Abstract
The manganese (Mn)-oxidizing protein (MopA) from Erythrobacter sp. strain SD21 is part of a unique enzymatic family that is capable of oxidizing soluble Mn(II). This enzyme contains two domains, an animal heme peroxidase domain, which contains the catalytic site, followed by a C-terminal calcium binding domain. Different from the bacterial Mn-oxidizing multicopper oxidase enzymes, little is known about MopA. To gain a better understanding of MopA and its role in Mn(II) oxidation, the 238-kDa full-length protein and a 105-kDa truncated protein containing only the animal heme peroxidase domain were cloned and heterologously expressed in Escherichia coli. Despite having sequence similarity to a peroxidase, hydrogen peroxide did not stimulate activity, nor was activity significantly decreased in the presence of catalase. Both pyrroloquinoline quinone (PQQ) and hemin increased Mn-oxidizing activity, and calcium was required. The Km for Mn(II) of the full-length protein in cell extract was similar to that of the natively expressed protein, but the Km value for the truncated protein in cell extract was approximately 6-fold higher than that of the full-length protein, suggesting that the calcium binding domain may aid in binding Mn(II). Characterization of the heterologously expressed MopA has provided additional insight into the mechanism of bacterial Mn(II) oxidation, which will aid in understanding the role of MopA and Mn oxidation in bioremediation and biogeochemical cycling.
INTRODUCTION
Manganese (Mn)-oxidizing bacteria and fungi are known to play an important role in the biogeochemical cycling of Mn and other elements by catalyzing the oxidation of soluble Mn(II) to Mn(III, IV) oxides. The Mn oxides formed can oxidize reduced metals, sulfides, and carbon compounds. In addition, the amorphous structure of the biogenic oxides can sequester heavy metals, reducing their toxicity (1–4). Mn-oxidizing bacteria have the potential to increase the rate of Mn oxidation by orders of magnitude over abiotic Mn oxidation, and this capability is found in many diverse phylogenetic groups (4). Yet, understanding the enzymes involved in Mn oxidation by these diverse bacteria has been difficult to achieve due to low levels of native expression and often unstable activity, which has prevented the purification and investigation of the enzymes under native conditions (5–7).
Multicopper oxidase (MCO) enzymes have been implicated in several of the model Mn-oxidizing strains (8). MCOs use molecular oxygen as the oxidant, with electrons being transferred from the substrate to molecular oxygen via multiple copper atoms in the enzyme. These enzymes are capable of oxidizing a broad range of substrates, including organic compounds, phenolic compounds, and various metals, such as Fe(II) and Mn(II) (9). They have been identified in Pseudomonas putida (10), Bacillus species (11, 12), Leptothrix discophora (13), and Pedomicrobium (14). Recently, two of the bacterial Mn-oxidizing MCO enzymes from Bacillus species (15, 16) and an MCO, CueO, from Escherichia coli (17) have been heterologously expressed, purified, and biochemically characterized. These unique MCO enzymes oxidize Mn(II) by two one-electron transfers to form an Mn(IV) oxide. The CotA protein from Bacillus pumilus WH4 and the CueO from E. coli can catalyze Mn(II) oxidation with a single subunit, yet the MnxG multicopper oxidase from Bacillus sp. strain PL-12 requires coexpression of the accessory proteins MnxDEF for activity (16).
In addition to the MCOs, another class of bacterial Mn-oxidizing proteins has been described. These proteins have sequence similarity to animal heme peroxidases of the peroxidase-cyclooxygenase superfamily of peroxidases (18). This class of enzymes includes myeloperoxidases (MPO) and lactoperoxidases (LPO), which use hydrogen peroxide as the oxidant and, as their names suggest, contain a heme cofactor. These animal heme peroxidases generally consist of the peroxidase domain and another nonenzymatic domain (18, 19). The Mn-oxidizing proteins (MopAs) of this type have been identified in Erythrobacter sp. strain SD21 and Aurantimonas manganoxydans strain SI85-9A1 by protein sequencing of Mn-oxidizing SDS-PAGE bands (5). These Mn-oxidizing proteins are in the peroxicin subfamily of the peroxidase-cyclooxygenase superfamily (18, 20). This subfamily has one or more animal heme peroxidase domains, with an additional domain containing multiple RTX-type calcium binding motifs (Fig. 1). Several proteins in this family, including the Mn-oxidizing protein from A. manganoxydans SI85-9A1, have repeated domains (18). RTX proteins are found in many Gram-negative bacteria and contain glycine and aspartate-rich nonapeptide repetitions for the binding of calcium ions (21). Although the functions of many animal heme peroxidases and RTX proteins are unknown, there are a variety of known functions for both of these types of proteins, including antibacterial activity for many animal heme peroxidases and protease activity and S-layer formation (21) for RTX-type proteins. The Mn-oxidizing proteins of this type oxidize Mn(II) to Mn(III) (5, 6). The Mn(III) formed from these organisms may form Mn(IV) oxides via another enzyme, such as an MCO, or by disproportionation. Mn(III) may also remain as Mn(III) bound to a ligand, as it has recently been identified in the environment (8, 22). The partially purified MopA protein from Aurantimonas manganoxydans SI85-9A1 has been shown to contain a heme cofactor via heme staining of an SDS-PAGE gel and to have Mn-oxidizing activity stimulated by hydrogen peroxide (5). In vitro studies on the Mn-oxidizing protein from Erythrobacter sp. SD21 did not show stimulation in the presence of hydrogen peroxide, but this activity was stimulated by PQQ and NAD+/NADH, and PQQ was found to copurify with the partially purified Mn(II)-oxidizing protein (6). The activity in native cell extract was also inhibited by peroxidase inhibitors cyanide, azide, and phenylhydrazine (6). The inability to purify the native active enzymes from these strains has significantly hindered their characterization and further understanding of this unique bacterial process.
FIG 1.
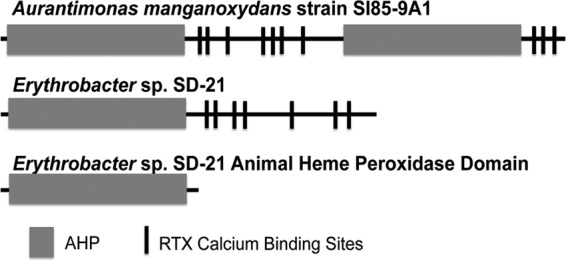
Protein diagram of the manganese-oxidizing peroxidase/protein (MopA). MopA, identified in Aurantimonas manganoxydans strain SI85-9A1 and Erythrobacter sp. strain SD21, has an animal heme peroxidase domain(s) and an RTX calcium binding domain.
Mn-oxidizing bacteria and their enzymes play an important role in the environment. Both the manganese-oxidizing peroxidase (Mop) type and the MCOs have been identified in Mn-oxidizing environmental samples (23), suggesting that both types of enzymes are likely to be relevant in situ. Although recent studies on MCOs have started to reveal the mechanism of this reaction, little is known about Mop enzymes. To better understand these newly identified animal heme peroxidase-type bacterial Mn-oxidizing enzymes, we have cloned the gene encoding MopA and the animal heme peroxidase (AHP) domain of MopA from Erythrobacter sp. SD21 and heterologously expressed this enzyme in E. coli. Characterization of the heterologously expressed enzyme begins to tell the story of how this unique enzyme may function in Mn oxidation.
MATERIALS AND METHODS
Cloning of the manganese(II)-oxidizing protein.
Erythrobacter sp. SD21 genomic DNA was isolated using standard techniques, and the mopA gene was amplified with Pfu Turbo (Agilent) using the forward primer 5′-ATGAAAAAACTGCTGTTCGCGATTCCGCTGGTGGTGCCGTTCTATAGCCATAGCATGGCCGTCAAACTCAACAAGC-3′, which contains a gIII signal sequence, and the reverse primer 5′-ATGATGATGATGATGATGGTCGACGGCGCTATTGAAGAGGAAGGTTTCG AAGAGTCC-3′, encoding a histidine tag. The 6.4-kb amplicon was cloned into the pSpeedET vector using the polymerase incomplete primer extension (PIPE) method (24). The pSpeedET vector used contains an inducible arabinose promoter for overexpression, a histidine tag for purification, and a kanamycin resistance gene for selection. The amplified vector used for PIPE cloning of the full-length mopA gene did not include the N-terminal histidine tag but had an added C-terminal histidine tag. The pSpeedET-Mop construct was designed for a 6× C-terminal histidine tag, but sequencing of different clones revealed some of the pSpeedET-Mop constructs had 12× and 18× histidine tags, thus allowing us to see if the histidine tag length could affect activity and purification.
A truncated version of the mopA gene encoding the catalytic AHP domain (nucleotides [nt] 1 to 2787) was cloned into the same pSpeedET expression vector by using PIPE cloning with the standard N-terminal 6×His tag. The AHP domain of mopA was amplified with the forward primer 5′-CTGTACTTCCAGGGCATGGCCGTCAAACTCAACAAGC-3′ and the reverse primer 5′-AATTAAGTCGCGTTAATTCAGCACTTCGTCATCGTGTTGC-3′. The AHP domain of mopA was also cloned into the pBAD/gIII expression vector (Life Technologies) by PIPES cloning. The vector was amplified using 5′-GAACAAAAACTCATCTCAGAAGAGGATCTGAATAGGCCG-3′ and 5′-GGTGCTATGGCTATAGAACGGCACCACCAGCGGAATCGCGAACCAGC-3′. The AHP domain of mopA was amplified for cloning into the pBAD/gIII vector using 5′-TTCTATAGCCATAGCACCATGGCCGTCAAACTCAACAAGC-3′ and 5′-GATGAGTTTTTGTTCATTCAGCACTTCGTCATCGTGTTGC-3′. All plasmids were cloned into chemically competent E. coli HK100 (24) for purification and sequencing. The resulting pSpeedET-Mop, pSpeedET-AHP, and pBAD/gIII-AHP recombinant plasmids were transformed into chemically competent E. coli Rosetta 2 (Novagen) cells carrying the pRARE plasmid that encodes rare tRNAs and contains a chloramphenicol resistance gene.
Overexpression of Mop and preparation of cell extract.
E. coli Rosetta 2 cells transformed with the pSpeedET-Mop and the pSpeedET-AHP plasmid were inoculated into LB medium supplemented with 60 μg/ml kanamycin and 30 μg/ml chloramphenicol and incubated overnight at 37°C and 250 rpm. A total of 3 ml of the overnight culture was added to 100 ml of LB medium with the same antibiotics under the same conditions. Once the cultures reached an optical density at 600 nm (OD600) of ∼0.5, they were induced with 0.02% l-arabinose and incubated for 4 h. The harvested cells were resuspended in 50 mM HEPES, pH 7.7, and lysed with a French press (16,000 lb/in2). The soluble cell extract was obtained by centrifugation at 9,100 × g for 20 min at 4°C. SDS-PAGE (25) and Western blotting were used to confirm expression. Western blots using the HisProbe-horseradish peroxidase (HRP) and SuperSignal West HisProbe kit (Thermo Scientific) were used according to the manufacturer's suggestions.
Mn oxidation assay.
Mn-oxidizing activity was determined as previously described (6). A 500-μl assay contained approximately 1.7 mg cell extract, 1 mM MnCl2, 10 mM CaCl2, 10 μM PQQ, 87.6 mM NaCl, and 43.8 mM HEPES (pH 8). The assay was incubated for 24 h with shaking (250 rpm). All assays were incubated at room temperature, similar to the optimal growth temperature for Erythrobacter sp. SD21 (26). Product formation increased linearly with time for more than 24 h, as determined by a time course. Catalase (5 μM), superoxide dismutase (50 μM), and hemin (0.5 μM) were added, as indicated. All assays were performed in triplicate. Oxidized Mn [Mn(III) or Mn(IV)] was quantified by adding 50 μl of the assay to 250 μl of 0.014% or 0.04% leucoberbelin blue (3), incubating for 15 min at room temperature, centrifuging (20,000 × g, 5 min, room temperature) to remove precipitated protein, and determining the A620. Potassium permanganate served as the standard.
For determination of kinetic parameters, Mn(II) was added at 0 mM, 0.1 mM, 0.4 mM, 0.6 mM, 1 mM, 1.5 mM, 2.0 mM, 5.0 mM, and 10 mM. Oxidized Mn was measured every 1 to 2 h for approximately 12 h to determine the rate of oxidation. Km and Vmax values were determined with the graphing software Kaleidograph (Synergy Software) fitted to the Michaelis-Menten kinetic equation.
Protein quantification, chemicals, and reagents.
Total protein was determined using the Coomassie Plus assay reagent (Pierce) with bovine serum albumin as the standard. All chemicals were reagent grade from Fisher or Sigma-Aldrich.
RESULTS AND DISCUSSION
Heterologously expressed MopA and the AHP domain catalyze Mn(II) oxidation.
In order to increase expression of and aid in purification of the Mn-oxidizing protein as well as to determine the domain necessary for activity, mopA and the AHP domain of mopA (Fig. 1) were cloned and heterologously expressed in E. coli Rosetta 2. The operon containing the mopA gene in Erythrobacter sp. SD21 has not been characterized, and mopA may be the only gene in its operon. Upstream of mopA, there is a two-component system response regulator that has currently not been linked to Mn oxidation. Therefore, no additional genes were expressed with mopA. The full-length mopA gene was cloned into the modified pSpeedET expression vector carrying a C-terminal His tag and a gIII N-terminal signal sequence. Several constructs with the His tag on the N or C terminus, different lengths of the His tag, and different expression vectors were tested for activity, with the most stable Mn-oxidizing activity occurring when expressed with an 18× C-terminal His tag and the N-terminal gIII signal sequence in the pSpeedET vector. The AHP domain was cloned into the pSpeedET expression vector and expressed with an N-terminal His tag. It was also cloned into the pBAD/gIII vector with a C-terminal His tag and an N-terminal gIII signal sequence. Activity and expression were relatively similar for the two constructs (0.184 nmol · min−1 · mg cell extract−1 for pSpeedET expression and 0.082 nmol · min−1 · mg cell extract−1 for pBAD/gIII); thus, the N-terminal His-tagged protein expressed from the pSpeedET vector was used for further studies. For both the AHP domain and the full-length MopA, the presence of the gIII signal did not help to purify an active protein from the periplasm.
An active MopA was expressed in E. coli cell extract, indicating that MopA does indeed catalyze Mn(II) oxidation. E. coli cell extract is unable to oxidize Mn in the absence of these expressed proteins, and assays containing heat-denatured protein or no protein did not oxidize Mn. The Mn-oxidizing activity of these heterologously expressed proteins strongly builds on their identification in Mn-oxidizing SDS-PAGE gels (5). The full-length 225-kDa MopA was expressed at a level comparable to expression in the native host (27) and ran as two bands at 150 and 250 kDa. These two bands have previously been seen with the native system (27). Affinity, ion exchange, gel filtration, and hydrophobic interaction chromatography were performed using a variety of conditions to purify the heterologously expressed enzyme, but Mn-oxidizing activity was lost. In addition, E. coli proteins that may stabilize, interact with, or stimulate Mn-oxidizing activity were investigated, but adding back E. coli cell extract did not rescue the lost Mn-oxidizing activity, nor did the addition of heme or pyrroloquinoline quinone (PQQ) (data not shown). Thus, a purified, active Mn-oxidizing enzyme could not be obtained, a problem previously encountered with purification from the native host (6). Therefore, the heterologously expressed enzyme was studied in cell extract.
The AHP domain was also actively expressed in E. coli. As indicated in Fig. 2, the 105-kDa AHP domain is expressed at much higher levels than the full-length protein. Similar to the full-length MopA, it does not run true to size on SDS-PAGE. The activity of the AHP domain alone suggests it is the catalytic domain, as has been found in other animal heme peroxidases with additional domains (19). This finding is intriguing in this case, since the calcium binding domain is found in concert with the AHP domain in both Erythrobacter sp. SD21 and Aurantimonas manganoxydans SI85-9AI and in several other organisms. Mn-oxidizing Pseudomonas putida GB-1 contains an MopA protein containing two AHP domains with calcium binding domains similar to those of A. manganoxydans SI85-9A1, although this protein has not been implicated in Mn oxidation (10). Leptothrix cholodnii SP6, another Mn-oxidizing strain, also has an animal heme peroxidase protein with a calcium binding domain (28), but the role of this protein in Mn oxidation is not known at this time. MopA-like proteins (with AHP and calcium binding domains) are not unique to known Mn oxidizers, as they can also be found in the genome sequences of Rhodopseudomonas palustris BisB5, Methylobacterium chloromethanicum CM4, and other species that have not been implicated in Mn oxidation.
FIG 2.
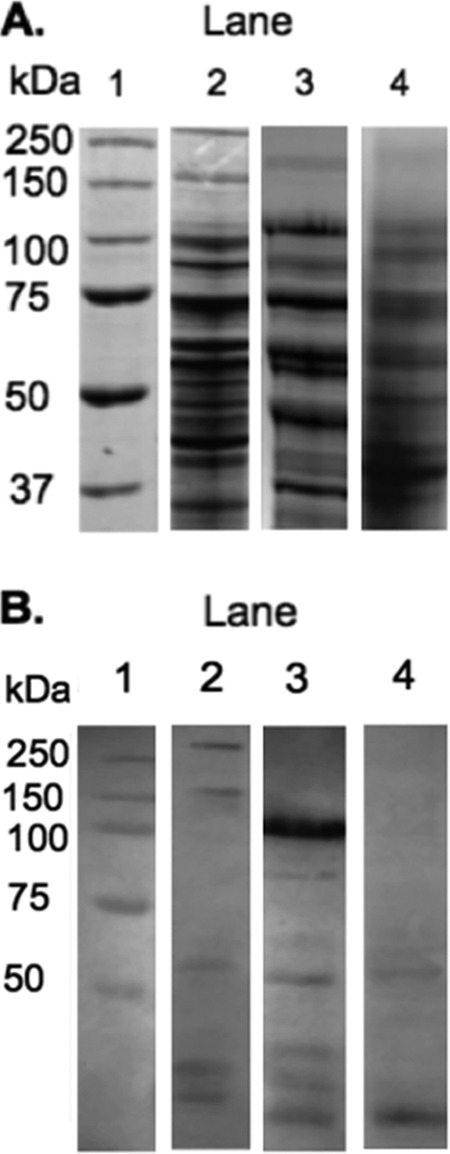
Heterologous expression of MopA and the AHP domain in E. coli cell extract: SDS-PAGE stained with Coomassie blue (A) and Western blot of the expression of the full-length MopA and the AHP domain from MopA (B) in E. coli cell extract. Lane 1, protein standard; lane 2, expression of the full-length protein; lane 3, expression of the AHP domain; lane 4, E. coli cell extract. Lanes shown were extracted from nonadjacent lanes or different gels and aligned using the protein standard.
H2O2 is not likely a substrate for Mn-oxidizing activity of MopA.
Because of the presence of the AHP domain and the sequence similarity to enzymes known to use hydrogen peroxide as a substrate, the effect of hydrogen peroxide was determined on enzyme activity for both the full-length enzyme and the AHP domain. Hydrogen peroxide (1 to 1,000 μM) was added to the enzyme assay, and there was no stimulatory effect on activity. In fact, high levels of hydrogen peroxide (>50 μM) decreased activity, likely due to reducing the oxidized Mn formed in the reaction (29). Catalase is likely present in the E. coli cell extract (30), but it may not be highly active, since addition of hydrogen peroxide decreased Mn oxidation. Therefore, additional catalase was also added to the enzyme assay, as it should inhibit any reactions requiring hydrogen peroxide. As shown in Fig. 3, catalase had only a small effect on Mn-oxidizing activity, providing further evidence that hydrogen peroxide is not likely a substrate. Studies on MPO indicate that hydrogen peroxide interacts with the histidine distal to the heme and an arginine at the catalytic site (31). Both residues are conserved in MopA. If H2O2 is a substrate of the Mn oxidation reaction, it may be difficult to determine, as the presence of H2O2 can affect the abundance of the product (H2O2 reacts directly with Mn oxides), but it does not appear to be required.
FIG 3.
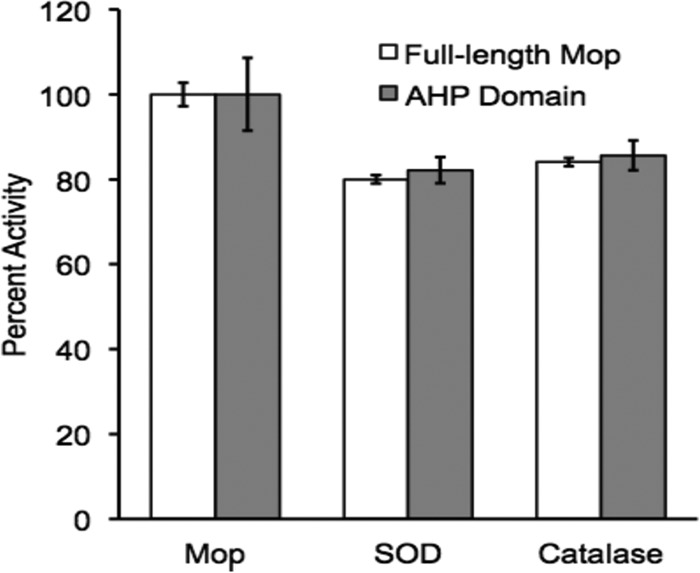
Superoxide and hydrogen peroxide are not substrates for MopA. The addition of 50 μM superoxide dismutase (SOD) and 5 μM catalase did not abolish Mn(II) oxidation of the full-length and the AHP domain in cell extract. Percent activity is compared to activity in the absence of SOD and catalase. Error bars represent standard deviations from triplicates.
Recently, Mn oxidation in Roseobacter sp. strain AzwK-3b was found to occur as a result of enzymatic superoxide formation (32). To assess if superoxide may be a substrate for MopA or a product of MopA that then catalyzes Mn oxidation, superoxide dismutase (SOD) was added to the assay (Fig. 3). SOD is also likely present in E. coli cell extract (33), which would prevent the accumulation of superoxide; additional SOD was added to be certain no superoxide was present in the assay and potentially oxidizing Mn(II). SOD addition did cause a slight decrease in activity, but not to the extent one would expect if superoxide acted as a substrate for the Mn oxidation reaction.
The natively expressed protein was inactive in the absence of oxygen (H. A. Johnson and B. M. Tebo, data not shown), suggesting that oxygen may be the terminal electron acceptor for MopA-catalyzed Mn oxidation. Coupling Mn oxidation to oxygen, or any other compound except hydrogen peroxide, seems very unusual for a protein with significant sequence similarity to peroxidases. MopA may oxidize Mn using a very novel mechanism. A purified protein will be required to further probe the mechanism of action.
Heme and PQQ stimulate MopA Mn oxidation.
Animal heme peroxidases are known to require a heme prosthetic group. This heme group is covalently attached to aspartate and glutamate residues in LPO (34) and to an additional methionine in MPO (31). The heme is also hydrogen bonded to a proximal histidine which interacts with an essential asparagine (35, 36). The aspartate and glutamate residues binding the heme are conserved among animal heme peroxidases and are present in MopA. For both the full-length and the truncated protein in cell extract, there is an increase in activity with the addition of 0.5 μM hemin (see Table 2). This increase in activity suggests that heme is important for activity. It also suggests that heme may be limited during expression, but addition of hemin (770 μM) or the heme precursor, 5-aminolevulinic acid (100 μM), during protein expression did not consistently increase activity (data not shown). The absence of an increase in activity upon addition of heme and 5-aminolevulinic acid during expression may be related to hydrogen peroxide not being a substrate, as covalent binding of heme has been shown to occur as a result of auto-oxidation of the enzyme with hydrogen peroxide. The reducing conditions of the E. coli expression system and the likely catalase activity (30) in whole cells could also make hydrogen peroxide limiting. A similar condition was reported (37) for LPO derived from a baculovirus expression system where covalent heme binding was increased by the addition of hydrogen peroxide. Hydrogen peroxide was also required for covalent binding of the heme to LspPOX, a bacterial peroxidase of this family (18).
TABLE 2.
Km and Vmax values of natively expressed MopA, heterologously expressed full-length MopA, and the heterologously expressed AHP domain in cell extracta
| Protein or domain | Km ± SD for Mn(II) (μM) | Vmax ± SD (nM Mn oxide equivalents · minute−1 · mg cell extract−1) |
|---|---|---|
| MopA from native host | 204 ± 43 | 2.5 ± 0.12 (6) |
| Full-length MopA | 545 ± 278 | 2.6 ± 0.38 |
| AHP domain | 3,624 ± 741 | 792 ± 52 |
Values are from triplicates.
Studies on the native protein suggested that PQQ may play a role in Mn oxidation (6). The addition of 10 μM PQQ to cell extract assays increased Mn oxidation by 50% with both the full-length MopA and the AHP domain (Table 1). This increase in activity with the heterologously expressed protein is consistent with, but much less than, the 8-fold increase in activity seen with the natively expressed protein in cell extracts (6). Although there is an increase in Mn-oxidizing activity resulting from PQQ addition with both the native and heterologously expressed proteins, the role of PQQ still needs to be elucidated. In contrast to the native system, the addition of 1 mM NAD+ or NADH to the cell extract assay did not affect activity (data not shown).
TABLE 1.
Effect of PQQ, hemin, and calcium on Mn oxidation
| Protein or domain | Component | % activity ± SDa |
|---|---|---|
| MopA from native host | PQQ | 824 ± 10 (6) |
| Full-length MopA | PQQ | 150 ± 0.5 |
| AHP domain | PQQ | 160 ± 0.8 |
| Full-length MopA | Hemin | 108 ± 2.6 |
| AHP domain | Hemin | 136 ± 6.8 |
| Mop from native host | Calcium | 140 ± 10 (6) |
| Full-length MopA | Calcium | Required |
| AHP domain | Calcium | Required |
Values are from triplicates.
Calcium is required for Mn oxidation.
Although calcium stimulated activity with the natively expressed protein, it was not required for activity (5). In contrast, calcium proved to be essential for activity of the heterologously expressed MopA and the AHP domain (Table 2). The calcium requirement for activity of both MopA and the AHP domain which lacks the calcium binding domain suggests a role for calcium in the catalytic domain. In fact, the structures of MPO (36) and LPO (34) indicate a bound calcium ion is required for orienting the distal histidine that interacts with the heme cofactor (31). Like MopA and the AHP domain, calcium binding is critical for activity of animal heme peroxidases (38). The residues that bind calcium in MPO and LPO are conserved in MopA and the AHP domain (39).
Determination of kinetic parameters in cell extract.
We determined the Km for Mn(II) and Vmax kinetic parameters for the heterologously expressed MopA and the AHP domain in cell extract. The Km for heterologously expressed MopA in cell extract was similar to that of the natively expressed protein (6) in cell extract (Table 1). Although the cell extract environments of the natively expressed protein and the heterologously expressed protein are likely different and therefore interpretation of the kinetic parameters has obvious limitations, it does provide evidence that we are examining the same protein catalyzing Mn-oxidizing activity. The Km for the AHP domain was significantly higher than that for the full-length MopA (Table 2). Although the calcium binding domain is not required for activity, it appears to significantly increase the affinity for Mn. It is irresistible to conjecture that the multiple calcium binding sites on this domain may bind Mn(II) to be delivered to the active site. The Km of the full-length MopA is significantly lower than the Km determined for the heterologously expressed MCOs CotA and CueO (14.85 ± 1.17 mM [15] and 17.33 ± 3.63 mM [17], respectively), suggesting that MopA activity is likely to be more relevant for biogeochemical cycling in most natural systems.
The Vmax values for MopA in the native host (6) and when expressed in E. coli were similar, approximately 2.5 nM Mn oxide equivalents · minute−1 · mg−1 (Table 2). One of the great advantages of heterologous expression is the increase in expression of a desired protein, but for the full-length MopA, based on qualitative SDS-PAGE analysis, the protein was expressed at similar levels in the native host and recombinantly, which is consistent with the similar Vmax values. The AHP domain had a >300-fold-higher Vmax than MopA (Table 2), likely due to the significant increase in expression (Fig. 2).
The animal heme peroxidase-type Mn-oxidizing enzymes likely play an important role in Mn cycling. This class of Mn-oxidizing enzymes was recently identified (5), and there are many questions regarding how they may function. The experiments described here provide important clues on the enzyme mechanism, which will assist future studies to purify and characterize this intriguing enzyme.
ACKNOWLEDGMENTS
We thank the National Science Foundation (MCB-1021187) and Research Corporation for financial support. This work was also supported by Associated Students Inc., with CSUF grants to K.N., K.C., A.L., J.R., and M.M. and a CSUF Junior Faculty Grant to H.A.J.
We also thank Justin Davis and Moussa Omidvar for experiment assistance.
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Spiro TG, Bargar JR, Sposito G, Tebo BM. 2010. Bacteriogenic manganese oxides. Acc. Chem. Res. 43:2–9. 10.1021/ar800232a. [DOI] [PubMed] [Google Scholar]
- 2.Miyata N, Tani Y, Sakata M, Iwahori K. 2007. Microbial manganese oxide formation and interaction with toxic metal ions. J. Biosci. Bioeng. 104:1–8. 10.1263/jbb.104.1. [DOI] [PubMed] [Google Scholar]
- 3.Tebo BM, Clement BG, Dick GJ. 2007. Biotransformations of manganese, p 1223–1238 In Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach LD. (ed), Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 4.Tebo BM, Bargar JR, Clement BG, Dick GJ, Murray KJ, Parker D, Verity R, Webb SM. 2004. Biogenic manganese oxides: properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 32:287–328. 10.1146/annurev.earth.32.101802.120213. [DOI] [Google Scholar]
- 5.Anderson CR, Johnson HA, Caputo N, Davis RE, Torpey JW, Tebo BM. 2009. Mn(II) oxidation is catalyzed by heme peroxidases in “Aurantimonas manganoxydans” strain SI85-9A1 and Erythrobacter sp. strain SD-21. Appl. Environ. Microbiol. 75:4130–4138. 10.1128/AEM.02890-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson HA, Tebo BM. 2008. In vitro studies indicate a quinone is involved in bacterial Mn(II) oxidation. Arch. Microbiol. 189:59–69. 10.1007/s00203-007-0293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams LF, Ghiorse WC. 1987. Characterization of extracellular Mn2+-oxidizing activity and isolation of an Mn2+-oxidizing protein from Leptothrix discophora SS-1. J. Bacteriol. 169:1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geszvain K, Butterfield C, Davis RE, Madison AS, Lee SW, Parker DL, Soldatova A, Spiro TG, Luther GW, Tebo BM. 2012. The molecular biogeochemistry of manganese(II) oxidation. Biochem. Soc. Trans. 40:1244–1248. 10.1042/BST20120229. [DOI] [PubMed] [Google Scholar]
- 9.Solomon EI, Sundaram UM, Machonkin TE. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96:2563–2605. 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 10.Geszvain K, McCarthy JK, Tebo BM. 2013. Elimination of manganese(II,III) oxidation in Pseudomonas putida GB-1 by a double knockout of two putative multicopper oxidase genes. Appl. Environ. Microbiol. 79:357–366. 10.1128/AEM.01850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick GJ, Torpey JW, Beveridge TJ, Tebo BM. 2008. Direct identification of a bacterial manganese(II) oxidase, the multicopper oxidase MnxG, from spores of several different marine Bacillus species. Appl. Environ. Microbiol. 74:1527–1534. 10.1128/AEM.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Waasbergen LG, Hildebrand M, Tebo BM. 1996. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J. Bacteriol. 178:3517–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corstjens PLAM, De Vrind JPM. 1997. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol. J. 14:91–108. 10.1080/01490459709378037. [DOI] [Google Scholar]
- 14.Ridge JP, Lin M, Larsen EI, Fegan M, McEwan AG, Sly LI. 2007. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ. Microbiol. 9:944–953. 10.1111/j.1462-2920.2006.01216.x. [DOI] [PubMed] [Google Scholar]
- 15.Su J, Bao P, Bai T, Deng L, Wu H, Liu F, He J. 2013. CotA, a multicopper oxidase from Bacillus pumilus WH4, exhibits manganese-oxidase activity. PLoS One 8:e60573. 10.1371/journal.pone.0060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butterfield CN, Soldatova AV, Lee SW, Spiro TG, Tebo BM. 2013. Mn(II,III) oxidation and MnO2 mineralization by an expressed bacterial multicopper oxidase. Proc. Natl. Acad. Sci. U. S. A. 110:11731–11735. 10.1073/pnas.1303677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su J, Deng L, Huang L, Guo S, Liu F, He J. 2014. Catalytic oxidation of manganese(II) by multicopper oxidase CueO and characterization of the biogenic Mn oxide. Water Res. 56:304–313. 10.1016/j.watres.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Auer M, Gruber C, Bellei M, Pirker KF, Zamocky M, Kroiss D, Teufer SA, Hofbauer S, Soudi M, Battistuzzi G, Furtmuller PG, Obinger C. 2013. A stable bacterial peroxidase with novel halogenating activity and an autocatalytically linked heme prosthetic group. J. Biol. Chem. 288:27181–27199. 10.1074/jbc.M113.477067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daiyasu H, Toh H. 2000. Molecular evolution of the myeloperoxidase family. J. Mol. Evol. 51:433–445. [DOI] [PubMed] [Google Scholar]
- 20.Zamocky M, Jakopitsch C, Furtmuller PG, Dunand C, Obinger C. 2008. The peroxidase-cyclooxygenase superfamily: reconstructed evolution of critical enzymes of the innate immune system. Proteins 72:589–605. 10.1002/prot.21950. [DOI] [PubMed] [Google Scholar]
- 21.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. 2010. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34:1076–1112. 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison AS, Tebo BM, Mucci A, Sundby B, Luther GW., III 2013. Abundant porewater Mn(III) is a major component of the sedimentary redox system. Science 341:875–878. 10.1126/science.1241396. [DOI] [PubMed] [Google Scholar]
- 23.Anderson CR, Davis RE, Bandolin NS, Baptista AM, Tebo BM. 2011. Analysis of in situ manganese(II) oxidation in the Columbia River and offshore plume: linking Aurantimonas and the associated microbial community to an active biogeochemical cycle. Environ. Microbiol. 13:1561–1576. 10.1111/j.1462-2920.2011.02462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klock HE, Koesema EJ, Knuth MW, Lesley SA. 2007. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins 71:982–994. 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Francis CA, Co E, Tebo BM. 2001. Enzymatic manganese(II) oxidation by a marine α-proteobacterium. Appl. Environ. Microbiol. 67:4024–4029. 10.1128/AEM.67.9.4024-4029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Francis CA, Tebo BM. 2002. Enzymatic manganese(II) oxidation by metabolically dormant spores of diverse Bacillus species. Appl. Environ. Microbiol. 68:874–880. 10.1128/AEM.68.2.874-880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Do SH, Batchelor B, Lee HK, Kong SH. 2009. Hydrogen peroxide decomposition on manganese oxide (pyrolusite): kinetics, intermediates, and mechanism. Chemosphere 75:8–12. 10.1016/j.chemosphere.2008.11.075. [DOI] [PubMed] [Google Scholar]
- 30.Switala J, Loewen PC. 2002. Diversity of properties among catalases. Arch. Biochem. Biophys. 401:145–154. 10.1016/S0003-9861(02)00049-8. [DOI] [PubMed] [Google Scholar]
- 31.Furtmuller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. 2006. Active site structure and catalytic mechanisms of human peroxidases. Arch. Biochem. Biophys. 445:199–213. 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Learman DR, Voelker BM, Vazquez-Rodriguez AI, Hansel CM. 2011. Formation of manganese oxides by bacterially generated superoxide. Nat. Geosci. 4:95–98. 10.1038/ngeo1055. [DOI] [Google Scholar]
- 33.Keele BB, Jr, McCord JM, Fridovich I. 1970. Superoxide dismutase from Escherichia coli B. A new manganese-containing enzyme. J. Biol. Chem. 245:6176–6181. [PubMed] [Google Scholar]
- 34.Singh AK, Singh N, Sharma S, Singh SB, Kaur P, Bhushan A, Srinivasan A, Singh TP. 2008. Crystal structure of lactoperoxidase at 2.4 Å resolution. J. Mol. Biol. 376:1060–1075. 10.1016/j.jmb.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Carpena X, Vidossich P, Schroettner K, Calisto BM, Banerjee S, Stampler J, Soudi M, Furtmuller PG, Rovira C, Fita I, Obinger C. 2009. Essential role of proximal histidine-asparagine interaction in mammalian peroxidases. J. Biol. Chem. 284:25929–25937. 10.1074/jbc.M109.002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng J, Fenna RE. 1992. X-ray crystal structure of canine myeloperoxidase at 3 Å resolution. J. Mol. Biol. 226:185–207. 10.1016/0022-2836(92)90133-5. [DOI] [PubMed] [Google Scholar]
- 37.DePillis GD, Ozaki S, Kuo JM, Maltby DA, Ortiz de Montellano PR. 1997. Autocatalytic processing of heme by lactoperoxidase produces the native protein-bound prosthetic group. J. Biol. Chem. 272:8857–8860. 10.1074/jbc.272.14.8857. [DOI] [PubMed] [Google Scholar]
- 38.Shin K, Hayasawa H, Lonnerdal B. 2001. Mutations affecting the calcium-binding site of myeloperoxidase and lactoperoxidase. Biochem. Biophys. Res. Commun. 281:1024–1029. 10.1006/bbrc.2001.4448. [DOI] [PubMed] [Google Scholar]
- 39.Santamaria-Hernando S, Krell T, Ramos-Gonzalez MI. 2012. Identification of a novel calcium binding motif based on the detection of sequence insertions in the animal peroxidase domain of bacterial proteins. PLoS One 7:e40698. 10.1371/journal.pone.0040698. [DOI] [PMC free article] [PubMed] [Google Scholar]


