Abstract
Methanotrophic Verrucomicrobia have been found in geothermal environments characterized by high temperatures and low pH values. However, it has recently been hypothesized that methanotrophic Verrucomicrobia could be present under a broader range of environmental conditions. Here we describe the isolation and characterization of three new species of mesophilic acidophilic verrucomicrobial methanotrophs from a volcanic soil in Italy. The three new species showed 97% to 98% 16S rRNA gene identity to each other but were related only distantly (89% to 90% on the 16S rRNA level) to the thermophilic genus Methylacidiphilum. We propose the new genus Methylacidimicrobium, including the novel species Methylacidimicrobium fagopyrum, Methylacidimicrobium tartarophylax, and Methylacidimicrobium cyclopophantes. These mesophilic Methylacidimicrobium spp. were more acid tolerant than their thermophilic relatives; the most tolerant species, M. tartarophylax, still grew at pH 0.5. The variation in growth temperature optima (35 to 44°C) and maximum growth rates (µmax; 0.013 to 0.040 h−1) suggested that all species were adapted to a specific niche within the geothermal environment. All three species grew autotrophically using the Calvin cycle. The cells of all species contained glycogen particles and electron-dense particles in their cytoplasm as visualized by electron microscopy. In addition, the cells of one of the species (M. fagopyrum) contained intracytoplasmic membrane stacks. The discovery of these three new species and their growth characteristics expands the known diversity of verrucomicrobial methanotrophs and shows that they are present in many more ecosystems than previously assumed.
INTRODUCTION
Methane oxidation is a microbial process which limits the amount of methane released to the atmosphere (1). Microbial methane oxidation can occur via different routes, including either anaerobically (2, 3, 4) or aerobically (5, 6, 7). The diversity of the biochemistry of methane oxidation is also reflected in the diversity of organisms performing these processes. For a long time, Alpha- and Gammaproteobacteria were considered to be the only organisms to perform aerobic methane oxidation. However, in 2007, methane-oxidizing Verrucomicrobia were discovered (genus Methylacidiphilum [8, 9, 10]); also, intra-aerobic bacteria belonging to the NC10 candidate phylum (3, 11) were recently described to oxidize methane using internally produced oxygen. Like most other aerobic methane oxidizers, the Verrucomicrobia use particulate methane monooxygenase (pMMO) to catalyze the first step of the methane oxidation. Unlike most proteobacterial methanotrophs (7), but like Methylomirabilis oxyfera (belonging to the NC10 phylum) (12), M. fumariolicum SolV (13) and M. infernorum V4 (14) grow as autotrophs, using only carbon dioxide as the carbon source via the Calvin cycle.
All verrucomicrobial methanotrophs known to date have been enriched from geothermal environments (7, 15, 16), and 16S rRNA gene surveys indicated their presence to be mainly limited to such environments (16). The three thermophilic strains belonging to the genus Methylacidiphilum share low pH optima (2 to 3.5) and high temperature optima (55 to 60°C) (7). In addition, 16S rRNA gene sequences from different geothermal sites showed identities of only 95% to 99% to the 16S rRNA gene sequence of M. fumariolicum SolV (8, 14, 16). This indicated that a larger diversity in verrucomicrobial methanotrophs might exist in geothermal environments. Geothermal environments are often characterized not only by extreme conditions but also by local and temporal fluctuations in pH, temperature, and methane concentrations. The flux and composition of geothermal gasses, including methane and hydrogen sulfide, differ considerably over time (17). The hot gasses escape directly as fumaroles or seep through soil layers, via routes that may change continuously. Hydrogen sulfide is oxidized by microbes into sulfuric acid, leading to very low pH values. It is likely that the pH is also influenced by rainfall.
During the isolation of the first (thermophilic) verrucomicrobial methanotrophs (8, 9), mesophilic isolates were also encountered. A mesophilic verrucomicrobial enrichment culture was obtained during the isolation of M. infernorum V4 (9). Recently, strain LP2A was isolated in pure culture from this mesophilic enrichment culture (16). Strain LP2A has a growth optimum temperature of 30°C, and its 16S rRNA gene sequence identity to the Methylacidiphilum genus is only 89.6%. In our search for methane-oxidizing species that are more acid tolerant and mesophilic, we started enrichment cultures from volcanic soil. Here we describe the isolation, physiology, phylogeny, and morphology of three new highly acid-tolerant, methanotrophic mesophilic verrucomicrobial strains.
MATERIALS AND METHODS
Enrichment and isolation.
Soil samples were taken from various spots at the Solfatara crater, which is at the center of the Campi Flegrei caldera, near Naples (Italy). Spots were chosen at least 20 m away from the hot area which surrounds the central mud pot. Their temperatures as judged manually were well below 50°C, and pH values ranged between 1 and 1.5. pH values were measured at the spots with indicator strips and confirmed by slurry measurements (about 0.3 ml g−1 soil). The rather dry top layer of about 0.5 to 1 cm in thickness was first removed, and samples of the humid layer just below were sampled into 50-ml sterile plastic tubes. The tubes were closed, and within 10 h, 15 ml of samples was mixed with 15 ml of store-bought mineral water. The mixtures were shaken by hand for 2 min in order to extract microorganisms from the soil. After the mixtures were allowed to settle for a few minutes, 1 ml from the liquid phase was transferred to a dilution series at 1 ml each time into 19 ml of medium in 60-ml bottles. The growth medium (8) was supplemented with 20% filtered (using a 0.22-μm-pore-size filter) liquid from the central mud pot and adjusted to pH 2.5 (using H2SO4). Bottles had a gas phase of 5% carbon dioxide and 10% methane in air and were incubated at 29°C with slow shaking (100 rpm) for 4 months. Bottles that showed growth were transferred three times and subsequently serially diluted (up to 10−8) to extinction. Serial dilutions were incubated for growth or immediately transferred onto floating membrane filters and incubated at 30°C as described previously (8). Single colonies that appeared on the membranes were transferred to liquid medium. The composition of the enrichments was investigated by light microscopy and fluorescence in situ hybridization (FISH; EubIII). The purity of the strains was investigated via 16S rRNA gene sequence analysis.
Growth.
Temperature and pH optima for the different strains were determined on the basis of growth rates in medium as described before (18). The growth rate was determined by following the optical density at 600 nm (OD600) of batch cultures under conditions of excess (>2%) methane and fast (350-rpm) shaking.
The pH range at which growth occurs was tested by gradually changing the pH when cultures were transferred (steps of 1 pH unit between pH 2 and pH 6; below pH 1, steps were only 0.1 pH unit). For pH 3 and below, the medium pH was adjusted by addition of sulfuric acid, which also acted as a buffer. For pH 3 and above, 50 mM MES (morpholineethanesulfonic acid) was included as a buffer (adjusted by addition of NaOH), which did not influence the growth rates. After growth, only pH values above 3 were different from the starting value but never differed by more than 0.1 pH unit. The ability of the strains to grow on methanol was tested by adding 10 mM methanol to the growth media in the absence of methane.
The ability of the strains to grow autotrophically was tested by incubation with labeled methane. 13C-labeled CH4 experiments were done in batch cultures (in duplicate for each of the three strains) as described previously (13), using 150-ml serum bottles containing 10 ml of culture medium and 40% of CO2 in air. 13C-labeled CH4 (Sigma-Aldrich) (99 atom% 13C; 3 to 5.5 ml) was added. Initial and final gas concentrations and amounts as well as mass ratios for CO2 were verified by gas chromatography-mass spectrometry (GC-MS) analysis at the start and after growth (when at least 90% of the added CH4 had been consumed) in order to calculate the recovery of 13C from CH4 in CO2. At the end of the experiment, the 13C/12C ratio of the biomass was determined by isotope ratio mass spectrometry (IRMS) as described before (13).
Phylogenetic and genomic analysis.
The draft genomes of strains 3B and 4AC were assembled from Illumina sequencing runs using CLCBio software with standard settings. This resulted in 604 and 314 contigs for strains 3B and 4AC, respectively. The contigs were submitted to the RAST server for annotation (19). The draft genome for strain 3C was assembled and annotated as part of a JGI Bioproject (PRJNA199166). The draft genomes of the three strains and the published genome of M. fumariolicum SolV (8, 20) were used to obtain scores corresponding to average nucleotide identity using BLAST [ANI(b)] and JSpecies with standard settings (21). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5 (22). The 16S rRNA gene sequences used for the analysis were full length. The 16S rRNA gene similarity percentage analysis was performed via the Jukes-Cantor model (23).
Morphological investigation by EM using different sample preparation techniques.
High-pressure freezing, freeze-substitution, Epon embedding, and sectioning were performed as described previously (24). For high-pressure freezing, cells were taken at one time point. Freeze-substitution was performed in acetone containing 2% osmium tetroxide, 0.2% uranyl acetate, and 1% H2O. Respectively, 66, 180, 88, and 173 images containing 1 to 50 typical cells were obtained by transmission electron microscopy (TEM) for M. fumariolicum SolV and strains 3B, 3C, and 4AC.
For freeze-etching (FE), cryo-scanning electron microscopy (cryoSEM), and negative staining, cells were taken from batch cultures at two time points. Freeze-etching on the three mesophilic strains of verrucomicrobial methanotrophs and on M. fumariolicum SolV was performed as described previously (25). Respectively, 73, 60, 79, and 66 typical images containing 1 to 4 freeze-etched cells per image were obtained using a CM12 transmission electron microscope (TEM) (FEI, Eindhoven, the Netherlands) for M. fumariolicum SolV and strains 3B, 3C, and 4AC.
Negative staining was performed by applying a cell suspension (concentrated by centrifugation for 4 min at 12,000 × g) on a (Formvar) carbon-coated copper grid. The grid was then immediately washed in two drops of MilliQ and subsequently incubated for 15 to 20 s on 2% uranyl acetate in MilliQ. The cells were then investigated by the use of a 1010 TEM (Jeol, Tokyo, Japan). Per strain, 24 electron-dense (ED) particles were examined, accounting for 27 cells for strains 3B and 4AC, 20 cells for strain 3C, and 24 cells for M. fumariolicum SolV.
CryoSEM was performed on plunge-frozen cells from the four strains as described previously (25) using a 6301F EDS FESEM instrument (Jeol, Tokyo, Japan). A total of 100 typical nondividing cells of each strain were analyzed. The length and width at the broadest part of the cell were measured for each cell.
A polysaccharide stain was performed to investigate if the electron-light particles identified in the three mesophilic strains were polysaccharide inclusions, as described for M. fumariolicum SolV (26). The stain and matching negative-control procedures were performed as described previously (24) on ultrathin sections of high-pressure-frozen cells (freeze-substituted in acetone containing 2% osmium tetroxide, 0.2% uranyl acetate, and 1% H2O) of the mesophilic strains and visualized using a TEM 1010 instrument (Jeol, Tokyo, Japan). In this method, electron-dense silver albumin aggregates indicate the presence of polysaccharide molecules.
Energy-dispersive X-ray (EDX) analysis was performed on all four strains to investigate the contents of the electron-dense particles. For this purpose, cells were high-pressure frozen, freeze-substituted in 2% osmium tetroxide, and sectioned as described previously (24). The ultrathin sections (ca. 60 nm thick) were visualized (without poststaining) in STEM mode in a JEM 2100 instrument (Jeol, Tokyo, Japan). Qualitative maps were made in which an enrichment of certain elements in specific locations of the cell could be investigated. Maps were acquired after 600 s of measurement time using the Bruker Quantax 200 esprit 1.9.4 software package. For each strain, 10 cells were analyzed in this fashion.
Deposition of cultures.
Preservation of the strains in any other state than as living cultures proved difficult. We are currently testing methods to generate viable frozen stocks. In the meantime, live cultures are available from us by request.
Nucleotide sequence accession numbers.
The genome data of strain 3C have been deposited under BioProject PRJNA165235 and those for strains 3B and 4AC under BioProject PRJNA255456. The pmoA and 16S rRNA sequences can be found in GenBank under the following numbers: KM210549 (3C_pmoA1), KM210550 (3B_pmoA1), KM210551 (3B_pmoA2), KM210552 (4AC_pmoA1), KM210553 (3C_16S), KM210554 (4AC_16S), and KM210555 (3B_16S).
RESULTS AND DISCUSSION
Enrichment and isolation.
Samples from the bare soil area of the Solfatara were taken at spots at least 20 m away from hot fumaroles or the central mud pot. The temperatures of these soils range from 25 to 40°C (27). Soil samples were extracted with water, serially diluted in medium, and incubated with methane for over 4 months. Four of eight soil sample enrichments showed methane consumption and growth at up to a 104 dilution. Lack of growth might be due to the sudden change in acidity that occurred when soils (pH 1) were extracted with water and diluted into medium (pH 2.5). Such pH shocks were found later to prevent the onset of growth of isolates from the enrichments. The positive enrichments contained almost exclusively verrucomicrobia, as judged by FISH microscopy using verrucomicrobium-specific probe EubIII (28) (data not shown). The positive 101 and 104 dilutions were chosen for further cultivation and isolation. After three transfers into new liquid medium, serial dilutions of the final cultures were transferred to floating membrane filters. After 4 weeks at 30°C, the highest dilutions (107) showed isolated colonies that were either small, yellowish, and shiny or larger and off-white. Different colony types were picked and purified by streaking on floating filters. Finally, this resulted in the isolation of strain 4AC (from the 101 dilution) and strains 3C and 3B (both from the 104 dilution). We observed that strain 4AC was sensitive to oxygen and showed a growth rate at an oxygen concentration of 5% that was 2-fold higher than that seen at the ambient oxygen concentration. Especially when shaken at an ambient oxygen concentration, cultures of strain 4AC appeared to have a long lag phase or even failed to grow. This may have prohibited the appearance of strain 4AC in higher serial dilutions. Strains 4AC and 3B consisted of rod-shaped bacteria as investigated with light microscopy. Cells of strain 3C were bigger and morphologically less homogenous than those of strains 4AC and 3B. Both long and short rod-shaped cells were observed as well as cells with a shape reminiscent of buckweed seeds, some of which were connected to each other at distances of up to a few micrometers. For all three isolates, only one rRNA operon was obtained upon genome sequencing (see below) and the isolates were considered to be a pure culture.
Genome properties.
For further characterization, the draft genomes of the new strains were assembled and annotated (see Materials and Methods). The GC content was 60.9% for both strain 3C and strain 4AC and 63.8% for strain 3B. The draft genome sizes (and numbers of identified protein-encoding genes) were 2.77 Mb (2,945 open reading frames [ORFs]), 2.75 Mb (2,804 ORFs), and 2.44 Mb (2,511 ORFs) for strains 3C, 3B, and 4AC, respectively. In general, the metabolic machinery was similar to that seen with thermophilic strains SolV (20) and V4 (29) and mesophilic strain LP2A (16). The annotated genomes were used to compile a table with the key predicted methylotrophy genes (see Table S1 in the supplemental material). The three new isolates possessed only one complete pmoCAB operon and thus differ from the thermophilic strains, which have three complete operons (20, 29). The soluble methane monooxygenase (sMMO)-encoding genes were absent in all strains. Other features of the draft genomes are included in the descriptions below.
Growth conditions.
Acid tolerance of the isolated strains was tested by gradually changing the acid concentration in batch cultures (with an OD of between 0.1 and 0.4). For strains 3C and 3B, growth was optimal between pH 1.5 and 3. Strain 4AC was the most acid tolerant; its optimum growth range extended down to pH 1, and it exhibited the lowest pH allowing growth, namely, pH 0.5. This was lower than for the thermophilic verrucomicrobial methanotrophs (pH 0.8) (7). This makes strain 4AC the most acidophilic methanotroph isolated thus far. Above pH 3, the growth rate of all strains dropped gradually. The maximum pH allowing growth was between pH 5 and 6.
The temperature and pH optima and ranges for these three mesophilic strains (Table 1) reflected the conditions in the acid soils (pH ranging from 1.5 to below 1) and indicated that they occupy different niches in their natural environment. The fact that these strains were enriched from the same soils indicates that local conditions are variable. This is evident for ecosystems such as that of the Solfatara, where heterogeneity in soil conditions occurs because of the irregular rainfall and the various routes the volcanic gasses take through the soil.
TABLE 1.
Conditions for optimal growth of the three mesophilic verrucomicrobial methanotroph strains as tested in batch culturesa
| Strain | Optimum temp (°C) (maximum temp) | μmax (h−1) (doubling time [h]) | Optimum pH range | Lowest pH (% of μmax) |
|---|---|---|---|---|
| 3B | 44 (49) | 0.042 (16) | 1.5–3 | 0.6 (30) |
| 4AC | 38 (43) | 0.035 (20) | 1–3 | 0.5 (40) |
| 3C | 35 (39) | 0.013 (53) | 1.5–3 | 0.6 (35) |
Because of the oxygen sensitivity of strain 4AC, this organism was grown at 5% O2 compared to 17% O2 for the other two strains. μmax = ln2/doubling time.
Like the thermophilic verrucomicrobial methanotrophs (7), the three mesophilic strains were able to grow on methanol. All strains were strictly dependent on the addition of mud pot water to the growth medium. This water could be replaced by cerium (III), which is one of the lanthanides that can serve as a metal cofactor in methanol dehydrogenase of M. fumariolicum SolV (30). All mesophilic strains contain two different xoxF genes encoding methanol dehydrogenases (see Table S1 in the supplemental material). All genes possess the typical lanthanide (III) coordination-specific aspartate located two amino acids downstream of the catalytic aspartate (31).
Most proteobacterial methanotrophs use methane both as an energy source and as a carbon source. Carbon is assimilated at the formaldehyde oxidation level via the ribulosemonophosphate (RuMP) pathway or serine pathway (32). In contrast, M. fumariolicum SolV (13) and M. infernorum V4 (14) and employ carbon dioxide as a carbon source using the Calvin cycle and therefore have an autotrophic lifestyle. Since all of the three new strains contain the genes involved in the Calvin cycle (including the two subunits of the key enzyme RuBisCO) (see Table S1 in the supplemental material), they were predicted to grow as autotrophs. The cbbL genes encoding the RuBisCO large subunit of the three strains were previously shown to form a phylogenetically distinct cluster closely related to M. fumariolicum SolV and M. infernorum V4 (13). The ability to grow autotrophically was tested by culturing the strains on 13C-labeled methane (2% to 3% of the headspace) added to batch cultures that contained a high percentage (40%) of unlabeled 12CO2 in the gas phase to serve as a trap for the produced 13CO2. After growth, 94% to 102% of the 13C label from methane was recovered as 13CO2. The finding that only 4% to 5% of the biomass produced was labeled with 13C, matching the average 13C percentage of the CO2 in the bottles during growth (which increased from 1.2% to 7% to 10%), confirmed that CO2 was the actual carbon source. This showed that these strains do not use intermediates of methane oxidation for their carbon supply as in the case of the serine or RuMP pathway (which would lead to, respectively, 50% or 100% 13C from methane incorporated into the biomass [5, 32]). The key genes of the serine and RuMP pathways were indeed absent from the draft genomes (see Table S1). With the finding that the three mesophilic strains are autotrophic, five of five studied methanotrophic Verrucomicrobia have been proven to be autotrophs. Furthermore, the genome of strain LP2A indicates that this organism too is an autotroph (16). This suggests that all methanotrophic Verrucomicrobia have an autotrophic lifestyle. For the methanotrophic proteobacteria, thus far only Methylococcus capsulatus was shown to possess Calvin cycle genes (33), although physiological evidence for an active Calvin cycle is lacking.
Description of morphology.
The morphology of the three mesophilic strains was studied in detail using multiple electron microscopy (EM) techniques. For comparison, the thermophilic verrucomicrobial methanotroph M. fumariolicum SolV was included in this study. The shape and dimensions of the cells were investigated using cryoSEM. This method showed the whole cells (in contrast to sections) in a near-native state (in contrast to negative staining, where the cells can collapse during the procedure). The cells of all four studied microorganisms were rod shaped, with one of the cell poles being broader than the other pole (Fig. 1D, 2D, 3D, and 4D). Using freeze-etching (FE), replicas of whole or fractured cells show the overall morphology and especially the appearance of the cell surface. FE showed the same morphology of the cells as cryoSEM (Fig. 1 to 4). No S-layers were observed; instead, the cell surface was smooth, as demonstrated by both FE and cryoSEM. Cells of strain 3C exhibited a wider variety in shapes than those of the other strains, as was also observed by light microscopy. In cryoSEM, cells of strain 3C were occasionally attached to each other via a pilus-like structure. The dimensions of all strains were similar (approximately 1.3 μm by 0.6 μm) (see Table S2 in the supplemental material). The width (measured at the broadest pole) was clearly greater for strain 3C, as was also obvious from the length/width (L/W) ratios (see Table S2). This leads to the resemblance of these cells to buckwheat seeds.
FIG 1.
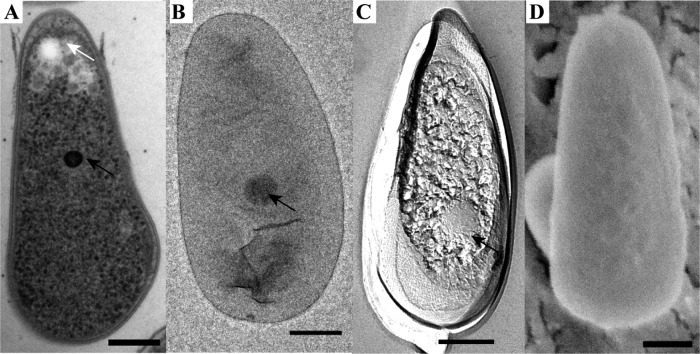
Morphology of the mesophilic verrucomicrobial methanotroph strain 3B as visualized by the use of thin sections of high-pressure-frozen and freeze-substituted cells (A), negative staining (B), FE (C), and cryoSEM (D). The cells are rod shaped with one broader cell pole and contain electron-dense (black arrows) and electron-light (white arrow) particles. Scale bars, 200 nm.
FIG 2.
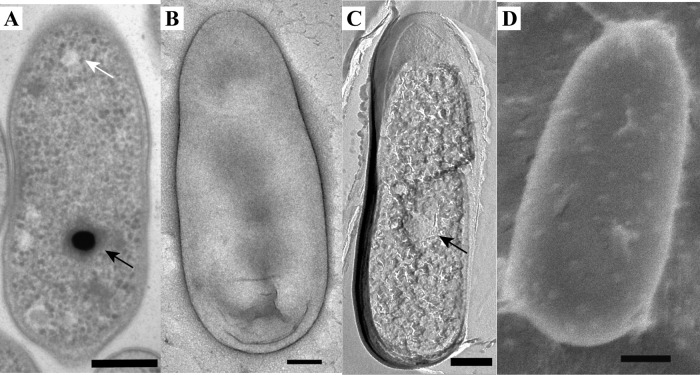
Morphology of the mesophilic verrucomicrobial methanotroph strain 4AC as visualized by the use of thin sections of high-pressure-frozen and freeze-substituted cells (A), negative staining (B), FE (C), and cryoSEM (D). The cells are rod shaped with one broader cell pole and contain electron-dense (black arrows) and electron-light (white arrow) particles. Scale bars, 200 nm.
FIG 3.
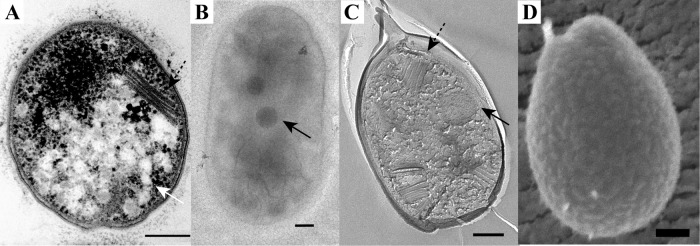
Morphology of the mesophilic verrucomicrobial methanotroph strain 3C as visualized by the use of thin sections of high-pressure-frozen and freeze-substituted cells (A), negative staining (B), FE (C), and cryoSEM (D). The cells are rod shaped with one broader cell pole and contain membrane stacks (dashed arrow) and electron-dense (black arrows) and electron-light (white arrow) particles. Scale bars, 200 nm.
FIG 4.
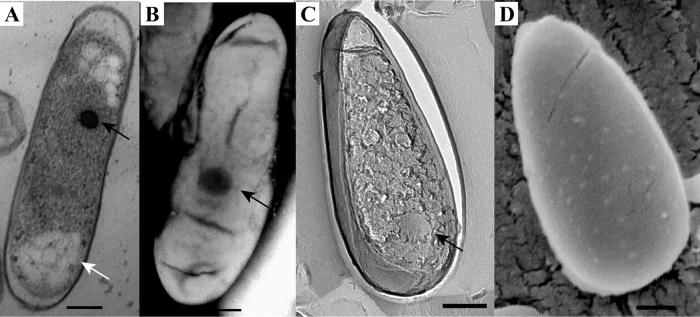
Morphology of the thermophilic verrucomicrobial methanotroph M. fumariolicum SolV as visualized by the use of thin sections of high-pressure-frozen and freeze-substituted cells (A), negative staining (B), FE (C), and cryoSEM (D). The cells are rod shaped with one broader cell pole and contain electron-dense (black arrows) and electron-light (white arrow) particles. Scale bars, 200 nm.
Thin sections of high-pressure-frozen and freeze-substituted cells were studied to get an insight into the ultrastructure of the cells. Both M. fumariolicum SolV and the three mesophilic strains feature Gram-negative cell envelopes, as two membranes can be visualized in thin sections (Fig. 1 to 4). There have been reports that multiple verrucomicrobial species (Verrucomicrobium spinosum, Prosthecobacter dejongeii, Chthoniobacter flavus, and Pedosphaera parvula strain Ellin514) have a Planctomycetes-Verrucomicrobia-Chlamydia (PVC)-specific cell plan in which the outermost membrane is in fact a cytoplasmic membrane (34). This is not apparent from our ultrastructural study of the four studied verrucomicrobial methanotrophs. In addition to the Gram-negative cell envelope, all four strains feature a typical-looking cytoplasm containing ribosomes. No condensed DNA has been observed in the thin sections of the four strains.
In strain 3C, intracytoplasmic membrane (ICM) stacks orthogonal to the membrane were observed in many sections (Fig. 3A), after FE (Fig. 3C), and during cryoSEM (data not shown). Although ICMs have been observed in the verrucomicrobial methanotroph Methylacidiphilum infernorum in rare occurrences (9), strain 3C is the first verrucomicrobial methanotroph described where membrane stacks are present in most cells. In the proteobacterial methanotrophs, a complex ICM system consisting of membrane stacks is a widespread characteristic and the membranes are thought to harbor the pMMO enzyme. The appearance and orientation of ICMs of type I methanotrophs differ from the appearance and orientation of ICMs of type II methanotrophs (5). The membranes occur as layers lining the periphery of the cell in type II methanotrophs; in type I methanotrophs, the membranes form stacks that are aligned more or less orthogonally to the cytoplasmic membrane. The membrane stacks of strain 3C resemble type I ICMs. It remains to be established if the pMMO enzymes are also localized in these membranes of strain 3C. Furthermore, the location of pMMO in the other verrucomicrobial methanotrophs that seem to lack internal membranes needs to be investigated.
In all strains, both electron-dense (ED) and electron-light (EL) particles were observed in the thin sections (Fig. 1A, 2A, 3A, and 4A) and after FE. The EL particles were smaller and more numerous than the ED particles. The ED particles were typically located near the center of the cell, and the EL particles seemed to localize preferentially to the cell poles. EL particles were observed in the majority of cells, and they resembled the glycogen storage particles in M. fumariolicum SolV (26). A polysaccharide stain verified that the EL particles contained glycogen (Fig. 5). Accordingly, the genomes of all studied verrucomicrobial methanotrophs harbor the key genes for glycogen synthesis and glycogen degradation (see Table S1 in the supplemental material).
FIG 5.
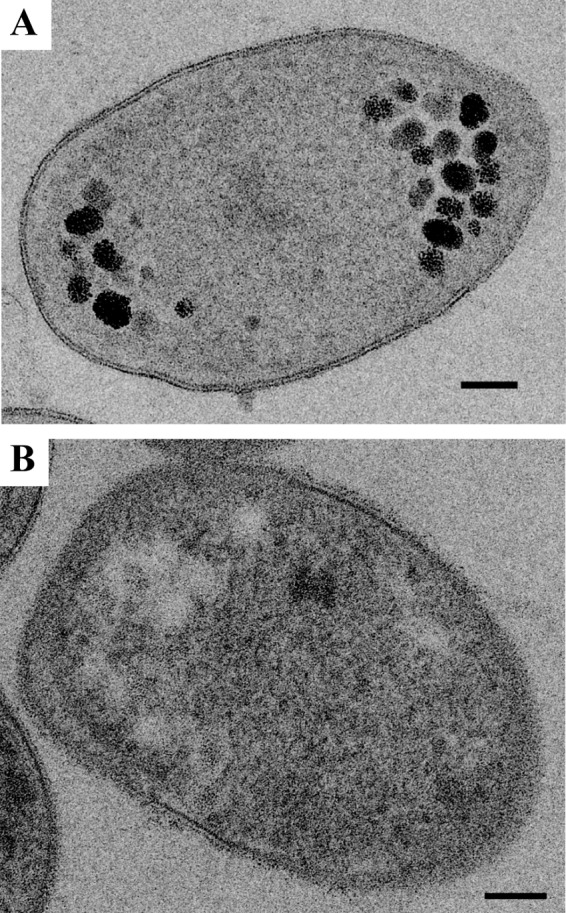
A polysaccharide stain on thin sections of high-pressure-frozen and freeze-substituted cells of the mesophilic verrucomicrobial methanotroph strain 4AC indicates that polysaccharides are present in the electron-light particles. Electron-light particles, appearing as white spots in the negative control (B), show clear staining in panel A (black silver aggregates) because of the polysaccharide content. Similar results were obtained for strains 3B and 3C (data not shown).
The amount and the size of the ED particles were studied via negative staining. The amount of ED bodies was on average close to one per cell (see Table S3 in the supplemental material). Two ED bodies per cell were observed only in some cells of strain 3C and in many dividing cells. The average diameters of the ED particles were similar for all strains and were slightly lower than 200 nm (see Table S3).
To investigate the composition of the cells and of the ED particles in particular, EDX was performed on thin sections of cryo-sectioned, freeze-substituted cells. Qualitative maps showed enrichments of specific elements throughout the cell and made clear that multiple elements were enriched in the electron-dense particles. For M. fumariolicum SolV (Fig. 6) and strains 3C and 3B, the ED particles were enriched in phosphorus and oxygen (see Table S3 in the supplemental material). This suggests that (poly)phosphate is present in the ED particles. Genes encoding polyphosphate kinase and exopolyphosphatase were present in both mesophilic and thermophilic strains (see Table S1). Polyphosphate-encompassing vesicles have been found in other organisms, where they are known as acidocalcisomes (35). Because polyphosphate has a negative charge, positive counterions are often found in the vicinity of these molecules. In the case of M. fumariolicum SolV and strain 3C, magnesium ions seemed to have this role. In strain 4AC, a minority of the ED particles showed a phosphorus signal. In addition, these particles showed sulfur enrichment. The role for sulfur, however, remains puzzling, since no genes for sulfur storage/utilization have been detected thus far in the draft genome (data not shown).
FIG 6.
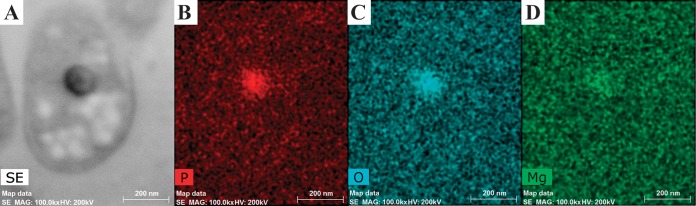
Thin section of M. fumariolicum SolV analyzed by EDX shows the ED particle (dark particle in the sectioned cell in STEM mode [A]) to be enriched in phosphorus (B), oxygen (C), and magnesium (D). SE, secondary electron detector; MAG: 100.0kx, magnification 100,000 times; HV, high voltage.
Phylogenetic and genomic analysis.
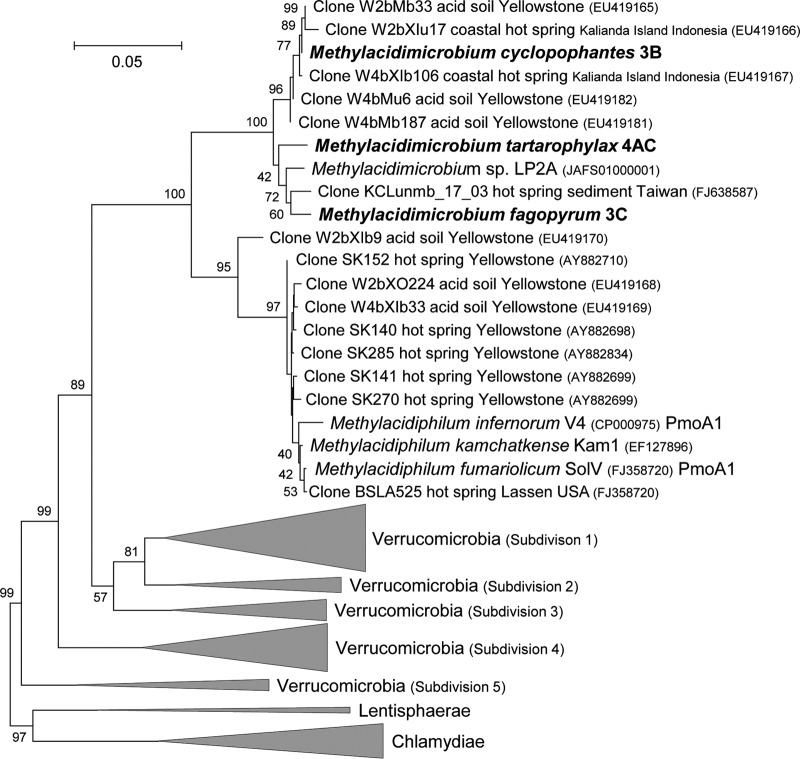
Phylogenetic analysis of the 16S rRNA gene of the three mesophilic strains showed them to be affiliated with the previously described (16) mesophilic LP2A strain (Table 2). The cluster of mesophiles separates clearly from the thermophilic cluster, with which they share only just below 90% identity (Fig. 7). The pmoA-based phylogeny agreed well with the 16S rRNA-based phylogeny, with strain LP2A as the closest cultivated neighbor (see Table S1 in the supplemental material). The pmoA genes of new isolates showed about 92% identity to those of strain LP2A at the amino acid level and about 70% to 72% amino acid identity to the pmoA1 and pmoA2 genes of the thermophilic strains. The same clustering was obtained using the mxaF (xoxF) (31) gene. It was previously asserted that verrucomicrobial 16S rRNA genes putatively belonging to methanotrophs cluster in three distinct groups (16). Analysis of the 16S rRNA genes that also included the environmental operational taxonomic units (OTUs) described previously (16) showed that the newly described mesophilic isolates belong to the same group as LP2A (data not shown).
TABLE 2.
Distance analysis (complete deletion) of 16S rRNA genes showing the similarities between the three mesophilic verrucomicrobial strains and the previously described mesophilic LP2A and thermophilic M. fumariolicum SolV strains
| Strain no. | Strain name | % similarity with strain: |
||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 1 | Methylacidiphilum fumariolicum SolV | |||||
| 2 | Methylacidimicrobium fagopyrum 3C | 89.1 | ||||
| 3 | Methylacidimicrobium tartarophylax 4AC | 89.6 | 97.3 | |||
| 4 | Methylacidimicrobium cyclopophantes 3B | 89.7 | 97.6 | 97.1 | ||
| 5 | LP2A | 89.6 | 98.1 | 97.3 | 98.1 | |
FIG 7.
16S rRNA gene-based phylogenetic tree of methanotrophic and other Verrucomicrobia showing that the methanotrophic mesophilic strains cluster separately from the methanotrophic thermophilic species. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the branch length sum of 4.46361805 is shown. The percentage of replicate trees (values above 40%) in which the associated taxa clustered in the bootstrap test (10,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths shown in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method and are in the units of the number of base substitutions per site. The analysis involved 128 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1,647 positions in the final data set. Evolutionary analyses were conducted in MEGA5.
To investigate if the three new strains represent three strains of the same species or multiple species, analyses based on the 16S rRNA gene and the average nucleotide identity using BLAST [ANI(b)] (with draft genomes) were performed. M. fumariolicum SolV was included in both analyses, and LP2A was included in the 16S rRNA gene sequence analysis. The three mesophilic strains (3B, 3C, and 4AC) share 89.1% to 98.1% 16S rRNA gene sequence identity (Table 2). They would therefore be characterized as three different species by the use of 16S rRNA gene analysis with the strict species cutoffs for similarity of 98.5% (36) or 98.7 to 99.1% (37, 38) (Table 2). Using the same species cutoffs, mesophilic strain LP2A should be classified as a fourth species, since it shares 89.6% to 98.1% 16S rRNA gene sequence identity with the three mesophiles. However, multiple studies have shown that 16S rRNA gene analysis is often not fit for resolution on the species level and that the ANI technique is better suited to such analyses (36, 39, 40). Therefore, we have also analyzed the ANI(b) values for the three mesophilic strains and M. fumariolicum SolV. The three strains share ANI(b) values between 77.3% and 87.8% (Table 3) and are therefore proposed to belong to three different species using the standard species identity cutoff value of 95% (41). Based on the ANI(b) and 16S rRNA gene similarity values (using a strict species cutoff), it should be concluded that each strain represents a distinct species.
TABLE 3.
ANIb analysis results show the similarities between the draft genomes of the three mesophilic verrucomicrobial strains and that of the thermophilic M. fumariolicum SolV strain
| Strain no. | Strain name | % similarity with strain: |
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 1 | Methylacidiphilum fumariolicum SolV | 63.7 | 63.2 | 63.7 | |
| 2 | Methylacidimicrobium fagopyrum 3C | 63.8 | 87.8 | 77.3 | |
| 3 | Methylacidimicrobium tartarophylax 4AC | 62.9 | 87.6 | 77.6 | |
| 4 | Methylacidimicrobium cyclopophantes 3B | 63.4 | 77.6 | 77.7 | |
A minimal 95% identity of the 16S rRNA gene sequences is widely accepted to affiliate two strains to the same genus (42, 43). Because of the 16S rRNA gene sequence identity of 89.1% to 89.7% (Table 2) between the four mesophilic species and M. fumariolicum SolV, it should be concluded that the four species belong to a different genus than M. fumariolicum SolV. The four mesophilic species have 16S rRNA gene sequence identities between 97.3% and 98.1% and therefore belong to the same genus. For this genus, we propose the name Methylacidimicrobium. The proposed names for the three species described in this article are as follows: Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium fagopyrum 3C, and Methylacidimicrobium cyclopophantes 3B. We do not propose a species name for strain LP2A but propose referring to this strain as Methylacidimicrobium strain LP2A.
Here we described three new species of a new genus of methanotrophic Verrucomicrobia isolated from a geothermal environment. The three species featured in this study indicate that a wide variety of methanotrophic verrucomicrobial species exist and that they are quite well adapted to different niches present in geothermal environments with respect to both temperature and acidity. The previously described LP2A strain (16), which, according to our analysis, also belongs to the genus Methylacidimicrobium, has a lower growth temperature range than the three Methylacidimicrobium species described here. The Methylacidiphilum species described before have a higher growth temperature range than all Methylacidimicrobium species. Although the optimum pH range for growth is essentially the same for all three Methylacidimicrobium species (1.5 to 3), these new mesophilic strains are more acid tolerant, with strain 4AC even growing at pH 0.5. The pH range for growth of Methylacidimicrobium strain LP2A is similar but slightly higher (1 to 5.2) (16), and the same is true for the Methylacidiphilum species (0.8 to 6) (7). It would be very interesting to study whether verrucomicrobial methanotrophs exist that live at higher pHs or if they are outcompeted by proteobacterial methanotrophs in these environments. It is therefore important to isolate and characterize additional verrucomicrobial methanotrophs.
So far, all studies in acidic, volcanic environments have exclusively yielded verrucomicrobial enrichments. No proteobacteria have been isolated from these environments to date (see, e.g., reference 16). This suggests that Verrucomicrobia dominate these ecosystems, although additional (especially metagenomic) surveys would be needed to verify this hypothesis.
It appears that many methanotrophic species are adapted to slightly different conditions, which would indicate that even when circumstances change, methanotrophy occurs in the ecosystem. The diversification of methanotrophs in the ecosystem may thus lead to a more stable ecosystem. As has been shown before in other ecosystems with a high number of microbial species per functional group, the amount of biomass and the density were more stable (44). High numbers of species that are adapted to slightly different conditions are also expected to stabilize ecosystem process rates in response to changes in environments (45).
The recent findings are just the beginning of understanding the diversity in verrucomicrobial methanotrophs. Much more research is needed to verify that an even broader methanotrophic diversity can exist, as is suggested from a study of operational taxonomic units (OTUs) that were retrieved from multiple geothermal sites (16). For the organisms that these OTUs represented, it has to be investigated whether they indeed perform methane oxidation and to find out which circumstances they need for their growth. Additional studies investigating the ecosystems are necessary to understand the interplay between the different genera and species of verrucomicrobial methanotrophs and the role they play in different environments.
Description of Methylacidimicrobium gen. nov.
Methylacidimicrobium (Me.thyl.a.ci.di.mi.cro′bi.um. N.L. n. methyl, the methyl group; N.L. n. acidum, acid [from L. adj. acidus, sour]; N.L. n. microbium, microbe; N.L. n. Methylacidimicrobium, methyl-using microbe living in an acid environment).
Gram-negative, rod-shaped bacteria with a broadening at one of the two cell poles. Cells occur as single cells or as small groups; do not form rosettes. Reproduce by binary fission. Nonmotile. Produce intracellular glycogen granules and additional intracellular (electron-dense) particles. Except for the type species M. fagopyrum 3C, no ICM were observed. sMMO is not present in the genomes. Representatives are extremely acidophilic and mesophilic. No growth occurs without lanthanides in the medium. Growth is also possible on methanol. Carbon is fixed via the Calvin cycle. The G-C content is 60.9% to 63.8%. Belongs to the Verrucomicrobia; the closest described methanotrophic bacterial genus is the verrucomicrobial thermo- and acidophilic genus Methylacidiphilum (7). Contains the type species Methylacidimicrobium fagopyrum 3C and Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium cyclopophantes 3B, and Methylacidimicrobium strain LP2A (16) as additional species. Habitat is acidic soil of elevated temperature, particularly in volcanic mud pots or near fumaroles.
Description of Methylacidimicrobium fagopyrum sp. nov.
Methylacidimicrobium fagopyrum (fa.go′py.rum. N.L. neuter n. fagopyrum, buckwheat; referring to the shape of the cell).
Description as for the genus plus the following traits. Cells are ca. 1.2 μm long and 0.7 μm wide. An ICM system was observed (Fig. 3) that consists of membrane stacks orthogonal to the cell wall. One or two particles (0.16 ± 0.08 μm in diameter) containing phosphorus, oxygen, magnesium, and nitrogen are present in most cells. The optimum temperature for growth is 35°C; no growth occurs above 39°C. Growth occurs at or above pH 0.6, with an optimum range of 1.5 to 3.0. The type strain is strain 3CT, which was isolated from soil of the Solfatara, at Pozzuoli, near Naples, Italy.
Description of Methylacidimicrobium tartarophylax sp. nov.
Methylacidimicrobium tartarophylax (tar.ta.ro′phy.lax. L. masc. n. Tartarus, underworld; Gr. masc. n. phylax, guardian; N.L. adj. tartarophylax, guardian of the underworld; referring to the enrichment location which in Roman times was believed to be in the vicinity of an entrance to the underworld).
Description as for the genus plus the following traits. Cells are ca. 1.4 μm long and 0.9 μm wide. No ICM system was observed (Fig. 2). One particle (0.15 ± 0.03 μm in diameter) containing sulfur, oxygen, and in some cases phosphorus is present in most cells. The optimum temperature for growth is 38°C; no growth occurs above 43°C. Growth occurs at and above pH 0.5, with pH 1 to 3 as optimum. Growth is inhibited by oxygen (growth at 5% oxygen is about two times faster than at ambient oxygen concentration). The type strain is strain 4ACT, which was isolated from soil of the Solfatara, at Pozzuoli, near Naples, Italy.
Description of Methylacidimicrobium cyclopophantes sp. nov.
Methylacidimicrobium cyclopophantes (cy.clo.po.phan′tes. L. masc. cyclops, cyclops; Gr. adj. suffix -phantes, resembling; N.L. n. adj. cyclopophantes, resembling a Cyclops; referring to the single large electron-dense particle which is present in each cell).
Description as for the genus plus the following traits. Cells are ca. 1.2 μm long and 0.6 μm wide. No ICM system was observed (Fig. 1). One particle (0.12 ± 0.04 μm in diameter) containing phosphorus and oxygen is present in most cells. The optimum temperature for growth is 44°C; no growth occurs above 49°C. Growth occurs at or above pH 0.6, with an optimum range of 1.5 to 3.0. The type strain is strain 3BT, which was isolated from soil of the Solfatara, at Pozzuoli, near Naples, Italy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Geert-Jan Janssen, Rob Mesman, Elly van Donselaar, Willie Geerts, Reinhard Rachel, and Andreas Klingl for advice on and assistance with numerous aspects of the ultrastructural research. We thank Jelle Eygensteyn for performing the 13C analysis. Daan Speth is acknowledged for help with the ANI(b) analysis. We thank Claudia Lüke for help with ARB. We thank Janric van Rookhuijzen for advice concerning the correct ancient Greek and Latin grammatical construction of the names.
This research was supported by ERC 232937; ERC 339880, and Gravitation Grant SIAM OCW/NWO 024.002.002. L.V.N. is supported by NWO VENI grant 863.09.009.
Footnotes
Published ahead of print 29 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01838-14.
REFERENCES
- 1.Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1:285–292. 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 2.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334. 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 3.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp, HJM. Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 4.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 5.Hanson RS, Hanson TE. 1996. Methanotropic Bacteria. Microbiol. Rev. 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol. Rev. 34:496–531. 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 7.Op den Camp, HJM. Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306. 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 8.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. 2007. Methanotrophy below pH1 by a new Verrucomicrobia species. Nature 450:874–878. 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 9.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882. 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 10.Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland N-K. 2008. Methane oxidation at 55 °C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U. S. A. 105:300–304. 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damsté JS, Op den Camp HJ, Jetten MS, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 12.Rasigraf O, Kool DM, Jetten MSM, Sinninghe Damsté JS, Ettwig KF. 2014. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph Methylomirabilis oxyfera. Appl. Environ. Microbiol. 80:2451–2460. 10.1128/AEM.04199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs K-J, Stunnenberg HG, Jetten MSM, Op den Camp HJM. 2011. Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol. 193:4438–4446. 10.1128/JB.00407-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp CE, Stott MB, Dunfield PF. 2012. Detection of autotrophic verrucomicrobial methanotrophs in a geothermal environment using stable isotope probing. Front. Microbiol. 3:303. 10.3389/fmicb.2012.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp CE, Op den Camp, HJM. Tamas I, Dunfield PF. 2013. Unusual members of the PVC superphylum: the methanotrophic Verrucomicrobia genus “Methylacidiphilum,” p 211–227 In Fuerst JA. (ed), Planctomycetes: cell structure, origins and biology. Humana Press (Springer), New York, NY. [Google Scholar]
- 16.Sharp CE, Smirnova AV, Graham JM, Stott MB, Khadka R, Moore TR, Grasby SE, Strack M, Dunfield PF. 18 April 2014. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 10.1111/1462-2920.12454. [DOI] [PubMed] [Google Scholar]
- 17.Etiope G, Klusman RW. 2002. Geologic emissions of methane to the atmosphere. Chemosphere 49:777–789. 10.1016/S0045-6535(02)00380-6. [DOI] [PubMed] [Google Scholar]
- 18.Khadem AF, Pol A, Jetten MSM, Op den Camp HJM. 2010. Nitrogen fixation by the verrucomicrobial methanotroph “Methylacidiphilum fumariolocum” SolV. Microbiology 156:1052–1059. 10.1099/mic.0.036061-0. [DOI] [PubMed] [Google Scholar]
- 19.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khadem AF, Wieczorek AS, Pol A, Vuilleumier S, Harhangi HR, Dunfield PF, Kalyuzhnaya MG, Murrel JC, Francoijs K-J, Stunnenberg HG, Stein LY, DiSpirito AA, Semrau JD, Lajus A, Médigue C, Klotz MG, Jetten MSM, Op den Camp HJM. 2012. Draft genome sequence of the volcano-inhibiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J. Bacteriol. 194:3729–3730. 10.1128/JB.00501-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131. 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jukes TH, Cantor CR. 1969. Evolution of protein molecules, p 21–132 In Munro HN. (ed), Mammalian protein metabolism. Academic Press, New York, NY. [Google Scholar]
- 24.van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Webb RI, Fuerst JA, Verkleij AJ, Jetten MSM, Strous M. 2008. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190:708–717. 10.1128/JB.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu ML, van Teeseling MCF, Willems MJR, van Donselaar EG, Klingl A, Rachel R, Geerts WJC, Jetten MSM, van Niftrik L. 2012. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera,” a novel polygon-shaped bacterium. J. Bacteriol. 194:284–291. 10.1128/JB.05816-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khadem AF, van Teeseling MCF, van Niftrik L, Jetten MSM, Op den Camp, HJM. Pol A. 2012. Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph “Ca. Methylacidiphilum fumariolicum” SolV. Front. Microbiol. 3:345. 10.3389/fmicb.2012.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merucci L, Bogliolo MP, Buongiorno MF, Teggi S. 2006. Spectral emissivity and temperature maps of the Solfatara crater from DAIS hyperspectral images. Ann. Geophys. 49:235–244. 10.4401/ag-3174. [DOI] [Google Scholar]
- 28.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444. 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 29.Hou S, Makarova KS, Saw JHW, Senin P, Ly BV, Zhou Z, Ren Y, Wang J, Galperin MY, Omelchenko MV, Wolf YI, Yutin N, Koonin EV, Stott MB, Mountain BW, Crowe MA, Smirnova AV, Dunfield PF, Feng L, Wang L, Alam M. 2008. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 3:26. 10.1186/1745-6150-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp HJM. 2014. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 16:255–264. 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 31.Keltjens JT, Pol A, Reimann J, Op den Camp HJM. 13 May 2014. PQQ-dependent methanol dehydrogenases: rare earth elements make a difference. Appl. Microbiol. Biotechnol. 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 32.Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. 2009. The expanding world of methylotrophic metabolism. Annu. Rev. Microbiol. 63:477–499. 10.1146/annurev.micro.091208.073600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Methé B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonasen I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K-C, Webb RI, Janssen PH, Sangwan P, Romeo T, Staley JT, Fuerst JA. 2009. Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiol. 9:5. 10.1186/1471-2180-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Docampo R, Moreno SNJ. 2001. The acidocalcisome. Mol. Biochem. Parasitol. 114:151–159. 10.1016/S0166-6851(01)00246-8. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt E. 2011. Molecular taxonomic parameters. Microbiol. Aust. 32:59–61. [Google Scholar]
- 37.Mende DR, Sunagawa S, Zeller G, Bork P. 2013. Accurate and universal delineation of prokaryotic species. Nat. Methods 10:881–884. 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- 38.Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152–155. [Google Scholar]
- 39.Chan JZ-M, Halachev MR, Loman NJ, Constantinidou C, Pallen MJ. 2012. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol. 12:302. 10.1186/1471-2180-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10:504–509. 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81–91. 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 42.Adékambi T, Shinnick TM, Raoult D, Drancourt M. 2008. Complete rpoB gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int. J. Syst. Evol. Microbiol. 58:1807–1814. 10.1099/ijs.0.65440-0. [DOI] [PubMed] [Google Scholar]
- 43.Schloss PD, Handelsman J. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686–691. 10.1128/MMBR.68.4.686-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naeem S, Li S. 1997. Biodiversity enhances ecosystem reliability. Nature 390:507–509. 10.1038/37348. [DOI] [Google Scholar]
- 45.Hooper DU, Chapin FS, III, Ewel JJ, Hector A, Inchausti P, Lavorel S, Lawton JH, Lodge DM, Loreau M, Naeem S, Schmid B, Setälä H, Symstad AJ, Vandermeer J, Wardle DA. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75:3–35. 10.1890/04-0922. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.