Abstract
Cyanobacteria are photosynthetic bacteria that are currently being developed as biological production platforms. They derive energy from light and carbon from atmospheric carbon dioxide, and some species can fix atmospheric nitrogen. One advantage of developing cyanobacteria for renewable production of biofuels and other biological products is that they are amenable to genetic manipulation, facilitating bioengineering and synthetic biology. To expand the currently available genetic toolkit, we have demonstrated the utility of synthetic theophylline-responsive riboswitches for effective regulation of gene expression in four diverse species of cyanobacteria, including two recent isolates. We evaluated a set of six riboswitches driving the expression of a yellow fluorescent protein reporter in Synechococcus elongatus PCC 7942, Leptolyngbya sp. strain BL0902, Anabaena sp. strain PCC 7120, and Synechocystis sp. strain WHSyn. We demonstrated that riboswitches can offer regulation of gene expression superior to that of the commonly used isopropyl-β-d-thiogalactopyranoside induction of a lacIq-Ptrc promoter system. We also showed that expression of the toxic protein SacB can be effectively regulated, demonstrating utility for riboswitch regulation of proteins that are detrimental to biomass accumulation. Taken together, the results of this work demonstrate the utility and ease of use of riboswitches in the context of genetic engineering and synthetic biology in diverse cyanobacteria, which will facilitate the development of algal biotechnology.
INTRODUCTION
Eukaryotic and prokaryotic species of algae are currently being developed as a renewable resource for the production of biofuels and other commercially important products (1). Because algae are photosynthetic and derive carbon from atmospheric carbon dioxide, they provide a means to produce biofuels and biological products with lower net greenhouse gas emissions. Furthermore, algae can be grown in low-quality water sources and on nonarable land, so they would not compete with resources required for food crop production. A major hurdle in the implementation of alga-based production methods is the current high cost of production. Genetic engineering plays a major role in efforts to improve efficiency at all steps of production (2, 3). As the field of algal bioengineering and synthetic biology grows, methods for the precise regulation of gene expression will be invaluable.
Cyanobacteria, commonly called blue-green algae, are photosynthetic bacteria that are promising candidates for the renewable production of valuable products such as biofuels (4, 5). As a phylum, cyanobacteria are quite diverse. They inhabit a range of habitats and possess a wide range of metabolic capabilities (6, 7). This diversity can be exploited by the isolation of production strains with desired traits and the bioengineering of exogenous metabolic pathways, stress tolerance, and growth enhancements obtained from other cyanobacterial or bacterial strains. One major advantage of using cyanobacteria in biotechnological production is that they are easier to manipulate genetically (8) than eukaryotic algae. Many strains can accept exogenous DNA through conjugation, and some strains can take up exogenous DNA by natural transformation. Cyanobacteria can integrate homologous DNA into their genomes for the expression of genetic constructs or the generation of knockout mutations. They can also harbor multicopy plasmids for the expression of desired gene products. These characteristics provide an attractive platform for genetic engineering, and expanding the genetic toolkit for cyanobacteria will advance this field. Existing tools for regulating gene expression in cyanobacteria include natural and heterologous promoters that can be regulated by various inputs (9). One widely used induction system originally developed in Escherichia coli is LacI repression of isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoters. However, in cyanobacteria, this system can have unwanted baseline expression in the absence of the inducer and poor induction levels (3, 10). Another nonnative promoter used in cyanobacteria is the tetracycline repressor system, which has recently been improved to produce a wider dynamic range of induction in a Synechocystis species (11). Endogenous regulated promoters from cyanobacteria include the high-light-responsive psbA promoter, the copper-regulated petE promoter, the nitrate/nitrite-inducible nirA promoter, and the nickel-regulated nrsA promoter (9). Although these promoters are functional in cyanobacteria, induction of these regulatory systems would also affect the endogenous cognate regulon and could produce unwanted effects. Therefore, synthetic or exogenous systems for regulating gene expression are preferred because they are less likely to affect natural regulatory pathways.
Riboswitches are naturally occurring mRNA elements found in all domains of life (12). They are a versatile tool for genetic engineering because they are small and modular systems that regulate gene expression without a requirement for the production of a protein transcription factor. Riboswitches are composed of an aptamer sequence that binds its cognate ligand, and the expression platform sequence that regulates gene expression through the adoption of alternative secondary-structure conformations. In eukaryotes, riboswitches can alter mRNA splicing to produce different protein isoforms, introduce premature stop codons, or affect mRNA stability (13). In bacteria, they are generally located in 5′ untranslated regions of mRNA transcripts. They can regulate transcription through the formation or disruption of transcriptional terminators. Other riboswitches can regulate translation by altering the accessibility of the ribosome-binding site. They often bind metabolites and regulate the expression of genes to maintain the homeostasis of metabolic pathways. To engineer riboswitches as a means to regulate gene expression, a set of synthetic riboswitches was recently developed through screening and rational design (14). These riboswitches respond to theophylline, a caffeine analog, and function through a translation initiation mechanism. They were originally evaluated in eight different bacterial species and have since been shown to function in other diverse bacterial species, including Mycobacterium species (15), Francisella novicida (16), Streptomyces coelicolor (17), and Synechococcus elongatus (18).
Riboswitches are a promising method for controlling gene expression in the field of algal bioengineering. To evaluate riboswitches in cyanobacteria, we characterized a set of six theophylline-responsive riboswitches (14) in a diverse set of cyanobacterial species by using a yellow fluorescent protein (YFP) reporter. We demonstrate that these riboswitches function effectively in these cyanobacterial strains and offer regulation of gene expression superior to that of traditional IPTG induction methods. We also demonstrate the use of riboswitches for regulating the expression of the toxic protein SacB as a model for the controlled expression of toxic products that may be required for certain industrial production schemes. This work shows that riboswitches can be used to regulate gene expression in diverse species of cyanobacteria for synthetic biology applications and the production of industrially important products.
MATERIALS AND METHODS
Cyanobacterial strains and cultivation.
All of the cyanobacterial strains were cultivated in BG11 medium and grown while shaking at 30°C under continuous illumination at 70 to 150 μmol photons m−2 s−1. For selection of integrative or replicative plasmids, BG11 agar plates were supplemented with 2 μg/ml spectinomycin (Sigma-Aldrich) and 2 μg/ml streptomycin (Sigma-Aldrich) and BG11 liquid medium was supplemented with 1 μg/ml spectinomycin and 1 μg/ml streptomycin. For induction assays, the growth medium was supplemented with vehicle controls, with theophylline from 200 mM stock solutions prepared in dimethyl sulfoxide (DMSO), or with IPTG from a 100 mM stock solution prepared in water.
Strain construction.
All of the plasmids used in this study (Table 1) were constructed by Seamless cloning (Life Technologies). For quantification of riboswitch induction, a conII promoter (19) was cloned upstream of a riboswitch-regulated myc-yfp reporter gene, followed by an rrnB transcriptional terminator. These reporters were subcloned into cyanobacterial vectors. For S. elongatus, integrative plasmids pAM5044 to pAM5049 and pAM5051 containing regions homologous to neutral site 1 were constructed and then introduced by natural transformation (20). RSF1010-based replicative plasmids pAM5052 to pAM5057 and pAM5059 harboring a mobA Y25F mutation (21, 22) were introduced into the remaining cyanobacterial strains by conjugation from E. coli strain AM1359 (23, 24) carrying the cargo plasmid. Strain AM1359 contains plasmids pRL443 for conjugation and pRL623 for methylation of donor DNA (25). For cloning of unstable YFP (pAM5061), codons encoding AAV were added via primers to the C terminus of myc-yfp. For evaluation of IPTG induction of Ptrc (pAM5062 to pAM5065), the conII promoter was replaced with lacIq and Ptrc, or Ptrc alone, which were amplified from pAM2255 (26). For riboswitch regulation, plasmids contained riboswitch F regulating myc-yfp, and for controls, riboswitch regulation was eliminated by deletion of the aptamer sequence. For sucrose sensitivity assays, vectors pAM5066 and pAM5067 were constructed with riboswitch F regulating sacB (27).
TABLE 1.
Plasmids used in this studya
| Plasmid(s) | Description |
|---|---|
| pAM5044 to pAM5049 | Integrative-vector series for insertion into neutral-site NS1 of PCC 7942 with aadA cassette and conII promoter driving expression of theophylline riboswitch variants A to F with myc-yfp reporter |
| pAM5051 | pAM5049 (riboswitch F) with EcoRI restriction site in place of myc-yfp, vector control |
| pAM5052 to pAM5057 | RSF1010 mobA Y25F replicative plasmid series with aadA cassette and conII promoter driving expression of theophylline riboswitch variants A to F with myc-yfp reporter |
| pAM5059 | pAM5057 (riboswitch F) with EcoRI restriction site in place of myc-yfp, vector control |
| pAM5061 | pAM5049 (riboswitch F) with insertion of C-terminal AAV degradation tag |
| pAM5062 | pAM5049 (riboswitch F) with lacIq repressor and Ptrc in place of PconII |
| pAM5063 | pAM5062 with deletion of theophylline aptamer |
| pAM5064 | pAM5062 (riboswitch F) with deletion of lacIq repressor gene |
| pAM5065 | pAM5062 with deletion of theophylline aptamer and lacIq repressor gene |
| pAM5066 | pAM5051 (riboswitch F) with sacB insert |
| pAM5067 | pAM5059 (riboswitch F) with sacB insert |
See Table S1 in the supplemental material for promoter and riboswitch sequences.
Induction assays.
Unless otherwise noted, cyanobacterial cultures were prepared in the following manner to assay reporter expression and phenotype. Cultures were pregrown for 4 to 6 days before induction assays. Cultures were normalized to an optical density at 750 nm (OD750) of 0.2, and 2-ml samples were treated with theophylline (Sigma-Aldrich), a DMSO vehicle control, or IPTG (Apex BioResearch Products). These cultures were grown for 19 to 24 h while shaking at 30°C under continuous illumination at 70 to 150 μmol photons m−2 s−1 and then assayed for induction. All assays were performed in triplicate with independent strains. To measure YFP expression, cultures were normalized by OD750 and 100-μl volumes of normalized cultures were added to the wells of a black-walled 96-well plate (Greiner). YFP expression was measured with an Infinite M200 Pro microplate reader (Tecan), with excitation at 490/9 nm and emission at 535/20 nm. Background YFP values were obtained from vector control strains and subtracted from YFP expression values. YFP induction in unicellular cyanobacteria was also quantified with a BD FACSort flow cytometer (BD Biosciences) equipped with an argon laser (488-nm laser line). Autofluorescence of cells was measured with FL3 (670-nm low-pass filter), and YFP expression was measured with FL1 (530/30 nm). Approximately 50,000 cells were analyzed per sample.
Microscopy.
All samples were mounted on 1.2% agarose pads before imaging. Light microscopy images were taken with an Axioskop microscope (Zeiss) through a 63× oil immersion objective equipped with differential interference contrast sliders and a Spot Pursuit color charge-coupled device camera (Spot Imaging Solutions). Images were acquired and processed with Spot Advanced 5.1 software. Fluorescence microscopy images were acquired with a DeltaVision Core system (Applied Precision, General Electric Healthcare) with an Olympus IX71 inverted microscope and an Olympus Plan Apochromat 100× oil immersion objective. Images of autofluorescence were acquired with 555/28-nm excitation filters and 617/73-nm emission filters. YFP images were acquired with 500/20-nm excitation filters and 535/30-nm emission filters. Images were processed with softWoRx workstation software. Brightness and contrast were scaled identically for each species.
Sucrose sensitivity assays.
Cultures were pregrown for 4 to 6 days before induction assays. All assays were performed with triplicate biological replicates. For solid-medium assays, cultures were concentrated to an OD750 of 2.0 and 5-fold serial dilutions were prepared. Five-microliter volumes of cells were spotted onto BG11 agar containing various concentrations of sucrose and 1 mM theophylline or 0.5% DMSO. Plates were incubated at 30°C under continuous illumination at 70 to 150 μmol photons m−2 s−1 for 3 days and then imaged. For liquid culture assays, 2-ml cultures were prepared as for the induction assay and sucrose was added to a final concentration of 1%. A 100-μl volume of culture was sampled daily, and OD750 was determined with an Infinite M200 Pro microplate reader (Tecan). Cultures were imaged on day 3.
Phylogenetic tree.
16S rRNA sequences of Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn were aligned with 119 reference cyanobacterial sequences (7). Sequence alignment was performed with the multiple-sequence alignment program MAFFT (28) by the E-INSI method with default parameters. The positions that can be reliably used in phylogenetic analysis were extracted with Gblocks (29) at settings that allowed the most relaxed selection of blocks and covered E. coli positions 24 to 1442. The phylogenetic tree was constructed with the maximum-likelihood method of PhyML (30) by using Shimodaira-Hasegawa (SH)-like branch support and based on a GTR+I+G model using four categories of substitution rate. The proportion of invariant sites and gamma distribution parameter was estimated by PhyML from the data set. The phylogenetic tree was trimmed to display representative sequences.
RESULTS
Theophylline-responsive riboswitches provide robust induction of gene expression in diverse cyanobacteria.
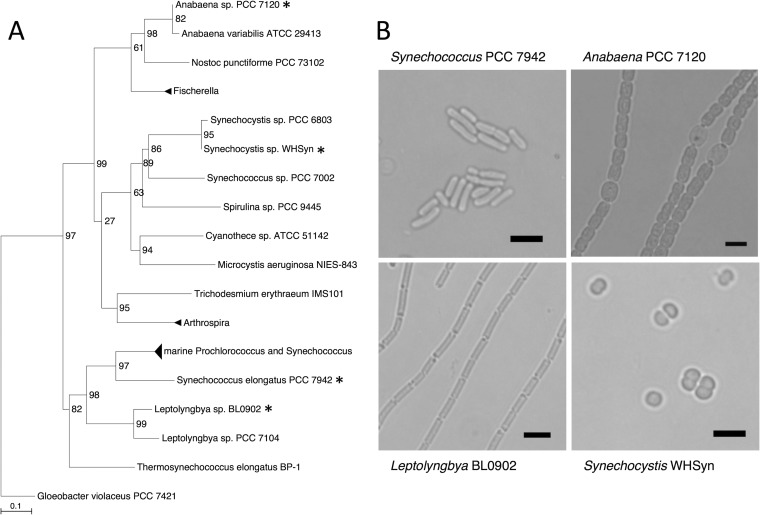
A promising strategy for controlling gene expression in cyanobacteria is the use of synthetic riboswitches. We evaluated the set of six theophylline-responsive riboswitches developed by Topp et al. (14) in four phylogenetically diverse cyanobacterial strains (Fig. 1A). The strains encompass a range of morphologies (Fig. 1B), metabolic capabilities, and growth characteristics. S. elongatus PCC 7942 is a model laboratory strain in which photosynthesis, circadian rhythms, and synthetic biology are studied. This strain has superior genetic manipulation capabilities, such as natural transformation and high rates of homologous recombination, making it an excellent model system for metabolic engineering. Anabaena sp. strain PCC 7120 is a model laboratory strain that can fix atmospheric nitrogen after differentiation of specialized cells called heterocysts that are spaced along filaments. Because supplying combined nitrogen is a significant cost for algal crop production (31), cyanobacterial strains that are capable of fixing nitrogen are of potential interest. Leptolyngbya sp. strain BL0902 is a filamentous cyanobacterial strain that was isolated as a contaminant in an algal production pond (24). This strain has documented outdoor-growth capabilities, is tolerant of a range of stress conditions, and is amenable to genetic manipulation. Another attractive production strain, Synechocystis sp. strain WHSyn, was isolated from an estuary in Woods Hole, MA, and can grow in a wide range of salinities (22).
FIG 1.
Diversity of cyanobacterial strains used in this study. (A) Phylogenetic tree of select cyanobacterial species. Species used in this study are marked with asterisks. SH-like branch support values are shown. The evolutionary distance between two sequences is depicted by the sum of the horizontal branches connecting them (scale bar represents 0.1 mutation per position). Gloeobacter violaceus PCC 7421 was used to root the tree. (B) Light micrographs of the cyanobacterial species used in this study. Top left, S. elongatus PCC 7942. Top right, Anabaena sp. strain PCC 7120. Bottom left, Leptolyngbya sp. strain BL0902. Bottom right, Synechocystis sp. strain WHSyn. Scale bars, 5 μm.
To assay functional characteristics of the set of six riboswitches in the selected cyanobacterial species, we constructed integrative and replicative shuttle plasmids (Table 1). The constructs contain the constitutive conII promoter, a synthetic E. coli consensus promoter shown to function in many cyanobacterial species (22), driving the expression of each of the theophylline riboswitches cloned upstream of a yellow fluorescent protein (YFP) reporter. To assay riboswitch function in S. elongatus PCC 7942, constructs were chromosomally integrated, while Anabaena sp. strain PCC 7120, Leptolyngbya sp. strain BL0902, and Synechocystis sp. strain WHSyn constructs were introduced on a derivative of RSF1010, a broad-host-range, low-copy-number replicative plasmid (22, 32). The use of RSF1010-based plasmids allows the same constructs to be characterized in several different strains and mimics a common way in which the riboswitch devices are likely to be used. Both genome and plasmid copy numbers can vary substantially between cyanobacterial strains and under different growth and genetic conditions (32–34). The genome copy number can range from monoploid to polyploid for different strains and can vary between different growth phases (33). Plasmid RSF1010 has a copy number of 10 in E. coli, and different RSF1010-based plasmids have 10 to 30 copies per cell in Synechocystis sp. strain PCC 6803 (32). However, although the copy number of empty shuttle plasmids may be relatively low and stable under different growth conditions, shuttle vectors containing genes that are a metabolic burden on cells can have variable copy numbers (34). Therefore, absolute gene expression levels cannot be reliably compared between different strains or growth phases but rather only between different constructs and induction conditions in the same strain under the same growth conditions.
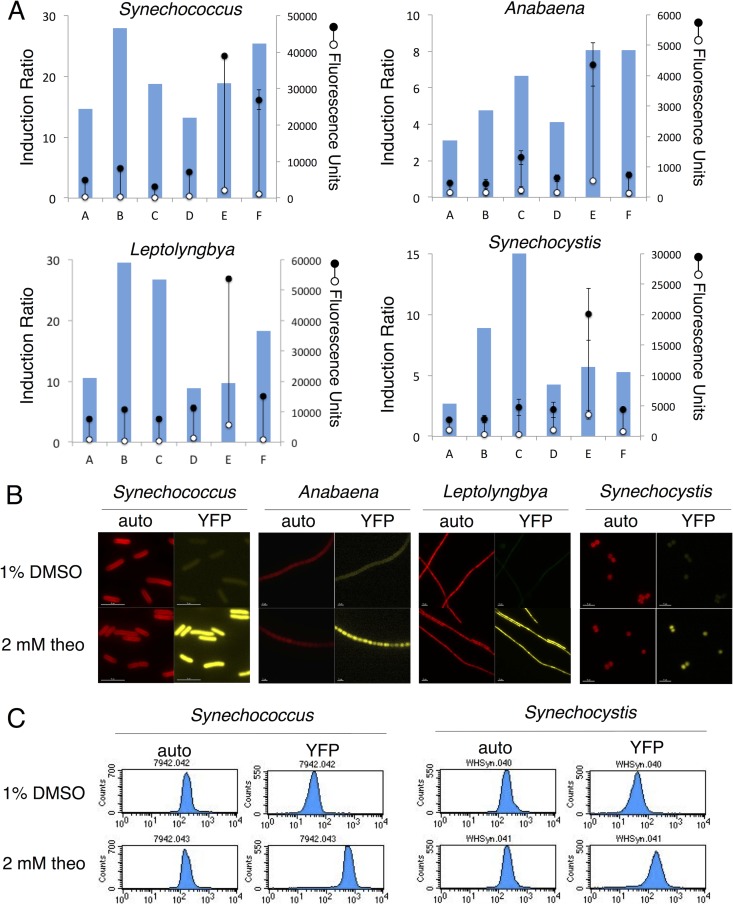
Six different riboswitches were characterized in each of four diverse cyanobacterial strains (Fig. 2). After treatment with 2 mM theophylline or a 1% DMSO vehicle control for 1 day, YFP expression was quantified with a fluorescence plate reader (Fig. 2A). An “induction ratio” provides a metric to gauge riboswitch induction and is determined by calculating the ratio of YFP expression in induced cultures to that in uninduced cultures after subtraction of the background fluorescence. These values were calculated for each of the riboswitches in each of the cyanobacterial strains. All riboswitch variants provided some level of regulation of YFP expression in all of the species tested but varied in their uninduced baseline expression level and their degree of YFP expression after induction (Fig. 2). Overall, induction ratios and expression levels after induction were higher in S. elongatus PCC 7942 and Leptolyngbya sp. strain BL0902 than in Synechocystis sp. strain WHSyn and Anabaena sp. strain PCC 7120. Riboswitch C had low baseline expression for most of the species tested, which then gave a high induction ratio even though it produced only low-to-moderate levels of expression after induction. The exception to this trend is Anabaena sp. strain PCC 7120, in which riboswitch C showed moderate baseline expression and moderate expression after induction. Riboswitch E produced a high expression baseline in all of the cyanobacterial strains tested but also produced the highest levels of expression after induction with theophylline. This could be due in part to its strong ribosome-binding site sequence and its optimal spacing before the start codon for cyanobacteria. For the strains that generally produced higher induction levels, S. elongatus PCC 7942 and Leptolyngbya sp. strain BL0902, both riboswitches B and F performed well. Riboswitch B provided low baseline expression and moderate induction and produced the highest induction ratios for these strains. Although riboswitch F (referred to as E* in references 14 and 18) allowed some baseline YFP expression in uninduced cultures, the level of YFP expression under inducing conditions was higher than that obtained with riboswitch B.
FIG 2.
Performance of theophylline-responsive riboswitch variants regulating YFP expression in model laboratory strains S. elongatus PCC 7942 and Anabaena sp. strain PCC 7120 and in model production strains Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn. Strains were treated with 2 mM theophylline or a 1% DMSO negative control for 1 day. (A) Fluorescence units (scale on the right) of YFP expression in induced (closed circles) and uninduced (open circles) cultures. The values shown are averages of three biological replicates ± the standard deviations. Columns depict induction ratios, calculated as induced values divided by uninduced values (scale on the left). Measurements were made with a fluorescence plate reader after normalization of cultures by OD750. (B) Fluorescence micrographs of induced and uninduced cultures of riboswitch F reporter strains. Images on the left show autofluorescence (auto) from photosynthetic pigments, and images on the right show YFP fluorescence. Scale bars, 5 μm. (C) Flow cytometry analysis of YFP induction in S. elongatus PCC 7942 and Synechocystis sp. strain WHSyn with riboswitch F regulation of YFP. Histograms with signal intensities on a log scale (x axis) versus the number of cells (y axis) show the distribution of autofluorescence (left, FL3), and the distribution of YFP expression (right, FL1).
Induction of YFP expression was verified through fluorescence microscopy imaging (Fig. 2B; see Fig. S1 and S2 in the supplemental material), which allowed us to examine YFP expression in individual cells and filaments. Unicellular S. elongatus PCC 7942 and Synechocystis sp. strain WHSyn showed little cell-to-cell variability in YFP expression after induction. Flow cytometry analysis of these strains showed population-wide shifts in YFP expression after induction, consistent with induction at the single-cell level (Fig. 2C; see Fig. S3 and S4 in the supplemental material). This low variability is comparable to induction in Leptolyngbya sp. strain BL0902, which had only moderate variability between cells within a single filament and between different filaments. In Anabaena sp. strain PCC 7120, there was also only moderate variability between cells within a single filament and between different filaments, potentially because of plasmid copy number variations. However, reporter expression measured at the population level (Fig. 2A) typically showed a low standard deviation between replicate samples for all four strains.
Characterization of riboswitch F in S. elongatus PCC 7942.
We further characterized riboswitch F in S. elongatus PCC 7942 because it supported high levels of expression after induction with relatively low baseline expression. To examine the range of protein expression from riboswitch F, we measured YFP expression at theophylline levels of 0.5 to 3.0 mM. YFP expression showed a dose-dependent increase with a linear response from 0.5 to 1.5 mM theophylline (Fig. 3A). We also examined the timing of induction by using both YFP and a destabilized YFP, which was constructed by appending a C-terminal AAV degradation tag that had been previously used in Synechocystis sp. strain PCC 6803 (10). The YFP-AAV reporter was included in this experiment to potentially produce a more accurate measure of the kinetics of induction. For both reporters, after induction with 2 mM theophylline, fluorescence increased steadily for 6 h before plateauing. The YFP and destabilized YFP-AAV reporters showed similar kinetics, but YFP-AAV reached only half the level of the unmodified YFP (Fig. 3B). These expression levels were maintained at 12 h and were similar to expression levels after 24 h (data not shown).
FIG 3.
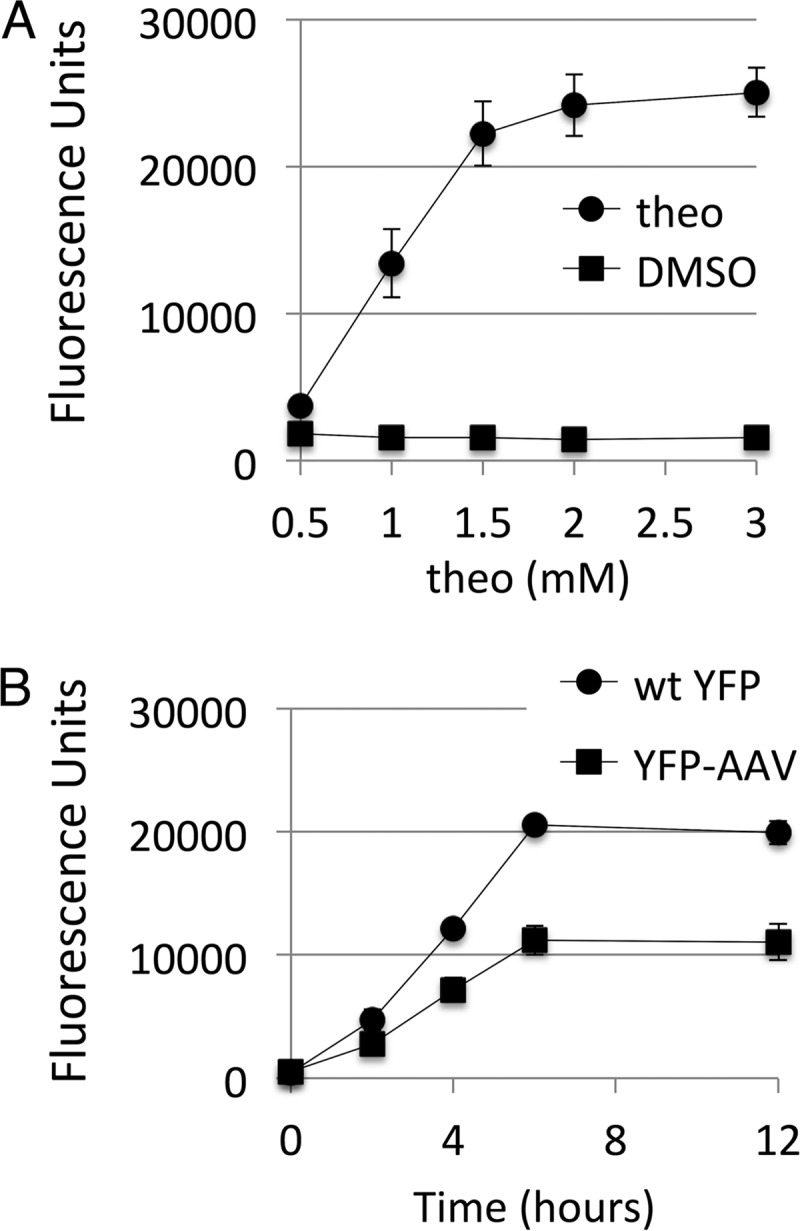
Effects of theophylline concentration and induction time. (A) S. elongatus PCC 7942 harboring riboswitch variant F regulating YFP expression was treated with a range of theophylline (theo) concentrations or equivalent volumes of a DMSO control. YFP fluorescence was measured 1 day after induction after normalization of cultures by OD750. (B) Time course of induction with YFP and destabilized YFP-AAV. Cultures were treated with 2 mM theophylline and harvested at 2, 4, 6, and 12 h for measurement of YFP expression. The values shown are averages of three biological replicates ± the standard deviations.
Comparison of theophylline/riboswitch, IPTG/LacI, and a hybrid regulatory system.
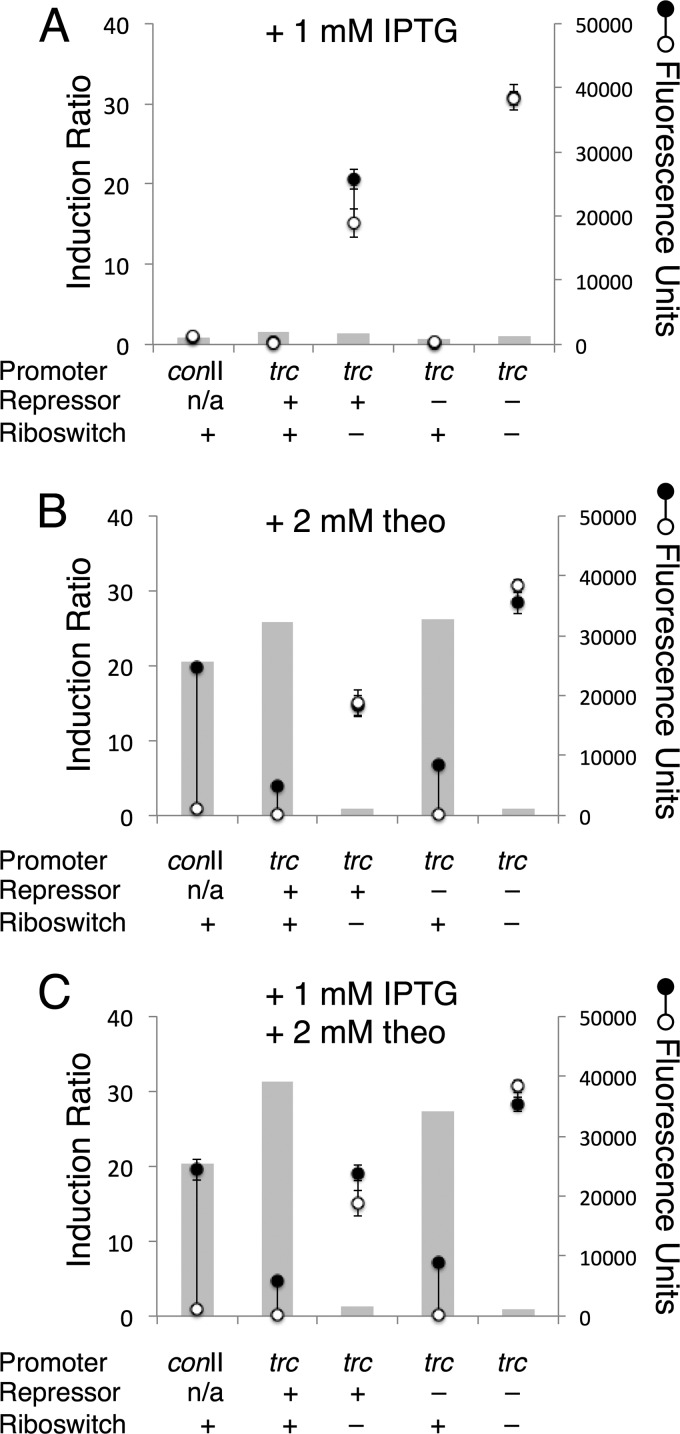
To compare the induction of gene expression with riboswitches to current inducible systems used in cyanobacteria, we examined theophylline induction of riboswitch F and IPTG induction of a Ptrc promoter, which is a hybrid of the E. coli trp and lacUV5 promoters. We constructed vectors to compare YFP expression driven from a trc promoter with and without regulation by riboswitch F and the LacI repressor produced by the lacIq gene. We found that in the absence of induction with theophylline or IPTG (Fig. 4, open circles), strains containing riboswitch F had low baseline expression of YFP. However, strains that relied on only LacI for repression of transcription had high baseline levels of YFP expression, a commonly known characteristic of IPTG induction. We observed some repressive activity conferred by LacI by comparing expression in strains that contain lacIq to strains that do not (Fig. 4, third and fifth columns).
FIG 4.
Comparison of riboswitch regulation, IPTG induction, and a hybrid regulatory system in S. elongatus PCC 7942. Reporter strains containing Ptrc driving expression of YFP were cloned with and without the lacIq repressor gene and with and without riboswitch F. Strains were induced with 1 mM IPTG (A), 2 mM theophylline (theo) (B), or 1 mM IPTG and 2 mM theophylline (C) (closed circles), and uninduced control cultures were treated with 1% DMSO (open circles). After induction for 1 day, cultures were normalized by OD750 and YFP expression was quantified with a fluorescence plate reader (scale on the right). The values shown are averages of three biological replicates ± the standard deviations. Gray columns depict induction ratios calculated by dividing induced values by uninduced values (scale on the left). n/a, not applicable.
We observed that the performance of riboswitch F induced with theophylline was superior to that of lacIq-Ptrc induced with IPTG. In the strain containing lacIq-Ptrc but lacking riboswitch F, we observed induction ratios in the presence of IPTG of 1.25× to 1.6× (Fig. 4A and C, third column), which are comparable to previous work (10, 18). This induction was not observed in strains also containing riboswitch F (Fig. 4A, second column) because the riboswitch effectively suppressed translation in the absence of theophylline. In strains containing riboswitch F, there was robust induction of YFP expression in the presence of theophylline (Fig. 4B, second and fourth columns). The induction ratios of Ptrc-driven expression regulated by riboswitch F were slightly higher than that for PconII-driven expression (Fig. 4B, first column), although the overall level of YFP expression after induction was lower, reaching a maximum of ∼24% of the YFP expression from strains lacking both the Lac repressor and riboswitch F (Fig. 4B, fifth column). The function of riboswitch F is potentially affected by the lac operator sequence present on the transcript (see Table S1 in the supplemental material). This impairment was not observed with riboswitch F transcripts from PconII, which reached ∼85% of YFP expression after induction compared to a construct lacking the aptamer sequence (see Fig. S5 in the supplemental material).
When we treated cells with both IPTG and theophylline, we saw a small increase in expression in the strain containing both lacIq-Ptrc and riboswitch F (Fig. 4C, second column), with an induction ratio of 31 rather than the 26 obtained by induction with theophylline alone (Fig. 4B, second column). Although combining the translational regulation of riboswitch F with transcriptional regulation by LacI/IPTG provided an incremental increase in induction, the contribution of IPTG-mediated transcriptional induction was minimal.
Regulated expression of toxic protein SacB.
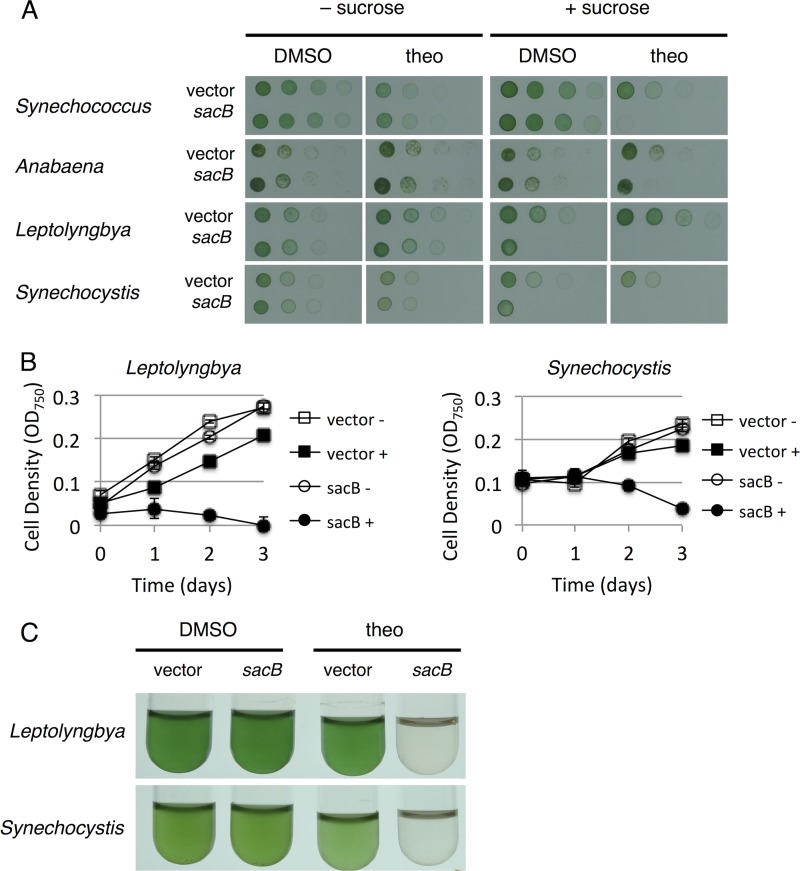
During the production of biofuels and other biological products, the formation of end products or their intermediates can be toxic to the algal crop. In this situation, a two-phase production strategy could be used in which an initial growth phase for accumulating biomass would be followed by a production phase during which induction of biosynthetic genes would occur. We expressed sacB as a model toxic gene under the regulation of riboswitch F in the four diverse cyanobacterial strains. The sacB gene encodes levansucrase, which produces toxic polysaccharide polymers, conferring sensitivity to sucrose and allowing counterselection of sacB in genetic experiments (27, 35). We tested the set of cyanobacterial species for regulated sucrose sensitivity in both solid-medium (Fig. 5A) and liquid-medium (Fig. 5B and C) assays.
FIG 5.
Regulation of toxic protein SacB with riboswitch variant F. (A) Solid-medium assay for sucrose sensitivity conferred by SacB expression. Sucrose concentrations were 5% for S. elongatus PCC 7942 and Anabaena sp. strain PCC 7120, 0.06% for Leptolyngbya sp. strain BL0902, and 0.25% for Synechocystis sp. strain WHSyn. BG11 agar plates contained a 0.5% DMSO vehicle control or 1 mM theophylline (theo). Plates were spotted with 5 μl of a cell suspension from a 5-fold dilution series and incubated for 3 days under standard conditions. (B) Liquid culture assay showing cell density over time for Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn. Cultures were treated with 1% sucrose and either 1% DMSO (open symbols) or 2 mM theophylline (closed symbols). The cell densities of 100-μl samples were measured with a microplate reader. The values shown are averages of three biological replicates ± the standard deviations. (C) Images of liquid cultures on day 3.
Overall, all of the cyanobacterial species displayed theophylline-dependent sucrose sensitivity in solid-medium assays, although there were differences between the different species. In the absence of sucrose, there was no difference in growth between vector controls and strains containing sacB on plates containing the DMSO vehicle or theophylline. However, when comparing growth on DMSO plates with that on theophylline plates (Fig. 5A), we observed some sensitivity to theophylline itself, particularly in S. elongatus PCC 7942 and Synechocystis sp. strain WHSyn. On BG11 agar plates, conditional sucrose toxicity was observed in S. elongatus PCC 7942 and Anabaena sp. strain PCC 7120 at 5% sucrose under inducing conditions with theophylline. With the DMSO vehicle alone, vector control strains and strains encoding sacB showed comparable growth (third column, first and second rows). Under theophylline-inducing conditions, S. elongatus PCC 7942 and Anabaena sp. strain PCC 7120 encoding sacB displayed reduced growth (fourth column, first and second rows). Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn were more sensitive to the toxic effects of SacB. On solid medium under inducing conditions, they displayed sucrose sensitivity at 0.06 and 0.25% sucrose, respectively (fourth column, third and fourth rows). At the same sucrose concentrations under noninducing conditions, Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn also displayed some sucrose sensitivity (third column, third and fourth rows). This suggests that the low-level baseline expression from riboswitch F allowed sufficient SacB production to confer toxicity to these strains even at low sucrose concentrations.
This high sensitivity of Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn to SacB toxicity in the presence of sucrose could also be observed in liquid culture assays (Fig. 5B and C). At 1% sucrose under inducing conditions, cultures of strains expressing SacB showed decreasing cell density and were cleared by day 3. Under noninducing conditions, these strains grew at rates comparable to those of the vector-only control strains. We observed slightly reduced growth associated with 2 mM theophylline in the strains containing vector controls. The solid-medium assay appeared to be more robust than the liquid-medium assay because it required lower sucrose concentrations. S. elongatus PCC 7942 and Anabaena sp. strain PCC 7120 were not as sensitive as Leptolyngbya sp. strain BL0902 and Synechocystis sp. strain WHSyn in solid-medium assays and did not display toxicity in the liquid-medium assay even at higher sucrose and theophylline concentrations (data not shown). Taken together, these experiments show effective riboswitch regulation of the model toxic protein SacB.
DISCUSSION
Cyanobacteria have great potential as a production platform for renewable resources because they derive energy from light and carbon from atmospheric carbon dioxide, such that they can produce biofuels and other commercially important products with a reduced carbon footprint. Because production costs are a major hurdle in the development of cyanobacteria for renewable production of biological products, devising methods to improve efficiency at every step of production is a major goal of cyanobacterial researchers. Improved methods for regulating gene expression in cyanobacteria, such as the use of riboswitches, can help advance this field because many steps of production can benefit from the regulated expression of genes. If production of the desired product is detrimental to cellular health or growth, an initial growth phase to maximize biomass could be followed by a production phase during which biosynthesis genes would be turned on by the addition of an inducer ligand. Another step of production that could be regulated is harvesting, such that genes that cause flocculation or floatation could be induced. Lysis of cells could also be induced to release end products for downstream processing. Misregulation of any one of these steps could potentially reduce biomass accumulation and decrease product formation, ultimately resulting in lower yields of the desired end products. Therefore, tight regulation of genes is a desirable trait for bioengineering applications. Our systematic evaluation of six theophylline-responsive riboswitches in four diverse cyanobacterial strains provides a useful guide for riboswitch selection. Different riboswitches could be used under different circumstances, such as conditions that require very low background levels or very high levels of induction. Additionally, our work with SacB shows that riboswitches can provide effective regulation of toxic gene products in cyanobacteria.
Riboswitches are increasingly being used for applied bioengineering (36) and are also being developed for use in eukaryotic cells (37, 38). While this particular set of theophylline-responsive riboswitches has proven to be effective in many different bacterial species, the potential for developing novel riboswitches is significant. In addition to selecting aptamers against exogenously added ligands, riboswitches that respond to endogenous signals generated within the cell can be developed to form feedback circuits within the cell (39). The modular nature and small size of riboswitches make them attractive targets for engineering or evolving riboswitches with new properties. The aptamer sequence binds its cognate ligand, while the expression platform sequence regulates gene expression dependent on the ligand occupancy of the aptamer. Aptamers that bind a ligand of choice can be obtained through in vitro evolution methods, and expression platform function can be obtained through in vivo screening. Because we observed some toxicity toward cyanobacteria at 2 mM theophylline, the potential to develop novel riboswitches responsive to alternative, less toxic ligands is a key benefit of using riboswitches to regulate gene expression (40). While positive selection markers such as antibiotic resistance genes can be used for screening of novel riboswitches, negative selection markers can also be useful. Expression of SacB in Leptolyngbya sp. strain BL0902 or Synechocystis sp. strain WHSyn could be used to screen for riboswitches with new properties because toxicity can be precisely tuned by varying the ligand and sucrose concentrations.
Riboswitches provide a powerful tool for synthetic biologists, and their use in genetic engineering of cyanobacteria is quite promising. Apart from basic transcription and translational machinery, riboswitches require only ligand-aptamer interaction to function. This obviates the need for exogenous proteins to be correctly made and then correctly interact with endogenous proteins, as would be required for heterologous transcription factors for inducible promoters. The simplicity of riboswitches makes this type of regulatory system more likely to function well among many different bacterial species, a notion supported by the diverse set of cyanobacterial strains used in this study. This is particularly meaningful to the field of algal biotechnology because riboswitches are likely to function in novel production strains of cyanobacteria.
The theophylline-responsive riboswitches in this work function through a translation initiation mechanism and can be paired with transcriptional regulation to provide an additional level of regulation. By combining different transcriptional regulatory systems with different riboswitches, the ability to fine tune the expression of target genes is increased, providing better control over metabolic and biosynthetic pathways. Although our experiments that combined IPTG induction with riboswitch regulation showed minimal benefits, this was due to a high baseline of transcription under noninducing conditions and a low induction ratio in the presence of IPTG. The low ability of IPTG to permeate cyanobacterial cells could play a role in the low induction ratio. In contrast, the theophylline-responsive riboswitches showed much better induction ratios, which may be partially due to better membrane permeability of theophylline, which is small and only weakly polar. An effective transcriptional regulatory system, such as the tetracycline-inducible system recently improved for use in cyanobacteria (11), could presumably provide the tight transcriptional control required for synergistic regulation of gene expression.
Through bioengineering and synthetic biology, microbes have been used for the production of a wide variety of products, such as industrial enzymes, vaccine components, antibody fragments and nanobodies, and therapeutics. Additionally, a variety of fuel molecules produced by microbes, including alkanes and alkenes, fatty acid methyl esters, and alcohols, are currently being developed as next-generation biofuels (41). An advantage of using photosynthetic cyanobacteria for commercial production over using heterotrophic microbes such as E. coli and Saccharomyces is that cyanobacteria potentially can be grown at very large industrial scales more cheaply. Some therapeutic proteins have already been engineered for production in eukaryotic algae (42). Arthrospira species of cyanobacteria (commonly referred to as Spirulina) are currently cultivated on an industrial scale as a nutritional supplement. The use of nitrogen-fixing cyanobacteria can eliminate the need for nitrogen fertilizer, a major cost and energy input into algal crop farming (31). Cyanobacteria have been engineered to produce fuel molecules and precursors, such as alcohols and triacylglycerols, and they naturally secrete overproduced free fatty acids (4, 43). With greater understanding of photosynthetic and metabolic processes (44), cyanobacteria can be engineered for optimal production of desired molecules. The relatively small genome size and genetic manipulability of cyanobacteria make them well suited for genetic engineering. The use of riboswitches for the versatile and robust regulation of gene expression in cyanobacteria will advance the field of algal biotechnology and facilitate the production of renewable next-generation biomolecules.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Department of Energy (DE-EE0003373) and the California Energy Commission Initiative for Large Molecule Sustainable Fuels (agreement 500-10-039). C.M.S. was supported by the Amgen Scholars Program. A.T.M. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
We thank Arnaud Taton for assistance with strain construction, providing the RSF1010 mobA Y25F plasmid, and phylogenetic tree analysis; Brian Palenik and Bianca Brahamsha for use of their light microscope and for Synechocystis sp. strain WHSyn; Emy Daniels for assistance with flow cytometry; and Joris Beld, Susan Cohen, and Mizuho Ota for critical reading of the manuscript.
Footnotes
Published ahead of print 22 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01697-14.
REFERENCES
- 1.Georgianna DR, Mayfield SP. 2012. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488:329–335. 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- 2.Gimpel JA, Specht EA, Georgianna DR, Mayfield SP. 2013. Advances in microalgae engineering and synthetic biology applications for biofuel production. Curr. Opin. Chem. Biol. 17:489–495. 10.1016/j.cbpa.2013.03.038. [DOI] [PubMed] [Google Scholar]
- 3.Berla BM, Saha R, Immethun CM, Maranas CD, Moon TS, Pakrasi HB. 2013. Synthetic biology of cyanobacteria: unique challenges and opportunities. Front. Microbiol. 4:246. 10.3389/fmicb.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducat DC, Way JC, Silver PA. 2011. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 29:95–103. 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Rosgaard L, de Porcellinis AJ, Jacobsen JH, Frigaard NU, Sakuragi Y. 2012. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J. Biotechnol. 162:134–147. 10.1016/j.jbiotec.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Gupta V, Ratha SK, Sood A, Chaudhary V, Prasanna R. 2013. New insights into the biodiversity and applications of cyanobacteria (blue-green algae)—prospects and challenges. Algal Res. 2:79–97. 10.1016/j.algal.2013.01.006. [DOI] [Google Scholar]
- 7.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, Herdman M, Sivonen K, Coursin T, Laurent T, Goodwin L, Nolan M, Davenport KW, Han CS, Rubin EM, Eisen JA, Woyke T, Gugger M, Kerfeld CA. 2013. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 110:1053–1058. 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruffing AM. 2011. Engineered cyanobacteria: teaching an old bug new tricks. Bioeng. Bugs 2:136–149. 10.4161/bbug.2.3.15285. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Wang J, Zhang W, Meldrum DR. 2012. Application of synthetic biology in cyanobacteria and algae. Front. Microbiol. 3:344. 10.3389/fmicb.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang HH, Camsund D, Lindblad P, Heidorn T. 2010. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 38:2577–2593. 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang HH, Lindblad P. 2013. Wide-dynamic-range promoters engineered for cyanobacteria. J. Biol. Eng. 7:10. 10.1186/1754-1611-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol. Cell 43:867–879. 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wachter A. 2010. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 7:67–76. 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 14.Topp S, Reynoso CM, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, Scott JR, Gallivan JP. 2010. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 76:7881–7884. 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeliger JC, Topp S, Sogi KM, Previti ML, Gallivan JP, Bertozzi CR. 2012. A riboswitch-based inducible gene expression system for mycobacteria. PLoS One 7(1):e29266. 10.1371/journal.pone.0029266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynoso CM, Miller MA, Bina JE, Gallivan JP, Weiss DS. 2012. Riboswitches for intracellular study of genes involved in Francisella pathogenesis. mBio 3(6):e00253–12. 10.1128/mBio.00253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudolph MM, Vockenhuber MP, Suess B. 2013. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology 159:1416–1422. 10.1099/mic.0.067322-0. [DOI] [PubMed] [Google Scholar]
- 18.Nakahira Y, Ogawa A, Asano H, Oyama T, Tozawa Y. 2013. Theophylline-dependent riboswitch as a novel genetic tool for strict regulation of protein expression in cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol. 54:1724–1735. 10.1093/pcp/pct115. [DOI] [PubMed] [Google Scholar]
- 19.Elledge SJ, Davis RW. 1989. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 3:185–197. 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- 20.Clerico EM, Ditty JL, Golden SS. 2007. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol. Biol. 362:155–171. 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- 21.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. 2007. The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J. Mol. Biol. 366:165–178. 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taton A, Unglaub F, Wright NE, Zeng WY, Paz-Yepes J, Brahamsha B, Palenik B, Peterson TC, Haerizadeh F, Golden SS, Golden JW. 29 July 2014. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon H, Golden JW. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935–938. 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 24.Taton A, Lis E, Adin DM, Dong G, Cookson S, Kay SA, Golden SS, Golden JW. 2012. Gene transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts. PLoS One 7(1):e30901. 10.1371/journal.pone.0030901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. 2003. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J. Biol. Chem. 278:19102–19110. 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 27.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577. 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 31.Peccia J, Haznedaroglu B, Gutierrez J, Zimmerman JB. 2013. Nitrogen supply is an important driver of sustainable microalgae biofuel production. Trends Biotechnol. 31:134–138. 10.1016/j.tibtech.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Ng WO, Zentella R, Wang Y, Taylor JS, Pakrasi HB. 2000. phrA, the major .sp. strain PCC 6803 codes for a cyclobutane-pyrimidine-dimer-specific DNA photolyase. Arch. Microbiol. 173:412–417. 10.1007/s002030000164. [DOI] [PubMed] [Google Scholar]
- 33.Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323:124–131. 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Huang XZ, Wang L, Risoul V, Zhang CC, Chen WL. 2013. Phenotypic variation caused by variation in the relative copy number of pDU1-based plasmids expressing the GAF domain of Pkn41 or Pkn42 in Anabaena sp. PCC 7120. Res. Microbiol. 164:127–135. 10.1016/j.resmic.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Ried JL, Collmer A. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239–246. 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- 36.Weigand JE, Suess B. 2009. Aptamers and riboswitches: perspectives in biotechnology. Appl. Microbiol. Biotechnol. 85:229–236. 10.1007/s00253-009-2194-2. [DOI] [PubMed] [Google Scholar]
- 37.Bocobza SE, Aharoni A. 2008. Switching the light on plant riboswitches. Trends Plant Sci. 13:526–533. 10.1016/j.tplants.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Verhounig A, Karcher D, Bock R. 2010. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc. Natl. Acad. Sci. U. S. A. 107:6204–6209. 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fowler CC, Li Y. 2014. Construction and application of riboswitch-based sensors that detect metabolites within bacterial cells. Methods Mol. Biol. 1103:177–197. 10.1007/978-1-62703-730-3_14. [DOI] [PubMed] [Google Scholar]
- 40.Wittmann A, Suess B. 2012. Engineered riboswitches: expanding researchers' toolbox with synthetic RNA regulators. FEBS Lett. 586:2076–2083. 10.1016/j.febslet.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 41.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556–563. 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Rasala BA, Muto M, Lee PA, Jager M, Cardoso RM, Behnke CA, Kirk P, Hokanson CA, Crea R, Mendez M, Mayfield SP. 2010. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 8:719–733. 10.1111/j.1467-7652.2010.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaiser BK, Carleton M, Hickman JW, Miller C, Lawson D, Budde M, Warrener P, Paredes A, Mullapudi S, Navarro P, Cross F, Roberts JM. 2013. Fatty aldehydes in cyanobacteria are a metabolically flexible precursor for a diversity of biofuel products. PLoS One 8(3):e58307. 10.1371/journal.pone.0058307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver JW, Atsumi S. 2014. Metabolic design for cyanobacterial chemical synthesis. Photosynth. Res. 120:249–261. 10.1007/s11120-014-9997-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.