Abstract
Exploring processes of coevolution of microorganisms and their hosts is a new imperative for life sciences. If bacteria protect hosts against pathogens, mechanisms facilitating the intergenerational transmission of such bacteria will be strongly selected by evolution. By disentangling the diversity of bacterial strains from the uropygium of hoopoes (Upupa epops) due to genetic relatedness or to a common environment, we explored the importance of horizontal (from the environment) and vertical (from parents) acquisition of antimicrobial-producing symbionts in this species. For this purpose, we compared bacterial communities among individuals in nonmanipulated nests; we also performed a cross-fostering experiment using recently hatched nestlings before uropygial gland development and some nestlings that were reared outside hoopoe nests. The capacity of individuals to acquire microbial symbionts horizontally during their development was supported by our results, since cross-fostered nestlings share bacterial strains with foster siblings and nestlings that were not in contact with hoopoe adults or nests also developed the symbiosis. Moreover, nestlings could change some bacterial strains over the course of their stay in the nest, and adult females changed their bacterial community in different years. However, a low rate of vertical transmission was inferred, since genetic siblings reared in different nests shared more bacterial strains than they shared with unrelated nestlings raised in different nests. In conclusion, hoopoes are able to incorporate new symbionts from the environment during the development of the uropygium, which could be a selective advantage if strains with higher antimicrobial capacity are incorporated into the gland and could aid hosts in fighting against pathogenic and disease-causing microbes.
INTRODUCTION
Symbiotic relationships are widespread in nature and are considered a crucial driving force in evolution (1). They deeply affect the diversity of life by expanding ecological and evolutionary opportunities for speciation and specialization of both hosts and symbionts (2, 3) and have profound effects on ecosystem functioning (4).
The vast majority of symbioses described in eukaryotes involve bacteria (5), which are considered an important selective force shaping the evolution of animals (4, 6). Although research on symbiotic interactions in nature has long focused on parasites, the study of nonpathogenic microbiota that live in close association with eukaryotes and benefit the ecology, fitness, and evolution of their hosts has recently received increased attention (reviewed in reference 7). Among other features, most bacteria produce compounds that inhibit the growth of potential competing microorganisms. Animals may benefit from hosting antibiotic-producing bacteria that combat potential pathogens, so these alliances should be widespread in nature (8). Tight mutualistic symbioses between animals and bacteria that confer protection against pathogens have been discovered and studied, mainly in invertebrates such as crustaceans (9), squids (10), aphids (11), or flies (12) but also in vertebrates such as salamanders (13) and European hoopoes (Upupa epops) (14). Bacteria with outstanding antibiotic properties have been isolated from the uropygial gland (referred to here as the UG) of hoopoes (15, 16) and wood hoopoes (Phoeniculus purpureus) (17), two closely related bird species (order Coraciiformes). The UG is a holocrine secretory gland located at the base of the tail (18) that produces a secretion with a variety of hydrocarbons, fatty acids, and esters (19). In hoopoes, beneficial effects of antibiotic-producing bacteria have been associated with increased hatching success (14, 20) and reduced feather degradation (21).
Host traits facilitating relationships with beneficial bacteria would represent a selective advantage due to the fitness benefits conferred by the mutualistic symbionts with antimicrobial production (22). Hosts should thus first ensure the acquisition of those symbionts, for which several mechanisms have been described (23). The acquisition of symbionts can be vertical, from parents to offspring (24, 25), or can be horizontal, from environment to offspring, including unrelated host individuals (26). The fitness of vertically transmitted symbionts is closely related to that of their hosts (e.g., 27, 28, 29, 30), favoring intimate coevolutionary processes (31) and enhancing the effectiveness of symbionts (29). Horizontal transmission leads to more-diffuse coevolutionary processes (32), implying that the effectiveness of mutualistic symbionts may depend on the availability of symbiont genotypes in the environment (29). Horizontally transmitted symbionts may allow hosts to exploit a wider range of environmental conditions or locations if symbionts acquired at different locations confer protection against common pathogenic microbes for each situation (23). In this sense, knowing how bacterial communities are acquired by new generations, and knowing the importance of vertical transmission of antibiotic-producing symbionts relative to that of horizontal transmission, is essential for understanding the symbiotic associations and therefore the coevolutionary relationship between counterparts.
We studied whether acquisition of bacteria by nestling hoopoes occurs early or later in their development, which may clarify the importance of vertical transmission relative to that of horizontal transmission of bacterial communities. Symbiotic bacteria are present in the dark secretions of the UG (referred to here as the UGS) of hoopoes, which are produced by incubating/brooding adult females and nestlings, but have not been found in the white secretions produced by males and nonbreeding adult females (14, 33). Bacterial symbionts are likely responsible for particularities such as brown color, malodor, and antimicrobial properties (14, 33, 34). The UG microbial community in adult females could be fixed during their lives, or, alternatively, new symbionts could colonize their UG, which would result in the presence of different microbial communities in the course of successive breeding seasons. On the other hand, nestlings may acquire symbionts vertically from mothers and/or horizontally from nest remains of previous reproduction events of con- or heterospecifics. Previous studies have shown that the dominant group of cultivable bacteria in the UGS of hoopoes belongs to the genus Enterococcus (14, 35, 36) and that some isolated strains from hoopoes produce compounds with strong antagonistic activity against other bacterial species (15, 16). Enterococci are common bacteria that can be found in soil and plants (37, 38) or in the gastrointestinal tract of birds (39, 40) and could therefore be easily acquired from different sources. Moreover, it is known that the antimicrobial properties of different Enterococcus bacterial strains harbored by hoopoes differ and that those with broader antimicrobial spectra are the most common in the UGS of hoopoes (35, 36). The mechanisms responsible for acquisition of symbionts with the highest potential benefits are unknown, and thus it is an ideal system for exploring the importance of vertical transmission relative to that of horizontal transmission in explaining the symbiotic microbiota.
We studied the mode of acquisition of UG symbiotic bacteria in hoopoes by means of two experiments. The first consisted of cross-fostering recently hatched nestlings between pairs of hoopoe nests. We predicted that if offspring acquired their uropygial bacteria in the egg or at hatching (hypothesis 1; Table 1), their bacterial community should resemble that of the nest of origin (prediction 1.1 [P1.1]; Table 1), and that the diversity and prevalence of bacterial genotypes and species should be more similar among genetically related nestlings that grew up in different nests than among unrelated nestlings sharing a nest (P1.2; Table 1). In addition, the bacterial community of cross-fostered nestlings should be more similar to that of their biological mother than to that of their foster mother (P1.3; Table 1). Moreover, if bacteria are acquired and established at early stages and no other bacteria can incorporate during development, the community of symbionts should remain constant over time (P1.4; Table 1), while the levels of bacterial diversity should be higher for experimental nests (i.e., with nestlings from two different mothers) than for control nests if the two communities are mixed (P1.5; Table 1), or they should be similar if they compete and if only one community is established in the gland (P1.6; Table 1).
TABLE 1.
Framework of predictions tested to elucidate the mode of acquisition of bacterial symbionts from the hoopoe's uropygial glanda
| Parameter | Prediction |
||
|---|---|---|---|
| Early transmission (H1) | Late transmission (H2) | Both transmissions (H3) | |
| Relative influence of nests in bacterial community acquisition by cross-fostered nestlings | Nest of origin most influential (P1.1) | Nest of rearing most influential (P2.1) | Nest of origin and nest of rearing equally influential (P3.1) |
| Similarity of bacterial communities among nestlings | Higher with biological siblings reared in different nests (P1.2) | Higher with foster siblings reared in the same nest (P2.2) | Similar with biological and foster siblings (P3.2) |
| Similarity of bacterial communities among nestlings and adult females | Higher to that of the biological mother than to that of the foster mother (P1.3) | Higher to that of the foster mother than to that of the biological mother (P2.3) | Bacteria shared with both mothers (P3.3) |
| Bacterial community change with time | Would remain constant (P1.4) | Could be modified (P2.4) | Could obtain bacteria from different environments and keep the bacterial community from the original nest (P3.4) |
| Diversity of strains and species in nests | Higher in cross-fostering nests if the two communities are mixed together (P1.5) or similar if the two communities compete and only one can become established (P1.6) | Similar in natural and exptl nests (P2.5) | Higher in cross-fostering nests if the two communities are mixed together (P3.5) or similar if the two communities compete and only one can become established (P3.6) |
H, hypothesis; P, prediction.
Alternatively, if nestlings acquire symbionts of the UG later during growth (hypothesis 2), the bacterial community of cross-fostered nestlings should represent that of the nest of rearing (P2.1; Table 1). In this case, symbiont communities of genetic and foster siblings reared in the same nest should be similar to each other (P2.2; Table 1) and to that of the rearing mother (P2.3; Table 1). Moreover, if bacteria colonize the UG of hoopoes throughout the nesting period, bacterial communities should change with age (P2.4; Table 1), while bacterial diversities in experimental and control nests should be similar if cross-fostered nestlings do not take with them bacteria acquired before the experiment (P2.5; Table 1). In addition, the two transmissions could also occur simultaneously (hypothesis 3; Table 1).
Previous scenarios assume that hoopoes acquire symbiotic bacteria from nest environments, but, due to bacterial ubiquity, it is also possible that innate properties of the UG of the nestlings allow the establishment of the symbiotic bacteria when the nestlings are reared under artificial conditions. To test this assumption, we performed another experiment which consisted of preventing contact of hatchlings with mothers, siblings, or nests (i.e., experimental incubation in the laboratory). We expected that the UG of nestlings reared outside the hoopoe nests would also be colonized by bacteria similar to those found for nestlings in natural hoopoe nests. Those approaches together will help to elucidate the symbiotic relationship between hoopoes and the bacteria living in their uropygial glands.
MATERIALS AND METHODS
Field work and bacterial sampling.
The study was performed during the 2003, 2005, and 2006 breeding seasons in the Hoya de Guadix (southeastern Spain), where around 400 nest boxes (interior height, 350 mm; interior width, 180 mm; interior depth, 210 mm; hole height, 240 mm; hole diameter, 55 mm) had been installed to follow the whole reproduction cycle of hoopoes (see reference 33 for more details) among other bird species.
The UGS of nestling hoopoes were sampled at the age of ringing (19 to 21 days after hatching). To explore possible variations in relation with age, some individual nestlings (n = 15) were also sampled during the first days of producing secretion (i.e., 8 to 11 days after hatching). Adult females were sampled at the beginning of incubation, after laying ended (n = 20). To estimate whether the composition of the bacterial community in adult females changes over the course of their stay in the nest, we also sampled five of them at the end of the incubation period. In four cases, we sampled the UGS of a particular adult female from different reproductive attempts, either in the same breeding season (1 case) or in two different years (3 cases). With these samples, we investigated the variation in bacterial communities hosted during different breeding attempts.
UGS was extracted by inserting the tip of a micropipette (Finpipette; 1 to 10 μl) directly through the UG external opening and pipetting several times until the papilla in which the secretion is stored was emptied (33). To reduce the probability of contamination, we wore new latex gloves for each extraction. We had previously wiped the feathers and skin around the UG with 96% ethanol and let the ethanol dry before inserting the micropipette. Then, each UGS sample was placed in a sterile Eppendorf tube (1.5 ml) and stored at 4°C until used.
In 2006, we performed a cross-fostering experiment in which two experimental nestlings that were 3 to 4 days old (with no secretion and the UG starting development) were exchanged with another two nestlings of the same age from another nest (n = 22). In addition to the cross-fostering experiment, in 2003 and 2006, we collected 9 hoopoe eggs from 8 clutches and incubated them artificially in the laboratory. These were placed in the incubator 2 days before the estimated hatching date. Within the first 3 h of life, these 9 hatchlings were introduced into nests of great tits (Parus major) with the rest of the tit nestlings, where they were fed by tit parents until fledging.
Finally, 5 nestlings (4 that were 2 to 6 days old from 1 nest and 1 that was 1 or 2 days old from another) without completely developed UG that were from partially depredated nests were reared by researchers in a nest box (similar to those in the field, but this box had not been used before) in the laboratory. Hoopoes do not build nests and do not take material to the boxes, so we did not put any material into the nest box. UGS samples were collected 15 days later.
Colony isolation and clustering.
UGS was diluted 1:5 in sterilized distilled water, and 5 μl of the solution was spread on tryptic soy agar (TSA; Scharlau, Barcelona, Spain). Plates were incubated aerobically at 32 to 37°C for 24 h. After growing, five colonies taken randomly from TSA were isolated from each sample and transferred to brain heart infusion (BHI; Scharlau, Barcelona, Spain) liquid medium and incubated for 24 h at 37°C. Then, 1 ml of the culture was centrifuged and the pellet was kept at −20°C until DNA extraction. A total of 644 colonies were isolated from 134 hoopoe individuals; 123 colonies belonged to 20 adult females from 24 clutches and 521 colonies to 114 nestlings from 46 (24 control and 22 experimental) nests.
Bacterial genomic DNA from pure cultures was extracted following a modification of the “salting-out” procedure (41). To type the isolates from each sample, the randomly amplified polymorphic DNA (RAPD) method was used (42). The RAPD-PCR technique is used as a rapid and reliable method for intra- and interspecific differentiation of bacterial groups. This has been found to be an efficient method for typing large numbers of isolated colonies to the species and strain levels (see, e.g., reference 43). Although there is some variability in the fingerprints obtained by RAPD technique, good reproducibility is achieved under controlled laboratory conditions (44). Our molecular analyses were all performed under identical laboratory conditions. Reactions were performed in a final volume of 50 μl, which included 5 μl of 10× Taq reaction buffer [75 mM Tris HCl (pH 9.0), 50 mM KCl, 20 mM (NH4)2SO4], 5 μl of 3 mM MgCl2, 2 μl of 400 μM deoxynucleoside triphosphates (dNTPs), 5 μl of 1 μM M13 primer (5′-GAGGGTGGCGGTTCT-3′), 1 U of MBL DNA polymerase, and 5 μl of template DNA. Amplifications were made in an iCycler 170-8720 thermocycler (Bio-Rad, Hercules, CA), and the reaction conditions were as follows: a denaturing step of 94°C for 1 min, 35 cycles of 94°C for 1 min, 40°C for 20 s (0.6°C/s ramp), and 72°C for 80 s, and a final elongation step at 72°C for 5 min.
Amplified DNA fragments were analyzed by electrophoresis through 1.5% agarose gels at 30 V for 16 h in 1× TAE buffer (40 mM Tris-acetate, 2.5 mM EDTA, pH 8) and revealed in ethidium bromide (0.5 μg/ml). A 1-kbp ladder (Biotools, Madrid, Spain) was used as a molecular size standard. Gels were photographed on a UV transillumination table (VilberLourmat, Marne-la-Vallée, France). Fingerprint pattern images were analyzed with Fingerprinting II Informatix Software 2000 (Bio-Rad, Hercules, CA). To cluster all the strains in RAPD groups, we used similarity coefficients based on the Pearson's product moment correlation coefficient to build dendrograms based on the unweighted-pair group method using average linkages (UPGMA). RAPD and cluster analyses were repeated with 34 randomly selected samples to establish the level of strength identity (i.e., RAPD group) and to check for variations due to the use of different RAPD gels. All the isolates with more than the 80% similarity were clustered in the same RAPD group and were thus considered to represent the same strain. This threshold is broadly used because it correlates with strain or species identity (e.g., 44, 45, 46).
Identification of isolates.
At least one isolate from each RAPD group was selected for sequencing, although in some groups more than one isolate was selected. The sequenced fragment was around 700 bp of the 16S rRNA gene, including variable regions V1 to V4, which allows the molecular identification of bacterial strains. The PCR was carried out using a 50-μl (total volume) mixture containing 5 μl of 10× Taq reaction buffer, 10 μl of 5× TaqEnhancer, 3 μl of 25 mM Cl2Mg, 2 μl of 0.4 mM dNTPs, 2 μl of 20 pM primer WO1 (5′-AGAGTTTGATC[A/C]TGGCTC-3′) and the same amount of primer WO12 (5′-TACGCATTTCACC[G/T]CTACA-3′), 1 U of Eppendorf Master Taqpolymerase, and 1 μl of template DNA. The amplification consisted of an initial denaturing step of 94°C for 4 min followed by amplification using 30 cycles of 30 s at 94°C, 30 s at 50°C, and 60 s at 72°C and a final extension at 72°C for 2 min. PCR products were purified with a Perfectprep gel cleanup kit (Eppendorf, Hamburg, Germany). The sequence of the 16S rRNA gene was determined by using CEQ 2000 dye terminator cycle sequencing with a Quick Start kit (Beckman Coulter, Brea, CA) according to the manufacturer's instructions. The resultant sequence was analyzed with a CEQ DNA analysis system (version 4.0). To identify the species of isolated strains, a search for homology to the DNA sequence was made using the BLAST algorithm (47) available at the National Center for Biotechnology Information (NCBI), from the “16S ribosomal DNA sequences (Bacteria and Archaea)” database, optimized for the “highly similar sequences” (Megablast). A target sequence was assigned to the species with the highest identity value (>98% in all the cases). A total of 172 sequences resulted, and they were deposited in the GenBank database (see below for accession numbers).
Statistical analyses.
To identify factors explaining the variability of the bacterial communities of the UG of hoopoes (predictions 1.1, 2.1, and 3.1; Table 1), binary pairwise distance matrices were constructed for strains and species identities as follows: 0 (i.e., no distance) indicated that two samples belonged to the same strain (i.e., more than 80% similarity) or species, and 1 (i.e., maximum distance) indicated that they belonged to different strains or species. Both types of matrices were considered dependent variables. We also built binary pairwise distance matrices for all the individuals and nests (i.e., with a value of 0 assigned when the two isolates belonged to the same individual or nest and a value of 1 assigned when the two isolates did not belong to the same individual or nest), which were used as the explanatory variables in the analyses. In the case of the isolates from experimental nests, two types of matrices were constructed, one for the original and the other for the rearing nest. Analyses were performed separately for adult females, nestlings that grew in control nests, and nestlings in cross-fostering nests. The association among matrices was analyzed by multiple regression on distance matrices (MRM) with the ecodist package (48) in R environment 2.13.1 (49). Significance was estimated by the Monte Carlo procedure after 10,000 permutations.
Individual differences in bacterial communities with respect to their UGS (predictions 1.2, 1.3, 2.2, 2.3, 3.2, and 3.3; Table 1) were estimated by using the function vegdist in the “vegan” package (50) implemented in R environment 2.13.1 (49), which calculates a dissimilarity index for pairs of individuals by considering coincidences in the prevalences of strains and species. The index values ranged from 0 to 1 and were transformed to similarity values (1 minus the distance value) before the analyses. Comparisons of similarity values of bacterial communities were performed with nonparametric analysis (Mann-Whitney U tests) in Statistica 7.0 (Dell Software).
The Shannon diversity indices of strains and species for individuals and nests (predictions 1.5, 1.6, 2.5, 3.5, and 3.6; Table 1) were also estimated in the R “vegan” package (47) as follows:
| (1) |
where pi is the proportion (p) of species or strains (i) related to all the isolates of each individual/nest and S is the number of species or strains. Differences in Shannon's diversity values estimated for individuals within different groups were also explored by means of nonparametric statistics (Kruskal-Wallis tests).
In addition, to assess whether the frequencies of each of the detected bacterial species differed among ages (for adult females and nestlings) (predictions 1.4, 2.4, and 3.4; Table 1), we used log-linear analyses with χ2 tests. Also, we estimated the overall probabilities associated with nestlings and adult females in terms of differences in frequencies of bacterial species by using χ2 test results and the associated P values and the following formula (52):
| (2) |
where k is 2 times the number of performed statistical analyses and p is the P value of the i analysis. Furthermore, we applied the false-discovery-rate (FDR) correction to establish the appropriate Q values, which were the calculated P values after the FDR correction (53).
Nucleotide sequence accession numbers.
The 172 sequences determined in this work were deposited in the GenBank database under accession numbers KC481273 to KC481322 and KF303342 to KF303460.
RESULTS
Prevalences of strains and species.
The 644 microbial colonies from hoopoes were clustered into 105 RAPD groups (i.e., strains). The number of isolates within each RAPD group was highly variable (range, 1 to 74; median, 3; standard deviation [SD], 10.86). In 78 RAPD groups, the clustered colonies were only from nestlings (n = 275); 12 RAPD groups included only colonies from adult females (n = 34), and the remaining 15 RAPD groups included isolates from adult females (n = 89) as well as nestlings (n = 246). Notably, those 15 (14.29%) RAPD groups, which included colonies isolated from UGS of adult females and nestlings, comprised 52% (n = 335 colonies) of the isolates.
We were able to identify most of the 644 isolates, 93.17% to the species level and 0.93% more to the genus level. The vast majority of the isolates identified belonged to the Enterococcus genus (91.77%), the most frequently identified species being E. faecalis (grouping 37 strains) followed by E. faecium (8 strains), E. avium (4 strains), E. durans (5 strains), E. mundtii (13 strains), E. casseliflavus (13 strains), and E. gallinarum (4 strains) (Table 2). The 15 additional isolates were identified as Stenotrophomonas rhizophila (3 strains), Aerococcus urinaeequi (4 strains), Streptococcus salivarius (1 strain), and Staphylococcus saprophyticus (1 strain) (Table 2).
TABLE 2.
Frequencies of identified bacterial species and nonidentified Enterococcus spp. in adult females and nestlings and comparisons among thema
| Bacterial species | Adult females (n = 115) |
Nestlings (n = 491) |
Difference and FDR correction |
||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | χ2 | P | Q | |
| Enterococcus faecalis | 49 | 42.6 | 222 | 45.2 | 0.26 | 0.61 | 0.61 |
| Enterococcus faecium | 4 | 3.5 | 85 | 17.3 | 14.23 | 0.0002 | 0.001 |
| Enterococcus avium | 46 | 40.0 | 19 | 3.9 | 127.02 | <0.0001 | 0.0001 |
| Enterococcus durans | 2 | 1.7 | 61 | 12.4 | 11.42 | 0.0007 | 0.002 |
| Enterococcus mundtii | 6 | 5.2 | 44 | 9.0 | 1.73 | 0.19 | 0.26 |
| Enterococcus casseliflavus | 1 | 0.9 | 39 | 7.9 | 7.56 | 0.006 | 0.011 |
| Enterococcus gallinarum | 0 | 0 | 7 | 1.4 | 1.18 | 0.28 | 0.31 |
| Stenotrophomonas rhizophila | 3 | 2.6 | 4 | 0.8 | 2.63 | 0.11 | 0.17 |
| Aerococcus urinaeequi | 0 | 0 | 5 | 1.0 | 1.18 | 0.28 | 0.31 |
| Streptococcus salivarus | 2 | 1.7 | 0 | 0 | 8.57 | 0.003 | 0.006 |
| Staphylococcus saprophyticus | 1 | 0.9 | 0 | 0 | 4.28 | 0.003 | 0.006 |
| Enterococcus spp. | 1 | 0.9 | 5 | 1.0 | |||
n and %, number and percentage of individuals in which each species is present, respectively. Results of the χ2 tests and associated P values, as well as the adjusted P values (Q) after the FDR corrections (significant values are in bold), are given for all the known species.
At the genus level, Enterococcus species were the most frequent in isolates from adult females (94.78%, n = 123) and nestlings (98.17%, n = 521). At the species level, the frequencies of bacterial species in samples from nestlings and adult females differed (χ2df = 22 = 101.62, P < 0.001), supporting hypothesis 2 (prediction 2.4; Table 1), i.e., the hypothesis that communities of bacterial strains vary among age groups. These differences were due mainly to the frequencies of E. faecium, E. avium, E. durans, E. casseliflavus, S. salivarius, and S. saprophyticus, whereas those of E. faecalis, E. mundtii, E. gallinarum, S. rhizophila, and A. urinaeequi in adult female and nestling samples did not differ after FDR correction (Table 2).
E. faecalis was also the most commonly detected species (76.09%) at the nest level, followed by E. faecium (34.78%), E. mundtii (32.61%), E. casseliflavus (26.09%), E. durans (23.91%), E. avium (17.39%), E. gallinarum (10.87%), A. urinaeequi (10.87%), and S. rhizophila (6.52%).
Factors explaining variations in frequencies of bacterial strains and species.
The communities of bacterial strains and species hosted in the glands of adult females differed among individuals (Table 3). In the case of nestlings in control nests, the identities corresponding to both the nests and the individuals from which colonies were isolated explained a significant proportion of the variations in the frequencies of bacterial strains; i.e., colonies belonging to the same individuals or nests grouped within the same strain with higher probability than those from different individuals or nests (Table 3). However, the nest but not the individual identity explained a significant proportion of variance of bacterial species identity, suggesting that nestlings of the same nest usually shared bacterial species but not necessarily the same strains.
TABLE 3.
Results of analysis of multiple regression on distance matrices for adult females and nestlings involved or not in cross-fostering experimentsa
| Hoopoe category | Bacterial category | Regression coefficient |
R2 | F | n | ||
|---|---|---|---|---|---|---|---|
| Indiv. | R-Nest | O-Nest | |||||
| Adult females | Species | 1.06** | NA | NA | 0.07 | 155.26** | 66 |
| Strains | 0.93** | NA | NA | 0.33 | 1209.47** | 70 | |
| Nestlings from control nests | Species | 0.27 | 0.59** | NA | 0.03 | 214.88** | 160 |
| Strains | 0.84** | 0.10** | NA | 0.16 | 1,416.07** | 172 | |
| Nestlings from cross-fostering nests | Species | 0.94** | 0.17** | 0.05* | 0.01 | 174.79** | 273 |
| Strains | 0.72** | 0.15** | −0.005 | 0.04 | 563.81** | 290 | |
Regression coefficient data were obtained from comparisons of the identifications of species or strains to the individual (Indiv.), nest-of-rearing (R-Nest), and nest-of-origin (O-Nest, only in cross-fostering experiments) matrices. The rest of the statistical coefficients (R2 and F) correspond to the overall model, while n data represent the sizes of the samples (i.e., the number of isolated colonies in each category). NA, not applicable. A tendency to significance is marked with a single asterisk (*) (P = 0.067), while significant P values are marked with double asterisks (**) (all P < 0.0001).
In experimental nests, the identity of nestling individuals within the same nest explained a significant proportion of variance at the levels of both the bacterial species and the bacterial strains isolated from their glands, supporting hypothesis 2 (prediction 2.1; Table 1). The identity of the nest of origin tended to explain a significant proportion of additional variance in species diversity (P = 0.067; Table 3) but not in strain diversity, in partial support of hypothesis 3 (prediction 3.1; Table 1). These results suggest that the bacterial communities in the UGS of foster siblings reared in the same nest tend to be similar and that siblings developing in different nests tend to share similar bacterial species.
Diversity of bacterial strains and species.
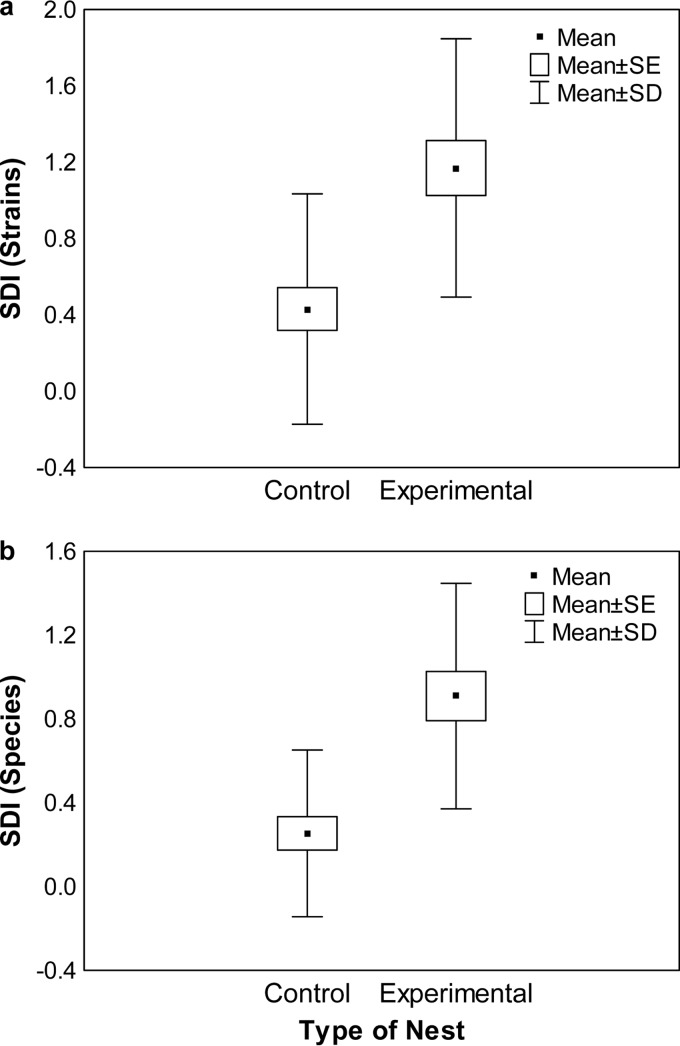
The Shannon diversity indices of bacteria did not differ significantly between the UGS of adult females and the UGS of nestlings from control nests with respect to either species (Kruskal-Wallis [K-W] test H1,65 = 0.43; P = 0.51) or strains (K-W H1,65 = 3.56, P = 0.06). Interestingly, the UGS of all nestlings growing in cross-fostered nests had greater species diversity than those of the control (K-W H1,119 = 4.45; P = 0.03), although there were no differences for bacterial strains (K-W H1,119 = 0.14; P = 0.71). Within experimental nests, fostered and native individuals did not differ in the diversities of bacterial strains (K-W H1,75 = 0.16; P = 0.69) or species (K-W H1,119 = 0.08; P = 0.77) found in their UGS. Finally, experimental nests (considering all nestlings in each nest) were more diverse than control nests for both strains (K-W H1,51 = 12.54; P < 0.001) and species (K-W H1,46 = 15.63; P < 0.001) (Fig. 1), suggesting that the inclusion of foreign nestlings increases the whole bacterial diversity of the nests (hypothesis 1 and 3, predictions 1.5 and 3.5; Table 1).
FIG 1.
Shannon diversity index (SDI) data for bacterial strains (a) and species (b) from uropygial secretions of hoopoes in control and experimental (cross-fostering) nests, including both exchanged and native nestlings. SE, standard error.
Similarity among bacterial communities in cross-fostered nests.
Experimental nestlings did not differ in similarity indices for bacterial communities in comparisons of those that shared the nest of hatching (i.e., siblings that hatched in the same nests) or of those that shared the nest of rearing (unrelated nestlings that grew in the same nest) (Mann-Whitney U test; Z = 0.05, P = 0.96, n = 94 comparisons) (Fig. 2). The similarity indices estimated for nestlings that shared just one nest (origin or rearing) and for those that shared both nests (siblings that hatched and were reared in the same nest) (Mann-Whitney U test; Z = 0.40, P = 0.69, n = 150 comparisons) (Fig. 2) did not differ. However, the values corresponding to the similarity of bacterial communities of UGS of nestlings that had never been in contact with each other were lower than the values estimated for nestlings that shared nest of origin or nest of rearing or both (Mann-Whitney U test; Z = −5.15, P < 0.001, n = 1,998 comparisons) (Fig. 2). These results support hypothesis 3 (bacterial acquisition before and after the cross-fostering experiment) and prediction 3.2 (Table 1).
FIG 2.
Similarity index (means ± standard errors of the means) data from comparisons among cross-fostered nestlings' bacterial communities in the UGS with that of other nestlings that shared the nest of origin (biological siblings), the rearing nest (foster siblings), or both nests (biological siblings that were cross-fostered to the same rearing nest) or of those with which they were never in contact.
Similarity among bacterial communities of adult females and nestlings.
In 11 (5 control and 6 experimental) nests, we recorded data from the adult female and the offspring. The mother and biological offspring harbored bacteria from the same species in 8 (3 control and 5 experimental) nests. For experimental nests, adult females and adopted nestlings shared bacterial species in one nest (prediction 2.3; Table 1). Bacterial strains detected in adult females were also detected in biological offspring in five experimental nests, but no coincidences were found with adopted nestlings (see Table 4 for more details). Cross-fostered nestlings and their biological mother had no coincidence when both strains and species of bacterial isolates were considered (n = 4 nests).
TABLE 4.
Coincidence of bacterial strains and species among nestlings and females in each nesta
| Nest | Type | Control nestlings (biological sons) |
Cross-fostered nestlings (adopted) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of nestlings | No. of bacterial colonies | % coincident bacterial strains | % coincident bacterial species | No. of nestlings | No. of bacterial colonies | % coincident bacterial strains | % coincident bacterial species | ||
| 1 | C | 2 | 10 | 0 | 0 | ||||
| 2 | E | 4 | 17 | 0 | 0 | 2 | 16 | 0 | 0 |
| 3 | E | 6 | 16 | 0b | 40 | 2 | 8 | 0b | 12.5 |
| 4 | E | 4 | 17 | 47.06 | 70.59 | ||||
| 5 | C | 5 | 22 | 4.54 | 100 | ||||
| 6 | C | 1 | 5 | 0 | 100 | ||||
| 7 | E | 3 | 13 | 0 | 88.89 | 1 | 2 | 0 | 0 |
| 8 | C | 1 | 5 | 0 | —c | ||||
| 9 | C | 1 | 5 | 100 | 100 | ||||
| 10 | E | 3 | 11 | 18.18 | 54.54 | 2 | 8 | 0 | 0 |
| 11 | E | 2 | 11 | 45.45 | 54.54 | 1 | 9 | 0 | 0d |
The quantities of autochthonous and adopted nestlings, as well as the numbers of bacterial colonies isolated, are given. Nest types: C, control; E, experimental.
Totals of 31.25% and 12.5% of the isolates from biological and foster sons belonged to RAPD groups, respectively, with similarity to isolates from females of 79%.
—, the bacterial species of the females were unidentified.
A total of 5 colonies from 1 nestling were unidentified at the species level.
Changes over a breeding attempt.
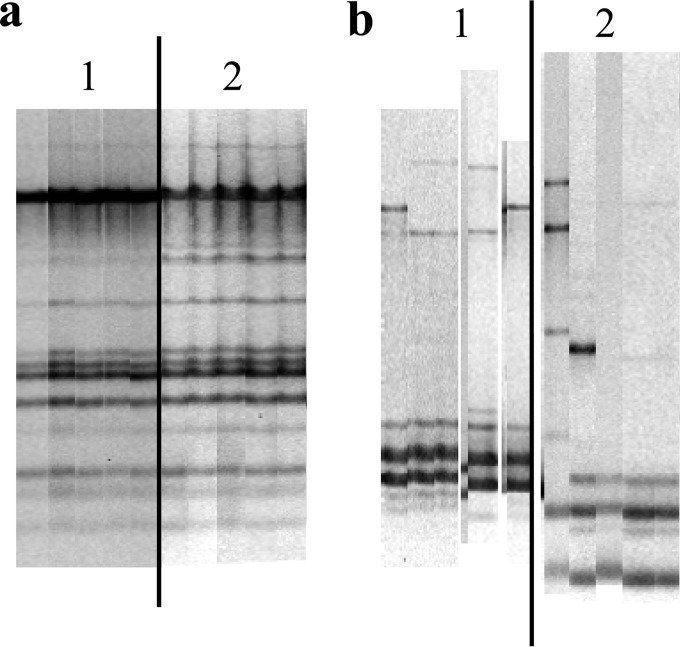
In four of the five adult females with UGS sampled twice during the same reproductive attempt (at the beginning and end of incubation), isolated colonies belonged to the same species and strains (Fig. 3a) (prediction 1.4; Table 1). The strain isolated from the fifth adult female had changed from the first to the second sampling, but the two strains were clustered very close to each other (similarity >72%) and belonged to the same species.
FIG 3.
Results of RAPD gel analyses of isolates from two adult females (a and b), with 5 isolates from each of the two sampling events (lanes 1 and 2). In adult female a, colonies were isolated from the same reproductive attempt (at the beginning and at the end of the incubation period), and the similarity among the RAPD profiles from the two sampling events is >95%. In adult female b, samples were taken in two consecutive years (and thus represent different reproductive attempts), and the similarities among the RAPD profiles from the two sampling events are <20%.
For nestlings that were sampled twice during their stay in the nest (n = 15), the detected bacterial species were the same in nine individuals, while the strains did not differ in seven nestlings in the two samplings (Table 5). However, no coincidences were found for four nestlings (Table 5). Thus, nestlings tend to keep the same strains, but new strains and species can colonize the UG (prediction 3.4; Table 1).
TABLE 5.
Coincidence between two sampling events in the same nestlings during their stay in the nesta
| Individual | Type | No. of bacterial colonies isolated |
No. (%) of coincident bacterial strains | No. (%) of coincident bacterial species | |
|---|---|---|---|---|---|
| Sampling 1 | Sampling 2 | ||||
| 1 | E | 5 | 1 | 0 | 0 |
| 2 | E | 5 | 1 | 0 | 0 |
| 3 | E | 5 | 5 | 0 | 5 (100) |
| 4 | C | 5 | 3 | 1 (33.33) | 1 (33.33) |
| 5 | E | 5 | 5 | 0 | 4 (80) |
| 6 | C | 2 | 5 | 0 | 0 |
| 7 | C | 5 | 1 | 1 (100) | 1 (100) |
| 8 | E | 4 | 5 | 4 (80) | 4 (80) |
| 9 | C | 5 | 1 | 0 | —b |
| 10 | E | 5 | 5 | 4 (80) | 4 (80) |
| 11 | E | 5 | 5 | 3 (66.66) | 5 (100) |
| 12 | C | 3 | 5 | 0 | 0 |
| 13 | C | 1 | 5 | 0 | — |
| 14 | E | 2 | 5 | 5 (100) | 5 (100) |
| 15 | C | 1 | 5 | 1 (20) | 2 (40) |
The first sampling (sampling 1) was performed around day 10 after the hatching date (when the birds start to produce the UGS), and the second (sampling 2) was performed around day 20 (before the birds fledged). The columns of the coincidences reflect the numbers and percentages (in parentheses) of the bacterial strains and species found in the second sampling that were also found in the first. Nestling types: C, control (nestlings that were not changed from the nest); E, experimental (adopted nestlings).
—, the bacterial species from the first sampling were not identified.
Changes between breeding attempts.
The strains detected in the UGS of three of four adult females (one of them bred in the same nest) during different reproductive attempts changed (Fig. 3b) (prediction 2.4; Table 1). Two of five (40%) detected strains appeared during two consecutive years (in different nests) in the UG of the four analyzed adult females. At the level of bacterial species, two of the four adult females harbored exactly the same species during the 2 years of sampling; one female maintained the species in two of five isolates (40%), and the community in the fourth had completely changed.
Bacterial community of nestlings that did not grow in hoopoe nests.
The UGS of nestlings that were never in contact with hoopoes (n = 9) apparently had the same characteristics (i.e., color and odor) as those from nestlings growing in hoopoe nests. Moreover, UGS of nestlings from artificially incubated eggs also harbored bacteria, all of those detected (42 isolates) being of the genus Enterococcus (E. faecalis, 59.52%; E. faecium, 19.05%; E. durans, 11.9%; E. mundtii, 2.38%; 3 Enterococcus spp.).
However, the isolated bacterial strains of nestlings that did not hatch and develop in hoopoe nests did not coincide with those in the isolates from their siblings that stayed in original nests. The 25 isolates from the UGS of the 5 nestlings that were reared in the laboratory belonged to the species E. faecalis, and only one nestling did not share the strains with the rest. In the other 4 nestlings, 19 isolates belonged to 2 different strains, and 1 isolate was different from the rest. Curiously, the bacterial community in the nestling that came from a different nest (see Materials and Methods) matched the communities in the isolates from the 5 foster siblings. Due to the predation of their relatives, we could not compare their bacterial communities.
DISCUSSION
The data show that hoopoes acquire and incorporate UG bacteria throughout the reproductive cycle and across years throughout their lives, given the following findings: (i) the frequencies of different bacterial species of adult females differed from those estimated for nestlings; (ii) the bacterial communities of some individuals did not remain constant throughout the nesting period; and (iii) the communities of adult females changed between reproductive events. Furthermore, the results also indicated the capacity of hoopoes to acquire colonies horizontally from the environment, because (i) the identity of the rearing nest instead of that of the nest of origin explained the variations of symbiont strains of cross-fostered nestlings and (ii) nestlings reared outside the hoopoe nests also hosted Enterococcus spp. which did not coincide with those of their relatives. Thus, our results show that acquisition of some symbiotic bacteria took place after the cross-fostering experiment was performed and that communities of adult females can also change over time. Given that nestling glands develop several days after hatching, the results suggest that symbionts can colonize the UG from external sources once developed. In the case of adult females, the UG undergoes drastic morphological and physiological changes at the beginning of each breeding attempt (30). Our results indicate that the bacterial communities in the UG differ among different breeding seasons.
Also, we found some evidence suggesting that the UG is colonized by bacteria before being completely developed and open to the outside environment, given that (i) the bacterial diversity in the UGS of nestlings was higher in experimental nests than in control nests (implying that cross-fostered nestlings carried some symbiont strains from their original nests), (ii) data corresponding to the nest of origin tended to explain variations in the frequencies of symbiont strains of cross-fostered nestling, and (iii) the indices of similarity between biological and foster siblings did not differ but were significantly higher than those of nonrelated individuals.
Below, we discuss such results in scenarios of vertical and horizontal transmission of symbionts, which may potentially be adjusted to particular environmental conditions.
Horizontal versus vertical transmission.
The cross-fostering experiment indicated that foster nestlings can obtain their bacteria several days after hatching from external sources, such as nest mates, foster mother, or the new nest, which could be interpreted as horizontal transmission since symbionts do not originate from the biological mother. Environmental influence in bacterial community assemblages has been previously found in cloacal samples (54, 55). In a natural scenario (nestlings growing in their original nests), nestlings could acquire bacteria from the genetic mother through nest contamination by strains from the adult female (56). Actually, some of the breeding behavior of adult female hoopoes such as egg besmearing with UGS (14, 33) or touching offspring with the bill impregnated with secretion may increase the probability of vertical transmission. Such behavior should result in enterococcal symbionts from their gland being abundant in the nest environment, thus easily reaching the developing UG of hatchlings. Accordingly, some nestlings shared bacterial strains and species with the adult females (see Table 4). However, adult females are not the only source of bacteria in nests, and symbionts may be shared if both adult females and nestlings acquired them from nest material instead, implying horizontal transmission. In addition, the host genotype could also influence bacterial assemblages (57), of which related individuals would tend to host similar strains. Cross-fostering experiments accompanied by manipulations of the availability of bacteria in nests are necessary to distinguish among these possibilities.
In any case, nestlings that were never in contact with adults or nests of hoopoes developed the same type of UGS as wild chicks, and Enterococcus spp. prevailed in their bacterial community. In great tit nests, there are many possible sources of Enterococcus, such as the nest material or nestling excrement. In addition, adult tits could also harbor bacteria in their UG and disseminate them in the nest environment (e.g., by besmearing the eggs), which would result in the transmission to hoopoe nestlings.
Thus, vertical transmission from parents after hatching is not necessary for hoopoes to acquire bacterial symbionts in their UG. Although we cannot rule out the possibility that the bacteria pass through the eggshell and infect the embryo before egg removal from hoopoe nests, this finding indicates the possibility that the UG is adapted to host enterococci, which are otherwise quite abundant in nature (e.g., 37, 38, 58).
Adaptable symbiont communities.
The antimicrobial effectiveness of symbionts in hoopoe glands depends on their genotypes (36). Bacterial symbionts of adult females changed from 1 year to another; thus, the adult females may be able to collect from the environment the bacterial strain most appropriate to particular conditions, given that the pathogenic environments may differ among years and/or nests (29). This plasticity in acquisition of symbionts may be advantageous for hoopoes.
We can only speculate on the mechanisms of possible selection of symbionts (59), but the differences that we detected between the bacterial communities of adult females and those of nestlings may have been due to differences in UG characteristics, hormone concentrations, or other unknown factors. In accordance with a possible process of symbiont selection and the effect of environments determining their bacterial community, we found that E. faecalis, the species with the greatest antibiotic activity (35), is the culturable bacterium that is prevalent in hoopoe glands (36). Thus, it is possible that the community of symbionts in the UG of hoopoes is a plastic trait conditioned by particularities of the nest environment each year.
The relatively low variation of symbiont taxa found indicates the possibility of coevolution between hoopoes and symbionts, leading to a better alignment of interests between the counterparts (60). Despite our having used a nonselective and highly nutritive medium (TSA), some 97% of bacteria cultured from hoopoe UG belonged to just one genus (Enterococcus), while no enterococci were detected in bacteria isolated in TSA from the plumage of birds (61). Although we cannot rule out the possible presence of bacteria that cannot be cultured, our results imply a certain specialization in hosting enterococci. Earlier studies have shown the relationship of bacteriocin production to the antimicrobial capacity of isolated colonies and also the association between genotypes and antimicrobial properties (15, 35, 36). Moreover, we know that hoopoes derive fitness advantages from harboring these symbionts (14, 21), suggesting a mutualistic relationship between enterococci and hoopoes. Accordingly, here we show that hoopoe nestlings reared under laboratory conditions or in nests of great tits managed to acquire antimicrobial enterococci in their UG.
Final remarks.
There are several examples in the animal kingdom of mutualistic relationships in which symbionts of relatively low diversity are acquired from the environment, as in the cases of nematodes, squids, and annelids (reviewed in reference 23). One of the leading issues in evolutionary biology is how microbiomes are established and maintained within their hosts due to the multitude of benefits they receive from symbiotic microbiota (23, 62). The reduced variability in symbiotic bacteria detected in the above-mentioned invertebrate taxa, as well as in nestlings and adult female hoopoes, could result from individuals acquiring a single or a few bacteria but also from selection acting on a more diverse bacterial community within individual hosts. Hosts, by modifying the environment of the more or less specialized places where symbionts develop, may favor the growth of beneficial bacteria and restrict the proliferation of pathogenic ones (59). Beneficial bacteria may be resistant to antimicrobial defenses of hosts, which may fuel interference competition among symbiotic bacteria, favoring the recruitment of the most effective antibiotic-producing strains (59). Indeed, bacteria isolated from hoopoe UGS produce bacteriocins with a wide inhibition spectrum (15, 35). These compounds establish several interaction networks and help maintain the biodiversity of microbial communities (63). Thus, hoopoes may harbor bacterial competition within the uropygial gland, resulting in communities with major antimicrobial properties (16, 35). In this study, we detected a strong environmental effect determining the bacterial community, suggesting that some of these mechanisms explain the relatively low diversity of the culturable bacterial communities of hoopoes.
Our results therefore suggest that hoopoes start to acquire bacteria soon after hatching and continue to do so during their stay in the nest and that, although bacterial colonization and establishment of the symbiosis with enterococci are innate in this species, special characteristics of the UGS of each individual could shape the bacterial community. Moreover, we show experimental evidence of horizontal and vertical transmission of bacterial strains. This mixed mode of transmission to nestlings could serve to gain simultaneously the advantages provided by horizontal transmission (23) and vertical transmission (22, 29) of beneficial microorganisms.
ACKNOWLEDGMENTS
This work was funded by the Spanish national government (Ministerio de Ciencia e Innovación), FEDER (CGL2010-19233–C03-01/BOS and CGL2010-19233-C03-0), and by Junta de Andalucía (P09-RNM-4557). M.R.-R. was funded by a postdoctoral fellowship from Junta de Andalucía (project RNM-02177) and by the JAE program.
We also acknowledge the three anonymous referees for their comments, which helped to improve the manuscript. David Nesbitt corrected the English grammar of the manuscript.
Footnotes
Published ahead of print 29 August 2014
REFERENCES
- 1.Maynard-Smith J, Szathmáry E. 1995. The major transitions in evolution. Oxford University Press, New York, NY. [Google Scholar]
- 2.Margulis L. 1993. Origins of species - acquired genomes and individuality. Biosystems 31:121–125. 10.1016/0303-2647(93)90039-F. [DOI] [PubMed] [Google Scholar]
- 3.Douglas AE. 1994. Symbiotic associations. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 4.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A. 110:3229–3236. 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindquist N, Barber PH, Weisz JB. 2005. Episymbiotic microbes as food and defence for marine isopods: unique symbioses in a hostile environment. Proc. Biol. Sci. 272:1209–1216. 10.1098/rspb.2005.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFall-Ngai MJ. 2002. Unseen forces: the influence of bacteria on animal development. Dev. Biol. 242:1–14. 10.1006/dbio.2001.0522. [DOI] [PubMed] [Google Scholar]
- 7.Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114:185–190. 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soler JJ, Martín-Vivaldi M, Peralta-Sánchez JM, Ruiz-Rodríguez M. 2010. Antibiotic-producing bacteria as a possible defence of birds against pathogenic microorganisms. Open Ornithol. J. 3:93–100. 10.2174/1874453201003010093. [DOI] [Google Scholar]
- 9.Gil-Turnes MS, Hay ME, Fenical W. 1989. Symbiotic marine-bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246:116–118. 10.1126/science.2781297. [DOI] [PubMed] [Google Scholar]
- 10.Barbieri E, Paster BJ, Hughes D, Zurek L, Moser DP, Teske A, Sogin ML. 2001. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (Cephalopoda:Loliginidae). Environ. Microbiol. 3:151–167. 10.1046/j.1462-2920.2001.00172.x. [DOI] [PubMed] [Google Scholar]
- 11.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. U. S. A. 100:1803–1807. 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215. 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 13.Banning J, Weddle A, Wahl GWI, Simon MA, Lauer A, Walters RL, Harris RN. 2008. Antifungal skin bacteria, embryonic survival, and communal nesting in four-toed salamanders, Hemydactylium scutatum. Oecologia 156:423–429. 10.1007/s00442-008-1002-5. [DOI] [PubMed] [Google Scholar]
- 14.Soler JJ, Martín-Vivaldi M, Ruiz-Rodríguez M, Valdivia E, Martín-Platero AM, Martínez-Bueno M, Peralta-Sánchez JM, Méndez M. 2008. Symbiotic association between hoopoes and antibiotic-producing bacteria that live in their uropygial gland. Funct. Ecol. 22:864–871. 10.1111/j.1365-2435.2008.01448.x. [DOI] [Google Scholar]
- 15.Martín-Platero AM, Valdivia E, Ruiz-Rodríguez M, Soler JJ, Martín-Vivaldi M, Maqueda M, Martínez-Bueno M. 2006. Characterization of antimicrobial substances produced by Enterococcus faecalis MRR 10–3, isolated from the uropygial gland of the hoopoe (Upupa epops). Appl. Environ. Microbiol. 72:4245–4249. 10.1128/AEM.02940-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín-Vivaldi M, Pena A, Peralta-Sánchez JM, Sánchez L, Ananou S, Ruiz-Rodríguez M, Soler JJ. 2010. Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc. Biol. Sci. 277:123–130. 10.1098/rspb.2009.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law-Brown J, Meyers PR. 2003. Enterococcus phoeniculicola sp. nov., a novel member of the enterococci isolated from the uropygial gland of the Red-billed Woodhoopoe, Phoeniculus purpureus. Int. J. Syst. Evol. Microbiol. 53:683–685. 10.1099/ijs.0.02334-0. [DOI] [PubMed] [Google Scholar]
- 18.Jacob J, Ziswiler V. 1982. The uropygial gland, p 359–362 In Farner DS, King JR, Parker KC. (ed), Avian biology, vol II Academic Press, London, United Kingdom. [Google Scholar]
- 19.Jacob J. 1978. Hydrocarbon and multibranched ester waxes from uropygial gland secretion of grebes (Podicipediformes). J. Lipid Res. 19:148–153. [PubMed] [Google Scholar]
- 20.Martín-Vivaldi M, Soler JJ, Peralta-Sánchez JM, Arco L, Martín-Platero AM, Martínez-Bueno M, Ruiz-Rodríguez M, Valdivia E. 30 April 2014. Special structures of hoopoe eggshells enhance the adhesion of symbiont-carrying uropygial secretion that increase hatching success. J. Anim. Ecol. 10.1111/1365-2656.12243. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Rodríguez M, Valdivia E, Soler JJ, Martín-Vivaldi M, Martín-Platero AM, Martínez-Bueno M. 2009. Symbiotic bacteria living in the hoopoe's uropygial gland prevent feather degradation. J. Exp. Biol. 212:3621–3626. 10.1242/jeb.031336. [DOI] [PubMed] [Google Scholar]
- 22.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83. 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 23.Chaston J, Goodrich-Blair H. 2010. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol. Rev. 34:41–58. 10.1111/j.1574-6976.2009.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran NA, Wernegreen JJ. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321–326. 10.1016/S0169-5347(00)01902-9. [DOI] [PubMed] [Google Scholar]
- 25.Darby AC, Douglas AE. 2003. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 69:4403–4407. 10.1128/AEM.69.8.4403-4407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. U. S. A. 97:10231–10235. 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herre EA. 1993. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science 259:1442–1445. 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- 28.Frank SA. 1996. Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 263:339–344. 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- 29.Douglas AE. 1998. Host benefit and the evolution of specialization in symbiosis. Heredity 81:599–603. [Google Scholar]
- 30.Poulsen M, Bot ANM, Currie CR, Nielsen MG, Boomsma JJ. 2003. Within-colony transmission and the cost of a mutualistic bacterium in the leaf-cutting ant Acromyrmex octospinosus. Funct. Ecol. 17:260–269. 10.1046/j.1365-2435.2003.00726.x. [DOI] [Google Scholar]
- 31.Cafaro MJ, Currie CR. 2005. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can. J. Microbiol. 51:441–446. 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- 32.Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu. Rev. Entomol. 49:71–92. 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 33.Martín-Vivaldi M, Ruiz-Rodríguez M, Soler JJ, Peralta-Sánchez JM, Méndez M, Valdivia E, Martín-Platero AM, Martínez-Bueno M. 2009. Seasonal, sexual and developmental differences in hoopoe Upupa epops preen gland morphology and secretions: evidence for a role of bacteria. J. Avian Biol. 40:191–205. 10.1111/j.1600-048X.2009.04393.x. [DOI] [Google Scholar]
- 34.Soler JJ, Martín-Vivaldi M, Peralta-Sánchez JM, Arco L, Juárez-García-Pelayo N. 11 July 2014. Hoopoes color their eggs with antimicrobial uropygial secretions. Naturwissenschaften. 10.1007/s00114-014-1201-3. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Rodríguez M, Martínez-Bueno M, Martín-Vivaldi M, Valdivia E, Soler JJ. 2013. Bacteriocins with a broader antimicrobial spectrum prevail in enterococcal symbionts isolated from the hoopoe's uropygial gland. FEMS Microbiol. Ecol. 85:495–502. 10.1111/1574-6941.12138. [DOI] [PubMed] [Google Scholar]
- 36.Ruiz-Rodríguez M, Valdivia E, Martín-Vivaldi M, Martín-Platero AM, Martínez-Bueno M, Méndez M, Peralta-Sánchez JM, Soler JJ. 2012. Antimicrobial activity and genetic profile of enteroccoci isolated from hoopoes uropygial gland. PLoS One 7:e41843. 10.1371/journal.pone.0041843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller T, Ulrich A, Ott EM, Muller M. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91:268–278. 10.1046/j.1365-2672.2001.01373.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen YS, Yanagida F, Shinohara T. 2005. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbiol. 40:195–200. 10.1111/j.1472-765X.2005.01653.x. [DOI] [PubMed] [Google Scholar]
- 39.Potti J, Moreno J, Yorio P, Briones V, Garcia-Borboroglu P, Villar S, Ballesteros C. 2002. Bacteria divert resources from growth for magellanic penguin chicks. Ecol. Lett. 5:709–714. 10.1046/j.1461-0248.2002.00375.x. [DOI] [Google Scholar]
- 40.Moreno J, Briones V, Merino S, Ballesteros C, Sanz JJ, Tomas G. 2003. Beneficial effects of cloacal bacteria on growth and fledging size in nestling Pied Flycatchers (Ficedula hypoleuca) in Spain. Auk 120:784–790. 10.1642/0004-8038(2003)120[0784:BEOCBO]2.0.CO;2. [DOI] [Google Scholar]
- 41.Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M. 2007. Fast, convenient and economical method for isolating genomic DNA from lactic acid bacteria using a modification of the protein “salting-out” procedure. Anal. Biochem. 366:102–104. 10.1016/j.ab.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 42.Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6535. 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzsimons NA, Cogan TM, Condon S, Beresford T. 2001. Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J. Appl. Microbiol. 90:600–608. 10.1046/j.1365-2672.2001.01285.x. [DOI] [PubMed] [Google Scholar]
- 44.Rossetti L, Giraffa G. 2005. Rapid identification of dairy lactic acid bacteria by M13-generated, RAPD-PCR fingerprint databases. J. Microbiol. Methods 63:135–144. 10.1016/j.mimet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Morandi S, Brasca M, Andrighetto C, Lombardi A, Lodi R. 2006. Technological and molecular characterisation of enterococci isolated from north-west Italian dairy products. Int. Dairy J. 16:867–875. 10.1016/j.idairyj.2005.09.005. [DOI] [Google Scholar]
- 46.Psoni L, Kotzamanides C, Andrighetto C, Lombardi A, Tzanetakis N, Litopoulou-Tzanetaki E. 2006. Genotypic and phenotypic heterogeneity in Enterococcus isolates from Batzos, a raw goat milk cheese. Int. J. Food Microbiol. 109:109–120. 10.1016/j.ijfoodmicro.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22:1–19 http://www.jstatsoft.org/v22/i07/paper. [Google Scholar]
- 49.R Core Team. 2013. R: a language and environment for statistical computing. R foundation for statistical computing. http://www.r-project.org/.
- 50.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHM, Wagner H. 2011. Vegan: community ecology R package, version 1.17-10 http://cran.r-project.org/web/packages/vegan/index.html.
- 51. Reference deleted.
- 52.Sokal RR, Rohlf FJ. 1995. Biometry. W. H. Freeman, San Francisco, CA. [Google Scholar]
- 53.Pike N. 2011. Using false discovery rates for multiple comparisons in ecology and evolution. Methods Ecol. Evol. 2:278–282. 10.1111/j.2041-210X.2010.00061.x. [DOI] [Google Scholar]
- 54.Lucas FS, Heeb P. 2005. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caerulus nestlings. J. Avian Biol. 36:510–516. 10.1111/j.0908-8857.2005.03479.x. [DOI] [Google Scholar]
- 55.Klomp JE, Murphy MT, Bartos Smith S, McKay JE, Ferrera I, Reysenbach AL. 2008. Cloacal microbial communities of female spotted towhees Pipilo maculatus: microgeographic variation and individual sources of variability. J. Avian Biol. 39:530–538. 10.1111/j.0908-8857.2008.04333.x. [DOI] [Google Scholar]
- 56.Banks JC, Cary SC, Hogg ID. 2009. The phylogeography of Adelie penguin faecal flora. Environ. Microbiol. 11:577–588. 10.1111/j.1462-2920.2008.01816.x. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Rodríguez M, Lucas FS, Heeb P, Soler JJ. 2009. Differences in intestinal microbiota between avian brood parasites and their hosts. Biol. J. Linn. Soc. Lond. 96:406–414. 10.1111/j.1095-8312.2008.01127.x. [DOI] [Google Scholar]
- 58.Marcinek H, Wirth R, Muscholl-Silberhorn A, Gauer M. 1998. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 64:626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheuring I, Yu DW. 2012. How to assemble a beneficial microbiome in three easy steps. Ecol. Lett. 15:1300–1307. 10.1111/j.1461-0248.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herre EA, Knowlton N, Mueller UG, Rehner SA. 1999. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14:49–53. 10.1016/S0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 61.Burtt EH, Ichida JM. 1999. Occurrence of feather-degrading bacilli in the plumage of birds. Auk 116:364–372. 10.2307/4089371. [DOI] [Google Scholar]
- 62.Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn A, Solan M, van der Gast CJ, Young J. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384–392. 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 63.Maróti G, Kereszt A, Kondorosi E, Mergaert P. 2011. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 162:363–374. 10.1016/j.resmic.2011.02.005. [DOI] [PubMed] [Google Scholar]