Abstract
The aim of this study was to investigate the phenotypic and genotypic antibiotic resistance profiles of pseudomonads isolated from surfaces of a goat and lamb slaughterhouse, which were representative of areas that are possible sources of meat contamination. Mesophilic (85 isolates) and psychrotrophic (37 isolates) pseudomonads identified at the species level generally were resistant to sulfamethoxazole, erythromycin, amoxicillin, ampicillin, chloramphenicol, trimethoprim, rifampin, and ceftazidime (especially mesophiles), as well as colistin and tetracycline (especially psychrotrophes). However, they generally were sensitive to ciprofloxacin, gentamicin, imipenem, and kanamycin regardless of species identity. Worryingly, in the present study, we found multidrug resistance (MDR) to up to 13 antibiotics, which was related to intrinsic and acquired resistance mechanisms. Furthermore, a link between various antimicrobial resistance genes was shown for beta-lactams and tetracycline, trimethoprim, and sulfonamides. The distribution and resistome-based analysis of MDR pseudomonads in different slaughterhouse zones indicated that the main sources of the identical or related pseudomonad strains were the animals (feet and wool) and the slaughterhouse environment, being disseminated from the beginning, or entrance environment, to the environment of the finished meat products. Those facts must be taken into consideration to avoid cross-contamination with the subsequent flow of mobile resistance determinants throughout all slaughterhouse zones and then to humans and the environment by the application of adequate practices of hygiene and disinfection measures, including those for animal wool and feet and also the entrance environment.
INTRODUCTION
The genus Pseudomonas belongs to the bacterial family Pseudomonadaceae of the class Gammaproteobacteria and is considered the most heterogeneous group of Gram-negative bacteria, including aerobic rods and motile, catalase-positive, and non-spore-forming bacteria (1). Their oxygen requirement can be changed under anaerobic conditions by using an alternative electron acceptor, such as nitrate. These bacteria are ubiquitous because of their simple nutritional requirements and their high metabolic versatility, having been isolated from a variety of sources, like soil, fresh water, humans, plant and animal surfaces, cosmetics, medical products and instruments, and foods of animal and vegetal origins. Thus, Pseudomonas belongs to a group of organisms of great ecological importance as opportunistic pathogens causing a variety of infectious diseases in animals and humans, since they are part of the normal bacterial flora of the pharynx, mucous membranes, and skin of humans (2). They also play a role as phytopathogens (3, 4) and as spoilage organisms. In this sense, pseudomonads may cause off-flavor (5–7), especially in proteinaceous foods with high water activity, like meat and fresh cheese, browning of minimally processed vegetables because of their pectinolytic activity (8, 9), off-flavor in fish products due to the production of volatile compounds and degradation of amino acids (10–13), and lipolysis and proteolysis of processed milk due to the production of thermotolerant enzymes (6, 14). Furthermore, they are of great concern in chilled food spoilage because of their psychrotrophic conditions, especially Pseudomonas fragi, Pseudomonas putida, and Pseudomonas fluorescens (15), which causes bitterness, putrefaction, rancid odor, liquefaction, gelatinization of curd, and slime and mucous formation on cheese surfaces.
Pseudomonads as spoilage or as pathogenic bacteria could inhabit vastly different ecological niches where the key factors driving to the emergence of resistance may be present (antibiotics and antibiotic resistance, or AR, genes) (16). The spread of multiple-drug-resistant (MDR) pseudomonads from different sources to humans and also to the environment implies the frequent spread of resistance genes by horizontal gene transfer (HGT), since many of them are located on plasmids, integrons, or transposons (17). The evolution and dissemination of AR genes among pseudomonads and among environments globally, which were enhanced by their genetic flexibility and versatility, is an increasing problem in infectious diseases. In this way, gene transfer crosses species and genus barriers (18); thus, genes flow to and from Gram-positive and Gram-negative bacteria in different environments.
The prevalence of MDR pseudomonads and enterobacteria in slaughterhouses, including swine and poultry environments, has been reported largely in several studies (19–22), creating a growing concern about their impact on animal and human health. At the slaughter and processing plant and the farm, it is difficult to reduce risks related to pathogens normally present in the gut of healthy animals and the teat, so microorganisms present in animal foods and their processing environment may cause a great challenge for human health in terms of their pathogenic power and their role as a potential reservoir of AR genes (20, 23, 24). Indeed, it is interesting that commensal bacteria, considered free of health risk, also could be vehicles of AR genes, although food-borne pathogens are the main reservoirs (25, 26). The principal factor linked to the emergence of microbial resistance is the extensive use or, rather, misuse of antibiotics in different areas, such as bacterial infection treatment, animal husbandry, and agriculture (27–32), which may generate an enormous worldwide selective pressure (16, 33).
In the present study, we report for the first time the prevalence of multiple-antibiotic-resistant pseudomonads in a goat and lamb slaughterhouse. This study involved the analysis of phenotypic and genotypic antibiotic resistance profiles of 122 mesophilic and psychrotrophic pseudomonads isolated from slaughterhouse surfaces throughout the chain of meat production and also from the end products. Mesophilic (growth at 30°C for 72 h) and psychrotrophic (growth at 7°C for 10 days) pseudomonads were isolated as described by Lavilla Lerma et al. (34) in King agar and tryptone soya agar, respectively. Furthermore, we evaluated the relationship between environmental pseudomonads and the end products with the aim of elucidating if they share an antibiotic resistome.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Thirty-seven psychrotrophic and 85 mesophilic isolates of antibiotic-resistant pseudomonads were used in the present study. The strains were obtained from different surfaces (entrance [E], sacrifice room [SR], cold room, cutting room [CR], freezing tunnel [FrT], and white room [WR]) of a local goat and lamb slaughterhouse in Jaén (Spain) and also from five meat products from different shops in Jaén, as described previously by Lavilla Lerma et al. (34). All strains were maintained and stored in tryptone soya broth (TSB; Scharlab, Barcelona, Spain) containing 20% glycerol at −80°C. For routine use, mesophilic and psychrotrophic pseudomonads were cultivated on TSB at 22°C for 24 to 48 h.
Antimicrobial agents.
The antimicrobial agents used in this study included various antibiotics used in the clinical area, such as penicillins (amoxicillin [AMX] and ampicillin [AMP]), a cephalosporin (ceftazidime [CAZ]), a fluoroquinolone (ciprofloxacin [CIP]), miscellaneous drugs (chloramphenicol, [CHL], rifampin [RIF], sulfamethoxazole [SMZ], and trimethoprim [TMP]), a macrolide (erythromycin [ERY]), aminoglycosides (gentamicin [GEN], kanamycin [KAN], and streptomycin [STR]), a carbapenem (imipenem [IPM]), lipopeptides (colistin [CL] and polymyxin B [PB]), and tetracycline (TET). Stock solutions of all antibiotics used in the present study (Tables 1 and 2) were prepared and diluted according to guidelines of the Clinical and Laboratory Standards Institute (38).
TABLE 1.
MIC distribution of 16 antibiotics for mesophilic pseudomonads isolated throughout the chain of meat production
| Antibiotic and species | No. of isolates in each MIC rangee (μg/ml) |
MIC breakpoint (μg/ml) | No. of resistant strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥1-<2 | ≥2-<4 | ≥4-<8 | ≥8-<16 | ≥16-<32 | ≥32-<64 | ≥64-<128 | ≥128-<256 | ≥256-<512 | ≥512 | |||
| Amoxicillin | ||||||||||||
| P. putida | 1 | 1 | ≥32a | 2 | ||||||||
| P. lundensis | 4 | 1 | 2 | 1 | 11 | 1 | 4 | 7 | 24 | |||
| P. fluorescens | 1 | 2 | 2 | |||||||||
| P. alkylphenolia | 1 | 1 | 2 | |||||||||
| Ampicillin | ||||||||||||
| P. putida | 1 | 1 | ≥32a | 2 | ||||||||
| P. lundensis | 4 | 1 | 1 | 2 | 9 | 1 | 4 | 9 | 25 | |||
| P. fluorescens | 1 | 1 | 1 | 3 | ||||||||
| P. alkylphenolia | 1 | 1 | 2 | |||||||||
| Ceftazidime | ||||||||||||
| P. putida | 1 | 1 | ≥32b | 2 | ||||||||
| P. lundensis | 2 | 2 | 5 | 2 | 4 | 2 | 14 | 22 | ||||
| P. fluorescens | 1 | 1 | 1 | 2 | ||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Imipenem | ||||||||||||
| P. putida | 1 | 1 | ≥16b | 0 | ||||||||
| P. lundensis | 10 | 7 | 9 | 4 | 1 | 1 | ||||||
| P. fluorescens | 1 | 1 | 1 | 1 | ||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Gentamicin | ||||||||||||
| P. putida | 2 | ≥16b | 0 | |||||||||
| P. lundensis | 7 | 10 | 8 | 2 | 1 | 3 | 6 | |||||
| P. fluorescens | 2 | 1 | 0 | |||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Kanamycin | ||||||||||||
| P. putida | 1 | 1 | ≥64a | 0 | ||||||||
| P. lundensis | 11 | 0 | 6 | 8 | 2 | 3 | 1 | 4 | ||||
| P. fluorescens | 1 | 2 | 0 | |||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Streptomycin | ||||||||||||
| P. putida | 1 | 1 | ≥64a | 0 | ||||||||
| P. lundensis | 1 | 1 | 4 | 12 | 5 | 2 | 4 | 2 | 8 | |||
| P. fluorescens | 2 | 1 | 0 | |||||||||
| P. alkylphenolia | 1 | 1 | 1 | |||||||||
| Rifampin | ||||||||||||
| P. putida | 2 | ≥32d | 2 | |||||||||
| P. lundensis | 2 | 1 | 3 | 16 | 5 | 3 | 1 | 25 | ||||
| P. fluorescens | 2 | 1 | 3 | |||||||||
| P. alkylphenolia | 2 | 2 | ||||||||||
| Sulfamethoxazole | ||||||||||||
| P. putida | 2 | ≥512b | 2 | |||||||||
| P. lundensis | 31 | 31 | ||||||||||
| P. fluorescens | 3 | 3 | ||||||||||
| P. alkylphenolia | 2 | 2 | ||||||||||
| Trimethoprim | ||||||||||||
| P. putida | 1 | 1 | ≥16a | 1 | ||||||||
| P. lundensis | 2 | 7 | 1 | 1 | 2 | 1 | 17 | 22 | ||||
| P. fluorescens | 2 | 1 | 3 | |||||||||
| P. alkylphenolia | 2 | 2 | ||||||||||
| Colistin | ||||||||||||
| P. putida | 1 | 1 | ≥8b | 0 | ||||||||
| P. lundensis | 7 | 4 | 2 | 1 | 1 | 4 | 1 | 11 | 20 | |||
| P. fluorescens | 1 | 1 | 1 | 1 | ||||||||
| P. alkylphenolia | 1 | 1 | 0 | |||||||||
| Polymyxin B | ||||||||||||
| P. putida | 1 | 1 | ≥8b | 0 | ||||||||
| P. lundensis | 10 | 6 | 1 | 1 | 13 | 15 | ||||||
| P. fluorescens | 2 | 1 | 0 | |||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Erythromycin | ||||||||||||
| P. putida | 2 | >4c | 2 | |||||||||
| P. lundensis | 4 | 1 | 1 | 9 | 9 | 7 | 27 | |||||
| P. fluorescens | 1 | 2 | 3 | |||||||||
| P. alkylphenolia | 2 | 2 | ||||||||||
| Chloramphenicol | ||||||||||||
| P. putida | 2 | ≥32b | 2 | |||||||||
| P. lundensis | 3 | 8 | 9 | 5 | 1 | 5 | 20 | |||||
| P. fluorescens | 1 | 1 | 1 | 2 | ||||||||
| P. alkylphenolia | 1 | 1 | 2 | |||||||||
| Ciprofloxacin | ||||||||||||
| P. putida | 2 | ≥4b | 0 | |||||||||
| P. lundensis | 29 | 1 | 1 | 2 | ||||||||
| P. fluorescens | 3 | 0 | ||||||||||
| P. alkylphenolia | 2 | 0 | ||||||||||
| Tetracycline | ||||||||||||
| P. putida | 2 | ≥16b | 0 | |||||||||
| P. lundensis | 10 | 3 | 7 | 6 | 1 | 2 | 1 | 1 | 11 | |||
| P. fluorescens | 1 | 1 | 1 | 0 | ||||||||
| P. alkylphenolia | 2 | 2 | ||||||||||
In the case of nondescribed antibiotics, we considered the breakpoint values suggested by CLSI (35) for E. coli.
The microbiological breakpoint values according to CLSI (35) for Pseudomonas sp. are given.
The microbiological breakpoint values according to Bruchmann et al. (36) for Pseudomonas sp. are given.
The microbiological breakpoint values according to Tribuddharat and Fennewald (37) for Pseudomonas sp. are given.
Resistant strains with a MIC higher than the breakpoints described in the table are indicated in boldface.
TABLE 2.
MIC distribution of 18 antibiotics for psychrotrophic pseudomonads isolated throughout the chain of meat production
| Antibiotic and species | No. of isolates with the following MIC rangee (μg/ml) |
MIC breakpoint (μg/ml) | No. of resistant strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥1-<2 | ≥2-<4 | ≥4-<8 | ≥8-<16 | ≥16-<32 | ≥32-<64 | ≥64-<128 | ≥128-<256 | ≥256-<512 | ≥512 | |||
| Amoxicillin | ||||||||||||
| P. fragi | 1 | 1 | 2 | ≥32a | 4 | |||||||
| P. putida | 2 | 2 | 1 | 2 | 7 | |||||||
| P. lundensis | 3 | 3 | ||||||||||
| Ampicillin | ||||||||||||
| P. fragi | 2 | 2 | ≥32a | 4 | ||||||||
| P. putida | 1 | 4 | 2 | 7 | ||||||||
| P. lundensis | 2 | 1 | 3 | |||||||||
| Ceftazidime | ||||||||||||
| P. fragi | 3 | 1 | ≥32b | 1 | ||||||||
| P. putida | 2 | 1 | 1 | 2 | 1 | 3 | ||||||
| P. lundensis | 3 | 0 | ||||||||||
| Imipenem | ||||||||||||
| P. fragi | 3 | 1 | ≥16b | 1 | ||||||||
| P. putida | 4 | 2 | 1 | 1 | ||||||||
| P. lundensis | 3 | 0 | ||||||||||
| Gentamicin | ||||||||||||
| P. fragi | 2 | 1 | 1 | ≥16b | 0 | |||||||
| P. putida | 4 | 1 | 1 | 1 | 1 | |||||||
| P. lundensis | 3 | 0 | ||||||||||
| Kanamycin | ||||||||||||
| P. fragi | 2 | 1 | ≥64a | 0 | ||||||||
| P. putida | 3 | 2 | 1 | 1 | 1 | |||||||
| P. lundensis | 3 | 0 | ||||||||||
| Streptomycin | ||||||||||||
| P. fragi | 3 | 1 | ≥64a | 1 | ||||||||
| P. putida | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |||||
| P. lundensis | 1 | 2 | 0 | |||||||||
| Rifampin | ||||||||||||
| P. fragi | 3 | 1 | ≥32d | 1 | ||||||||
| P. putida | 1 | 3 | 1 | 1 | 1 | 3 | ||||||
| P. lundensis | 3 | 0 | ||||||||||
| Sulfamethoxazole | ||||||||||||
| P. fragi | 2 | 2 | ≥512b | 2 | ||||||||
| P. putida | 2 | 5 | 5 | |||||||||
| P. lundensis | 1 | 2 | 2 | |||||||||
| Trimethoprim | ||||||||||||
| P. fragi | 1 | 1 | 1 | 1 | ≥16a | 3 | ||||||
| P. putida | 1 | 1 | 5 | 5 | ||||||||
| P. lundensis | 3 | 3 | ||||||||||
| Colistin | ||||||||||||
| P. fragi | 1 | 2 | 1 | ≥8b | 1 | |||||||
| P. putida | 1 | 3 | 1 | 1 | 1 | 3 | ||||||
| P. lundensis | 1 | 1 | 1 | 2 | ||||||||
| Polymyxin B | ||||||||||||
| P. fragi | 3 | 1 | ≥8b | 1 | ||||||||
| P. putida | 1 | 4 | 1 | 1 | 2 | |||||||
| P. lundensis | 2 | 1 | 1 | |||||||||
| Erythromycin | ||||||||||||
| P. fragi | 3 | 1 | >4c | 4 | ||||||||
| P. putida | 1 | 2 | 1 | 3 | 7 | |||||||
| P. lundensis | 1 | 1 | 1 | 3 | ||||||||
| Chloramphenicol | ||||||||||||
| P. fragi | 2 | 2 | ≥32b | 4 | ||||||||
| P. putida | 2 | 1 | 4 | 5 | ||||||||
| P. lundensis | 1 | 2 | 3 | |||||||||
| Ciprofloxacin | ||||||||||||
| P. fragi | 4 | ≥4b | 0 | |||||||||
| P. putida | 7 | 0 | ||||||||||
| P. lundensis | 3 | 0 | ||||||||||
| Tetracycline | ||||||||||||
| P. fragi | 3 | 1 | ≥16b | 1 | ||||||||
| P. putida | 1 | 3 | 3 | 3 | ||||||||
| P. lundensis | 1 | 2 | 2 | |||||||||
In the case of nondescribed antibiotics, we considered the breakpoint values suggested by CLSI (35) for E. coli.
The microbiological breakpoint values according to CLSI (35) for Pseudomonas sp. are given.
The microbiological breakpoint values according to Bruchmann et al. (36) for Pseudomonas sp. are given.
The microbiological breakpoint values according to Tribuddharat and Fennewald (37) for Pseudomonas sp. are given.
Resistant strains with a MIC higher than the breakpoints described in the table are indicated in boldface.
Molecular identification of pseudomonads. (i) DNA extraction.
Total DNA was extracted from cultures by the method described by de Los Reyes-Gavilan et al. (39). This DNA preparation was used in further PCRs.
(ii) ERIC-PCR fingerprinting of Pseudomonas strains.
Enterobacterial repetitive intergenic consensus (ERIC)-PCR fingerprinting of Pseudomonas isolates was done as described by Martín-Platero et al. (40). DNA was amplified with primer ERIC1-R in 35 cycles (94°C for 3 min; 35 cycles of 94°C for 30 s, 48°C for 60 s, and 72°C for 5 min; and 72°C for 7 min). Reactions were carried out in a total volume of 25 μl containing 2.5 μl of 10× Taq reaction buffer, 3 mM MgCl2, 400 μM deoxynucleoside triphosphates (dNTPs), 1 μM ERIC1-R primer (5′-ATGTAAGCTCCTGGGGATTCAC-3′), 1 U of Taq DNA polymerase (GE Healthcare), and 1 μl of template DNA. Amplification products were separated by electrophoresis on a 1.8% (wt/vol) agarose gel in 1× TBE buffer (0.45 mM Tris-HCl, 0.45 mM boric acid, 1 mM EDTA, pH 8.3) during 16 h at 46 V. The gels were stained in ethidium bromide and photographed on a UV transilluminator. Photopositives were scanned into a computer and subsequently analyzed using Bionumerics software, version 2.5 (Applied Maths, Kortrijk, Belgium). The grouping of the ERIC-PCR patterns was performed by means of the Pearson product moment correlation coefficient and cluster analysis with the unweighted-pair group method using arithmetic average linkages (UPGMA).
(iii) Identification of Pseudomonas sp. strains at species level.
Once the fingerprinting analysis was done, representative strains of each cluster were selected for their genetic identification by sequencing of rpoD and gyrB genes amplified by PCR using the primers described by Yamamoto et al. (41), and the nucleotide sequences were deposited in GenBank under accession numbers KM364994 to KM365013 and KM370331 to KM370332. A search for homology of the DNA sequences was done using the BLAST algorithm available at the National Centre for Biotechnology Information (NCBI).
To confirm the identity of strains, a multiplex species-specific PCR of the carbamoyl phosphate synthase gene small subunit (carA) was done as described by Ercolini et al. (42) to detect P. lundensis and P. putida.
Antimicrobial susceptibility testing.
The MICs of the above-mentioned antibiotics were measured in a concentration range from 2 to 4,096 μg/ml for all antibiotics except imipenem, which ranged from 1 to 4,096 μg/ml. After incubation, the MIC was read as the lowest concentration of each antimicrobial agent that inhibited the visible growth of the strain. All of the MIC determinations of each antimicrobial for each strain were carried out in triplicate, and reliable results were taken if at least two out of three replicates were in agreement. The microbiological breakpoints of most antibiotics tested were those defined by CLSI (35). Concerning beta-lactams (amoxicillin and ampicillin), kanamycin, streptomycin, and trimethoprim, we used the microbiological breakpoint proposed by CLSI (35) for Escherichia coli, since the official ECOFF (epidemiological cutoff) for Pseudomonas spp. has not been designated by the same international organization. The microbiological breakpoints of erythromycin and rifampin were those proposed by Bruchmann et al. (36) and Tribuddharat and Fennewald (37), respectively.
Molecular screening of resistance determinants.
PCR amplifications of well-known structural genes associated with resistance to beta-lactams (blaOXA, blaCTX, blaSHV-1, and blaTEM), chloramphenicol (catA1, catA2, catA3, and catB3), macrolides (ereA, ereB, ermA, ermB, msrA and mrsB, and mefA), tetracycline (tetA, tetB, tetO, and tetQ), aminoglycosides [aad(E), aphA-3, aac(6′)-Ie-aph(2′)-Ia, aph(2′)-Ib, aph(2′)-Ic, aph(2′)-Id, aph(3′)-IIIa, and ant(4′)-Ia], trimethoprim (dfrA and dfrD), and sulfonamide (sulI, sulII, and sulIII) were performed by following methods described elsewhere (43–50) and using primers listed in Table S1 in the supplemental material. Efflux pumps mediating multiple antibiotic resistance also were included in this study (see Table S1), such as AcrA, AcrB, TolC, MexAB, MexCD, and MexXY (51, 52).
To investigate whether observed resistance to rifampin was due to mutations in the rpoB gene, PCR of the partial rpoB gene fragment (nucleotides 1524 to 2159) was done as described by Hosokawa et al. (53) using the primers listed in Table S1 in the supplemental material. The nucleotide sequences were deposited in GenBank under accession numbers KM370326 to KM370330.
Analysis of integrons.
Class 1, 2, and 3 integrons were detected as described by White et al. (54) and Ploy et al. (55) (see Table S1 in the supplemental material).
Statistical analysis.
Statistical analysis of data was accomplished using Excel 2007 and XLSTAT 2014 (trial version 2014.1.03; Addinsoft, France), and the correlation between all slaughterhouse variables (zones, antibiotics, and population type) and phenotypic resistance was determined by principal component analysis (PCA).
To identify the source of multiple antibiotic resistance, agglomerative hierarchical cluster analysis was performed using XLSTAT 2014 (trial version 2014.1.03; Addinsoft, France) according to Ward's method for clustering and the square Euclidean distance as a measure of distance grouping to measure population similarities between sampling zones in pseudomonad resistomes, which was based on the incidence of resistance determinants.
Nucleotide sequence accession numbers.
Nucleotide sequences determined during this work were deposited in GenBank under the following accession numbers: KM364994 to KM365013, KM370326 to KM370330, and KM370331 to KM370332.
RESULTS
Fingerprinting and identification of antibiotic-resistant mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces and meat products.
A collection of 122 isolates of antibiotic-resistant pseudomonads (85 and 37 mesophilic and psychrotrophic isolates, respectively) isolated by Lavilla Lerma et al. (34) were reduced to 52 (38 mesophilic and 14 psychrotrophic) strains by ERIC-PCR analysis, since isolates with identical ERIC-PCR patterns were considered the same strain (see Fig. S1 in the supplemental material). The genetic diversity of mesophilic pseudomonads was studied by ERIC-PCR, and the dendrogram generated using Pearson correlation demonstrated the existence of one main cluster, G1 (80% was used as the cutoff for defining the cluster), subdivided in two subclusters: subcluster G1A, with 26 strains, and G1B, with 12 strains (see Fig. S1A). Similarly, psychrotrophic pseudomonads showed one main cluster, G1 (37 strains), subdivided in two subclusters, G1A and G1B, with 11 and 3 strains, respectively (see Fig. S1B). The identification of representative strains of each group in the dendrograms revealed that mesophilic pseudomonads were represented mainly by P. lundensis (82%), followed by a small proportion of P. fluorescens (8%), P. alkylphenolia (5%), and P. putida (5%) (see Fig. S1A). However, psychrotrophic pseudomonads were represented by P. putida (50%), P. fragi (29%), and P. lundensis (21%) (see Fig. S1B). The analysis of ERIC-PCR dendrograms displaying the distances between the 122 strains revealed that both mesophilic and psychrotrophic pseudomonads showed low degrees of heterogeneity (similarity coefficient, ~80.4 to 82%).
On the other hand, the analysis of ERIC-PCR dendrograms showed that strains isolated from the end products (the same clone) also were detected on surfaces throughout the chain of meat production. Furthermore, isolates detected at the entrance (E) were the same as those detected in the end products (i.e., P. lundensis 1K04.1 and P. lundensis 1K13.1 from meat product were the same clones as P. lundensis M1K10.2 and P. lundensis M1K06.2, respectively, from the entrance).
Antibiotic susceptibility assays and MIC distributions.
The MIC determination of the different antibiotics was performed with 52 pseudomonads identified at the species level in the present study. The results obtained (Tables 1 and 2 and Fig. 1 and 2) indicated that resistance to 16 antibiotics was detected in almost all pseudomonads tested depending on the antibiotic used, the species analyzed (P. fragi, P. alkylphenolia, P. fluorescens, P. lundensis, and P. putida), the population type, and the sampling zone.
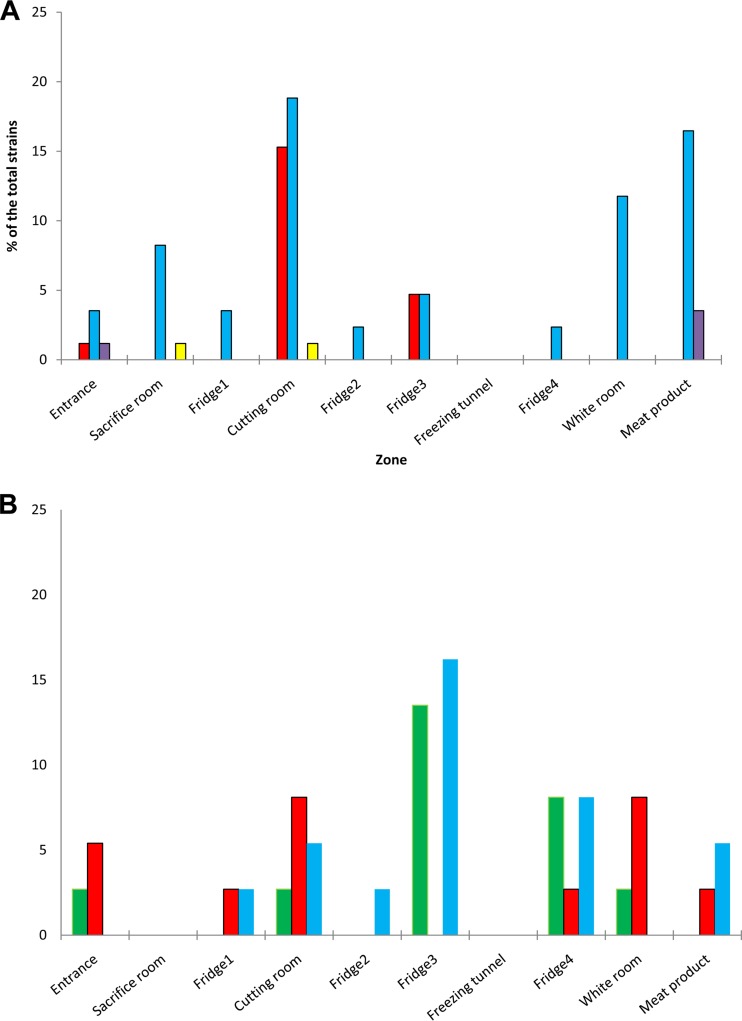
FIG 1.
Resistance to antibiotics of mesophilic (A) and psychrotrophic (B) pseudomonads isolated from slaughterhouse surfaces throughout the chain of meat production and from the end products according to species identity. P. alkylphenolia, yellow; P. fluorescens, purple; P. lundensis, blue; P. putida, red; P. fragi, green.
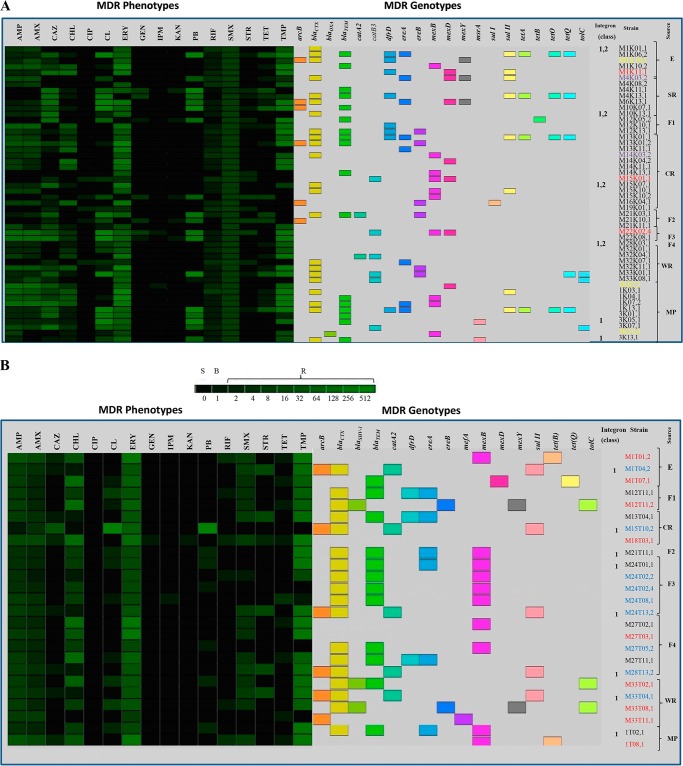
FIG 2.
Heat-map summary of MDR phenotypes and genotypes, proteolytic activity, and the sources of mesophilic (A) and psychrotrophic (B) pseudomonads isolated from slaughterhouse surfaces throughout the chain of meat production and from the end products. The level of resistance is indicated by a green scale (R, resistant; S, susceptible; B, breaking point). Antimicrobial abbreviations: AMP, ampicillin; AMX, amoxicillin; CAZ, ceftazidime; CHL, chloramphenicol; CIP, ciprofloxacin; CL, colistin; ERY, erythromycin; GEN, gentamicin; IPM, imipenem; KAN, kanamycin; PB, polymixin B; RIF, rifampin; SMX, sulfamethoxazole; STR, streptomycin; TET, tetracycline; TMP, trimethoprim. P. alkylphenolia, purple; P. fluorescens, yellow; P. lundensis, black; P. putida, red; P. fragi, blue.
Analysis of antibiotic resistance according to population type. (i) Mesophilic pseudomonads.
Generally, mesophilic pseudomonads were resistant to sulfamethoxazole (100%); ampicillin, rifampin, and erythromycin (81 to 100%); amoxicillin, ceftazidime (except P. alkylphenolia), trimethoprim, and chloramphenicol (50 to 100%); and tetracycline (100% of P. alkylphenolia strains) regardless of the species analyzed (Table 1). However, sensitivity was shown to colistin (all P. putida and P. alkylphenolia strains), streptomycin and tetracycline (all P. putida and P. fluorescens strains), polymyxin B (all strains were sensitive except for 48% of P. lundensis strains), imipenem (97 to 100%, except for 33% of P. fluorescens strains), and ciprofloxacin, kanamycin, and gentamicin (all strains were sensitive except for 6, 13, and 19%, respectively, of P. lundensis strains).
(ii) Psychrotrophic pseudomonads.
High resistance of psychrotrophic pseudomonads was shown to amoxicillin, ampicillin, and erythromycin (100% of species), as well as to chloramphenicol and trimethoprim (71 to 100%). Nevertheless, higher sensitivity was obtained with ciprofloxacin (100% of all species), gentamicin and kanamycin (for all species except 14% of P. putida strains), imipenem (except for P. fragi), and streptomycin and rifampin (100% of P. lundensis strains). For the rest of the antibiotics, intermediate resistance was shown for all species (Table 2).
In most cases, both sensitive mesophilic and psychrotrophic pseudomonads showed unimodal MIC distributions in the low-intermediate range of concentrations (Tables 1 and 2), while the resistant pseudomonads showed bi- or multimodal MIC distributions in the intermediate-high range of concentrations, allowing the differentiation of two or three subpopulations: one sensitive and one or two resistant subpopulations. The distinction between intrinsic and acquired resistance was determined for resistant pseudomonads, which displayed bi- or multimodal MIC distributions. In this sense, acquired resistance was detected in mesophilic P. lundensis (Table 1) to all antibiotics to which they showed resistance, except sulfamethoxazole and ciprofloxacin. However, psychrotrophic P. lundensis showed acquired resistance to sulfamethoxazole, colistin, polymyxin B, erythromycin, and tetracycline (Table 2). Regarding mesophilic P. putida, acquired resistance was shown to all antibiotics except rifampin, sulfamethoxazole, erythromycin, and chloramphenicol, while in psychrotrophic P. putida, acquired resistance to several antibiotics, except gentamicin, rifampin, and tetracycline, was shown. With respect to other species, acquired resistance was detected in mesophilic P. fluorescens and P. alkylphenolia and also in psychrotrophic P. fragi (Tables 1 and 2).
Analysis of antibiotic resistance according to sampling zone.
The analysis of the distribution of resistant strains throughout the chain of meat production revealed that mesophilic pseudomonads were the most heterogeneous group, being highly represented by P. lundensis (82%) along the different zones of the meat processing plant (Fig. 1A). Moreover, mesophilic P. lundensis was isolated with high frequency in the cutting room (CR) and from end products (MP) (18.8 and 16.5%, respectively), followed by the white room (WR; 11.8%) and sacrifice room (SR; 8.2%) samples (Fig. 1A). Similarly, psychrotrophic P. lundensis organisms were isolated from different zones of the meat processing plants and also from the end products, being isolated frequently from the refrigerators F3 (16.2%) and F4 (8.1%), CR and MP (5.4%), and refrigerators F2 and F1 (2.7%) (Fig. 1B).
Mesophilic P. putida was detected at high levels in CR (15.3%), as well as in F3 (4.7%) and E (1.2%) (Fig. 1A). However, psychrotrophic P. putida strains were distributed in different slaughterhouse zones and end products, being isolated from WR and CR (8.1%), the entrance (5.4%), and MP, F1, and F4 (2.7%).
Concerning the rest of the species, a few strains of mesophilic P. fluorescens (1 to 3.5%) and P. alkylphenolia (1%) were isolated from E and MP (P. fluorescens) as well as SR and CR (P. alkylphenolia) samples. Psychrotrophic P. fragi was distributed throughout the meat processing plant to the end products, being isolated mostly from F3 (13.5%), F4 (8.1%), and also E, CR, and WR (2.7%) samples. Moreover, neither mesophilic nor psychrotrophic pseudomonads were detected in the freezing tunnel (FrT), and no psychrotrophic pseudomonads were isolated from the SR (Fig. 1).
Analysis of antibiotic resistance according to the type of antibiotic.
Almost all mesophilic pseudomonad strains showed resistance to sulfamethoxazole, erythromycin, rifampin, amoxicillin, ampicillin, ceftazidime (except P. alkylphenolia), chloramphenicol, and trimethoprim regardless of their identity at the species level (Fig. 2A). Similarly, psychrotrophic pseudomonads showed the same antibiotic resistance pattern against all drugs except for rifampin and ceftazidime, to which only P. putida and P. fragi showed resistance, and the strains also were resistant to colistin and tetracycline (Fig. 2B). However, all or almost all pseudomonads were very sensitive to imipenem, kanamycin, ciprofloxacin, and gentamicin (Fig. 2).
MDR phenotypes and genotypes.
Multidrug resistance (defined as resistance to 3 or more different antimicrobials) was observed in all mesophilic and psychrotrophic pseudomonads displaying resistance to 4 to 13 antibiotics (Fig. 2). Furthermore, about 65 mesophilic and psychrotrophic pseudomonads were resistant to at least 8 to 13 antibiotics (Fig. 2).
To identify resistance determinants responsible for the MDR phenotypes observed, all antibiotic-resistant mesophilic and psychrotrophic pseudomonads were screened by PCR for the presence of known resistance genes as described above. The analysis of antibiotic resistance in all strains indicated that phenotypic and genotypic resistance was linked in most cases, since specific and nonspecific resistance determinants were detected (Fig. 2). However, the analysis of aminoglycoside-resistant pseudomonads showed that the genes [aad(E), aphA3, aac(6′)-Ie-aph(2′)-Ia, aph(2′)-Ib, aph(2′)-Ic, aph(2′)-Id, aph(3′)-IIIa, and ant(4′)-Ia] encoding transferases involved in gentamicin, kanamycin, or streptomycin resistance were not detected. On the other hand, both mesophilic and psychrotrophic pseudomonad-resistant strains frequently exhibited the following resistance determinants: blaCTX > blaTEM as beta-lactam resistance genes, sulII > sulI as sulfonamide resistance genes, and ereA > ereB > msrA. mefA was detected in one strain only of psychrotrophic P. putida as an erythromycin resistance gene, and catA2 > catB3 was detected in psychrotrophic pseudomonads, while mesophilic pseudomonads showed the opposite situation for chloramphenicol resistance genes. dfrD was detected as a trimethoprim resistance gene, and tetQ > tetO-tetA was detected in mesophilic pseudomonads (Fig. 2A) and tetB > tetQ in psychrotrophic pseudomonads (Fig. 2B). (“blaCTX > blaTEM” indicates that blaCTX has greater incidence than blaTEM, etc.)
On the other hand, the analysis of the partial rpoB gene sequences revealed that neither of the rifampin-resistant pseudomonads possessed a point mutation in the Rif region. Furthermore, in the case of resistant strains which did not exhibit specific resistance determinants to the corresponding antibiotics, efflux pumps as unspecific mechanisms responsible for the MDR phenotype were detected, such as mexB > mexD > mexY genes, coding for MexAB-OprM, MexCD-OprJ, and MexXY-OprN efflux pumps, respectively. Furthermore, acrB and tolC genes of the AcrAB-TolC efflux system also were detected in both mesophilic and psychrotrophic pseudomonads. However, few resistant pseudomonad strains harbored any of the resistance determinants described above.
Regarding horizontal gene transfer (HGT), integron class 1 was detected in some mesophilic and psychrotrophic pseudomonads (P. lundensis and P. fragi) isolated throughout the meat processing plant and from end products (Fig. 2). Furthermore, integron class 2 also was detected in mesophilic pseudomonads (Fig. 2A).
Statistical analysis of resistance. (i) PCA of multidrug resistance in pseudomonads.
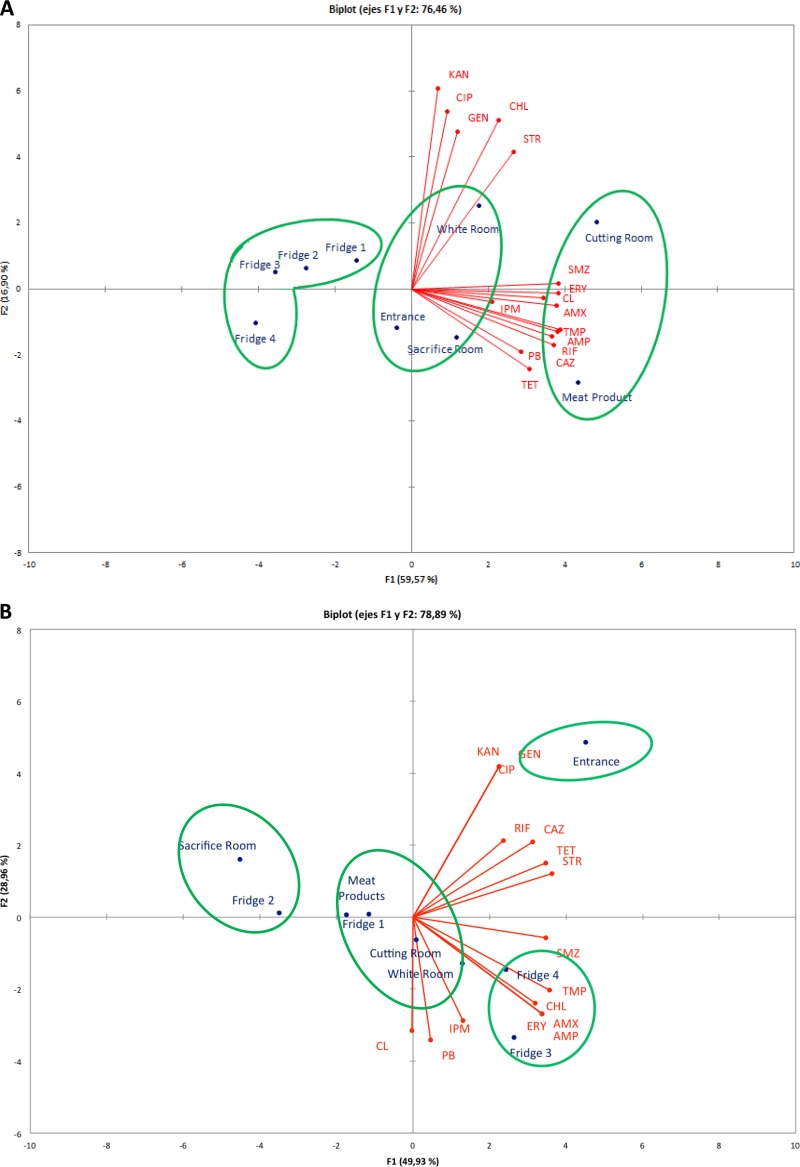
Figure 3 shows the biplot graph of the relationship between the antibiotics tested (scores) and strain variables (population type, sampling zones, and loads). Principal component analysis (PCA) of the data from the phenotypic antibiotic resistance of mesophilic and psychrotrophic pseudomonads in different slaughterhouse zones and end products resulted in three clusters for mesophiles and four clusters for psychrotrophs (Fig. 3). As shown in Fig. 3A, the first cluster represented the most resistant mesophilic pseudomonads, being isolated from CR and MP; however, they exhibited the opposite antibiotic resistance profile, since CHL, GEN, and STR were the most relevant antibiotics in CR and PB-TET-CAZ-RIF were the most relevant in MP. The second cluster was formed by the less resistant mesophilic pseudomonads, which were isolated from all refrigerators (F1, F2, F3, and F4), with CIP-GEN-KAN-PB being the most relevant antibiotics. However, mesophilic pseudomonads from E, SR, and WR (third cluster) occupied an intermediate position between the other clusters, exhibiting resistance to a wide range of antibiotics, with CIP-KAN-GEN-CHL being the most relevant antibiotics in the WR and several antibiotics being the most relevant in the SR (Fig. 3A).
FIG 3.
Biplot of the simultaneous evaluation of the relationship of scores (antibiotics) and sample variables (sampling zone and population type. (A) Mesophilic pseudomonads; (B) psychrotrophic pseudomonads. Antibiotic abbreviations: AMP, ampicillin; AMX, amoxicillin; CAZ, ceftazidime; CIP, ciprofloxacin; CL, colistin; CHL, chloramphenicol; ERY, erythromycin; GEN, gentamicin; IPM, imipenem; KAN, kanamycin; PB, polymyxin B; RIF, rifampin; SMZ, sulfamethoxazole; STR, streptomycin; TET, tetracycline; TMP, trimethoprim.
On the other hand, PCA of phenotypic antibiotic resistance in psychrotrophic pseudomonads (Fig. 3B) showed that E (first cluster) and F3 and F4 (second cluster) represented clusters with higher levels of resistance, with KAN and GEN being the most relevant antibiotics in E and ERY-AMX-APM-TMP being the most relevant antibiotics in F3 and F4. Of the remaining two clusters, one was represented by the less resistant pseudomonads found in SR and F2 samples, and one occupied an intermediate position and was found in MP, F1, CR, and WR samples. The resistance of the last two clusters was determined by a wide range of antimicrobials, and the differences were less noticeable than those in other zones.
(ii) Similarity analysis of multidrug-resistant pseudomonads.
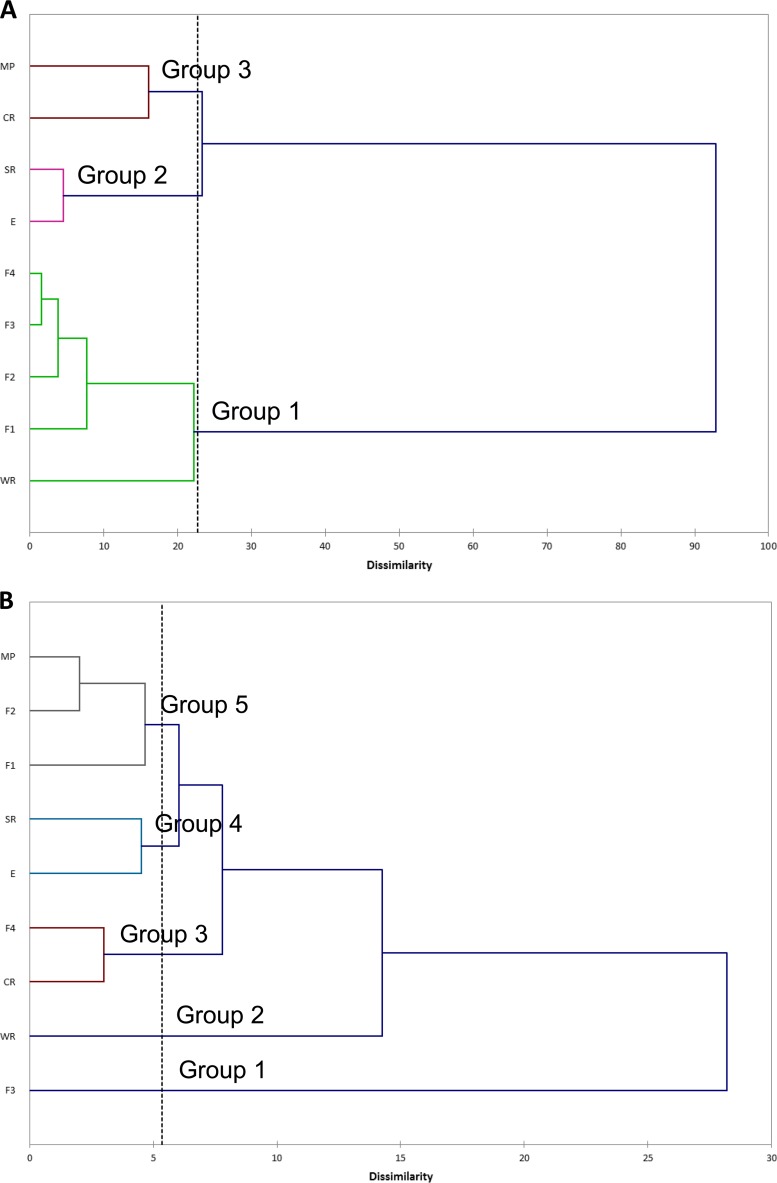
Figure 4 shows the dendrogram obtained when resistant populations of the sampling zones and end products based on the genotypic resistance profiles was analyzed. According to this clustering (Fig. 4A), all refrigerators (F1, F2, F3, and F4) and WR (group 1) had a resistant mesophilic population of pseudomonads clustered separately from SR and E (group 2), which clustered together with CR and MP (group 3). However, resistome-based clustering of psychrotrophic pseudomonads (Fig. 4B) showed that refrigerators were distributed throughout almost all groups: F3 (group 1), F4 (group 3), and F1, F2, and MP (group 5).
FIG 4.
Dendrogram showing mesophilic (A) and psychrotrophic (B) pseudomonad population clusters of goat and lamb slaughterhouse sampling zones from the entrance area and sacrifice room to the end product stage, typed by resistomes. Hierarchical cluster analysis for multiple antibiotic resistance patterns was performed by using the Ward method for clustering and the square Euclidean distance for distance measures.
DISCUSSION
The slaughterhouse is considered an ideal environment for spreading antimicrobial-resistant zoonotic pathogens that contaminate surfaces, meat products, and wastewater (56, 57). The spread of antimicrobial resistance genes throughout the food chain increases the resistance gene pool from which both pathogens and commensals can pick up resistance determinants, including those that pose a potential threat to public health and ecological balance (21, 58, 59). The main microorganisms recovered from hides, carcasses, butchered meat, and meat processing plant surfaces (60, 61) include a wide spectrum of Gram-negative bacteria, with P. fluorescens and the psychrotrophic P. fragi, P. lundensis, and P. putida being the most relevant spoilage agents of fresh meat stored aerobically (42, 62–65). In the present study, MDR pseudomonads (100% of strains isolated in this study) were isolated from all slaughterhouse zones (77%), except the freezing tunnel, and from the end products (23%), represented mainly by P. lundensis (65%) and P. putida (17%), followed by P. fragi (8%), P. fluorescens (6%), and P. alkylphenolia (4%). The high genetic relatedness of mesophilic and psychrotrophic pseudomonads and the fact that some isolates showed identical or highly similar ERIC-PCR profiles, although they were isolated from different zones (entrance, sacrifice room, refrigerators, cutting room, and white room) or even from the end products, suggest that the main sources of the identical or related pseudomonad strains were the animals (feet and wool) and the slaughterhouse environment. Thus, carcasses contaminated with environmental bacteria may spread pseudomonads throughout all of the slaughterhouse zones, including the end products. For this reason, the isolation of pseudomonads from living animals and comparison to slaughterhouse strains deserves further studies.
MDR phenotypes (resistance to 4 to 13 antibiotics) were detected in both mesophilic and psychrotrophic pseudomonads, being resistant to sulfamethoxazole, erythromycin, amoxicillin, ampicillin, chloramphenicol, trimethoprim, rifampin, and ceftazidime (especially mesophiles), as well as colistin and tetracycline (especially psychrotrophs), regardless of species identity. However, a low percentage of resistant pseudomonads was obtained with ciprofloxacin, gentamicin, imipenem, and kanamycin. The most resistant mesophilic pseudomonads, especially P. lundensis strains, were frequently isolated from the cutting room and meat products. However, resistant psychrotrophic pseudomonads were isolated at high levels from refrigerators (F3 and F4). The multidrug resistance of pseudomonads is due to multiple intrinsic or acquired mechanisms, such as the low permeability of the outer membrane (20, 66–68), the production of beta-lactamases, and the presence of multidrug efflux pumps with wide substrate profiles (66, 69). Worryingly, in the present study we found multidrug resistance for up to 13 antibiotics (65% of pseudomonads were resistant to 8 to 13 antibiotics) caused by practically all known mechanisms of antimicrobial resistance. The increase in antibiotic resistance of pseudomonads isolated from the slaughterhouse environment can be due to several reasons, such as the use of antimicrobials (biocides and antibiotics) that could enhance gene transfer and recombination through the activation of the SOS system (70, 71), temperature of storage, growth in biofilm, and the presence of pathogens as potential reservoirs of resistance genes.
The high resistance of pseudomonads to beta-lactams (ampicillin, amoxicillin, and ceftazidime) was related to the presence of plasmid-mediated extended-spectrum beta-lactamases (ESBLs) encoded by blaCTX, blaTEM, blaSHV-1, and blaOXA genes. Thus, the observed acquired resistance of mesophilic and psychrotrophic pseudomonads reflected by the bi- or multimodal MIC distributions was due to the acquisition in most cases of blaCTX and blaTEM genes by horizontal gene transfer (insertion sequences, class 1 integrons, transposons, and plasmids). However, only some strains exhibited the presence of class 1 and 2 integrons. It should be noted that the independent acquisition of mobile elements carrying a bla gene, blaCTX or blaTEM, can lead to the simultaneous occurrence of more than one gene in the same strain.
Concerning chloramphenicol, catA2 and catB3 resistance genes prevalent in psychrotrophic and mesophilic pseudomonads, respectively, are widespread in many bacteria (44), suggesting that the observed acquired resistance was due to horizontal gene transfer. On the other hand, the acquisition of the sulfamethoxazole (sulII) resistance gene, which is found predominantly on plasmids and associated with class 1 integrons (72, 73), via horizontal gene transfer by pseudomonads was reported in enteric bacteria isolated from healthy food animals and humans (74, 75), with sulII being the most prevalent gene in the absence of clinical selection pressure (76). Here, the genes catA2 and sulII acquired by psychrotrophic P. fragi in the entrance environment probably was due to the presence of class 1 integron in those strains acquired from other microorganisms of the environment or the animals and may be responsible for the spread of those genes throughout the chain of meat production.
Resistance to tetracycline (tetA, tetB, tetO, and tetQ genes), trimethoprim (dfrD gene), and erythromycin (ereA, ereB, msrA, and mefA genes) was due partially to the presence of the corresponding resistance genes, with ereA and tetQ genes being the most prevalent. The genes tetO and tetQ were found to be predominant in the gastrointestinal tracts of pigs and steers and also in the manure (77), often being associated with conjugative transposons (77, 78). In the present study, the occurrence of acquired tetQ and ereA genes in P. putida from the entrance environment suggests that the source of those genes is related to animals or the entrance environment.
Genetic linkage of sulII, dfrD, and tet genes to determinants such as blaCTX and blaTEM, conferring resistance to beta-lactams, was due in part to the presence of class 1 integrons (17%), which are implicated in the carriage and the genetic mobility of resistance genes (79, 80); thus, the beta-lactams still commonly used might help explain the persistence of those genes and the increase in MDR of pseudomonads in the slaughterhouse. Accordingly, Tadesse et al. (81) established a link between sulfonamide resistance genes and determinants conferring resistance to tetracycline and streptomycin. MDR of pseudomonads lacking specific genetic resistance determinants may be due to other mechanisms, such as drug efflux pumps with a wide spectrum of activity (MexAB-OprM, MexCD-OprJ, MexXY-OprM, and AcrAB-TolC; 52% of strains harbored such unspecific mechanisms), with mexB, mexD, and acrB genes being detected at high levels. Those efflux pumps act synergistically with the permeability barrier to result in significant intrinsic resistance to many antibiotics (82–85).
The correlation of resistance in pseudomonads using PCA of sampling zones and end products determined that CR and MP are considered the main sources of antibiotic-resistant mesophilic pseudomonads, although they showed the opposite behavior concerning the relevance of antibiotics to determine resistance. The most resistant psychrotrophic pseudomonads were isolated from F3, F4, and E. However, resistome-based clustering did not support this conclusion, with mesophilic pseudomonads from CR and MP (group 3) being highly related and sharing the same source of resistance determinants with E and SR (group 2), as occurred with psychrotrophic pseudomonads from F3 (group 1), F4 (group 3), and F1 and F2 (group 5). Those data suggested a clear divergence between phenotypic and genotypic resistance of pseudomonads in a slaughterhouse environment, since specific and unspecific mechanisms induced by a wide range of antibiotics may occur in different zones. Often, more than one gene was associated with a given phenotypic resistance.
In summary, we revealed, for the first time, a high prevalence of pseudomonads with MDR to commonly used antibiotics on goat and lamb slaughterhouse surfaces, which may reflect the misuse or abuse of the antimicrobial agents in animals and the environment. Furthermore, the high similarity between different slaughterhouse surfaces and end products regarding phenotypic and molecular antibiotic resistance profiles of MDR pseudomonads isolated in this study suggested that meat products play a role as a reservoir of resistance determinants to be spread to human pathogens. Following the high slaughterhouse surface contamination with MDR pseudomonads, it must be assumed that these highly resistant microorganisms also can be directly transmitted to humans by transport, transaction, and food preparation. The relationship between environmental microorganisms and human pathogens is not clear; however, recent reports showed that soil bacteria and human pathogens shared an antibiotic resistome (86), as did animals and farm workers (59, 87, 88). Considering that the entrance environment (the first zone in a goat and lamb slaughterhouse; animals should be kept there for a determined time period before sacrifice) shared several resistance determinants with end products implicated in resistance to several antibiotics (about 6), we can suggest that the entrance, where some antibiotic resistance determinants were detected for the first time (catA2 and sulII), is the key zone in antibiotic resistance spreading throughout different slaughterhouse zones, including the end products. Furthermore, other zones, such as the cutting room and refrigerators, where most MDR pseudomonads were isolated, should be exhaustively controlled, especially F1 (about 6 resistance determinants acquired), which was located between the SR and CR. This fact must be taken into consideration to avoid cross-contamination with the subsequent flow of mobile resistance determinants throughout all slaughterhouse zones and to avoid the spread of resistance to humans and the environment by the application of adequate practices of hygiene and disinfection measures, including animal wool and feet and also the entrance environment. Practical strategies could be applied in slaughterhouses, including good husbandry practices to prevent disease and good hygiene of animals before access to entrance into a pre-entrance room, which could be created with the aim of applying a brief shower to eliminate the majority of microorganisms from wool and feet.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AGL2009-08921 and P08-AGR-4295, Plan Propio de la Universidad de Jaén, and Campus de Excelencia Internacional Agroalimentario CeiA3. L.L.L. was the beneficiary of a fellowship from the Spanish Ministry of Education and Science.
Footnotes
Published ahead of print 29 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01998-14.
REFERENCES
- 1.Brady MT, Feigin RD. 1998. Pseudomonas and related species, p 1401–1413 In Feigin RD, Cherry JD. (ed), Textbook of pediatric infectious diseases. W.B. Saunders, Philadelphia, PA. [Google Scholar]
- 2.Prasad G, Minakshi 2007. Normal microbial flora of human body and host parasite relationship, p 1–23 In Immunology and medical microbiology. National Science Digital Library, Boulder, CO. [Google Scholar]
- 3.Ridgway HF, Safarik J. 1990. Identification and catabolic activity of well-derived gasoline-degrading bacteria from a contaminated aquifer. Appl. Environ. Microbiol. 56:3565–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender CL, Alarcon-Chaidez F, Gross DC. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeth HC, Fitz-Gerald CH. 1983. Lipolytic enzymes and hydrolytic rancidity in milk and milk products, p 195–239 In Fox PF. (ed), Developments in dairy chemistry. Applied Science, London, United Kingdom. [Google Scholar]
- 6.Reddy MC, Bills DD, Lindsey RC, Libbey LM. 1968. Ester production by Pseudomonas fragi. I. Identification and quantification of some esters produced in milk cultures. J. Dairy Sci. 51:656–659. [Google Scholar]
- 7.Ternstrom A, Lindberg A-M, Molin G. 1993. Classification of the spoilage flora of raw and pasteurized bovine milk, with special reference to Pseudomonas and Bacillus. J. Appl. Bacteriol. 75:25–34. 10.1111/j.1365-2672.1993.tb03403.x. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen-the C, Carlin F. 1994. The microbiology of minimally processed fresh fruits and vegetables. Crit. Rev. Food Sci. Nutr. 34:371–401. 10.1080/10408399409527668. [DOI] [PubMed] [Google Scholar]
- 9.Riva M, Franzetti L, Galli A. 2001. Microbiological quality of shelf-life modelling of ready to eat cicorino. J. Food Prot. 64:228–234. [DOI] [PubMed] [Google Scholar]
- 10.Molin G, Ternstrom A. 1986. Phenotypically based taxonomy of psychrotrophic Pseudomonas isolated from spoiled meat, water and soil. Int. J. Syst. Bacteriol. 36:257–274. 10.1099/00207713-36-2-257. [DOI] [Google Scholar]
- 11.Miller A, III, Scanlan RA, Lee JS, Libbey LM. 1973. Identification of volatile compounds produced in sterile fish muscle (Sebastes melanops) by Pseudomonas fragi. Appl. Microbiol. 25:952–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tryfinopoulou P, Tsakalidon E, Nychas G-JE. 2002. Characterization of Pseudomonas sp. associated with spoilage of gilt-head sea bream stored under various conditions. Appl. Environ. Microbiol. 68:65–72. 10.1128/AEM.68.1.65-72.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Lopez I, Otero A, Garcia-Lopez M-L, Santos JA. 2004. Molecular and phenotypic characterization of nonmotile gram-negative bacteria associated with spoilage of freshwater fish. J. Appl. Microbiol. 96:878–886. 10.1111/j.1365-2672.2004.02214.x. [DOI] [PubMed] [Google Scholar]
- 14.Wiedmann M, Weilmeier D, Dineen SS, Ralyea RM, Boor KJ. 2000. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66:2085–2095. 10.1128/AEM.66.5.2085-2095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaut-Rollier I, Vauterin L, De Vos P, Massart DL, Devriese LA, De Zutter L, Van Hoof J. 1999. A numerical taxonomic study of the Pseudomonas flora isolated from poultry meat. J. Appl. Microbiol. 87:15–28. 10.1046/j.1365-2672.1999.00785.x. [DOI] [PubMed] [Google Scholar]
- 16.Levy SB. 1997. Antibiotic resistance: an ecological imbalance. Ciba Found. Symp. 207:1–9. [DOI] [PubMed] [Google Scholar]
- 17.Breidenstein EBM, de la Fuente-Nuñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19:419–426. 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 18.DeFlaun MF, Levy SB. 1989. Genes and their varied hosts, p 1–32 In Levy SB, Miller RV. (ed), Gene transfer in the environment. McGraw-Hill, New York, NY. [Google Scholar]
- 19.Miko A, Pries K, Schroeter A, Helmuth R. 2005. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 56:1025–1033. 10.1093/jac/dki365. [DOI] [PubMed] [Google Scholar]
- 20.De Oliveira, KMP. Pericles DDS, Grisolia AB. 2013. Antimicrobial susceptibility profile of Pseudomonas spp. isolated from a swine slaughterhouse in Dourados, Mato Grosso do Sul State, Brazil. Rev. Argent. Microbiol. 45:57–60. [PubMed] [Google Scholar]
- 21.Gregova G, Kmetova M, Kmet V, Venglovsky J, Feher A. 2012. Antibiotic resistance of Escherichia coli isolated from a poultry slaughterhouse. Ann. Agric. Environ. Med. 19:75–77. [PubMed] [Google Scholar]
- 22.Schwaiger K, Huther S, Hölzel C, Kämpf P, Bauer J. 2012. Prevalence of antibiotic-resistant Enterobacteriaceae isolated from chicken and pork meat purchased at the slaughterhouse and at retail in Bavaria, Germany. Int. J. Food Microbiol. 154:206–211. 10.1016/j.ijfoodmicro.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Aarestrup FM, Wegener HC, Collignon P. 2008. Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti-Infect. Ther. 6:733–750. 10.1586/14787210.6.5.733. [DOI] [PubMed] [Google Scholar]
- 24.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74:417–433. 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durán GM, Marshall DL. 2005. Ready-to-eat shrimp as an international vehicle of antibiotic-resistant bacteria. J. Food Prot. 68:2395–2401. [DOI] [PubMed] [Google Scholar]
- 26.Wang HH, Manuzon M, Lehman M, Wan K, Luo HL, Wittum TE, Yousef A, Bakaletz LO. 2006. Food comensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 254:226–231. 10.1111/j.1574-6968.2005.00030.x. [DOI] [PubMed] [Google Scholar]
- 27.Wegener HC. 2003. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 6:439–445. 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Munsch-Alatossava P, Alatossava T. 2007. Antibiotic resistance of raw milk associated psychrotrophic bacteria. Microbiol. Res. 162:115–123. 10.1016/j.micres.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Dixon B. 2000. Antibiotics as growth promoters: risks and alternatives. ASM News 66:264–265. [Google Scholar]
- 30.Feinman SE. 1999. Antibiotics in animal feeds—drug resistance revisited. ASM News 64:24–29. [Google Scholar]
- 31.SCAN. 1996. Report of the Scientific Committee for Animal Nutrition (SCAN) on the possible risk for humans on the use of avoparcin as feed additive. Office for EC Publications, Luxemburg City, Luxemburg. [Google Scholar]
- 32.SCAN. 1998. Opinion of the Scientific Committee for Animal Nutrition (SCAN) on the immediate and long-term risk to the value of streptogramins in human medicine posed by the use of virginiamycin as an animal growth promoter. Office for EC Publications, Luxemburg City, Luxemburg. [Google Scholar]
- 33.Levy SB. 1998. The challenge of antibiotic resistance. Sci. Am. 278:22–39. [DOI] [PubMed] [Google Scholar]
- 34.Lavilla Lerma L, Benomar N, Gálvez A, Abriouel H. 2013. Prevalence of bacteria resistant to antibiotics and/or biocides on meat processing plant surfaces throughout meat chain production. Int. J. Food Microbiol. 161:97–106. 10.1016/j.ijfoodmicro.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Bruchmann J, Kirchen S, Schwartz T. 2013. Sub-inhibitory concentrations of antibiotics and wastewater influencing biofilm formation and gene expression of multi-resistant Pseudomonas aeruginosa wastewater isolates. Environ. Sci. Pollut. Res. 20:3539–3549. 10.1007/s11356-013-1521-4. [DOI] [PubMed] [Google Scholar]
- 37.Tribuddharat C, Fennewald M. 1999. Integron-mediated rifampin resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:960–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 15th informational supplement, vol 29 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 39.de Los Reyes-Gavilan CG, Limsowtin GKY, Tailliez P, Séchaud L, Acholas JP. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M. 2009. Characterization and safety evaluation of enterococci isolated from Spanish goats' milk cheeses. Int. J. Food Microbiol. 132:24–32. 10.1016/j.ijfoodmicro.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394. [DOI] [PubMed] [Google Scholar]
- 42.Ercolini D, Russo F, Blaiotta G, Pepe O, Mauriello G, Villani F. 2007. Simultaneous detection of Pseudomonas fragi, P. lundensis, and P. putida from meat by use of a multiplex PCR assay targeting the carA gene. Appl. Environ. Microbiol. 73:2354–2359. 10.1128/AEM.02603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birkett C, Ludlam H, Woodford N, Brown D, Brown N, Roberts M, Milner N, Curran M. 2007. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum β-lactamases. J. Med. Microbiol. 56:52–55. 10.1099/jmm.0.46909-0. [DOI] [PubMed] [Google Scholar]
- 44.Kim M, Kwon TH, Jung SM, Cho SH, Jin SY, Park NH, Kim CG, Kim JS. 2013. Antibiotic resistance of bacteria isolated from the internal organs of edible snow crabs. PLoS One 8:e70887. 10.1371/journal.pone.0070887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, Hildebrandt B, Müller-Bertling S, Witte W, Goossens H. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus, and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59:900–912. 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 46.Knapp CW, Zhang W, Sturm BS, Graham DW. 2010. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions. Environ. Pollut. 158:1506–1512. 10.1016/j.envpol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Ng L-K, Martin I, Alfo M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistance genes. Mol. Cell. Probes 15:209–215. 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]
- 48.Pei R, Kim S-C, Carlson KH, Pruden A. 2006. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 40:2427–2435. 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 49.Spiliopoulou I, Petinaki E, Papandreou P, Dimitracopoulos G. 2004. erm(C) is the predominant genetic determinant for the expression of resistance to macrolides among methicillin-resistant Staphylococcus aureus clinical isolates in Greece. J. Antimicrob. Chemother. 53:814–817. 10.1093/jac/dkh197. [DOI] [PubMed] [Google Scholar]
- 50.Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh H, Stenhoff J, Jalal S, Wretlind B. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 9:323–328. 10.1089/107662903322762743. [DOI] [PubMed] [Google Scholar]
- 52.Swick MC, Morgan-Linnell SK, Carlson KM, Zechiedrich L. 2011. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob. Agents Chemother. 55:921–924. 10.1128/AAC.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosokawa K, Park N-H, Inaoka T, Itoh Y, Ochi K. 2002. Streptomycin-resistant (rpsL) or rifampicin-resistant (rpoB) mutation in Pseudomonas putida KH146-2 confers enhanced tolerance to organic chemicals. Environ. Microbiol. 4:703–712. 10.1046/j.1462-2920.2002.00348.x. [DOI] [PubMed] [Google Scholar]
- 54.White PA, McIver CJ, Deng YM, Rawlinson WD. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265–269. 10.1111/j.1574-6968.2000.tb08906.x. [DOI] [PubMed] [Google Scholar]
- 55.Ploy MC, Denis F, Courvalin P, Lambert T. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of an hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684–2688. 10.1128/AAC.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsola E, Drosinos EH, Zoiopoulos P. 2008. Impact of poultry slaughter house modernisation and updating of food safety management systems on the microbiological quality and safety of products. Food Control 19:423–431. 10.1016/j.foodcont.2007.05.003. [DOI] [Google Scholar]
- 57.Romanova NA, Gawande PV, Brovko LY, Griffiths MW. 2007. Rapid methods to assess sanitizing efficacy of benzalkonium chloride to Listeria monocytogenes biofilms. J. Microbiol. Methods 71:231–237. 10.1016/j.mimet.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Hammerum A, Heuer O. 2009. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 48:916–921. 10.1086/597292. [DOI] [PubMed] [Google Scholar]
- 59.Van den Bogaard A, London N, Driessen C, Stobberingh E. 2001. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47:763–771. 10.1093/jac/47.6.763. [DOI] [PubMed] [Google Scholar]
- 60.European Food Safety Authority. 2010. The Community Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from animals and food in the European Union in 2004–2007. EFSA J. 8:1309. [Google Scholar]
- 61.European Food Safety Authority. 2010. Community summary reports on trend and sources of zoonoses, zoonotic agents, antimicrobial resistance and food borne outbreaks in the European Union 2008. EFSA J. 8:1496. [Google Scholar]
- 62.Labadie J. 1999. Consequences of packaging on bacterial growth. Meat is an ecological niche. Meat Sci. 52:299–305. [DOI] [PubMed] [Google Scholar]
- 63.Gill CO. 2003. Active packaging in practice: meat, p 378–396 In Ahvenainem H. (ed), Novel food packaging technology. Woodhead Publishing Limited and CRC Press LLC, Boca Raton, FL. [Google Scholar]
- 64.Gennari M, Dragotto F. 1992. A study of the incidence of different fluorescent Pseudomonas species and biovars in the microflora of fresh and spoiled meat and fish, raw milk, cheese, soil and water. J. Appl. Bacteriol. 72:281–288. 10.1111/j.1365-2672.1992.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 65.Stanbridge LH, Davies AR. 1998. The microbiology of chill-stored meat, p 175–177 In Davies A, Board R. (ed), Microbiology of meat and poultry. Blackie Academic & Professional, London, United Kingdom. [Google Scholar]
- 66.Nikaido H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382–388. 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 67.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Juan C, Zamorano L, Mena A, Albertí S, Pérez JL, Oliver A. 2010. Metallo-β-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J. Antimicrob. Chemother. 65:474–478. 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- 69.Henwood CJ, Livermore DM, James D, Warner M. 2001. Antimicrobial susceptibility of Pseudomonas aeruginosa: results of a UK survey and evaluation of the British Society for Antimicrobial Chemotherapy disc susceptibility test. J. Antimicrob. Chemother. 47:789–799. 10.1093/jac/47.6.789. [DOI] [PubMed] [Google Scholar]
- 70.Couce A, Blazquez J. 2009. Side effects of antibiotics on genetic variability. FEMS Microbiol. Rev. 33:531–538. 10.1111/j.1574-6976.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- 71.Guerin E, Cambray G, Da Re S, Mazel D, Ploy MC. 2010. The SOS response controls antibiotic resistance by regulating the integrase of integrons. Med. Sci. 1:28–30. 10.1051/medsci/201026128. [DOI] [PubMed] [Google Scholar]
- 72.Radstrom P, Swedberg G, Skold O. 1991. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 35:1840–1848. 10.1128/AAC.35.9.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784. 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 74.Enne VI, Livermore DM, Stephens P, Hall LMC. 2001. Persistence of sulfonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328. 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 75.Kozak GK, Pearl DL, Parkman J, Reid-Smith RJ, Deckert A, Boerlin P. 2009. Distribution of sulfonamide resistance genes in Escherichia coli and Salmonella isolates from swine and chickens at abattoirs in Ontario and Quebec, Canada. Appl. Environ. Microbiol. 75:5999–6001. 10.1128/AEM.02844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Enne VI, Bennett PM, Livermore DM, Hall LM. 2004. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 53:958–963. 10.1093/jac/dkh217. [DOI] [PubMed] [Google Scholar]
- 77.Zhu YG, Johnson TA, Su J-Q, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 110:3435–3440. 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leng Z, Riley DE, Berger RE, Krieger JN, Roberts MC. 1997. Distribution and mobility of the tetracycline resistant determinant tetQ. J. Antimicrob. Chemother. 40:551–559. 10.1093/jac/40.4.551. [DOI] [PubMed] [Google Scholar]
- 79.Lynne AM, Kaldhone P, David D, White DG, Foley SL. 2009. Characterization of antimicrobial resistance in Salmonella enterica serotype Heidelberg isolated from food animals. Foodborne Pathog. Dis. 6:207–215. 10.1089/fpd.2008.0172. [DOI] [PubMed] [Google Scholar]
- 80.Gaze WH, Abdouslam N, Hawkey PM, Wellington EM. 2005. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 49:1802–1807. 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF. 2012. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 18:741–749. 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poole K, Krebes K, McNally C, Neshat S. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X-Z, Livermore DM, Nikaido H. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38:1732–1741. 10.1128/AAC.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zgurskaya HI, Nikaido H. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219–225. 10.1046/j.1365-2958.2000.01926.x. [DOI] [PubMed] [Google Scholar]
- 85.Aeschlimann JR. 2003. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 23:916–924. [DOI] [PubMed] [Google Scholar]
- 86.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111. 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith TC, Gebreyes WA, Abley MJ, Harper AL, Forshey BM, Male MJ, Martin HW, Molla BZ, Sreevatsan S, Thakur S, Thiruvengadam M, Davies PR. 2013. Methicillin-resistant Staphylococcus aureus in pigs and farm workers on conventional and antibiotic-free swine farms in the USA. PLoS One 8:e63704. 10.1371/journal.pone.0063704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang XY, Ding LJ, Yue J. 2009. Occurrence and characteristics of class 1 and class 2 integrons in resistant Escherichia coli isolates from animals and farm workers in northeastern China. Microb. Drug Resist. 15:323–328. 10.1089/mdr.2009.0020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.