huang WE. 2012. A culture-independent approach to unravel uncultured bacteria and functional genes in a complex microbial community. PLoS One 7:e47530. 10.1371/journal.pone.0047530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin F, Malagnoux L, Violet F, Jakoncic J, Jouanneau Y. 2013. Diversity and catalytic potential of PAH-specific ring-hydroxylating dioxygenases from a hydrocarbon-contaminated soil. Appl. Microbiol. Biotechnol. 97:5125–5135. 10.1007/s00253-012-4335-2. [DOI] [PubMed] [Google Scholar]
- 19.Demaneche S, Meyer C, Micoud J, Louwagie M, Willison JC, Jouanneau Y. 2004. Identification and functional analysis of two aromatic ring-hydroxylating dioxygenases from a Sphingomonas strain degrading various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714–6725. 10.1128/AEM.70.11.6714-6725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aury JM, Cruaud C, Barbe V, Rogier O, Mangenot S, Samson G, Poulain J, Anthouard V, Scarpelli C, Artiguenave F, Wincker P. 2008. High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies. BMC Genomics 9:603. 10.1186/1471-2164-9-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi H, Taniguchi T, Itoh T. 2008. Metageneannotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA res. 15:387–396. 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glemet E, Codani JJ. 1997. LASSAP, a LArge Scale Sequence compArison Package. Comput. Appl. Biosci. 13:137–143. [DOI] [PubMed] [Google Scholar]
- 24.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuler L, Chadhain SMN, Jouanneau Y, Meyer C, Zylstra GJ, Hols P, Agathos SN. 2008. Characterization of a novel angular dioxygenase from fluorene-degrading Spingomonas sp. strain LB126. Appl. Environ. Microbiol. 74:1050–1057. 10.1128/AEM.01627-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jouanneau Y, Meyer C, Jakoncic J, Stojanoff V, Gaillard J. 2006. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45:12380–12391. 10.1021/bi0611311. [DOI] [PubMed] [Google Scholar]
- 27.Jouanneau Y, Meyer C. 2006. Purification and characterization of an arene cis-dihydrodiol dehydrogenase endowed with broad substrate specificity toward polycyclic aromatic hydrocarbon dihydrodiols. Appl. Environ. Microbiol. 72:4726–4734. 10.1128/AEM.00395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, Jin HM, Lee HJ, Kim JM, Jeon CO. 2012. Complete genome sequence of the BTEX-degrading bacterium Pseudoxanthomonas spadix BD-a59. J. Bacteriol. 194:544. 10.1128/JB.06436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jouanneau Y, Micoud J, Meyer C. 2007. Purification and characterization of a three-component salicylate 1-hydroxylase from Sphingomonas sp. strain CHY-1. Appl. Environ. Microbiol. 73:7515–7521. 10.1128/AEM.01519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828–3841. 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parales RE, Lee K, Resnick SM, Jiang HY, Lessner DJ, Gibson DT. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641–1649. 10.1128/JB.182.6.1641-1649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones MD, Crandell DW, Singleton DR, Aitken MD. 2011. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ. Microbiol. 13:2623–2632. 10.1111/j.1462-2920.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinyakong O, Habe H, Omori T. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J. Gen. Appl. Microbiol. 49:1–19. 10.2323/jgam.49.1. [DOI] [PubMed] [Google Scholar]
- 34.Romine MF, Stillwell LC, Wong KK, Thurston SJ, Sisk EC, Sensen C, Gaasterland T, Fredrickson JK, Saffer JD. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinyakong O, Habe H, Yoshida T, Nojiri H, Omori T. 2003. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem. Biophys. Res. Commun. 301:350–357. 10.1016/S0006-291X(02)03036-X. [DOI] [PubMed] [Google Scholar]
- 36.Kweon O, Kim SJ, Freeman JP, Song J, Baek S, Cerniglia CE. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1:e00135–10. 10.1128/mBio.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu CL, Liu W, Ferraro DJ, Brown EN, Parales JV, Ramaswamy S, Zylstra GJ, Gibson DT, Parales RE. 2007. Purification, characterization, and crystallization of the components of a biphenyl dioxygenase system from Sphingobium yanoikuyae B1. J. Ind. Microbiol. Biotechnol. 34:311–324. 10.1007/s10295-006-0199-8. [DOI] [PubMed] [Google Scholar]
- 38.Schuler L, Jouanneau Y, Chadhain SM, Meyer C, Pouli M, Zylstra GJ, Hols P, Agathos SN. 2009. Characterization of a ring-hydroxylating dioxygenase from phenanthrene-degrading Sphingomonas sp. strain LH128 able to oxidize benz[a]anthracene. Appl. Microbiol. Biotechnol. 83:465–475. 10.1007/s00253-009-1858-2. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa K, Suenaga H, Goto M. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189–5196. 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinyakong O, Habe H, Supaka N, Pinpanichkarn P, Juntongjin K, Yoshida T, Furihata K, Nojiri H, Yamane H, Omori T. 2000. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiol. Lett. 191:115–121. 10.1111/j.1574-6968.2000.tb09327.x. [DOI] [PubMed] [Google Scholar]
- 41.Seo JS, Keum YS, Hu Y, Lee SE, Li QX. 2007. Degradation of phenanthrene by Burkholderia sp. strain C3: initial 1,2- and 3,4-dioxygenation and meta- and ortho-cleavage of naphthalene-1,2-diol. Biodegradation 18:123–131. [DOI] [PubMed] [Google Scholar]
- 42.Moreno-Ruiz E, Hernaez MJ, Martinez-Perez O, Santero E. 2003. Identification and functional characterization of Sphingomonas macrogolitabida strain TFA genes involved in the first two steps of the tetralin catabolic pathway. J. Bacteriol. 185:2026–2030. 10.1128/JB.185.6.2026-2030.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.