ABSTRACT
Cancer cells are susceptible to oncolytic viruses, albeit variably. Human adenoviruses (HAdVs) are widely used oncolytic agents that have been engineered to produce progeny within the tumor and elicit bystander effects. We searched for host factors enhancing bystander effects and conducted a targeted RNA interference screen against guanine nucleotide exchange factors (GEFs) of small GTPases. We show that the unfolded protein response (UPR), which is readily inducible in aggressive tumor cells, enhances melanoma or epithelial cancer cell killing upon HAdV infection. UPR was triggered by knockdown of Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF-1) or the GBF-1 inhibitor golgicide A (GCA) and stimulated HAdV infection. GBF-1 is a GEF for ADP ribosylation factors (Arfs) regulating endoplasmic reticulum (ER)-to-Golgi apparatus and intra-Golgi apparatus membrane transport. Cells treated with GCA enhanced HAdV-induced cytopathic effects in epithelial and melanoma cancer cells but not normal cells, if the drug was applied several hours prior to HAdV inoculation. This was shown by real-time label-free impedance measurements using the xCELLigence system. GCA-treated cells contained fewer incoming HAdVs than control cells, but GCA treatment boosted HAdV titers and spreading in cancer cells. GCA enhanced viral gene expression or transgene expression from the cytomegalovirus promoter of B- or C-species HAdVs but did not enhance viral early region 1A (E1A) expression in uninfected cell lines or cells transfected with plasmid reporter DNA. The UPR-enhanced cell killing required the nuclease activity of the UPR sensor inositol-requiring enzyme 1 (IRE-1) and X box binding protein 1 (XBP-1), which alleviate ER stress. The collective results show that chemical UPR induction and viruses boost tumor cell killing by enhancing oncolytic viral efficacy.
IMPORTANCE Cancer is difficult to combat. A wide range of oncolytic viruses show promise for killing cancer cells, yet the efficacy of oncolytic killing is low. We searched for host factors enhancing adenovirus cancer cell killing and found that the knockdown of Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF-1) or chemical inhibition of GBF-1 enhanced adenovirus infection by triggering the IRE-1/XBP-1 branch of the unfolded protein response (UPR). IRE-1/XBP-1 promote cell survival and enhanced the levels of the adenoviral immediate early gene product E1A, virus spreading, and killing of cancer cells. Aggressive tumor cells depend on a readily inducible UPR and, hence, present prime targets for a combined strategy involving adenoviruses and small chemicals inducing UPR.
INTRODUCTION
Cancer is a devastating multifactorial disease and difficult to combat owing to genomic instability, uncontrolled proliferation, dissemination, and poor immunologic control (for reviews, see references 1 and 2). Treatment with oncolytic viruses is an emerging therapeutic practice (reviewed in references 3 and 4). Oncolytic viral therapy takes advantage of the fact that many enveloped and nonenveloped viruses destroy host cells as part of their replication strategy. Oncolytic viruses include herpesvirus, measles virus, vesicular stomatitis virus, influenza A virus, Newcastle disease virus, vaccinia virus, poliovirus, parvovirus, and adenovirus. Currently, human adenoviruses (HAdVs) are the most widely used oncolytic agents that have been engineered to produce progeny within the tumor and kill tumor rather than normal cells (5).
Oncolytic viruses directly kill cancer cells and may trigger an immune response against cancer-specific or viral epitopes presented on major histocompatibility complex class I protein to immune cells. This poses the problem that an oncolytic virus can be eliminated by the immune system before reaching full efficacy, for example, if the host is not tolerant against immune-dominant viral antigens. Since immune tolerance against dominant viral antigens is rare, other ways to enhance the oncolytic efficacy of viruses have been explored. For example, treatments with biological agents or chemicals or the physical induction of stress sensitizes tumor cells to be killed by oncolytic viruses (6, 7). In some instances, stress induction leads to the inhibition of virus replication; for example, radiation therapy attenuates vaccinia virus infection (8). Alternatively, inhibition of cell stress can enhance oncolysis; for example, blockage of endoplasmic reticulum (ER) stress augments rhabdovirus oncolysis (9).
Here, we report that chemical or genetic inhibition of Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor 1 (GBF-1) activates the unfolded protein response (UPR) from the ER and enhances gene expression from HAdV species C, type 5 (HAdV-C5), and HAdV species B, type 3 (HAdV-B3). GBF-1 inhibition boosts HAdV-induced cell killing and viral dissemination in human lung epithelial or melanoma-derived cancer cells. GBF-1 is a cis-Golgi guanine nucleotide exchange factor (GEF) for ADP ribosylation factors (Arfs) and regulates ER-Golgi apparatus and intra-Golgi apparatus membrane traffic (10, 11). It is widely expressed in human cells and controls the dynamics of Arfs and COP-I at the ER-Golgi apparatus interface (12, 13). Notably, GBF-1 depletion by RNA interference induces the UPR by locating site-specific protease 1 (SP1) and SP2 from the Golgi apparatus to the ER and proteolytic activation of activating transcription factor 6 (ATF-6) (14).
HAdVs are widespread nonenveloped DNA viruses causing mild, self-limiting infections in immunocompetent individuals (15). Species B and C HAdVs target the urogenital and respiratory tracts and have been extensively developed into vectors for clinical therapy (5). They attach to host cells via the coxsackievirus adenovirus receptor (CAR), CD46, or desmoglein 2 and in most cases integrin secondary receptors (16). This triggers initial steps of virus uncoating, internalization, and endosomal membrane rupture by pH-independent mechanisms (17–19). Cytosolic viruses are transported by dynein/dynactin and microtubules to the nuclear pore complex, where kinesin-mediated virus disassembly and disruption of the nuclear pore complex occur and viral DNA is imported into the nucleus (20–23). Expression of the early region 1A (E1A) genes from episomal viral DNA controls a range of host and viral genes (24, 25). E1A starts the viral gene expression and genome replication programs, which drive viral immune escape and, ultimately, the release of progeny viruses from the nucleus upon cell lysis (26), yet the clinical oncolytic efficacy of adenoviruses and other virus-derived oncolytic vectors has been modest (27, 28). This is paralleled by a recent observation from 2-dimensional cell cultures showing that inefficient viral transmission correlates with low-level lytic infection events (29). The results presented here show that induction of the UPR through the inositol-requiring enzyme 1 (IRE-1) sensor and the X box binding protein 1 (XBP-1) transcription factor leads to enhanced viral cytotoxicity in primary human cancer cells. This is a hitherto unknown pathway leading from ER stress and the host transcriptional response to enhanced viral gene expression and oncolysis.
MATERIALS AND METHODS
Cells and viruses.
Cells and viruses were grown as described before (17, 30–32). A549 cells (American Type Culture Collection [ATCC]) are human lung epithelial carcinoma cells, HeLa-ATCC cells are cells of a HeLa cell clone obtained from the American Type Culture Collection, 911 cells are human embryonic retinoblasts containing base pairs 79 to 5789 of the HAdV-C5 genome (33), HEK293T cells are human embryonic kidney cells containing base pairs 1 to 4344 of the HAdV-C5 genome (34, 35), and WI38 is a human diploid cell line derived from normal embryonic lung tissue (American Type Culture Collection). XBP-1−/− and wild-type (wt) control mouse embryonal fibroblasts (MEFs; obtained from Laurie Glimcher, Weill Cornell Medical College) were maintained as described previously (36). Replicating wt HAdV-C5 (HAdV-C5_wt) and green fluorescent protein (GFP)-tagged HAdV-C2-dE3B (HAdV-C2-dE3B_GFP) were grown in A549 cells as described previously (32, 37, 38). Nonreplicating HAdV-C5-dE1_GFP (30, 32) and HAdV-B3-dE1_GFP (39) containing GFP under the control of the cytomegalovirus (CMV) major immediate early promoter were grown in 911 cells. The formation of HAdV-C2-dE3B_GFP progeny was as described previously (29, 40). Cell viability was measured by resazurin as described previously (41). The infectious titer of HAdV-C5_wt and HAdV-C2-dE3B_GFP was determined on A549 cells, and that of HAdV-C5-dE1_GFP was determined on 911 cells.
Transfections and infections.
Knockdown experiments were performed in 96-well imaging plates (Greiner Bio-one) using reverse transfection. Small interfering RNA (siRNA; 1 pmol/well diluted in 5 μl double-distilled H2O; ON-TARGET Plus SMARTPool; Dharmacon) or siRNA pools (siP RNAs; siPools; siTools Biotech GmbH, Martinsried, Germany) (42) were mixed with Lipofectamine RNAiMax reagent (0.2 μl/well; Invitrogen) in 19.8 μl Opti-MEM medium (Invitrogen) and incubated at room temperature for 5 min, and 6,000 A549 cells/well were added in 75 μl of growth medium. The medium was changed on the next day to fresh growth medium, and at 48 h posttransfection, the cells were infected with HAdV-C5_wt (multiplicity of infection [MOI], 0.09) or HAdV-C5-dE1_GFP (MOI, 0.07) for 18 h, fixed, and stained with 4′,6-diamidino-2-phenylindole (DAPI) or, in the case of HAdV-C5_wt infection, immunostained with rabbit anti-protein VI antibody (17) and secondary Alexa Fluor 488-conjugated antirabbit antibody (Life Technologies). DAPI stain was used to mark the cell nucleus, and a custom-made script (Matlab; Mathworks, USA) or a custom-made CellProfiler (version 2.0) pipeline (as described previously [43]) was used to quantify the average nuclear intensity of the GFP and protein VI signals, which were used as measures of infection efficiency. The spreading of HAdV-C2-dE3B_GFP was analyzed by time-lapse fluorescence microscopy as described previously (29). A detailed description of the imaging procedures is available on request. Human rhinovirus species A, type A1 (HRV-A1A), infections were analyzed at 7 h postinfection (p.i.) as described previously (44).
In golgicide A (GCA; Sigma) experiments, the cells were pretreated with the drug or its solvent (dimethyl sulfoxide [DMSO]) for 5 h. The GCA concentration in all experiments was 20 μM. To determine virus progeny formation from control versus GCA-treated cells, HAdV-C2-dE3B_GFP (0.008 infectious unit/cell) was added to confluent A549 cells and progeny were collected from the clarified culture medium and from cells by Freon extraction at 40 h p.i. Samples were titrated on HeLa-ATCC cells grown on 96-well imaging plates using serial 10-fold dilutions of cell extracts or culture supernatants, at 18 h postinfection samples were fixed and DAPI stained, and GFP-positive cells from wells that had less than 100% infection were counted. One GFP-positive cell was scored as one infectious particle.
Cell impedance measurements by xCELLigence and cell counting.
The xCELLigence system (Roche Applied Science and ACEA Biosciences) consists of four components, an analyzer, a device station, a control unit with software, and E plates (disposable E plates 16). The device was used as described by the supplier (45). E plates have a gold-plated sensor array at the bottom which measures the electrical impedance across the plate. Impedance is recorded in terms of a dimensionless quantity termed the cell index (CI). For background measurements, 100 μl of culture medium was added to the E-plate well, equilibrated at 37°C, and supplemented with 6,500 or 25,000 cells at 37°C. Forty-eight hours later, inhibitor was added to the cells for 5 h, followed by the addition of virus. Impedance was recorded every 15 min until the CI value reached background levels. Regression analyses and graphs were rendered in GraphPad Prism software (GraphPad Software Inc.). To quantify the cytopathogenicity of HAdV infection, CI values were plotted against time, and the time point when the CI of infected cells had decreased to 50% of the maximum CI for noninfected cells (ΔCIT50) was determined.
Differential interference contrast (DIC) images of A549 cells seeded on 96-well optical plates were obtained with a bright-field microscope equipped with an AxioCam MRc5 camera (Carl Zeiss). For segmentation, images were enhanced using a band-pass filter and contrast enhanced in Image J software. Manual counting of cells was performed using the built-in plug-in cell counter of ImageJ software.
Immunoblot assays.
A549 cells grown in a 24-well plate were infected with HAdV-C5_wt (2 μg/sample) at 37°C for 5 or 9 h, and cell extracts were prepared in hot SDS-PAGE sample buffer, boiled, sheared through a 20-gauge needle, and analyzed on a 15% polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride membrane by semidry blotting (Hoefer TE 77; Amersham Biosciences). The membrane was blocked in 5% milk powder and incubated with M73 anti-E1A (catalog number 05-599; Millipore), anti-GBF-1 (catalog number 612116; BD Biosciences), anti-IRE-1 (catalog number 3294; Cell Signaling Technology), anti-XBP-1 (catalog number 7160; Santa Cruz), anti-protein VI (17), or anti-β-tubulin (Amersham) antibodies, washed with Tris-buffered saline with 0.1% Tween 20, and incubated with horseradish peroxidase (HRP)-coupled secondary antibody (Cell Signaling Technology), followed by chemiluminescence development using an Amersham Hyperfilm ECL kit (GE Healthcare).
XBP-1 splicing.
cDNA was synthesized from total RNA extracts, and reverse transcription-PCR was carried out with primers spanning XBP-1, as described previously (46). PCR products were restriction digested with PstI and analyzed by 2% agarose gel electrophoresis.
GCA effects on gene expression from transfected or integrated DNA.
HEK293T cells were treated with GCA for 5 h, and E1A levels were scored from cell extracts by immunoblotting with M73 anti-E1A antibody. pMAX-EGFP plasmid DNA (Amaxa) driving GFP expression from the CMV promoter was transfected into A549 cells using a Neon transfection system (Life Technologies). After 24 h, the transfected cells were treated with GCA for 5 h, washed, stained with Hoechst 33342, and live imaged by automated high-throughput microscopy 5 or 21 h after drug washout, and single cells were analyzed for GFP expression by use of CellProfiler software and a nuclear DAPI mask extended by 5 pixels to account for cytoplasmic GFP.
Effect of GCA on HAdV-C5 binding to cells.
Cells grown on alcian blue-coated coverslips were treated with GCA (20 μM) for 12 or 0.5 h, followed by inoculation with atto565-labeled HAdV-C5_wt in the cold for 30 min, washing, and a 5-min pulse at 37°C. Confocal maximum-intensity projections were analyzed with a custom-made Matlab script, in which the cell outline was manually segmented using a boosted DAPI channel. The number of virus particles per cell within the cell outlines was counted, and the results were analyzed using GraphPad Prism software and the Mann-Whitney test for statistical analyses.
Statistical analyses.
The results of the infection assays are shown as the means for 3 parallel wells, unless otherwise indicated, and experiments were repeated 3 to 4 times.
RESULTS
Inhibition of GBF-1 enhances postentry steps in HAdV infection.
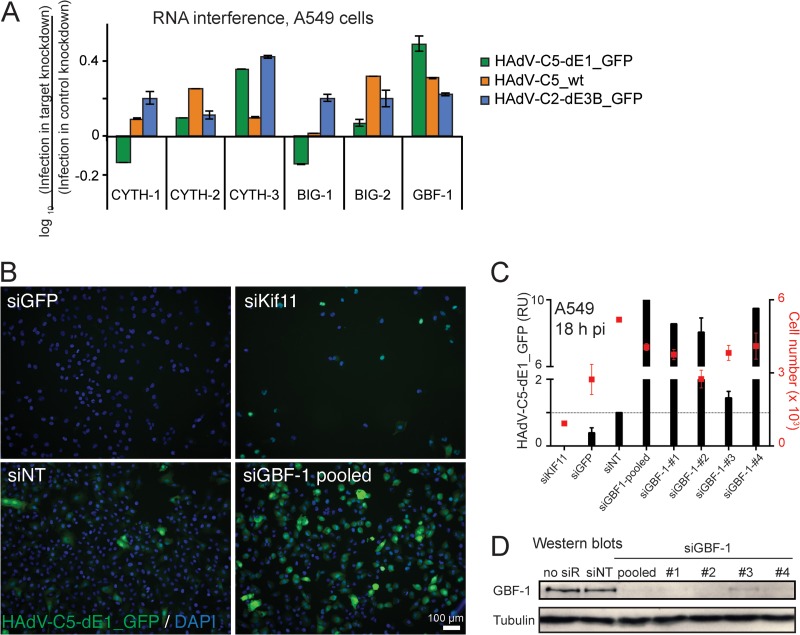
In the search for host factors enhancing bystander effects and cell killing upon viral infection, we conducted an RNA interference screen against guanine nucleotide exchange factors (GEFs) of small GTPases implicated in secretion. The knockdown of GBF-1 stimulated adenovirus infection, measured with HAdV-C5_wt, replication-defective HAdV-C5-dE1_GFP, and replication-competent HAdV-C2-dE3B_GFP (Fig. 1A). In both HAdV-C5-dE1_GFP and HAdV-C2-dE3B_GFP, GFP was under the control of the major early CMV promoter. The extent of the infection boost was between 3-fold and 10-fold in different independent experiments, and the boost with different GBF-1 siRNAs correlated with GBF-1 knockdown (Fig. 1B to D).
FIG 1.
Knockdown of GBF-1 enhances HAdV infections. (A) RNA interference miniscreen against Arf GEFs identifies GBF-1 knockdown as an enhancer of HAdV-C5-dE1_GFP, HAdV-C5_wt, and HAdV-C2-dE3B_GFP infections. Cells were reverse transfected with pooled siRNAs (1 pmol/well) against cytohesin 1 (CYTH-1), CYTH-2, CYTH-3, brefeldin A-inhibited guanine nucleotide exchange protein 1 (BIG-1), BIG-2, or GBF-1 for 48 h, infected as indicated, fixed at 18 h p.i., and analyzed for infection. Results are expressed as the log10 ratio of the mean nuclear intensity of GFP in infected cells normalized to that in control cells transfected with nontargeting siRNA. (B to D) Knockdown of GBF-1 siRNA (siGBF-1) enhances HAdV-C5-dE1_GFP infection in A549 cells. Single or pooled GBF-1 siRNAs, along with control nontargeting siRNA (siNT), kinesin family member protein 11 siRNA (siKif11), and GFP siRNA (siGFP), were reverse transfected into A549 cells, and cells were infected at 48 h posttransfection. At 18 h postinfection, the cells were fixed, stained with DAPI, and analyzed for infection. (B) Representative images. (C) Quantification of the GFP signal in cells transfected with the indicated siRNAs. RU, relative units, representing the mean nuclear GFP signal from three parallel samples ± SDs. The cell toxicity of the siRNAs was measured by use of the cell number shown on the secondary x axis. (D) Western blots. no siR, no siRNA.
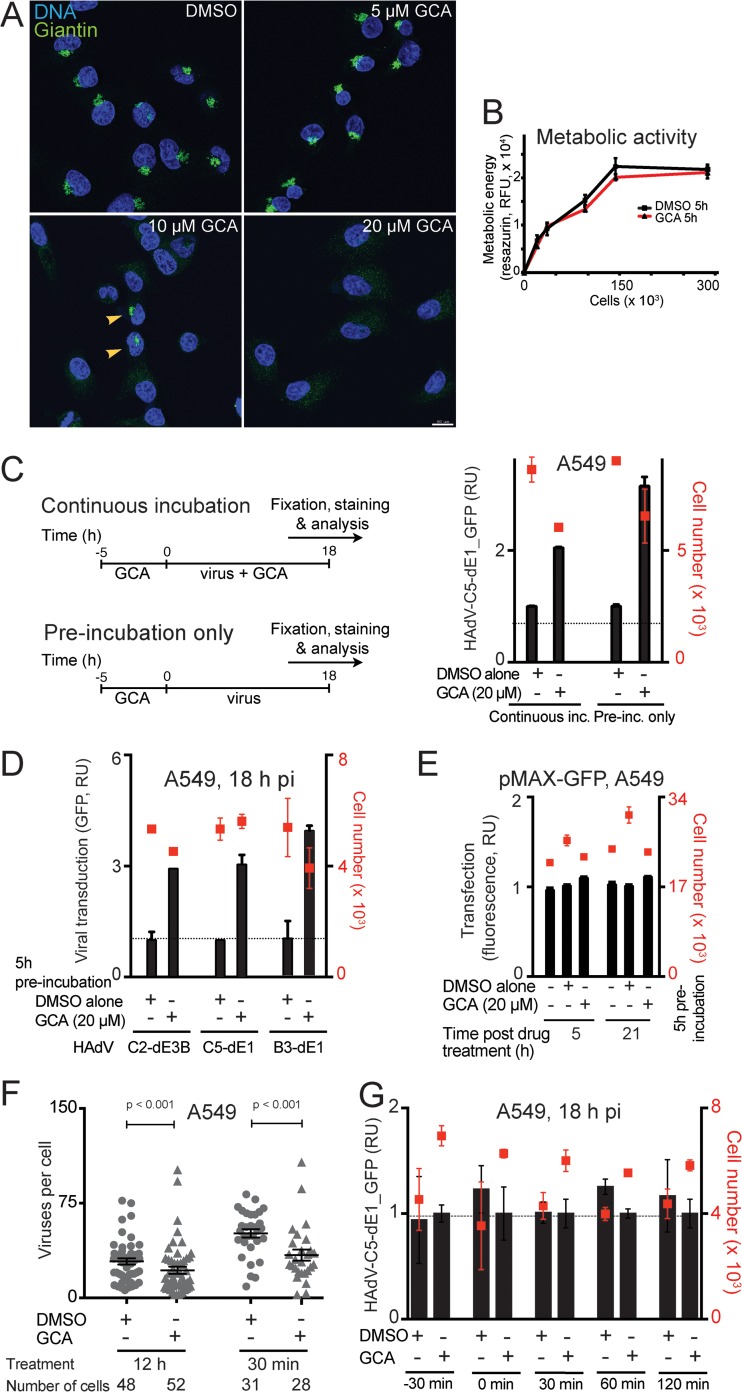
We next employed a specific inhibitor of GBF-1, golgicide A (GCA). GCA stabilizes GBF-1 on ER-Golgi apparatus membranes, interferes with ER-Golgi apparatus and intra-Golgi apparatus transport, and disperses the Golgi apparatus (47). Twenty micromolar GCA dispersed the Golgi apparatus in A549 cells but had no strong effects on metabolic cell activity, shown by immunostaining of giantin and resazurin measurements, respectively (Fig. 2A and B). GCA treatment of A549 cells for at least 5 h prior to infection enhanced infection with replicating and nonreplicating HAdV-B and -C, as measured by GFP transgene expression, but did not affect CMV promoter-driven GFP expression from transfected plasmid DNA (Fig. 2C to E). This suggested that the enhancement was not merely due to a promoter effect. The enhancement was also not due to an increased virus association with cells, since quantification of atto565-labeled HAdV-C5 on A549 cells after 30 min virus binding in the cold and after a 5-min warm-up at 37°C indicated less efficient virus binding to GCA-treated cells than to control cells (Fig. 2F). If GCA was added shortly before or after virus was added, essentially no boosting effect was observed, strongly suggesting that GBF-1 is not acutely involved in infection but, rather, is involved through a mechanism that takes hours to build up (Fig. 2G). In further experiments, we used a 5-h GCA pretreatment to induce the infection-enhancing effect.
FIG 2.
Prolonged inhibition of GBF-1 enhances HAdV infection in A549 cells but not virus binding to cells. (A) Dispersal of the Golgi apparatus upon treatment of A549 cells with the GBF-1 inhibitor GCA for 30 min. Cells were fixed and immunostained with antibodies directed against the Golgi apparatus-associated protein giantin (green), and nuclei (blue) were stained with DAPI. Samples were imaged by confocal fluorescence microscopy. Images show maximum projections of confocal sections. Note that control DMSO-treated cells showed normal perinuclear Golgi apparatus staining. Five micromolar GCA had no effect, 10 μM caused incomplete disruption of the Golgi apparatus, and 20 μM induced efficient disruption of the Golgi apparatus in all cells. Bar, 20 μm. (B) Minor effect of GCA on metabolic activity of A549 cells. Cells were treated with GCA or control DMSO for 5 h, and the metabolic activity in cells was measured by resazurin fluorescence assay (RFU, relative fluorescence units). (C) A 5-h preincubation with GCA is sufficient to enhance HAdV-C5-dE1_GFP infection of A549 cells. Cells were preincubated with GCA or DMSO for 5 h and inoculated with HAdV-C5-dE1_GFP, and infection was carried out in the presence or absence of GCA. Cells were fixed at 18 h p.i., and the mean nuclear intensity of GFP was used to score infection efficiency. (Left) Graphical representation of the experiment; (right) experimental results with mean values from three parallel experiments ± SD. inc., incubation. (D) A 5-h preincubation with GCA enhances HAdV-C5-dE1_GFP, HAdV-C2_dE3B_GFP, and HAdV-B3-dE1_GFP infection in A549 cells. (E) GCA has no effect on a plasmid-mediated CMV promoter-driven GFP expression. A549 cells were transfected with plasmid pMAX-GFP, at 24 h posttransfection were treated with GCA for 5 h, and were analyzed for GFP expression at 5 or 21 h after drug removal. (F) Decrease in atto565-labeled HAdV-C5 attachment to A549 cells upon GCA treatment. Cells were treated with GCA for 12 or 0.5 h, followed by inoculation with atto565-labeled HAdV-C5_wt in the cold for 30 min, washing, and a 5-min pulse at 37°C. The number of virus particles in individual cells was determined from maximum-intensity projections of confocal stacks using a custom-made Matlab script. Each symbol represents one cell. Error bars represent the means ± SEMs, and P values were calculated using the Mann-Whitney test for statistics. (G) Acute inhibition of GBF-1 upon virus addition does not enhance HAdV-C5-dE1_GFP infection in A549 cells. GCA or DMSO was added to the cells at 30 min prior to virus infection or at the indicated times postinfection, and incubation was continued until 18 h p.i., when the cells were analyzed.
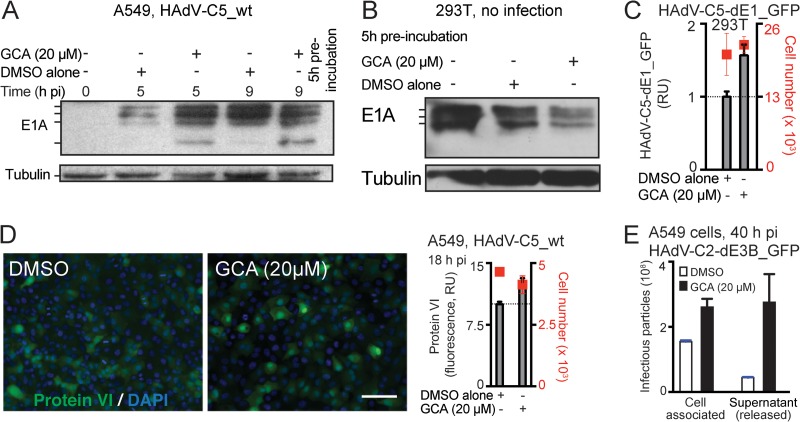
Figure 3 shows the effect of GCA on HAdV-C gene expression and progeny formation. E1A, the first viral gene expressed after delivery of the HAdV genome into the nucleus, produces five different mRNAs through differential splicing. The abundance of these transcripts is temporally regulated, with the largest two transcripts (13S and 12S) dominating early in infection and the smallest transcript (9S) being more abundant late in infection, after viral DNA replication (see, for example, references 48 and 49). Furthermore, both positive- and negative-feedback mechanisms control the expression of E1A (50), and thus, the E1A protein levels in infected cells become stabilized after an initial increase. GCA enhanced the expression of the HAdV-C5 13S and 12S protein products at 5 h p.i. and accelerated the appearance of the 9S product at 5 and 9 h p.i. (Fig. 3A). In contrast, E1A levels in HEK293T cells, which harbor an integrated copy of the E1A region (34, 35), were not enhanced but, rather, were reduced by GCA, yet GCA boosted HAdV-C5-dE1_GFP infection in these cells, albeit less efficiently than it did in A549 cells (Fig. 3B and C). Further, GCA boosted the expression of the late viral protein protein VI in HAdV-C5-infected A549 cells by about 30% at 18 h p.i. compared to the level of expression in untreated cells (Fig. 3D). GCA treatment resulted in enhanced viral titers at 40 h p.i. within both cells and the extracellular medium (Fig. 3E). Together, the data indicate that in the context of viral infection GCA enhances early and late viral gene products from extrachromosomal DNA rather than chromosomal DNA and accelerates the formation of viral progeny.
FIG 3.
Inhibition of GBF-1 by GCA enhances HAdV-C early and late gene expression, as well as virus production in A549 cells. (A) A 5-h preincubation with GCA accelerates E1A expression from HAdV-C5_wt in A549 cells, as indicated by Western blotting of infected cell lysates. (Top) E1A forms encoded by the differentially spliced E1A transcripts are indicated. (Bottom) Results for the β-tubulin loading control. (B) GCA does not increase E1A levels in uninfected HEK293T cells, which express E1A from a chromosomal copy. Cell extracts were prepared after 5 h of incubation with DMSO or GCA, and E1A levels were determined by immunoblotting using β-tubulin as a loading control. (C) GCA boosts HAdV-C5-dE1_GFP infection in HEK293T cells. Cells were preincubated with GCA for 5 h, inoculated with virus, and analyzed for GFP expression 18 h p.i. (D) GBF-1 inhibition enhances expression of the late protein protein VI in A549 cells. Cells were preincubated with GCA for 5 h, infected with HAdV-C5_wt, and analyzed for protein VI expression at 18 h p.i. (Left) Representative images. Green, protein VI signal; blue, DAPI signal. Bar, 100 μm. (Right) Quantification of average nuclear protein VI signal. (E) Inhibition of GBF-1 accelerates the production and release of HAdV-C2-dE3B_GFP in A549 cells 40 h p.i. Cells were preincubated with DMSO or GCA for 5 h and inoculated with the virus (MOI, 0.008), and at 40 h p.i., progeny particles were collected from the cells and culture supernatants. Titers of the cell-associated and supernatant fractions were determined on HeLa-ATCC cells by counting the number of GFP-positive cells at 18 h p.i.
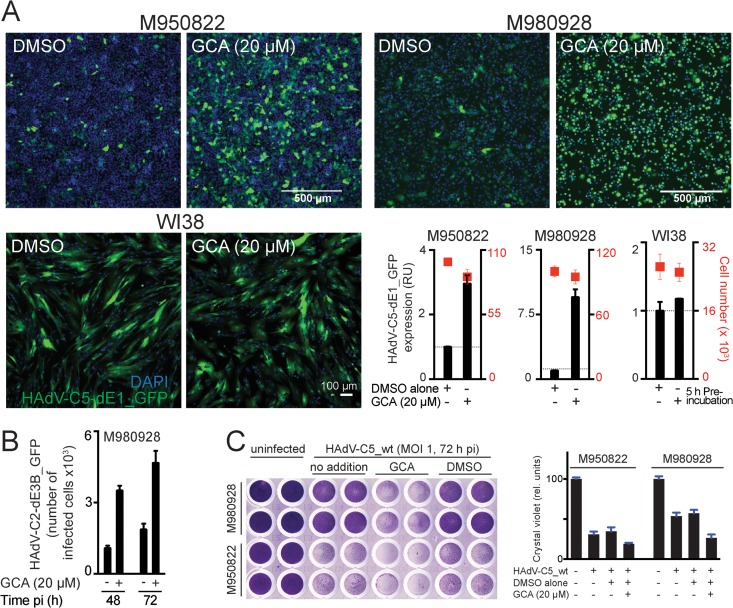
GCA enhances cancer cell killing.
We next tested whether inhibition of GBF-1 boosts HAdV infection of cultures of primary tumor stage III melanoma cells, M950822 and M980928, both of which are CAR positive (32, 51). GCA enhanced HAdV-C5-dE1_GFP infection severalfold in both cases but had few effects in normal human WI38 fibroblasts (Fig. 4 A). GCA boosted the HAdV-C2-dE3B_GFP infection spread in M980928 cells, measured by an increase in the number of infected cells at 48 h p.i. compared to the number at 72 h p.i. in a live cell assay (Fig. 4B). Crystal violet cell staining of HAdV-C5_wt-infected M950822 and M980928 cells at 72 h p.i. also indicated that virus cytotoxicity was enhanced by the GCA pretreatment (Fig. 4C).
FIG 4.
GBF-1 inhibition enhances adenovirus infection of melanoma cells. (A) Inhibition of GBF-1 by GCA enhances HAdV-C5-dE1_GFP infection of M950822 and M980928 melanoma cells but not normal human WI38 fibroblasts. Cells were preincubated with DMSO or GCA for 5 h, inoculated with the virus, and analyzed at 18 h p.i. Shown are representative images and quantification of the mean nuclear GFP signal. (B) Inhibition of GBF-1 enhances HAdV-C2-dE3B_GFP spreading in melanoma-derived M980928 cells. The cells were preincubated with DMSO or GCA for 5 h and inoculated with the virus (MOI, ∼0.00016). The data are from a live experiment in which recordings were made every 4 h to 5 h. Shown is the number of GFP-positive cells at 48 h and 72 h p.i. (C) Inhibition of GBF-1 enhances HAdV-C5_wt-induced killing of M950822 and M980928 cells. The cells were preincubated with DMSO or GCA for 5 h, inoculated with HAdV-C5_wt (MOI, 1), and stained with crystal violet at 72 h p.i. (left). (Right) Quantification of crystal violet staining, which is proportional to cell numbers. rel. units, relative units.
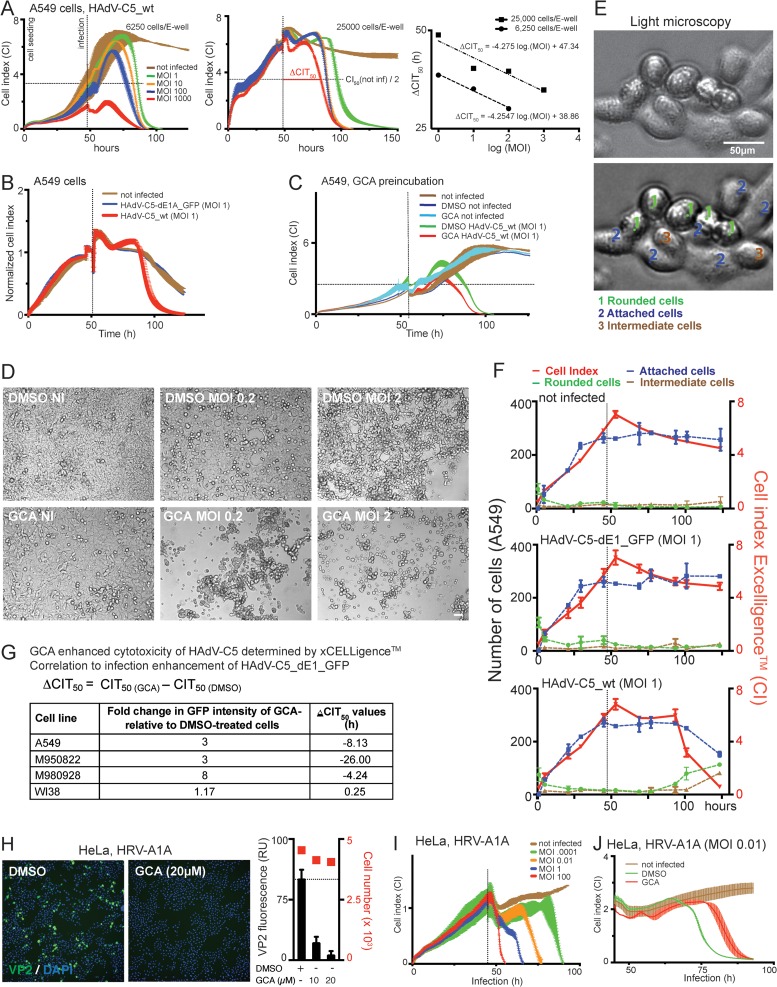
To corroborate these results, we measured virus-induced changes to cell phenotypes by recording the impedance at the cell-substrate interface using an xCELLigence system. Changes in impedance, also dubbed the cell index (CI), can be measured in real time and a label-free manner and are proportional to parameters like cell adhesion, proliferation, cell-cell interactions, and cytotoxicity (45, 52). The CI dropped as a linear function of the log MOI upon infection with HAdV-C5_wt but was independent of the initial number of cells on the substrate (Fig. 5A). The replication-defective HAdV-C5_dE1_GFP had fewer effects on CI than HAdV-C5_wt, and GCA pretreatment reduced the CI of HAdV-C5_wt-infected A549 cells but had essentially no effects on the CI of uninfected cells (Fig. 5B and C). The CI measurements were corroborated by differential interference contrast (DIC) microscopy, which showed intact monolayers of uninfected cells and progressively rounded cells upon infection with HAdV-C5_wt and GCA treatment (Fig. 5D to F). Note that the increase in the number of attached cells at 0 to 40 h postseeding correlated with an increase in CI values (compare the red and blue lines in Fig. 5F). The numbers of attached cells leveled off at 40 h postseeding, but the CI values continued to rise up to 50 h postseeding. Both the number of attached cells and the CI values remained relatively constant from 75 h onwards in the case of uninfected cells or cells infected with the nonreplicating HAdV-C5-dE1_GFP. In the case of HAdV-C5_wt infection, both the CI and the amount of attached cells decreased after 85 h. When the CI value at 50 h is taken to be 100%, a 50% reduction in the CI was reached at ∼100 h.
FIG 5.
Inhibition of GBF-1 enhances HAdV-induced cytopathic effects but blocks rhinovirus infection. (A) CI profiles from impedance measurements of A549 cells infected with HAdV-C5_wt indicate cytopathic effects. Impedance was recorded every 15 min using an xCELLigence system. Each point represents the average value from two replicates with SDs. The times on the x axis indicate the times after cell seeding. Vertical lines show the time of infection, and horizontal lines refer to 50% of the maximum CI of noninfected cells. (Right) Regression fit of ΔCIT50 values, where each point represents a single ΔCIT50 value. Note that the CI profile of HAdV-C5_wt infection is MOI dependent but not cell density dependent. (B) A549 cells infected with HAdV-C5_wt (red) or replication-deficient HAdV-C5-dE1_GFP (blue) yield significantly different CI profiles. The profile of HAdV-C5-dE1_GFP-infected cells is similar to that of noninfected control cells (brown). (C) A 5-h preincubation with GCA enhanced HAdV-C5_wt-induced cytotoxicity in A549 cells. Data points represent the means from two samples per condition ± SDs. (D) DIC images of control and GCA-treated (5 h preincubation) uninfected and HAdV-C5_wt-infected A549 cells at 72 h p.i. Bar, 50 μm. (E, F) Comparison of CI values with cell appearance in DIC images. (E) Representative DIC images of A549 cells classified as rounded (1, green), attached (2, blue), or in an intermediate state (3, brown). The upper image is unprocessed, whereas the lower image shows an example of images that were filtered through a band-pass filter and contrast enhanced using ImageJ software. The latter images were used for cell classification. (F) Comparison of the CI profiles of uninfected, HAdV-C5-dE1_GFP-infected (MOI, 1), and HAdV-C5_wt-infected (MOI, 1) A549 cells with the number of rounded, attached, and intermediate cells in DIC images of corresponding parallel samples. (G) Summary of GCA-mediated infection enhancement for HAdV-C5-dE1_GFP and ΔCIT50 values (h) for HAdV-C5_wt. Negative values in the ΔCIT50 column indicate that the CI of GCA-treated cells reached 50% of the maximum CI of noninfected cells earlier than control DMSO-treated cells. (H) GCA inhibits HRV-A1A infection of HeLa-Ohio cells, as indicated by anti-VP2 immunostaining. Cells were infected with HRV-A1A (MOI, 0.01) in the presence of 20 μM GCA and analyzed for VP2 expression at 7 h p.i. (Left) Representative images (green, VP2; blue, DAPI); (right) quantification of the cytoplasmic VP2 signal. (I) CI profile of HeLa-Ohio cells infected with HRV-A1A at different MOIs. Values are the averages, including SDs, from two replicates. (J) The GBF-1 inhibitor GCA reduced the HRV-A1A-induced cytopathic effect in HeLa-Ohio cells. Cells were infected with HRV-A1A (MOI, 0.01) in the presence of 20 μM GCA. Data represent the means ± SDs for 2 samples per condition.
In good agreement with the findings presented above, manual counting indicated an equal distribution of rounded-off and attached cells at ∼125 h. The sensitivity of cell viability measurements depends on the slope of the dip in the CI profile, a measure for attached cells. A steep slope of the CI profiles compared to a gradual slope obtained by cell counting indicated a higher sensitivity of the xCELLigence system than microscopic inspection for measurement of the cytopathic effects. Importantly, the CI values of HAdV-C5_wt infection were strongly reduced in GCA-treated cancer cells, including M950822 and M980928 melanoma cells, but not in normal WI38 cells compared to the CI values of control infected cells, as indicated by the negative ΔCIT50 values for A549 and melanoma cells in Fig. 5G. Notably, the ΔCIT50 values correlated well with the HAdV-C5-dE1_GFP infection phenotype (Fig. 5G) and confirmed the strong acceleration of HAdV infection by GCA.
Contrary to HAdV infection, GCA increased the CIT values from human rhinovirus (HRV) A1A infections, indicating fewer cytopathic effects, in good correlation with infection reduction (Fig. 5H to J). This result was in agreement with the notion that GCA blocks the replication of HRV-related enteroviruses by dissociating Arf1 and COP-I from Golgi apparatus membranes (53) and that HRV-A1A requires lipid flux between the ER and Golgi apparatus for replication (54).
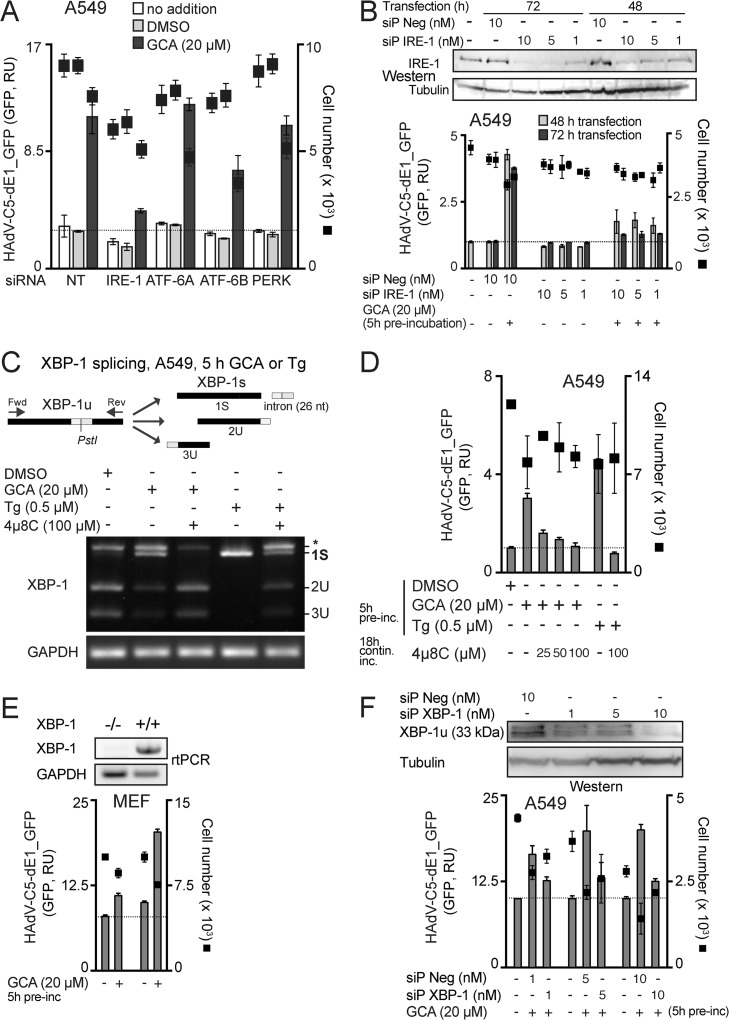
IRE-1 and XBP-1 are required for GCA enhancement of HAdV infection.
UPR enhances the protein folding capacity in the ER under stress conditions, such as physical, chemical, or biological insults, developmental processes, or cancer (55). ER stress activates three signaling arms of UPR, IRE-1/XBP-1, protein kinase R (PKR)-like ER kinase (PERK), and ATF-6 (56). This can lead to cell protection or cell death. The former is favored by the IRE-1 branch, and the latter is favored by the ATF-6 and PERK arms (56). Since knockdown of GBF-1 has been shown to induce the ATF-6 arm of the UPR (14), we tested whether the UPR was linked to the GCA boost of HAdV infection. The knockdown of IRE-1alpha with a pool of four siRNAs had the strongest effect on blunting GCA enhancement, although ATF-6B knockdown reduced the boost as well (Fig. 6 A). We confirmed these results with a pool of 30 synthetic siRNAs (siP RNAs), which were reported to have no detectable off-target effects (42). The siP IRE-1 RNAs blocked the infection boost and reduced the IRE-1 protein levels (Fig. 6B). The treatment of cells with GCA triggered the IRE-1/XBP-1 branch of the UPR, similar to the ER stress activator thapsigargin, as indicated by the activation of cytoplasmic splicing of XBP-1 mRNA, and this splicing was inhibited by the IRE-1 nuclease inhibitor 4μ8C (Fig. 6C) (57). Similar to GCA, thapsigargin boosted HAdV-C5-dE1_GFP infection and 4μ8C blunted both the GCA and thapsigargin boosts (Fig. 6D). For HAdV-C5-dE1_GFP, the GCA infection boost was strongly but not completely reduced in XBP-1-knockout mouse embryo fibroblasts (36), akin to the findings for XBP-1 siP RNA-treated A549 cells (Fig. 6E and F), suggesting a major, though not exclusive, role of XBP-1 in boosting infection.
FIG 6.
GCA enhances adenovirus infection through IRE-1 and XBP-1. (A) Effect of knockdown of ER stress sensors on GCA-mediated infection boost. A549 cells were reverse transfected with control nontargeting siRNAs (NT) or pooled siRNAs against ER stress sensors ATF-6A, ATF-6B, PERK, and IRE-1alpha. At 43 h posttransfection, cells were preincubated with DMSO or GCA for 5 h (no addition indicates no pretreatment) and inoculated with HAdV-C5-dE1_GFP, and the average nuclear GFP signal was analyzed at 18 h p.i. (B) IRE-1 knockdown by siP RNAs reduces the HAdV-C5-dE1_GFP infection boost in GCA-treated A549 cells. siP Neg, nontargeting negative-control siP RNAs. Intracellular IRE-1 levels were determined by Western blotting using β-tubulin as a loading control. (C) GBF-1 inhibition by GCA induces ER stress and activates IRE-1 nuclease and splicing of XBP-1 mRNA. A549 cells were treated with GCA or the ER stress activator thapsigargin (Tg) for 5 h, and IRE-1 activation was analyzed by PstI digestion of XBP1 cDNA amplicons. The spliced XBP-1 cDNA amplicon lacks a PstI site (1S), whereas the unspliced one retains the site and is cleaved into two fragments (2U and 3U) upon PstI digestion. The uppermost band (*) is a spliced/unspliced XBP-1 hybrid amplicon (69). XBP-1 splicing was inhibited by the IRE-1 nuclease inhibitor 4μ8C in GCA- and thapsigargin-treated cells. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) cDNA amplicons were used as a loading control. Fwd, forward; Rev, reverse; nt, nucleotides. (D) GCA-induced HAdV-C5-dE1_GFP infection boost in A549 cells requires IRE-1 endonuclease activation. Cells were preincubated with GCA or thapsigargin for 5 h with or without 4μ8C, inoculated with HAdV-C5-dE1_GFP, and analyzed for GFP expression at 18 h p.i. contin. inc., continuous incubation. (E) Reduced GCA infection boost in XBP-1−/− mouse embryo fibroblasts. XBP-1+/+ or XBP-1−/− MEFs were preincubated with GCA, inoculated with HAdV-C5-dE1_GFP, and analyzed for GFP expression at 18 h p.i. (F) XBP-1 knockdown by siP RNAs reduces the HAdV-C5-dE1_GFP infection boost in GCA-treated A549 cells. Cells were reverse transfected with siP RNAs against XBP-1 or nontargeting negative-control siP RNA (siP Neg), at 72 h posttransfection preincubated with GCA or DMSO for 5 h, inoculated with HAdV-C5-dE1_GFP, and analyzed for GFP expression at 18 h p.i. Levels of knockdown of the unspliced XBP-1 protein (XBP-1u) were controlled by Western blotting using β-tubulin as a loading control.
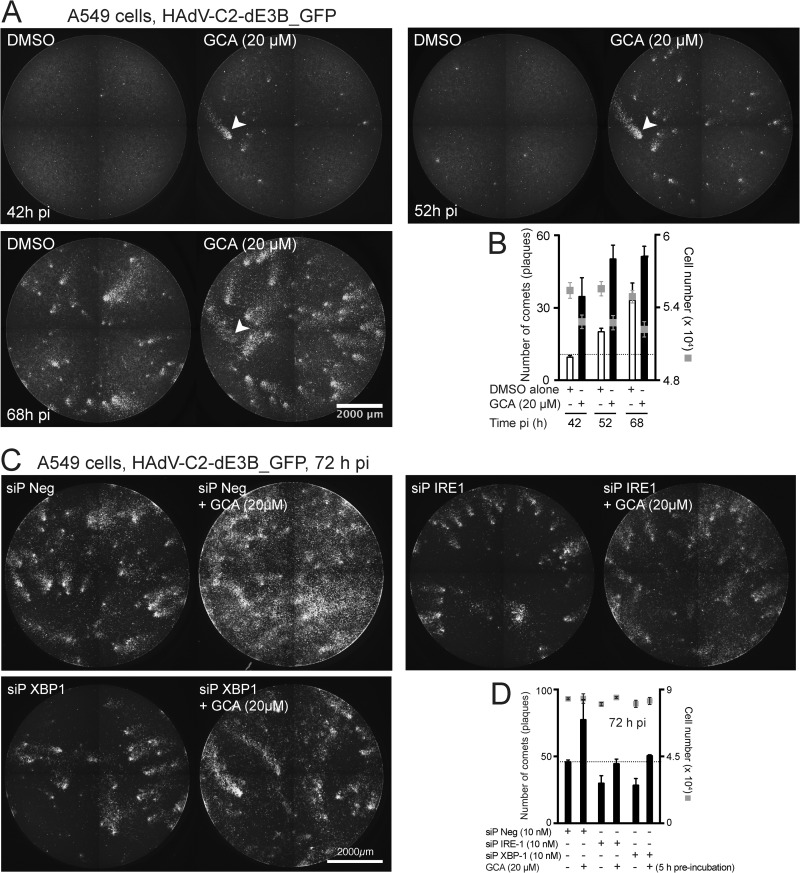
The involvement of the IRE-1/XBP-1 branch in the GCA infection boost was further tested by live cell assays measuring the spread of infection, where comets of infected cells are formed by replicating HAdV-C2-dE3B_GFP (29). Comets are the equivalent of plaques but are obtained in the absence of gelling medium. They consist of dozens of GFP-positive cells in an elongated arrangement due to the convection flow of cell-free viruses from lytic infected cells. GCA enhanced the formation of comets in a time-dependent manner (Fig. 7A, B). Importantly, siP RNAs against IRE-1 or XBP-1 blunted the GCA enhancement of comet formation (Fig. 7C and D). Notably, these siP RNAs also reduced the number of comets without GCA treatment, implying that IRE-1 and XBP-1 contribute to the replication of HAdV in the absence of exogenous UPR stimulation.
FIG 7.
Inhibition of GBF-1 by GCA enhances spreading of virus infection via IRE-1 and XBP-1. (A, B) Confluent A549 cells were preincubated with GCA for 5 h and infected with replication-competent HAdV-C2-dE3B_GFP (MOI, 0.00016), and spreading of infection was analyzed by time-lapse fluorescence microscopy. (A) Spreading of infection is manifested by the typical comet phenotypes of infected GFP-positive cells. Arrowhead, one of the many comets which increase in size as infection proceeds. (B) Quantification of the number of comets. The data are from two parallel experiments. (C, D) A549 cells were reverse transfected with the negative-control siP RNA (siP Neg) or siP RNA against IRE-1 or XBP-1. At 72 h posttransfection, cells were preincubated with GCA or DMSO for 5 h and inoculated with HAdV-C2-dE3B_GFP (MOI, 0.008) and spreading of infection was analyzed by time-lapse fluorescence microscopy. (C) Images from the recordings obtained at 72 h p.i. (D) Quantification of the comets at 72 h p.i. The data in panel D were from two parallel experiments.
DISCUSSION
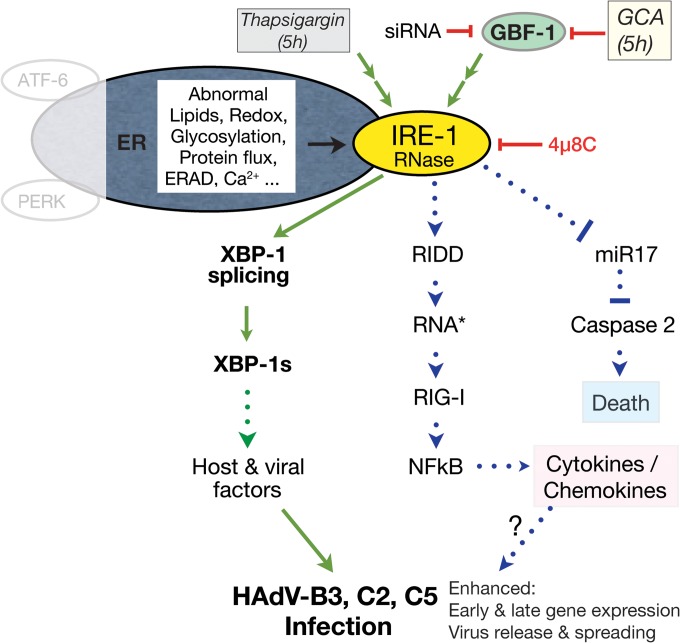
IRE-1 senses unfolded proteins in the ER lumen, and this activates the IRE-1 cytosolic kinase and endonuclease functions and leads to splicing of XBP-1 mRNA, yielding a transcript which encodes the active XBP-1 transcription factor (58). XBP-1 alleviates ER stress. We show here that the inhibition of the Arf activator GBF-1 by GCA induces the IRE-1/XBP-1 branch of the UPR (summarized in a model presented in Fig. 8). Chemical induction of the UPR boosted HAdV-C5 or HAdV-B3 infection specifically in cancer cells. This may have therapeutic relevance, since the CAR-tropic HAdV-C5 is used to treat metastatic tumors and the CD46/DSG-2-tropic HAdV-B3 is used to treat late-stage cancers with downregulated CAR, for example, prostate cancer (59). Importantly, UPR is readily inducible in aggressive tumor cells and promotes survival, angiogenesis, autophagy, the epithelial-mesenchymal transition (EMT), or adaptation to hypoxic conditions. While angiogenesis, EMT, and hypoxia adaptation depend on the PERK branch, the XBP-1 branch supports some forms of breast cancer (60–62). We expect that cells with fast or strong induction of IRE-1/XBP-1 will enhance HAdV infection particularly well, while cells with low or slow induction will show a lesser infection boost upon chemical UPR induction. In both cases, however, cancer cells can be killed by HAdV.
FIG 8.
Model for the UPR-induced HAdV infection boost by GCA. ER stress can be induced by abnormal ER-associated lipids, redox potential, glycosylation, or protein flux through the secretory pathway or by the depletion of calcium ions, for example, by the Ca2+ ATPase inhibitor thapsigargin (70). Cells sense ER stress by three major pathways, the activating transcription factor 6 (ATF-6), the PKR-like ER kinase (PERK), and the inositol-requiring enzyme 1 (IRE-1) pathways. Here we show that the small compound golgicide A (GCA), which blocks the guanine nucleotide exchange factor GBF-1 of Arf GTPases, implicated in ER-Golgi apparatus membrane transport, induces the IRE-1/X-box binding protein 1 (XBP-1) branch of the UPR (green arrows). GCA and, in some cases, the knockdown of GBF-1 by siRNA enhance infection of cancer cells with both replicating and nonreplicating HAdV of the C and B species. This requires the RNase activity of IRE-1 and leads to the splicing of the XBP-1 mRNA in the cytoplasm, yielding an mRNA which encodes the active XBP-1 transcription factor (also dubbed XBP-1s). XBP-1 splicing can be blocked by the IRE-1 RNase inhibitor 4μ8C (57). XBP-1s may enhance the transcription of both host and viral genes and thereby boost early and late viral gene expression, virus release from infected cells, and viral spreading. Besides the XBP-1 branch, IRE-1 also signals through regulated IRE-1-dependent decay (RIDD), generating double-stranded RNA (RNA*) and leading to the activation of retinoic acid-inducible gene 1 (RIG-I), the nuclear factor kappa light chain enhancer of activated B cells (NF-κB), and downstream cytokines and chemokines, which may boost HAdV infection (31, 71–73). A third output from IRE-1 signaling is the degradation of microRNA 17 (miR17), an inhibitor of the apoptotic caspase 2 (74). If this pathway is triggered by GCA, it may enhance HAdV-induced cell killing. Green arrows, the GCA pathway activating the IRE-1/XBP-1 branch of the UPR and leading to HAdV infection boost, as shown in this work; green dotted arrow, possible activations downstream of GCA-induced UPR; blue dotted arrows, pathways relating to work previously published depicting two signaling branches downstream of IRE-1 that could contribute to the enhancement of HAdV infection or cancer cell killing.
On a mechanistic level, our results demonstrate that knockdown or inhibition of GBF-1 by GCA enhanced early viral and transgene expression from adenovirus, enhanced late gene expression, cytopathic effects, and the formation and release of progeny from infected cells, and enhanced virus spreading to neighboring cells. This cascade of effects boosted cancer cell killing. It was, to a large extent, dependent on the ability of GCA to induce the IRE-1/XBP-1 branch of the UPR. Interestingly, the GCA effects were not limited to wild-type HAdV but also occurred with replication-defective HAdV expressing a transgene under the control of the major immediate early CMV promoter. Like for HAdV-C5_wt, this enhancement required IRE-1/XBP-1, suggesting that this UPR branch acts on the E1A and the CMV promoters. However, promoter mechanisms are not sufficient to induce the viral infection boost, since GCA did not enhance gene expression from transfected plasmid DNA or chromosomally integrated viral E1 DNA. Furthermore, our results also indicate that GCA did not enhance virus binding to cells. We speculate that viral insults to cell integrity during entry enhance infection, together with elements of the IRE-1/XBP-1 pathway. This combined action may enhance nuclear import of the viral genome, affect the structure or protein composition of the viral genome in the nucleus, or involve epigenetic regulatory machineries (23, 63–65). Since UPR induction enhanced both the GFP expression from HAdV-C5-dE1_GFP and the E1A or GFP expression from a replicating HAdV-C, this suggests that some early event in the virus life cycle is targeted. However, whether this as-yet-unidentified early effect also fully explains the accelerated spreading of virus infection or whether infection is accelerated/enhanced by more than one mechanism is at present unclear. Furthermore, the exact molecular mechanism(s) by which IRE-1/XBP-1 enhance HAdV infections is still unclear. This could involve one or more cellular (or viral) genes induced by XBP-1.
The approach outlined here allows the identification of host pathways boosting infection with viral vectors in any cell type of interest. Specifically, the cell impedance measurements can score cytopathic effects of virus infections in real time and a label-free manner and can identify both infection-enhancing and infection-inhibiting compounds in a semi-high-throughput format without the need to construct specific reporter viruses. Boosters of viral infection are needed to enhance and tune the efficacy of oncolytic virotherapies. Potential signaling branches downstream of the IRE-1 UPR node triggered by the small chemical GCA to enhance cancer cell killing are discussed in Fig. 8. Oncolytic therapies kill cancer cells, lead to inflammation, and, ideally, present tumor-associated antigens to immune cells to mount immune responses against tumors (27, 66). Viral oncolysis also crucially depends on efficient intratumoral transmission of the oncolytic agents and the ability of the virus to overcome innate immunity (67). Notably, the spreading of HAdV-C occurs by cell-free viruses after lysis of infected cells, yet spreading and oncolysis are limited both in cell cultures and in organisms (29, 68). Our data raise the possibility that viral oncolysis can be chemically tuned by manipulating the UPR and that this can be applied for cancer treatment.
ACKNOWLEDGMENTS
We thank Laurie Glimcher and Ann-Hwee Lee (Weill Cornell Medical College, NY) for XBP-1 MEFs as well as Daria Mudrak and Bettina Cardel (University of Zurich, Zurich, Switzerland) and Wai-Ming Lee (University of Wisconsin, Madison, WI) for supplying HRV-A1A and anti-VP2 hybridoma antibody.
The work was supported by the Swiss National Science Foundation (grant 31003A_141222/1) and an Initial Training Network grant, AdVance, from the European Union to U.F.G.
We declare that we have no conflict of interest.
V.P., M.S., and M.P. designed and performed the experiments; V.P., M.S., A.Y., V.A., S.H., and U.F.G. interpreted the data; V.P., M.S., and U.F.G. wrote the manuscript; and U.F.G. coordinated the study.
Footnotes
Published ahead of print 3 September 2014
REFERENCES
- 1.Chen DS, Mellman I. 2013. Oncology meets immunology: the cancer-immunity cycle. Immunity 39:1–10. 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Bell J, McFadden G. 2014. Viruses for tumor therapy. Cell Host Microbe 15:260–265. 10.1016/j.chom.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell SJ, Peng KW, Bell JC. 2012. Oncolytic virotherapy. Nat. Biotechnol. 30:658–670. 10.1038/nbt.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. 2013. Gene therapy clinical trials worldwide to 2012—an update. J. Gene Med. 15:65–77. 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 6.Bovenberg MS, Degeling MH, Tannous BA. 2013. Advances in stem cell therapy against gliomas. Trends Mol. Med. 19:281–291. 10.1016/j.molmed.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Touchefeu Y, Franken P, Harrington KJ. 2012. Radiovirotherapy: principles and prospects in oncology. Curr. Pharm. Des. 18:3313–3320. 10.2174/1381612811209023313. [DOI] [PubMed] [Google Scholar]
- 8.Kyula JN, Khan AA, Mansfield D, Karapanagiotou EM, McLaughlin M, Roulstone V, Zaidi S, Pencavel T, Touchefeu Y, Seth R, Chen NG, Yu YA, Zhang Q, Melcher AA, Vile RG, Pandha HS, Ajaz M, Szalay AA, Harrington KJ. 2014. Synergistic cytotoxicity of radiation and oncolytic Lister strain vaccinia in (V600D/E)BRAF mutant melanoma depends on JNK and TNF-alpha signaling. Oncogene 33:1700–1712. 10.1038/onc.2013.112. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney DJ, Lefebvre C, Allan K, Brun J, Sanaei CA, Baird S, Pearce N, Gronberg S, Wilson B, Prakesh M, Aman A, Isaac M, Mamai A, Uehling D, Al-Awar R, Falls T, Alain T, Stojdl DF. 2011. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell 20:443–456. 10.1016/j.ccr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Claude A, Zhao B-P, Kuziemsky CE, Dahan S, Berger SJ, Yan J-P, Armold AD, Sullivan EM, Melançon P. 1999. GBF1: a novel Golgi-associated Bfa-resistant guanine nucleotide exchange factor that displays specificity for Adp-ribosylation factor 5. J. Cell Biol. 146:71–84. 10.1083/jcb.146.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, Claude A, Chun J, Shields D, Presley J, Melançon P. 2006. GBF1, a cis-Golgi and VTCs-localized Arf-GEF, is implicated in ER-to-Golgi protein traffic. J. Cell Sci. 119:3743–3753. 10.1242/jcs.03173. [DOI] [PubMed] [Google Scholar]
- 12.Mansour SJ, Herbrick JA, Scherer SW, Melancon P. 1998. Human GBF1 is a ubiquitously expressed gene of the sec7 domain family mapping to 10q24. Genomics 54:323–327. 10.1006/geno.1998.5563. [DOI] [PubMed] [Google Scholar]
- 13.Szul T, Garcia-Mata R, Brandon E, Shestopal S, Alvarez C, Sztul E. 2005. Dissection of membrane dynamics of the Arf-guanine nucleotide exchange factor GBF1. Traffic 6:374–385. 10.1111/j.1600-0854.2005.00282.x. [DOI] [PubMed] [Google Scholar]
- 14.Citterio C, Vichi A, Pacheco-Rodriguez G, Aponte AM, Moss J, Vaughan M. 2008. Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc. Natl. Acad. Sci. U. S. A. 105:2877–2882. 10.1073/pnas.0712224105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greber UF, Arnberg N, Wadell G, Benko M, Kremer EJ. 2013. Adenoviruses—from pathogens to therapeutics: a report on the 10th International Adenovirus Meeting. Cell. Microbiol. 15:16–23. 10.1111/cmi.12031. [DOI] [PubMed] [Google Scholar]
- 16.Wolfrum N, Greber UF. 2013. Adenovirus signalling in entry. Cell. Microbiol. 15:53–62. 10.1111/cmi.12053. [DOI] [PubMed] [Google Scholar]
- 17.Burckhardt CJ, Suomalainen M, Schoenenberger P, Boucke K, Hemmi S, Greber UF. 2011. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe 10:105–117. 10.1016/j.chom.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Moyer CL, Wiethoff CM, Maier O, Smith JG, Nemerow GR. 2011. Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J. Virol. 85:2631–2641. 10.1128/JVI.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suomalainen M, Luisoni S, Boucke K, Bianchi S, Engel DA, Greber UF. 2013. A direct and versatile assay measuring membrane penetration of adenovirus in single cells. J. Virol. 87:12367–12379. 10.1128/JVI.01833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bremner KH, Scherer J, Yi J, Vershinin M, Gross SP, Vallee RB. 2009. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6:523–535. 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strunze S, Engelke MF, Wang I-H, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. 2011. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 10:210–223. 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Suomalainen M, Nakano MY, Boucke K, Keller S, Stidwill RP, Greber UF. 1999. Microtubule-dependent minus and plus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657–672. 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang IH, Suomalainen M, Andriasyan V, Kilcher S, Mercer J, Neef A, Luedtke NW, Greber UF. 2013. Tracking viral genomes in host cells at single-molecule resolution. Cell Host Microbe 14:468–480. 10.1016/j.chom.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Dazard J-E, Zhang K, Sha J, Yasin O, Cai L, Nguyen C, Ghosh M, Bongorno J, Harter ML. 2011. The dynamics of E1A in regulating networks and canonical pathways in quiescent cells. BMC Res. Notes 4:160. 10.1186/1756-0500-4-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pelka P, Ablack JN, Fonseca GJ, Yousef AF, Mymryk JS. 2008. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J. Virol. 82:7252–7263. 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toth K, Wold WS. 2010. Increasing the efficacy of oncolytic adenovirus vectors. Viruses 2:1844–1866. 10.3390/v2091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miest TS, Cattaneo R. 2014. New viruses for cancer therapy: meeting clinical needs. Nat. Rev. Microbiol. 12:23–34. 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toth K, Dhar D, Wold WS. 2010. Oncolytic (replication-competent) adenoviruses as anticancer agents. Expert Opin. Biol. Ther. 10:353–368. 10.1517/14712590903559822. [DOI] [PubMed] [Google Scholar]
- 29.Yakimovich A, Gumpert H, Burckhardt CJ, Lutschg VA, Jurgeit A, Sbalzarini IF, Greber UF. 2012. Cell-free transmission of human adenovirus by passive mass transfer in cell culture simulated in a computer model. J. Virol. 86:10123–10137. 10.1128/JVI.01102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleischli C, Sirena D, Lesage G, Havenga MJ, Cattaneo R, Greber UF, Hemmi S. 2007. Species B adenovirus serotypes 3, 7, 11 and 35 share similar binding sites on the membrane cofactor protein CD46 receptor. J. Gen. Virol. 88:2925–2934. 10.1099/vir.0.83142-0. [DOI] [PubMed] [Google Scholar]
- 31.Lutschg V, Boucke K, Hemmi S, Greber UF. 2011. Chemotactic antiviral cytokines promote infectious apical entry of human adenovirus into polarized epithelial cells. Nat. Commun. 2:391. 10.1038/ncomms1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagel H, Maag S, Tassis A, Nestle FO, Greber UF, Hemmi S. 2003. The alphavbeta5 integrin of hematopoietic and nonhematopoietic cells is a transduction receptor of RGD-4C fiber-modified adenoviruses. Gene Ther. 10:1643–1653. 10.1038/sj.gt.3302058. [DOI] [PubMed] [Google Scholar]
- 33.Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, van Ormondt H, Hoeben RC, van der Eb AJ. 1996. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum. Gene Ther. 7:215–222. 10.1089/hum.1996.7.2-215. [DOI] [PubMed] [Google Scholar]
- 34.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis N, Evelegh C, Graham FL. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233:423–429. 10.1006/viro.1997.8597. [DOI] [PubMed] [Google Scholar]
- 36.Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greber UF, Webster P, Weber J, Helenius A. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 38.Greber UF, Willetts M, Webster P, Helenius A. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477–486. 10.1016/0092-8674(93)90382-Z. [DOI] [PubMed] [Google Scholar]
- 39.Sirena D, Ruzsics Z, Schaffner W, Greber UF, Hemmi S. 2005. The nucleotide sequence and a first generation gene transfer vector of species B human adenovirus serotype 3. Virology 343:283–298. 10.1016/j.virol.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puntener D, Engelke MF, Ruzsics Z, Strunze S, Wilhelm C, Greber UF. 2011. Stepwise loss of fluorescent core protein V from human adenovirus during entry into cells. J. Virol. 85:481–496. 10.1128/JVI.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jurgeit A, McDowell R, Moese S, Meldrum E, Schwendener R, Greber UF. 2012. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 8:e1002976. 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannus M, Beitzinger M, Engelmann JC, Weickert MT, Spang R, Hannus S, Meister G. 2014. siPools: highly complex but accurately defined siRNA pools eliminate off-target effects. Nucleic Acids Res. 42:8049–8061. 10.1093/nar/gku480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jurgeit A, Moese S, Roulin P, Dorsch A, Lotzerich M, Lee WM, Greber UF. 2010. An RNA replication-center assay for high content image-based quantifications of human rhinovirus and coxsackievirus infections. Virol. J. 7:264. 10.1186/1743-422X-7-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiegel M. 2009. Real-time and dynamic monitoring of virus-mediated cytopathogenicity. Biochemica 3:15–17. [Google Scholar]
- 46.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066–3077. 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. 2009. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5:157–165. 10.1038/nchembio.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu MC, Hsu MT. 1997. Adenovirus E1B 19K protein is required for efficient DNA replication in U937 cells. Virology 227:295–304. 10.1006/viro.1996.8349. [DOI] [PubMed] [Google Scholar]
- 49.Miller MS, Pelka P, Fonseca GJ, Cohen MJ, Kelly JN, Barr SD, Grand RJ, Turnell AS, Whyte P, Mymryk JS. 2012. Characterization of the 55-residue protein encoded by the 9S E1A mRNA of species C adenovirus. J. Virol. 86:4222–4233. 10.1128/JVI.06399-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida K, Higashino F, Fujinaga K. 1995. Transcriptional regulation of the adenovirus E1A gene. Curr. Top. Microbiol. Immunol. 199:113–130. [DOI] [PubMed] [Google Scholar]
- 51.Schmitz M, Graf C, Gut T, Sirena D, Peter I, Dummer R, Greber UF, Hemmi S. 2006. Melanoma cultures show different susceptibility towards E1A-, E1B-19 kDa- and fiber-modified replication-competent adenoviruses. Gene Ther. 13:893–905. 10.1038/sj.gt.3302739. [DOI] [PubMed] [Google Scholar]
- 52.Solly K, Wang X, Xu X, Strulovici B, Zheng W. 2004. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev. Technol. 2:363–372. 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
- 53.van der Linden L, van der Schaar HM, Lanke KH, Neyts J, van Kuppeveld FJ. 2010. Differential effects of the putative GBF1 inhibitors golgicide A and AG1478 on enterovirus replication. J. Virol. 84:7535–7542. 10.1128/JVI.02684-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roulin P, Lotzerich M, Tanner LB, Torta FT, van Kuppefeld F, Wenk MR, Greber UF. A phosphatidylinositol 4-phosphate and cholesterol counter-current model for the formation of rhinovirus replication compartments at ER-Golgi interface. Cell Host Microbes, in press. [DOI] [PubMed] [Google Scholar]
- 55.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 56.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318:944–949. 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. U. S. A. 109:E869–E878. 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tirasophon W, Welihinda AA, Kaufman RJ. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812–1824. 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, Schmitt LD, McCormick F. 2002. Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res. 62:3812–3818. [PubMed] [Google Scholar]
- 60.Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, Mai J, Shen H, Hu DZ, Adoro S, Hu B, Song M, Tan C, Landis MD, Ferrari M, Shin SJ, Brown M, Chang JC, Liu XS, Glimcher LH. 2014. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 508:103–107. 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, Gupta PB. 2014. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 4:702–715. 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 62.Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, Lambin P, van der Kogel AJ, Koritzinsky M, Wouters BG. 2010. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 120:127–141. 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berscheminski J, Groitl P, Dobner T, Wimmer P, Schreiner S. 2013. The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. J. Virol. 87:965–977. 10.1128/JVI.02023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. 2008. Adenovirus small e1a alters global patterns of histone modification. Science 321:1084–1085. 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schreiner S, Kinkley S, Bürck C, Mund A, Wimmer P, Schubert T, Groitl P, Will H, Dobner T. 2013. SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog. 9:e1003775. 10.1371/journal.ppat.1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mellman I, Coukos G, Dranoff G. 2011. Cancer immunotherapy comes of age. Nature 480:480–489. 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A, Greber UF. 2014. Innate immunity to adenovirus. Hum. Gene Ther. 25:265–284. 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemminki O, Diaconu I, Cerullo V, Pesonen SK, Kanerva A, Joensuu T, Kairemo K, Laasonen L, Partanen K, Kangasniemi L, Lieber A, Pesonen S, Hemminki A. 2012. Ad3-hTERT-E1A, a fully serotype 3 oncolytic adenovirus, in patients with chemotherapy refractory cancer. Mol. Ther. 20:1821–1830. 10.1038/mt.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han D, Lerner AG, Vande Walle L, Upton J-P, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. 2009. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138:562–575. 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hetz C, Martinon F, Rodriguez D, Glimcher LH. 2011. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 91:1219–1243. 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 71.Cho JA, Lee AH, Platzer B, Cross BC, Gardner BM, De Luca H, Luong P, Harding HP, Glimcher LH, Walter P, Fiebiger E, Ron D, Kagan JC, Lencer WI. 2013. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe 13:558–569. 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Fejer G, Drechsel L, Liese J, Schleicher U, Ruzsics Z, Imelli N, Greber UF, Keck S, Hildenbrand B, Krug A, Bogdan C, Freudenberg MA. 2008. Key role of splenic myeloid DCs in the IFN-alphabeta response to adenoviruses in vivo. PLoS Pathog. 4:e1000208. 10.1371/journal.ppat.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 186:323–331. 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, Papa FR, Oakes SA. 2012. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science 338:818–822. 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]