ABSTRACT
Membrane fusion in herpesviruses requires viral glycoproteins (g) gB and gH/gL. While gB is considered the actual fusion protein but is nonfusogenic per se, the function of gH/gL remains enigmatic. Crystal structures for different gH homologs are strikingly similar despite only moderate amino acid sequence conservation. A highly conserved sequence motif comprises the residues serine-proline-cysteine corresponding to positions 437 to 439 in pseudorabies virus (PrV) gH. The PrV-gH structure shows that proline438 induces bending at the end of an alpha-helix, thereby placing cysteine404 and cysteine439 in juxtaposition to allow formation of a strictly conserved disulfide bond. However, PrV vaccine strain Bartha unexpectedly carries a serine at this conserved position. To test the influence of this substitution, we constructed different gH chimeras carrying proline or serine at position 438 in gH derived from either PrV strain Kaplan or strain Bartha. Mutants expressing gH with serine438 showed reduced fusion activity in transient-fusion assays and during infection, with delayed penetration kinetics and a small-plaque phenotype which indicates that proline438 is important for efficient fusion. A more drastic effect was observed when disulfide bond formation was completely blocked by mutation of cysteine404 to serine. Although PrV expressing gHC404S was viable, plaque size and penetration kinetics were drastically reduced. Alteration of serine438 to proline in gH of strain Bartha enhanced cell-to-cell spread and penetration kinetics, but restoration of full activity required additional alteration of aspartic acid to valine at position 59.
IMPORTANCE The role of the gH/gL complex in herpesvirus membrane fusion is still unclear. Structural studies predicted a critical role for proline438 in PrV gH to allow the formation of a conserved disulfide bond and correct protein folding. Functional analyses within this study corroborated these structural predictions: mutation of this residue resulted in a drastic impairment of membrane fusion kinetics not only in vitro in transient transfection-fusion assays but also during virus infection. Elimination of formation of the disulfide bond yielded the same phenotype in transient assays but had a more drastic effect on virus replication. Thus, our studies add important information to structure-function analyses of herpesvirus gH.
INTRODUCTION
In contrast to many other known viral fusion systems which rely on one or two proteins, herpesvirus membrane fusion requires the concerted action of at least three viral glycoproteins (g). In the prototypical alphaherpesvirus, herpes simplex virus 1 (HSV-1), binding of gD to one of its specific host cell receptors induces a conformational change that, in turn, triggers the actual fusion process by signaling to the gH/gL heterodimer and/or gB (1–4). gB and gH/gL are essential for membrane fusion during entry as well as for direct viral cell-to-cell spread and are therefore considered the core fusion machinery of herpesviruses. While gB shows typical features of class III fusion proteins, it is not sufficient by itself to induce efficient membrane fusion but requires the presence of gH/gL. The function of this complex, whose structure does not resemble that of any known fusion protein (5–7), still remains elusive (reviewed in reference 8).
Herpesvirus gH molecules are type I transmembrane proteins with several N-glycosylation consensus sequences in the ectodomain and a short cytoplasmic tail following the C-terminal membrane anchor. Amino acid sequences are only moderately conserved across the Herpesviridae and are largely limited to several regions within the C-terminal half of the ectodomain (5, 9). One of the conserved sequence motifs corresponds to amino acids 437-SPCAVSLRRDL-447 in gH of the alphaherpesvirus pseudorabies virus (PrV), encompassing a highly conserved serine-proline-cysteine motif, and a second region close to the predicted transmembrane region, 620-LFPNGTV-626, comprising a conserved N-glycosylation site (9). The preservation of several other cysteine residues throughout the ectodomain already implied similarity of the tertiary structures (9), which was confirmed by comparison of the crystal structures of the herpes simplex virus 2 (HSV-2) and Epstein-Barr virus (EBV) gH/gL complexes as well as the PrV gH core domain (5–7).
All known gH homologs form a heterodimeric complex with gL, a small glycoprotein (156 amino acids [aa] in PrV) which depends on gH interaction for membrane association and virion incorporation (10–13). While in many herpesviruses gH also requires gL for correct processing and transport, PrV, bovine herpesvirus 4 (BoHV-4), and murine herpesvirus 4 (MuHV-4) gH is also incorporated into virions in the absence of gL (14–16). However, in contrast to BoHV-4 and MuHV-4, where gL is not strictly required for fusion, PrV requires gL for entry of free virions (15), as do all other herpesviruses studied so far. However, direct viral cell-to-cell transmission, which relies on a similar, but not identical, set of proteins, occurs in PrV even in the absence of gL, although at drastically decreased efficiency (15). This minimal capability for direct viral cell-to-cell spread has been used for serially passaging PrV-ΔgL in tissue culture cells, ultimately resulting in a rescuant, PrV-ΔgLPass, which efficiently replicated without gL (17). In this revertant, a hybrid protein is expressed from a fused gene consisting of the receptor binding domain of gD joined in-frame to an N-terminally truncated gH core fragment that lacks the putative gL binding domain (Fig. 1). This gDH hybrid protein is sufficient to induce membrane fusion in combination with gB and complements the defect of mutants simultaneously lacking gD, gH, and gL (17, 18), indicating that membrane fusion can be accomplished without gL and with a minimal set of two herpesviral proteins.
FIG 1.
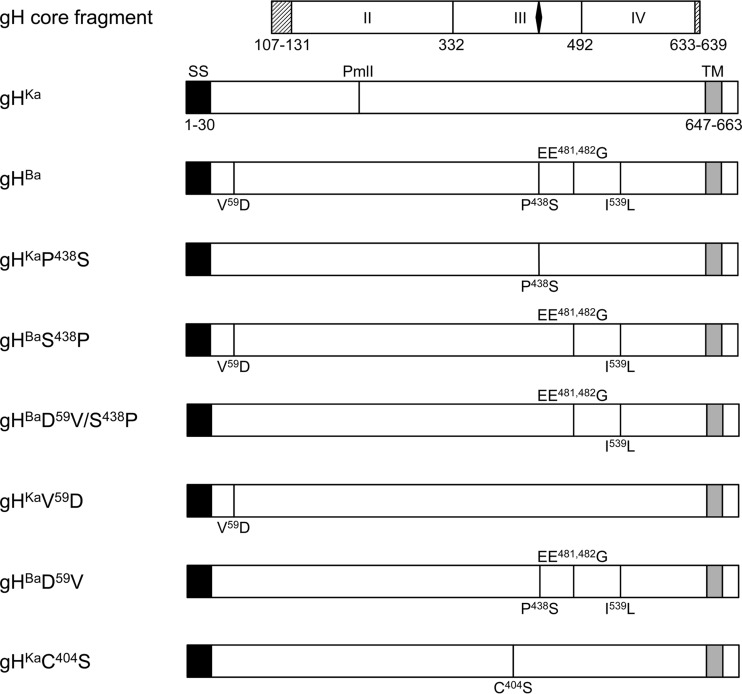
Schematic diagram of gH. The gH open reading frames are shown as rectangles. The core fragment of gH present in the gDH hybrid protein, which had been used for crystallization, is presented above. The domains are labeled with roman numbers with boundaries shown. The black diamond indicates the location of the SPC motif. The shaded boxes left and right indicate residues that were not resolved in the structure. The predicted signal sequence (SS; aa 1 to 30) is highlighted in black; the transmembrane domain (TM; aa 647 to 663) is indicated in gray. The location of the PmlI restriction site used for cloning is shown. Amino acid differences between PrV-Ba gH and PrV-Ka gH are given in one-letter code with the corresponding position.
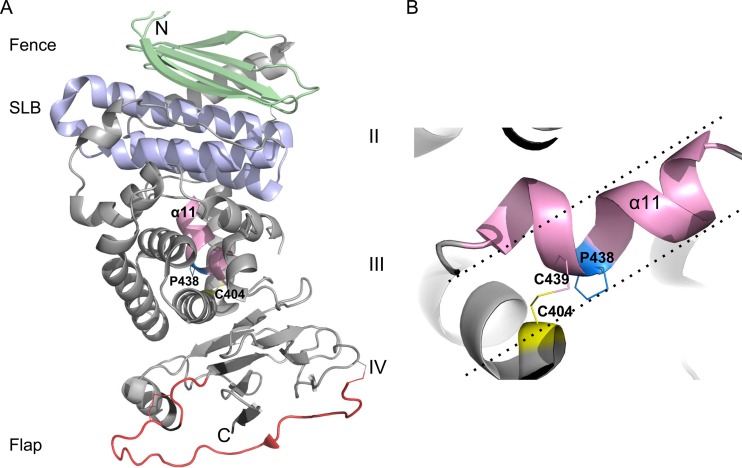
From the crystal structures of the ectodomains of HSV-2 and EBV gH/gL (6, 7), as well as the core fragment of PrV gH, which was derived from the gDH hybrid protein (5), four distinct domains could be defined. While domain I, which is involved in binding gL, is missing in the PrV gH structure (Fig. 1), domains II to IV are largely superimposable among the three proteins. Domain II contains two conserved structures, an antiparallel beta-sheet designated the “fence” and an elongated antiparallel 3-helix bundle which shows structural similarities to a domain of cellular syntaxins and has accordingly been designated the “syntaxin-like bundle” (SLB) (5) (Fig. 2). Domain III consists mainly of alpha-helices and harbors one of the conserved sequence motifs, including residues serine437, proline438, and cysteine439 (numbering for PrV gH), with proline and cysteine invariably present in all herpesvirus gH homologs. Proline438 induces bending at the end of an alpha-helix, so that cysteine439 is brought into proximity to cysteine404 and is able to form a strictly conserved disulfide bond with cysteine404 (Fig. 2) (5). The membrane-proximal domain IV, which shows the highest conservation, is composed of two four-stranded beta-sheets connected by the so-called “flap” region. This hydrophilic flap, which is followed by a conserved N-glycosylation site, covers a hydrophobic patch, which was shown to play an important role during fusion. It is speculated that movement of the flap, which masks the underlying hydrophobic patch, promotes membrane fusion (5, 19).
FIG 2.
PrV gH structural features. (A) Crystal structure of the PrV gH core domain (PDB code 2XQY) with the fence, SLB, and flap region colored in light green, blue, and red, respectively. The domains are labeled with roman numbers. The N and C termini of the molecule are designated with the letters N and C. Alpha-helix 11 (α11), located in domain III and carrying the SPC motif, is highlighted in pink. (B) Zoomed-in view of the SPC motif as seen from the back of the molecule shown in panel A. Replacing Pro438 with any other amino acid would result in an unbent alpha-helix 11, which would project as indicated by the dotted lines. Bending of the helix is clearly important for residues Cys439 and Cys404 to become juxtaposed for formation of the disulfide bond.
Recently, the complete genome sequence of PrV strain Bartha (PrV-Ba), an attenuated strain widely used as a vaccine against PrV infection in pigs, was deciphered and compared to wild-type strains PrV-Kaplan (PrV-Ka) and PrV-Becker (20). While the attenuating mutations in PrV-Ba were mapped earlier by marker rescue experiments (21–24) and were shown to be a result of a large deletion within the unique short region comprising genes encoding gI, gE, pUS9, and pUS2, as well as subtle amino acid changes in pUL21 and gC, comparison of the deduced amino acid sequences revealed a total of 46 open reading frames with nonsilent mutations (20). Among these mutations, an alteration in gH within the highly conserved sequence motif 437serine-proline-cysteine439 was identified. Interestingly, PrV-Ba gH contains a serine instead of the helix-bending proline (20).
Since proline438 is strictly conserved throughout other herpesvirus gH homologs, as is the disulfide bridge between cysteine404 and cysteine439 (5, 9), we were interested in analyzing the effect of this alteration on gH function. To this end, we generated PrV gH variants expressing either serine or proline at position 438 as well as a serine mutant at position 404 and analyzed them in transient transfection-fusion assays and, after rescue of the respective recombinant viruses in PrV-Ka, in one-step growth, penetration kinetics, and cell-to-cell transmission assays.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney cells (RK13) and RK13-gHgL cells (25) were grown in Dulbecco‘s modified Eagle's minimum essential medium (MEM) supplemented with 10% fetal calf serum at 37°C in the presence of 5% CO2. Recombinants used in this study were derived from pPrV-ΔgGG, comprising the PrV-Ka genomic DNA cloned as a bacterial artificial chromosome (BAC) with the mini-F plasmid sequence and a green fluorescent protein (GFP) expression cassette at the nonessential gG gene locus (19). In pPrV-ΔgHABF (for simplicity, here designated pPrV-ΔgH), gH codons from 51 to the end were deleted (S. Böhm, E. Eckroth, M. Backovic, B. G. Klupp, F. A. Rey, T. C. Mettenleiter, and W. Fuchs, unpublished data). pPrV-gHKa was generated by reinsertion of the wild-type Kaplan gH (gHKa) together with a downstream-located kanamycin resistance gene (kan) for selection in Escherichia coli and was used here as the wild-type control. For comparison, a BAC clone (here designated pPrV-Ba) derived from PrV-Bartha (pPrV-BaΔgGG [26]), generated after deletion and reinsertion of the Bartha gH gene (gHBa), including the downstream kan, was used. The gH deletion mutants were grown in RK13-gHgL cells, while all the other recombinants were propagated on RK13 cells.
Expression plasmids and gH mutagenesis.
Plasmids pcDNA-gB, pcDNA-gH, and pcDNA-gL express the full-length gB, gH, and gL of PrV-Ka, respectively, under the control of the human cytomegalovirus immediate early promoter/enhancer complex (18). pcDNA-gD expressing PrV-Ka gD was generated after PCR amplification using primers gD-fw and gD-rv (Table 1) and PrV-Ka genomic DNA as a template. The 1.2-kb PCR product was cloned into pcDNA3 (Invitrogen) after cleavage with EcoRI using the corresponding restriction sites added by the primers. pcDNA-gHBa was generated using primers DH-3 and DHrev1 (Table 1) on genomic PrV-Ba DNA (27). The 2.1-kb PCR product was cloned into pcDNA3 using the EcoRI and XbaI restriction enzyme sites provided by the primers. Correct amplification and insertion were verified by sequencing and indirect immunofluorescence assay after transfection (data not shown). For sequencing, a BigDye Terminator cycle sequencing kit (Applied Biosystems) was used, and products were run on an ABI 3130 genetic analyzer (Applied Biosystems).
TABLE 1.
Primers used in this study
| Primer | Sequencea (5′–3′) | Locationb (nt) |
|---|---|---|
| gD-fw | CAC AGA ATT CAC CTG CCA GCG CCA TGC | 121131–121148 |
| gD-rv | CAC AGA ATT CCA TCG ACG CCG GTA CTG C | 122370–122353 |
| DH-3 | AGA ATT CAA AGT TTG CCG TGC CCG TC | 60748–60766 |
| DHrev1 | CAT CTA GAC ACG CGC ACG CAG AGA GT | 62873–62856 |
| P438S-fw | CGA CGT CCT CTC GTC GTG CGC CGT CTC | 62068–62094 |
| P438S-rv | GAG ACG GCG CAC GAC GAG AGG ACG TCG | 62094–62068 |
| S438P-fw | CGA CGT CCT CTC GCC GTG CGC CGT CTC | 62068–62094 |
| S438P-rv | GAG ACG GCG CAC GGC GAG AGG ACG TCG | 62094–62068 |
| C404S-fw | CGC ACC ACG GCC ATG TCC ACC GCG GAG CGC GCC | 61964–61996 |
| C404S-rv | GGC GCG CTC CGC GGT GGA CAT GGC CGT GGT GCG | 61996–61964 |
To introduce mutations into the gH sequence, a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) was used as described recently (28). Specific primers (Table 1) derived from the genomic sequence of PrV-Ka (GenBank accession no. JQ809328 [29]) or PrV-Ba (GenBank accession no. JF797217 [20]) were applied to generate pcDNA-gHKaP438S, pcDNA-gHKaC404S, pcDNA-gHBaS438P, and pcDNA-gHBaD59V/S438P (Fig. 1). pcDNA-gHKa or pcDNA-gHBa was used as a template, and correct mutagenesis was verified by sequencing. To generate pcDNA-gHKaV59D and pcDNA-gHBaD59V, we cleaved pcDNA-gHKa and pcDNA-gHBa with PmlI and XbaI, and the resulting 1.4-kb fragments were ligated into the respective other plasmid. All plasmids were sequenced and tested for gH expression by indirect immunofluorescence and Western blotting (data not shown).
Fusion assays.
Fusion activity was measured after transient expression in RK13 cells. For each assay, approximately 1.8 × 105 RK13 cells per well in 24-well culture dishes were seeded the day before transfection. Cells were then transfected with 125 ng of each plasmid expressing one of the gH variants, full-length gB, gD, and gL, as well as pEGFP-N1 (Clontech) as a marker in 50 μl of serum-free MEM using 1 μl polyethylenimine (PEI). After incubation for 15 min at room temperature, the mixture was added to the cells. pcDNA3 was used as a negative control, substituting for the gH expression plasmid in the transfection assays. Forty-eight hours posttransfection, cells were washed with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde for 20 min, and washed twice with PBS. To quantify the fusion activity, we evaluated 10 randomly selected fields of view in each well by counting the number of green-fluorescing syncytia and measuring the polykaryocyte area using a Nikon Eclipse Ti-S fluorescence microscope and Nikon NIS-Elements imaging software (Nikon). Syncytia were defined as cells with three or more nuclei. Total fusion activity was calculated by multiplication of the number of syncytia by the mean value of the syncytium area in each assay. The experiment was repeated three times, and the average values (values for assays with pcDNA-gHKa set as 100%) of the results with the corresponding standard deviations are given.
Generation of gH recombinants.
The transfer plasmids used for reinsertion of authentic, heterologous, or mutated gH genes into PrV-Ka were based on the expression plasmid pcDNA-gHKDE (Böhm et al., unpublished), comprising the full-length gHKa followed by kan and a DrdI restriction site. A 1.7-kbp KpnI-BamHI fragment covering most of the gH gene (corresponding to codons 88 to 662) was replaced with the corresponding fragments of pcDNA-gHBa, pcDNA-gHKaP438S, and pcDNA-gHKaC404S to generate pPrV-gHBaD59V, pPrV-gHKaP438S, and pPrV-gHKaC404S. The transfer plasmid for pPrV-gHKaV59D was generated after substitution of a 0.5-kbp SanDI-BsrGI fragment (comprising codons 42 to 216) in pcDNA-gHKDE by the corresponding fragment from pcDNA-gHBa. The triplet encoding serine 438 in gHBa was mutated to a proline codon by site-directed mutagenesis with primers S438P-fw and S438P-rv (Table 1), resulting in the transfer plasmid for pPrV-gHBaS438P. By recloning of the 1.7-kb KpnI/BamHI fragment from this plasmid into pcDNA-gHKDE, the transfer plasmid for pPrV-gHBaD59V/S438P was obtained. All plasmids were digested with DrdI, and the 3.5-kbp fragments containing the complete gH gene followed by kan were isolated and subsequently used for Red-mediated mutagenesis of pPrV-ΔgH (Böhm et al., unpublished). PrV recombinants were characterized by restriction analyses and Southern blot hybridization of genomic DNA. Correct mutagenesis was verified by PCR amplification and sequencing of the complete gH open reading frame (data not shown).
Western blotting.
For immunoblotting, RK13 cells were infected at a multiplicity of infection (MOI) of 3. Twenty-four hours after infection, cells and supernatants were harvested, pelleted by centrifugation, and washed twice with PBS. The pellet was lysed in sodium dodecyl sulfate (SDS) sample buffer, and the proteins were separated in SDS–10% polyacrylamide gels. After blotting onto nitrocellulose and blocking with 5% skimmed milk, membranes were incubated with rabbit sera specific for gH (17), capsid triplex component pUL38 (unpublished data), or a monoclonal mouse anti-gE antibody (17). Bound antibodies were detected by incubation with peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Dianova). Blots were analyzed using an imager (VersaDoc; Bio-Rad) after incubation with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
One-step growth kinetics.
For analysis of replication properties, RK13 cells were infected at an MOI of 3 and incubated on ice for 1 h. Then the inoculum was replaced by prewarmed medium and the cells were further incubated at 37°C. After 1 h of incubation at 37°C, nonpenetrated extracellular virus was inactivated by low-pH treatment (30). The cells were harvested at different times thereafter by scraping into the supernatant and were stored at −80°C. After thawing, the supernatant was titrated on RK13 cells. This assay was repeated two times, and mean values with the corresponding standard deviations were calculated.
Plaque assays.
To determine the plaque size, RK13 cells grown in 12-well culture dishes were infected with the different recombinants with approximately 100 PFU under plaque assay conditions and fixed with 3% paraformaldehyde 2 days postinfection (p.i.). The fluorescent area of 30 plaques was measured microscopically with a Nikon Eclipse Ti-S fluorescence microscope using Nikon NIS-Elements imaging software and was compared to the area measured for pPrV-gHKa, which was set as 100%. Mean values with standard deviations from three independent experiments are shown.
Penetration kinetics.
The kinetics of penetration of the different viruses was analyzed in RK13 cells. Cells grown in 12-well culture dishes were infected on ice with approximately 150 PFU of the different recombinants. After 1 h, the inoculum was replaced by prewarmed medium and the cells were incubated at 37°C; after 0, 5, 10, 20, and 40 min, the remaining extracellular virus was inactivated by low-pH treatment in one well, while a parallel well was used as a 100% penetration control and washed with PBS only. Two days p.i., cells were fixed and stained with crystal violet, and plaques were counted. To calculate the penetration rate, we compared the number of PFU surviving the low-pH inactivation to the number of plaques formed in the control. Mean values with standard deviations from three independent assays are given.
RESULTS
Sequencing of PrV-Bartha gH.
Since the published genome sequences of PrV-Ka generated by Szpara et al. (20) and our laboratory (29) differ at several positions in the coding regions, we first verified whether the Bartha strain used by us (Bartha K61) (27) comprises the same mutations in the gH open reading frame as those previously reported (20). To this end, we amplified the complete gH gene of PrV-Ba genomic DNA and sequenced it after cloning. Comparison of the deduced amino acid sequences revealed changes in PrV-Ba gH identical to those previously reported (20), namely, V59D, P438S, EE481,482G, and I539L (Fig. 1), thus verifying the proline-to-serine mutation at position 438 within the highly conserved sequence motif (5).
Serine at position 438 in PrV gH reduces fusion activity in transient transfection-fusion assays.
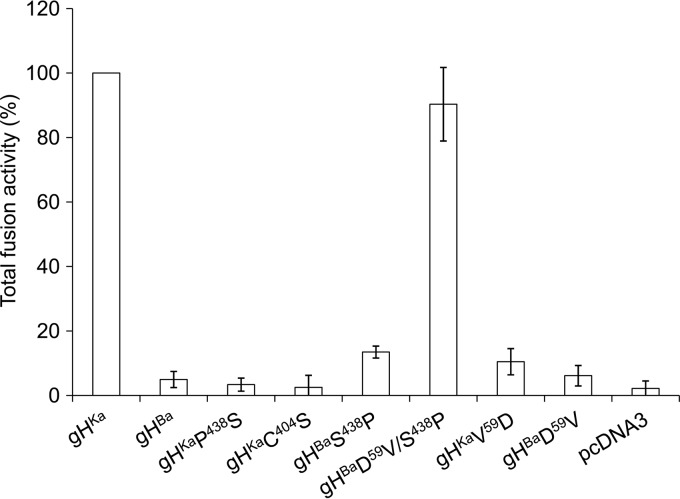
To analyze the influence of the mutation in gH in our transient transfection-fusion assay, RK13 cells were cotransfected with plasmids expressing full-length gB, gD, gL of PrV-Ka, and the different gH plasmids pcDNA-gHKa, pcDNA-gHBa, pcDNA-gHKaP438S, and pcDNA-gHBaS438P. pEGFP-N1 was included for easy visualization of transfected cells. Two days after transfection, syncytium formation was analyzed under a fluorescence microscope. Figure 3 shows the mean values for three independent assays with the corresponding standard deviations. Cotransfection of the plasmids expressing the native PrV-Ka glycoproteins resulted in approximately 60 syncytia in the 10 randomly selected fields of view, which was set as 100%. Substitution of proline438 with serine (gHKaP438S) led to a ca. 90% reduction of fusion activity resulting in activity similar to that observed in assays with gHBa and just slightly higher than in assays without any gH expression plasmid (pcDNA3), showing that proline at position 438 plays an important role for gH function during membrane fusion in these transient assays.
FIG 3.
Transient transfection-fusion assays. To test for fusion activity, RK13 cells were cotransfected with plasmids coding for gHKa, gHBa, or the respective gH mutants and plasmids encoding the full-length forms of gB, gD, and gL, as well as enhanced green fluorescent protein (EGFP) as a marker. The empty vector pcDNA3 served as a negative control, substituting for the gH expression plasmid. Total fusion activity (number of syncytia × syncytium area) in assays containing gHKa was set as 100%, and the corresponding standard deviations for three independent assays were calculated.
Bending of alpha-helix 11 in gH induced by proline438 was suggested to be important for formation of a highly conserved disulfide bond connecting cysteine404 and cysteine439 in PrV gH (Fig. 2) (5). To substantiate this assumption, we generated a Ka-gH expression plasmid with cysteine404 mutated to serine (gHKaC404S). In transient-fusion assays, gHKaC404S yielded reduced fusion activity similar to that of gHKaP438S (Fig. 3), suggesting that proline438 is indeed involved in mediating bridging between cysteine404 and cysteine439.
Restoring proline438 in gHBa (gHBaS438P) resulted in slightly enhanced fusion activity, which, however, did not reach the levels of fusion obtained with gHKa (Fig. 3). Thus, to test which other mutation(s) present in gHBa may influence its fusion activity, we altered other amino acids which differed between PrV-gHBa and PrV-gHKa (Fig. 1). The V59D substitution in gHKa (gHKaV59D) resulted in a drastic drop in activity, while restoring V59 in gHBa (gHBaD59V) showed only a slight increase in syncytium formation. However, when both positions in gHBa were altered to the wild-type PrV-Ka residues (gHBaD59V/S438P), fusion activity reached a level similar to that of native gHKa, indicating that besides proline438, valine59 plays an important role in the fusion process. Apparently, the other alterations (EE481,482G, I539L), which remained unchanged, had no significant effect on fusion (data not shown).
The glycosylation pattern is altered in gH mutants carrying serine438 or serine404.
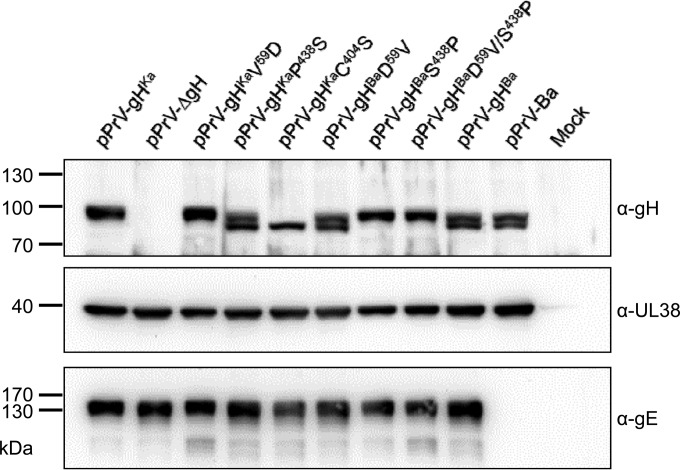
To test the influence of the amino acid substitutions during infection, we generated pPrV-Ka recombinants expressing the different gH proteins by rescuing pPrV-ΔgH. Expression of gH was tested by immunoblotting using monospecific anti-gH serum. As shown in Fig. 4, at 24 h after infection, in pPrV-gHKa-infected cells gH is detectable as a single protein species, indicating efficient processing by transfer through the Golgi apparatus, while in pPrV-Ba as well as in pPrV-gHBa, the PrV-Ka-derived mutant expressing gHBa, two gH species most likely representing immature, partially glycosylated and mature, completely glycosylated forms were detected. The same pattern was observed for all PrV-Ka mutants expressing gH with serine438 (gHKaP438S, gHBaD59V), while mutants with proline438 (gHKaV59D, gHBaS438P, and gHBaD59V/S438P) showed the wild-type pattern, indicating that serine438 influences protein processing. In pPrV-gHKaC404S-infected cells, only immature gH could be demonstrated, and no gH-specific signal was detected in pPrV-ΔgH-infected cells. It is notable that the mutations did not introduce an additional N-linked glycosylation site. The capsid protein pUL38, which served as a loading control, was present in similar amounts in all infected cell lysates, while gE was absent from the Bartha-derived recombinant pPrV-Ba.
FIG 4.
Western blotting. RK13 cells were infected with the indicated viruses for 24 h, harvested, and lysed. Proteins were separated on SDS–10% polyacrylamide gels, blotted, and incubated with monospecific antisera or monoclonal antibodies as indicated on the right. The locations and molecular masses (in kDa) of marker proteins are shown on the left.
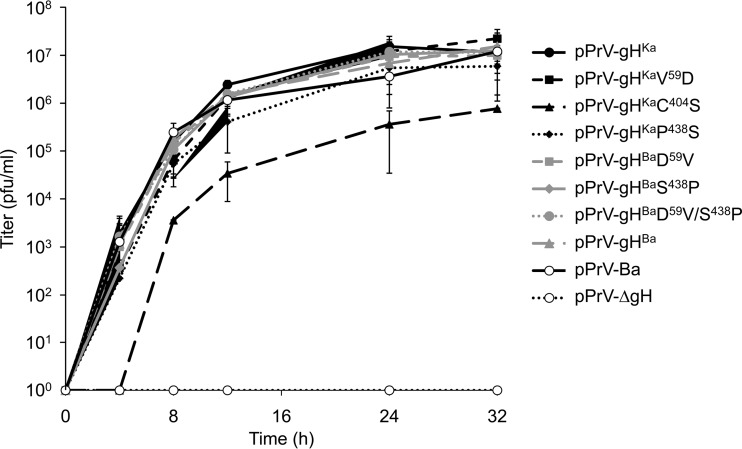
The proline438serine substitution in PrV gH has no influence on final viral titers.
To test for viral replication, RK13 cells were infected at an MOI of 3 with the different viruses. Cells and supernatants were harvested 0, 4, 8, 12, 24, and 32 h after infection, and titers were determined on RK13 cells. The mean values from two independent experiments are shown in Fig. 5. While the gH deletion mutant pPrV-ΔgH does not productively replicate on RK13 cells, titers of viruses expressing the different gH variants were comparable, demonstrating that the proline438serine substitution has no effect on production of infectious progeny. pPrV-gHKaC404S proved to be replication competent, but titers were approximately 10-fold lower, indicating that a complete block of formation of the disulfide bond has a greater effect than impairment by mutation of the neighboring proline.
FIG 5.
One-step growth kinetics. RK13 cells were infected with parental or mutant viruses at an MOI of 3. At the indicated times, cells and supernatants were harvested, and virus progeny titers on RK13 cells were determined. The mean values for two independent assays and the corresponding standard deviations are shown.
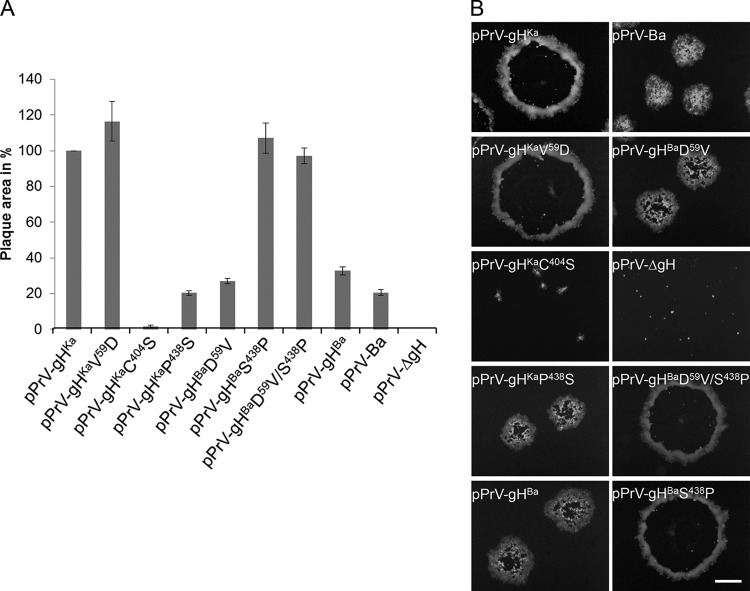
The proline438serine substitution in PrV gH affects direct viral cell-to-cell transmission.
Since gH is essential for direct viral cell-to-cell spread, plaque areas were measured 2 days after infection. PrV-Ba was already known to form small plaques, primarily due to a large deletion in the unique short region comprising, among others, the genes encoding gI and gE (22), which is similar for the BAC-derived mutant pPrV-Ba (Fig. 6). Expression of gHBa by PrV-Ka also resulted in a small-plaque phenotype. This was also found for all mutants expressing gH variants with serine438. In contrast, recombinants with proline438 formed plaques which were comparable to those of pPrV-gHKa, indicating that serine438 impairs, while proline at this position promotes, direct viral cell-to-cell spread. In contrast to the transient-fusion assays where valine at position 59 enhanced fusogenicity, no further increase could be observed in pPrV-gHBaD59V/S438P. pPrV-gHKaC404S produced only tiny foci of infected cells, congruent with reduced final viral titers after one-step replication. Infection of RK13 cells with pPrV-ΔgH resulted in only single fluorescent cells (Fig. 6B).
FIG 6.
Plaque formation of virus mutants. (A) Cell-to-cell spread was tested after infection of RK13 cells with the different viruses under plaque assay conditions. The plaque areas of 30 plaques for each virus were measured microscopically. The plaque size of PrV-Ka was set as 100%. Mean values and the corresponding standard deviations from three independent assays were plotted. (B) RK13 cells were infected with the indicated viruses. After 2 days, cells were fixed and fluorescent images were acquired with a fluorescence microscope (Nikon Eclipse Ti-S). In the negative control (pPrV-ΔgH), only single infected cells were observed. Bar = 400 μm.
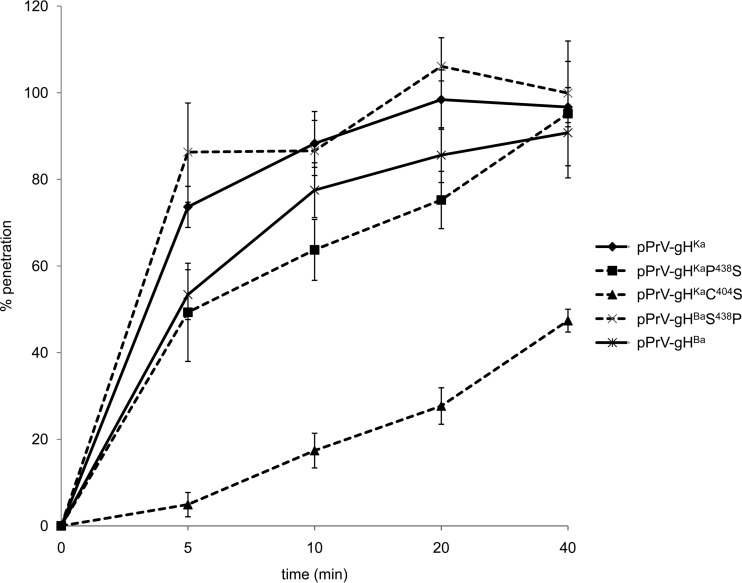
The gH proline438serine substitution delays penetration kinetics.
Penetration kinetics was analyzed to assess the influence of the proline438serine substitution on the rate of fusion during entry. While pPrV-gHKa entered cells extremely fast, with 80% of input virus protected from low-pH treatment after approximately 5 min, pPrV-gHBa reached this level only after approximately 20 min. The penetration kinetics of pPrV-gHKaP438S was as slow as that of pPrV-gHBa, indicating a significant influence of the proline438serine substitution on fusion kinetics during virus entry (Fig. 7). In contrast, the introduction of proline438 into gHBa restored penetration kinetics to rates comparable to those for pPrV-gHKa (Fig. 7). Penetration of pPrV-gHKaC404S was the slowest, but the virus was still able to enter cells and to initiate productive infection.
FIG 7.
Penetration kinetics. To analyze the rate of entry, RK13 cells were infected with approximately 150 PFU per well of the different viruses on ice. At 1 h p.i., the inoculum was replaced with prewarmed medium and the cells were incubated for the time periods indicated before inactivation of the remaining extracellular virus by low-pH treatment. Two days after infection, cells were fixed and stained with crystal violet. The rate of penetration was calculated as the number of PFU in low-pH-inactivated wells compared to that in control wells, which were treated with PBS only and set as 100%. Mean values with standard deviations from three independent assays are shown.
DISCUSSION
The gH/gL complex is an essential component of the herpesviral fusion machinery, but its role is not well understood. Although an active role in membrane fusion has been suggested for gH (31–33), analysis of the available crystal structures showed no signatures of known fusion proteins (5–7). More recent data indicate that the gH/gL complex plays a regulatory role, mediating the timely and efficient transition of gB into its fusion-active state (6). However, how this is accomplished remains unknown.
Amino acid sequence conservation is only moderate between the different herpesvirus gH molecules. However, the “SPC” motif located at positions 437 to 439 in PrV gH, with proline and cysteine invariably present in gH homologs of all herpesvirus subfamilies, was thought to be indispensable (5). It was, therefore, surprising that PrV strain Bartha, an attenuated vaccine strain, carries a serine at position 438 (20). To test the influence of this unexpected alteration on gH function, we mutated proline438 to serine in PrV-Ka gH and changed serine438 to proline in PrV-Ba gH.
In transient transfection-fusion assays, a drastic impairment in syncytium formation was evident when RK13 cells were cotransfected with expression plasmids for full-length gB, gD, and gL and plasmids expressing gH with the serine438 substitution. This indicates that proline438 plays a major role in supporting efficient cell-cell fusion. Since it was speculated that proline438 is necessary for the bending of alpha-helix 11 as a prerequisite for cysteine439 to come into the vicinity of and form a disulfide bridge with the conserved cysteine404 (Fig. 2) (5), we tested whether the replacement of cysteine404 with serine resulted in the same effect. Fusion assays with gHKaC404S also demonstrated a drastic drop in syncytium formation, similar to that for gHKaP438S, emphasizing the importance of these residues for protein function. While mutational analyses of the conserved proline have not been performed for any herpesvirus gH so far, mutation of the corresponding cysteines involved in disulfide bond formation in herpes simplex virus (HSV) (C5/C6 = cysteine554/cysteine589) and varicella-zoster virus (VZV) (cysteine540/cysteine575) rendered the proteins nonfunctional, pointing to an essential role for the disulfide bond (34, 35).
The proline438serine substitution also affected gH maturation, as demonstrated by the increased presence of immature gH (Fig. 4). It can be speculated that the presence of serine instead of proline in alpha-helix 11 (5) influences the folding of gH and that incorrectly folded gH molecules are retained in the endoplasmic reticulum (ER) and are not efficiently transported through the Golgi apparatus. Alternatively, improper folding may impair access to glycosylation sites during the transit of gH through the secretory pathway. Nevertheless, PrV-Ba gH, which contains this substitution, is functionally active to support viral replication, as is gHKaP438S, at least in cell culture. In cells expressing gHKaC404S, only immature gH was detected, in line with the importance of this disulfide bond for gH function in HSV and VZV.
Whether the proline438serine substitution affects virulence in vivo remains to be determined. However, it is important to note that full restoration of the virulence of PrV-Ba was achieved after repair of the deletion in the unique short region and point mutations in pUL21 (21, 23, 36). Thus, proline438 in gH is apparently not essential for PrV virulence.
Repair of proline438 in gHBa enhanced syncytium formation, but not to the same levels as gHKa, indicating that the other alterations present in gHBa also have a negative effect on membrane fusion. Mutation of aspartate59 to valine only slightly increased the fusion activity of gHBa, while introduction of aspartate59 into gHKa instead of valine drastically reduced fusogenicity, indicating that this amino acid position is also important for gH function. Since domain I, which also contains the gL binding site, was not part of the gH construct that was crystallized (Fig. 2), it is unclear whether this mutation might affect domain III directly or whether it influences fusogenicity by affecting gL binding. Unfortunately, the region encompassing V59 cannot be aligned with the structure of the other gH proteins. However, gHBa is functional in vitro and in vivo, indicating that this alteration does not abolish gL binding and gH/gL function. Restoration of both D59 and S438 in gHBa (gHBaD59V/S438P) resulted in syncytium formation comparable to that with native gHKa, demonstrating that the other differences between gHBa and gHKa do not play a role in membrane fusion.
To analyze the influence of the proline/serine variation during virus infection, we generated PrV-Ka recombinants expressing the different gH molecules. Despite the obvious impairment of syncytium formation in the transfection-fusion assays, the PrV recombinants expressing gH with the serine substitution at position 438 replicated to titers comparable to those of pPrV-gHKa and pPrV-gHBa, indicating that the mutation has no effect on virus production after high-MOI infection, which is in line with the notion that once inside the cell, gH is not required for continuing viral replication (25). In contrast, pPrV-gHKaC404S replicated to ca. 10-fold-reduced viral titers. Although this represents a significant defect, it clearly shows that, in contrast to HSV and VZV, PrV does not require the disulfide bond involving C404 for gH function. Our data suggest that the P438S mutation may impair protein folding such that only a fraction of the molecules folds correctly by a spontaneous tilt of alpha-helix 11, is able to leave the ER, and is properly processed. In contrast, another fraction is retained in the ER and remains in an immature state, as indicated by our protein analyses. Since correctly folded and properly processed gH will be incorporated into progeny virions, infectious particles will form but may contain less gH than the wild type. Maturation of gH is even more impaired by the complete inactivation of the disulfide bond in the C404S mutant, which correlates with the more severe phenotypic defects. However, even in the absence of the disulfide bond, PrV gH retains a residual capacity to function during entry.
Direct viral cell-to-cell spread as measured by plaque size was significantly reduced in RK13 cells infected with the serine438 variants, but plaque size could be restored to wild-type levels by serine438proline reversions. The strongest phenotype is shown by pPrV-gHKaC404S, which produces only tiny foci of infected cells, yet there is noticeable viral spread compared to infection of only single cells with the gH deletion mutant.
The molecular mechanism of direct cell-to-cell transmission of herpesviruses has remained unknown. While it relies on the same set of proteins as fusion of free virions in many herpesviruses, PrV can spread via direct cell-cell transmission in the absence of the receptor binding gD (37, 38) and also, partly, in the absence of gL (15), while gB and gH are absolutely required. This demonstrates that both processes depend on similar but not identical mechanisms. In addition, other proteins such as the gE/gI complex facilitate the direct transit of infectivity from infected to neighboring uninfected cells. It was suggested that gE is targeted to cell junctions, thereby helping virus spread (39). In consequence, restoration of the deleted unique short region to PrV-Ba resulted in significantly larger plaque sizes (36). In our experiments, plaque size could also be increased by just altering serine438 in PrV-Ba gH expressed in a PrV-Ka background. We are currently analyzing whether in a PrV-Ba background in the presence of the deletion in the unique short region, eliminating expression of gE and gI, alteration of these amino acids in gH has a similar effect.
Penetration kinetics showed that the velocity of virus entry is delayed in viruses expressing serine438 instead of proline. While 80% of pPrV-gHKa entered cells within approximately 5 min after temperature shift, all mutants expressing gH with serine438 required longer to reach the same value, indicating that either the fusion at the plasma membrane is less efficient or uptake might take place by a different mechanism, e.g., endocytosis. Thus, the transient transfection-fusion assay, which shows a distinct impairment of gH activity by the proline438serine alteration, correlates with the results from penetration kinetics and plaque size determination, but not one-step replication after high MOI infection and final virus titers. Penetration of pPrV-gHKaC404S is slowest, with only ca. 50% of the virus protected from acid inactivation after 40 min. Thus, inability to form this disulfide bond, which is essential in HSV and VZV, strongly impairs gH function in PrV but does not disable it completely. Therefore, despite the significant effect of the C404S mutation on PrV gH maturation as well as on viral cell-cell spread, penetration, and one-step replication, the mutated gH is still able to function, although at drastically lower efficiency. This unexpected result demonstrates that it is imperative to perform multiple assays with multiple viruses to unravel the functional consequences of specific alterations within this conserved protein.
In summary, alteration of the conserved proline at position 438 of PrV gH to serine influences the fusion activity of the protein in transient assays as well as cell-cell spread and penetration kinetics during infection. It also affects gH maturation. All these effects are mimicked, but increased, by abolition of the disulfide bond involving cysteine404. Thus, our data support the assumption that proline438 assists in, but is not essential for, formation of the cysteine404-cysteine439 disulfide bond by bending alpha-helix 11 and that formation of this disulfide bond is impaired but not eliminated by the amino acid exchange. Therefore, PrV-Ba retains replication competence despite the presence of the proline438serine mutation, which is a prime prerequisite for its use as a safe and efficacious PrV vaccine.
ACKNOWLEDGMENTS
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/11-1).
We thank Cindy Meinke and Maximilian Sell for expert technical assistance.
Footnotes
Published ahead of print 3 September 2014
REFERENCES
- 1.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299. 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, Eisenberg RJ. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825–3834. 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avitabile E, Forghieri C, Campadelli-Fiume G. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532–11537. 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653–1668. 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backovic M, DuBois RM, Cockburn JJ, Sharff AJ, Vaney MC, Granzow H, Klupp BG, Bricogne G, Mettenleiter TC, Rey FA. 2010. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc. Natl. Acad. Sci. U. S. A. 107:22635–22640. 10.1073/pnas.1011507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888. 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646. 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klupp BG, Mettenleiter TC. 1991. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology 182:732–741. 10.1016/0042-6822(91)90614-H. [DOI] [PubMed] [Google Scholar]
- 10.Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaye JF, Gompels UA, Minson AC. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693–2698. 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 12.Klupp BG, Baumeister J, Karger A, Visser N, Mettenleiter TC. 1994. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J. Virol. 68:3868–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu DX, Gompels UA, Nicholas J, Lelliott C. 1993. Identification and expression of the human herpesvirus 6 glycoprotein H and interaction with an accessory 40K glycoprotein. J. Gen. Virol. 74:1847–1857. 10.1099/0022-1317-74-9-1847. [DOI] [PubMed] [Google Scholar]
- 14.Gillet L, May JS, Colaco S, Stevenson PG. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J. Virol. 81:280–291. 10.1128/JVI.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp BG, Fuchs W, Weiland E, Mettenleiter TC. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687–7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lété C, Machiels B, Stevenson PG, Vanderplasschen A, Gillet L. 2012. Bovine herpesvirus type 4 glycoprotein L is nonessential for infectivity but triggers virion endocytosis during entry. J. Virol. 86:2653–2664. 10.1128/JVI.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klupp BG, Mettenleiter TC. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp BG, Nixdorf R, Mettenleiter TC. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760–6768. 10.1128/JVI.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs W, Backovic M, Klupp BG, Rey FA, Mettenleiter TC. 2012. Structure-based mutational analysis of the highly conserved domain IV of glycoprotein H of pseudorabies virus. J. Virol. 86:8002–8013. 10.1128/JVI.00690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 7:e1002282. 10.1371/journal.ppat.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp BG, Lomniczi B, Visser N, Fuchs W, Mettenleiter TC. 1995. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 212:466–473. 10.1006/viro.1995.1504. [DOI] [PubMed] [Google Scholar]
- 22.Lomniczi B, Blankenship ML, Ben-Porat T. 1984. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J. Virol. 49:970–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomniczi B, Watanabe S, Ben-Porat T, Kaplan AS. 1987. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J. Virol. 61:796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins AK, Ryan JP, Whealy ME, Enquist LW. 1989. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J. Virol. 63:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp B, Altenschmidt J, Granzow H, Fuchs W, Mettenleiter TC. 2008. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J. Virol. 82:6299–6309. 10.1128/JVI.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klingbeil K, Lange E, Teifke JP, Mettenleiter TC, Fuchs W. 2014. Immunization of pigs with an attenuated pseudorabies virus recombinant expressing the haemagglutinin of pandemic swine origin H1N1 influenza A virus. J. Gen. Virol. 95:948–959. 10.1099/vir.0.059253-0. [DOI] [PubMed] [Google Scholar]
- 27.Bartha A. 1961. Experimental reduction of virulence of Aujeszky's disease virus. Magy. Állatorv. Lapja 16:42–45 (In Hungarian.) [Google Scholar]
- 28.Passvogel L, Trübe P, Schuster F, Klupp BG, Mettenleiter TC. 2013. Mapping of sequences in pseudorabies virus pUL34 that are required for formation and function of the nuclear egress complex. J. Virol. 87:4475–4485. 10.1128/JVI.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimm KS, Klupp BG, Granzow H, Müller FM, Fuchs W, Mettenleiter TC. 2012. Analysis of viral and cellular factors influencing herpesvirus-induced nuclear envelope breakdown. J. Virol. 86:6512–6521. 10.1128/JVI.00068-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 31.Cole NL, Grose C. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13:207–222. 10.1002/rmv.377. [DOI] [PubMed] [Google Scholar]
- 32.Galdiero S, Falanga A, Vitiello M, Browne H, Pedone C, Galdiero M. 2005. Fusogenic domains in herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 280:28632–28643. 10.1074/jbc.M505196200. [DOI] [PubMed] [Google Scholar]
- 33.Kinzler ER, Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827–7837. 10.1128/JVI.79.12.7827-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairns TM, Landsburg DJ, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332:550–562. 10.1016/j.virol.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Vleck SE, Oliver SL, Brady JJ, Blau HM, Rajamani J, Sommer MH, Arvin AM. 2011. Structure-function analysis of varicella-zoster virus glycoprotein H identifies domain-specific roles for fusion and skin tropism. Proc. Natl. Acad. Sci. U. S. A. 108:18412–18417. 10.1073/pnas.1111333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Porat T, DeMarchi J, Pendrys J, Veach RA, Kaplan AS. 1986. Proteins specified by the short unique region of the genome of pseudorabies virus play a role in the release of virions from certain cells. J. Virol. 57:191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66:894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauh I, Mettenleiter TC. 1991. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 65:5348–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DC, Webb M, Wisner TW, Brunetti C. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821–833. 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]