ABSTRACT
Protein synthesis, the most energy-consuming process in cells, responds to changing physiologic priorities, e.g., upon mitogen- or stress-induced adaptations signaled through the mitogen-activated protein kinases (MAPKs). The prevailing status of protein synthesis machinery is a viral pathogenesis factor, particularly for plus-strand RNA viruses, where immediate translation of incoming viral RNAs shapes host-virus interactions. In this study, we unraveled signaling pathways centered on the ERK1/2 and p38α MAPK-interacting kinases MNK1/2 and their role in controlling 7-methyl-guanosine (m7G) “cap”-independent translation at enterovirus type 1 internal ribosomal entry sites (IRESs). Activation of Raf-MEK-ERK1/2 signals induced viral IRES-mediated translation in a manner dependent on MNK1/2. This effect was not due to MNK's known functions as eukaryotic initiation factor (eIF) 4G binding partner or eIF4E(S209) kinase. Rather, MNK catalytic activity enabled viral IRES-mediated translation/host cell cytotoxicity through negative regulation of the Ser/Arg (SR)-rich protein kinase (SRPK). Our investigations suggest that SRPK activity is a major determinant of type 1 IRES competency, host cell cytotoxicity, and viral proliferation in infected cells.
IMPORTANCE We are targeting unfettered enterovirus IRES activity in cancer with PVSRIPO, the type 1 live-attenuated poliovirus (PV) (Sabin) vaccine containing a human rhinovirus type 2 (HRV2) IRES. A phase I clinical trial of PVSRIPO with intratumoral inoculation in patients with recurrent glioblastoma (GBM) is showing early promise. Viral translation proficiency in infected GBM cells is a core requirement for the antineoplastic efficacy of PVSRIPO. Therefore, it is critically important to understand the mechanisms controlling viral cap-independent translation in infected host cells.
INTRODUCTION
Mammalian mRNAs have starkly diverse 5′ untranslated regions (UTRs), ranging from simple <50-nucleotide (nt) leaders preceding a single initiation AUG to vast (>1,000-nt), intricate structures with multiple upstream AUGs. The physiologic purpose of 5′-UTR complexity is to impede the scanning phase of protein synthesis initiation (1). Scanning occurs when 43S preinitiation complexes (PICs), containing 40S ribosomal subunits/eukaryotic translation initiation factor 3 (eIF3) and the eIF2–GTP–Met-tRNA ternary complex, connect with mRNA. The PIC-mRNA link is eIF4G, which binds PICs via eIF3, engages the translation initiation helicase eIF4A, and propels PICs toward the initiation AUG (2). Conventionally, eIF4G binds to eIF4E, tethering PICs to the 5′ 7-methyl-guanosine (m7G) cap of mRNAs. Alternatively, certain templates can recruit PICs to 5′ UTRs internally, independent of a 5′ end, m7G cap, or eIF4E (3). This requires an internal ribosomal entry site (IRES), a concept first described with enteroviruses (EVs) (4) and cardioviruses (5). EV (type 1) and cardiovirus (type 2) IRESs initiate translation by recruiting the eIF4G/4A translation initiation helicase (3, 6), a mechanism that may be shared by eukaryotic, IRES-competent mRNAs (7, 8).
Translation initiation at IRESs is tightly restricted in cells, as unbridled cap-independent PIC recruitment would derail global protein synthesis control. Such restraints affect viral pathogens that rely on IRESs, because translation of incoming viral genomes must occur promptly, prior to virus-induced host cell rearrangements that favor viral protein synthesis. Poliovirus (PV) IRES competence is reduced by point mutations in IRES stem-loop domain V of the live-attenuated (Sabin) vaccines (9) that impair eIF4G/PIC engagement (10, 11). Similarly, a deficit for PIC recruitment in PVSRIPO, the type 1 live-attenuated PV (Sabin) vaccine containing an HRV2 IRES (12, 13), maps to human rhinovirus type 2 (HRV2) IRES stem-loop domains V/VI (14) in a region harboring the eIF4G/4A helicase complex footprint (10). PV is sensitive to such IRES impediments, specifically in neuron-like cells (15), e.g., in the primate central nervous system (CNS) (16), but is unaffected in less differentiated cancer cells (17). This suggests fundamentally different conditions for IRES competency in postmitotic neurons versus mitotically active tumor cells.
Protein synthesis responds to phosphoinositol 3-kinase (PI3K)/mTOR and Raf-ERK1/2 signal transduction pathways that converge on translation machinery. Thus, inappropriate cooption of such signaling, a hallmark of cancer, may enable unfettered IRES activity. For example, PVSRIPO translation defects in (neuron-like) HEK293 cells (15) are fully restored upon transformation with oncogenic Ras, due to activation of Raf–ERK1/2–mitogen-activated protein kinase (MAPK)-interacting kinase (MNK) signaling (18). MAPK-mediated activation of MNK and simultaneous protein kinase C (PKC)-Raf-ERK1/2 signals to eIF4G (19) lead to MNK-eIF4G binding (20) and phosphorylation of eIF4E(S209) (21). Although MNK facilitates mitogen-induced protein synthesis, tumorigenesis (22), and tumor chemoresistance (23), the mechanisms of MNK-mediated posttranscriptional gene regulation remain obscure.
In this work, we deciphered MNK-centered signaling networks that control PVSRIPO IRES-mediated translation, cytotoxicity, and cancer cell killing. Two genes give rise to MNK1 and -2 isoforms that occur in two splice variants each (MNK1a and -b/MNK2a and -b). The “b” isoforms lack MAPK activation domains and nuclear export signals, and only the “a” isoforms respond to upstream MAPK signals (thus, “MNK1/2” here refer to the latter). Our studies revealed novel MNK functions, independent of MNK-eIF4G binding/phosphorylation of eIF4E(S209), that substantiate major unrecognized roles for MNK in posttranscriptional gene control. Our findings suggest that MNK regulates Ser/Arg (SR)-rich protein kinase (SRPK) and its prime substrates, the SR-rich proteins, key factors of constitutive and alternative splicing, mRNA export, stability, and translation (24), including translation initiation at type 1 picornavirus IRESs (25). These signals permit rampant IRES activity, viral translation, and tumor-specific cytotoxicity of PVSRIPO in cancerous cells with constitutively active Raf-ERK1/2 MAPK signal transduction networks.
MATERIALS AND METHODS
Cell lines, expression plasmids, viruses, stimulants, and inhibitors.
Du54, 43, and U87 glioma cells and HeLa R19 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). Stable doxycycline (Dox)-inducible HeLa (8) and HEK293 (20) cell lines were grown in DMEM supplemented with 10% FBS, blasticidin S (2.5 μg/ml; Sigma-Aldrich), and hygromycin B (100 μg/ml; Invitrogen). PVSRIPO was described previously (16). CHICO virus was cloned as follows: the hepatitis C virus (HCV) IRES was amplified from a full-length HCV genomic cDNA (26) using primer pair 1/2 (Table 1), digested with MluI and SacI, and ligated into corresponding sites of an infectious coxsackievirus B3 (CBV3) cDNA (27); virus was derived as previously described (27). PVSRIPO, PV type 1 (Sabin), CBV3, and CHICO were propagated in HeLa cells as described previously (18) and concentrated by centrifugation through a 100-kDa-cutoff spin column (Millipore). HRV16 was obtained from Y. Bochkov, University of Wisconsin, and propagated as described previously (28). 12-O-Tetradecanoylphorbol-13-acetate (TPA) (Tocris) was dissolved in dimethyl sulfoxide (DMSO). Inhibitors of MEK (UO126; Promega), MNK (CGP57380; Tocris), and SRPK (SRPin340; Millipore) were dissolved in DMSO and used as described in the figures and figure legends. Dox (Sigma) was dissolved in sterile water and used at a final concentration of 1 μg/ml. Unless otherwise indicated, 5 × 105 cells were seeded in 6-well plates the day before infection and treated with TPA (200 nM), UO126 (20 μM), or CGP57380 (10 and 30 μM) 30 to 60 min prior to infection or with SRPin340 (1 to 10 μM) at the time of infection. After the growth medium was replaced with prewarmed serum-free DMEM (Invitrogen) containing virus at a multiplicity of infection (MOI) of 5, inhibitors were added to maintain the signal blockade, whereas TPA was removed with the addition of virus.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide name | No. | Sequence |
|---|---|---|
| chico-5′ | 1 | 5′-AGCACGCGTCCCCTGTGAGGAACTACTG-3′ |
| chico-3′ | 2 | 5′-CGCCGAGCTCCATATGTTGTGAGACCTTTGATGACCTTACCCAAATTACGCG-3′ |
| deltaCMV-5′ | 3 | 5′-TTACGCGTGGGCGGTAGGCGTGTACG-3′ |
| deltaCMV-3′ | 4 | 5′- CCAAGCTTAAGTTTAAACGCTAGAG-3′ |
| TO-5′ | 5 | 5′-phos-TCGACTCCCTATCAGTGATAGAGATCTCCCTATCAGTGATAGAGATCG-3′ |
| TO-3′ | 6 | 5′-phos-TCGACGATCTCTATCACTGATAGGGAGATCTCTATCACTGATAGGGAG-3′ |
| MNK(T334D)-1-5′ | 7 | 5′-TCCCCGATCCGCAAGTCCTCCAG-3′ |
| MNK(T334D)-1-3′ | 8 | 5′-GATAATGCGGCCGCTCAGATGCTGTGGGCGGG-3′ |
| MNK(T334D)-2-5′ | 9 | 5′-GAACCTGAGGTAGTGGAGGTCTTCACGG-3′ |
| MNK(T334D)-2-3′ | 10 | 5′-GCGGATCGGGGAGTCCCTTTTC-3′ |
| eIF4E(S209A)-5′ | 11 | 5′-TACTAAGAGCGGCGCCACCACTAA-3′ |
| eIF4E(S209A)-3′ | 12 | 5′-TTAGTGGTGGCGCCGCTCTTAGTA-3′ |
| Myc-eIF4G1(ΔMNK)-5′ | 13 | 5′-GCGGTACCCACAGAAAGCACAGATAATAG-3′ |
| Myc-eIF4G1(ΔMNK)-3′ | 14 | 5′-GCCTCGAGGGCTGTGACAGATTTAAGGGCC-3′ |
Viral proliferation and cytotoxicity assays.
Viral progeny/one-step growth curves were determined as previously described (20), except that a 30-min virus attachment step at room temperature was replaced by incubation with prewarmed medium at 37°C for 60 min. For ATP release assays, 5 × 104 glioma cells (U87, Du54, and 43) were seeded in 24-well plates, mock or PVSRIPO infected, and treated as described in the figure legends. Supernatants were harvested at the designated intervals after infection (MOI = 2 for U87/Du54; MOI = 4 for 43) and analyzed with the Enliten ATP Assay System (Promega) according to the manufacturer's instructions.
siRNA knockdown/reconstitution assays and MNK expression assays.
For small interfering RNA (siRNA) transfections, 1 × 105 cells were seeded in 35-mm dishes and transfected the following day. All-Stars nontargeting control siRNA (siCtrl) or siRNAs targeting MNK1, eIF4E, Dap5, SRPK1/2 (Qiagen), or hnRNP A1 (Thermo-Dharmacon) were transfected (50 pmol/35-mm dish) using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. The cells were used 60 to 72 h (all but SRPK1/2 depletions) or 84 h (SRPK1/2 depletions) posttransfection for further assays. For MNK/eIF4E reconstitution after siRNA transfection, Dox was added 8 h or 48 h prior to infection, respectively.
IPs, cell fractionation, IF assays, and immunoblots.
Immunoprecipitations (IPs) were carried out using anti-Flag-conjugated agarose beads (Sigma) as previously described (19). eIF4G1 knockdown/knock-in cells were plated out in 150-mm dishes, treated with Dox (96 h), and lysed with polysome lysis buffer (20). Dap5/eIF4G1(682-1600)-expressing cells were Dox induced (24 h), and co-IPs were performed as described previously (20). For fractionation, cells were processed with the NE-PER fractionation kit (Thermo Pierce); whole-cell lysates were collected by lysing cells in LDS buffer (Invitrogen) containing benzonase (Sigma) and 5% β-mercaptoethanol (Sigma). For immunofluorescence (IF) assays, cells were grown on poly-d-lysine (Invitrogen)-coated, UV cross-linked coverslips; transfected with siRNA or treated with inhibitors as described above; and fixed using methanol at −20°C (15 min). After blocking with 5% goat serum (1 h; Invitrogen) in PBS-Triton X-100 (0.3%) buffer, SRPK1 or SF2 antibodies (1:500) were added in PBS-Triton with 1% bovine serum albumin (BSA) and incubated (1 h) at room temperature. The cells were washed and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse (1:250; Invitrogen) and tetramethyl rhodamine isocyanate (TRITC)-conjugated goat anti-rabbit (1:250; Invitrogen) diluted in blocking buffer (1 h). The cells were washed, Progold anti-fade DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) mounting medium was used to mount coverslips, and the slides were photographed using an Olympus IX71 fluorescence microscope. Immunoblots were performed as previously described (29) using antibodies specific to poliovirus 2C/2BC (18), CBV3 3D/3CD (a generous gift from K. Klingel, University of Tübingen, Tübingen, Germany), phosphorylated ERK1/2 (p-ERK1/2), ERK1/2, p-RSK(S380), RSK, p-S6K(T389), S6K, MNK1, p-eIF4E(S209), eIF4E, eIF4A, hemagglutinin (HA), hnRNP A1, eIF4G1, eIF3A, poly(ADP-ribose) polymerase (PARP), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and Dap5 (Cell Signaling); tubulin and myc (Sigma-Aldrich); SRPK1 and SRPK2 (BD Biosciences); and SF2 (Novus). All immunoblot quantitations were performed on a Li-Cor Odyssey Fc.
Expression plasmids and stable cell lines.
To reduce spurious exogenous MNK expression before Dox induction and/or blatant MNK overexpression, the pcDNA5-FRT/TO vector (Invitrogen) was modified as follows. First, a MluI-HinDIII fragment containing the cytomegalovirus (CMV) promoter was replaced with its promoterless counterpart generated by PCR with primers 3 and 4 (Table 1). Second, 4 added tetracycline (Tet) operator sequences (TetO) were inserted into a SalI site adjacent to the existing TetO2 element using complementary oligonucleotides 5 and 6 (Table 1), generating pcDNA5/FRT/TO6. The T334D mutation in HA-tagged MNK1 was introduced by PCR using primer pairs 7/8 and 9/10 (Table 1) and wild-type (wt) MNK1 as a template. The resulting overlapping fragments were fused by PCR using primers 8 and 9 (Table 1), digested with Bsu36I and NotI, and ligated into the Bsu36I/NotI-digested pcDNA3.1 wt MNK1 plasmid. HA-tagged MNK1, MNK1(D191A), MNK1(ΔMAPK), MNK2 (20), and MNK1(T334D) fragments were subcloned into pcDNA5/FRT/TO6 using AflII and XhoI sites. A myc-tagged wt eIF4E expression plasmid (30) was used to generate myc-eIF4E(S209A) with primer pair 11/12 (Table 1) and a Quick-Change kit 2 (Stratagene). Both wt and S209A myc-tagged eIF4E sequences were subcloned into pcDNA5/FRT/TO. The myc-eIF4G1(ΔMNK)-Flag construct lacks 15 amino acids of the C terminus and was cloned by PCR using primer pair 13/14 (Table 1) and myc-eIF4G1-Flag (29) as a template. The resulting fragment was cleaved with Acc65I/XhoI and ligated into the Acc65I/XhoI-digested myc-eIF4G1-Flag vector. The (endogenous) eIF4G1 knockdown/(exogenous) knock-in cell line system (31) was used for reconstitution with pcDNA5 (mock), wt myc-eIF4G1-Flag, or myc-eIF4G1(ΔMnk)-Flag. The pCI-flag-Dap5 vector (a generous gift from Martin Holcik, University of Ottawa, Ottawa, Canada) was digested with XhoI, followed by treatment with Klenow fragment and NotI digestion, and the resulting Flag-Dap5 fragment was inserted into EcoRV/NotI sites of pcDNA5/FRT/TO. The Dox-inducible Dap5-expressing cell line was established in Flip-In HEK293 cells as described previously (20). MNK, eIF4E, and eIF4G expression lines in the HeLa Flip-In background were generated as described previously (8).
RNA reporter constructs and assays.
Capped β-globin reporter RNA was synthesized in vitro using the T7 Message Machine (Ambion), and uncapped viral IRES-driven reporters were synthesized using the T7 Megascript kit (Ambion) (32). In vitro-transcribed RNA was purified using the RNeasy kit (Qiagen) and analyzed by agarose gel electrophoresis and NanoDrop UV spectrophotometry. For reporter translation assays, 0.5 μg of Renilla luciferase (rluc; IRES driven) RNA and 0.5 μg of capped firefly luciferase (fluc) RNA were cotransfected per well using DMRIE-C following the manufacturer's protocol (Invitrogen). Four hours posttransfection, the cells were harvested with passive lysis buffer (Promega), and rluc/fluc values were measured using the dual-luciferase assay kit (Promega) (18). rluc values were divided by fluc values to correct for transfection efficiency. For reporter assays with Dox-induced MNK overexpression, 3 × 105 cells/well were plated in 6-well plates (with or without Dox); the fold stimulation was determined by dividing values obtained with Dox by those without Dox.
Statistics.
Quantitated immunoblot/reporter data were normalized between experiments as described in the figure legends and are represented as averages and standard errors of the mean (SEM). JMP10 (SAS) was used to perform statistical analysis using either Student's t test or an analysis of variance (ANOVA)-protected Student t test (enabling multiple comparisons within a data set). Significance was defined as a P value of <0.05, and the specific tests used for each experiment are described in the figure legends.
RESULTS
PVSRIPO translation and cytotoxicity in GBM cells respond to the MEK-ERK1/2 status.
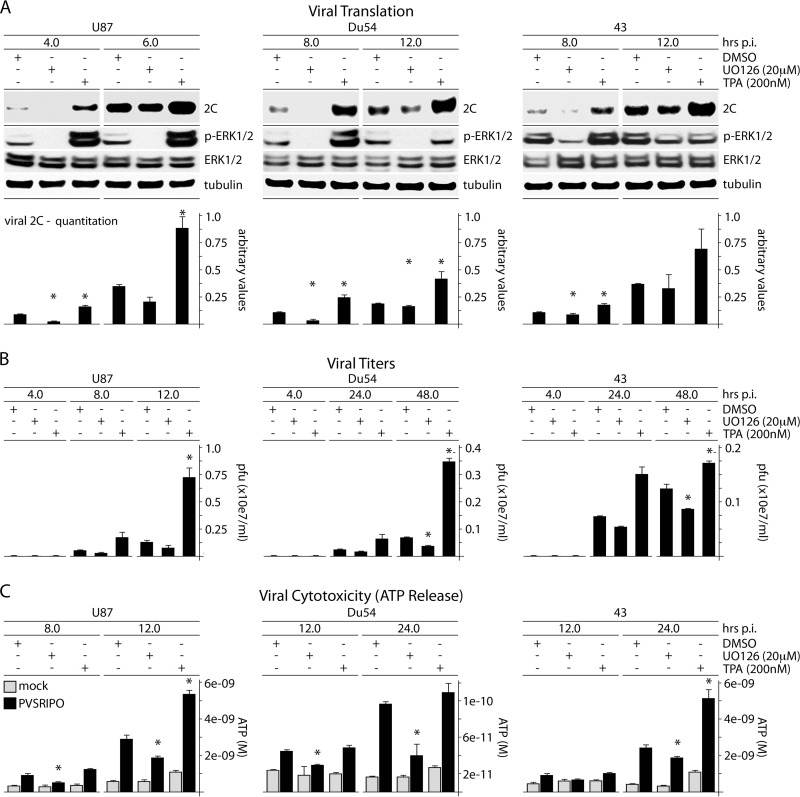
PV host cell killing is irrevocably linked to viral protein synthesis, because instant translation of incoming viral RNA yields a set of highly cytotoxic viral proteins. To mechanistically unravel the role of Raf-MEK-ERK1/2 signals in PVSRIPO tumor-specific translation, we tested PVSRIPO translation, propagation, and cytotoxicity in glioblastoma (GBM) cells treated with the MEK inhibitor UO126 or the phorbol ester TPA (Fig. 1); the signaling schema is shown in Fig. 2A). Since PSVRIPO has similar growth characteristics in all GBM cell lines tested (17), we chose 3 well-studied, representative models at random: Du54 (33) (ex vivo passage 18), 43 (34) (ex vivo passage 23), and U87 (35) (ex vivo passage >100). The cells were not serum starved and displayed significant intrinsic p-ERK1/2. UO126 (added 30 min before infection and throughout the assay) diminished p-ERK1/2 levels; TPA (added 30 min before infection and removed with the addition of virus) increased p-ERK1/2 for at least 8 h (Fig. 1A). The cells were lysed at the indicated intervals postinfection (p.i.) to assess viral translation (Fig. 1A), propagation (Fig. 1B), and cytotoxicity (ATP release) (Fig. 1C). These parameters were reduced in UO126-treated GBM cells and enhanced in TPA-stimulated cultures (correlating with the p-ERK1/2 status), suggesting that MEK-ERK1/2 signals drive PVSRIPO translation, and thus cytotoxicity, in GBM.
FIG 1.
ERK1/2 signals control PVSRIPO translation, proliferation, and cytotoxicity in GBM cells. (A) U87, Du54, or 43 GBM cells were treated with DMSO (mock), UO126 (20 μM), or TPA (200 nM) (30 min); infected with PVSRIPO; and harvested at the designated intervals. UO126 was maintained after infection; TPA was not. The immunoblots track viral protein (2C); the quantitation represents the average of 3 experiments normalized to the first control value for each series. (B) Supernatants from cells treated and infected as described for panel A were collected to determine viral progeny (PFU); the averages of two experiments are shown. (C) ATP release was measured in supernatants from the experiments in panel B. The ATP concentration was determined using a standard curve; the average of two assays is shown. The error bars represent SEM; the asterisks indicate significant ANOVA-protected t tests.
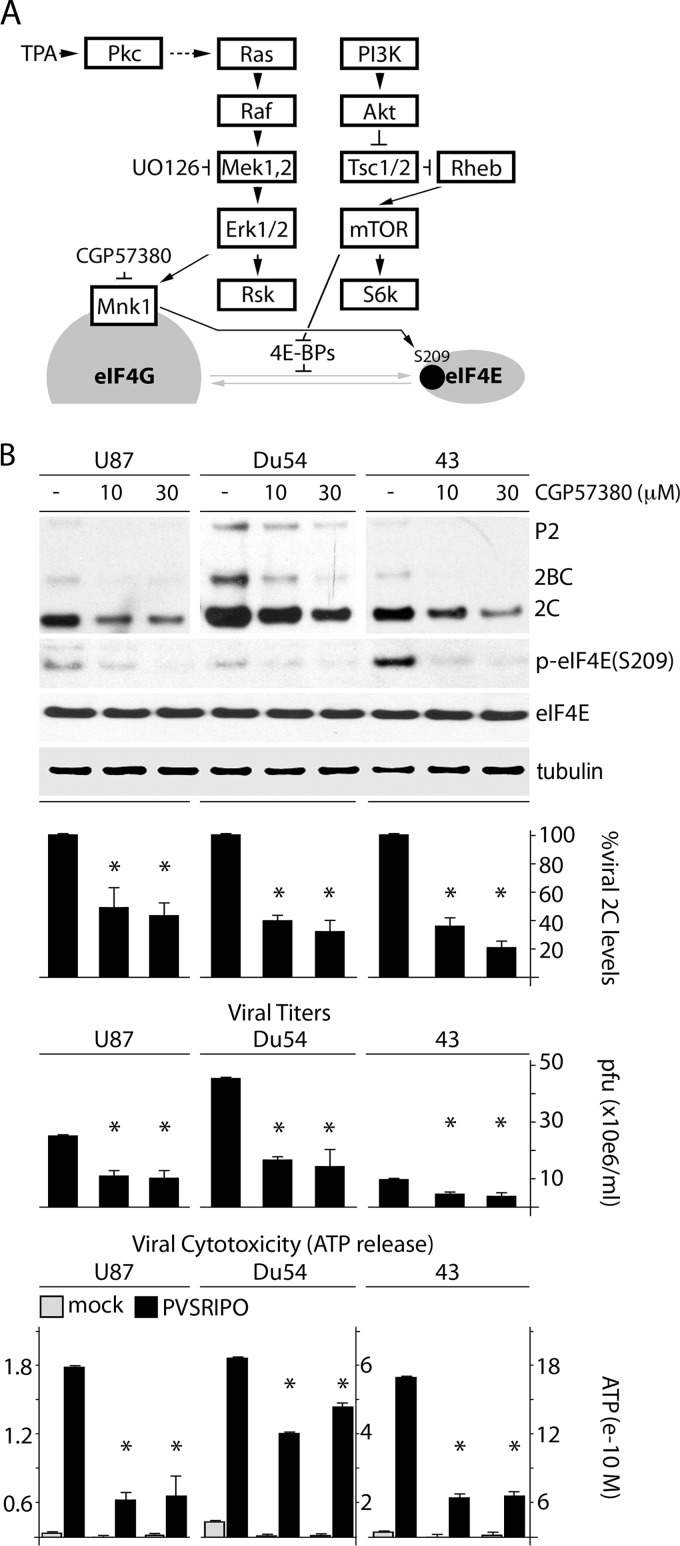
FIG 2.
MNK inhibition reduces PVSRIPO translation, proliferation, and cytotoxicity in GBM cells. (A) Schema of signaling pathways to translation initiation factors investigated in this study. (B) U87, Du54, and 43 GBM cell lines were pretreated (1 h) with DMSO or CGP57380 (10 or 30 μM), followed by infection in the presence of DMSO/CGP57380. Cells were harvested (6 h p.i.), and viral protein (2C) was assessed by immunoblotting. Supernatants were collected (12 h p.i.) to determine viral progeny and ATP release, as indicated. The bars represent the average of 3 experiments normalized to the control (DMSO) values, and the error bars represent SEM. The asterisks denote significant ANOVA-protected t tests.
ERK1/2-responsive PVSRIPO translation depends on MNK.
Since it has been implicated in PVSRIPO IRES competence previously (18), we investigated whether MNK controls TPA-responsive PVSRIPO tumor cytotoxicity. MNK inhibition with CGP57380 (Fig. 2A) decreased p-eIF4E(S209) and reduced viral translation, propagation, and cytotoxicity in GBM cell lines (Fig. 2B). To buttress our results with a rigorous MNK depletion/reconstitution assay, we combined MNK1 depletion with Dox-inducible reconstitution of several MNK variants. We depleted only MNK1 (and not MNK2) because it is more abundant then MNK2 (36) and because MNK2 lacks autoinhibitory structural features (37), resulting in high basal activity in the absence of ERK1/2 signaling (38). Thus, MNK2 is less likely to be involved in TPA-induced PVSRIPO tumor cytotoxicity.
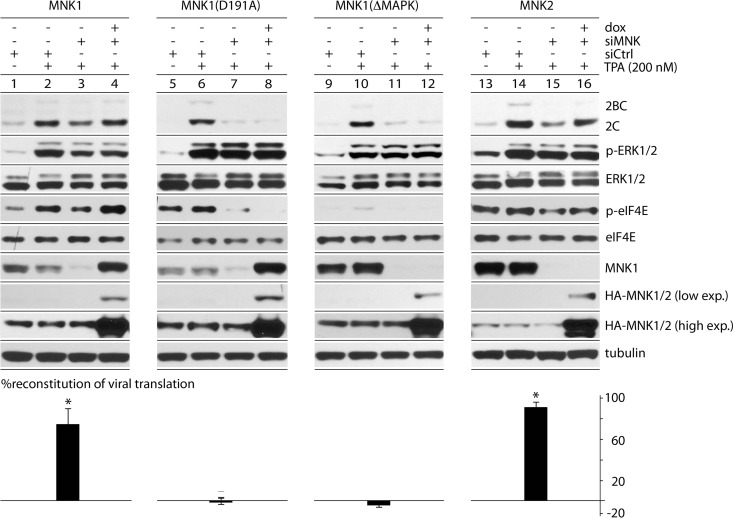
We reconstituted with exogenous, HA-tagged wt MNK1, kinase-dead MNK1(D191A) (20), non-MAPK-responsive MNK1(ΔMAPK), or wt MNK2 (Fig. 3). This achieved efficient endogenous MNK1 depletion with reconstitution of HA-tagged MNK1 (Fig. 3). MNK1 depletion reduced eIF4E phosphorylation and PVSRIPO translation (Fig. 3, lanes 3, 7, 11, and 15). Reconstitution with wt HA-MNK1 compensated for the MNK1 depletion effect on both p-eIF4E- and TPA-stimulated PVSRIPO translation (Fig. 3, lanes 1 to 4). Reconstitution with kinase-dead (Fig. 3, lanes 5 to 8) or non-MAPK-responsive (Fig. 3, lanes 9 to 12) MNK1 did not. The D191A mutation (in the metal-binding coordinating site) abolishes MNK1 kinase activity, but not MAPK activation. Therefore, MNK1(D191A) responds to MAPK activation with constitutive eIF4G binding (20). In contrast, MNK1(ΔMAPK) cannot assume a MAPK-induced conformation required for eIF4G binding (20). Failed reconstitution with MNK1(D191A) indicates that MNK binding to eIF4G is not sufficient for TPA stimulation of PVSRIPO translation. Exogenous MNK2 rescued TPA stimulation of PVSRIPO in MNK1-depleted cells (Fig. 3, lanes 13 to 16), suggesting that MNK1 and -2 exert similar effects on PVSRIPO translation. PVSRIPO translation was consistently more resistant to MNK1 depletion in mock-induced cells harboring wt MNK1/2 transgenes (Fig. 3, compare lanes 2 and 3 and lanes 14 and 15 with lanes 6 and 7 and lanes 10 and 11), likely because of leaky HA-MNK1/2 expression in the absence of Dox. Our findings suggest that TPA-mediated stimulation of PVSRIPO translation and cytotoxicity rely on catalytically active MNK1.
FIG 3.
TPA-mediated stimulation of viral translation depends on catalytically active MNK. Dox-inducible cells expressing wt HA-tagged MNK1, MNK1(D191A), MNK1(ΔMAPK), or MNK2 (see the text for details) were used for mock or TPA stimulation after siCtrl (mock) or siMNK (MNK1) depletion. Dox-induced expression (exp.) of the corresponding HA-tagged MNK (lanes 4, 8, 12, and 16) was detected by HA immunoblotting. The MNK1 antibody used does not recognize MNK1(ΔMAPK) or MNK2. Viral translation was assayed at 4 h p.i. The percent reconstitution of viral translation was calculated as follows: (siMNK + Dox + TPA − siMNK + TPA) divided by (siCtrl + TPA − siMNK + TPA), representing the average of 3 experiments. The error bars represent SEM, and the asterisks indicate significance (P < 0.05 by Student's t test compared to 0% reconstitution).
MNK broadly stimulates viral cap-independent translation.
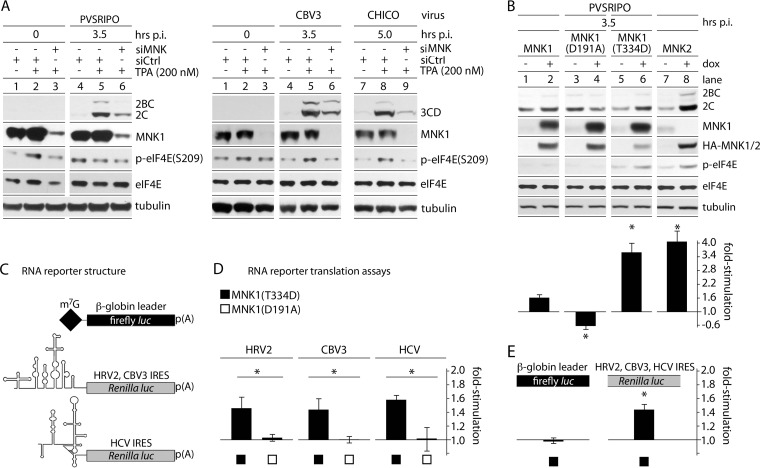
MNK could affect host-virus interactions that precede translation, i.e., entry. We previously found that Raf-MEK-ERK1/2 activation does not alter transfer of viral RNA into the host cytoplasm (18). Subsequent analyses (see Fig. 9 and our companion paper [39]) implicated a signal transduction network with preeminent roles in posttranscriptional gene regulation and no plausible involvement in PV entry processes. Also, MNK could influence HRV2 IRES competency specifically, e.g., through sequence-specific RNA-binding trans-acting factors, or generally, by broadly promoting generic conditions for cap-independent initiation. We tested these possibilities, assessing MNK's effect on several IRESs in diverse viral contexts (Fig. 4A) and conducting translation assays by transfecting corresponding viral 5′-UTR-driven RNA reporters (Fig. 4B to E). MNK-dependent TPA stimulation occurred equally with PV type 1 (Sabin) (the PVSRIPO backbone) and with HRV16 (representing an HRV IRES in an authentic context [data not shown]). We observed MNK-dependent TPA stimulation of viral translation equally with CBV3 and with CBV3(CHICO) containing a foreign HCV IRES (Fig. 4A).
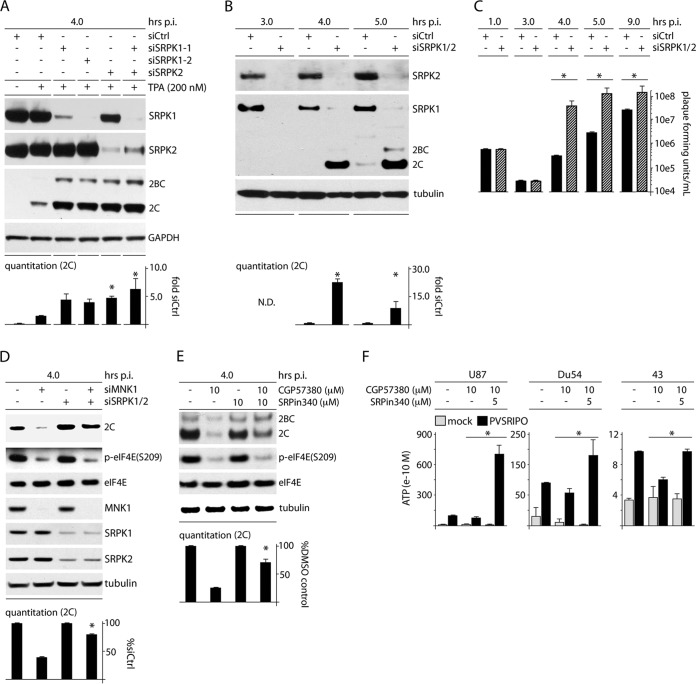
FIG 9.
SRPK depletion enhances PVSRIPO translation and propagation, counters the MNK depletion/inhibition effect, and increases viral translation and cytotoxicity in GBM. (A) HeLa cells were treated with siCtrl (with or without TPA) or siRNA targeting SRPK1 and/or SRPK2 (all plus TPA) prior to infection with PVSRIPO (4 h). The quantitation represents the average viral protein 2C levels normalized for the siCtrl-plus-TPA sample from 2 assays; asterisks denote ANOVA-protected t test. (B) HeLa cells were treated with siCtrl or siRNA targeting SRPK1 and -2 and assessed for viral 2C expression by immunoblotting at 4 and 5 h p.i. Average quantitations from 3 tests, normalized for the control values for each interval, are shown. (C) Viral titers from HeLa cells treated as for panel B and infected at an MOI of 10 were determined for the designated intervals; the average of two experiments is shown, normalizing between experiments using the siCtrl values. (D) HeLa cells were cotransfected with siCtrl or siRNA targeting SRPK1/2 followed by transfection with siCtrl or MNK1 siRNA as shown and infected as for panel B. The quantitation represents the average viral protein 2C levels normalized between experiments by setting siCtrl values to 1. (E) HeLa cells were treated with DMSO (mock), CGP57380 (10 μM), SRPin340 (10 μM), or combined inhibitors coincident with PVSRIPO infection (4 h p.i.). Cells were harvested and analyzed for viral protein by immunoblotting. Quantitation of viral protein 2C is shown for 3 assays normalized by setting the controls (without CGP57380) to 1. (F) U87, Du54, and 43 GBM cells were treated with DMSO, CGP57380, or CGP57380 plus SRPin340 at the concentrations indicated at the time of infection. The supernatants were harvested (12 h p.i.), and the average ATP concentration for 3 (Du54) or 2 (U87 and 43) assays was determined. (B to F) The asterisks denote significant paired Student's t tests; error bars represent SEM.
FIG 4.
MNK activity selectively enhances viral IRES-mediated translation. (A) HeLa cells were treated with control (siCtrl) or MNK1-targeting (siMNK) siRNA 24 h prior to TPA/mock stimulation. PVSRIPO (left) or CBV3/CHICO (right) infection and subsequent analyses were carried out as for Fig. 1. The assays were repeated 3 times, and representative series are shown. (B) Cells with Dox-inducible expression of wt MNK1, MNK1(D191A), MNK1(T334D), or MNK2 were mock/Dox induced (12 h) and infected with PVSRIPO. Viral translation was quantitated, and the fold stimulation of viral translation was calculated by dividing the Dox-induced value by the mock-induced value for 3 independent tests. (C) Structure of RNA reporters used (32). (D) Uncapped, in vitro-transcribed rluc RNA reporters driven by the HRV2, CBV3, or HCV 5′ UTR were cotransfected with m7G-capped β-globin leader fluc reporters into Dox-/mock-induced MNK1(T334D)- or MNK1(D191A)-expressing cells (4 h). IRES-driven (rluc) values were divided by β-globin 5′-UTR firefly luciferase values to correct for transfection differences. Dox-induced values were then divided by mock-induced values for each cell line to calculate the fold stimulation of IRES-mediated translation due to MNK1(T334D)/MNK1(D191A) expression. The data represent 3 independent assays done in triplicate for each cell line and 5′ UTR. (E) Pooled β-globin 5′-UTR fluc values and IRES-driven rluc values in MNK1(T334D)-expressing cells. (B, D, and E) The error bars represent SEM, and the asterisks indicate significance (P < 0.05 by Student's t test).
To confirm a role for MNK catalytic activity in PVSRIPO translation without the possible off-target effects of TPA or siRNA transfections, we used Dox-inducible expression of (MAPK-dependent) HA-MNK1 and -MNK2 or kinase-dead MNK1(D191A) compared to (MAPK-independent) constitutively active MNK1(T334D) (Fig. 4B to E). Expression of wt MNK1 and MNK1(D191A) failed to induce eIF4E(S209) phosphorylation (Fig. 4B, lanes 2 and 4). In contrast, expression of MNK1(T334D) and MNK2 enhanced p-eIF4E(S209) levels (Fig. 4B, lanes 6 and 8), due to their constitutive or intrinsic basal activity, respectively. Increased p-eIF4E(S209) in the absence of Dox is likely due to cryptic expression of MNK(T334D)/MNK2 (Fig. 4B, lanes 5 and 7). Wt MNK1 expression alone (in the absence of MAPK stimulation) did not increase PVSRIPO translation, due to the lack of catalytic MNK activity (Fig. 4B). In accordance with eIF4E(S209) phosphorylation (indicating MNK catalytic activity), at 3.5 h p.i., PVSRIPO translation was stimulated ∼3.5- to 4-fold with MNK1(T334D)/MNK2 expression.
To assess MNK's influence on viral IRES competency directly, we conducted studies with transfected in vitro-transcribed RNA reporters (Fig. 4C to E). As was done with PVSRIPO infection (Fig. 4B), we compared reporter RNA translation in cells expressing Dox-inducible, kinase-dead MNK1(D191A) or constitutively active MNK1(T334D). We tested a firefly luciferase (fluc)-expressing reporter controlled by the (m7G cap-dependent) β-globin leader and reporters expressing Renilla luciferase (rluc) driven by the HRV2, CBV3, or HCV IRES (Fig. 4C). The β-globin leader is an unstructured 38-nt stretch preceding a single initiation AUG in an ideal Kozak context and represents an unencumbered translation template with no scanning impediment. HeLa cells were each cotransfected with the β-globin leader (internal control for transfection efficiency and for unencumbered m7G cap-dependent translation) and one of the IRES-driven constructs. Expression of MNK1(D191A) did not change the ratio of IRES to β-globin leader efficiency for any of the viral IRESs (Fig. 4D). In contrast, MNK1(T334D) induction consistently favored cap-independent translation at all IRESs (Fig. 4D). To test whether IRES induction was linked to repression of conventional cap-dependent translation, the β-globin leader values (fluc) from all MNK1(T334D) samples were pooled (dividing values with Dox by those without Dox) and compared to all corresponding, pooled IRES/rluc ratios (pooling was used to reach the highest statistical power possible). Our data suggest that induced IRES efficiency is the sole contributor to the increased IRES-to-m7G cap initiation ratio observed after MNK(T334D) expression (Fig. 4E).
MNK stimulation of PVSRIPO translation is independent of eIF4E(S209) phosphorylation.
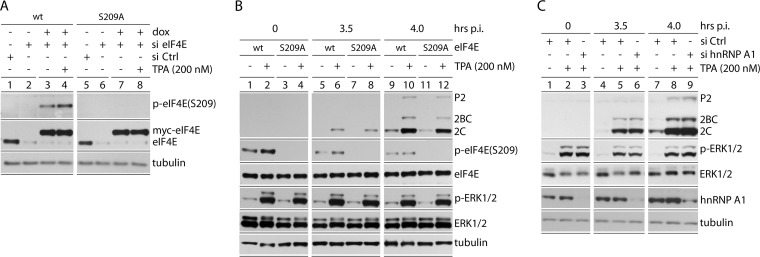
MNK control over viral IRES-mediated translation could implicate its “classic” substrate, eIF4E(S209). While viral IRES-mediated translation does not require eIF4E, the effects of eIF4E phosphorylation on translation machinery at large could affect PIC recruitment to IRESs. To assess this possibility, siRNA-mediated depletion of endogenous eIF4E was combined with Dox-induced reconstitution of myc-tagged wt eIF4E or mutant eIF4E(S209A) (Fig. 5). This created knockdown/knock-in cell lines with nearly exclusive expression of myc-tagged wt eIF4E or eIF4E(S209A) at physiological levels (Fig. 5A). Basal and TPA-stimulated eIF4E(S209) phosphorylation occurred with wt eIF4E (Fig. 5A, lanes 3 and 4), but not with the S209A mutant (Fig. 5A, lanes 7 and 8). The PVSRIPO infection/TPA stimulation assay showed equal stimulation of viral translation caused by TPA, excluding eIF4E(S209) phosphorylation as a factor in PVSRIPO translation competence (Fig. 5B). We also evaluated a possible involvement of hnRNP A1, because it is a proposed HRV IRES trans-acting factor (40) and MNK substrate (41). siRNA-mediated depletion of hnRNP A1 had no influence on PVSRIPO translation upon TPA stimulation (Fig. 5C).
FIG 5.
eIF4E(S209A) substitution and hnRNP A1 depletion do not affect TPA stimulation of PVSRIPO translation. (A) Dox-inducible cell lines expressing wt myc-eIF4E or myc-eIF4E(S209A) were treated with siCtrl (lanes 1 and 5) or eIF4E-targeting siRNA (lanes 2 to 4 and 6 to 8). Dox induction reconstituted wt eIF4E or eIF4E(S209A) to roughly endogenous levels. (B) The assay conditions from panel A, lanes 3 and 4 and lanes 7 and 8, were used to track viral translation after TPA stimulation in the presence of wt myc-eIF4E or myc-eIF4E(S209A) at 3.5 and 4 h p.i. (C) HeLa cells were mock or TPA stimulated following siCtrl or hnRNP A1 siRNA, as shown. The cells were infected, and viral protein was analyzed by immunoblotting (3.5 and 4 h p.i.). The experiments were performed in triplicate (B) or duplicate (C); representative series are shown.
MNK stimulation of PVSRIPO translation does not require MNK binding to eIF4G.
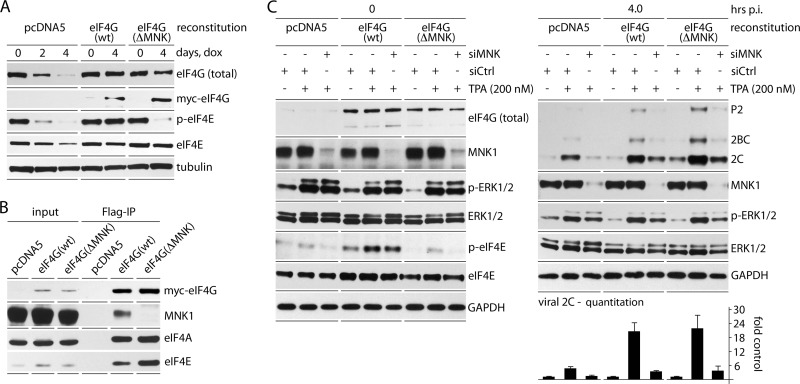
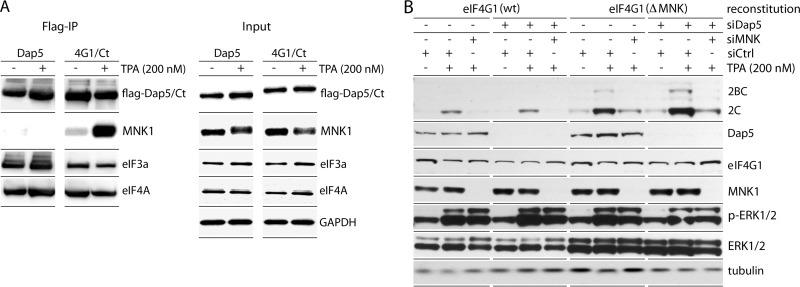
To reach its substrate, eIF4E, MNK must bind to eIF4G (20, 42) (Fig. 2A). Since eIF4E(S209) phosphorylation is not involved in MNK stimulation of PVSRIPO, we tested if MNK-eIF4G binding is required. We used a Dox-inducible (endogenous) eIF4G1 knockdown/(exogenous) myc-eIF4G1-Flag knock-in system described previously (31). Dox induction (96 h) yielded >90% depletion of endogenous eIF4G1 with reconstitution to roughly native levels (Fig. 6A). Mammalian cells express 3 eIF4G isoforms, eIF4G1, eIF4G2 (UniProt name, eIF4G-3), and death-associated protein 5 (Dap5; UniProt name, eIF4G-2), which were all reported to bind MNK (42). We focused on MNK binding to eIF4G1 because it is significantly more abundant than eIF4G2 (∼70:1 in HeLa cells [36]) and because we have evidence that Dap5 does not bind MNK (Fig. 7A). Dox-inducible expression of Flag-tagged Dap5 or eIF4G1(682-1600), the structural homolog of Dap5, followed by Flag IP led to co-IP of endogenous MNK1 with eIF4G1(682-1600), but not with Dap5 (Fig. 7A). Our findings are at variance with earlier reports (42), which may be due to significant MNK1 and Dap5 overexpression in that study. Confirming a dominant role for eIF4G1 in MNK binding, there was almost complete abolition of p-eIF4E(S209) upon eIF4G1 depletion alone (Fig. 6A and C).
FIG 6.
MNK1 stimulation of viral translation does not require MNK1 binding to eIF4G1. (A) Dox-inducible endogenous eIF4G1 knockdown with simultaneous mock reconstitution (pcDNA5) or reconstitution with wt myc-eIF4G1-Flag or myc-eIF4G1(ΔMNK)-Flag. (B) Flag IP of lysates from the three cell lines after Dox induction (96 h). MNK1 co-IP occurred only with wt eIF4G1 reconstituted cells. (C) MNK1/mock depletion, TPA/mock stimulation, and PVSRIPO infection of the three cell lines as shown in Fig. 4A. Prior to infection, all cells were Dox induced (96 h), followed by siCtrl/siMNK1 treatment and TPA/mock stimulation, as shown. Note deficient eIF4E(S209) phosphorylation in mock-/eIF4G(ΔMNK)-reconstituted cells. Viral protein 2C levels were quantitated (4 h p.i.), and the values represent the average of 3 experiments normalized using the corresponding siCtrl-mock lanes. The error bars indicate SEM.
FIG 7.
Dap5 does not interact with MNK and does not affect MNK-dependent PVSRIPO translation competence. (A) HEK293 cells with Dox-inducible expression of Dap5-Flag or eIF4G1(682-1600)-Flag (Ct) (8) were treated with Dox (24 h) and subsequently mock (DMSO) or TPA (200 nM) stimulated (2 h). Flag IP of Dap5/eIF4G1(682-1600) and co-IP of MNK1, eIF4A, and eIF3a are shown. The experiment was repeated 10 times; a representative series is shown. A shift in MNK1 electrophoretic mobility after TPA stimulation likely reflects phosphorylation. (B) HeLa (endogenous) eIF4G1 knockdown/(exogenous) wt eIF4G1 or eIF4G1(ΔMNK) knock-in cells were treated as for Fig. 5. In addition, the cells were treated with siCtrl or siDap5 (72 h) prior to infection. PVSRIPO translation was assayed for each condition alongside relevant controls. The assay was repeated twice, and a representative series is shown.
We generated cell lines reconstituted with pcDNA5 (mock), wt myc-eIF4G1-Flag (reconstitution to the native state), or myc-eIF4G1(ΔMNK)-Flag (reconstitution with eIF4G1 incapable of binding MNK) (Fig. 6C). eIF4G1(ΔMNK) lacks 15 C-terminal amino acids and cannot bind MNK (19); Flag co-IP confirmed eIF4G1-MNK1 interaction only in wt reconstituted cells (Fig. 6B). The absence of eIF4G1-MNK binding in ΔMNK reconstituted cells is evident as decreased p-eIF4E(S209) with and without TPA stimulation (compare p-ERK1/2 to p-eIF4E) at levels similar to mock reconstituted cells (Fig. 6C). eIF4G1 depletion reduced viral translation by ∼75%, reflecting eIF4G's critical role in picornavirus IRES-mediated initiation (Fig. 6C). The response of viral translation to TPA was indistinguishable in wt eIF4G1 and eIF4G1(ΔMNK) reconstituted cells (Fig. 6C). This shows that MNK-eIF4G1 binding is not required for the TPA stimulatory effect on PVSRIPO translation. Codepletion of Dap5 in eIF4G1(ΔMNK) reconstituted cells did not affect PVSRIPO translation, either (Fig. 7C), confirming that Dap5 (which does not bind MNK [Fig. 7A]) does not compensate for eIF4G1(ΔMNK) in our assay. In accordance with other assays in this study, PVSRIPO translation in wt eIF4G1 and eIF4G1(ΔMNK) reconstituted cells dropped ∼8-fold in response to MNK1 depletion (Fig. 6C and 7B). In aggregate, these results suggest a role for MNK1 in protein synthesis control independent of its relationship with eIF4G and eIF4E.
MNK1 activation alters SRPK activity.
Surprisingly, our data suggest that neither of MNK's direct links to translation factors accounts for its effects on PVSRIPO translation and tumor cytotoxicity. We identified a possible mechanism, however, by examining a recently proposed regulatory relationship of MNK2 with SRPK (43). There are two homologous, ubiquitously expressed SRPK isoforms in mammals, which are constitutively active (44) and are controlled by subcellular partitioning and heat shock protein association. Cytoplasmic retention of SRPK in complexes with molecular chaperones (45) is reversed by upstream signals that result in nuclear translocation. Reversible phosphorylation of the SRPK's principal substrates, the SR proteins, is essential to their defining roles in posttranscriptional gene regulation, including translation control (46).
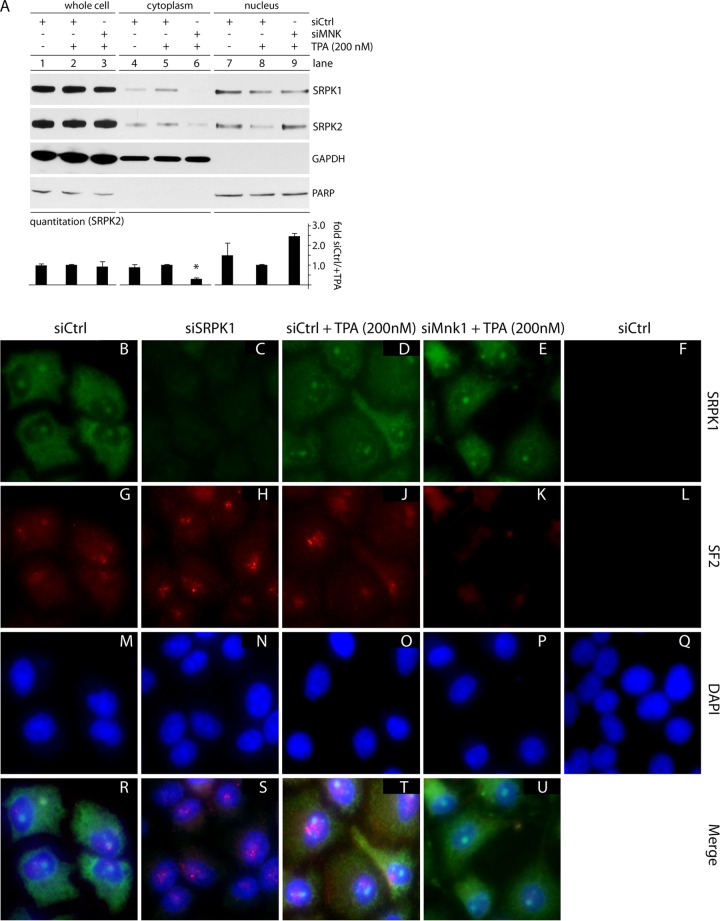
Considering its mode of regulation, we first assessed SRPK partitioning upon MNK activation (by TPA) or MNK1 depletion (with TPA stimulation). HeLa cells were TPA stimulated, MNK1 depleted, and harvested to generate fractionated extracts. TPA and/or MNK1 depletion did not alter total SRPK1/2 levels but did change their nucleocytoplasmic partitioning (Fig. 8A). TPA led to cytoplasmic accumulation and diminished nuclear presence of SRPK1/2, while MNK1 depletion (with TPA stimulation) had the opposite effect, reducing cytoplasmic SRPK2 ∼5-fold while increasing its nuclear abundance (Fig. 8A). MNK1 depletion in TPA-stimulated cells altered SRPK partitioning patterns beyond baseline (without TPA) levels, indicating a role for MNK1 in basal control of SRPK function. To confirm these results, we performed indirect IF studies in fixed cells (Fig. 8B to U). SRPK partitioning is difficult to test directly, because the effects are subtle (Fig. 8A) (47). A function of SRPK activity is dissociation of nuclear speckles (48), which is readily detected. Therefore, we tested the SRPK substrate and speckle component SF2 (SRSF1, or ASF) by indirect IF (Fig. 8G to L). SF2 nuclear-speckle disassembly occurs with SRPK overexpression, suggesting that SRPK activity inversely correlates with SF2 nuclear-speckle intensity (48, 49). Corroborating these findings, SRPK1 depletion (Fig. 8C) intensified nuclear-speckle signals (Fig. 8H and S). A similar effect was achieved with TPA stimulation (Fig. 8J and T). MNK1 depletion, in the presence of TPA, abolished speckle signal in the sample (Fig. 8K and U), in accordance with nuclear accumulation of SRPK1/2 (Fig. 8A and E). Our findings suggest that TPA stimulation leads to SRPK1/2 cytoplasmic retention, resulting in speckle stabilization. This effect is mediated by MNK1, because MNK1 depletion (in the presence of TPA) favors SRPK1/2 nuclear influx and speckle dissociation.
FIG 8.
MNK1 depletion causes SRPK nuclear accumulation and SF2 nuclear-speckle dissociation. (A) Whole-cell, cytoplasmic, and nuclear lysates were prepared from cells treated with siCtrl with and without TPA (1 h) or siMNK plus TPA (1 h). Immunoblots for SRPK2 were quantitated, and average SRPK2 values for each fraction from two experiments are shown; the asterisk denotes an ANOVA-protected t test. The error bars indicate SEM. (B to U) Indirect IF using cells treated with siCtrl (with or without TPA; 1 h), siMNK (plus TPA; 1 h), or siSRPK1 were stained for SRPK1 (green), nuclear-speckle marker SF2 (red), and DAPI (blue), as labeled. Individual staining for each group (B to Q) and merged tricolor staining (R to U) are shown. A negative-staining (no primary antibody added) control for SRPK1 and SF2 is shown on the right (F, L, and Q). The indirect IF experiment was performed three times, and representative images are shown.
MNK1 acts on PVSRIPO translation via SRPK.
Nucleocytoplasmic shuttling SR proteins have been broadly implicated in translation (50), and SRp20 has been specifically linked to PV IRES competence (25). Therefore, SRPK-mediated phosphorylation of SR proteins, leading to their nuclear import (51) and reduced affinity for RNA (50), may negatively affect PVSRIPO translation and cytotoxicity. MNK1 activity leads to nuclear exclusion and cytoplasmic accumulation of SRPK and restricts nuclear SRPK activities, e.g., speckle dissociation. If such restrictive effects of MNK on SRPK are indeed responsible for PVSRIPO tumor cytotoxicity, then outright SRPK depletion should enhance viral translation and cytotoxicity. To test this, we assessed viral translation in SRPK-depleted cells (Fig. 9).
SRPK1/2 depletion significantly elevated viral protein synthesis in the presence of TPA, ∼4- to 5-fold beyond levels achieved with TPA stimulation alone (Fig. 9A). Depletion of either SRPK1 or SRPK2 enhanced PVSRIPO translation almost as well as codepletion of both (Fig. 9A), supporting recent evidence that proper SRPK activity relies on both kinase isoforms (47). One-step growth curve assays in SRPK1/2-depleted HeLa cells revealed ∼20-fold and ∼7-fold increases of PVSRIPO protein at 4 h and 5 h p.i., respectively (Fig. 9B). Viral progeny levels were ∼200-fold and ∼120-fold elevated at the corresponding intervals (Fig. 9C). SRPK1/2 codepletion had no effect on PVSRIPO adsorption to cells (even recovery of PFU at 1 h p.i.) or on the eclipse of bound particles (at 3 h p.i.) but profoundly altered the efficiency of PVSRIPO translation and progeny production (Fig. 9C).
To link the observed role of SRPK in control over viral translation and cytotoxicity to the effects of MNK catalytic activity, we first tested PVSRIPO translation with combined MNK1 and SRPK1/2 codepletion (Fig. 9D). At 4 h p.i., MNK1 depletion reduced viral translation to ∼40% (Fig. 9D). This effect was mitigated by codepletion of SRPK1/2, although the SRPK1/2 knockdown efficiency was reduced in this assay (likely due to triple depletion in a single sample) (Fig. 9D). Next, we tested PVSRIPO translation in cells with combined MNK (CGP57380) and SRPK (SRPin340) blockade (Fig. 9E). As is the case for SRPK1/2 codepletion (Fig. 9D), combining SRPin340 inhibition reversed the effect of MNK1 inhibition with CGP57380 on PVSRIPO translation. We recapitulated this effect in the GBM cytotoxicity experiment (ATP release assay) (Fig. 2B) by combining the repressive effects of CGP57380 with SRPK inhibition (Fig. 9F). SRPin340 restored the loss of PVSRIPO cytotoxicity with MNK inhibition in 43, Du54, and U87 GBM cells (Fig. 9E). Our findings are in line with a suppressive role of SRPK activity on picornavirus type 1 IRES-mediated translation initiation, which is alleviated through MNK activity.
DISCUSSION
Translation machinery is under the tight control of Raf-MEK-ERK1/2 and PI3K-AKT-mTOR signal transduction networks in cells. Advanced invasive and treatment-refractory cancers invariably exhibit inappropriate cooption of such signaling cascades, contributing to an environment of broadly unhinged protein synthesis control. The mechanistic understanding of translation regulation in cancer is incomplete at best. Targeting GBM (or other cancer types) with PVSRIPO is based on the observation that type 1 IRES-driven translation is favored in a cancer setting (17). Unfettered IRES-driven translation of PV polypeptides in malignant cells produces rapid and drastic cytotoxicity, leading to the presentation of combined pathogen- and danger-associated molecular patterns with a potential for recruiting host immunogenic responses (52). Because IRES competency is a decisive factor in PVSRIPO oncolytic efficacy, we set out to unravel the signal transduction network that controls viral m7G cap-independent translation and viral cytotoxicity in tumor cells.
The present and previous (18) investigations pointed toward a role for Raf-MEK-ERK1/2 signals and the ERK1/2-activated protein kinase MNK, in particular. Much evidence links MNK to important gene-regulatory processes, e.g., oncogenesis (22). However, few of MNK's functions are mechanistically understood. We discovered that MNK affects viral IRES-mediated translation independent of its best-known roles as an eIF4G binding partner or the eIF4E(S209) kinase. Rather, our data suggest that MNK broadly influences posttranscriptional gene regulation via SRPK, a “master” regulator of the SR proteins, key factors in alternative and constitutive splicing, mRNA export, mRNA stability, and translation (53). Activation of MNK was associated with SRPK cytoplasmic retention and reduced SRPK nuclear activity, consistent with a recent report that MNK2 activity prevents eIF4G(S1148) phosphorylation downstream of SRPK (43). SRPK depletion had substantial stimulatory effects on PVSRIPO replication, even in HeLa cells that naturally support rampant viral IRES-mediated translation and propagation. The fact that siRNA-mediated SRPK depletion enhanced viral dynamics eliminates the possibility of off-target suppression due to the RNA transfection procedure or host viability effects, which can skew analyses of host factor involvement in virus susceptibility studies. Combined SRPK depletion/inhibition reversed the repressive effects of MNK depletion/inhibition on PVSRIPO translation competence and cytotoxic potential in HeLa cells or GBM cells.
A role for SRPK in control over viral IRES-mediated translation very likely involves its prime substrates, the SR proteins. This is because most SR proteins shuttle rapidly and continuously (54) between the nucleus and cytoplasm, associate with translating ribosomes, and stimulate translation of reporter mRNAs in vitro and in vivo (50). They induce translation of spliced and intronless mRNAs equally, indicating that their effect on translation is independent of a prior splicing event (50). Most importantly, a particular shuttling SR protein (SRp20) was previously implicated in PV translation (25). The mechanism of SRp20 involvement in EV IRES-mediated translation is unknown, but it may occur through contacts with the poly(rC) binding protein 2 (PCBP2) (25), a host IRES trans-acting factor (ITAF) implicated in PIC recruitment (3) and EV IRES-mediated translation initiation (3, 55). A possible explanation for their contribution to alternative initiation may involve SR protein-aided tethering of PICs to mRNA. This could be particularly important for templates that lack conventional recruitment of PICs to the 5′ m7G cap by eIF4E, e.g., PVSRIPO RNA. The broad effects of SRPK on cap-independent translation shared between very diverse IRESs of HCV and EVs may be explained by the relatively degenerate RNA sequences recognized by SR proteins and/or by the common regulation of all SR proteins by SRPK. Although HCV IRES-mediated initiation is fundamentally different from that of type 1 picornaviral IRESs, because PIC recruitment is independent of eIF4F (56), it similarly involves PCBP2 (57).
MNK1-mediated effects on SRPK partitioning/activity, evident as increased nuclear-speckle intensity and cytoplasmic retention, is in accordance with previously described effects of SRPK on the SR proteins' role in cytoplasmic functions, e.g., translation. For example, SRPK-mediated SR protein phosphorylation causes them to lose RNA affinity (50) and enhances their nuclear import (51). A key question emerging from our studies is the mechanism underlying MNK-mediated control over SRPK, since SRPK is not a plausible MNK substrate. This question is addressed in a companion report, which revealed important information on an unexpected broad and deep involvement of MNK in mitogenic signal transduction networks impinging on posttranscriptional gene regulation (39). Given their apparently critical role as host pathogenesis factors, elucidating the precise mechanism for the SR protein/SRp20 involvement in IRES-mediated translation is required; however, this is beyond the scope of this study.
Our research provides mechanistic support for a new experimental cancer therapy demonstrating early promise in clinical trials in recurrent-GBM patients (52). PVSRIPO exerts forceful tumor cytotoxicity of a type and range difficult to achieve with other therapeutic modalities. In targeting ERK1/2-MNK signaling, PVSRIPO cytotoxicity is enabled by a basal, homeostatic survival mechanism that is critical for mitogenic-, genotoxic-, metabolic-, or hypoxic-stress resilience and therapy resistance of many cancers (58). Targeting such fundamental aspects of the malignant phenotype is key for the therapy of notoriously treatment-resistant and genetically heterogeneous cancers, such as recurrent GBM.
ACKNOWLEDGMENTS
We thank A. Palmenberg and Y. Bochkov (University of Wisconsin) for the HRV16 infectious clone, M. Holcik (University of Ottawa, Ottawa, Canada) for the Dap5 cDNA, and K. Klingel (University of Tübingen, Tübingen, Germany) for CBV3 3D antibodies. We thank M. Dobrikov (Duke University) for critical reading of the manuscript.
This research was supported by PHS awards CA124756 (M.G.) and 5T32CA009111 (M.C.B.), by the National Center for Advancing Translational Sciences of the NIH (UL1TR001117), and by the Southeastern Brain Tumor Foundation.
Footnotes
Published ahead of print 3 September 2014
REFERENCES
- 1.Kozak M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903. 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136:447–460. 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J. 33:76–92. 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU. 2011. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acid Res. 39:4851–4865. 10.1093/nar/gkr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hundsdoerfer P, Thoma C, Hentze MW. 2005. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. U. S. A. 102:13421–13426. 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser C, Dobrikova EY, Bradrick SS, Shveygert M, Herbert JT, Gromeier M. 2008. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA 14:2170–2182. 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wimmer E, Hellen CU, Cao X. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353–436. 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 10.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U. S. A. 106:9197–9202. 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochs K, Zeller A, Saleh L, Bassili G, Song Y, Sonntag A, Niepmann M. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77:115–122. 10.1128/JVI.77.1.115-122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merrill MK, Dobrikova EY, Gromeier M. 2006. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 80:3147–3156. 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merrill MK, Gromeier M. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 80:6936–6942. 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell SA, Lin J, Dobrikova EY, Gromeier M. 2005. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 79:6281–6290. 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrikova EY, Goetz C, Walters RW, Lawson SK, Peggins JO, Muszynski K, Ruppel S, Poole K, Giardina SL, Vela EM, Estep JE, Gromeier M. 2012. Attenuation of neurovirulence, biodistribution, and shedding of a poliovirus:rhinovirus chimera after intrathalamic inoculation in Macaca fascicularis. J. Virol. 86:2750–2759. 10.1128/JVI.06427-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gromeier M, Lachmann S, Rosenfeld MR, Gutin PH, Wimmer E. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. U. S. A. 97:6803–6808. 10.1073/pnas.97.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goetz C, Everson RG, Zhang LC, Gromeier M. 2010. MAPK signal-integrating kinase controls cap-independent translation and cell type-specific cytotoxicity of an oncolytic poliovirus. Mol. Ther. 18:1937–1946. 10.1038/mt.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobrikov M, Dobrikova E, Shveygert M, Gromeier M. 2011. Phosphorylation of eukaryotic translation initiation factor 4G1 (eIF4G1) by protein kinase C{alpha} regulates eIF4G1 binding to Mnk1. Mol. Cell. Biol. 31:2947–2959. 10.1128/MCB.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shveygert M, Kaiser C, Bradrick SS, Gromeier M. 2010. Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen-activated protein kinase occurs through modulation of Mnk1-eIF4G interaction. Mol. Cell. Biol. 30:5160–5167. 10.1128/MCB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16:1909–1920. 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda T, Sasaki M, Elia AJ, Chio II, Hamada K, Fukunaga R, Mak TW. 2010. Combined deficiency for MAP kinase-interacting kinase 1 and 2 (Mnk1 and Mnk2) delays tumor development. Proc. Natl. Acad. Sci. U. S. A. 107:13984–13990. 10.1073/pnas.1008136107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grzmil M, Huber RM, Hess D, Frank S, Hynx D, Moncayo G, Klein D, Merlo A, Hemmings BA. 2014. MNK1 pathway activity maintains protein synthesis in rapalog-treated gliomas. J. Clin. Invest. 124:742–754. 10.1172/JCI70198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, Coso OA, Srebrow A. 2005. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 12:1037–1044. 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 25.Bedard KM, Daijogo S, Semler BL. 2007. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 26:459–467. 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradrick SS, Walters RW, Gromeier M. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acid Res. 34:1293–1303. 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobrikova E, Florez P, Bradrick S, Gromeier M. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. U. S. A. 100:15125–15130. 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall DJ, Bates ME, Guar L, Cronan M, Korpi N, Bertics PJ. 2005. The role of p38 MAPK in rhinovirus-induced monocyte chemoattractant protein-1 production by monocytic-lineage cells. J. Immunol. 174:8056–8063. 10.4049/jimmunol.174.12.8056. [DOI] [PubMed] [Google Scholar]
- 29.Dobrikov MI, Dobrikova EY, Gromeier M. 2013. Dynamic regulation of the translation initiation helicase complex by mitogenic signal transduction to eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 33:937–946. 10.1128/MCB.01441-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradrick SS, Gromeier M. 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4:e7030. 10.1371/journal.pone.0007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobrikov MI, Shveygert M, Brown MC, Gromeier M. 2014. Mitotic phosphorylation of eukaryotic initiation factor 4G1 (eIF4G1) at Ser1232 by Cdk1:cyclin B inhibits eIF4A helicase complex binding with RNA. Mol. Cell. Biol. 34:439–451. 10.1128/MCB.01046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradrick SS, Dobrikova EY, Kaiser C, Shveygert M, Gromeier M. 2007. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosome entry sites. RNA 13:1582–1593. 10.1261/rna.556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullard DE, Schold SC, Jr, Bigner SH, Bigner DD. 1981. Growth and chemotherapeutic response in athymic mice of tumors arising from human glioma-derived cell lines. J. Neuropathol. Exp. Neurol. 40:410–427. 10.1097/00005072-198107000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, Ballman KV, Kitange GJ, Guha A, Pandita A, James CD. 2006. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin. Cancer Res. 12:2264–2271. 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 35.Ponten J, Macintyre EH. 1968. Long term culture of normal and neoplastic human glia. Acta Pathol. Microbiol. Scand. 74:465–486. [DOI] [PubMed] [Google Scholar]
- 36.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7:548. 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jauch R, Cho MK, Jakel S, Netter C, Schreiter K, Aicher B, Zweckstetter M, Jackle H, Wahl MC. 2006. Mitogen-activated protein kinases interacting kinases are autoinhibited by a reprogrammed activation segment. EMBO J. 25:4020–4032. 10.1038/sj.emboj.7601285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheper GC, Morrice NA, Kleijn M, Proud CG. 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21:743–754. 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown MC, Dobrikov MI, Gromeier M. 2014. Mitogen-activated protein kinase-interacting kinase regulates mTOR/AKT signaling and controls the serine/arginine-rich protein kinase-responsive type 1 internal ribosome entry site-mediated translation and viral oncolysis. J. Virol. 88:13149–13160. 10.1128/JVI.01884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. 2007. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell 18:1302–1311. 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buxade M, Parra JL, Rousseau S, Shpiro N, Marquez R, Morrice N, Bain J, Espel E, Proud CG. 2005. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23:177–189. 10.1016/j.immuni.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18:270–279. 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu SI, Katz M, Chin S, Qi X, Cruz J, Ibebunjo C, Zhao S, Chen A, Glass DJ. 2012. MNK2 inhibits eIF4G activation through a pathway involving serine-arginine-rich protein kinase in skeletal muscle. Sci. Signal 5:ra14. 10.1126/scisignal.2002466. [DOI] [PubMed] [Google Scholar]
- 44.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. 2005. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell 20:77–89. 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 45.Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. 2009. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 23:482–495. 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA, Fu XD. 2006. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell 17:876–885. 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, Fu XD. 2012. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47:422–433. 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gui JF, Lane WS, Fu XD. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678–682. 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 49.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 50.Sanford JR, Gray NK, Beckmann K, Caceres JF. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755–768. 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai MC, Lin RI, Tarn WY. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A. 98:10154–10159. 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown MC, Dobrikova EY, Dobrikov MI, Walton RW, Gemberling SL, Nair SK, Desjardins A, Sampson JH, Friedman HS, Friedman AH, Tyler DS, Bigner DD, Gromeier M. 17 June 2014. Oncolytic polio virotherapy of cancer. Cancer. 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Y, Steitz JA. 2005. SRprises along a messenger's journey. Mol. Cell 17:613–615. 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 54.Caceres JF, Screaton GR, Krainer AR. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55–66. 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93:11115–11120. 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959–1962. 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Jeng KS, Lai MM. 2011. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J. Virol. 85:7954–7964. 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebolt-Leopold JS, Herrera R. 2004. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4:937–947. 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]