ABSTRACT
Equine hepacivirus (EHcV) has been identified as a closely related homologue of hepatitis C virus (HCV) in the United States, the United Kingdom, and Germany, but not in Asian countries. In this study, we genetically and serologically screened 31 serum samples obtained from Japanese-born domestic horses for EHcV infection and subsequently identified 11 PCR-positive and 7 seropositive serum samples. We determined the full sequence of the EHcV genome, including the 3′ untranslated region (UTR), which had previously not been completely revealed. The polyprotein of a Japanese EHcV strain showed approximately 95% homology to those of the reported strains. HCV-like cis-acting RNA elements, including the stem-loop structures of the 3′ UTR and kissing-loop interaction were deduced from regions around both UTRs of the EHcV genome. A comparison of the EHcV and HCV core proteins revealed that Ile190 and Phe191 of the EHcV core protein could be important for cleavage of the core protein by signal peptide peptidase (SPP) and were replaced with Ala and Leu, respectively, which inhibited intramembrane cleavage of the EHcV core protein. The loss-of-function mutant of SPP abrogated intramembrane cleavage of the EHcV core protein and bound EHcV core protein, suggesting that the EHcV core protein may be cleaved by SPP to become a mature form. The wild-type EHcV core protein, but not the SPP-resistant mutant, was localized on lipid droplets and partially on the lipid raft-like membrane in a manner similar to that of the HCV core protein. These results suggest that EHcV may conserve the genetic and biological properties of HCV.
IMPORTANCE EHcV, which shows the highest amino acid or nucleotide homology to HCV among hepaciviruses, was previously reported to infect horses from Western, but not Asian, countries. We herein report EHcV infection in Japanese-born horses. In this study, HCV-like RNA secondary structures around both UTRs were predicted by determining the whole-genome sequence of EHcV. Our results also suggest that the EHcV core protein is cleaved by SPP to become a mature form and then is localized on lipid droplets and partially on lipid raft-like membranes in a manner similar to that of the HCV core protein. Hence, EHcV was identified as a closely related homologue of HCV based on its genetic structure as well as its biological properties. A clearer understanding of the epidemiology, genetic structure, and infection mechanism of EHcV will assist in elucidating the evolution of hepaciviruses as well as the development of surrogate models for the study of HCV.
INTRODUCTION
The Flaviviridae family is composed of four genera: Flavivirus, Pestivirus, Pegivirus, and Hepacivirus. Flaviviridae family viruses are enveloped and contain a single-stranded, positive-sense RNA genome, which encodes a single large precursor polyprotein composed of approximately 2,800 to 3,000 amino acids. The genus Hepacivirus had included only two species, hepatitis C virus (HCV) and GB virus B (GBV-B), until 2010. GBV-B was isolated from serum samples obtained from laboratory tamarins by 11 passages of serum obtained from a human patient with idiopathic hepatitis (1). Although GBV-B experimentally infects tamarins and common marmosets, but not chimpanzees, in vivo (2, 3), the natural host of GBV-B has not yet been clarified. Several hepacivirus species were recently detected in dogs, horses, bats, and rodents and tentatively designated nonprimate hepaciviruses (NPHVs). Bat hepaciviruses have been isolated from some species of bats in Kenya (4), while rodent hepaciviruses have been isolated from several species of rodents in Germany, the Netherlands, South Africa, and Namibia (5, 6). GBV-B is phylogenetically more similar to rodent hepacivirus than to HCV (5). Several strains of equine hepacivirus (EHcV) have been isolated from domestic horses in the United States, the United Kingdom, and Germany (5, 7, 8). The canine hepacivirus was isolated from dogs in the United States (9) but has not yet been genetically or serologically detected in any dogs other than those from the first report (5, 7, 8). The polypeptides of canine hepacivirus show approximately 95% amino acid homology to those of the EHcV strains, suggesting that canine hepacivirus may belong to the same species as EHcV and that infections may be rare in dogs (5, 7, 8, 10). Recent phylogenetic analyses identified EHcV as the most closely related viral homologue of HCV among the reported NPHV strains; however, epidemiological and virological information on EHcV is limited. The open reading frames of EHcV strains show approximately 95% homology to one another, suggesting that previously reported EHcV strains may be classified into one species. Several genome sequences of rodent hepacivirus have already been completely determined (5). The 3′ untranslated region (UTR) of HCV was found to include three stem-loop (SL) structures, while variable stem-loop structures were found in that of rodent hepacivirus and GBV-B (5). However, the nucleotide sequence of the EHcV 3′ UTR has not yet been determined completely because the adenine-rich [(A)-rich] sequence downstream of the stop codon in the EHcV genome interrupts an ordinary 3′-rapid amplification of cDNA ends (RACE) reaction (8). The RNA secondary structure of the hepacivirus 3′ UTR may indicate species specificity (5).
On the basis of amino acid similarities among the polyproteins of NPHVs and HCV, the N-terminal one-fourth of the NPHV polyprotein has been predicted to be cleaved by signal peptidase into mature structural proteins and a viroporin (core, E1, E2, and p7), while the C-terminal three-fourths has been predicted to be cleaved by viral proteases into maturated nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (6). Core, E1, and E2 have been predicted to form viral particles with host lipids, although it remains unclear whether p7 is incorporated into a viral particle. Signal peptide peptidase (SPP) was shown to further cleave the C-terminal transmembrane region of HCV and GBV-B core protein after signal peptidase-dependent cleavage (11, 12). However, whether SPP cleaves the C-terminal transmembrane region of the NPHV core protein remains unknown.
The mature core proteins of HCV and GBV-B are localized mainly on lipid droplets (LDs) (13, 14). The core proteins of dengue virus are also localized on LDs but are not cleaved by SPP (15), suggesting that localization of the core protein on LDs may be one of the common characteristics of the Flaviviridae family. The HCV core protein is known to be partially localized in the detergent-resistant membrane (DRM), which originates from lipid raft-like membranes (16, 17). The DRM is composed of cholesterol and sphingolipids, which are included in the replication compartment known as the membranous web (18, 19). Therefore, LDs and DRM are considered to be the intracellular compartments for the replication and viral assembly of HCV, but it is currently unknown whether NPHV core proteins are localized on LDs and DRM.
Epidemiological information on EHcV is still limited. The results of the present study demonstrated that Japanese-born domestic horses were infected with EHcV, which showed high homology to the reported strains on the basis of its nucleotide and amino acid sequences. We predicted the RNA secondary structures around the 5′ and 3′ UTRs of the EHcV genome and analyzed the biological properties of the EHcV core protein in relation to the HCV core protein.
MATERIALS AND METHODS
Samples.
Serum samples 1 to 13 were collected from Japanese-born domestic horses raised on one farm, farm A, located in Hokkaido, Japan, while groups of serum samples numbered 14 to 18 and 19 to 31 were from horses on farms B and C, respectively, located in Tokyo, Japan (Fig. 1). The distance between Hokkaido and Tokyo is about 1,000 km. All sample collections conformed to guidelines for the care and use of laboratory animals (Yamanashi University) and were approved by the Institutional Committee of Laboratory Animal Experimentation (Yamanashi University). All samples were divided into small aliquots and stored at −80°C until nucleic acid extraction.
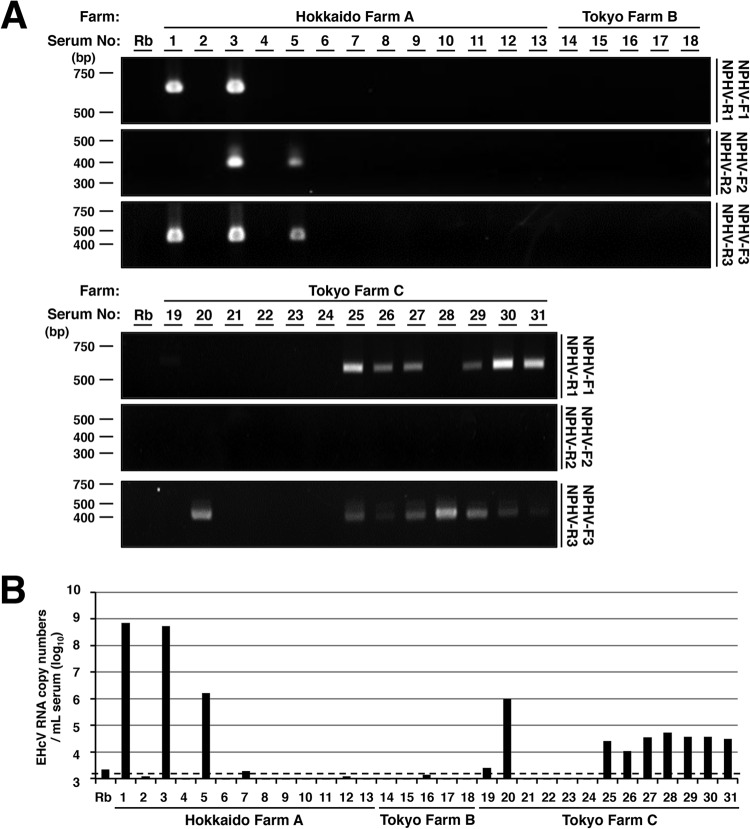
FIG 1.
Detection and genetic analyses of NPHV genomic RNA in sera of Japanese domestic horses. (A) Total RNAs extracted from 31 equine sera and normal rabbit serum (Rb) as a negative control were subjected to RT-PCR analysis. Hokkaido Farm A, Tokyo Farm B, and Tokyo Farm C indicate the farms where the individual horses were reared. Three sets of primers, NPHV-F1 and NPHV-R1, NPHV-F2 and NPHV-R2, and NPHV-F3 and NPHV-R3, were used to amplify NPHV-specific gene regions. The PCR products were electrophoresed and stained with ethidium bromide. (B) Total RNAs were isolated from sera, reverse-transcribed, and estimated as a copy number per ml. Normal rabbit serum was used as a negative control. The dashed line indicates the cutoff level.
RT-PCR.
Total RNAs were prepared from horse sera using a Qiagen viral RNA extraction kit (Qiagen, Valencia, CA). RNAs were converted to cDNA using a PrimeScript reverse transcription-PCR (RT-PCR) kit (TaKaRa, Shiga, Japan) with random primers. The viral gene was amplified by PCR using PuReTaq Ready-To-Go PCR beads (GE Healthcare, Piscataway, NJ) with three pairs of primers: NPHV-F1 (5′-TGTCACCTACTATCGGGG-3′) and NPHV-R1 (5′-TCAAGCCTATACAGCAAAGG-3′), NPHV-F2 (5′-ATCATTTGTGATGAGTGCC-3′) and NPHV-R2 (5′-CATAAGGGCGTCCGTGGC-3′), and NPHV-F3 (5′-GTGGTCGCCACGGATGCC-3′) and NPHV-R3 (5′-ACCCTATGAAGACGCTCTCC-3′). PCR was carried out as follows: one cycle at 92°C for 5 min; 35 repeats of one cycle at 94°C for 0.5 min, 58°C for 0.5 min, and 72°C for 0.5 min, in that order; and one cycle at 72°C for 1 min followed by holding at 4°C. The PCR products were electrophoresed on 1.5% agarose gels, stained with ethidium bromide, and visualized using the BioDoc-It imaging system (UVP, Upland, CA).
Determination of the EHcV genomic sequence.
The viral genome of EHcV was segmentally amplified by PCR using the primers listed in Table 1. The PCR products were cloned into T vectors prepared from pBluescript II SK(−) (20). The DNA sequences of the PCR products were determined using an ABI Prism BigDye Terminator version 1.1 cycle sequencing kit and an ABI Prism 310 genetic analyzer (Life Technologies, Tokyo, Japan). More than three colonies were picked up among the transformants of Escherichia coli with regard to the accuracy of the sequence. The nucleotide sequences of the PCR products were determined in forward and reverse directions. The junction of two adjacent PCR products was confirmed by PCR using primers that overlapped two close regions. The 5′-terminal sequence upstream of the open reading frame was determined with a 5′-RACE core set (TaKaRa) using the 5′ phospholyrated RT primer for the NPHV 5′ UTR (5′-CATCCTATCAGACCG-3′). The 3′-terminal region downstream of the (A)-rich region was determined by the 3′-RACE method (21, 22), modified as follows: Total RNAs were prepared from horse serum using TRIzol LS reagent (Invitrogen, Carlsbad, CA) with 40 μg of glycogen (Nacalai Tesque, Kyoto, Japan). The poly(U) tail was added to the 3′ end of the RNA preparation using Escherichia coli poly(U) polymerase (New England BioLabs, Ipswich, MA) and was incubated for 45 min at 37°C. The resulting preparation was reverse transcribed by the SuperScript First-Strand Synthesis system (Life Technologies) using an oligo(dA) adapter primer (5′-TTGCGAGCACAGAATTAATACGACTCACAAAAAAAAAAAAVN-3′). The sequence of each region was determined by sequencing more than 3 clones. The primers for PCR amplification and the RACE methods are listed in Table 1. The whole sequence of the EHcV strain isolated from serum sample 3 (GenBank accession number AB863589) was determined by the method described above. The EHcV strain was designated JPN3/JAPAN/2013 in this study. The partial NS5B-coding regions and 3′ UTRs were amplified from serum samples 5 and 1. The nucleotide sequences of samples 5 and 1 (GenBank accession numbers AB921150 and AB921151, respectively) were determined by the method described above. The neighbor-joining trees of the nucleotide sequences from the NPHV, HCV, and GBV-B strains were predicted by the method of Saitou et al (23). Trees were constructed by the maximum composite likelihood method calculated by using the program MEGA5 (24) (see Fig. 3). The secondary protein structures were predicted by the method of Garnier et al. (25) (see Fig. 6). Hydrophobicity plots of the EHcV and HCV core proteins were prepared by the method of Kyte and Doolittle (26) and drawn using the software Genetyx (Nihon Genetyx, Tokyo, Japan) (see Fig. 5).
TABLE 1.
List of PCR primers used in this study
| Primer | Nucleotide position | Genome location | F or Ra | Sequence (5′→3′) |
|---|---|---|---|---|
| Primer for cloning of the NPHV genome | 92–111 | 5′ UTR | F | ATGTGTCACTCCCCCTATGG |
| 367–386 | 5′ UTR | R | CTATGGTCTACGAGACCGGC | |
| 268–285 | 5′ UTR | F | AGCCGAAATTTGGGCGTG | |
| 1207–1224 | E1 | R | AAACAGAAGCCATAGCGG | |
| 1116–1132 | E1 | F | AGTGCTTGTTGGGTGCC | |
| 1697–1713 | E2 | R | GTCCTTTGCACTTCGGG | |
| 1605–1623 | E2 | F | ACTGTTAAGCAGATGTGGG | |
| 2103–2121 | E2 | R | CACAGAGTTGGTAAGTAGC | |
| 2007–2023 | E2 | F | AAGCAGTGTGGTGCTCC | |
| 2526–2545 | E2 | R | AAACAGAACCAGAGAATTGC | |
| 2375–2392 | E2 | F | CCCTGCCTTCACTACTGG | |
| 2898–2913 | NS2 | R | CGAGATAGCGCCAAGC | |
| 2847–2867 | NS2 | F | TTTATGCTAGTAAAGTGGTGG | |
| 3396–3415 | NS2 | R | GGTGATAAAAGTCTCCATCC | |
| 3318–3334 | NS2 | F | ATCCTCCATGGCTTGCC | |
| 3819–3835 | NS3 | R | GGGCCACCTGAACTACC | |
| 3732–3750 | NS3 | F | ACCAGGACGGGTCAGGTCG | |
| 4254–4270 | NS3 | R | ATAATGTCATAAGCACC | |
| 4177–4195 | NS3 | F | CTAGTTGCAAGACAACGGG | |
| 4682–4700 | NS3 | R | AGTGTTGCAGTCAGTGACG | |
| 4574–4591 | NS3 | F | TGTCACCTACTATCGGGG | |
| 5199–5218 | NS3 | R | TCAAGCCTATACAGCAAAGG | |
| 4574–4591 | NS3 | F | TGTCACCTACTATCGGGG | |
| 5199–5218 | NS3 | R | TCAAGCCTATACAGCAAAGG | |
| 5134–5152 | NS3 | F | CTCCCAGCAAAGATGAACG | |
| 5997–6014 | NS4B | R | AGCACCCACACCAACAGC | |
| 5919–5934 | NS4B | F | AAGATCTTGAGTGGTG | |
| 6651–6632 | NS5A | R | GCCGATAACTCTGACAGC | |
| 6547–6564 | NS5A | F | ACACCTGGAAAACAGCCG | |
| 7293–7310 | NS5A | R | AGATTCCGTGGCCGAAGG | |
| 7235–7252 | NS5A | F | AGCTCTCGTTTCCGGGTG | |
| 7573–7590 | NS5B | R | TAGCTGACGCTGTTGTGG | |
| 7511–7527 | NS5B | F | ACGCCACCCTATAGGCC | |
| 8027–8046 | NS5B | R | GTTGACGGGGAGTGTATTGG | |
| 7926–7943 | NS5B | F | ATCGTTTACCCCGATTTG | |
| 8528–8545 | NS5B | R | CAAGATGTTATCTGCTCC | |
| 8457–8474 | NS5B | F | CGTGACTTCACTAATGCC | |
| 9069–9086 | NS5B | R | GTCAATCGAGTTTACGCC | |
| Primer for 5′ RACE | 235–252 | 5′ UTR | F | AATCGCGGCTTGAACGTC |
| 213–230 | 5′ UTR | R | TGTACTCACGGATTCACG | |
| Primer for 3′ RACE | 8979–8999 | NS5B | F | CTTAAAGTACGTGGTGGTCGC |
| Adapter primer | R | GCGAGCACAGAATTAATACGAC |
F, forward; R, reverse.
FIG 3.
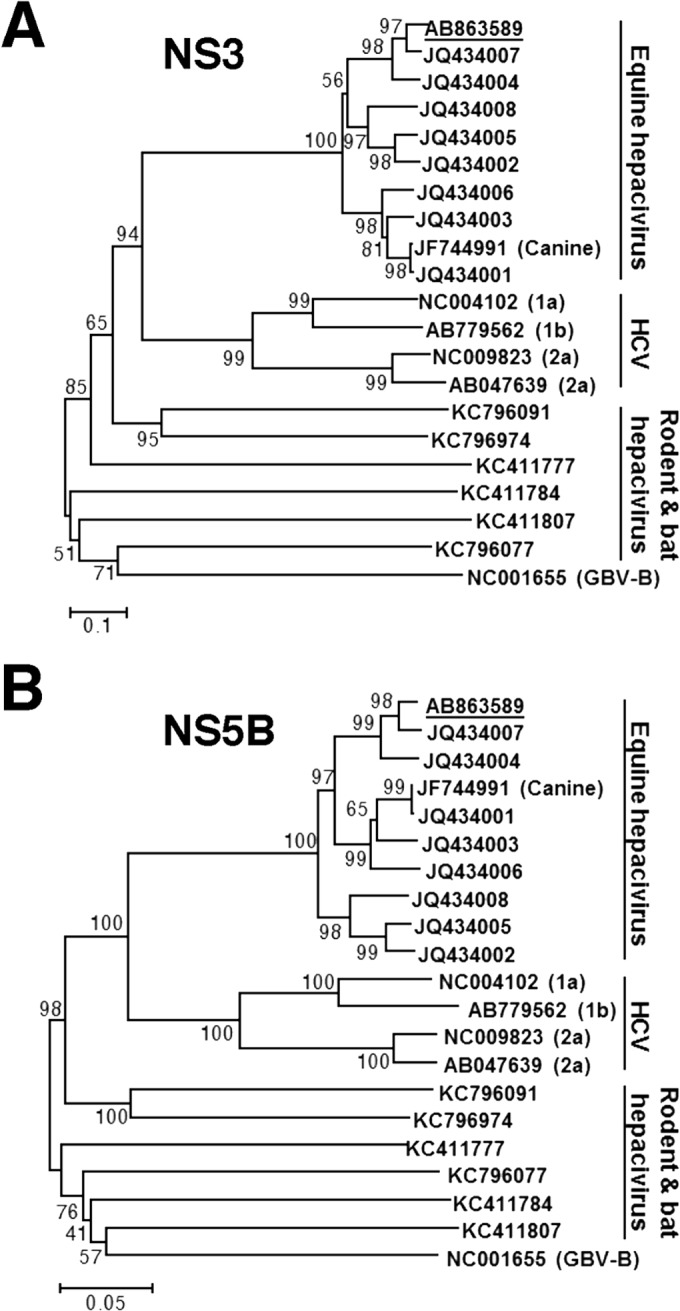
Phylogenetic analysis of the EHcV gene. Neighbor-joining trees of the nucleotide sequences from the NS3 (A) and NS5B (B) regions of the NPHV, HCV, and GBV-B strains are shown (23). Trees were constructed by the maximum composite likelihood method calculated using the program MEGA5 (24). The percentage of replicate trees in which the associated taxa were clustered together in the bootstrap test (1,000 replicates) is indicated next to the branches. Analyses were carried out using 10 strains of EhcV, JPN3/JAPAN/2013, A6-066 (GenBank accession no. JQ434003), B10-022 (GenBank accession no. JQ434004), F8-068 (GenBank accession no. JQ434005), G1-073 (GenBank accession no. JQ434002), G5-077 (GenBank accession no. JQ434006), H3-011 (GenBank accession no. JQ434008), H10-094 (GenBank accession no. JQ434007), NZP1 (GenBank accession no. JQ434001), and AAK-2011 (canine hepacivirus; GenBank accession no. JF744991); 4 strains of HCV, H77 (genotype 1a; GenBank accession no. NC004102), LyHCV (genotype 1b; GenBank accession no. AB779562), HC-J6CH (genotype 2a; GenBank accession no. NC009823), and JFH1 (genotype 2a; GenBank accession no. AB047639); 3 strains of bat hepacivirus, PDB-112 (GenBank accession no. KC796077), PDB-445 (GenBank accession no. KC796091), and PDB-829 (GenBank accession no. KC796074); 3 strains of rodent hepacivirus, RMU10-3382 (GenBank accession no. KC411777), NLR-AP-70 (GenBank accession no. KC411784), and SAR-46 (GenBank accession no. KC411807); and another primate hepacivirus, GBV-B (GenBank accession no. NC001655). The Japanese strain JPN3/JAPAN/2013 (GenBank accession no. AB863589) is underlined.
FIG 6.
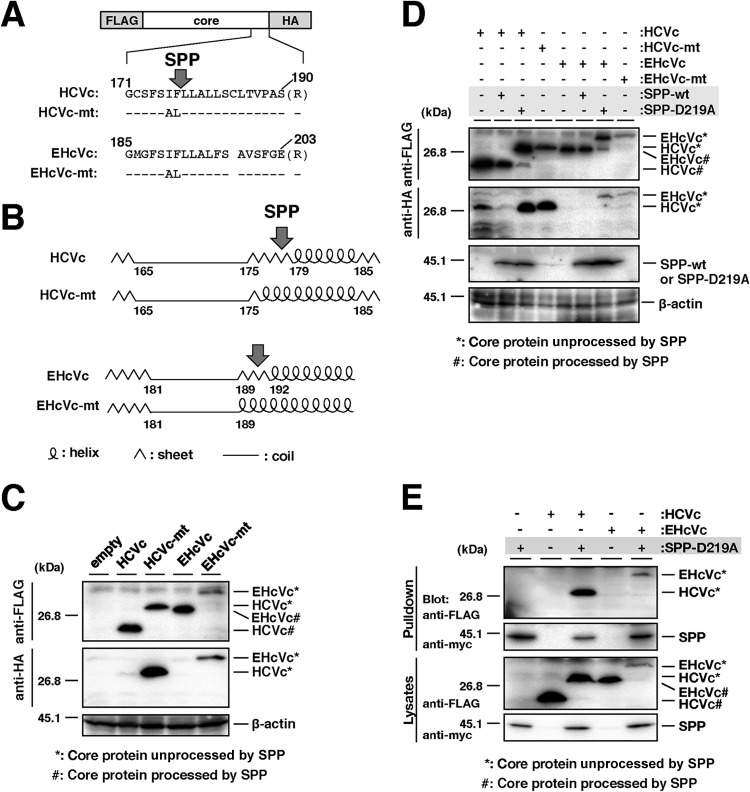
Intramembrane processing of the EHcV core protein by SPP. (A) The plasmids encoding HCVc, HCVc-mt, EHcVc, and EHcVc-mt are shown as a schematic diagram. Their C-terminal regions (171 to 190, HCV core protein; 185 to 203, EHcV core protein) were aligned. The C-terminal Ala of each core protein was replaced with Arg (R) to prevent signal peptidase-dependent cleavage for the detection of the SPP-uncleaved core protein with the anti-HA antibody. Bars indicate the amino acids that were the same as those of the wild-type residues. (B) The secondary protein structures in the C-terminal transmembrane regions of the HCV and EHcV core proteins and mutants were predicted by the method of Garnier et al. (25). Arrows indicate putative SPP cleavage sites. (C) HCVc, HCVc-mt, EHcVc, and EHcVc-mt were expressed in 293FT cells and immunoblotted with the anti-FLAG and -HA antibodies. (D) HCVc or EHcVc was expressed with SPP-wt or SPP-D219A in the 293FT cell line. HCVc-mt and EHcVc-mt were expressed in the absence of SPP-wt and SPP-D219A as uncleavable controls. (E) HCVc or EHcVc was coexpressed with or without SPP-D219A. SPP-D219A was pulled down with Ni beads. Coprecipitated proteins were immunoblotted with the anti-FLAG antibody.
FIG 5.
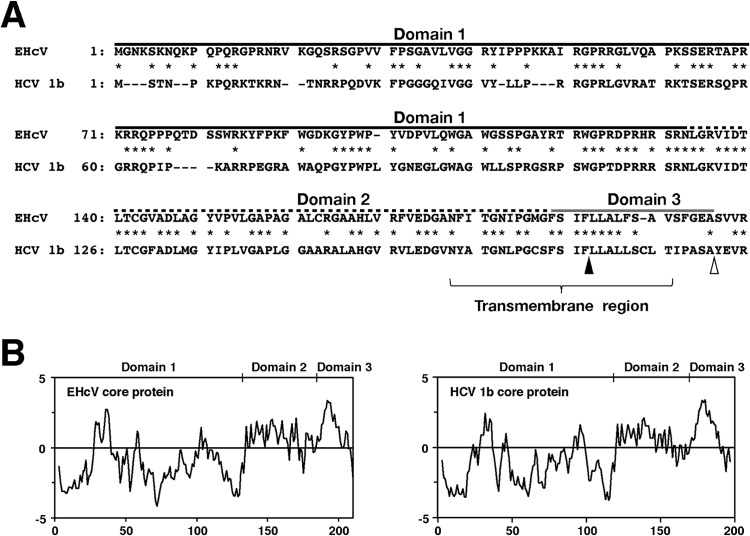
Amino acid alignment and hydrophobicity of EHcV and HCV core proteins. (A) Alignment of the core proteins of EHcV (JPN3/JAPAN/2013) and HCV genotype1b (Con1; GenBank accession number AJ238799). Asterisks indicate identical amino acid residues. Bars indicate gaps to achieve maximum amino acid matching. The black and white arrowheads indicate the predicted cleavage site of the core protein of HCV by SPP and signal peptidase, respectively. The EHcV core protein was composed of three domains, domain 1 (a black line, residues 2 to 132), domain 2 (a broken black line, residues 133 to 187), and domain 3 (a gray line, residues 188 to 204), relative to those of the HCV core protein (42). (B) Hydrophobicity plots of the EHcV and HCV core proteins were prepared by the method of Kyte and Doolittle (26). The horizontal and vertical axes represent amino acid position and hydrophobicity, respectively.
Quantification of viral genomic RNAs in horse sera.
Total RNA was prepared from equine serum using a Qiagen viral RNA extraction kit and was then reverse transcribed into cDNA by using a PrimeScript RT-PCR kit with random primers. The amount of targeted viral RNA was estimated using SYBR GreenER qPCR SuperMix (Life Technologies) and the ABI StepOnePlus real-time PCR system (Life Technologies). The region encoding NS3 was targeted with the primer pair NPHV-F3 (5′-GTGGTCGCCACGGATGCC-3′) and NPHV-R3 (5′-ACCCTATGAAGACGCTCTCC-3′). Total RNAs extracted from conventional rabbit serum were used as a negative control to determine the analytical threshold line. The in vitro-transcribed RNA of EHcV was utilized for the standard curve.
Prediction of RNA secondary structures.
The 5′-UTR sequences of EHcV strains were aligned with the MUSCLE program and subjected to a manual search for covariant nucleotide substitutions. The RNA folding structure upstream of domain III in the 5′ UTR was predicted using the Mfold web server (27) with conventional phylogenetic conservation analysis due to the lack of sufficient homology to the 5′ UTR sequences of HCV strains. The NS5B-coding regions and 3′ UTRs of EHcV strains were aligned with the program MUSCLE. Conserved secondary structures were predicted as described above. The secondary structures of the 3′ UTR in EHcV were predicted by the Mfold web server without confirming phylogenetic data because of the absence of additional available sequences of the EHcV 3′ UTR and the lack of sufficient homology to the HCV X-tail sequences.
Plasmids.
The PCR product encoding the EHcV core protein was amplified from serum sample 3 and was then cloned into the BamHI and XhoI sites of pcDNA3.1-Flag/HA, which encodes the FLAG and hemagglutinin (HA) epitope tags, as reported previously (28). Ala204 was replaced with Lys to prevent signal peptidase-dependent cleavage. The translated EHcV core protein was added to the FLAG and HA epitope tags at the N and C termini (EHcVc), respectively. A point mutation was generated using a KOD mutagenesis kit (Toyobo, Osaka, Japan). The PCR products encoding EHcVc or the mutant in which Ile190 and Phe191 were replaced with Ala and Leu (EHcVc-mt), respectively, were introduced into the AflII and EcoRV sites of pCAGGS using an In-Fusion HD cloning kit (TaKaRa). The introduced fragments of all plasmids were confirmed by sequencing using an ABI Prism 310 genetic analyzer (Applied Biosystems). The plasmid encoding the N-terminally FLAG-tagged and C-terminally HA-tagged HCV core protein (HCVc) and the mutant in which Ile176 and Phe177 were replaced with Ala and Leu (HCVc-mt), respectively, were described previously (28). The gene encoding human signal peptide peptidase (SPP) or its mutant was introduced into pcDNA3.1-myc/His C (Invitrogen) instead of the plasmids described previously (28). The resulting plasmids encoded C-terminally myc-His×6-tagged wild-type SPP (SPP-wt) or the mutant protein in which Asp219 was replaced with Ala (SPP-D219A).
Cell culture and transfection.
The human embryonic kidney cell line 293FT and the human hepatoma cell line Huh7OK1 (29) were maintained in Dulbecco's modified Eagle's minimal essential medium (DMEM) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, nonessential amino acids (Sigma, St. Louis, MO), sodium pyruvate (Sigma), and 10% fetal bovine serum (FBS) and were then cultured at 37°C under the conditions of a humidified atmosphere and 5% CO2. Plasmids were transfected into cell lines using XtremeGene 8 (Roche) according to the manufacturer's protocol.
Western blot analysis.
293FT cells were cultured in 6-well plates and transfected with the appropriate plasmids. The transfected cells were harvested at 18 h posttransfection, washed with cold phosphate-buffered saline (PBS), and suspended in 50 μl of the lysis buffer consisting of 20 mM Tris-HCl (pH 7.5), 135 mM NaCl, 10% glycerol, 1% Triton X-100, and protease inhibitor cocktail (Merck Bioscience, Calbiochem, San Diego, CA). The lysates were centrifuged at 19,000 × g for 5 min at 4°C. The supernatants were mixed with 16 μl of 4× SDS sample buffer and then boiled at 60°C for 20 min. The resulting mixtures were subjected to SDS-PAGE. The proteins in a gel were transferred to polyvinylidene difluoride (PVDF) membranes and incubated with mouse anti-FLAG antibodies (Sigma), mouse anti-HA antibodies (Covance, Princeton, NJ), mouse anti-c-myc antibodies (BD Pharmingen, San Diego, CA), or mouse anti-beta-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and were then incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies. Immunocomplexes were visualized with SuperSignal West Femto substrate (Thermo Scientific, Rockford, IL) and detected using an LAS-4000 Mini image analyzer (GE Healthcare, Buckinghamshire, United Kingdom).
Detection of antibodies against EHcV.
To detect anti-EHcV antibodies in horse sera, we subjected lysates prepared from 293FT cells expressing EHcVc, which is an N-terminally FLAG-tagged and C-terminally HA-tagged EHcV core protein (a positive reference), or cells transfected with an empty plasmid (a negative reference) to Western blotting, as described above. The resulting PVDF membranes were incubated with Blocking One solution (Nacalai Tesque) for blocking at room temperature for 30 min and then incubated with 1,000-fold-diluted horse serum in 10-fold-diluted Blocking One. Mouse anti-FLAG or rabbit anti-EHcV core antibody was used as a positive serum control. The resulting membrane was incubated with an HRP-conjugated antibody to mouse, rabbit, or horse IgG (Abcam, Cambridge, UK) at room temperature for 1 h. Protein bands with a molecular mass of 28 kDa were detected in the positive reference, but not in the negative reference, using positive serum or an antibody to the FLAG epitope tag or EHcV core protein. The rabbit polyclonal antibody against the EHcV core protein was generated by immunization using peptides of the residues from 2 to 15, GNKSKNQKPQPQRG (Scrum Inc., Tokyo, Japan).
Pulldown assay for SPP binding.
Human embryonic kidney 293FT cells expressing EHcVc or HCVc with or without SPP-D219A were harvested at 18 h posttransfection, washed with cold PBS, suspended in 100 μl of the lysis buffer, and centrifuged at 14,000 ×g for 5 min at 4°C. Twenty microliters of the lysate was mixed with 20 μl of 2× SDS sample buffer. The remaining lysate was adjusted to 250 μl with the lysis buffer and incubated for 2 h at 4°C after the addition of 20 μl of His-Select nickel affinity gel (Sigma) equilibrated 50% (vol/vol) with lysis buffer. The nickel beads that included SPP-wt or SPP-D219K were washed five times with 500 μl of lysis buffer by centrifugation at 5, 000 × g for 1 min at 4°C and then suspended in 40 μl of 1× SDS sample buffer. After being boiled at 60°C for 20 min, the supernatant was subjected to Western blotting to detect the coprecipitated core proteins.
Immunofluorescence microscopy.
Huh7OK1 cells were incubated with fresh DMEM containing Bodipy 558/568 (2 μg/ml; Molecular Probes) for 1 h at 37°C to visualize lipid droplets (LDs). The cells were washed once with prewarmed DMEM and incubated for 30 min at 37°C. The treated cells were then fixed in 4% paraformaldehyde for 30 min at room temperature. After two washes with PBS, the cells were permeabilized with permeabilization buffer containing 0.1% saponin (eBioscience, San Diego, CA) for 30 min at 37°C and blocked with PBS containing 2% FBS (blocking buffer) for 30 min at room temperature. The cells were incubated with an appropriate antibody, as indicated in the figure legends. The cells were washed three times with PBS. The mounted cells were observed with a FluoView FV1000 laser scanning confocal microscope (Olympus, Tokyo, Japan). Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Flotation assay.
A flotation assay was carried out according to the method described previously (17). Briefly, 293FT cells expressing EHcVc or EHcVc-mt were cultured on a 10-cm dish. The transfected cells were washed once with cold PBS at 18 h posttransfection and harvested using a cell scraper. The cells were suspended in 1.2 ml of 25 mM Tris-HCl (pH 7.5) containing 150 mM NaCl, 5 mM EDTA, and protease inhibitor cocktail (Merck, Calbiochem) (TNE buffer) and were then homogenized by 10 passes through a 26-gauge needle. Each 0.6-ml aliquot of the homogenates was incubated for 30 min on ice with or without 1% Triton X-100 and was then mixed with 0.4 ml of OptiPrep (Axis-Shield, Oslo, Norway). An appropriate concentration of OptiPrep was adjusted with TNE buffer. This mixture was overlaid with 1.2 ml of 30% OptiPrep, 1.2 ml of 25% OptiPrep, and 0.8 ml of 5% OptiPrep, in that order, and was centrifuged at 42,000 rpm for 5 h at 4°C in an SW50.1 rotor (Beckman Coulter, Fullerton, CA). Each fraction, with a volume of 0.4 ml, was collected from the top of the centrifugation tube and was then precipitated by mixing with 4 volumes of cold acetone at −30°C. The resulting pellet was resolved in 50 μl of 1× sample buffer and then subjected to Western blot analysis using a mouse anti-FLAG antibody (Sigma), a rabbit anti-calreticulin antibody (Sigma), and a rabbit anti-caveolin-1 antibody (Sigma). The fractions containing calreticulin in the absence and presence of Triton X-100 were defined as the membrane and detergent-soluble membrane fractions, respectively. In the presence of the detergent, fractions 3 to 5, which contained caveolin-1 but only small amounts of calreticulin, were defined as the detergent-resistant membrane fractions.
Nucleotide sequence accession numbers.
The whole sequence of the EHcV strain isolated from serum sample 3 was deposited in GenBank under accession number AB863589. The nucleotide sequences of the partial NS5B-coding regions and 3′ UTRs from samples 5 and 1 were registered as AB921150 and AB921151, respectively.
RESULTS
Detection of the EHcV genome and antibody to EHcV in sera of Japanese-born horses.
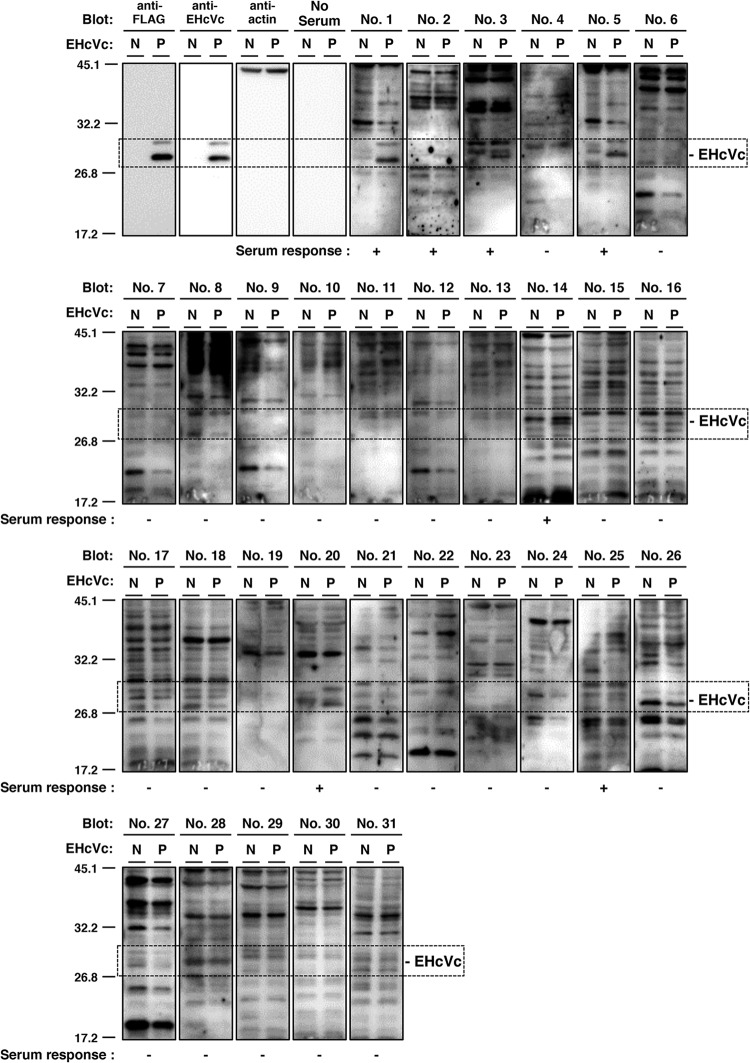
To clarify whether NPHVs were distributed in Japan, we collected 31 horse serum samples and examined them in order to detect the EHcV genome and antibody to the core protein. We prepared total RNAs from horse sera and screened them using RT-PCR analyses with three sets of PCR primers (NPHV-F1/NPHV-R1, NPHV-F2/NPHV-R2, and NPHV-F3/NPHV-R3) that targeted the NS3-coding region that is relatively conserved among NPHVs. Total RNA prepared from conventional rabbit serum was used as a negative control. PCR products with the expected sizes were found in horse serum samples 1, 3, 25, 26, 27, and 29 to 31 using NPHV-F1/NPHV-R1, in horse serum samples 3 and 5 using NPHV-F2/R2, and in horse serum samples 1, 3, 5, 20, and 25 to 31 using NPHV-F3/R3 (Fig. 1A). The EHcV genome was detected in 11 of 31 (35%) serum samples by RT-PCR (Fig. 1A and B). Copy numbers of the EHcV genome in horse sera varied from 104 to 109 copies per ml of sera (Fig. 1B). Although a PCR product was slightly amplified from serum sample 19 by PCR using the primer pair NPHV-F1/R1, the copy number of the virus genome in serum sample 19 was estimated to be low, at a level similar to that of the negative control. Thus, we could not determine whether serum sample 19 included a viral genome. We then immunologically surveyed horse sera by Western blotting. Western blotting analyses using horse sera to detect antibodies to the EHcV core protein (Fig. 2) showed that the sera of samples 1, 2, 3, 5, 14, 20, and 25 were immunoreactive to the EHcV core protein (7 positive serum samples of a total of 31 samples; 22.6%). The sera of samples 1, 3, 5, and 20 were PCR positive and seropositive. Serum samples 2 and 14 were PCR negative and seropositive, whereas samples 26 to 31 were PCR positive and seronegative. These results suggest that EHcV has infected Japanese-born domestic horses.
FIG 2.
Serological screening of Japanese-born domestic horses. Lysates of 293FT cells transfected with an empty plasmid (a negative reference, N) or the plasmid encoding EHcVc (a positive reference, P) were subjected to Western blotting using serum from each horse. The serum response “+” indicates that the protein band with the same molecular size as the EHcV core protein was specifically detected in the “P” lane, but not in the “N” lane, while the serum response “−” indicates that the protein band with the same molecular size as the EHcV core protein was detected in neither the “P” lane nor the “N” lane. Both antibodies to the FLAG tag and to the EHcV core protein were used as serum positive controls, while protein amounts were standardized with blotting using the antibody to beta-actin. “No serum” indicates the membrane was incubated without primary antibodies but with HRP-conjugated anti-horse IgG antibodies as a background of the secondary antibody.
Genetic analysis of EHcV.
PCR products corresponding to the 5′ UTR and the open reading frame were segmentally amplified from serum sample 3 by 5′ RACE and RT-PCR, respectively. In the present study, we successfully determined the 3′-terminal sequence downstream of a stop codon using the 3′-RACE method with poly(U) polymerase. We determined the nucleotide sequence of the putative full genome, which was designated JPN3/JAPAN/2013 (GenBank accession number AB863589). The full-length genome of strain JPN3/JAPAN/2013 is composed of 9,355 nucleotides, consisting of the 5′ UTR with a nucleotide length of 389, the 3′ UTR with a nucleotide length of 134, and an open reading frame with a nucleotide length of 8,832. The open reading frame encodes 2,943 amino acids. Table 2 summarizes the amino acid homology of the JPN3/JAPAN/2013 polyprotein with the polyproteins of the other EHcV strains. The polyprotein of JPN3/JAPAN/2013 shared more than 94% homology with the other EHcV polyproteins and exhibited the highest homology, 97.8%, with NPHV-H10-094 (GenBank accession number JQ434007), which was isolated from a horse in the United States (8). The NS3- and NS5B-coding regions of the EHcV strains were phylogenetically analyzed by the neighbor-joining method. The phylogenetic trees of the NS3 (Fig. 3A) and NS5B regions (Fig. 3B) showed that JPN3/JAPAN/2013 was included in the clade comprising the U.S. strains NPHV-H10-094 (GenBank accession number JQ434007) and B10-022 (GenBank accession number JQ434004).
TABLE 2.
Amino acid sequence homologies of the polyproteins
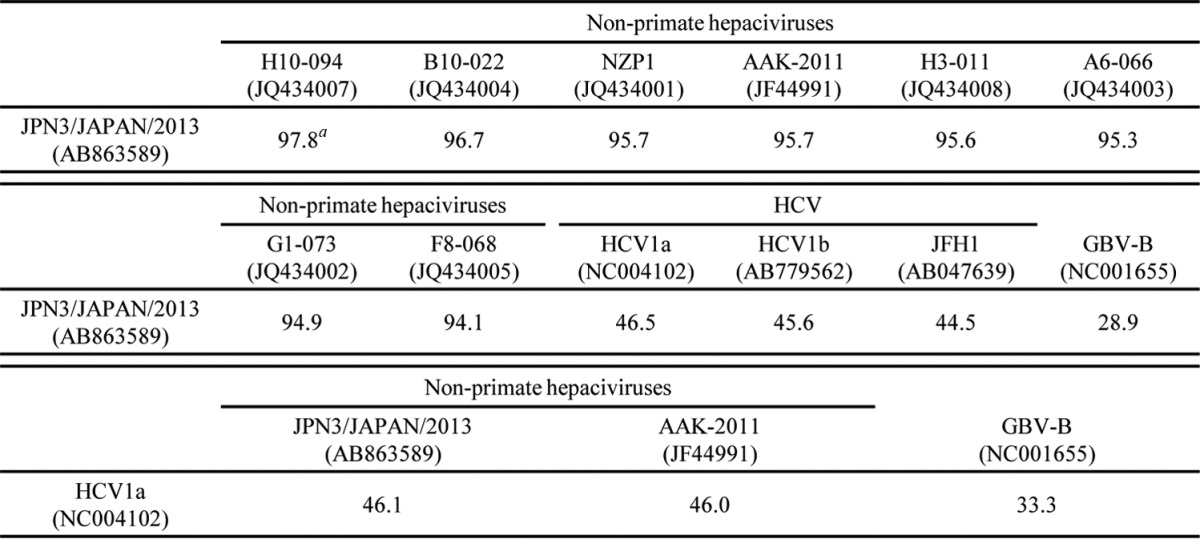
, percent identity.
Putative RNA secondary structures around the UTRs of EHcV.
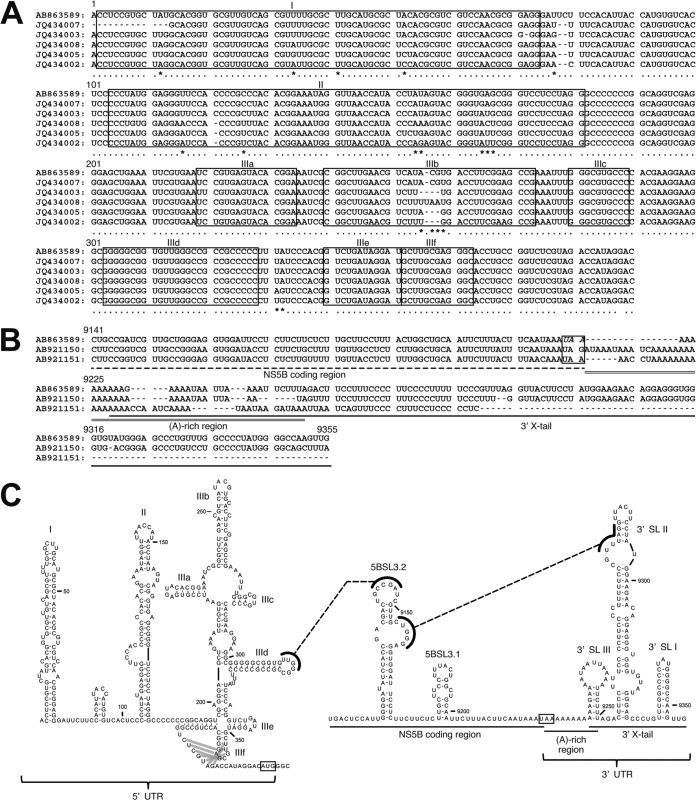
The 5′-terminal region of JPN3/JAPAN/2013 was compared with those of the EHcV genomes (Fig. 4A). The HCV internal ribosome entry site (IRES)-like structure was embedded in the 5′ UTRs of NPHVs (5, 6). The 5′-UTR region was well conserved among the EHcV strains and showed a mean diversity of approximately 4% among the EHcV strains (Fig. 4A). The 3′-terminal sequence downstream of the (A)-rich region in the EHcV genome had not been reported because the (A)-rich region downstream of the stop codon of EHcV interrupted the reaction in the ordinary 3′-RACE method (5, 6). In the present study, we determined the nucleotide sequences downstream of the (A)-rich region from serum sample 3 (JPN3/JAPAN/2013; GenBank accession number AB863589), sample 5 (JPN5/JAPAN/2015; GenBank accession number AB921150), and sample 1 (JPN1/JAPAN/2015; GenBank accession number AB921151) by the modified 3′-RACE method using poly(U) polymerase, although the region in serum sample 1 was incompletely amplified (Fig. 4B). The regions downstream of the (A)-rich region were conserved between serum samples 3 and 5, whereas the (A)-rich regions varied among the three strains (Fig. 4B).
FIG 4.
RNA structure analysis of the 5′ and 3′ ends of EHcV. (A) Alignment of the 5′ UTRs of the EHcV strains. Asterisks and black lines indicate variable residues and miRNA-targeting regions, respectively. SL structures are enclosed in rectangles. GenBank numbers are listed in the legend to Fig. 3. (B) Alignment of the 3′ UTRs of the EHcV strains. The 3′-terminal sequences with GenBank numbers AB921150 and AB921151 were determined using serum samples 5 and 1, respectively, and were aligned with that of JPN3/JAPAN/2013 (GenBank accession no. AB863589). (C) The secondary structures of the 5′ UTR, NS5B-coding region, and 3′ UTR were predicted on the basis of minimum free energy predictions. The stem loops in the 5′ UTR (left) were designated according to the stem loops of the HCV 5′-UTR structures (30). A gray line, double line, and single line indicate a pseudoknot structure, miR-122-target region, and complementary sequence, respectively. The stem loops in the NS5B-coding region and 3′ UTR (right) were designated to correspond to the stem loops embedded in the HCV 3′-terminal region (32, 33). The NS5B-coding region, (A)-rich region, and 3′-X-tail sequence are indicated under the schematic structure. Start and stop codons are enclosed by rectangles.
The secondary structure of 5′ UTR in strain JPN3/JAPAN/2013 was predicted according to the method described previously (8) (Fig. 4C). The stem-loops in the 5′ UTR were designated according to the stem-loops of the HCV 5′-UTR structures (30). Stem-loops (SLs) I, II, IIIa to IIIf, and the pseudoknot interaction were predicted within the 5′ UTR of strain JPN3/JAPAN/2013. These structures were the same as that of the strain reported previously (9), although several nucleotide insertions and deletions were more predominant in the apical loop of subdomain IIIb than in the other strains reported previously (Fig. 4A and C). Two seed sites of the microRNA miR-122 (Fig. 4A and C) were found in the 5′ UTR of strain JPN3/JAPAN/2013 at nucleotide residues 81 to 89 (UCCACAUUA) and 98 to 103 (CACUCC), which also corresponded to the predicted miR-122 seed sites in the 5′ UTRs of the other EHcV strains (9).
The HCV 3′ UTR, which is generally 200 to 300 nucleotides in length, consists of a short variable region, the poly(U/UC) stretch sequence, and the 3′-X-tail region, in that order (31–33). Although the EHcV 3′ UTR, which is composed of 138 nucleotides, is shorter than the HCV 3′ UTR, the 3′ UTR of EHcV consists of the (A)-rich sequence and 3′-X-tail region, in that order. The (A)-rich sequence of EHcV may vary in length (Fig. 4B). We subsequently predicted the secondary structure of the EHcV 3′ UTR. Although the EHcV 3′ UTR, which is composed of 138 nucleotides, is shorter than the HCV 3′ UTR, the 3′ UTR includes three predicted SL structures (Fig. 4C). Based on the SL structures in the HCV 3′ X-tail, these SL structures in the EHcV 3′ UTR were designated 3′SL I, 3′SL II, and 3′SL III, in that order from the 3′ terminus (Fig. 4C). Interestingly, the (A)-rich sequence was partially incorporated into the 3′SL III, although the poly(U/UC) stretch sequence in the HCV 3′ UTR is separated from any 3′SL structures (31–33). Furthermore, the two SL structures in the 3′ side of the EHcV NS5B-coding region were predicted to correspond to 5BSL3.2 and 5BSL3.3 in the NS5B-coding region of HCV. HCV 5BSL3.2 was previously shown to interact with 3′SL II to form the kissing-loop interaction, which is required for HCV replication (33). The secondary structure prediction shown in Fig. 4C suggests that the kissing-loop interaction may be conserved between 5BSL3.2 and the 3′SL II of the EHcV genome through their complementary sequences. The long-range RNA-RNA interaction between the apical loop of subdomain IIId in HCV IRES and the bulge of 5BSL3.2 supports IRES-dependent translation and viral RNA replication (34–36). In the case of the EHcV genome, the complement sequences were detected in the apical loops of subdomain and the 5BSL3.2-like subdomain (Fig. 4C), suggesting that the long-range RNA-RNA interaction may reside in the EHcV genome. These results indicated that HCV-like RNA secondary structures may be conserved around both UTRs of the EHcV genome.
Cleavage of the EHcV core protein by SPP.
The C-terminal transmembrane region of the HCV core protein was previously shown to be cleaved by SPP following the cleavage of the core-E1 junction by signal peptidase (11, 28, 37). The core protein is known to be released from the precursor polyprotein embedded in the endoplasmic reticulum (ER) membrane, and it then moves mainly to lipid droplets (LDs) (13, 14). Although SPP-dependent cleavage and LD translocation of the capsid protein are features common to HCV and GBV-B (13), it currently remains unknown whether the EHcV core protein shows these properties. The EHcV core protein shared 49.5% amino acid homology with the HCV core protein (genotype 1b) (Fig. 5A) and exhibited a hydrophobic/hydrophilic pattern similar to that of the HCV core protein (Fig. 5B). The EHcV core protein was predicted to be composed of domains 1, 2, and 3 relative to the HCV core protein. The transmembrane region of the EHcV core protein was predicted to span from Asn177 to Val199 by TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The transmembrane region of the EHcV core protein was 65% identical to that of the HCV core proteins (Fig. 5A). The C-terminal residue of the mature HCV core protein was found to be Phe177 in human and insect cell lines (17, 38). Our previous findings suggest that Ile176 and Phe177 of the HCV core protein may be responsible for SPP-dependent cleavage, because the replacement of Ile176 and Phe177 with Ala and Leu, respectively, abrogated intramembrane cleavage by SPP and impaired virus production (17, 28, 39). Weihofen et al. reported that SPP cleaved a peptide bond of the alpha-helix-breaking structure in a transmembrane region of the membrane protein (40). The replacement of Ile176 and Phe177 with Ala and Leu, respectively, in the HCV core protein converted the beta-sheet structure (alpha-helix-breaking structure) to an alpha-helix structure in the transmembrane region, as reported previously (28) (Fig. 6A and B). Ile190 and Phe191 of the EHcV core protein, which correspond to Ile176 and Phe177, respectively, of the HCV core protein, reside in the alpha-helix-breaking structure of the transmembrane region (Fig. 6A and B). In contrast, the replacement of Ile190 and Phe191 with Ala and Leu, respectively, in the EHcV core protein were predicted to convert the beta-sheet to an alpha-helix structure in a manner similar to that for the HCV core protein (Fig. 6A and B). To investigate the involvement of SPP in the maturation of the EHcV core protein, we expressed EHcVc or HCVc in 293FT cells with an SPP or SPP mutant. These core proteins were expected to be resistant to signal peptidase-dependent processing because the C-terminal residue Ala of both core proteins was replaced with Arg, resulting in the detection of an immature core protein by the anti-HA antibody (Fig. 6A) (28). The core proteins with molecular masses of 23 kDa and 28 kDa were detected mainly with the anti-FLAG antibody in 293FT cells expressing HCVc and HCVc-mt, respectively (Fig. 6C, lanes 2 and 3); however, the 23-kDa band was not detected with the anti-HA antibody (Fig. 6C, lane 2). When EHcVc was expressed in 293FT cells, it was detected at a molecular mass of 27 kDa with the anti-FLAG antibody, but not with the anti-HA antibody (Fig. 6C, lane 4). In contrast, EHcV-mt, in which the 190th and 191st residues were Ala and Leu instead of Ile and Phe, respectively, was detected mainly at a molecular mass of 30 kDa with the anti-FLAG and anti-HA antibodies (Fig. 6C, lane 5). A loss-of-function SPP mutant (SPP-D219A) in which the 219th residue was Ala instead of Asp was shown to have a dominantly negative effect on SPP-dependent cleavage of the HCV core protein and to be coprecipitated with the immature core protein (28). HCVc had a molecular mass of 23 kDa in the presence of wild-type SPP (SPP-wt) and was detected with the anti-FLAG antibody, but not with the anti-HA antibody (Fig. 6D, lane 2), suggesting that the 23-kDa protein band may be a mature core protein. HCVc mainly had a molecular mass of 28 kDa in the presence of SPP-D219A and was detected with the anti-FLAG and anti-HA antibodies (Fig. 6D, lane 3), which corresponds to the mobility of HCVc-mt (Fig. 6D, lane 4). These results suggest that SPP-D219A may abrogate the intramembrane cleavage of HCVc. In a manner similar to that for the HCV core protein, EHcVc was detected mainly at a molecular mass of 27 kDa in the presence of SPP-wt with the anti-FLAG antibody, but not with the anti-HA antibody (Fig. 6D, lane 6). EHcVc was detected mainly at a molecular mass of 30 kDa in the presence of SPP-D219A with anti-FLAG and anti-HA antibodies (Fig. 6D, lane 7), corresponding to the mobility of EHcVc-mt (Fig. 6D, lane 8). When SPP-D219A was coexpressed with either HCVc or EHcVc, immature HCVc and EHcVc were coprecipitated with SPP-D219A (Fig. 6E, lanes 3 and 5). These results suggest that the EHcV core protein may be cleaved by SPP and that Ile190 and Phe191 of the EHcV core protein are critical for SPP-dependent cleavage.
The intracellular localization of the hepacivirus core protein.
The HCV core protein is known to be localized mainly on the surface of LDs and is partially fractionated in the detergent-resistant membrane (DRM) close to the budding sites on the ER (13, 14, 16, 17). The core protein is considered to encompass the viral genome on the ER membrane, followed by budding into the lumen side (13, 14, 16, 17). To examine the intracellular localization of the EHcV core protein, we expressed HCVc or EHcVc in the Huh7OK1 cell line and stained the core proteins with the anti-FLAG antibody after staining LDs. Consistent with the findings of previous studies (14, 41, 42), HCVc was localized mainly on LDs (Fig. 7, row 3), whereas HCV-mt was not (Fig. 7, row 4). In a manner similar to that for the HCV core protein, EHcVc was localized mainly on LDs (Fig. 7, top row), whereas EHcVc-mt was not (Fig. 7, second row). These results suggest that the EHcV core protein may be localized mainly on LDs after SPP-dependent cleavage.
FIG 7.
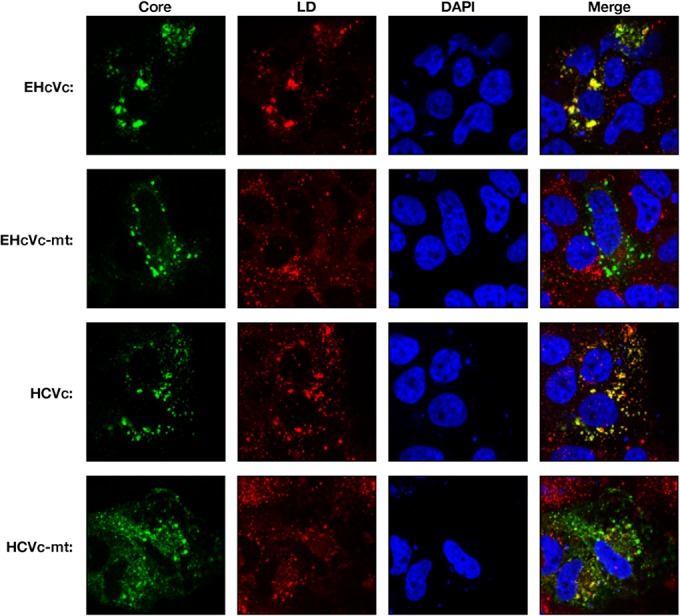
Intracellular localization of hepacivirus core proteins. HCVc, HCVc-mt, EHcVc, or EHcVc-mt was expressed in the Huh7OK1 cell line. The resulting cells were stained with Bodipy 558/568 (red) and then fixed with 4% paraformaldehyde at 24 h posttransfection, permeabilized, and subjected to indirect immunofluorescence staining. Each core protein was detected using mouse anti-FLAG antibodies and then Alexa 488-conjugated anti-mouse IgG (green). Cell nuclei were stained with DAPI after fixation (blue).
The DRM is defined as the cholesterol/sphingolipid-rich microdomain, which is resistant to nonionic detergents such as Triton X-100, considered to be a characteristic of lipid rafts. HCV was previously shown to be propagated in lipid raft-like compartments, including the membranous web (43–45). Furthermore, the HCV core protein is known to be associated with lipid raft-like compartments as well as LDs (16, 17, 41, 42). Therefore, we determined whether the EHcV core protein could be detected in the DRM fractions. EHcVc or EHcVc-mt was expressed in 293FT cells. The resulting cells were lysed on ice in the presence or absence of 1% Triton X-100. The DRM fractions were separated from the soluble proteins by a flotation assay with a stepwise density gradient in the presence or absence of Triton X-100. Serial fractions were collected after ultracentrifugation and were then subjected to Western blot analysis after being concentrated. EHcVc and EHcVc-mt were fractionated broadly from fractions of samples 3 to 11 without Triton X-100, and the fraction from sample 8 contained the largest amount of the core protein (Fig. 8, left panels). The distributions of the core proteins were roughly consistent with that of calreticulin, a marker protein of the ER membrane. When the cells expressing EHcVc were lysed in the presence of Triton X-100, a large amount of the core protein was localized in fractions 9 to 11 (Fig. 8, top three panels on the right). These fractions were enriched in calreticulin, corresponding to the detergent-soluble fractions (Fig. 8, fractions 7 to 11, top three panels on the right). However, EHcVc was partially detected in fractions 3 to 6 together with caveolin-1, a marker protein of the lipid raft (Fig. 8, fractions 3 to 6, top three panels on the right), suggesting that the EHcV core protein may have been partially distributed in the DRM fractions. In contrast, EHcVc-mt was localized in the detergent-soluble fractions (Fig. 8, fractions 9 to 11, bottom three panels on the right), but not in the DRM fractions (Fig. 8, fractions 3 to 6, bottom three panels on the right), in the presence of Triton X-100. EHcVc-mt was resistant to SPP-dependent processing, as described above (Fig. 6). These results suggest that the EHcV core protein may have been partially localized in the DRM and also that SPP-dependent processing may be required for DRM localization of the EHcV core protein.
FIG 8.
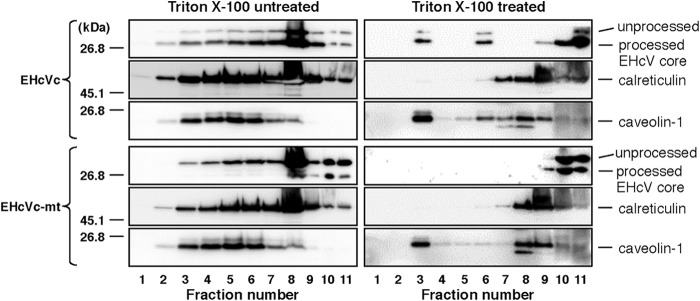
The EHcV core protein partially migrated to the DRM fraction after SPP-dependent processing. 293FT cells expressing either EHcVc or EHcVc-mt were homogenized with or without 1% Triton X-100 and then subjected to a flotation assay. Proteins in each fraction were concentrated with cold acetone and then subjected to Western blotting using the anti-FLAG, anti-calreticulin, or anti-caveolin-1 antibody.
DISCUSSION
The results of the present study indicate that EHcV infects Japanese-born domestic horses. Previous studies suggested that EHcV infected mainly horses and rarely dogs (5, 7–9). Our results demonstrate that EHcV commonly infects Japanese-born domestic horses (35.6% PCR positive and 22.6% seropositive). Several groups reported a prevalence of less than 10% PCR positivity in horses raised in the United States, the United Kingdom, and Germany (5, 7–9). Although the infection route of EHcV remains unknown, horses that were previously imported to Japan may be highly infected with EHcV. The serological prevalence in the present study appeared to be lower than that reported previously (8). A specific signal of the viral protein may be selected by Western blotting, used herein, rather than by the luciferase immunoprecipitation system, as reported previously (8), since the serum of each horse reacted to different proteins irrespective of the EHcV core protein (Fig. 2). The predicted full sequence of the EHcV strain amplified from serum sample 3 had high homology to those of the previously reported strains (Table 2). The polyproteins of previous strains had approximately 95% amino acid homology to one another irrespective of the area in which the horses originated, suggesting that these strains may belong to the same virological species. The parents of horse number 3 were born in Japan, while its grandparents were imported from the United States and Canada. Unfortunately, the sera of the parents and grandparents were not obtained in the present study. The EHcV strains obtained from Japanese-born horses may have originated from the United States or Canada. Another possibility is that one species of EHcV may have recently been distributed worldwide.
The primary and secondary structures of both UTRs are conserved among HCV strains and are essential for replication and translation. Four major stem-loop (SL) motifs have been detected in the 5′ UTR of the HCV genome, three SL structures of which are known to be required for IRES activity (46). Domain IIId plays a crucial role in anchoring of the 40S ribosome for IRES activity (47). Domain IIIb and the four-way helical junction of domains IIIa, IIIb, and IIIc bind eukaryotic initiation factor 3 (eIF3) and form a ternary complex, thereby forming the 48S preinitiation complex on HCV RNA (48). Moreover, domain II is known to be required to enhance eIF5-mediated GTP hydrolysis and the release of eIF2 from the 48S complex (48). These equivalent motifs were observed in the predicted secondary structures of the 5′ UTR of the reported EHcV strains (49), as well as in strain JPN3/JAPAN/2013 in the present study (Fig. 4C). A recent study demonstrated that the EHcV 5′ UTR exhibited IRES-dependent translation activity (50); however, further studies are needed to fully understand the IRES activity of the EHcV 5′UTR.
SL motifs embedded in the NS5B-coding region and UTRs of the HCV genome are known to be associated with viral replication. Several studies found that the mutational disruption of the complement sequence between 5BSL3.2 and 3′SL2 inhibited HCV RNA replication (33, 51). Additionally, the apical loop of domain IIId in the HCV 5′ UTR was shown to interact with the bulge of 5BSL3.2, supporting IRES-dependent translation and viral RNA replication (34–36). The RNA secondary structures of the 3′ UTR in the EHcV genome remained unknown due to limited information on its nucleotide sequence. 3′ RACE using poly(U) polymerase was employed in the present study because the ordinary 3′-RACE reaction using poly(A) polymerase was stopped at the (A)-rich region of the EHcV 3′ UTR. The nucleotide sequence of the EHcV 3′ UTR was determined, and its RNA secondary structure was then predicted (Fig. 4B and C). The results of the present study revealed that the 3′ UTR of the EHcV genome consists of the (A)-rich sequence and relatively shorter 3′-X-tail sequence. The three SL structures of the EHcV 3′ UTR were similar to those of the HCV 3′ UTR but were markedly different from the 3′ UTRs of GBV-B and rodent hepaciviruses. Drexler et al. described the structural characteristics of the 5′ and 3′ UTRs in rodent hepacivirus as well as phylogenetic information, liver tropism, and the pathogenicity of the virus (5). The SL motifs embedded in the 3′ X-tails of rodent hepacivirus and GBV-B varied. The structures of the 3′ UTRs appeared to correspond to the phylogenetic relationship of the hepaciviruses (5). Figure 4C shows that there were two stem-loop structures within the NS5B-coding region of EHcV corresponding to 5BSL3.2 and 5BSL3.3 of HCV RNA. Complementary regions were observed between the 5BSL3.2-like domain and 3′SL2, as well as between the 5BSL3.2-like domain and domain IIId of the EHcV genome (Fig. 4B and C). The kissing-loop and long-range RNA-RNA interactions may be structurally conserved between EHcV and HCV. Functional analyses of the cis-acting elements of the EHcV genome will contribute to the establishment of an EHcV infection system.
The mature HCV core protein was previously shown to be generated from the viral precursor polyprotein by signal peptidase followed by SPP-dependent processing of the transmembrane region (52). The core proteins of HCV and GBV-B are known to be cleaved by SPP (12, 37). The transmembrane regions of both the HCV and EHcV core proteins were found to be structurally conserved, based on their amino acid sequences and hydrophobicity plots (Fig. 5A and B and Fig. 6B). The replacement of Ile190 and Phe191 with Ala and Leu, respectively, in the EHcV core protein abrogated the intramembrane processing of the EHcV core protein (Fig. 6C). The loss-of-function mutant of SPP inhibited intramembrane processing of the EHcV core protein (Fig. 6D). Furthermore, the loss-of-function mutant of SPP specifically interacted with an uncleaved form of the EHcV core protein (Fig. 6E). These results indicate that the transmembrane region of the EHcV core protein may have been cleaved by SPP. The mature HCV core protein is known to be translocated into LDs and partially on lipid raft-like membranes. Previous studies reported that the HCV core protein on the LDs may be recruited near the replication complex in the membranous web, which consists of cholesterol- and sphingolipid-rich lipid components (43–45). Viral assembly was shown to occur on the ER membrane close to LDs and the membranous web (14). In addition to the HCV core protein, the nonstructural proteins and viral RNA of HCV were detected in the DRM fractions. The HCV RNA polymerase NS5B was previously reported to interact with sphingomyelin (53). Furthermore, a serine palmitoyltransferase inhibitor suppressed HCV replication by disrupting the replication complex (53, 54). These findings indicate that the DRM is provided as a scaffold for the formation of the HCV replication complex (45, 54). In the present study, we showed that the mature EHcV core protein was localized mainly on LDs and partially on the DRM (Fig. 7 and 8). A mutational analysis of the EHcV core protein indicated that SPP-dependent cleavage may be required for the localization of the EHcV core protein on LDs and the DRM in a manner similar to that for the HCV core protein. In addition, the assembly mechanism of EHcV may be similar to that of HCV.
In conclusion, the results of the present study show that EHcV shares common features with the HCV genomic structure and the biological properties of the capsid protein. In vivo and ex vivo infection systems for EHcV have not yet been successfully established. SCID mice carrying chimeric human livers are currently employed as a small animal model for in vivo infection with HCV (55) but are not suitable for studies on immunity and pathogenicity due to an immunodeficiency. Chimpanzees are not yet available for in vivo HCV research. Further studies on the mechanisms underlying EHcV infection will contribute to the development of an in vivo surrogate model system for studying HCV immunity and pathogenicity.
ACKNOWLEDGMENTS
We thank M. Furugori for her secretarial work, I. Katoh for helpful discussions, and C. Endoh for technical assistance.
This work was supported by Grants-in-Aid from the Ministry of Health, Labor, and Welfare, Japan (H24-Kanen-008 and H25-Kanen-002 and -008); the Ministry of Education, Culture, Sports, Science, and Technology, Japan; and the Japan Science and Technology Agency (JST) (Houga-24659204).
Footnotes
Published ahead of print 10 September 2014
REFERENCES
- 1.Simons JN, Leary TP, Dawson GJ, Pilot-Matias TJ, Muerhoff AS, Schlauder GG, Desai SM, Mushahwar IK. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1:564–569. 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 2.Beames B, Chavez D, Lanford RE. 2001. GB virus B as a model for hepatitis C virus. ILAR J. 42:152–160. 10.1093/ilar.42.2.152. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J, Apgar CL, Govindarajan S, Purcell RH. 2001. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 65:694–697. 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- 4.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 110:8194–8199. 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 9:e1003438. 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. 2013. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio 4(2):e00216–13. 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons S, Kapoor A, Sharp C, Schneider BS, Wolfe ND, Culshaw G, Corcoran B, McGorum BC, Simmonds P. 2012. Nonprimate hepaciviruses in domestic horses, United Kingdom. Emerg. Infect. Dis. 18:1976–1982. 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 86:6171–6178. 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. 2011. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 108:11608–11613. 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Laan LJ, de Ruiter PE, van Gils IM, Fieten H, Spee B, Pan Q, Rothuizen J, Penning LC. 5 June 2014. Canine hepacivirus and idiopathic hepatitis in dogs from a Dutch cohort. J. Viral Hepat. 10.1111/jvh.12268. [DOI] [PubMed] [Google Scholar]
- 11.Hüssy P, Langen H, Mous J, Jacobsen H. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93–104. 10.1006/viro.1996.0510. [DOI] [PubMed] [Google Scholar]
- 12.Targett-Adams P, Schaller T, Hope G, Lanford RE, Lemon SM, Martin A, McLauchlan J. 2006. Signal peptide peptidase cleavage of GB virus B core protein is required for productive infection in vivo. J. Biol. Chem. 281:29221–29227. 10.1074/jbc.M605373200. [DOI] [PubMed] [Google Scholar]
- 13.Hope RG, Murphy DJ, McLauchlan J. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261–4270. 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 14.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 15.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matto M, Rice CM, Aroeti B, Glenn JS. 2004. Hepatitis C virus core protein associates with detergent-resistant membranes distinct from classical plasma membrane rafts. J. Virol. 78:12047–12053. 10.1128/JVI.78.21.12047-12053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto K, Mori Y, Komoda Y, Okamoto T, Okochi M, Takeda M, Suzuki T, Moriishi K, Matsuura Y. 2008. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation. J. Virol. 82:8349–8361. 10.1128/JVI.00306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461. 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 19.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984. 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchuk D, Drumm M, Saulino A, Collins FS. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19:1154. 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajima S, Takasaki T, Matsuno S, Nakayama M, Kurane I. 2005. Genetic characterization of Yokose virus, a flavivirus isolated from the bat in Japan. Virology 332:38–44. 10.1016/j.virol.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 22.Tilgner M, Shi PY. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 78:8159–8171. 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnier J, Osguthorpe DJ, Robson B. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97–120. 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto K, Moriishi K, Miyamura T, Matsuura Y. 2004. Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein. J. Virol. 78:6370–6380. 10.1128/JVI.78.12.6370-6380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25:5015–5025. 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honda M, Brown EA, Lemon SM. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. U. S. A. 96:2291–2295. 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blight KJ, Rice CM. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392. 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lourenço S, Costa F, Debarges B, Andrieu T, Cahour A. 2008. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and trans-acting viral factors. FEBS J. 275:4179–4197. 10.1111/j.1742-4658.2008.06566.x. [DOI] [PubMed] [Google Scholar]
- 35.Cristina J, del Pilar Moreno M, Moratorio G. 2007. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Res. 127:185–194. 10.1016/j.virusres.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 36.Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. 2006. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 80:11579–11588. 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980–3988. 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino T, Fukuda H, Imajoh-Ohmi S, Kohara M, Nomoto A. 2004. Membrane binding properties and terminal residues of the mature hepatitis C virus capsid protein in insect cells. J. Virol. 78:11766–11777. 10.1128/JVI.78.21.11766-11777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kopp M, Murray CL, Jones CT, Rice CM. 2010. Genetic analysis of the carboxy-terminal region of the hepatitis C virus core protein. J. Virol. 84:1666–1673. 10.1128/JVI.02043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215–2218. 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 41.Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, Brechot C. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. U. S. A. 94:1200–1205. 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913–1925. [DOI] [PubMed] [Google Scholar]
- 43.Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480–3488. 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487–5492. 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160–4168. 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. 2009. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 37:1141–1151. 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9:370–374. 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 49.Locker N, Easton LE, Lukavsky PJ. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26:795–805. 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart H, Walter C, Jones D, Lyons S, Simmonds P, Harris M. 2013. The non-primate hepacivirus 5′ untranslated region possesses internal ribosomal entry site activity. J. Gen. Virol. 94:2657–2663. 10.1099/vir.0.055764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 82:9008–9022. 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. 2004. Structural biology of hepatitis C virus. Hepatology 39:5–19. 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 53.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 8:e1002860. 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katsume A, Tokunaga Y, Hirata Y, Munakata T, Saito M, Hayashi H, Okamoto K, Ohmori Y, Kusanagi I, Fujiwara S, Tsukuda T, Aoki Y, Klumpp K, Tsukiyama-Kohara K, El-Gohary A, Sudoh M, Kohara M. 2013. A serine palmitoyltransferase inhibitor blocks hepatitis C virus replication in human hepatocytes. Gastroenterology 145:865–873. 10.1053/j.gastro.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927–933. 10.1038/90968. [DOI] [PubMed] [Google Scholar]