ABSTRACT
The four dengue virus (DENV) serotypes (DENV serotype 1 [DENV-1] to DENV-4) are transmitted by Aedes aegypti and A. albopictus mosquitoes, causing up to 390 million DENV infections worldwide each year. We previously reported a clade replacement of the DENV-2 Asian-American genotype NI-1 clade by the NI-2B clade in Managua, Nicaragua. Here, we describe our studies of the replicative ability of NI-1 and NI-2B viruses in an A. aegypti cell line (Aag2) and A. aegypti mosquitoes reared from eggs collected in Managua. In coinfection experiments, several different pairs of NI-1 and NI-2B clinical isolates were used to infect Aag2 cells or blood-fed A. aegypti mosquitoes. Results consistently showed a significant replicative advantage of NI-2B over NI-1 viruses early after infection in vitro, and in mosquitoes, NI-2B viruses attained a higher replicative index than NI-1 isolates 3 to 7 days postinfection (dpi). At 7 dpi, NI-2B viruses displayed a significantly higher replicative index in legs and salivary glands; however, this advantage was lost by 14 and 21 dpi. We also found that the percentage of mosquitoes in which NI-2B viruses were dominant was significantly higher than that in which NI-1 viruses were dominant on day 7 but not at later time points. Taken together, these data demonstrate that clade NI-2B holds a replicative advantage over clade NI-1 early in infection that wanes at later time points. This early fitness advantage of NI-2B viruses over NI-1 viruses in the native vector, A. aegypti, suggests a shorter extrinsic incubation period for NI-2B viruses, which could have contributed to the clade replacement event in Managua.
IMPORTANCE Dengue virus (DENV), one of the most medically important arthropod-borne viruses, is transmitted to humans by Aedes aegypti and A. albopictus mosquitoes in tropical and subtropical regions worldwide. Dengue epidemics continue to increase in frequency, geographic range, and severity and are a major public health concern. This is due to globalization, unplanned urbanization, and climate change, as well as host genetics and immune responses and viral genetic changes. DENV consists of four serotypes, in turn composed of genotypes and genetically distinct clades. What drives the frequent replacement of a previously circulating DENV clade by another is unclear. Here, we investigate the replicative fitness of two clades of DENV serotype 2 in Aedes aegypti cells and mosquitoes collected from the region where the viruses circulated and conclude that increased replicative fitness could have contributed to a DENV clade replacement event in Nicaragua. These findings provide insight into vector-driven evolution of DENV epidemics.
INTRODUCTION
Dengue virus (DENV) is an arbovirus of global importance comprised of four phylogenetically related serotypes, DENV serotype 1 (DENV-1) to DENV-4. DENV is transmitted by Aedes aegypti and A. albopictus mosquitoes, which are typically found in tropical and subtropical regions of the world. However, due to a complex combination of factors that include migration, globalization, and climate change (1, 2), considerable expansion in the habitat range of the mosquito vectors that transmit DENV and in the geographic reach and number of dengue epidemics has recently occurred. DENV now causes an estimated 390 million annual human infections worldwide (3), a quarter of which manifest as an acute, debilitating fever (dengue fever) that can progress to life-threatening manifestations with vascular leak (dengue hemorrhagic fever and dengue shock syndrome) (4), severe bleeding, and/or organ damage, collectively known as severe dengue (5).
Risk factors that contribute to dengue severity include preexisting immunity and viral genetics (6), as well as host genetic factors. A previous infection with a virus of a different serotype has been shown in some instances to generate a cross-reactive enhancing effect instead of a protective immune response (7–11). More severe disease is postulated to result from cross-reactive T cells (12, 13) and/or antibody-dependent enhancement that increases uptake of the virus into Fcγ receptor-bearing target cells (14).
The introduction of new DENV serotypes and genotypes and frequent lineage replacements of closely related clades are factors that are important for understanding DENV evolutionary dynamics. These replacements may begin with a cloud of closely related virus strains (i.e., intrahost viral diversity), generated in part by the error-prone RNA-dependent RNA polymerase of DENV (15, 16). The intrahost diversity of viruses is typically described in chronic infections, such as those caused by HIV and hepatitis C virus (17–19), but has also been found in acute infections with DENV and other RNA viruses (20–23). During arbovirus infections, such populations are subject to natural selection in both the mosquito and the human host. For example, an increase in viral fitness, i.e., faster replication, speed of dissemination, or evasion of acquired or innate immunity, may lead to positive selection in mosquitoes or humans. A replicative advantage could result in a shorter extrinsic incubation period (EIP), the time taken for an infected mosquito to become infectious to a human host, which could in turn increase the likelihood and the rate of virus transmission to humans (24). Alternatively, stochastic events leading to a genetic bottleneck event within or among hosts may also lead to the emergence of new genetic variants that could compete with existing viral populations and may ultimately form new genetic clades. Such replacement events are not unique to DENV and have also been reported for other flaviviruses, such as Japanese encephalitis and West Nile viruses (25–27).
Previously, a clade replacement within the Asian-American genotype of DENV-2 was described in Managua, Nicaragua, in which the NI-1 clade was replaced by the NI-2B clade over three epidemic seasons (6). These clades are distinguishable from one another by nine nonsynonymous mutations in the coding region and four mutations in the untranslated regions (UTRs). The replacement event occurred simultaneously with an increase in disease severity among participants in two prospective studies in Managua. The increased severity of disease was found to result from a dynamic interplay between changing viral genetics and preexisting serotype-specific immunity (6). Children with DENV-1-specific immunity experienced more severe disease when infected by the NI-1 clade of DENV-2, whereas children previously exposed to DENV-3 manifested more severe disease when infected by DENV-2 NI-2B. In addition, it was found that NI-2B viruses displayed a replicative advantage over NI-1 viruses in in vitro experiments with C6/36 mosquito cells and human dendritic cells.
Yet to be defined, however, is the role of the mosquito vector in the DENV-2 clade replacement in Nicaragua. Here, we present data on the replicative fitness of the two DENV clades in cultured Aag2 cells derived from A. aegypti mosquitoes, the natural host for DENV. In addition, we extended our studies to F2 and F3 generation A. aegypti mosquitoes hatched from eggs collected in Managua, Nicaragua. Overall, our results suggest that NI-2B viruses replicate more efficiently than NI-1 viruses under pressure not only in cultured cells but also in competition assays in A. aegypti mosquitoes.
MATERIALS AND METHODS
Viruses and virus isolation.
The panel of viruses used in the work presented here was collected as part of two prospective pediatric studies of dengue in Nicaragua: a community-based cohort study and a hospital-based study (6). Both studies were reviewed and approved by the Institutional Review Boards (IRBs) of the University of California, Berkeley, and the Nicaraguan Ministry of Health. In addition, the cohort study protocol was reviewed and approved by the IRB of the International Vaccine Institute in Seoul, South Korea. Parents or legal guardians of all subjects in both studies provided written informed consent, and subjects 6 years of age and older provided assent. In the cohort study, participants were seen at the study health center, the Health Center Sócrates Flores Vivas (HCSFV), for all medical episodes and were encouraged to come to the HCSFV at the first signs of fever or dengue-like signs or symptoms. In the hospital-based study, subjects suspected of having dengue were enrolled upon presentation to the national pediatric reference hospital, the Hospital Infantil Manuel de Jesús Rivera. In both studies, blood was drawn on the day of presentation and at convalescence (14 to 21 days after the onset of symptoms) and subjected to molecular biological, virological, and serological diagnostic tests for dengue. In the acute-phase samples, DENV infection was confirmed by a nested reverse transcription-PCR (RT-PCR) assay which distinguishes among the four DENV serotypes using primers directed against the capsid gene (28, 29). Virus isolation was performed on RT-PCR-positive samples as previously described (30). In brief, serum from DENV RT-PCR-positive samples was diluted 1:20 in sterile phosphate-buffered saline (PBS) and used to inoculate A. albopictus C6/36 cells, which were then incubated at 34°C for 10 days in minimal essential medium (MEM; Gibco-BRL) containing Earle's salts, l-glutamate, nonessential amino acids, 0.11% sodium bicarbonate, 105 U/ml of penicillin, 75 U/ml of streptomycin, and 2% fetal bovine serum (FBS; Denville Scientific). Successful isolation was confirmed and serotype information was obtained by RT-PCR of the culture supernatant.
Full-length sequences of viral RNA genomes were obtained from the Broad Institute, as described by OhAinle et al. (6). Phylogenetic analysis has been described previously; briefly, full-length genomic sequences were used to construct maximum likelihood and Bayesian maximum clade credibility trees (6). This analysis defined two viral clades, NI-1 and NI-2. NI-2 was subcategorized into NI-2A and 2B. NI-2A was transient, and the vast majority of NI-2 viruses were classified as NI-2B (see http://www.broadinstitute.org/annotation/viral/Dengue/GlobalPopulationStructure.html for a description of the Nicaraguan sample sets). When a full-length sequence was not obtained, the clade was determined using a genotyping assay (6).
Cells and in vitro infections.
C6/36 cells (a gift from Paul Young, University of Queensland, Brisbane, Australia) were used for virus propagation (30). In brief, cells were inoculated with virus at a multiplicity of infection (MOI) of 0.5 and incubated at 28°C in M199 medium (Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin plus 100 μg/ml streptomycin (PenStrep), and 1× HEPES buffer. All viral isolates were passaged 1 to 4 times in C6/36 cells; for the experiments, viral stocks from passages 2 to 4 were used. Virus titers were determined using a quantitative RT-PCR (qRT-PCR) assay, described below. Single-infection and coinfection experiments (see below) were performed in A. aegypti (Aag2) cells (a gift from Carol Blair, Colorado State University), which were maintained at 28°C in Schneider's Drosophila medium (Invitrogen) supplemented with 8% FBS, PenStrep, and 1× MEM nonessential amino acids (Gibco). For single infections and coinfections with NI-1/NI-2B viruses, 5 × 104 Aag2 cells were plated in each well of a 96-well plate 24 h prior to infection. Bordering wells were filled with sterile water to prevent gas exchange. A viral inoculum containing 2.5 × 108 genome equivalents (GE) of each clade (single infections) or both clades combined (coinfections) was added to each well, and virus adsorption was performed in infection medium (Schneider's Drosophila medium with 2% FBS and PenStrep) for 2 h at 28°C. Cells were washed with PBS to remove unadsorbed virus, and cells were maintained in 200 μl of infection medium. All infections were performed in duplicate. Supernatants were collected at the time points postinfection indicated on the figures. For infections using mycophenolic acid (MPA), the 2-h viral adsorption period was followed by 2 washes with PBS, and cells were maintained in Schneider's Drosophila medium containing either 2 μg/ml MPA (Sigma) in methanol or the same volume of methanol without MPA.
Plaque-forming titers were determined by inoculating a confluent monolayer of BHK-21 cells (clone 15) in 12-well plates with virus supernatant serially diluted in alpha-MEM (Invitrogen) supplemented with 5% FBS and PenStrep in a total volume of 500 μl. After a 2-h incubation, wells were overlaid with 2× MEM and 2× low-melting-point agarose (Sigma), mixed 1:1. At 7 days postinfection (dpi), the wells were fixed with 10% formalin solution. After the agarose plugs were removed, the wells were stained using crystal violet (1% crystal violet in 30% ethanol) and rinsed with 30% ethanol. The plaques were counted, and the number of PFU/ml was calculated.
Mosquito husbandry and infections.
A. aegypti egg rafts were collected in Managua, Nicaragua, in 2010. Mosquito husbandry (egg hatching and mosquito rearing) and mosquito infections were performed in the Arbovirus Laboratory at the Wadsworth Center, Slingerlands, NY. Mosquitoes were reared in 30.5-cm3 cages placed within a controlled chamber that was maintained at a temperature of 28°C, a relative humidity of 50 to 65%, and a photoperiod of 12 h of light and 12 h of dark.
For each experiment, 300 F2 or F3 adult mosquitoes (100 males and 200 females) were sorted and allowed to mate in mesh-top 3.8-liter paper cartons for 5 to 7 days. During this period, the mosquitoes were fed using cotton pads saturated with a 10% sucrose solution. The mosquitoes were then switched to a water-only feed for a 24-h period prior to infection. On the day of infection, female mosquitoes were moved to 0.6-liter cups (∼50 females per cup) that were fitted with Hemotek membrane feeders (Discovery Workshops), sausage casings, or pledgets loaded with a blood meal that had been warmed to 37°C. Mosquitoes were fed on defibrinated rabbit or bovine blood (Rockland) containing 2.5% sucrose, 0.2 μM ATP, and fresh viral supernatant (viral titers were determined by qRT-PCR 16 h prior to infection; see below). On the day after feeding indicated below, mosquitoes were anesthetized using CO2, and engorged females were sorted into cups (∼20 to 50 mosquitoes per cup) and provided with continued access to 10% sucrose solution. At the time points indicated on the figures, live mosquitoes were collected, euthanized using triethylamine (Sigma), and handled as follows. On days 3 to 6, the whole bodies of mosquitoes were collected. On days 7 to 21, salivary glands were dissected and legs were separated from the carcass. The carcasses, legs, and salivary glands were placed in 2-ml microcentrifuge tubes filled with 1 ml of mosquito diluent (20% heat-inactivated FBS, Dulbecco's PBS, 50 U/ml penicillin, 50 μg/ml streptomycin, 50 μg/ml gentamicin, and 2.5 μg/ml amphotericin B [Fungizone]) containing one 5-mm metal bead (Daisy) for homogenization. Homogenization was performed for 30 s at 24 Hz in a Mixer Mill MM301 mixer (Retsch) (31). Samples were frozen at −80°C and processed as described below.
Viral RNA quantitation.
Viral RNA was extracted from cellular supernatants using a QIAamp viral RNA kit (Qiagen). RNA extraction from mosquito homogenates was automated using a QIAamp virus BioRobot MDx kit (Qiagen) and a BioRobot MDx workstation. qRT-PCR with a primer/probe combination targeting a 100-nucleotide region of the DENV NS5 gene was used to measure the levels of total DENV RNA in single infections. The numbers of viral RNA GE were extrapolated from a standard curve generated from a predesigned reference standard (6), using primers and probes specific to the Asian-American DENV-2 genotype directed to a region that is identical between NI-1 and NI-2B (forward primer, 5′ACAAGTCGAACAACCTGGTCCAT; reverse primer, 5′GCCGCACCATTGGTCTTCTC; probe, 5′FAM-TGGGATTTCCTCCCATGATTCCACTGG-TAMRA, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine). To assess the relative levels of NI-1 and NI-2B viruses in coinfection experiments, viral RNA was amplified as described previously (6) using a primer set designed to amplify a 950-nucleotide sequence in the NS3 gene that contains 10 single nucleotide polymorphisms (SNPs) from viruses of either clade. Amplicons were separated on a 1% agarose gel, and positive RT-PCR products were sequenced on an ABI 3730xl DNA analyzer. The relative levels of NI-1 and NI-2B viruses were determined using polySNP software (32) from ABI chromatogram peak height ratios at polymorphic sites, and 5 to 8 known SNP loci in the sequenced PCR products were averaged for each determination.
RESULTS
Growth of clades NI-1 and NI-2B in Aag2 cells.
We first analyzed the growth of clades NI-1 and NI-2B in A. aegypti Aag2 cells, an RNA interference (RNAi)-competent line derived from A. aegypti mosquitoes, the predominant vector for DENV transmission in Nicaragua. In contrast, C6/36 cells derived from A. albopictus mosquitoes are deficient in the RNAi pathway, which is an important antiviral function in invertebrates. We measured viral growth by determination of the number of focus-forming units (FFU) and were unable to detect statistically significant differences between the clades in single infections under normal growth conditions in Aag2 cells at any of the time points tested (Fig. 1). As previously established, however, differences in fitness (or growth) can be demonstrated by alternative assays, such as coinfection assays, that may identify subtle effects of genetic changes on replication kinetics (33–35). Furthermore, these data are not entirely surprising because (i) both clades were individually successful in nature in infecting mosquitoes and in causing illness in humans (6), and (ii) in vitro infections can fail to accurately represent the complexity of natural transmission cycles.
FIG 1.
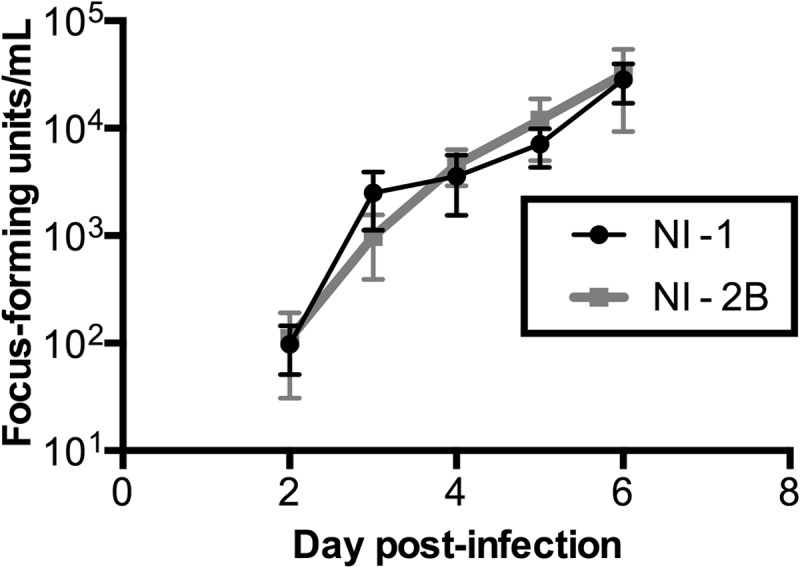
Growth curves of single-clade infections in Aag2 mosquito cells. Equal numbers of GE of five viral isolates from each clade were used to infect Aag2 cells, and supernatants were collected on days 2 to 6 postinfection. Supernatants were analyzed by determination of the number of focus-forming units on BHK cell monolayers, using anti-DENV monoclonal antibody 4G2. Plotted are the mean and standard error for the isolates at each time point. An unpaired t test revealed no statistically significant difference between the clades on any given day: day 2, P > 0.999; day 3, P = 0.318; day 4, P = 0.532; day 5, P = 0.944; and day 6, P = 0.667.
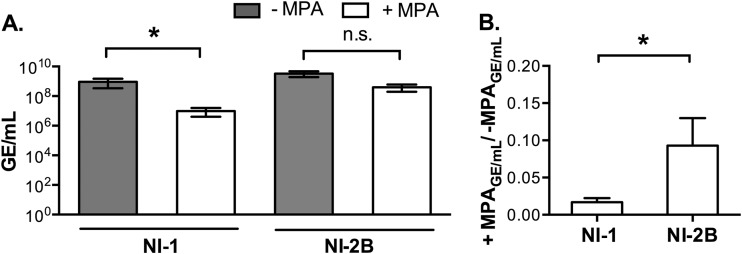
Since differences in replicative fitness were not detected in single infections, we considered that differences between the two clades could be detected under different forms of selective pressure. One means of challenging DENV replication is to treat cells with mycophenolic acid (MPA), which inhibits the de novo pathway of guanosine nucleotide synthesis in the host cell and hinders DENV replication (36). Aag2 cells were infected with NI-1 or NI-2B isolates (five from each clade) in the presence of MPA or an equal volume of the methanol diluent alone. We first performed a dose-response titration and selected a sublethal concentration of MPA (2 μg/ml) for subsequent experiments (data not shown). No significant difference in the ratio of the number of genome equivalents (GE)/number of PFU of clade NI-1 and NI-2B isolates grown in either Aag2 or C6/36 cells was observed (data not shown); therefore, we measured the titer as the number of GE for these experiments. The mean titer for five NI-1 isolates was significantly decreased in the presence of MPA (Wilcoxon rank sum test, P < 0.0001), whereas NI-2B virus replication was not significantly affected by MPA (Fig. 2A). Moreover, when we compared the reduction in replication of the clades to one another, the replication of NI-1 isolates decreased significantly in the presence of MPA compared to that of NI-2B isolates (Fig. 2B). Given that a comparison of single infections showed no statistically significant differences in virus replication (Fig. 1) and that under pressure a selective advantage for clade NI-2B viruses was apparent (Fig. 2), we concentrated our efforts on assays that would detect differences in the replication of the DENV-2 clades under conditions of stress.
FIG 2.
Replication of DENV-2 clades under selective pressure by MPA treatment. Equal numbers of GE of five viral isolates from each DENV-2 clade were used to infect Aag2 cells with the addition of either MPA or an equivalent amount of methanol alone following a 2-h adsorption period. The data are plotted as the mean and standard error of the mean. (A) Number of GE/ml for five isolates at 3 dpi in the presence or absence of MPA, as indicated. Statistical significance was determined by a paired t test: for NI-1, P < 0.001 (*); for NI-2B, P = 0.0810 (no significant difference [n.s.]). (B) Reduction in viral titer for each clade in the presence versus absence of MPA according to the formula number of GE with MPA/number of GE without MPA. Significance was determined using the Mann-Whitney test (*, P = 0.037).
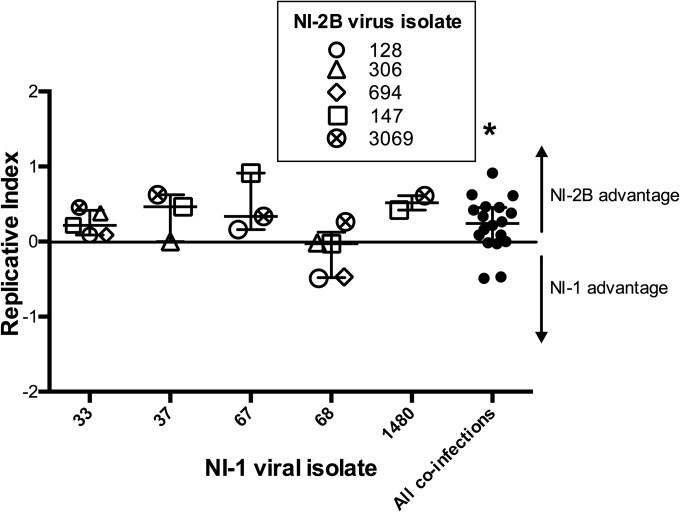
Another strategy to apply selective pressure to viruses in culture is by coinfection; this serves as a means to detect a replicative advantage between closely related viruses (6, 33–35). In the competition experiments presented here, the relative amounts of each clade are reported using a replicative index, defined as ln[(NI-2B/NI-1 ratio on day x)/(NI-2B/NI-1 ratio on day 0)] (34, 37). This approach adjusts for small errors in inputs, as it divides the NI-1/NI-2B ratio obtained on a given day (the NI-1/NI-2B ratio on day x) by the NI-1/NI-2B ratio input (the NI-1/NI-2B ratio on day 0). We coinfected Aag2 cells with one of five NI-1 isolates and a comparable amount of a series of NI-2B viruses in separate experiments. At 4 days postinfection (dpi), the supernatants were collected and frozen. We amplified the RNA extracted from supernatants by RT-PCR using primers specific for the NS3 region, capturing 10 known SNPs that differentiate NI-1 from NI-2B. DNA amplification was confirmed by gel electrophoresis, and the PCR amplicons were sequenced using the Sanger method. The resulting chromatograms were analyzed using the polySNP program (see Materials and Methods) to determine the relative amount of each clade in each sample. Each of the DENV isolates in this study originated from a single patient. As the genetic diversity of the virus population differs among patients, testing of various low-passage isolates represents the breadth of infections that exists in the Nicaraguan studies. For this reason, when relevant, each isolate was analyzed separately. NI-2B viruses replicated to higher levels than NI-1 viruses in four out of five of the competition assays performed at day 4 postinfection (Fig. 3). NI-1 isolate 68 was unusual, as it outcompeted some of the NI-2B isolates, suggesting that strain 68 was different from the other NI-1 isolates. On comparing the genomic sequence of strain 68 to the sequences of the other NI-1 isolates, several additional mutations (amino acid mutation H224Y in the NS1 protein and nucleotide mutations A → C10368 and G → A10415 in the 3′ UTR) which could contribute to the observed phenotypic difference of NI-1 isolate 68 were revealed. Overall, isolate 68 is not more closely related to the NI-2B strains than the other NI-1 isolates; in fact, of the isolates used here, isolate 68 falls in the middle of the cluster on a phylogenetic tree of the sequenced Nicaraguan NI-1 isolates (data not shown). The functional difference must be due to one or several particular mutations and warrants further investigation that is beyond the scope of this paper.
FIG 3.
Coinfection of NI-1 and NI-2B DENV-2 isolates in Aag2 mosquito cells. Aag-2 cells were infected at an MOI of 0.01 PFU/cell. The cells were infected with comparable numbers of GE of clinical isolates of NI-1 and NI-2B viruses mixed as indicated. Each NI-1 strain (indicated by the numbers on the x axis) was mixed with one of up to five different NI-2B viruses (denoted by the key) in three separate experiments. Cell supernatants were collected at 4 dpi. The ratio of the viral inoculum and viral growth on a given day in all coinfections was determined by sequencing. The data were analyzed using the replicative index, which was derived according to the formula ln[(NI-2B/NI-1 ratio on day x)/(NI-2B/NI-1 ratio on day 0)]. This parameter measures the relative log change of each clade on a given day from its input value. The neutral expectation, 0, shows no change in relative proportion from the input and is indicated by the horizontal line. The Wilcoxon signed rank test comparing the experimental results to the baseline value of 0 was used to determine significance. Error bars show the 25th and 75th percentiles. The line in the middle indicates the median. *, P= 0.027.
Single infection and coinfection of clades NI-1 and NI-2B in Nicaraguan A. aegypti mosquitoes.
We began our in vivo studies by infecting A. aegypti mosquitoes reared from eggs collected in Managua, Nicaragua, using a single virus from each clade. Detection of virus in each mosquito tissue indicates attainment of distinct milestones in arbovirus infection. The presence of virus in the mosquito carcass signifies infection of the mosquito midgut, whereas the presence of virus in the legs indicates that the virus has escaped the midgut barrier and disseminated to other organs. Consistent with the results obtained in Aag2 cells, we observed no statistically significant difference in titer between the clades at day 7 and later time points (Fig. 4A and data not shown). Another means of establishing a fitness advantage by a virus would be a superior ability to infect mosquitoes. Therefore, we calculated the infectivity of each clade in single infections by tallying the percentage of blood-fed mosquitoes that were infected, as determined by a positive RT-PCR result, but no significant difference between the clades in the carcasses or in the legs was observed at day 7 (Fig. 4B) or at later time points (data not shown). Similar results were obtained in the salivary glands, which were tested at day 21 only (data not shown).
FIG 4.
Titer and infectivity of DENV-2 clades in single infections in A. aegypti mosquitoes. (A) Titer (GE) of NI-1 or NI-2B viruses in Nicaraguan A. aegypti mosquito tissues on day 7 in single infections. Statistical significance was determined by the Mann-Whitney unpaired t test. P value for carcasses, 0.7379; P value for legs, 0.2081. (B) Infectivity calculated from the number of infected mosquitoes in a group (determined by RT-PCR) divided by the number of blood-fed mosquitoes in the same group. A two-sided Z-test comparing the proportions of the two groups was used to determine significance: for carcasses, P = 0.841; for legs, P = 0.575.
As shown in Fig. 1 to 3 and reported previously (33–35), coinfection of closely related RNA viruses provides a more sensitive means of detecting a superior replicative ability; thus, given the very limited number of low-generation Nicaraguan mosquito eggs and low-passage viral isolates, we decided to perform coinfection experiments in mosquitoes rather than to pursue the single-infection experiments. A. aegypti mosquitoes were offered a blood meal containing comparable amounts of GE of a viral isolate from each clade prepared from fresh viral supernatants from C6/36 cells, yielding a combined titer of 1 × 1010 GE/ml. Blood meal-engorged mosquitoes were separated into groups of 20 to 50, depending on feeding rates, and were collected on the days postinfection indicated below for dissection. Mosquitoes collected at 3, 5, and 6 dpi were not dissected, whereas for mosquitoes collected on days 7 to 21 dpi, legs and salivary glands were removed from euthanized mosquitoes and processed separately from their carcasses. As mentioned above, detection of virus in each mosquito tissue indicates attainment of distinct milestones in infection. In addition to testing the carcass and legs in coinfection experiments, we also measured the presence of virus in the salivary glands as an indication of virus dissemination to the saliva and, consequently, of transmission potential (26). Thus, we used detection of virus in salivary glands, which we reliably detected after 7 dpi, to demonstrate transmission potential.
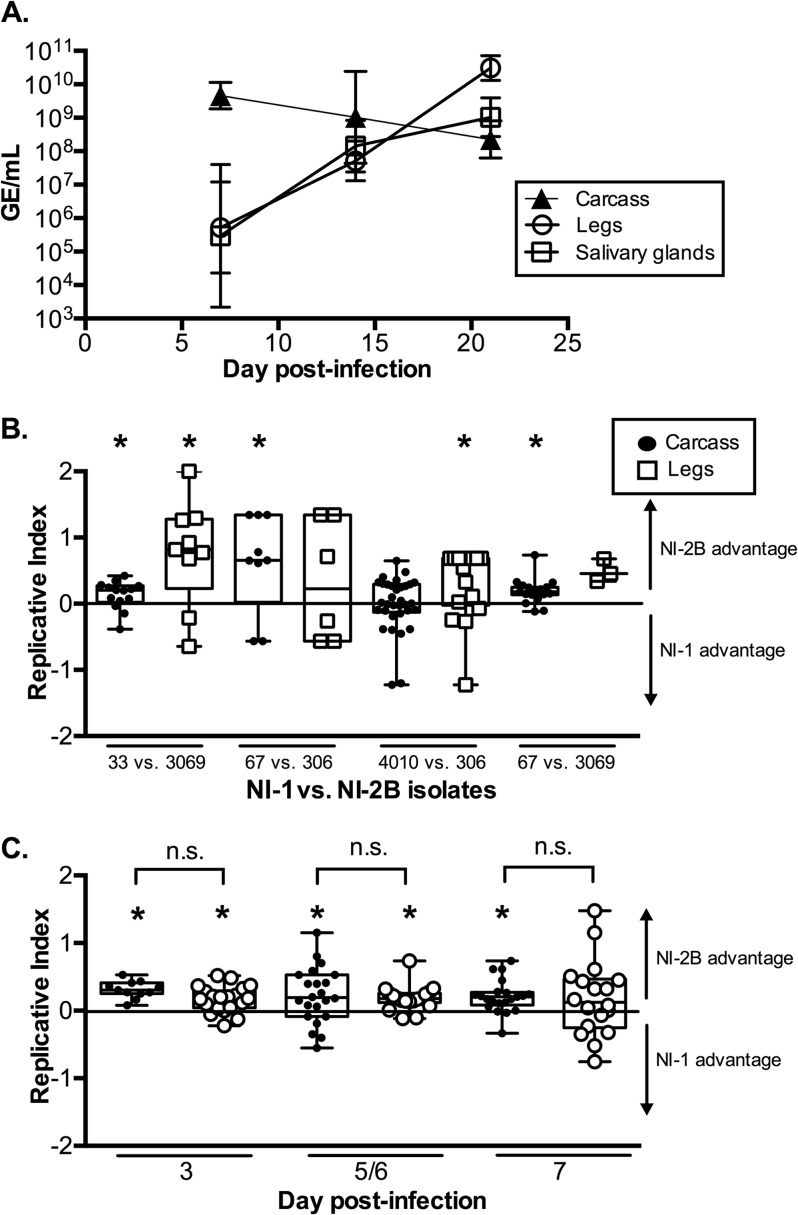
First, we confirmed that the mosquitoes had undergone an active DENV infection by measuring total RNA levels using qRT-PCR at different time points and in different tissues in five mosquitoes coinfected with isolates 67 and 3069. GE titers increased in the legs and salivary glands over time, indicating that a productive infection had taken place (Fig. 5A). As expected, the viral titer in the carcass dropped over time as virus disseminated out of the midgut and into the legs and salivary glands (38). Next, four separate pairings of NI-1 and NI-2B isolates were competed; one pair, isolate 67 versus isolate 3069, was competed twice, for a total of five in vivo competition experiments.
FIG 5.
Coinfections of DENV-2 clades in Nicaraguan A. aegypti mosquitoes. (A) At 7, 14, and 21 dpi, RNA was extracted from tissues from five mosquitoes infected with DENV-2 isolates 67 and 3069. The RNA from mosquitoes previously identified to be DENV infected by RT-PCR was amplified by qRT-PCR (see Materials and Methods). Plotted is the geometric mean titer with the 95% confidence interval. (B) Mosquitoes that fed on a blood meal containing comparable numbers of GE of a viral isolate from each of the two DENV-2 clades were harvested at 7 dpi. The replicative index was calculated as described in the legend to Fig. 3. The middle line is the median, the top and bottom of the box represent the 25th and 75th percentiles, respectively, and the whiskers represent the minimum and maximum values in each data set. Plotted are the results for the carcasses and legs from four separate competition assays. Statistical significance was determined as described in the legend to Fig. 3. The P values and numbers of infected mosquitoes for each competition assay are as follows, with significant values indicated in bold below and with an asterisk in the figure: for viral isolate 33 versus viral isolate 3069, P = 0.035 for carcasses (n = 14) and P = 0.021 for legs (n = 9); for isolate 67 versus isolate 306, P = 0.020 for carcasses (n = 9) and P = 0.343 for legs (n = 6); for isolate 4010 versus isolate 306, P = 0.272 for carcasses (n = 14) and P = 0.020 for legs (n = 17); and for isolate 67 versus isolate 3069, P < 0.001 (n = 19) for carcasses and P = 0.109 (n = 3) for legs. (C) One clade competition assay (isolate 67 versus isolate 3069) was also performed at earlier time points with different NI-1/NI-2B ratios of 0.4:0.6 and 0.6:0.4. The replicative index for viruses in carcasses is shown. Black circles, NI-1/NI-2B ratio of 0.4:0.6; white circles, NI-1/NI-2B ratio of 0.6:0.4. The P values (with significant values indicated in bold below and with an asterisk in the figure) and numbers of mosquitoes assayed for each infection are as follows: for an NI-1/NI-2B ratio of 0.4:0.6, P = 0.001 (n = 21) on day 3, P = 0.002 (n = 11) on day 5/6, and P < 0.001 (n = 19) on day 7; for an NI-1/NI-2B ratio of 0.6:0.4, P = 0.003 (n = 11) on day 3, P = 0.026 (n = 22) on day 5/6, and P = 0.023 (n = 18) on day 7. No statistically significant difference (n.s.) was found between the coinfections of each isolate pair on a given day.
Although different clinical isolates from the two clades were competed against each other and some variation was found, a reproducible pattern was observed among pairs of coinfected viruses during competition. The replicative index (as described above) of the viruses in the carcasses and legs of infected mosquitoes at 7 dpi in four separate coinfection experiments is shown in Fig. 5B. In all competition experiments, the median replicative index of clade NI-2B viruses was significantly higher than that of NI-1 viruses in either the legs or the carcass. The replicative advantage of NI-2B viruses at 7 dpi suggested that differences in replicative ability might also be observed at earlier time points. To investigate this, we conducted two coinfection experiments in which blood-fed mosquitoes were collected at three early time points: 3, 5/6, and 7 dpi. In all coinfection experiments at early time points, NI-2B viruses demonstrated a replicative advantage over NI-1 viruses (Fig. 5C).
We also competed one pair of isolates (isolates 67 and 3069) twice, using different input ratios of NI-1/NI-2B. The levels of NI-1 viruses in the blood meal of the first coinfection were higher than those of NI-2B viruses (NI-1/NI-2B, 0.6:0.4), while the second coinfection had a higher NI-2B input (NI-1/NI-2B, 0.4:0.6). No statistically significant significance between the replicative indices of this isolate pairing was observed on a given day, suggestive of the reproducibility of the assay. Furthermore, the analysis of the replicative indices of these two coinfections with reciprocal input NI-1/NI-2B ratios demonstrated a significant replicative advantage of NI-2B viruses at the earliest time point of infection, 3 dpi, that continued until the last shared time point in the these coinfections, 7 dpi (Fig. 5C).
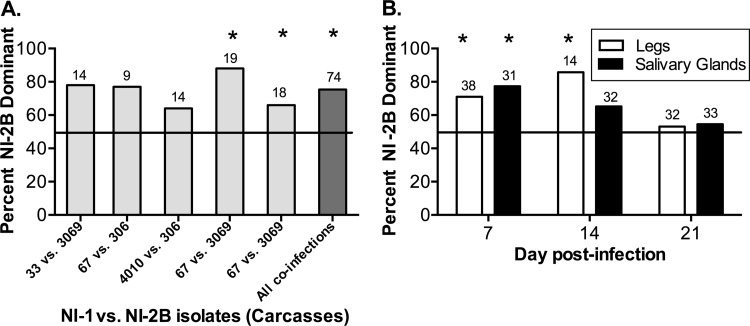
An alternative approach is to determine the proportion of mosquitoes with higher levels of NI-2B viruses than NI-1 viruses. Accordingly, we tallied the number of mosquito carcasses in which clade NI-2B was more abundant than clade NI-1, defined as >50% NI-2B in a given mosquito. In the carcasses of mosquitoes in each of the five coinfections, NI-2B was found at higher levels than NI-1, with percentages ranging from 60 to 90% among the coinfections (Fig. 6A). The same analysis was performed for the legs and salivary glands as a function of time, combining data from all coinfection experiments (Fig. 6B). In the legs, more mosquitoes in which NI-2B was dominant were observed at 7 and 14 dpi. The salivary glands displayed more mosquitoes in which NI-2B was dominant at 7 dpi; however, this replicative advantage decreased and disappeared at later time points.
FIG 6.
Proportion of mosquitoes with the dominant NI-2B clade during coinfection. The number of mosquitoes in which NI-2B virus was found to be more abundant than NI-1 virus, defined as greater than 50% NI-2B isolates in a given mosquito, was tallied and reported as a percentage of the total number of infected mosquitoes in each group. (A) The percentage of mosquitoes in which the NI-2B clade was dominant (i.e., more NI-2B viruses than NI-1 viruses) in day 7 carcasses in five coinfection experiments (virus pairs listed on the x axis) is shown in light gray, with the number of mosquitoes indicated above each bar; the last bar represents the sum tally for all five coinfections. A two-sided exact binomial test was used to determine significance, using a hypothetical probability of success of 0.5 (i.e., assuming that both clades have equivalent replicative fitness). The P values (with significant values indicated in bold below and with an asterisk in the figure) and numbers of mosquitoes assayed for each infection are as follows: for NI-1/NI-2B, for isolate 33 versus isolate 3069, P = 0.057 (n = 14); for isolate 67 versus isolate 306, P = 0.180 (n = 9); for isolate 4010 versus isolate 306, P = 0.424 (n = 14); for isolate 67 versus isolate 3069, P < 0.001 (n = 19); for isolate 67 versus isolate 3069, P= 0.031 (n = 18); for all coinfections, P < 0.001 (n = 74). (B) Percentage of mosquitoes in which clade NI-2B was dominant in legs and salivary glands. Samples from all coinfection assays, collected on the day indicated below, were tallied together and are presented as a function of time (day 7, day 14, and day 21). Significance was determined as described above for panel A. The P values (with significant values indicated in bold below and with an asterisk in the figure) and numbers of tissues assayed for each infection are as follows: for legs, day 7, P = 0.014 (n = 38); day 14, P = 0.013 (n = 14); and day 21, P = 0.860 (n = 32); for salivary glands, day 7, P = 0.003 (n = 31); day 14, P = 0.600 (n = 32); and day 21, P = 0.729 (n = 33).
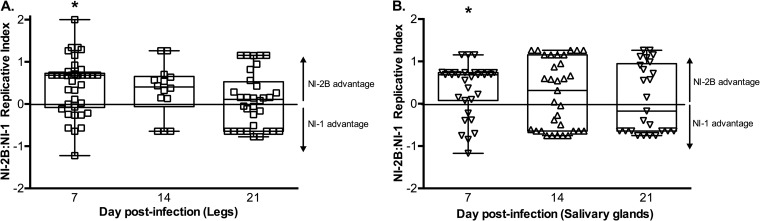
The dissemination and transmission potential of each clade were measured by analyzing the dissected legs and salivary glands, respectively, of mosquitoes coinfected with NI-1 and NI-2B viruses. As seen in the carcasses of these mosquitoes, the median replicative index in legs and salivary glands demonstrated significantly higher levels of NI-2B at day 7 postinfection (Fig. 7). However, in both tissue types, this replicative advantage was lost by days 14 and 21. Combined with the NI-2B dominance disappearing at later time points, these data support the hypothesis that NI-2B has a shorter EIP, the time for an infected mosquito to become infectious, than NI-1.
FIG 7.
Replicative advantage of NI-2B viruses in mosquito legs and salivary glands. Replicative index values for legs (A) and salivary glands (B) of all coinfection experiments pooled by day are shown. The P values (with significant values indicated in bold below and with an asterisk in the figure) and numbers of mosquitoes assayed for each infection are as follows: for legs, day 7, P < 0.001 (n = 41); day 14, P = 0.159 (n = 14); and day 21, P = 0.055 (n = 32); for salivary glands, day 7, P = 0.012 (n = 31); day 14, P = 0.136 (n = 33); and day 21, P = 0.325 (n = 25).
DISCUSSION
In recent years, understanding the role of genotypes and clades in lineage replacements of DENV as well as of genotype- and clade-specific immunity has become increasingly important. For example, the DENV-2 clade replacement event in Managua, Nicaragua, between 2005 and 2007 was associated with an increase in disease severity within the human population that was attributed to a complex interaction between serotype-specific immunity and genetic changes in the virus (6). These earlier experiments also suggested that the clade replacement could be explained by the increased fitness of NI-2B viruses in human primary dendritic cells as well as in A. albopictus mosquito cells. However, since A. aegypti mosquitoes are the most prevalent vectors for DENV in Nicaragua, experimental evaluation of virus replication in this species would better approximate the natural conditions for studying the role of viral fitness in the Nicaraguan clade replacement event. For this reason, in the present study, we tested both A. aegypti cultured cells and live mosquitoes to more fully characterize the role of the vector in clade replacement and to obtain a better approximation of relative fitness in vivo (39).
Our initial experiments in Aag2 cells, challenging virus replication by using an inhibitor (MPA) or by coinfection of DENV-2 clades, showed a reproducible advantage of NI-2B isolates over NI-1 isolates. Consistent with previous findings presented in the literature (33–35), we did not find significant differences between clades in single infections in cultured cells or in mosquito tissues. Significantly, competition experiments in A. aegypti mosquitoes revealed that NI-2B also displayed a similar advantage at the organismal level, as measured by several parameters. In coinfection experiments with several pairs of low-passage isolates in A. aegypti mosquitoes, NI-2B isolates were found to attain a higher replicative index than NI-1 isolates at the earliest times of viral detection, 3 to 7 dpi. At 7 dpi, the first day of virus detection in legs and salivary glands, NI-2B viruses had a significantly higher replicative index in the legs and salivary glands. However, this advantage was lost by days 14 and 21 dpi. We also found that the percentage of mosquitoes in which NI-2B was dominant was significantly higher than the percentage of mosquitoes in which NI-1 was dominant on day 7 but not at later time points. Taken together, these data show that NI-2B has a replicative advantage over NI-1 early in infection and that this advantage wanes at later time points. These results suggest a shorter EIP of NI-2B viruses than NI-1 viruses in A. aegypti mosquitoes.
NI-1 and NI-2B viruses exhibit nine amino acid differences in their proteins: K97R in the capsid protein (C), M492V in the envelope protein (E), R94K in nonstructural protein 1 (NS1), L279F in NS1, T245P in NS3, N245S in NS4B, K200Q in NS5, T290I in NS5, and R401K in NS5. In addition, there are four nucleotide variations in the 5′ and 3′ UTRs. The higher replicative fitness of NI-2B viruses than NI-1 viruses could potentially have resulted from genetic differences influencing one or more steps in the DENV life cycle, such as virus binding/endocytosis, uncoating, membrane fusion, translation, RNA replication, and/or viral egress. For example, a single amino acid change in the DENV-2 envelope protein of an Asian genotype at position 390 compared to that found in the American DENV-2 genotype (Asn to Asp, Ser, or Aln) was associated with decreased replication in monocyte-derived macrophages and dendritic cells (40, 41). In the context of the Nicaraguan DENV-2 isolates, the mutations in the NS3 helicase and/or NS5 methyltransferase and RNA-dependent RNA polymerase in NI-1 compared with the sequences of the NI-2B enzymes could presumably have an effect on viral RNA synthesis; the ability of these mutations to affect the biochemical function of these enzymes is under investigation. In addition, genetic determinants in the DENV-2 3′ UTR have been associated with a decrease in translation efficiency and, consequently, reduced DENV infectivity in cultured cells (42). Alternatively, genetic determinants in coding and noncoding regions can also influence host antiviral defense mechanisms. For example, differences in the production or activity of the short flaviviral noncoding RNAs (sfRNA) can influence viral interactions with the interferon and RNAi innate immune pathways (43, 44). The DENV and West Nile virus sfRNAs are generated by the secondary structure of the 3′ UTR blocking the host cell exonuclease XRN1 (45), and sfRNA has been shown to interfere with Dicer, thus preventing cleavage of double-stranded RNA (43). NI-1 and NI-2B differ at several nucleotide positions in the sfRNA region of the 3′ UTR; these differences could potentially result in more effective interference with antiviral host defenses, resulting in a replicative advantage for NI-2B viruses.
Numerous studies have demonstrated the importance of defining DENV genotype/clade replacement events (46). Replacement of the DENV-2 American genotype with the Asian-American genotype in the Americas was associated with increased disease severity (47). Likewise, clade replacement in DENV-3 was associated with an increase in disease severity in Sri Lanka after 1989 (48, 49). Genotype specificity has also been suggested to be a potential cause for the lack of protection against DENV-2 observed during the phase IIb proof-of-concept trial of Sanofi Pasteur's tetravalent CYD23 vaccine in Thailand (50). Genetic differences between the DENV-2 strain included in the tetravalent vaccine and the virus that circulated in the subjects at the time of the vaccine trial could have contributed to the vaccine's reduced efficacy against DENV-2 (50). Thus, genetic differences in the virus can influence disease manifestations, transmission potential, and possibly, vaccine efficacy.
Tracking human health alone is insufficient to understand viral evolution and control outbreaks. The investigation of vectorial capacity, biting and breeding patterns, and geographic range driven by climate change (51) may be key in controlling the spread of arboviruses. In this regard, the Ross-MacDonald model (52), a mathematical model of mosquito-borne pathogen transmission estimating vectorial capacity related to the basic reproductive number, R0, can be adapted to assess DENV transmission potential. The model includes mosquito and human variables, such as mosquito density, biting rate, probability of successful infection in humans and in mosquitoes, recovery rate in humans, mosquito mortality rate, and the EIP. The EIP is expressed as an exponent; thus, a slight change in EIP could have a large impact on the transmission of DENV in a human population (24).
Here, we have shown that a DENV-2 clade replacement event in Nicaragua appears to have been driven by a replicative advantage of the replacing clade in the native mosquito vector, A. aegypti, rather than a stochastic event. Our data suggest a shorter EIP of clade NI-2B, which in turn could increase its epidemic potential. Functional studies such as these help to define the effect of DENV lineage replacements and can improve our understanding of DENV evolutionary dynamics in relation to disease severity and transmission potential.
ACKNOWLEDGMENTS
We thank Saira Saborio and Angel Balmaseda in the National Virology Laboratory of the Nicaraguan Ministry of Health for the virus isolates and Sonia Valle and Emperatriz Lugo in the Department of Entomology of the Nicaraguan Ministry of Health for collecting and preparing the A. aegypti eggs in Managua used to rear the mosquitoes used in these studies. We are grateful to Richard Paul for his thoughtful comments, to Hope Biswas for assistance in biostatistical support, and to Nikitha Mangalapally for technical assistance. C.A.Q. thanks S. D. Guzman.
This work was supported by NIH grant R01 GM087405 (to E.H.).
Footnotes
Published ahead of print 3 September 2014
REFERENCES
- 1.Gould EA, Higgs S. 2009. Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg. 103:109–121. 10.1016/j.trstmh.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle JL, Harris E. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 62:71–92. 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed. World Health Organization Press, Geneva, Switzerland. [Google Scholar]
- 5.World Health Organization. 2009. Dengue guidelines for diagnosis, treatment, prevention and control. New edition. World Health Organization Press, Geneva, Switzerland. [PubMed] [Google Scholar]
- 6.OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, Nunez A, Lennon NJ, Birren BW, Gordon A, Henn MR, Harris E. 2011. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci. Transl. Med. 3:114ra128. 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke DS, Nisalak A, Johnson DE, Scott RM. 1988. A prospective study of dengue infections in Bangkok. Am. J. Trop. Med. Hyg. 38:172–180. [DOI] [PubMed] [Google Scholar]
- 8.Graham RR, Juffrie M, Tan R, Hayes CG, Laksono I, Ma'roef C, Erlin Sutaryo, Porter KR, Halstead SB. 1999. A prospective seroepidemiologic study on dengue in children four to nine years of age in Yogyakarta, Indonesia. I. Studies in 1995-1996. Am. J. Trop. Med. Hyg. 61:412–419. [DOI] [PubMed] [Google Scholar]
- 9.Guzman MG, Alvarez M, Rodriguez-Roche R, Bernardo L, Montes T, Vazquez S, Morier L, Alvarez A, Gould EA, Kouri G, Halstead SB. 2007. Neutralizing antibodies after infection with dengue 1 virus. Emerg. Infect. Dis. 13:282–286. 10.3201/eid1302.060539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman MG, Kouri G, Valdes L, Bravo J, Vazquez S, Halstead SB. 2002. Enhanced severity of secondary dengue-2 infections: death rates in 1981 and 1997 Cuban outbreaks. Rev. Panam. Salud Publica 11:223–227. 10.1590/S1020-49892002000400003. [DOI] [PubMed] [Google Scholar]
- 11.Thein S, Aung MM, Shwe TN, Aye M, Zaw A, Aye K, Aye KM, Aaskov J. 1997. Risk factors in dengue shock syndrome. Am. J. Trop. Med. Hyg. 56:566–572. [DOI] [PubMed] [Google Scholar]
- 12.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, Sawasdivorn S, Duangchinda T, Dong T, Rowland-Jones S, Yenchitsomanus PT, McMichael A, Malasit P, Screaton G. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921–927. 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 13.Rothman AL. 2011. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 11:532–543. 10.1038/nri3014. [DOI] [PubMed] [Google Scholar]
- 14.Halstead SB. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60:421–467. 10.1016/S0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 15.Ciota AT, Kramer LD. 2010. Insights into arbovirus evolution and adaptation from experimental studies. Viruses 2:2594–2617. 10.3390/v2122594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eigen M. 1996. On the nature of virus quasispecies. Trends Microbiol. 4:216–218. 10.1016/0966-842X(96)20011-3. [DOI] [PubMed] [Google Scholar]
- 17.Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L, Diaz G, Balestrieri A, Purcell RH. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. U. S. A. 99:3081–3086. 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joos B, Trkola A, Fischer M, Kuster H, Rusert P, Leemann C, Boni J, Oxenius A, Price DA, Phillips RE, Wong JK, Hirschel B, Weber R, Gunthard HF. 2005. Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J. Virol. 79:9026–9037. 10.1128/JVI.79.14.9026-9037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan DG, Bruden D, Deubner H, McArdle S, Chung M, Christensen C, Hennessy T, Homan C, Williams J, McMahon BJ, Gretch DR. 2007. Hepatitis C virus dynamics during natural infection are associated with long-term histological outcome of chronic hepatitis C disease. J. Infect. Dis. 196:239–248. 10.1086/518895. [DOI] [PubMed] [Google Scholar]
- 20.Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. 2012. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J. Virol. 86:8546–8558. 10.1128/JVI.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerzak G, Bernard KA, Kramer LD, Ebel GD. 2005. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J. Gen. Virol. 86:2175–2183. 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin SR, Hsieh SC, Yueh YY, Lin TH, Chao DY, Chen WJ, King CC, Wang WK. 2004. Study of sequence variation of dengue type 3 virus in naturally infected mosquitoes and human hosts: implications for transmission and evolution. J. Virol. 78:12717–12721. 10.1128/JVI.78.22.12717-12721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thai KT, Henn MR, Zody MC, Tricou V, Nguyet NM, Charlebois P, Lennon NJ, Green L, de Vries PJ, Hien TT, Farrar J, van Doorn HR, de Jong MD, Birren BW, Holmes EC, Simmons CP. 2012. High-resolution analysis of intrahost genetic diversity in dengue virus serotype 1 infection identifies mixed infections. J. Virol. 86:835–843. 10.1128/JVI.05985-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tjaden NB, Thomas SM, Fischer D, Beierkuhnlein C. 2013. Extrinsic incubation period of dengue: knowledge, backlog, and applications of temperature dependence. PLoS Negl. Trop. Dis. 7:e2207. 10.1371/journal.pntd.0002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DW, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett AD. 2005. Phylogenetic analysis of North American West Nile virus isolates, 2001-2004: evidence for the emergence of a dominant genotype. Virology 342:252–265. 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am. J. Trop. Med. Hyg. 71:493–500. [PubMed] [Google Scholar]
- 27.Schuh AJ, Ward MJ, Leigh Brown AJ, Barrett AD. 2014. Dynamics of the emergence and establishment of a newly dominant genotype of Japanese encephalitis virus throughout Asia. J. Virol. 88:4522–4532. 10.1128/JVI.02686-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris E, Roberts TG, Smith L, Selle J, Kramer LD, Valle S, Sandoval E, Balmaseda A. 1998. Typing of dengue viruses in clinical specimens and mosquitoes by single-tube multiplex reverse transcriptase PCR. J. Clin. Microbiol. 36:2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balmaseda A, Sandoval E, Perez L, Gutierrez CM, Harris E. 1999. Application of molecular typing techniques in the 1998 dengue epidemic in Nicaragua. Am. J. Trop. Med. Hyg. 61:893–897. [DOI] [PubMed] [Google Scholar]
- 31.Ciota AT, Lovelace AO, Jia Y, Davis LJ, Young DS, Kramer LD. 2008. Characterization of mosquito-adapted West Nile virus. J. Gen. Virol. 89:1633–1642. 10.1099/vir.0.2008/000893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall GS, Little DP. 2007. Relative quantitation of virus population size in mixed genotype infections using sequencing chromatograms. J. Virol. Methods 146:22–28. 10.1016/j.jviromet.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciota AT, Payne AF, Ngo KA, Kramer LD. 2014. Consequences of in vitro host shift for St. Louis encephalitis virus. J. Gen. Virol. 95:1281–1288. 10.1099/vir.0.063545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland JJ, de la Torre JC, Clarke DK, Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novella IS, Ball LA, Wertz GW. 2004. Fitness analyses of vesicular stomatitis strains with rearranged genomes reveal replicative disadvantages. J. Virol. 78:9837–9841. 10.1128/JVI.78.18.9837-9841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diamond MS, Zachariah M, Harris E. 2002. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304:211–221. 10.1006/viro.2002.1685. [DOI] [PubMed] [Google Scholar]
- 37.Martinez MA, Carrillo C, Gonzalez-Candelas F, Moya A, Domingo E, Sobrino F. 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65:3954–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardy JL, Houk EJ, Kramer LD, Reeves WC. 1983. Intrinsic factors affecting vector competence of mosquitoes for arboviruses. Annu. Rev. Entomol. 28:229–262. 10.1146/annurev.en.28.010183.001305. [DOI] [PubMed] [Google Scholar]
- 39.Lambrechts L, Fansiri T, Pongsiri A, Thaisomboonsuk B, Klungthong C, Richardson JH, Ponlawat A, Jarman RG, Scott TW. 2012. Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J. Virol. 86:1853–1861. 10.1128/JVI.06458-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cologna R, Rico-Hesse R. 2003. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 77:3929–3938. 10.1128/JVI.77.7.3929-3938.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pryor MJ, Carr JM, Hocking H, Davidson AD, Li P, Wright PJ. 2001. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 65:427–434. [DOI] [PubMed] [Google Scholar]
- 42.Edgil D, Diamond MS, Holden KL, Paranjape SM, Harris E. 2003. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology 317:275–290. 10.1016/j.virol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 43.Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. 2012. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 86:13486–13500. 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, Silverman R, Akira S, Barton DJ, Diamond MS, Khromykh AA. 2012. West Nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J. Virol. 86:5708–5718. 10.1128/JVI.00207-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3:19–28. 10.1016/S1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 47.Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230:244–251. 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- 48.Kanakaratne N, Wahala WM, Messer WB, Tissera HA, Shahani A, Abeysinghe N, de-Silva AM, Gunasekera M. 2009. Severe dengue epidemics in Sri Lanka, 2003-2006. Emerg. Infect. Dis. 15:192–199. 10.3201/eid1502.080926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. 2003. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg. Infect. Dis. 9:800–809. 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 51.Ciota AT, Kramer LD. 2013. Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 5:3021–3047. 10.3390/v5123021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macdonald G. 1957. The epidemiology and control of malaria. Oxford University Press; London, United Kingdom. [Google Scholar]