Abstract
Identification of Salmonella serotypes is important for understanding the environmental diversity of the genus Salmonella. This study evaluates the diversity of Salmonella isolates recovered from 165 of 202 Central Florida surface water samples and investigates whether the serotype of the environmental Salmonella isolates can be predicted by a previously published multiplex PCR assay (S. Kim, J. G. Frye, J. Hu, P. J. Fedorka-Cray, R. Gautom, and D. S. Boyle, J. Clin. Microbiol. 44:3608–3615, 2006, http://dx.doi.org/10.1128/JCM.00701-06). Multiplex PCR was performed on 562 Salmonella isolates (as many as 36 isolates per water sample) to predict serotypes. Kauffmann-White serogrouping was used to confirm multiplex PCR pattern groupings before isolates were serotyped, analyzed by pulsed-field gel electrophoresis, and assayed for antimicrobial susceptibility. In 41.2% of the Salmonella-positive water samples, all Salmonella isolates had identical multiplex PCR patterns; in the remaining 58.8%, two or more multiplex PCR patterns were identified. Within each sample, isolates with matching multiplex PCR patterns had matching serogroups. The multiplex patterns of 495 isolates (88.1%) did not match any previously reported pattern. The remaining 68 isolates matched reported patterns but did not match the serotypes for those patterns. The use of the multiplex PCR allowed the number of isolates requiring further analysis to be reduced to 223. Thirty-three Salmonella enterica serotypes were identified; the most frequent included serotypes Muenchen, Rubislaw, Anatum, Gaminara, and IV_50:z4,z23:−. A majority (141/223) of Salmonella isolates clustered into one genotypic group. Salmonella isolates in Central Florida surface waters are serotypically, genotypically, and phenotypically (in terms of antimicrobial susceptibility) diverse. While isolates could be grouped as different or potentially the same using multiplex PCR, the multiplex PCR pattern did not predict the Salmonella serotype.
INTRODUCTION
Salmonella bacteria were traditionally considered zoonotic: they were believed to originate from the gastrointestinal tracts of animals, and outbreaks of salmonellosis were traced back to these reservoirs (1). More recently, the role of the nonhost environment as a source for Salmonella bacteria and food contamination has been explored (2–4). One of the nonhost reservoirs of Salmonella on which considerable effort has focused is surface water, which poses a potential public health risk through consumption, indirect consumption (for example, irrigation of edible horticultural crops), or direct contact. A number of studies have characterized the distribution of Salmonella isolates from different water sources (5–10). In evaluations of surface water for Salmonella isolates, multiple serotypes are often identified from a single environmental sample, indicating the diversity of this genus in the environment.
Salmonella serotypes isolated from surface waters are often not clinically common (4–12) and often include serotypes of Salmonella enterica subsp. enterica, Salmonella enterica subsp. arizonae, and Salmonella enterica subsp. houtenae (5, 8). Serotypes of Salmonella enterica subsp. enterica that have been isolated in more than one surface water survey include Anatum, Braenderup, Gaminara, Inverness, Muenchen, Newport, Rubislaw, Saintpaul, and Typhimurium (5–8). The number of different Salmonella serotypes isolated from one water sample can differ greatly among samples and may be influenced by the sample volume and enrichment scheme (7, 9); as many as 27 different Salmonella serotypes have been reported from a single surface water sampling site (9). The inclusion of multiple watersheds in studies may also lead to the isolation of a large number of different Salmonella serotypes (7–9).
A current pitfall of Salmonella classification is the length of time it takes to identify serotypes, commonly by serological discrimination with classification according to the Kauffmann-White scheme (13) or through any number of emerging genetic analysis methods (14–18). Many of the rapid methods proposed for Salmonella classification are based on, and tested against, the most clinically relevant serotypes (18), and their usefulness in predicting serotypes of environmental isolates is unknown. In order to begin to understand the diversity of Salmonella isolates in the nonhost environment, it is crucial to serotype multiple isolates from each sample (9, 10). The use of conventional serotyping for this purpose can be prohibitively expensive, laborious, and time-consuming, making a rapid method desirable. The diversity of 562 Salmonella isolates previously recovered from Central Florida surface water sites sampled over 12 months (19) is presented here, along with an evaluation of a previously published multiplex PCR method for rapid serotyping of the isolates (18).
MATERIALS AND METHODS
Water sample collection and Salmonella isolation.
Salmonella isolation from Central Florida surface waters has been described by McEgan et al. (19). Briefly, surface water samples were collected from 18 sites in Central Florida for 12 consecutive months (total, 202 water samples). All sites were in rural agricultural areas, away from animal-agricultural operations, and have been described by McEgan et al. (19). Sampling sites included two lakes, one pond, six creeks, two streams, one river, and six canals. Salmonella bacteria were isolated by using a Salmonella enrichment procedure modified from that in the U.S. Food and Drug Administration (FDA) Bacteriological Analytical Manual (BAM) (20). Salmonella bacteria were genetically confirmed by PCR of the invA and oriC genes (21); a total of 562 Salmonella isolates were collected (19) and were stored in tryptic soy broth (TSB; Difco, Becton Dickinson, Sparks, MD, USA) supplemented with 15% glycerol at −80°C.
Multiplex PCR analysis.
Multiplex PCR analysis was performed on all Salmonella isolates. Frozen (−80°C) isolates were streaked onto tryptic soy agar (TSA; Difco, Becton Dickinson) and were incubated for 24 ± 2 h at 35 ± 2°C. A single colony was then transferred to TSB and was incubated for 24 ± 2 h at 35 ± 2°C. From these cultures, DNA was extracted using the Mo Bio (Carlsbad, CA, USA) UltraClean DNA kit according to the manufacturer's instructions. Isolated DNA was then subjected to the multiplex PCR described by Kim et al. (18) with minor modifications. The assay uses the amplification of 10 specific gene sections (in two separate PCRs) that result in the production of a serotype-specific pattern of bands to differentiate 30 of the most clinically relevant serotypes of S. enterica subsp. enterica (18). The presence–absence patterns of the 10 gene sections were compared to the patterns reported by Kim et al. (18). The primers and reaction parameters used here were those described previously by Kim et al. (18), except that all the PCR reagents were obtained from the Fisher exACTGene Complete PCR kit (Fisher Scientific, Pittsburgh, PA, USA). DNA amplicons were separated by agarose (2.5%) gel electrophoresis and were visualized under UV light.
Serogrouping and serotyping.
Frozen (−80°C) isolates were streaked onto TSA and were incubated for 24 ± 2 h at 35 ± 2°C. A single colony from each isolate was applied to a glass slide with 5 μl sterile saline. Salmonella O Poly Antiserum (Becton Dickinson, Sparks, MD, USA) was applied, and autoagglutination was reported after 1 min of rocking of the glass slide. All isolates were serogrouped in order to determine if isolates that grouped together based on multiplex PCR patterns also belonged to the same serogroup. If isolates from the same water sample had identical presence–absence patterns for the 10 gene sections amplified during the multiplex PCR and belonged to the same serogroup, only one was selected for further analysis. The selected isolates were sent to the National Veterinary Laboratory Service (USDA, Ames, IA, USA) for serotyping.
PFGE.
Isolates sent for serotyping were analyzed using DNA fingerprinting conducted using XbaI DNA macrorestriction followed by pulsed-field gel electrophoresis (PFGE). PFGE was performed according to the standard laboratory protocol recommended by the Centers for Disease Control and Prevention PulseNet program (22). The PulseNet universal standard strain H9812 of Salmonella enterica serotype Braenderup was used as the reference marker.
Gel images were captured using a MultiDoc-It imaging system (UVP, Upland, CA, USA), and images were analyzed using Bionumerics software, version 6.6 (Applied Maths, NV, Austin, TX, USA). Similarity amongst PFGE patterns was assessed using the unweighted-pair group method with arithmetic averages, with 2.0% band position tolerances. Dice coefficients had 1.5% optimization values. PFGE patterns that were >80% similar were considered to be in the same genetic cluster; similarity coefficients were obtained by calculating Dice coefficients.
Antimicrobial susceptibility testing.
Isolates sent for serotyping and analyzed by PFGE were assayed for susceptibility to 15 antibiotics by the calibrated dichotomous sensitivity method (23). The following antibiotics were tested: amikacin (An), amoxicillin-clavulanic acid (Amc), ampicillin (Am), cefoxitin (Fox), ceftriaxone (Cro), cephalothin (Cf), chloramphenicol (C), ciprofloxacin (Cip), gentamicin (G), imipenem (Imp), kanamycin (K), nalidixic acid (Na), streptomycin (S), sulfamethoxazole-trimethoprim (STx), and tetracycline (Te). Antimicrobial susceptibility was determined according to the test standards presented by Bell et al. (23).
RESULTS
Salmonella isolates.
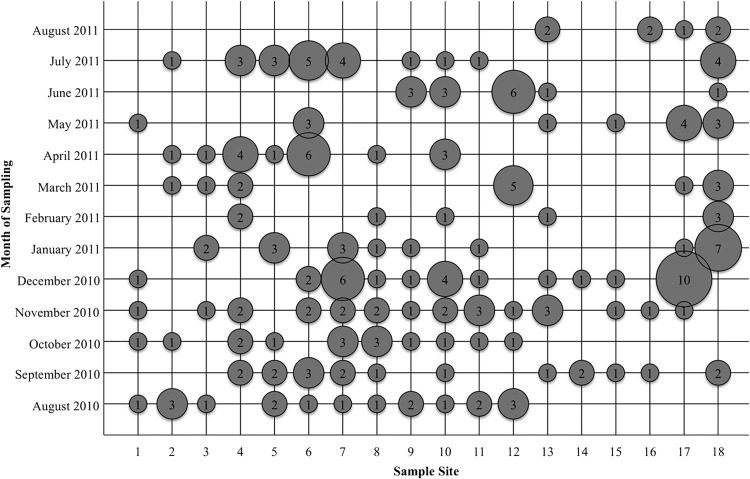
Salmonella isolates were collected from 165 (81.7%) of the 202 water samples analyzed (Fig. 1). All Salmonella isolates collected displayed typical morphology and growth characteristics on XLT4 agar or Salmonella CHROMagar. A total of 562 isolates were obtained; as many as 36 isolates were obtained and confirmed from a single sample.
FIG 1.
Unique Salmonella isolates recovered from 18 sampling sites over the 12-month survey (19). For each combination of sampling site and date, the number of unique isolates collected is given inside a bubble, whose size is relative to that number.
Multiplex PCR, serogrouping, and serotyping.
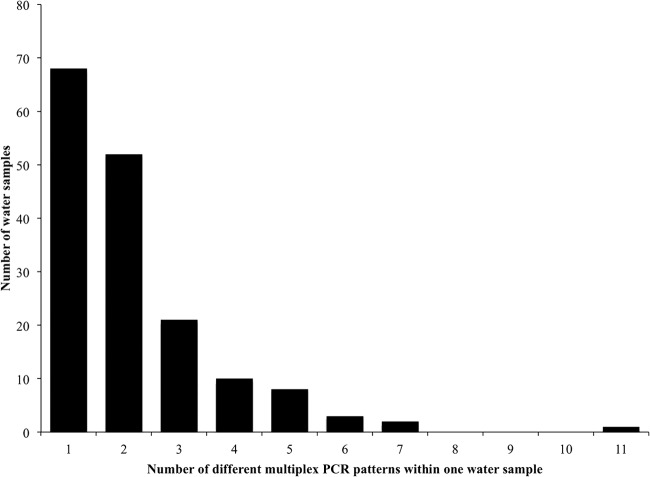
All 562 isolates were analyzed by multiplex PCR and serogrouping. Salmonella isolates from the same sample (water collected from the same site at the same time) with identical presence–absence multiplex PCR patterns were assumed to be potentially the same isolate; Salmonella isolates with different multiplex PCR patterns were assumed to be different. In 68 (41.2%) of the 165 water samples from which Salmonella isolates were recovered, all Salmonella isolates had identical multiplex PCR patterns, as shown in Fig. 2. In 52 (31.5%) water samples, two patterns were identified; in 21 (12.7%) water samples, three patterns were identified among the isolates. More than three patterns were identified in the remaining 24 (14.5%) water samples.
FIG 2.
Number of single surface water samples with the indicated number of unique multiplex PCR patterns (18). A single water sample comprises water that was collected from the same site at the same time.
If a multiplex pattern was previously reported by Kim et al. (18) as a specific serotype, that serotype was designated the “suggested serotype,” and the corresponding serogroup was designated the “suggested serogroup” (Table 1). Sixty-seven of the 562 isolates had multiplex patterns matching those reported by Kim et al. (18), while the multiplex patterns of 495 isolates (88.1%) did not match any pattern reported by Kim et al. For the 67 S. enterica isolates matching reported multiplex patterns (18), the suggested serotypes included Bovismorbificans (22 isolates), Braenderup (9 isolates), Chester (1 isolate), Derby (1 isolate), Hadar (3 isolates), Muenchen (4 isolates), Newport (2 isolates), Ohio (9 isolates), Oranienburg (4 isolates), Poona (3 isolates), Saintpaul (3 isolates), Thompson (5 isolates), and Typhimurium (1 isolate).
TABLE 1.
Suggested serotypes and corresponding serogroups of Salmonella isolates recovered from Central Florida surface waters compared with actual serogroupsa
| Suggested serotype | Suggested serogroup | No. of isolates | No. of isolates with the following actual serogroup: |
% matchingb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | ||||
| Bovismorbificans | B | 22 | 13 | 5 | 2 | 23 | ||||
| Braenderup | B | 9 | 7 | 1 | 1 | 78 | ||||
| Chester | A | 1 | 1 | 0 | ||||||
| Derby | A | 1 | 1 | 0 | ||||||
| Hadar | B | 3 | 1 | 2 | 0 | |||||
| Muenchen | B | 4 | 2 | 2 | 50 | |||||
| Newport | B | 2 | 2 | 0 | ||||||
| Ohio | B | 9 | 7 | 1 | 1 | 78 | ||||
| Oranienburg | B | 4 | 2 | 2 | 50 | |||||
| Poona | B | 3 | 1 | 2 | 0 | |||||
| Saintpaul | A | 3 | 2 | 1 | 67 | |||||
| Thompson | B | 5 | 5 | 100 | ||||||
| Typhimurium | A | 1 | 1 | 100 | ||||||
| Other | 494 | 57 | 88 | 46 | 6 | 9 | 5 | 17 | ||
Serotypes were suggested on the basis of multiplex PCR patterns corresponding to those reported by Kim et al. (18). The actual serogroups of the isolates were determined by autoagglutination.
Percentage of isolates for which the actual serogroup matched the suggested serogroup.
Serogroups were determined for all 562 S. enterica isolates and are reported in Table 1 as “actual serogroups.” In all 165 water samples from which Salmonella isolates were recovered, if isolates had matching multiplex patterns, as described above, the same isolates also had matching serogroups. However, not all isolates with matching serogroups had identical multiplex PCR patterns within one water sample. Only 31 of the 67 suggested serotypes (46.3%) based on the study of Kim et al. (18) matched the actual serogroup (Table 1).
To explore the possibility that the multiplex PCR could be an effective rapid method for determining serotypes of Salmonella strains collected from environmental sources, one representative Salmonella isolate from each water sample–multiplex PCR pattern combination was sent for serotyping. Of the 562 Salmonella isolates analyzed by multiplex PCR and serogrouping, 223 were sent for Kauffmann-White serotyping. Thirty-one Salmonella isolates were not recovered following frozen storage and could not be analyzed further. None of the “suggested serotypes” based on the multiplex PCR patterns of Kim et al. (18) matched the serotype determined by Kauffmann-White serotyping.
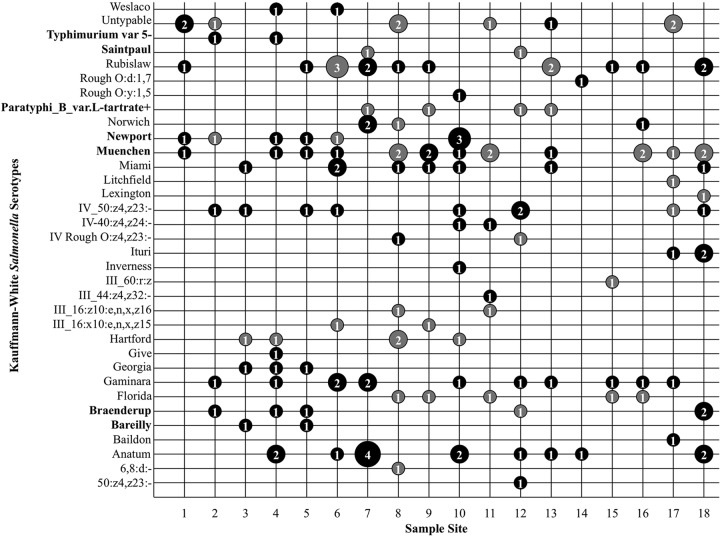
Kauffmann-White serotyping identified a total of 33 different serotypes for the 223 isolates sent for serotyping. Water samples were collected over a continuous 12-month time frame; the number of unique isolates from each water sample at each sampling time is given in Fig. 1. The number of different serotypes isolated from any individual sampling site over the course of the 12-month study ranged from 2 at site 14 to 10 at sites 8 and 10. The average number of serotypes isolated from a site was 6. The frequency of isolation for each serotype at each location across all sample times is shown in Fig. 3. No one serotype was isolated in all months of the study from any of the sampling sites. The most frequently isolated S. enterica serotypes were Muenchen (11.5% of isolates), Rubislaw (9.5%), Anatum (8.8%), Gaminara (8.8%), and IV_50:z4,z23:− (6.8%). When the 33 S. enterica serotypes isolated in this study were compared to the CDC's list of the 20 most common clinical serotypes, only 7 were found to be clinically common: serotypes Bareilly, Braenderup, Muenchen, Newport, Paratyphi B var. L-tartrate+, Saintpaul, and Typhimurium var. 5- (Fig. 3) (12). Salmonella serotypes isolated during the most months included serotypes Anatum (8 of 12 months), Muenchen (7 of 12 months), and Rubislaw (6 of 12 months). No temporal pattern of Salmonella serotype isolation was observed.
FIG 3.
Frequency of isolation of each serotype by location across all 12 sample times. For each combination of sampling site and serotype, the number of unique isolates collected across all sample dates is given inside a bubble, whose size is relative to that number. Black bubbles represent isolates in PFGE cluster A; gray bubbles represent isolates in all other PFGE clusters. Serotypes on the CDC's list of the 20 most common clinical serotypes are shown in boldface on the y axis.
PFGE analysis.
Isolates sent for Kauffmann-White serotyping (n = 223) were further analyzed using PFGE–XbaI macrorestriction banding patterns. Two isolates did not produce banding patterns by PFGE, even with the addition of thiourea: a Salmonella enterica serotype Bareilly strain, isolated from site 5 in August 2010, which also failed to produce a banding pattern by multiplex PCR, and a Salmonella enterica serotype Paratyphi B var. L-tartrate+ strain, isolated from site 12 in March 2011, which produced a banding pattern with multiplex PCR but did not match the other Salmonella enterica serotype Paratyphi B isolate that was recovered from the same site at the same time. Analysis of the PFGE patterns from 221 Salmonella isolates generated 11 major genotypic clusters (clusters A to K) (see Fig. S1 in the supplemental material), each comprising at least three isolates, with a Dice coefficient index cutoff point of 80% (Fig. 4 and 5). Analyzing the Salmonella isolates separately in two dendrograms (Fig. 4 and 5) caused individual isolates to move their positions from those in the complete dendrogram in Fig. S1 in the supplemental material. While the isolates within each cluster remained the same, the cluster labels became disordered (Fig. 5); for ease of interpretation, cluster labels were maintained as ordered in the complete dendrogram (see Fig. S1).
FIG 4.
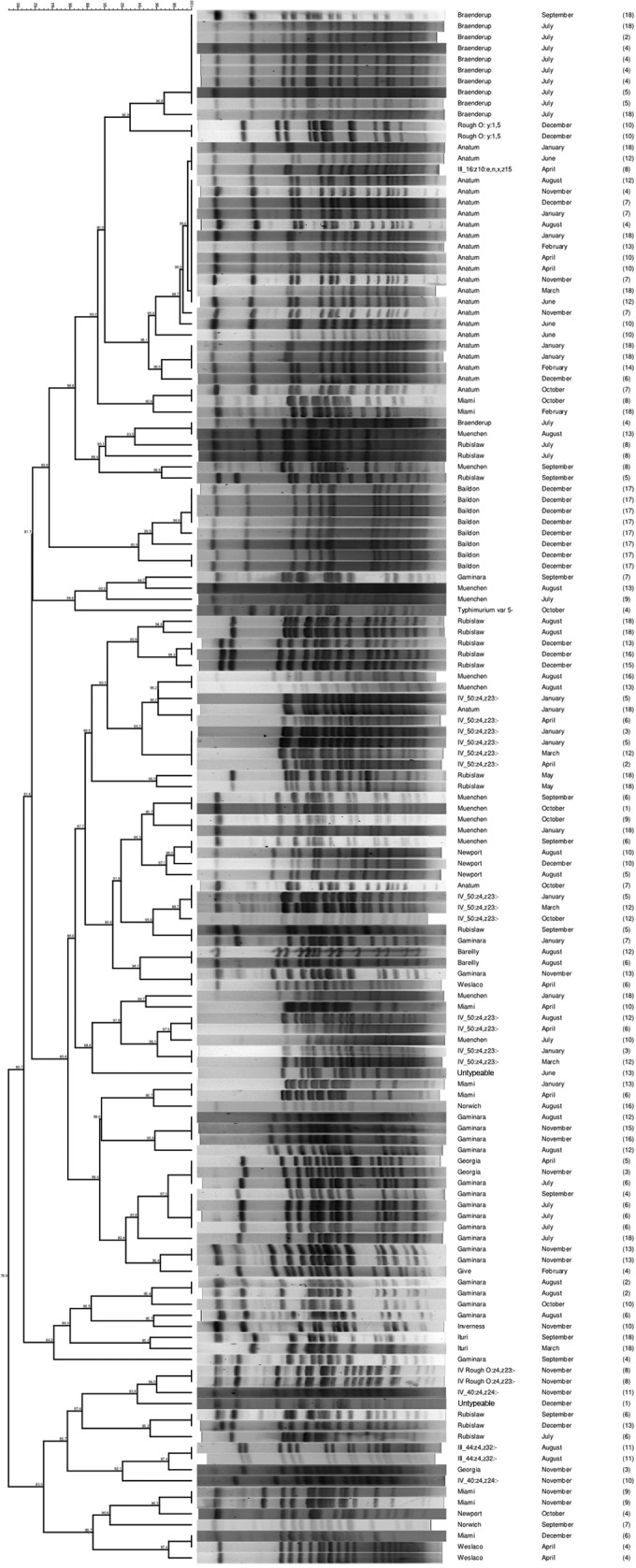
Dendrogram of Salmonella isolates recovered from Central Florida in cluster A, the largest cluster of isolates >80% similar to each other, determined by PFGE-XbaI fingerprinting. Additional information on nodal numbers can be found in Fig. S1 in the supplemental material.
FIG 5.
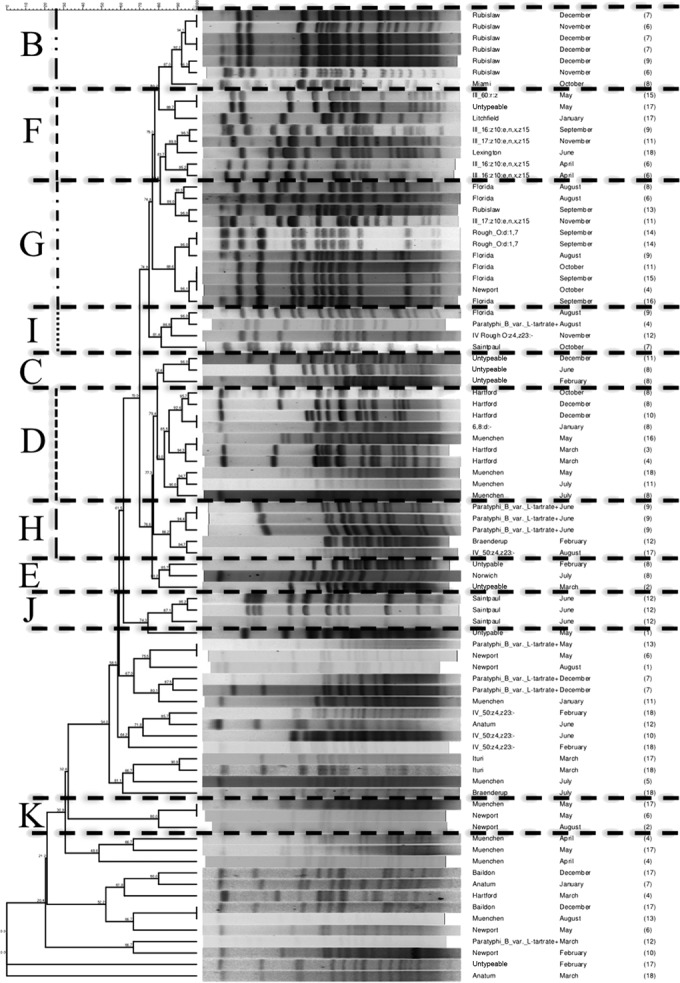
Dendrogram of Salmonella isolates recovered from Central Florida that are not grouped within cluster A, determined by PFGE-XbaI fingerprinting. All isolates that are >80% similar by PFGE are grouped into clusters. Clusters are labeled with letters from B to K, as determined by the complete dendrogram produced from all Salmonella isolates recovered from Central Florida (see Fig. S1 in the supplemental material). Lines connecting isolates are as follows: for group B, two short dashes followed by a long dash; for group D, all short dashes; for group F, all medium dashes; for group G, alternating short and medium dashes; for group H, long dashes; for group I, a dotted line. (Only groups with four or more isolates are labeled on the dendrogram.) Additional information on nodal numbers can be found in Fig. S1 in the supplemental material.
The majority (141/223) of isolates, including at least 1 isolate from each sample site, clustered into one genotypic group (cluster A) (Fig. 3 and 4); 23 of the 33 total serotypes are represented in this cluster. S. enterica serotypes not found within cluster A included Florida, Hartford, Lexington, Litchfield, Paratyphi B, Saintpaul, 6,8:d:−, III_60:r:z, and Rough_O:d:1,7 (Fig. 5). Sampling site 1 had the least genotypic diversity, with 2 isolates in cluster A and 3 isolates outside the major genotypic clusters (Fig. 3). Sampling sites 8, 9, 11, and 12 had the greatest genotypic diversity; each yielded isolates in 6 of the 11 genotypic clusters (Fig. 3). All typeable isolates of Salmonella enterica serotype Bareilly (n = 2) clustered as identical by PFGE in cluster A—the only occurrence of this among the 221 isolates evaluated by PFGE. All Salmonella enterica serotype Gaminara isolates (8.8% of isolates) were found in cluster A. Salmonella enterica serotype Muenchen isolates (11.5% of isolates) clustered into groups A, D, and K; Salmonella enterica serotype Rubislaw isolates (9.5%), into groups A, B, and G; Salmonella enterica serotype Anatum isolates (8.8%), into group A; and Salmonella enterica serotype IV_50:z4,z23:− isolates (6.8%), into groups A and H. Additional isolates of these serotypes were found outside of the major genotypic cluster groups.
Over the entire sampling period, there were 19 instances in which 2 to 10 isolates of the same serotype from different water samples had identical PFGE patterns (100% similarity). In six instances of isolates with identical PFGE patterns, some of the isolates were collected from the same sampling site and in the same sampling month but had different multiplex PCR patterns. Ten Salmonella Anatum isolates with identical PFGE patterns were isolated in seven different months and from six different sites. Of the 23 total Salmonella Anatum isolates (Fig. 3 and 4), 20 in cluster A had 89.6% similar PFGE patterns. Nine Salmonella Braenderup isolates with identical PFGE patterns came from sites 2, 4, 5, and 18 in September 2010 and July 2011 (including the four isolates from July 2011, site 4, with different multiplex PCR patterns). Of the 13 total Salmonella Braenderup isolates (Fig. 3 and 4), 11 in cluster A had 87.6% similar PFGE patterns. Twelve of the 19 instances in which identical PFGE patterns were reported involved only two isolates; in one case, Salmonella enterica serotype Newport isolates from samples collected at site 10 in August 2010 and December 2010 had identical patterns. Isolates with identical PFGE patterns were collected from different sites in the same month twice: Salmonella Bareilly from sites 4 and 5 in August 2010 and Salmonella Rubislaw from sites 13 and 16 in December 2010.
Antimicrobial susceptibility testing.
The 223 serotyped Salmonella isolates were tested for antimicrobial susceptibility. Twenty-four isolates were susceptible to all antimicrobials tested; 41 were resistant to streptomycin; and 50 were resistant to both streptomycin and kanamycin. Eighteen Salmonella isolates were resistant to more than five of the antibiotics tested; the antibiotic resistance profiles of these strains are shown in Table 2. None of the multidrug-resistant (MDR) Salmonella isolates were resistant to ceftriaxone or ciprofloxacin. Three of the 13 Salmonella Braenderup strains isolated and 3 of the 8 Salmonella enterica serotype Florida strains isolated were MDR. The Salmonella Braenderup isolates were recovered from three sampling sites (sites 2, 4, and 5) in July and were 87.6% similar by PFGE; the Salmonella Florida strains were isolated from sampling sites 4, 5, and 9 during August and October and were 80.8% similar by PFGE. Two isolates each of Salmonella Bareilly (of 4 total isolates), Salmonella Gaminara (of 13 total isolates), Salmonella enterica serotype Hartford (of 5 total isolates), and Salmonella Muenchen (of 17 total isolates) were MDR. The Salmonella Bareilly strains were isolated from sampling sites 4 and 5 during August 2010 and had identical PFGE patterns; the Salmonella Gaminara strains were isolated from sampling sites 15 and 16 during November 2010 and also had identical PFGE patterns. The Salmonella Hartford strains were isolated from sampling sites 3 and 4 during March, and the Salmonella Muenchen strains were isolated from sampling sites 5 and 11 during July 2011; these isolates had different PFGE patterns. Of the 18 MDR Salmonella isolates, 4 isolates were from sampling sites 4 and 5; the remaining sampling sites had only 1 or no MDR Salmonella isolates.
TABLE 2.
Antibiotic resistance profiles of Salmonella isolates from Central Florida surface watersa
| Serotype of MDR isolate | Site | Month | Antimicrobial resistance profile |
|---|---|---|---|
| Anatum | 13 | February | Amc Am Fox Cf S Te |
| Bareilly | 4 | August | Amc Am Fox Cf K S STx Te |
| Bareilly | 5 | August | Amc Am Fox Cf C K Na S STx Te |
| Braenderup | 2 | July | Amc Am Cf Imp K S |
| Braenderup | 4 | July | Amc Am Fox K Na S STx |
| Braenderup | 5 | July | Amc Am Fox Cf C K Na STx Te |
| Florida | 4 | August | Amc Am Fox Cf Imp K S |
| Florida | 5 | October | Amc Am Fox Cf K S |
| Florida | 9 | August | Amc Am Fox Cf K S |
| Gaminara | 15 | November | Amc Am Fox Cf C K Na S STx Te |
| Gaminara | 16 | November | Amc Am Fox Cf K S |
| Hartford | 3 | March | Amc Fox Cf K Na STe |
| Hartford | 4 | March | Amc Am Fox Cf C K Na STe |
| Muenchen | 5 | July | Amc Cf C K Na STx Te |
| Muenchen | 11 | July | Amc Am Cf C K Na STx Te |
| Rough_O:d:1,7 | 14 | September | Amc Am Fox Cf Gm S |
| Rubislaw | 7 | July | Am Fox Cf K S STx Te |
| Saintpaul | 12 | June | Amc Am Fox Cf K S |
Only isolates resistant to more than five of the antibiotics screened are listed.
DISCUSSION
Studies evaluating the prevalence of Salmonella bacteria in surface waters have reported different prevalences: 81.7% (165/202) in Central Florida (19), 96% (106/110) (1,000-ml samples) in North Florida (7), 79% (57/72) (555-ml samples) in Georgia (5), 62% (90/145) (Moore swabs suspended in the water for 3 to 5 days) in Ontario, Canada (9), 57% (42/74) (25-ml samples) in North Carolina (6), 11% (16/145) (100-ml samples) in New York State (8), and 7% (18/252) (100-ml samples or 3-day Moore swabs) in California (24). Even within one study of Central California coastal waterways, Salmonella prevalences ranged from 0 to 72% in 3,330-ml water samples (25). Differences in water sampling volumes and enrichment protocols complicate a direct comparison of the Salmonella prevalence reported here with those reported previously for other regions. To overcome this difficulty, Strawn et al. (10) isolated Salmonella strains from the produce environment (including 250-ml water samples) in two locations using identical sample collection, detection, and isolation schemes; Salmonella bacteria were isolated from 38% (15/40) of surface water samples in South Florida and 10% of surface water samples (36/354; including those reported in reference 8) in New York State. While it is unfortunate that the numbers of water samples (40 in South Florida versus 354 in New York State) are not the same, it is clear that differences in Salmonella prevalence exist on a regional level, independently of sample size or detection and isolation schemes. These differences may be due to other climatic factors, environmental pressures, wildlife, or agricultural practices. Future studies designed to specifically determine the exact factors that influence pathogen prevalence in surface waters in different regions are warranted.
As has been reported previously (5, 9, 10), more than one Salmonella serotype is frequently isolated from a single Salmonella-positive surface water sample. Considering only differences in Kauffmann-White serotyping, multiple serotypes were identified in 32/165 Salmonella-positive water samples. When multiple Salmonella serotypes were identified from a single water sample, the typical number of serotypes identified was 2; as many as 4 different serotypes were isolated from a single water sample. Similarly, of 72 water samples from Georgia, 15 samples yielded 2 serotypes each and 3 samples yielded 3 serotypes each (5). Strawn et al. (10) reported 13/97 water samples with multiple serotypes and 1 sample from South Florida with 3 different serotypes. The different numbers of isolates per sample or isolation scheme selected for further analysis complicate a direct comparison of the numbers of Salmonella serotypes from a single water sample. The potential for isolating multiple Salmonella serotypes from a single sample highlights the importance of using multiple enrichment and plating media to truly explore Salmonella diversity and to facilitate the identification of isolates with atypical characteristics (7, 9, 10).
As few as 7 (New York State) (8) and as many as 38 (Ontario, Canada) (9) different serotypes have been isolated during surface water surveys, indicating a tremendous diversity of Salmonella serotypes in the environment. Strawn et al., comparing Salmonella isolates from water samples in South Florida and New York State using the same isolation scheme, identified 9 and 12 Kaufmann-White serotypes, respectively, with 4 serovars found in water samples from both locations (10). Surface water surveys in the southeastern United States have reported 5, 8, and 13 different serotypes from North Carolina, North Florida, and Georgia surface waters, respectively (5–7). In addition to sample size and isolation differences, the diversity of serotypes reported here (33 serotypes) may reflect the study design (a greater number of sampling locations and watersheds sampled) or other environmental pressures (wildlife, agricultural practices, etc.).
Of the 33 Kauffmann-White serotypes of S. enterica isolated from Central Florida, 15 have been reported in other surface water studies (5–10). They include serotype Anatum (from Georgia and Ontario, Canada), serotype Baildon (from South Florida), serotype Bareilly (from Georgia), serotype Braenderup (from Georgia, North Florida, and South Florida), serotype Gaminara (from Georgia, North Carolina, and South Florida), serotype Give (from Ontario, Canada, and New York State), serotype Hartford (from Ontario, Canada), serotype Inverness (from North Carolina and North Florida), serotype Litchfield (from Ontario, Canada, and South Florida), serotype Miami (from North Carolina), serotype Muenchen (from Georgia, North Florida, and Ontario, Canada), serotype Newport (from North Carolina, North Florida, New York State, Ontario, Canada, and South Florida), serotype Rubislaw (from Georgia, North Florida, New York State, and South Florida), serotype Saintpaul (from Georgia and Ontario, Canada), and serotype Typhimurium (from New York State and Ontario, Canada). None of the 16 serotypes, determined by multilocus sequencing, from Central California coastal waterways (25) match serotypes in this study. Independently of differences in sampling volume and enrichment protocols, 15/33 serotypes recovered here were also recovered during previous surveys of surface water, suggesting more-frequent presence or greater fitness in surface waters for these serotypes than for other Salmonella serotypes. Of the 11 Salmonella serotypes isolated in Central Florida and from at least two other locations, only 5 (serotypes Braenderup, Muenchen, Rubislaw, Saintpaul, and Typhimurium) are also reported on the CDC's list of top 20 clinical isolates (12). The frequency, over multiple surface water surveys, of Salmonella serotypes that are not often linked to human cases of salmonellosis suggests that the diversity of Salmonella strains in the environment includes many serotypes not commonly seen or clinically relevant. Future studies to evaluate potential differences in environmental fitness, associations with hosts, and virulence among surface water Salmonella isolates from different regions are justified.
The number of Salmonella serotypes differed significantly among the 18 sites tested. The smallest number of serotypes isolated from a single site over the 12 months of our study was 2 from site 14, a canal in a 55% agricultural land usage watershed (19). Two streams and one creek in watersheds with 32 to 33% agriculture (sites 10, 4, and 6) had the highest numbers of serotypes (11, 10, and 9 serotypes, respectively) isolated over the 12 months. Salmonella serotypes Anatum, Gaminara, Muenchen, and Newport were common to sites 10, 6, and 4 and were also reported in North Florida (7) and South Florida (10) surface waters. The repeated isolation of these serotypes from Florida surface waters may reflect increased fitness in this environment.
The multiplex PCR described by Kim et al. (18) could not serotype Salmonella isolates from Central Florida surface waters. Jean-Gilles Beaubrun et al. (26) report contradictory results, where 76.2% (48/63) of environmental Salmonella isolates from tomato farms in Virginia were correctly identified by the multiplex PCR (18) as S. Newport, S. Braenderup, and S. Typhimurium. However, different patterns were reported for S. enterica serotypes Berta, Derby, Hadar, Infantis, Montevideo, Muenchen, Newport, and Paratyphi, and more than one multiplex pattern was reported for serotypes Senftenberg and Tennessee (26). While the multiplex PCR (18), did not predict the serotype of Salmonella isolates here, the use of multiplex patterns to group isolates collected from one water sample decreased the number of isolates for further characterization by more than half, decreasing the cost and time required to screen the numerous isolates. The use of the multiplex patterns (18) to group isolates introduces the risk of identifying isolates as identical when they are in fact different serotypes, while allowing a greater number of isolates to be screened in a timely manner.
PFGE analysis revealed 19 sets of Salmonella isolates with identical banding patterns from multiple months or sampling sites. Salmonella Anatum isolates with identical patterns were isolated from sampling site 7 in November, December, and January; the 3-month isolation of Salmonella Anatum from one sample site may indicate its survival in the surface water or repetitive introduction from the same source. Repeated surface water samples in which Salmonella Anatum isolates with identical PFGE patterns were identified have been seen previously in North Carolina, in an area with a known influence from swine agriculture (6). Repeated isolation of Salmonella enterica serotype Heidelberg strains with identical PFGE patterns from stream water sampled from May through October has been reported in Ontario, Canada (9). Salmonella isolates subjected to genotypic clustering by PFGE using only one enzyme (XbaI), as performed here and by Patchanee et al. (9), may be further differentiated by repeating PFGE using a second enzyme or by alternate analysis. As such, the matches described here may be an artifact of the method of analysis selected, supported by the occurrence of more than one multiplex PCR pattern for isolates with identical PFGE patterns.
The majority (115/223) of Central Florida isolates were susceptible to all antimicrobials tested or were resistant only to kanamycin and/or streptomycin, both of which are secondary metabolites of Streptomyces spp., common soil microbes that can be found in the environment; this type of resistance, therefore, is not uncommon among environmental isolates (27, 28). Previously, Salmonella isolates from watersheds influenced by pig farms had increased antimicrobial resistance (6). Here, sites 4 and 5 had the greatest number of MDR Salmonella isolates; while sampling sites selected in this study were not immediately adjacent to animal agriculture, the high prevalence of antimicrobial-resistant isolates may indicate an animal agriculture influence on the watershed.
As in previous studies (5–10, 25), a diverse group of Salmonella serotypes was isolated from Central Florida surface waters. Although the isolates could not be serotyped rapidly using the multiplex PCR (18), this technique allowed isolates from the same sample to be grouped as different or potentially the same, decreasing the effort and expense of subsequent typing. However, while the diversity of isolates was also reflected in the genetic profile determined by PFGE using XbaI, the occurrence of more than one multiplex PCR pattern for isolates with identical PFGE patterns indicates the need for more-discriminatory typing tools.
Supplementary Material
ACKNOWLEDGMENTS
We thank the USDA CSREES (Specialty Crops Research Initiative 2008-51180-04846) and the University of Florida Alumni Fellowships Fund for financial support of this project.
We thank Gwen Lundy, Luis Martinez, and Brian Buzzie for technical assistance.
Footnotes
Published ahead of print 29 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02191-14.
REFERENCES
- 1.D'Aoust JY. 1989. Salmonella, p 327–445 In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, New York, NY. [Google Scholar]
- 2.Jackson GJ, Langford CF, Archer DL. 1991. Control of salmonellosis and similar foodborne infections. Food Control 2:26–34. 10.1016/0956-7135(91)90115-D. [DOI] [Google Scholar]
- 3.Ijabadeniyi OA, Debusho LK, Vanderlinde M, Buys EM. 2011. Irrigation water as a potential preharvest source of bacterial contamination of vegetables. J. Food Saf. 31:452–461. 10.1111/j.1745-4565.2011.00321.x. [DOI] [Google Scholar]
- 4.Greene SK, Daly ER, Talbot EA, Demma LJ, Holzbauer S, Patel NJ, Hill TA, Walderhaug MO, Hoekstra RM, Lynch MF, Painter JA. 2008. Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 136:157–165. 10.1017/S095026880700859X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haley BJ, Cole DJ, Lipp EK. 2009. Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl. Environ. Microbiol. 75:1248–1255. 10.1128/AEM.01648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patchanee P, Molla B, White N, Line DE, Gebreyes WA. 2010. Tracking Salmonella contamination in various watersheds and phenotypic and genotypic diversity. Foodborne Pathog. Dis. 7:1113–1120. 10.1089/fpd.2010.0572. [DOI] [PubMed] [Google Scholar]
- 7.Rajabi M, Jones M, Hubbard M, Rodrick G, Wright AC. 2011. Distribution and genetic diversity of Salmonella enterica in the Upper Suwannee River. Int. J. Microbiol. 2011:461321. 10.1155/2011/461321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strawn LK, Fortes ED, Bihn EA, Nightingale KK, Grohn YT, Worobo RW, Wiedmann M, Bergholz PW. 2013. Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl. Environ. Microbiol. 79:588–600. 10.1128/AEM.02491-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas JL, Slawson RM, Taylor WD. 2013. Salmonella serotype diversity and seasonality in urban and rural streams. J. Appl. Microbiol. 114:907–922. 10.1111/jam.12079. [DOI] [PubMed] [Google Scholar]
- 10.Strawn LK, Danyluk MD, Worobo RW, Wiedmann M. 2014. Distribution of Salmonella subtypes differ between two U.S. produce-growing regions. Appl. Environ. Microbiol. 80:3982–3991. 10.1128/AEM.00348-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perch M, Braden CR, Bishop R, Fields P, Plikaytis R, Tauxe RV. 2003. Salmonella surveillance summary, 2003. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 12.Centers for Disease Control and Prevention. 2009. National Salmonella surveillance annual summary, 2009. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncezid/dfwed/PDFs/SalmonellaAnnualSummaryTables2009.pdf. [Google Scholar]
- 13.Bopp CA, Brenner FW, Fields PI, Wells JG, Stockbine NA. 2003. Escherichia, Shigella, and Salmonella, p 645–671 In Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. (ed), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC. [Google Scholar]
- 14.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S. 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 8:e1002776. 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leader BT, Frye JG, Hu J, Fedorka-Cray PJ, Boyle DS. 2009. High-throughput molecular determination of Salmonella enterica serovars by use of multiplex PCR and capillary electrophoresis analysis. J. Clin. Microbiol. 47:1290–1299. 10.1128/JCM.02095-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida C, Franklin K, Konczy P, McQuiston JR, Fields PI. 2007. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J. Microbiol. Methods 70:261–271. 10.1016/j.mimet.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 45:3323–3334. 10.1128/JCM.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Frye JG, Hu J, Fedorka-Cray PJ, Gautom R, Boyle DS. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp enterica. J. Clin. Microbiol. 44:3608–3615. 10.1128/JCM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEgan R, Mootian G, Goodridge LD, Schaffner DW, Danyluk MD. 2013. Predicting Salmonella populations from biological, chemical, and physical indicators in Florida surface waters. Appl. Environ. Microbiol. 79:4094–4105. 10.1128/AEM.00777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. Food and Drug Administration. 2011. Salmonella. In Bacteriological analytical manual U.S. Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm Accessed 15 June 2011. [Google Scholar]
- 21.Malorny B, Hoorfar J, Bunge C, Helmuth R. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290–296. 10.1128/AEM.69.1.290-296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribot E, Fair M, Gautom R, Cameron D, Hunter S, Swaminathan B, Barrett T. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 23.Bell SM, Gatus BJ, Pham JN, Rafferty DL. 2013. Antibiotic susceptibility testing by the CDS method: a manual for medical and veterinary laboratories, 7th ed. Antibiotic Reference Laboratory, Prince of Wales Hospital, Randwick, NSW, Australia: http://web.med.unsw.edu.au/cdstest/. [Google Scholar]
- 24.Gorski L, Parker CT, Liang A, Cooley MB, Jay-Russell MT, Gordus AG, Atwill ER, Mandrell RE. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734–2748. 10.1128/AEM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters SP, Gonzalez-Escalona N, Son I, Melka DC, Sassoubre LM, Boehm AB. 2013. Salmonella enterica diversity in central Californian coastal waterways. Appl. Environ. Microbiol. 79:4199–4209. 10.1128/AEM.00930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jean-Gilles Beaubrun J, Cheng C-M, Chen K-S, Ewing L, Wang H, Agpaoa MC, Huang M-CJ, Dickey E, Du JM, Williams-Hill DM, Hamilton B, Micallef SA, Rosenberg Goldstein RE, George A, Joseph SW, Sapkota AR, Jacobson AP, Tall BD, Kothary MH, Dudley K, Hanes DE. 2012. The evaluation of a PCR-based method for identification of Salmonella enterica serotypes from environmental samples and various food matrices. Food Microbiol. 31:199–209. 10.1016/j.fm.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Turpin P, Dhir V, Maycroft K, Rowlands C, Wellington E. 1992. The effect of Streptomyces species on the survival of Salmonella in soil. FEMS Microbiol. Ecol. 101:271–280. 10.1111/j.1574-6968.1992.tb05784.x. [DOI] [Google Scholar]
- 28.Egan S, Wiener P, Kallifidas D, Wellington E. 2001. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie Van Leeuwenhoek 79:127–133. 10.1023/A:1010296220929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.