Abstract
Several yeast strains have been engineered to express different cellulases to achieve simultaneous saccharification and fermentation of lignocellulosic materials. However, successes in these endeavors were modest, as demonstrated by the relatively low ethanol titers and the limited ability of the engineered yeast strains to grow using cellulosic materials as the sole carbon source. Recently, substantial enhancements to the breakdown of cellulosic substrates have been observed when lytic polysaccharide monooxygenases (LPMOs) were added to traditional cellulase cocktails. LPMOs are reported to cleave cellulose oxidatively in the presence of enzymatic electron donors such as cellobiose dehydrogenases. In this study, we coexpressed LPMOs and cellobiose dehydrogenases with cellobiohydrolases, endoglucanases, and β-glucosidases in Saccharomyces cerevisiae. These enzymes were secreted and docked onto surface-displayed miniscaffoldins through cohesin-dockerin interaction to generate pentafunctional minicellulosomes. The enzymes on the miniscaffoldins acted synergistically to boost the degradation of phosphoric acid swollen cellulose and increased the ethanol titers from our previously achieved levels of 1.8 to 2.7 g/liter. In addition, the newly developed recombinant yeast strain was also able to grow using phosphoric acid swollen cellulose as the sole carbon source. The results demonstrate the promise of the pentafunctional minicellulosomes for consolidated bioprocessing by yeast.
INTRODUCTION
The demand for bioethanol has steadily increased over the past decade. In addition to the reduction of carbon footprint, the increased production and adoption of bioethanol has also reduced reliance on the volatile fossil fuel market, thus enhancing energy security (1). At present, corn and sugarcane represent the major feedstocks for the production of bioethanol. However, great efforts are being made to reduce reliance on these food sources and to produce ethanol from cellulosic waste materials such as corn stover, rice straw, and bagasse (1). Many commercial processes for the production of cellulosic ethanol involve chemical and enzymatic treatment of cellulosic substrates to release sugars, which are further converted to ethanol by anaerobic fermentation (2). The large amount of enzymes and high cost associated with the breakdown of recalcitrant lignocellulosic materials render cellulosic ethanol prohibitively expensive to compete with gasoline, thus hampering the widespread production of cellulosic ethanol (3). In view of these limitations, consolidated bioprocessing (CBP), which involves enzyme production, saccharification, and fermentation in a single step, has been proposed as a promising platform for cellulosic biofuel production (3, 4). Several studies have investigated the heterologous expression of cellulases in ethanol-producing organisms (5–10). In particular, the expression of cellulases in Saccharomyces cerevisiae has been widely examined (5–11). The cellulases that are commonly expressed in S. cerevisiae include endoglucanases (EGs), cellobiohydrolases (CBHs), and β-glucosidases (BGLs). The endo-acting EG mediates the intrachain hydrolysis of cellulose, generating reducing and nonreducing ends for exo-acting CBH. Dimeric cellobiose units released by the CBH are then converted to glucose by BGL. Apart from the heterologous expression of these cellulases, a variety of strategies have also been adopted in the engineering of cellulose-degrading S. cerevisiae strains. Some of these strategies include tethering secreted cellulases on yeast cell surfaces to maintain high local concentrations (5) and expressing nonenzymatic scaffolds for the docking and assembly of cellulases to form synergistic minicellulosomes (7–10). Although the proposed engineering strategies did lead to the improved production of ethanol, the current titers and productivities still need substantial enhancement to meet the industrial requirements.
On top of the commonly utilized cellulases, recent studies have found that the addition of lytic polysaccharide monooxygenases (LPMOs; formally known as family 61 glycoside hydrolases) and cellobiose dehydrogenases (CDHs) to enzyme cocktails of CBH, EG, and BGL led to significant improvements in cellulosic degradation (12–15). LPMO-encoding genes are abundant in biomass-converting microorganisms. However, their function and mechanism have only been elucidated in recent years. Owing to their flat substrate-binding surfaces, these enzymes can access and cleave tightly packed crystalline chains via an oxidative mechanism in the presence of nonenzymatic electron donors or enzymatic electron donors such as CDH (12, 16). The cleavage of crystalline regions by LPMO renders these tightly packed regions more accessible to EG and CBH, thus enabling synergistic degradation of cellulose.
Here, we examined the expression of GH61a (a LPMO) and CDH in addition to CBH, EG, and BGL to improve cellulolytic ability of S. cerevisiae. Apart from the cellulases, we also expressed and anchored pentameric scaffolds to the surfaces of the yeast cells for the assembly of pentafunctional minicellulosomes. The resultant recombinant yeast strain was able to simultaneously hydrolyze and ferment both amorphous and crystalline cellulose, yielding up to 2.7 g/liter ethanol when phosphoric acid swollen cellulose (PASC) was used as the substrate. This result represents a 60% improvement in titer compared to our previously reported strains with trifunctional minicellulosomes (7). With the addition of LPMO and CDH, the newly engineered strain was also able to grow using PASC as the sole carbon source, thus demonstrating the promise of this strategy in consolidated bioprocessing.
MATERIALS AND METHODS
Strains, media, and reagents.
S. cerevisiae EBY100 (Invitrogen, Carlsbad, CA) and HZ848 (MATα ade2-1 ade3Δ22 Δura3 his3-11,15 trp1-1 leu2-3,112 can1-100) were used for yeast cell surface display. The recombinant yeast strains in the present study are summarized in Table 1. Escherichia coli DH5α (Invitrogen) was used for recombinant DNA manipulation. S. cerevisiae EBY100 transformants were selected and maintained on SC-Trp, SC-Leu, or SC-Trp-Leu plates (0.167% yeast nitrogen base without amino acids and ammonium sulfate [Difco Laboratories, Detroit, MI], 0.5% ammonium sulfate, 2% glucose, 1.5% agar, and appropriate amino acid supplements) and were induced in YPG (1% yeast extract, 2% peptone, 2% galactose). S. cerevisiae HZ848 transformants were selected and maintained on SC-Ura plates. E. coli was cultured in Luria-Bertani medium (Fisher, Pittsburgh, PA) supplemented with ampicillin at 50 μg/ml. Clostridium thermocellum DSM1237 (ATCC, Manassas, VA) was cultured anaerobically in reinforced clostridia media (Difco) supplemented with 0.6% cellobiose. All restriction enzymes were obtained from New England BioLabs (Ipswich, MA). Unless otherwise indicated, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO).
TABLE 1.
Recombinant S. cerevisiae strains used in the cellulose hydrolysis studies
| Strain | Plasmid(s) | Phenotype |
|---|---|---|
| Control (EBY100) | pYD1-CipA3 and pRS425-insert | Galactose-inducible display of three-domain miniscaffoldins without cellulase activity |
| Three-enzyme minicellulosome (EBY100) | pYD1-CipA3-EG2 and pRS425-CBH2-BGL1 | Galactose-inducible display of trifunctional minicellulosomes with CBH2, EG2, and BGL1 activities |
| Five-enzyme minicellulosome (EBY100) | pYD1-CipA5-EG2-LPMO-CDH and pRS425-CBH2-BGL1 | Galactose-inducible display of pentafunctional minicellulosomes with CBH2, EG2, BGL1, LPMO, and CDH activities |
| Three-enzyme minicellulosome CipA5 (EBY100) | pYD1-CipA5-EG2 and pRS425-CBH2-BGL1 | Galactose-inducible display of trifunctional minicellulosomes with CBH2, EG2, and BGL1 activities |
| Control (HZ848) | pRS426-control | Control, no display |
| Three-enzyme minicellulosome (HZ848) | pRS426-CipA3-CBH2-EG2-BGL1 | Constitutive display of trifunctional minicellulosomes with CBH2, EG2, and BGL1 activities |
| Five-enzyme minicellulosome (HZ848) | pRS426-CipA5-CBH2-EG2-BGL1-LPMO-CDH | Constitutive display of pentafunctional minicellulosomes with CBH2, EG2, BGL1, LPMO, and CDH activities |
Plasmid construction.
The sequences of all PCR primers and templates used are listed in Table SA1 in the supplemental material. Primers were synthesized by Sigma-Aldrich (Singapore). Thermoascus aurantiacus GH61a (a LPMO) and Humicola insolens CDH genes were synthesized in accordance with previously reported sequences (17, 18) by GenScript (Piscataway, NJ). Plasmids pYD1-CipA3, pYD1-CipA3-EG2, and pRS425-CBH2-BGL1 were constructed in our previous work (7). To generate the remaining plasmids, DNA fragments were PCR amplified from the appropriate templates with 30- to 40-bp sequence homology to adjacent fragments. Plasmids pYD1-CipA5-EG2, pRS425-LPMO, pRS425-LPMO-CDH, pYD1-CipA5-EG2-LPMO-CDH, pRS425-TEF1p-CipA3-PGK1t, pRS425-PGK1t-TPI1p-CBH2-GPD1t, pRS425-GPD1t-ENO2p-EG2-TEF1t, pRS425-TEF1t-PDC1p-BGL1-HXT7t, pRS426-control, pRS426-CipA3-CBH2-EG2-BGL1, and pRS426-CipA5-CBH2-EG2-BGL1-LPMO-CDH were then constructed using either the DNA assembler method (19) or Gibson assembly (20) as described in Table SA1 in the supplemental material, with the exception of pYD1-CipA5 and pRS425-insert, which were constructed as follows. For the construction of pYD1-CipA5, a five-cohesin miniscaffoldin (cipA5) was amplified from C. thermocellum genomic DNA. Due to the sequence identity of the fourth and fifth cohesin subunits on CipA5, a gene was generated by ligating the gene encoding a four-cohesin miniscaffoldin, the cipA4 gene, and the fifth cohesin subunit, the 5thcoh2 gene. The plasmid was created by ligating NheI/PmeI-digested plasmid pYD1-CipA3, SacI/NheI-digested cipA4 gene, and the SacI/PmeI-digested 5thcoh2 gene and transforming the ligation product into E. coli cells. For the construction of pRS425-insert, the insert gene fragment was PCR amplified from S. cerevisiae genomic DNA, and the plasmid was created by ligating SgrDI/BsgI-digested plasmid pRS425-CBH2 and the SgrDI/BsgI-digested insert gene fragment and transforming the ligation product into DH5α cells. All plasmids were checked by diagnostic PCR and restriction digestions. The constructed plasmids were transformed into appropriate yeast strains using the lithium acetate-polyethylene glycol method as previously described (21).
Yeast surface display and flow cytometry analysis.
S. cerevisiae EBY100 clones transformed with different plasmid constructs were cultured, induced for 66 h, and analyzed by using flow cytometry as described elsewhere (22), except that only 2.5 × 106 cells were used in each staining assay. The primary monoclonal antibodies used in the assay were anti-V5, anti-His, anti-c-Myc, anti-Flag, anti-T7, and anti-HA. The primary monoclonal antibodies and the appropriate fluorescent conjugation kits were purchased from Abcam (Cambridge, United Kingdom), with the exception of the anti-V5 primary monoclonal antibodies purchased from Invitrogen. Approximately 10,000 cells were analyzed for each flow cytometry analysis.
Enzyme activity assay.
Phosphoric acid swollen cellulose (PASC) was generated from Avicel PH-101 crystalline cellulose as described elsewhere (23) and was washed with sterile double-distilled H2O at least 10 times to remove any soluble sugars. EBY100 transformants displaying different minicellulosomes on the cell surface were analyzed for their abilities to hydrolyze PASC. After induction in YPG, the cells were washed three times with water to prevent medium carryover and then resuspended in either sPASC medium (6.7 g/liter yeast nitrogen base, 10 g/liter PASC, 0.87 g/liter synthetic complete amino acids mix) to an optical density at 600 nm (OD600) of 1 or hydrolysis buffer (50 mM sodium acetate [pH 5.0]) supplemented with 0.1% PASC to an OD600 of 10. For some conditions, 100 mM methyl glyoxal was added to the hydrolysis buffer to inhibit utilization of the glucose released from PASC. Hydrolysis reactions were performed at 30°C with agitation at 100 rpm. The amount of glucose released from PASC was measured by using a glucose oxidase assay kit from Sigma-Aldrich.
Fermentation.
After induction in YPG, EBY100 transformants displaying different minicellulosomes were washed three times with water and resuspended in YP medium (1% yeast extract, 2% peptone) supplemented with 0.001% ergosterol, 0.042% Tween 80, and 1% PASC or Avicel to obtain an OD600 of ∼50. Fermentation was carried out in sealed serum bottles with 5% volumetric air headspace at 30°C with agitation at 250 rpm. Then, 500-μl samples were removed at the indicated time intervals. Ethanol concentration was determined by gas chromatography-mass spectroscopy using a Shimadzu GCMS-QP 2010 (Shimadzu Corp., Kyoto, Japan) and HP-INNOWAX column (Agilent, Inc., Palo Alto, CA) with helium as the carrier gas (flow rate, 1 ml/min). The temperature program used for compound separation was 51°C for 1 min, increased to 53°C at a rate of 0.5°C/min, increased to 80°C at 10°C/min, increased to 200°C at 25°C/min, and finally 200°C for 1 min.
Growth assay.
After overnight culture in SC-Ura, HZ848 transformants were washed three times with water, streaked onto sPASC plates (6.7 g/liter yeast nitrogen base, 10 g/liter PASC, 2% agar, 0.87 g/liter synthetic complete amino acid mix), and incubated at 30°C. The washed cells were also resuspended in sPASC medium to an OD600 of ∼1, or 5 × 107 cells/ml, as determined by hemocytometer and incubated at 30°C with an agitation of 250 rpm.
RESULTS
Design and construction of pentafunctional minicellulosomes for yeast surface display.
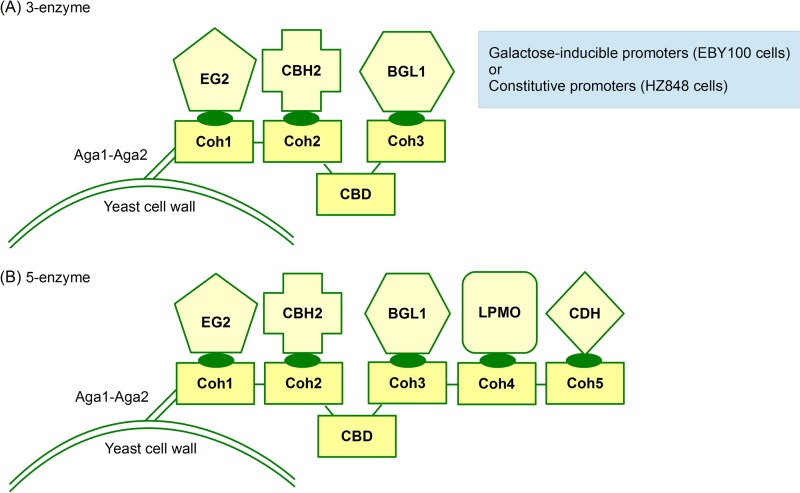
In our previous work, we designed trifunctional minicellulosomes containing three types of cellulases for cellulose hydrolysis (7). The trifunctional minicellulosomes consisted of trimeric miniscaffoldins, endoglucanases (EGs), cellobiohydrolases (CBHs), and β-glucosidases (BGLs), which were the minimum components required for cellulose utilization by S. cerevisiae. In the present study, pentafunctional minicellulosomes were designed so as to accommodate two additional enzymes, an LPMO (GH61a) and a cellobiose dehydrogenase (CDH), for enhanced synergistic degradation of cellulose (Fig. 1). The new miniscaffoldin, CipA5, was engineered based on the scaffold protein CipA from C. thermocellum. CipA5 contained a cellulose-binding domain (CBD) and five cohesin modules (Coh1-Coh2-CBD-Coh3-Coh4-Coh5), while CipA3 contained a cellulose-binding domain and three cohesin modules (Coh1-Coh2-CBD-Coh3). The fusion of the genes encoding CipA3 and CipA5 to the C terminus of the Aga2 protein enabled the respective miniscaffoldins to be tethered to the yeast α-agglutinin mating adhesion receptor and subsequently displayed on yeast cell surfaces.
FIG 1.
Design of yeast surface display for assembly of minicellulosomes. (A) Schematic representation of the three-enzyme strain. In this conventional minicellulosome system, EG2, CBH2, and BGL1 are secreted and docked onto the cohesin scaffolds to form trifunctional minicellulosomes. (B) Schematic representation of the five-enzyme strain. On top of the three cellulases, LPMO and CDH are secreted and docked to enhance cellulose degradation and utilization.
To allow secretion and display of the enzymes on yeast cell surface, different expression cassettes, each containing a promoter, a secretion signal peptide, an epitope tag, a cellulase enzyme, a dockerin module, and a transcription terminator, were designed. Two classes of promoters, galactose inducible and constitutive, were examined. The dockerin modules allowed the docking of the fusion cellulases on surface-displayed miniscaffoldins upon secretion, while the N-terminal epitope tags allowed detection of surface display through flow cytometry. These expression cassettes were assembled in pRS425, pRS426, or pYD1 plasmids with designs similar to those in our previous study (7). Of the enzymes used here, T. reesei EG2 and CBH2 and A. aculeatus BGL1 had been functionally expressed in S. cerevisiae in our earlier study (7). The genes encoding T. aurantiacus GH61a (an LPMO) and H. insolens CDH were synthesized in accordance with previously reported sequences (17, 18). The first 22 amino acids in the GH61a sequence were designated as the signal peptide and were replaced by the synthetic prepro-leader sequence (24) from S. cerevisiae while CDH was secreted using the α-factor signal peptide. Although there was no prior report for the expression of T. aurantiacus GH61a and H. insolens CDH in S. cerevisiae, Methanosarcina thermophila GH61a (25) with 53% amino acid sequence identity to T. aurantiacus GH61a and Myriococcum thermophilum CDH (26) with 60% sequence identity to H. insolens CDH had been functionally expressed in S. cerevisiae.
Yeast surface assembly of trifunctional and pentafunctional minicellulosomes.
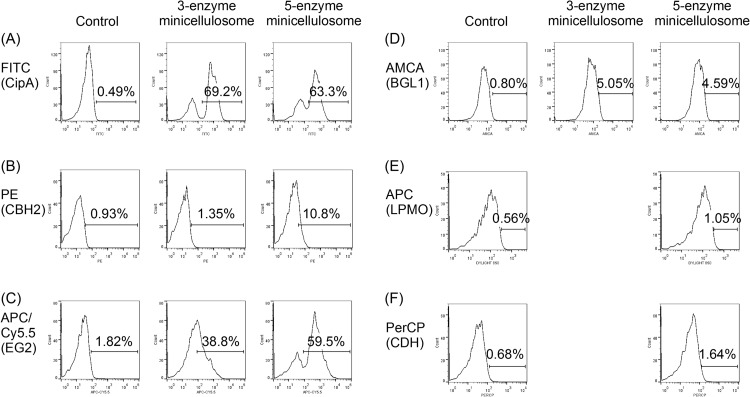
To examine the expression and secretion of fusion enzymes, followed by the surface display of the enzymes to form functional minicellulosomes, EBY100 cells were transformed with the different plasmids listed in Table 1 and induced with galactose for 66 h. As before, the anchorage of the miniscaffoldins on cell surfaces was verified by labeling the cells with anti-V5 antibody, followed by analysis by flow cytometry (Fig. 2A). As indicated by V5 epitope detection, both CipA3 and CipA5 miniscaffoldins, each harboring a single V5 epitope, were successfully displayed on the yeast cell surfaces. Percentages of V5-positive cells were fairly comparable for the three-enzyme and five-enzyme systems, with slightly higher levels of miniscaffoldins for the three-enzyme system. This could be attributed to better expression of the smaller CipA3 miniscaffoldins.
FIG 2.
Flow cytometry analysis of yeast cells displaying minicellulosomes. (A) Both the CipA3 and the CipA5 miniscaffoldins were successfully displayed on the yeast cell surface, as indicated by V5 epitope detection. (B) CBH2 was expressed, secreted, and docked onto the miniscaffoldins, as indicated by the FLAG epitope detection. (C) EG2 was expressed, secreted, and docked onto the miniscaffoldins, as indicated by the His epitope detection. (D) BGL1 was expressed, secreted, and docked onto the miniscaffoldins, as indicated by the c-Myc epitope detection. (E) LPMO was expressed, secreted, and docked onto the miniscaffoldins, as indicated by the HA detection. (F) CDH was expressed, secreted, and docked onto the miniscaffoldins, as indicated by the T7 detection.
Since the codisplay of different enzymes on the miniscaffoldins had already been demonstrated in our previous work (7), the induced yeast cells were only labeled with a single type of antibody in each flow cytometry experiment to minimize the effect of steric hindrance between bulky antibodies. The secretion and docking of CBH2, EG2 and BGL1 onto the surface-displayed CipA3 and CipA5 miniscaffoldins were verified by detection of the FLAG, His, and c-Myc tag epitopes, respectively (Fig. 2B, C, and D). The enhanced display of CBH2 and EG2 on CipA5 could be attributed to an increase in the overall number of cohesins. Despite the smaller number of scaffolds as shown by the V5 epitope detection, the overall number of cohesins was increased for the five-enzyme minicellulosomes compared to the three-enzyme minicellulosomes. However, the display levels for BGL1 were fairly comparable under both conditions, indicating that the docking of BGL1 on the cell surface might be limited by its expression and secretion instead of the availability of cohesins. The secretion and docking of LPMO and CDH were verified by HA and T7 epitope detection. The display levels of LPMO and CDH were comparable to those of CBH2 on the three-enzyme minicellulosome but appeared to be lower than those of the other cellulases. This could be due to lower expression levels or lower labeling efficiencies.
Functional analysis of LPMO and CDH in pentafunctional minicellulosomes.
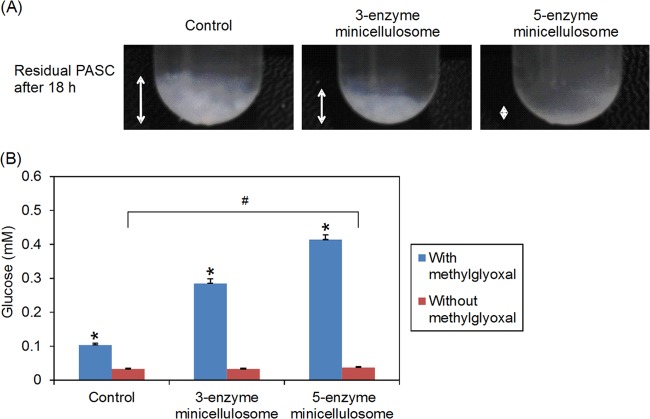
The function of the three-enzyme minicellulosome had already been demonstrated in our previous work (7). To evaluate whether the LPMO and CDH expressed and secreted by S. cerevisiae were able to enhance hydrolysis of cellulose, we cultured the control strain, the three-enzyme minicellulosome strain, and the five-enzyme minicellulosome strain in hydrolysis media containing 1% PASC. S. cerevisiae EBY100 strains containing galactose-inducible promoters were used for this analysis. After 18 h of culture in PASC-containing media, it was apparent that both the three-enzyme minicellulosome and the five-enzyme minicellulosome strains were able to hydrolyze cellulose, as shown by the reduction in residual amounts of insoluble PASC (Fig. 3A). The expression of LPMO and CDH enabled the five-enzyme strain to hydrolyze PASC to a greater extent, resulting in smaller amounts of residual PASC compared to the three-enzyme system. This demonstrated that the cell surface-displayed LPMO and CDH were functional and served to enhance cellulose hydrolysis. We further quantified the hydrolytic activities by measuring the amounts of residual glucose in the respective culture media after 4-h incubations in 0.1% PASC. In these studies, some of the cultures were supplemented with 100 mM methyl glyoxal to inhibit glucose utilization by the cells. In the absence of methyl glyoxal, the glucose levels in all three conditions were low due to cellular uptake of the generated glucose (Fig. 3B). When 100 mM methyl glyoxal was added to the hydrolysis buffers, there were clear differences in the glucose levels generated by hydrolysis of PASC, with the largest amounts of glucose observed for the five-enzyme strain. Under the test conditions, the PASC hydrolysis activity for the three-enzyme strain was 1.2 U, while the activity for the five-enzyme strain was 1.7 U, where 1 U of PASC hydrolysis activity was defined as the activity required to produce 1 μM glucose/min at 30°C. This represented an improvement of >40% in the cellulose hydrolysis activity.
FIG 3.
Hydrolytic activity of minicellulsomes. (A) The different strains were cultured in sPASC media (1% PASC) for 18 h at 30°C. The PASC degradation could be seen from the decrease in amounts of residual PASC in both three-enzyme and five-enzyme stains. The addition of LPMO and CDH in the five-enzyme strain enhanced cellulolytic activity of the cells. (B) The different strains were cultured in hydrolysis media (0.1% PASC) for 4 h at 30°C. The starting OD of the cultures was 10. For some of the cultures, 100 mM methyl glyoxal was added to inhibit glucose metabolism. In the absence of methyl glyoxal, any glucose produced by PASC hydrolysis was utilized by the cells resulting in low glucose concentrations. In the presence of methyl glyoxal, glucose utilization is inhibited and glucose produced by PASC hydrolysis is accumulated in the media. The five-enzyme strain had significantly higher cellulose hydrolysis activity, as indicated by the larger amounts of glucose released. Values represent the means of three samples. Error bars denote standard deviations. *, P < 0.05 (Student t test).
Direct conversion of cellulose to ethanol.
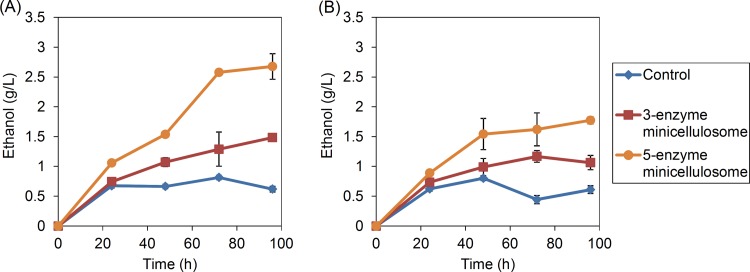
The five-enzyme strain containing galactose-inducible promoters was further compared to the three-enzyme strain for its ability to produce ethanol directly from both PASC and Avicel. The fermentations were carried out under microaerobic conditions instead of strict anaerobic conditions since oxygen was required for the function of LPMO enzymes. To achieve these conditions, the cells were cultured in the fermentation medium in serum bottles with 5% volumetric air headspaces. Under these conditions, the maximum titers of ethanol attained were 1.5 g/liter for the three-enzyme strain and 2.7 g/liter for the five-enzyme strain for PASC fermentation (Fig. 4A). As demonstrated in an earlier study by Wen et al., the ethanol produced by the control strain could be attributed to the fermentation of the YP medium rather than the fermentation of the PASC (7). When Avicel was used as the sole carbon source for fermentation, the maximum titers of ethanol attained were 1.0 g/liter for the three-enzyme strain and 1.8 g/liter for the five-enzyme strain (Fig. 4B). To investigate whether these enhancements were due to the presence of LPMO and CDH or the increased amount of surface-displayed cellulases, we further constructed a three-enzyme CipA5 strain which only expressed CBH2, EG2, and BGL1 enzymes but had CipA5 miniscaffoldins. When PASC was used as the carbon source, the final titer achieved with this three-enzyme CipA5 strain was 68% of the five-enzyme minicellulosome strain containing LPMO and CDH, even though the total amount of enzymes displayed on the cell surface should be similar (see Fig. SA1 in the supplemental material). This demonstrated that the expression of LPMO and CDH, in addition to CBH2, EG2, and BGL1, had a synergistic effect on cellulose degradation.
FIG 4.
Fermentation activities of minicellulosome-displaying cells. The strains were cultured under microaerobic conditions at 30°C in YP–1% PASC (A) or YP–1% Avicel (B). Samples were withdrawn at different time points and analyzed by gas chromatography-mass spectrometry. With the five-enzyme strain, it was possible to produce up to 2.7 g/liter ethanol from PASC and up to 1.5 g/liter ethanol from Avicel. Values represent means of three samples. Error bars denote the standard deviations.
Growth of minicellulosome-displaying yeast cells.
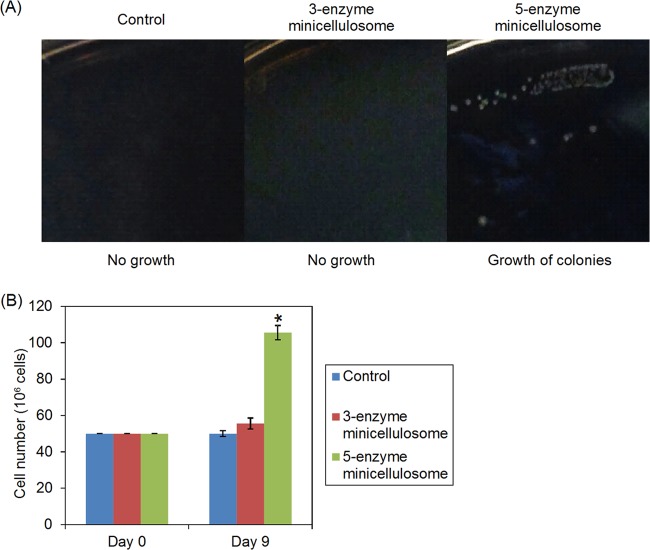
To investigate whether the engineered yeast cells were able to grow using PASC as the sole carbon source, constitutive promoters were used for the expression of cellulases and scaffolds in place of the galactose-inducible promoters. The HZ848 strain was used for the growth assay since it had higher growth rates than the EBY100 strain. HZ848 cells transformed with the different plasmids listed in Table 1 were streaked onto 1% sPASC plates after extensive washing to remove any residual sugars. After the plate was incubated for 10 days at 30°C, very small colonies appeared for the five-enzyme strain, while no colonies were formed for the control and three-enzyme strains. However, the colonies grew very slowly and remained small even after a 2-week incubation (Fig. 5A). These strains were also cultured in sPASC media containing 1% PASC. After culture for 9 days, no growth was observed for the control strain, and the cell numbers only increased slightly for the three-enzyme strain. In contrast, the five-enzyme strain exhibited a doubling of cell numbers (Fig. 5B).
FIG 5.
Growth of minicellulosome-displaying cells. (A) The different strains were grown at 30°C on the same agar plate with 1% PASC as the sole carbon source. Colonies were formed only for the five-enzyme strain after an incubation of 10 days. The colonies were photographed after an incubation of 2 weeks. (B) The different strains were grown in sPASC media containing 1% PASC. Cell numbers were determined by counting with a hemocytometer. Values represent the means of three samples. Error bars denote the standard deviations.
DISCUSSION
The high cost of cellulases and limited efficiencies of traditional enzyme cocktails have been the key roadblocks to the commercial success of cellulosic ethanol. In view of these limitations, consolidated bioprocessing presents great potential to replace existing technologies and to lower the costs required for the production of cellulosic ethanol (3). To this end, a number of studies have investigated the engineering of S. cerevisiae to achieve direct conversion of cellulose to ethanol (5–11). However, there is still plenty of room for improvement in the ethanol titers. As demonstrated by many recent studies, the addition of oxidative LPMOs to hydrolytic cellulase cocktails led to significant enhancement to the breakdown of cellulose (12–15). In a recent study published while the present manuscript was being prepared, the integration of bacterial LPMOs to cellulosomes in vitro resulted in 2.6-fold enhancement in the production of soluble sugars compared to free enzymes without the LPMOs (29). In the present study, we examined the expression and subsequent display of LPMO and CDH on functional minicellulosomes. The expression of these enzymes together with traditional cellulases improved ethanol titers from previously achieved levels of 1.8 g/liter to new levels of 2.7 g/liter. In previous works by Yamada et al. and Nakatani et al., the authors were able to achieve ethanol titers of 3.1 and 3.4 g/liter using cocktail delta integration of cellulase genes (8) and the expression of the expansin enzyme in addition to traditional cellulases (10). However, the fermentation conditions in these studies were different from the conditions used here. In the studies by Yamada et al. and Nakatani et al., a higher PASC content of 20 g/liter and cell loading of 200 g/liter wet cell mass were used. Furthermore, although several earlier studies have used the strategy of displaying cellulases on cell surfaces of S. cerevisiae (5, 7–10), this is the first study to demonstrate growth of recombinant S. cerevisiae with surface-displayed cellulases using cellulose as the sole carbon source.
Despite the promising results obtained here, there are several improvements that can be made to the engineered strain in follow-up studies. First, the flow cytometry data suggest that the display levels of the cellulases were not balanced. The display levels of EG2 were much higher than those of CBH2, BGL1, LPMO, and CDH. This could be a result of several factors. First, since the cellulases were labeled by different antibodies conjugated to different fluorophores, there might be differences in labeling efficiencies and fluorescence intensities. Second, there might be differences in the expression and display levels of the cellulases. The expression of the existing cellulases can be enhanced by codon optimization, and the display levels can be further improved by testing different signal peptides for secretion. To achieve greater synergy in the breakdown of cellulose, the optimal ratios of hydrolytic and oxidative enzymes can also be determined through in vitro experiments and regulated by different strategies such as varying promoter strengths for different cellulases. Alternatively, with the growth of our engineered strain on PASC plates, a high-throughput selection platform can even be set up for combinatorial engineering approaches such as COMPACTER (27). Using the COMPACTER method, the expression levels of different cellulases can be optimized simultaneously without going through the laborious process of determining optimal expression ratios in vitro. In addition to expression levels, there might be an unequal distribution of cellulases on individual miniscaffoldins due to the lack of specificity between cohesins and dockerins. This can be remedied by engineering the scaffolds to present unique binding sites (28).
While the traditional enzyme cocktails were comprised solely of hydrolytic enzymes, LPMO is an oxidative enzyme that requires the presence of oxygen for its function (12–15). In the present study, instead of adopting strict anaerobic conditions typically used in fermentation, we introduced oxygen by leaving a small headspace for air in our sealed tubes. This modification appeared to impose a penalty to the fermentative capacity of the S. cerevisiae cells, as demonstrated by the decline in titers from our previously reported levels of 1.8 g/liter to levels of 1.5 g/liter for the three-enzyme minicellulosome strain. However, the overall ethanol titer still went up to 2.7 g/liter, thus demonstrating that the low levels of oxygen was sufficient for some LPMO activity. The improvement in ethanol titers also corroborated the synergistic effects of adding LPMO and CDH to traditional cellulases, as reported in previous in vitro studies (12–15). Due to the current limitations of the experimental setup, the oxygen levels were not regulated and measured in the present study. To further improve ethanol titers, the effect of oxygen concentrations on consolidated bioprocessing can be more closely examined.
This is the first report of functional expression of both LPMO and CDH using S. cerevisiae. These enzymes were displayed on miniscaffoldins, together with traditional hydrolases, to construct synergistic minicellulosomes. With the addition of LPMO and CDH, we achieved enhanced cellulose degradation, as demonstrated by the increase in sugar and ethanol content. We were also able to achieve growth of yeast cells using cellulose as the sole carbon source. Although the growth rates of the engineered five-enzyme strain on PASC plates were low, the formation of colonies on the PASC plate opens up the possibility for high-throughput screening. With such unprecedented screening capabilities, different engineering strategies can be rapidly evaluated to further improve consolidated bioprocessing in S. cerevisiae.
Supplementary Material
Footnotes
Published ahead of print 22 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02070-14.
REFERENCES
- 1.Sahin Y. 2011. Environmental impacts of biofuels. Energy Education Sci. Technol. Part A Energy Sci. Res. 26:129–142. [Google Scholar]
- 2.Brown TR, Brown RC. 2013. A review of cellulosic biofuel commercial-scale projects in the United States. Biofuels Bioprod. Biorefin. 7:235–245. 10.1002/bbb.1387. [DOI] [Google Scholar]
- 3.Olson DG, McBride JE, Joe Shaw A, Lynd LR. 2012. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 23:396–405. 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 4.Hasunuma T, Kondo A. 2012. Consolidated bioprocessing and simultaneous saccharification and fermentation of lignocellulose to ethanol with thermotolerant yeast strains. Proc. Biochem. 47:1287–1294. 10.1016/j.procbio.2012.05.004. [DOI] [Google Scholar]
- 5.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. 2004. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 70:1207–1212. 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Den Haan R, Rose SH, Lynd LR, van Zyl WH. 2007. Hydrolysis and fermentation of amorphous cellulose by recombinant Saccharomyces cerevisiae. Metab. Eng. 9:87–94. 10.1016/j.ymben.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Wen F, Sun J, Zhao H. 2010. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76:1251–1260. 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada R, Taniguchi N, Tanaka T, Ogino C, Fukuda H, Kondo A. 2011. Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol. Biofuels 4:8. 10.1186/1754-6834-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan L-H, Zhang Z-J, Yu X-Y, Xue Y-X, Tan T-W. 2012. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. U. S. A. 109:13260–13265. 10.1073/pnas.1209856109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatani Y, Yamada R, Ogino C, Kondo A. 2013. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb. Cell Fact. 12:66. 10.1186/1475-2859-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X-Z, Sathitsuksanoh N, Zhu Z, Percival Zhang YH. 2011. One-step production of lactate from cellulose as the sole carbon source without any other organic nutrient by recombinant cellulolytic Bacillus subtilis. Metab. Eng. 13:364–372. 10.1016/j.ymben.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Horn S, Vaaje-Kolstad G, Westereng B, Eijsink VG. 2012. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 5:45. 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fushinobu S. 2014. Metalloproteins: a new face for biomass breakdown. Nat. Chem. Biol. 10:88–89. 10.1038/nchembio.1434. [DOI] [PubMed] [Google Scholar]
- 14.Podkaminer KK, Kenealy WR, Herring CD, Hogsett DA, Lynd LR. 2012. Ethanol and anaerobic conditions reversibly inhibit commercial cellulase activity in thermophilic simultaneous saccharification and fermentation (tSSF). Biotechnol. Biofuels 5:43. 10.1186/1754-6834-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cannella D, Jørgensen H. 2014. Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol. Bioeng. 111:59–68. 10.1002/bit.25098. [DOI] [PubMed] [Google Scholar]
- 16.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen J-CN, Johansen KS, Krogh KBRM, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. 2011. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U. S. A. 108:15079–15084. 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dotson W, Greenier J, Ding H. January 2006. Polypeptides having cellulolytic enhancing activity and polynucleotides encoding same. US patent 20,060,005,279.
- 18.Sweeney M, Vlasenko E, Abbate E. June 2011. Methods for increasing hydrolysis of cellulosic material in the presence of cellobiose dehydrogenase. US patent 20,100,159,536.
- 19.Shao Z, Zhao H, Zhao H. 2009. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37:e16. 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 21.Gietz RD, Woods RA. 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313:107–120. 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 22.Wen F, Esteban O, Zhao H. 2008. Rapid identification of CD4+ T-cell epitopes using yeast displaying pathogen-derived peptide library. J. Immunol. Methods 336:37–44. 10.1016/j.jim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhang YH, Cui J, Lynd LR, Kuang LR. 2006. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules 7:644–648. 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 24.Clements JM, Catlin GH, Price MJ, Edwards RM. 1991. Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences. Gene 106:267–271. 10.1016/0378-1119(91)90209-T. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Zhang X, Yoon J, Rao K, Grate JH, Baidyaroy D, Elgart DD. February 2013. Gh61 glycoside hydrolase protein variants and cofactors that enhance gh61 activity. US patent 20,130,052,698.
- 26.Sygmund C, Santner P, Krondorfer I, Peterbauer CK, Alcalde M, Nyanhongo GS, Guebitz GM, Ludwig R. 2013. Semi-rational engineering of cellobiose dehydrogenase for improved hydrogen peroxide production. Microb. Cell Fact. 12:38. 10.1186/1475-2859-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du J, Yuan Y, Si T, Lian J, Zhao H. 2012. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 40:e142. 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai S-L, Oh J, Singh S, Chen R, Chen W. 2009. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75:6087–6093. 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arfi Y, Shamshoum M, Rogachev I, Peleg Y, Bayer EA. 2014. Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc. Natl. Acad. Sci. U. S. A. 111:9109–9114. 10.1073/pnas.1404148111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.