ABSTRACT
Paramyxoviruses and other negative-strand RNA viruses encode matrix proteins that coordinate the virus assembly process. The matrix proteins link the viral glycoproteins and the viral ribonucleoproteins at virus assembly sites and often recruit host machinery that facilitates the budding process. Using a co-affinity purification strategy, we have identified the beta subunit of the AP-3 adapter protein complex, AP3B1, as a binding partner for the M proteins of the zoonotic paramyxoviruses Nipah virus and Hendra virus. Binding function was localized to the serine-rich and acidic Hinge domain of AP3B1, and a 29-amino-acid Hinge-derived polypeptide was sufficient for M protein binding in coimmunoprecipitation assays. Virus-like particle (VLP) production assays were used to assess the relationship between AP3B1 binding and M protein function. We found that for both Nipah virus and Hendra virus, M protein expression in the absence of any other viral proteins led to the efficient production of VLPs in transfected cells, and this VLP production was potently inhibited upon overexpression of short M-binding polypeptides derived from the Hinge region of AP3B1. Both human and bat (Pteropus alecto) AP3B1-derived polypeptides were highly effective at inhibiting the production of VLPs. VLP production was also impaired through small interfering RNA (siRNA)-mediated depletion of AP3B1 from cells. These findings suggest that AP-3-directed trafficking processes are important for henipavirus particle production and identify a new host protein-virus protein binding interface that could become a useful target in future efforts to develop small molecule inhibitors to combat paramyxoviral infections.
IMPORTANCE Henipaviruses cause deadly infections in humans, with a mortality rate of about 40%. Hendra virus outbreaks in Australia, all involving horses and some involving transmission to humans, have been a continuing problem. Nipah virus caused a large outbreak in Malaysia in 1998, killing 109 people, and smaller outbreaks have since occurred in Bangladesh and India. In this study, we have defined, for the first time, host factors that interact with henipavirus M proteins and contribute to viral particle assembly. We have also defined a new host protein-viral protein binding interface that can potentially be targeted for the inhibition of paramyxovirus infections.
INTRODUCTION
Hendra virus and Nipah virus are zoonotic paramyxoviruses belonging to the genus Henipavirus (1–3). Natural hosts for these viruses are pteropid fruit bats such as flying foxes, which suffer no apparent illness from the infections but act as reservoirs, allowing spillover transmissions that can be deadly to other animals and to people (4, 5). Hendra virus was first identified in Australia in 1994, after causing fatal infections in multiple horses and in one person who was exposed to an infected horse (6, 7). Numerous spillovers of Hendra virus to horses in Australia have occurred since that initial outbreak, and these have led to 7 human cases and 4 human fatalities to date (8, 9). Nipah virus was discovered after a Malaysian outbreak in 1998-1999, in which the virus was transmitted from bats to domesticated pigs. The virus circulated among the pigs and ultimately infected over 200 pig farmers, resulting in more than 100 fatalities (10). Like Hendra virus, Nipah virus has caused repeated spillovers in the years since its initial emergence, with many of the subsequent Nipah virus outbreaks occurring in Bangladesh and India (9).
Paramyxoviruses and other negative-strand RNA viruses encode matrix proteins that function to organize the assembly and release of virus particles (11). Once formed, the particles are membrane enveloped and covered with a layer of glycoprotein spikes, consisting of the viral attachment and fusion proteins. In addition, the particles contain negative-sense RNA genomes that are encapsidated by nucleocapsid proteins to form the viral ribonucleoprotein complexes (RNPs). During paramyxovirus assembly, the matrix (M) proteins accumulate at sites on cellular membranes from which the particles will bud and recruit other components to these locations, including the viral glycoproteins, the viral RNPs, and in many cases host budding machinery (12, 13).
The assembly and budding process that leads to the formation of enveloped virus particles can often be reconstituted in transfected cells, allowing the production of virus-like particles (VLPs), which resemble virions morphologically but lack viral genomes and many of the other viral components necessary for infectivity. For the paramyxoviruses, M protein expression in mammalian cells is necessary, and in many cases sufficient, to trigger the budding and release of VLPs with a size and shape that are consistent with authentic virions. For example, the M proteins of Sendai virus (14, 15), human parainfluenza virus type 1 (16), Newcastle disease virus (17), measles virus (18, 19), and Nipah virus (20–22) are sufficient to induce the formation and release of VLPs from transfected cells. In many cases, the viral glycoproteins and/or nucleocapsid proteins become incorporated into the VLPs if those proteins are coexpressed with M protein (13). In the cases of parainfluenza virus 5 (PIV5) (23) and mumps virus (24), efficient VLP release necessitates expression of viral glycoprotein and nucleocapsid protein, in addition to M protein.
Recruitment of host machinery via the matrix and Gag proteins of negative-strand RNA viruses and retroviruses is critical in many cases for proper formation and release of virus particles (25–28), yet for many paramyxoviruses, including the henipaviruses, M protein-host protein interactions remain largely unexplored. In this study, we identified the beta subunit of the AP-3 adapter protein complex, AP3B1, as a binding partner for the Nipah virus and Hendra virus M proteins. AP-3 complexes are known to play important roles during the trafficking of membrane proteins between various endosomal compartments within mammalian cells. Here, binding between viral M proteins and AP3B1 was mapped to the serine-rich and acidic Hinge domain of the AP3B1 protein. Budding of Nipah VLPs was significantly impaired upon small interfering RNA (siRNA)-mediated depletion of AP3B1 from cells. VLP budding could also be inhibited through expression of short M-binding polypeptides derived from the AP3B1 Hinge region. Our findings suggest that AP-3 directed trafficking processes are important during henipavirus particle formation and identify a new host-protein-virus protein binding interface that could prove useful as a target in future efforts aimed at developing therapeutics to treat these viral infections.
MATERIALS AND METHODS
Plasmids.
cDNA corresponding to the Nipah virus M protein was a kind gift from Paul Rota (Centers for Disease Control and Prevention, Atlanta, GA), and cDNA corresponding to the Hendra virus M protein was a kind gift from Chris Broder (Uniformed Services University, Bethesda, MD). These cDNAs were modified using PCR to encode tandem N-terminal Strep(II) and 6×His (Strep6His) tags (amino acid sequence, WSHPQFEKHHHHHH) or N-terminal Myc tags (amino acid sequence, EQKLISEEDL). The resulting cDNAs were subcloned into the eukaryotic expression vector pCAGGS (29) to generate pCAGGS-NiV M, pCAGGS-HeV M, pCAGGS-SH-NiV M, pCAGGS-SH-HeV M, pCAGGS-Myc-NiV M, and pCAGGS-Myc-HeV M. cDNA corresponding to Nipah virus M was also modified by PCR to encode an N-terminal Flag tag (amino acid sequence DYKDDDDK) and subcloned into the expression vector pcDNATM3.1/myc-His(-)A (Invitrogen, Carlsbad, CA) for use in fluorescence microscopy experiments (the pcDNA vector was used in this case because it results in a more moderate level of M protein expression, making M protein localization easier to define and visualize). cDNAs corresponding to the PIV5 and mumps virus M proteins, subcloned into pCAGGS vectors, have been described before (23, 24). cDNA corresponding to Sendai virus M protein (Z strain), subcloned into the pCAGGS vector, was a kind gift from Takemasa Sakaguchi.
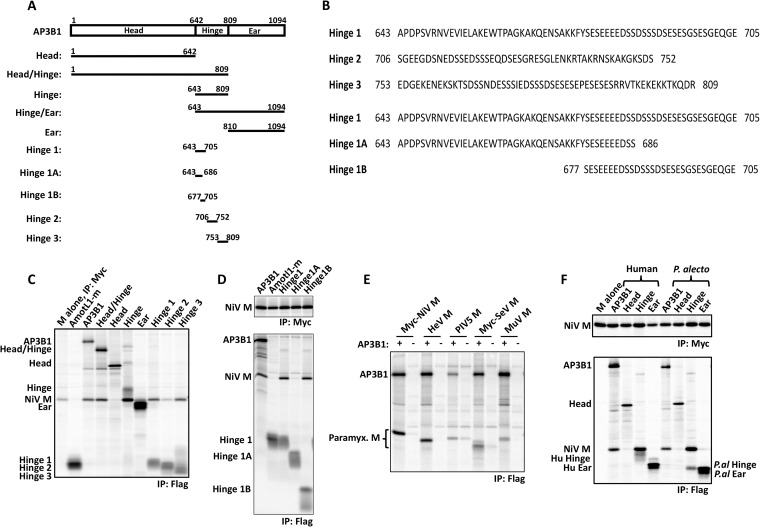
cDNA corresponding to full-length human AP3B1 was purchased from Open Biosystems (Thermo Fisher Scientific, Waltham, MA; clone ID 3914400). This sequence was modified using PCR to incorporate an N-terminal Flag tag and subcloned into the pCAGGS vector to generate plasmid pCAGGS-AP3B1. Subfragments of AP3B1, each with an N-terminal Flag tag, were generated by PCR using the full-length AP3B1 cDNA as the template and subcloned into pCAGGS, with boundaries as illustrated in Fig. 3. To obtain cDNA corresponding to full-length Pteropus alecto AP3B1, RNA was isolated from immortalized P. alecto kidney (PaKiT) cells, and cDNA was synthesized using the Superscript III first-strand synthesis system (Life Technologies, Grand Island, NY) according to the manufacturer's recommendations. The AP3B1 sequence was PCR amplified using primers designed based on the published Pteropus vampyrus sequence (30) (sense primer, 5′ ATGTCCAGTAACAGCTTCG 3′, and antisense primer, 5′ TTACCCCTGGGACAGGACAGG 3′). The sequence was modified to encode an N-terminal Flag tag and subcloned into the pCAGGS expression vector. Amino acid sequences of the human and P. alecto AP3B1 proteins were aligned using ClustalW2 (31) to define the Head, Hinge, and Ear domains. These subfragments of P. alecto AP3B1 were modified to encode N-terminal Flag tags and subcloned into the pCAGGS vector.
FIG 3.
Small AP3B1-derived polypeptides bind Nipah virus M protein. (A) Schematic representation of human AP3B1 and AP3B1-derived polypeptides. (B) Amino acid sequences of human AP3B1 Hinge-derived polypeptides. (C to F) 293T cells were transfected to produce Myc-tagged Nipah virus M protein (C, D, and F) or the indicated paramyxovirus M proteins (E) together with the indicated Flag-tagged AP3B1-derived polypeptides, and coimmunoprecipitation was carried out as described in the legend to Fig. 2. The bat (P. alecto)-derived AP3B1-derived polypeptides used for panel F are the equivalents of the corresponding human segments illustrated in panel A, based on ClustalW2 sequence alignment between the human and P. alecto AP3B1 proteins. The upper gels in panels D and F correspond to control immunoprecipitations of Myc-M protein using Myc antibody, while the lower gels correspond to coimmunoprecipitation experiments using Flag antibody.
The plasmid pCAGGS-AmotL1-m has been described previously (32). The SH-eGFP sequence was generated by PCR using the pEGFP-C1 vector (Clontech, Mountain View, CA) as the template and modified to append the N-terminal Strep6His tags. The resulting cDNA was subcloned into the pCAGGS vector to generate plasmid pCAGGS-SH-eGFP. cDNAs for host protein candidates EXOSC10 (clone ID 5505500), HERC5 (clone ID 9021584), HTATSF1 (clone ID 3504952), ILF2 (clone ID 2820505), NKRF (clone ID 5228666), RPS3 (clone ID 6160514), TCOF1 (clone ID 3616898), VPRBP (clone ID 4853730), YBX1 (clone ID 3914485), and ZC3HAV1 (clone ID 5418915) were all purchased from Open Biosystems (Thermo Fisher Scientific, Waltham, MA). Each cDNA was amplified and modified by PCR to append an N-terminal Flag tag and was subsequently subcloned into the pCAGGS vector.
Henipavirus M protein affinity purification and mass spectrometry.
For affinity purification of viral M proteins and M-interacting host factors, 293T cells (in groups of five 10-cm-diameter dishes) were transfected with pCAGGS plasmids corresponding to SH-M, SH-eGFP, or untagged M, at 3 μg/dish. At 24 h posttransfection (p.t.), cells were harvested and lysed in StrepTactin lysis buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40 [pH 8.0]). Cell lysates were clarified by centrifugation and further passed through 0.45-μm syringe filters to remove debris. For RNase-treated samples, clarified lysates were incubated with 200 μg/ml RNase A at room temperature for 30 min. RNase A-treated samples were clarified a second time by centrifugation before being passed through the syringe filter. Purification of M protein was done using the ÄKTAprime Plus fast protein liquid chromatography (FPLC) system (GE Life Sciences, Pittsburgh, PA) equipped with a 1-ml StrepTrap-HP column. Proteins were eluted from the column using a solution containing 100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 2.5 mM desthiobiotin, pH 8.0. Eluted proteins were concentrated using 500-3 U-Tube concentrators (Novagen, Madison, WI). Concentrated samples were resolved by SDS-PAGE using either 10% or 15% gels and stained with Coomassie brilliant blue (American Bioanalytical, Natick, MA) for gel excision. Additional SDS-PAGE gels run in parallel were stained with either Sypro orange or Lucy 506 (Bio-Rad Laboratories, Hercules, CA) for documentation of protein bands. Excised bands were submitted to the Taplin Mass Spectrometry Facility (Harvard Medical School, Boston, MA) for protein identification by liquid chromatography and tandem mass spectrometry (LC/LC-MS/MS).
Coimmunoprecipitation.
Coimmunoprecipitation of viral M proteins with AP3B1 and other host proteins was performed using modifications of methods that have been previously described (33). 293T cells grown in 6-cm-diameter dishes to 70 to 80% confluence in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), were transfected with pCAGGS plasmids encoding Myc-tagged viral M proteins (0.4 μg/dish) with or without pCAGGS plasmids encoding Flag-tagged candidate host proteins or AP3B1 derivatives (1.0 μg/dish). Cells were transfected with Lipofectamine-Plus reagents (Invitrogen, Carlsbad, CA) per the manufacturer's protocol. At 24 h p.t., cells were starved for 30 min in DMEM containing 2% FBS and 1/10 the normal amount of methionine and cysteine followed by labeling for 3 to 5 h in the same medium supplemented with 40 μCi of 35S-labeled Promix/ml (PerkinElmer, Waltham, MA). Cells were harvested and mixed with lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF] [pH 8.0]). The resulting cell lysates were clarified by centrifugation, followed by rocking for 2 h at 4°C in the presence of anti-Myc monoclonal antibody (Life Technologies, Grand Island, NY) or anti-Flag M2 magnetic beads (Sigma-Aldrich, St. Louis, MO). Immune complexes were collected by centrifugation after incubation with protein A Sepharose beads for 0.5 to 1 h and washed 3 times with lysis buffer. Proteins were separated by SDS-PAGE using 10%, 15%, or 17.5% gels and were detected using a Fuji FLA-7000 phosphorimager (Fujifilm Medical Systems, Stamford, CT).
Measurements of VLP production.
To generate VLPs, 293T cells grown in 6-cm dishes were transfected with pCAGGS plasmids encoding Myc-tagged Nipah virus M protein or Myc-tagged Hendra virus M protein (0.4 μg/dish), together with various plasmids encoding AP3B1-derived polypeptides (full-length AP3B1/Head/Hinge/Ear, 0.75 μg/dish; Hinge 1/Hinge 2/Hinge 3, 1 μg/dish; Hinge 1A/Hinge 1B, 1.5 μg/dish). Transfections were carried out in Opti-MEM using Lipofectamine-Plus reagents. At 24 h p.t., the culture medium was replaced with DMEM containing 2% FBS, 1/10 the normal amount of methionine and cysteine, and 40 μCi of 35S-labeled Promix/ml. After an additional 18 h, cell and medium fractions were collected. VLPs from the culture medium fractions were pelleted through 20% sucrose cushions, resuspended, floated to the tops of sucrose flotation gradients, pelleted again, and then resuspended in SDS-PAGE loading buffer containing 2.5% (wt/vol) dithiothreitol, as described previously (34). Cell lysate preparation and immunoprecipitation of proteins from the cell lysate fraction were carried out as described previously (23). Anti-Myc monoclonal antibody was used to immunoprecipitate viral M proteins, and anti-Flag M2 magnetic beads were used to immunoprecipitate AP3B1 and AP3B1-derived polypeptides. The precipitated proteins and VLPs were separated on 10% SDS gels and detected using a Fuji FLA-7000 phosphorimager. VLP production efficiency was calculated as the quantity of M protein in purified VLPs divided by the quantity of M protein in the corresponding cell lysate fraction, normalized to the value obtained in the positive-control experiment.
Membrane flotation assays to measure M protein membrane association.
293T cells in 10-cm dishes were transfected with pCAGGS plasmids encoding Nipah virus M protein (0.8 μg/dish) together with AP3B1-derived polypeptides (1.5 μg/dish). At 24 h p.t., cells were harvested, resuspended in 600 μl of hypotonic buffer (25 mM NaCl, 50 mM Na2HPO4 [pH 7.3], 1 mM phenylmethylsulfonyl fluoride), and incubated for 30 min at 4°C. The cells were subjected to 40 strokes of Dounce homogenization followed by microcentrifugation at 200 × g for 5 min to remove debris and nuclei. The resulting homogenates were mixed with 1.5 ml of 80% sucrose in NTE (0.1 M NaCl; 0.01 M Tris-HCl, pH 7.4; 1 mM EDTA). Layers of 50% sucrose (2.4 ml) and 10% sucrose (0.6 ml) in NTE were placed on top of the Dounce-homogenized mixtures, and samples were centrifuged at 160,000 × g for 4 h in a Sorvall AH650 swinging-bucket rotor. Six equal fractions were collected from the top of each gradient. Proteins from the gradient fractions were separated by SDS-PAGE using 10% gels and subjected to immunoblot analysis using a polyclonal antibody to Nipah virus M that has been described previously (24). Protein bands were detected and quantified using a Fuji FLA-7000 laser scanner. The fraction of membrane-bound M protein was calculated as the amount of M protein detected in the top three fractions of the gradient divided by the total amount of M protein detected in all six fractions.
RNA interference (RNAi).
Three 19-nucleotide siRNAs for human AP3B1 (SASI_Hs01_00018424, SASI_Hs01_00018425, and SASI_Hs01_00018426) were purchased from Sigma-Aldrich (St. Louis, MO), together with the universal negative-control siRNA. 293T cells in 6-cm dishes were cotransfected with pCAGGS plasmid encoding Myc-tagged Nipah virus M (0.4 μg per dish) and 100 nM siRNA (either the negative-control siRNA or a mixture of SASI_Hs01_00018424 (50 nM) plus SASI_Hs01_00018426 (50 nM). SASI_Hs01_00018424 targets the sequence CGAAUCUAGUUCAAUAGAA, and SASI_Hs01_00018426 targets the sequence GCAACAAAGAUUCUGCUAA. siRNA and plasmid cotransfection was performed using Lipofectamine-Plus reagents. VLPs were collected, and VLP production was calculated as described above. Additional transfections were carried out in parallel to measure the efficiency of AP3B1 depletion. 293T cells in 6-well plates were transfected with siRNA (100 nM) using Lipofectamine-Plus reagents as described above. At 24 h p.t., the culture medium was replaced with DMEM supplemented with 2% FBS. At 42 h p.t., cells were harvested and lysed in SDS-PAGE loading buffer containing 2.5% (wt/vol) dithiothreitol. Proteins were fractionated by SDS-PAGE using 10% gels, and the levels of endogenous AP3B1 protein were measured by immunoblot analysis using the AP3B1-specific polyclonal antibody AP3B1 13384–1-AP (Proteintech Group, Chicago, IL).
Immunofluorescence microscopy.
293T cells seeded on poly-d-lysine-coated glass coverslips and grown to 50% confluence were transfected with plasmid pcDNATM3.1-Flag-NiV M (50 ng/well) using Lipofectamine-Plus. At 24 h p.t., cells were washed three times with warm phosphate-buffered saline (PBS) for 10 min per wash, fixed with 4% paraformaldehyde in PBS for 15 min, and then washed three additional times. Cells were then permeabilized using 0.1% saponin, incubated in a blocking solution containing 1% bovine serum albumin (BSA) and 0.1% fish gelatin, and incubated with primary and secondary antibody solutions as described previously (35). Nipah virus M protein was visualized using anti-DDK monoclonal antibody specific to the Flag tag (Origene, Rockville, MD), and endogenous AP3B1 was visualized using anti-AP3B1 rabbit polyclonal antibody (Proteintech Group, Chicago IL). Secondary antibodies used were Alexa Fluor 594 goat anti-mouse IgG2a for detection of M protein and Alexa Fluor 488 goat anti-rabbit for detection of AP3B1 (Life Technologies). Washes were done after each antibody incubation, and cell nuclei were stained using ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Cells were visualized with a Zeiss AxioImager M1 fluorescence microscope (Carl Zeiss Inc., Thornwood, NY), and images were captured using an Orca R2 digital camera (Hamamatsu Photonics, Bridgewater, NJ). Images were deconvolved using iVision software (BioVision Technologies, Exton PA).
RESULTS
Identification of henipavirus M-associating host proteins by affinity purification and mass spectrometry.
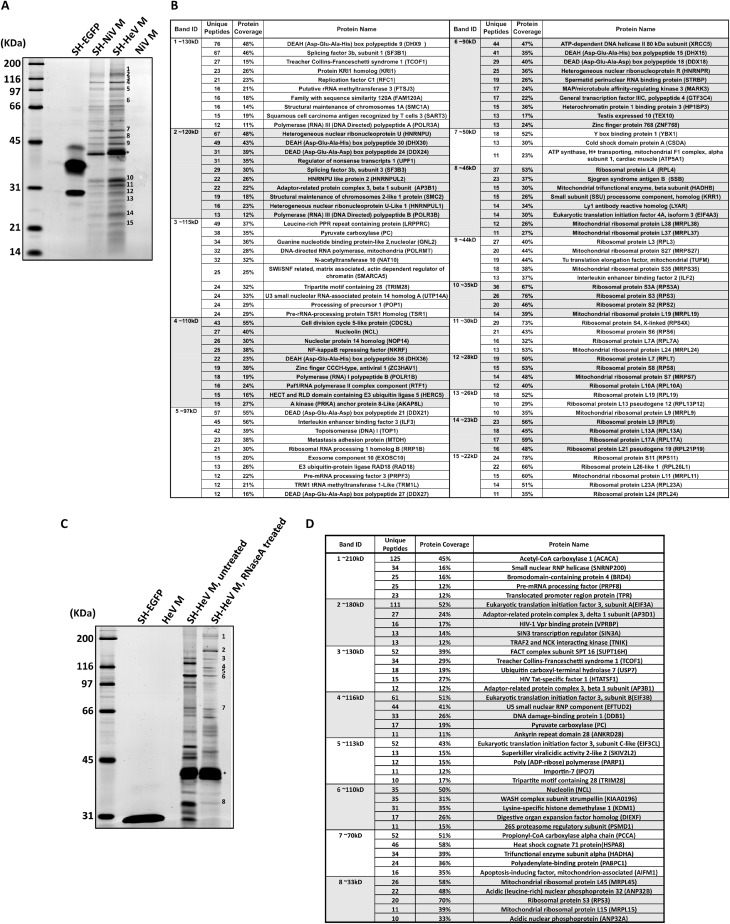
To define host factors involved in henipavirus particle formation, we affinity-purified Hendra virus M protein from the lysates of transfected cells and identified copurifying host factors using mass spectrometry. This approach employed a modified M protein, SH-HeV M, that harbors tandem N-terminal Strep(II) and 6×His tags. We found that placing these affinity tags at the N terminus of Hendra virus M protein had minimal effects on M protein stability and VLP production function, whereas addition of the same tags to the C-terminal end of M protein caused substantial stability and VLP production defects (data not shown). SH-HeV M protein was expressed in 293T cells using transient transfection, and M protein was affinity-purified from cell lysates by FPLC via the Strep(II) tag. SDS-PAGE and whole protein staining revealed a complex mixture of polypeptides which copurified with Hendra virus M protein (Fig. 1A). A parallel purification using the highly similar Nipah virus M protein (SH-NiV M) in place of the Hendra virus M protein led to a near-identical profile of copurifying polypeptides visualized by SDS-PAGE (Fig. 1A). These copurifying polypeptides were almost completely absent in control experiments using either tagged eGFP (SH-EGFP) or untagged Nipah virus M protein (Fig. 1A). Fifteen bands resulting from the Hendra virus M copurification procedure were selected for analysis. These were excised from the gel and subjected to in-gel trypsinization and multidimensional liquid chromatography and tandem mass spectrometry (LC/LC-MS/MS) for peptide identification. Results of this analysis revealed multiple distinct polypeptides associated with each of the excised bands (Fig. 1B). Several proteins involved in intracellular trafficking were identified (TCOF1, AP3B1, and EXOSC10), as well as some that have previously been linked to virus replication (ZC3HAV1, AP3B1, HERC5, and ILF2/3). More than half of the host proteins identified through this analysis are RNA-associated proteins, including RNA helicases, splicing factors, nuclear ribonucleoproteins, and ribosomal proteins.
FIG 1.
Identification of henipavirus M-associating host proteins by affinity purification and mass spectrometry. (A) 293T cells were transfected to express affinity-tagged henipavirus M proteins, untagged M protein, or affinity-tagged eGFP, as indicated. Cell lysates were prepared and subjected to FPLC using a StrepTrap-HP column. Eluted proteins were resolved by SDS-PAGE and visualized using Sypro orange. The asterisk denotes the migrations of SH.NiV M and SH.HeV M. Numbers indicate protein bands that were excised from a duplicate Coomassie blue-stained SDS gel for MS-based identification of polypeptides. (B) Proteins identified by MS from the 15 bands illustrated in panel A. Coverage indicates the percentage of amino acid residues within the protein that are present in at least one of the identified peptides. (C) Affinity-tagged Hendra virus M protein was purified from cell lysates using FPLC as in panel A, with an additional RNase A digestion step performed just prior to FPLC. (D) Proteins identified by MS from the 8 bands illustrated in panel C.
We speculated that the large proportion of RNA-associated proteins identified after M protein copurification might have been a consequence of an M protein interaction with cellular RNA. Under this scenario, a large number of proteins might copurify with M protein not because they interact with M protein itself but rather because they directly or indirectly interact with the cellular RNA that M protein has bound. Although the henipavirus M proteins have not previously been shown to bind RNA, matrix proteins from related viruses such as respiratory syncytial virus (36), Ebola virus (37), and influenza virus (38) have been shown to bind cellular RNA. We reasoned that treatment of cell lysates with RNase prior to the FPLC purification step would likely reduce or eliminate any potential RNA-directed interactions and might serve to facilitate the identification of proteins and/or protein complexes that bind directly to M protein. Indeed, RNase treatment markedly changed the profile of polypeptides copurifying with Hendra virus M protein as visualized by SDS-PAGE (Fig. 1C), and mass spectrometry analysis of excised bands revealed that only a minor fraction of these polypeptides correspond to RNA helicases, splicing factors, and other RNA-associated proteins (Fig. 1D).
AP3B1 interacts with henipavirus M proteins via its Hinge domain.
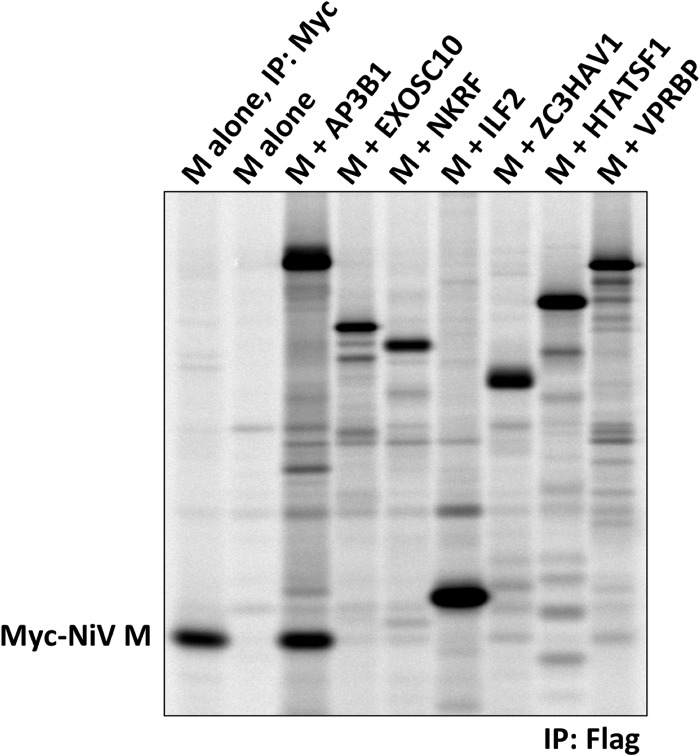
To further interrogate the abilities of Hendra virus M-copurifying host factors to interact with henipavirus M proteins, coimmunoprecipitation experiments were performed. Eleven candidate M-interacting host factors were selected for these experiments: AP3B1, EXOSC10, HERC5, HTATSF1, ILF2, NKRF, RPS3, TCOF1, VPRBP, YBX1, and ZC3HAV1. Several of these were selected because they are known to be involved in intracellular trafficking as noted above, and some were selected because they are known to be important for viral replication in other viral systems. cDNAs corresponding to these candidate proteins were obtained, and N-terminal Flag tags were appended. The Flag-tagged host proteins were expressed together with Myc-tagged Nipah virus M protein in transfected 293T cells. The host proteins were precipitated with Flag antibody, and coprecipitation of M protein was evaluated (Fig. 2 and data not shown). Strong coprecipitation of M protein was observed in the presence of AP3B1. This was the only candidate host factor for which substantial M protein coprecipitation was observed, although TCOF1, VPRBP, and ZC3HAV1 each led to a weak level of coprecipitation.
FIG 2.
Coimmunoprecipitation of Nipah virus M protein with candidate host proteins identified by MS. 293T cells were transfected to produce Myc-tagged Nipah virus M protein together with the indicated Flag-tagged candidate host factors. Proteins synthesized in the transfected cells were metabolically labeled, and cells were lysed in a solution containing 1% NP-40. Immunoprecipitation was carried out using Myc antibody (lane 1) or anti-Flag conjugated resin (lanes 2 to 9), and proteins were detected using a phosphorimager.
AP3B1 forms the beta subunit of tetrameric AP-3 adapter complexes, which act as cargo adapters during endosomal trafficking and sorting (39–41). Trafficking of HIV-1 Gag to multivesicular bodies is mediated in part by AP-3 complexes, and HIV-1 assembly is impaired in AP-3-deficient cells (42–44). Based on homology with the beta subunits of other adapter protein complexes, AP3B1 consists of three domains: an N-terminal Head domain that comprises approximately 60% of the protein, a C-terminal Ear domain (approximately 25% of the protein), and a serine-rich, acidic Hinge domain that separates the Head and Ear regions (Fig. 3A and B) (41, 45). To more clearly define the binding interface between AP3B1 and henipavirus M proteins, mapping studies were performed. A series of Flag-tagged human AP3B1 protein derivatives were constructed as illustrated in Fig. 3A, and these were used to coimmunoprecipitate Nipah virus M protein in transfected 293T cells (Fig. 3C and D). M-binding function was localized to the Hinge domain of the protein, as coimmunoprecipitation was observed with the full-length, Head/Hinge, and Hinge constructs but not with the Head or Ear constructs (Fig. 3C). AmotL1-m was used as a negative control in these experiments. This 83-amino-acid (aa) polypeptide binds to the M protein of another paramyxovirus, PIV5 (32), but fails to interact with henipavirus M proteins. To further localize the M-binding region within AP3B1, its Hinge domain was divided into three roughly equal segments, designated Hinge 1, Hinge 2, and Hinge 3 (Fig. 3A and B). Expression of the Hinge 1 segment led to M protein coimmunoprecipitation (Fig. 3C). Interestingly, expression of the Hinge 3 segment also led to a similar level of M protein coimmunoprecipitation. In contrast, Hinge 2 segment expression resulted in poor M protein coimmunoprecipitation. Both Hinge 1 and Hinge 3 are highly acidic and serine rich (Fig. 3B). The two sequences are nonoverlapping, but they both contain the 10-amino-acid sequence DSSSDSESES. The Hinge 1 sequence was further subdivided (Fig. 3A and B). Hinge 1A lacks DSSSDSESES and failed to bind M protein (Fig. 3D). The 29-amino-acid Hinge 1B contains DSSSDSESES and was sufficient for M protein binding (Fig. 3D). Thus, a short polypeptide derived from the Hinge region of AP3B1 was sufficient for interaction with Nipah virus M protein in coimmunoprecipitation assays.
The M proteins of other paramyxoviruses (Hendra virus, PIV5, Sendai virus, and mumps virus) were also tested for interaction with full-length AP3B1 in coimmunoprecipitation assays (Fig. 3E). We found the Hendra virus and Nipah virus M proteins to be virtually indistinguishable in these assays, as both exhibited highly efficient coprecipitation with AP3B1. The PIV5, Sendai virus, and mumps virus M proteins all coimmunoprecipitated with AP3B1 as well, but the efficiency of coimmunoprecipitation appeared to be less. Although it is difficult to assess relative binding affinities using this approach, our results suggest that a diverse group of paramyxovirus M proteins have the potential to interact with AP3B1.
Further binding experiments were performed using AP3B1 protein derived from the black flying fox, Pteropus alecto, which is a natural host reservoir species for both Nipah virus and Hendra virus. Flag-tagged versions of P. alecto AP3B1 were constructed (full-length, Head, Hinge, and Ear), and coimmunoprecipitation of Nipah virus M protein was evaluated (Fig. 3F). Strong coimmunoprecipitation of M protein with the full-length P. alecto AP3B1 was observed, similar to the result obtained with human AP3B1. The Hinge domain of P. alecto AP3B1 was able to coimmunoprecipitate M protein, but the Head and Ear domains failed to coimmunoprecipitate M protein, again consistent with results obtained using the human-derived AP3B1 constructs and suggesting that the human and P. alecto AP3B1 proteins are fundamentally similar to one another with respect to their interactions with henipavirus M proteins. Consistent with this idea, sequence comparisons revealed a high degree of conservation between the human and P. alecto AP3B1 proteins (90% amino acid identity overall, 27 out of 29 aa residues identical within the Hinge 1B region, and 10 out of 10 aa residues identical within the DSSSDSESES sequence).
Overexpression of M-binding, AP3B1-derived polypeptides blocks production of henipavirus VLPs.
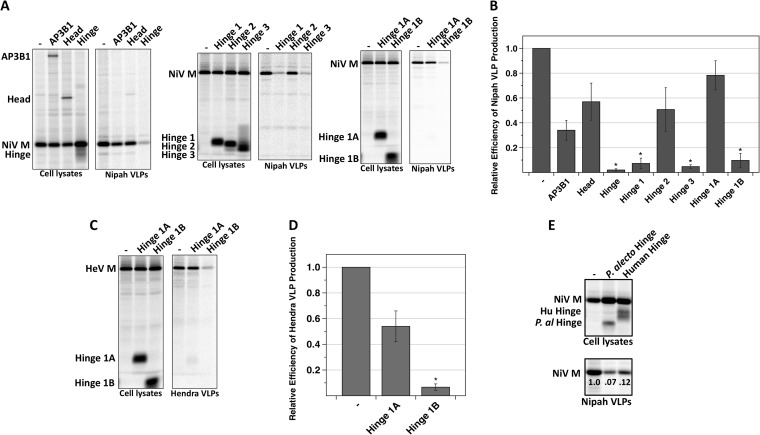
Short host factor-derived polypeptides that bind to viral Gag or M proteins can sometimes act as potent inhibitors of virus budding, either because they act as competitive inhibitors and prevent full-length endogenous host proteins from binding or because they otherwise interfere with budding function when they are bound to the viral proteins (32, 46–51). To test if AP3B1-derived polypeptides can inhibit henipavirus particle production, these polypeptides were expressed together with henipavirus M proteins in transfected 293T cells for production of VLPs. After metabolic labeling, VLPs were collected from the culture supernatants, pelleted through sucrose cushions, further purified by flotation on sucrose gradients, and analyzed on SDS gels (Fig. 4). We found that VLPs were abundantly produced when Nipah virus M protein was expressed in the absence of AP3B1-derived polypeptides (Fig. 4A), consistent with observations that have been reported before (20, 21). When the M-binding Hinge polypeptide was expressed together with M protein, VLP production was reduced more than 20-fold (Fig. 4A and B). Similar reductions in VLP production (between 10- and 20-fold) were observed upon expression of the M-binding polypeptides Hinge 1 and Hinge 3. Even the 29-amino-acid Hinge 1B polypeptide was inhibitory, resulting in VLP production that was 10-fold reduced from the normal level (Fig. 4A and B). In contrast, the AP3B1-derived polypeptides which did not bind to M protein in coimmunoprecipitation experiments (Head, Hinge 2, and Hinge 1A) affected VLP production less than 2-fold (Fig. 4A and B). This same pattern was also observed with Hendra VLP production. We found that expression of the Hendra virus M protein in the absence of any other viral proteins (and in the absence of any AP3B1-derived polypeptides) led to abundant VLP formation and release (Fig. 4C and D). Coexpression of the Hinge, Hinge 1, Hinge 3, and Hinge 1B polypeptides inhibited production of Hendra VLPs, while the Head, Hinge 2, and Hinge 1A polypeptides failed to inhibit (Fig. 4C and D and data not shown). Hence, Hendra virus was found to be quite similar to Nipah virus both in the requirements for VLP production (M protein alone is sufficient) and in the sensitivity to inhibition by AP3B1-derived polypeptides. Additional VLP release experiments were performed using the Nipah virus M protein expressed together with the bat (P. alecto) version of the AP3B1 Hinge domain (Fig. 4E). We found that expression of the P. alecto AP3B1 Hinge domain inhibited VLP production just as effectively as expression of the human AP3B1 Hinge domain. Overall, these experiments defined small, AP3B1-derived polypeptides that bind henipavirus M proteins and potently inhibit the production of henipavirus-like particles in transfected cells.
FIG 4.
Small AP3B1-derived polypeptides inhibit henipavirus VLP production. (A) 293T cells were transfected to produce Nipah virus M protein together with the indicated human AP3B1-derived polypeptides. After metabolic labeling of cells, lysates were prepared and M protein was immunoprecipitated using Myc antibody, while AP3B1-derived polypeptides were immunoprecipitated using anti-Flag-conjugated resin. VLPs from culture supernatants were purified by centrifugation through sucrose cushions followed by flotation on sucrose gradients. Purified VLPs were loaded directly onto SDS gels without immunoprecipitation, and proteins were visualized using a phosphorimager. (B) Three independent experiments were performed as described for panel A, and VLP production efficiencies were calculated as the amount of viral M protein detected in VLPs divided by the amount of M protein detected in the corresponding cell lysate fraction and were normalized to the values obtained in the absence of any polypeptide coexpression. Error bars indicate standard deviations. Differences from the values obtained in the absence of polypeptide coexpression were assessed for statistical significance by using a two-tailed Student t test, and P values of <0.01 are denoted by asterisks. (C) VLPs were produced as described for panel A but using Hendra virus M protein in place of Nipah virus M protein. (D) Efficiency of Hendra VLP production was calculated using data obtained from three independent experiments performed as for panel C. (E) Nipah VLPs were produced as described for panel A, with coexpression of either the human or the P. alecto version of the AP3B1 Hinge domain. Relative efficiencies of VLP production are indicated.
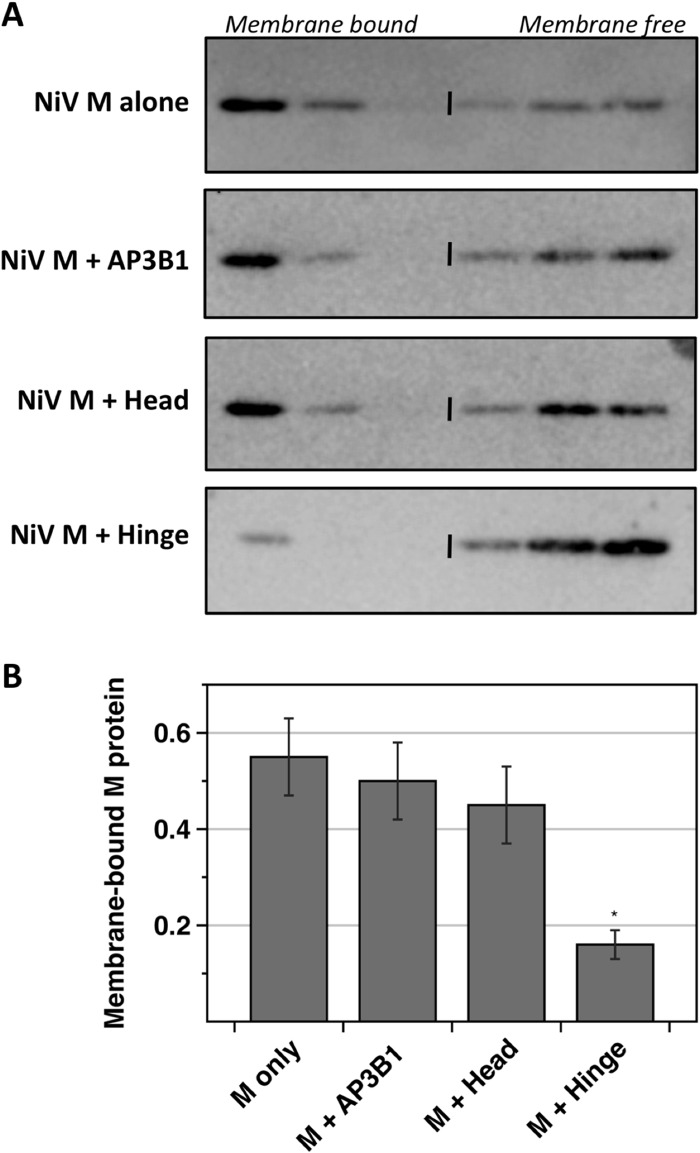
To gather insight into the mechanism of budding inhibition caused by AP3B1-derived polypeptides, the membrane-binding function of M protein was monitored. Detergent-free lysates were prepared from 293T cells transfected to express Nipah virus M protein together with various AP3B1-derived polypeptides, and membrane binding of M protein was assessed using sucrose flotation gradients (Fig. 5). Fifty to sixty percent of M protein was found in the membrane-bound (floated) fraction of the gradient when M was expressed alone, and this did not change significantly upon coexpression of either full-length AP3B1 or the AP3B1 Head polypeptide. However, coexpression of AP3B1 Hinge polypeptide reduced the fraction of membrane-bound M protein to less than 20% (Fig. 5). This result suggests a mechanism for inhibition in which M protein that is bound to AP3B1-derived polypeptides is subsequently unable to interact with cellular membranes as it normally would.
FIG 5.
AP3B1 Hinge polypeptide inhibits the membrane-binding ability of Nipah virus M protein. (A) 293T cells were transfected to produce Nipah virus M protein together with the indicated human AP3B1-derived polypeptides. Detergent-free cell lysates were prepared and overlaid with sucrose solutions to form flotation gradients. After ultracentrifugation, samples were collected from the tops of the gradients. Proteins from gradient fractions were resolved by SDS-PAGE, and Nipah virus M protein was detected by immunoblotting using M-specific polyclonal antibody. (B) Three independent experiments were performed as described for panel A. The percentage of M protein that was membrane bound (top three fractions of the gradients) was quantified using a phosphorimager. Results were plotted, with standard deviations indicated by error bars. Differences from the values obtained in the absence of polypeptide coexpression were assessed for statistical significance by using a two-tailed Student t test, and P values of <0.01 are denoted by asterisks.
Depletion of AP3B1 from cells impairs Nipah VLP production.
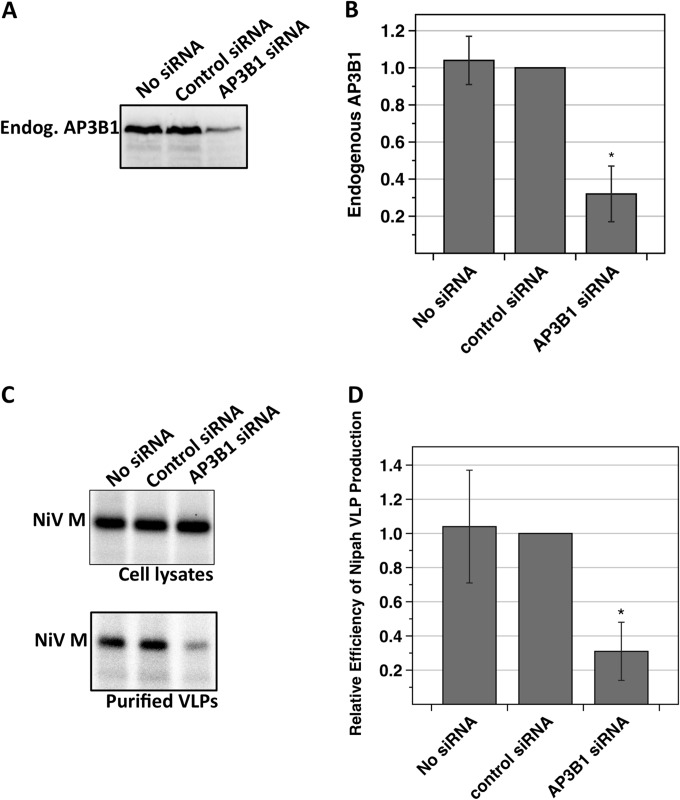
To investigate the importance of AP-3 complexes for Nipah virus M protein function and particle assembly, siRNAs were used to deplete AP3B1 protein from 293T cells. The efficiency of AP3B1 depletion was monitored in Western blot experiments (Fig. 6A and B). The quantity of endogenous AP3B1 was reduced to approximately 30% of its normal level in cells transfected with AP3B1-specific siRNAs. To measure the effect of AP3B1 depletion on Nipah VLP production, cells were simultaneously transfected with M-expressing plasmid and siRNA (Fig. 6C and D). VLP production was reduced 3-fold in AP3B1-depleted cells, relative to cells that had been transfected with a control siRNA. These results suggest that the ability of Nipah virus M protein to efficiently direct VLP formation is dependent upon the presence of physiological levels of AP3B1 within host cells.
FIG 6.
siRNA knockdown of endogenous human AP3B1 decreases Nipah VLP production. 293T cells were transfected with a plasmid encoding Nipah virus M protein together with 100 nM siRNA as indicated. (A and B) Endogenous AP3B1 was detected by immunoblotting using AP3B1-specific antibody, and protein bands were quantified using a laser scanner. Results from three independent experiments were plotted, with standard deviations indicated by error bars. (C and D) Nipah VLPs were purified and detected as described in the legend to Fig. 4. Results from three independent experiments were plotted, with standard deviations indicated by error bars. Differences from the values obtained in the presence of the control siRNA were assessed for statistical significance by using a two-tailed Student t test, and P values of <0.01 are denoted by asterisks.
Partial colocalization of Nipah virus M protein with endogenous AP3B1 in transfected cells.
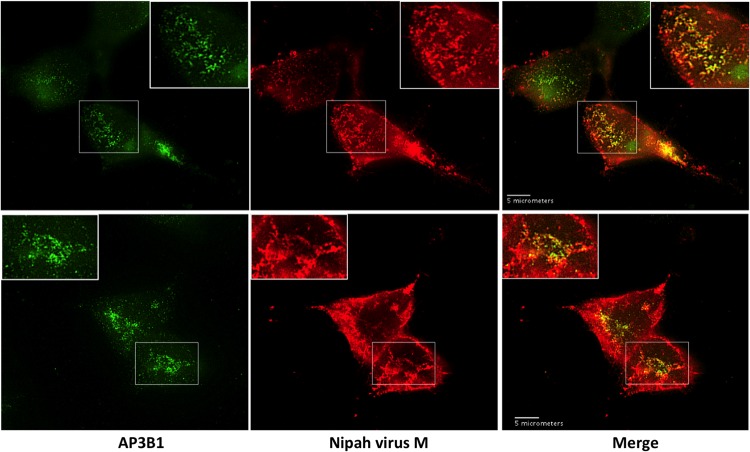
Further evidence for biological relevance of M protein interaction with AP3B1 was obtained by monitoring colocalization of these proteins in cells (Fig. 7). 293T cells were transfected to produce Nipah virus M protein, and colocalization between M protein and endogenous AP3B1 was observed by fluorescence microscopy. M protein was found mainly at the plasma membrane and in the cytoplasm (Fig. 7), and a smaller amount of M protein was often detected in the cell nucleus, consistent with previous observations (20, 52). The cytoplasmic M protein formed numerous small clusters, and many of these clusters exhibited substantial colocalization with endogenous AP3B1, indicating that a portion of M protein localizes to AP-3 trafficking compartments. It should be noted that this AP3B1-colocalized M protein represents only a minority of the total M protein in the cell, as some of the cytoplasmic M protein and virtually all of the plasma membrane-associated M protein are not AP3B1 colocalized. These observations are consistent with the possibility that M protein associates transiently with cellular compartments containing AP3B1 during its trafficking to the sites of virus assembly.
FIG 7.
Partial colocalization of Nipah virus M protein with endogenous AP3B1 in 293T cells. 293T cells on glass coverslips were transfected to produce Flag-tagged Nipah virus M protein, and subcellular localizations of M protein (red) and endogenous AP3B1 (green) were visualized by immunofluorescence microscopy at 24 h posttransfection. Two representative sets of images are shown in the top and bottom panels.
DISCUSSION
Here, we used a co-affinity purification approach to identify the beta subunit of the AP-3 adapter protein complex as a binding partner for the Nipah and Hendra virus M proteins. Binding to Nipah virus M protein was mapped to the serine-rich and acidic Hinge domain of AP3B1, and a 29-amino-acid polypeptide derived from the Hinge domain was found to be sufficient for M protein binding. Expression of AP3B1-derived, M-binding polypeptides prevented M protein association with cellular membranes and inhibited the budding of Nipah and Hendra VLPs, suggesting that the M protein-AP3B1 binding interface could prove useful as a target in future efforts to inhibit these viruses using small molecules. Significant colocalization between M protein and AP3B1 was observed in transfected mammalian cells, and siRNA-mediated depletion of AP3B1 impaired the production of Nipah VLPs, suggesting that the presence of functioning AP-3 complexes benefits the assembly and release of henipavirus particles.
AP-3 adapter protein complexes in mammalian cells direct the trafficking of membrane proteins from tubular sorting endosomes to late endosomes and lysosomes (40, 53, 54). A role for AP-3 complexes in enveloped virus assembly and release has already been well established in the case of HIV-1. HIV-1 Gag protein was shown to interact with the delta subunit of AP-3 (42), and siRNA-mediated depletion of AP-3 complexes impaired the assembly and release of HIV-1 particles (42). HIV-1 particle assembly was also impaired in cells derived from human patients with AP-3 deficiency, caused by mutations to AP3B1 (44). An N-terminal fragment of the AP-3 delta subunit (AP-3D-5′) was sufficient for binding to HIV-1 Gag, and this polypeptide could inhibit HIV-1 particle release and interfere with Gag protein trafficking to multivesicular bodies (42). Although interaction with AP-3 appears to be mediated through the MA component of Gag protein, as judged by GST pulldown assays (42), the molecular details of this interaction are not completely understood, and evidence for direct binding between MA and the protein interactive domain of the AP-3 delta subunit in vitro using NMR could not be obtained (55).
AP-3 interaction with henipavirus M proteins was mapped to the acidic, serine-rich Hinge domain of the AP-3 beta subunit, AP3B1. Interestingly, this AP3B1 Hinge region has been studied previously in the context of two different cellular binding partners: kinesin family member 3A (Kif3A) (45) and rabip4′ (56). The interaction between AP3B1 and Kif3A was identified in a yeast two-hybrid screen, using a Hinge-containing fragment of AP3B1 as bait (45). Here, the serine-rich Hinge region of AP3B1 had been characterized as a target for IP7-directed pyrophosphorylation, and the yeast two-hybrid investigation was carried out to determine if the pyrophosphorylation modification might impact protein-protein interactions. The screen identified Kif3A protein, and the Kif3A-AP3B1 interaction was confirmed in pulldown experiments (45). The interaction was found to be negatively regulated by AP3B1 pyrophosphorylation. In addition, the interaction was found to be important for HIV-1 particle release, as release of HIV-1 Gag VLPs could be inhibited either through siRNA depletion of Kif3A or by expression of a motorless Kif3A polypeptide that binds AP3B1 and presumably acts as a competitive inhibitor to block AP3B1 interaction with full-length, endogenous Kif3A (45). In a separate study, binding partners for the endosomal protein rabip4′ were isolated using an affinity pulldown procedure followed by mass spectrometry, and this identified an interaction between rabip4′ and AP3B1 (56). Mapping studies showed that the Hinge domain of AP3B1 bound to the FYVE domain of rabip4′, and knockdown studies suggested that AP-3:rabip4′ complexes are likely important for proper control of lysosome distribution within cells (56). Taken together with our results obtained with the henipavirus M proteins, these studies suggest that the AP3B1 Hinge domain is capable of directing protein interactions with a variety of partners. It will be important in future studies to determine whether these different viral and cellular proteins can bind simultaneously to AP3B1 or whether instead these proteins compete with one another for binding to the same site within the Hinge domain.
AP3B1-derived polypeptides were potent inhibitors of Nipah and Hendra VLP production. The shortest of these fragments, Hinge 1B, caused a 10-fold reduction in VLP production yet is only 29 amino acid residues in length. Consistent with these results, earlier studies found that short polypeptides derived from various host proteins that bind to viral Gag or M proteins are often potent inhibitors of virus budding. For example, expression of a Gag-binding fragment of Tsg101 (TSG-5′) caused a 60% reduction in HIV-1 particle production (46, 47), and expression of a Gag-binding fragment of Aip1/Alix caused a 5-fold reduction in HIV-1 particle production (48–51). Likewise, budding of the paramyxovirus PIV5 was reduced more than 3-fold upon expression of an M-binding polypeptide derived from the host protein AmotL1 (32). In the cases of Aip1/Alix and AmotL1, even overexpression of the full-length host proteins caused moderate negative effects on viral particle production (32, 48), drawing a further parallel with the results described here, in which full-length AP3B1 overexpression reduced Nipah VLP production to approximately 35% of its normal level. In this case, it is likely that the imbalance in AP-3 complex components caused by AP3B1 overexpression leads to some of the M protein binding to free AP3B1 that has not interacted with the other AP-3 components to form a functioning complex. Interestingly, although full-length AP3B1 overexpression moderately reduced VLP production, it did not have any noticeable effect on M protein membrane binding. Expression of AP3B1 Hinge polypeptide, in contrast, caused a more severe impairment in VLP production, along with significant impairment of M protein membrane binding. It is possible that Hinge and other AP3B1-derived polypeptides bind more strongly to M protein than full-length AP3B1 does and that the functional VLP assay is more sensitive at detecting these differences than the sucrose flotation-based membrane-binding assay. Another possibility is that the AP3B1-derived polypeptides and full-length AP3B1 could affect M protein in different ways, with the AP3B1-derived polypeptides disrupting the M protein conformation such that membrane-binding function is lost, while overexpressed full-length AP3B1 might cause moderate levels of inhibition merely by occupying M protein binding sites and preventing fully formed AP-3 complexes from interacting at these sites. Overall, the inhibition of M protein function by both AP3B1-derived polypeptides and overexpressed full-length AP3B1 supports the general concept that binding interfaces between host factors and budding-relevant viral proteins can be targeted for disruption as an effective antiviral approach (47, 57). Indeed, small-molecule budding inhibitors have recently been identified that target the PTAP-Tsg101 interface in the case of HIV-1 (58, 59) or the PPxY-Nedd4 interface in the cases of Ebola virus, Marburg virus, Lassa virus, and rabies virus (60). Similar approaches that target the AP3B1-M binding interface may prove useful in the identification of new therapeutics for paramyxovirus infections.
ACKNOWLEDGMENTS
We thank Chris Broder for helpful discussions and for providing Hendra virus cDNA reagents, and we are grateful to Paul Rota for providing Nipah virus M cDNA.
This work was supported in part by the Middle Atlantic Regional Center of Excellence (MARCE) for Biodefense and Emerging Infectious Disease Research NIH grant AI057168 to A.P.S.
Footnotes
Published ahead of print 10 September 2014
REFERENCES
- 1.Eaton BT, Broder CC, Middleton D, Wang LF. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 4:23–35. 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop KA, Broder CC. 2008. Hendra and Nipah viruses: lethal zoonotic paramyxoviruses, p 155–187 In Scheld WM, Hammer SM, Hughes JM. (ed), Emerging infections. ASM Press, Washington, DC. [Google Scholar]
- 3.Ksiazek TG, Rota PA, Rollin PE. 2011. A review of Nipah and Hendra viruses with an historical aside. Virus Res. 162:173–183. 10.1016/j.virusres.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 4.Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, Daszak P, Henipavirus Ecology Research Group 2011. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 85:946–951. 10.4269/ajtmh.2011.10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton BA, Wang LF, Marsh GA. 2013. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Health 60:69–83. 10.1111/j.1863-2378.2012.01501.x. [DOI] [PubMed] [Google Scholar]
- 6.O'Sullivan JD, Allworth AM, Paterson DL, Snow TM, Boots R, Gleeson LJ, Gould AR, Hyatt AD, Bradfield J. 1997. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet 349:93–95. 10.1016/S0140-6736(96)06162-4. [DOI] [PubMed] [Google Scholar]
- 7.Wong KT, Robertson T, Ong BB, Chong JW, Yaiw KC, Wang LF, Ansford AJ, Tannenberg A. 2009. Human Hendra virus infection causes acute and relapsing encephalitis. Neuropathol. Appl. Neurobiol. 35:296–305. 10.1111/j.1365-2990.2008.00991.x. [DOI] [PubMed] [Google Scholar]
- 8.Playford EG, McCall B, Smith G, Slinko V, Allen G, Smith I, Moore F, Taylor C, Kung YH, Field H. 2010. Human Hendra virus encephalitis associated with equine outbreak, Australia, 2008. Emerg. Infect. Dis. 16:219–223. 10.3201/eid1602.090552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aljofan M. 2013. Hendra and Nipah infection: emerging paramyxoviruses. Virus Res. 177:119–126. 10.1016/j.virusres.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435. 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 11.Liljeroos L, Butcher SJ. 2013. Matrix proteins as centralized organizers of negative-sense RNA virions. Front. Biosci. (Landmark Ed.) 18:696–715. 10.2741/4132. [DOI] [PubMed] [Google Scholar]
- 12.Takimoto T, Portner A. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133–145. 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int. J. Biochem. Cell Biol. 42:1416–1429. 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384–11391. 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10. 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 16.Coronel EC, Murti KG, Takimoto T, Portner A. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantua HD, McGinnes LW, Peeples ME, Morrison TG. 2006. Requirements for the assembly and release of Newcastle disease virus-like particles. J. Virol. 80:11062–11073. 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. 2007. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J. Gen. Virol. 88:1243–1250. 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- 19.Runkler N, Pohl C, Schneider-Schaulies S, Klenk HD, Maisner A. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell. Microbiol. 9:1203–1214. 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 20.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J. Virol. 80:12070–12078. 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patch JR, Crameri G, Wang LF, Eaton BT, Broder CC. 2007. Quantitative analysis of Nipah virus proteins released as virus-like particles reveals central role for the matrix protein. Virol. J. 4:1. 10.1186/1743-422X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patch JR, Han Z, McCarthy SE, Yan L, Wang LF, Harty RN, Broder CC. 2008. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol. J. 5:137. 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt AP, Leser GP, Waning DL, Lamb RA. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952–3964. 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. 2009. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J. Virol. 83:7261–7272. 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrison MS, Sakaguchi T, Schmitt AP. 2011. Paramyxovirus budding mechanisms, p 193–218 In Luo M. (ed), Negative strand RNA virus. World Scientific, Hackensack, NJ. [Google Scholar]
- 26.Lyles DS. 2013. Assembly and budding of negative-strand RNA viruses. Adv. Virus Res. 85:57–90. 10.1016/B978-0-12-408116-1.00003-3. [DOI] [PubMed] [Google Scholar]
- 27.Rossman JS, Lamb RA. 2013. Viral membrane scission. Annu. Rev. Cell Dev. Biol. 29:551–569. 10.1146/annurev-cellbio-101011-155838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 30.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt S, Johnson N, Juettemann T, Kahari AK, Keenan S, Kulesha E, Martin FJ, Maurel T, McLaren WM, Murphy DN, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ruffier M, Sheppard D, Taylor K, Thormann A, Trevanion SJ, Vullo A, Wilder SP, Wilson M, Zadissa A, Aken BL, Birney E, Cunningham F, Harrow J, Herrero J, Hubbard TJ, Kinsella R, Muffato M, Parker A, Spudich G, Yates A, Zerbino DR, Searle SM. 2014. Ensembl 2014. Nucleic Acids Res. 42:D749–D755. 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Pei Z, Bai Y, Schmitt AP. 2010. PIV5 M protein interaction with host protein angiomotin-like 1. Virology 397:155–166. 10.1016/j.virol.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei Z, Harrison MS, Schmitt AP. 2011. Parainfluenza virus 5 m protein interaction with host protein 14-3-3 negatively affects virus particle formation. J. Virol. 85:2050–2059. 10.1128/JVI.02111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt PT, Ray G, Schmitt AP. 2010. The C-terminal end of parainfluenza virus 5 NP protein is important for virus-like particle production and M-NP protein interaction. J. Virol. 84:12810–12823. 10.1128/JVI.01885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison MS, Schmitt PT, Pei Z, Schmitt AP. 2012. Role of ubiquitin in parainfluenza virus 5 particle formation. J. Virol. 86:3474–3485. 10.1128/JVI.06021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez L, Cuesta I, Asenjo A, Villanueva N. 2004. Human respiratory syncytial virus matrix protein is an RNA-binding protein: binding properties, location and identity of the RNA contact residues. J. Gen. Virol. 85:709–719. 10.1099/vir.0.19707-0. [DOI] [PubMed] [Google Scholar]
- 37.Gomis-Ruth FX, Dessen A, Timmins J, Bracher A, Kolesnikowa L, Becker S, Klenk HD, Weissenhorn W. 2003. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure 11:423–433. 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakefield L, Brownlee GG. 1989. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 17:8569–8580. 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newell-Litwa K, Seong E, Burmeister M, Faundez V. 2007. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J. Cell Sci. 120:531–541. 10.1242/jcs.03365. [DOI] [PubMed] [Google Scholar]
- 40.Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. 2004. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164:1065–1076. 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dell'Angelica EC, Ooi CE, Bonifacino JS. 1997. Beta3A-adaptin, a subunit of the adaptor-like complex AP-3. J. Biol. Chem. 272:15078–15084. 10.1074/jbc.272.24.15078. [DOI] [PubMed] [Google Scholar]
- 42.Dong X, Li H, Derdowski A, Ding L, Burnett A, Chen X, Peters TR, Dermody TS, Woodruff E, Wang JJ, Spearman P. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120:663–674. 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 43.Garcia E, Nikolic DS, Piguet V. 2008. HIV-1 replication in dendritic cells occurs through a tetraspanin-containing compartment enriched in AP-3. Traffic 9:200–214. 10.1111/j.1600-0854.2007.00678.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Sutton J, Woodruff E, Villalta F, Spearman P, Dong X. 2012. Defective HIV-1 particle assembly in AP-3-deficient cells derived from patients with Hermansky-Pudlak syndrome type 2. J. Virol. 86:11242–11253. 10.1128/JVI.00544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. 2009. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U. S. A. 106:21161–21166. 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demirov DG, Ono A, Orenstein JM, Freed EO. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. U. S. A. 99:955–960. 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed EO. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56–59. 10.1016/S0966-842X(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. 2005. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J. Biol. Chem. 280:40474–40480. 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- 49.Fujii K, Hurley JH, Freed EO. 2007. Beyond Tsg101: the role of Alix in ‘ESCRTing' HIV-1. Nat. Rev. Microbiol. 5:912–916. 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- 50.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414–12419. 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 282:3847–3855. 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 52.Wang YE, Park A, Lake M, Pentecost M, Torres B, Yun TE, Wolf MC, Holbrook MR, Freiberg AN, Lee B. 2010. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 6:e1001186. 10.1371/journal.ppat.1001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MS, Bonifacino JS. 2001. Adaptor-related proteins. Curr. Opin. Cell Biol. 13:444–453. 10.1016/S0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 54.Dell'Angelica EC. 2009. AP-3-dependent trafficking and disease: the first decade. Curr. Opin. Cell Biol. 21:552–559. 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 55.Kyere SK, Mercredi PY, Dong X, Spearman P, Summers MF. 2012. The HIV-1 matrix protein does not interact directly with the protein interactive domain of AP-3delta. Virus Res. 169:411–414. 10.1016/j.virusres.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivan V, Martinez-Sanchez E, Sima LE, Oorschot V, Klumperman J, Petrescu SM, van der Sluijs P. 2012. AP-3 and Rabip4′ coordinately regulate spatial distribution of lysosomes. PLoS One 7:e48142. 10.1371/journal.pone.0048142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harty RN. 2009. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res. 81:189–197. 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. 2008. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem. Biol. 3:757–764. 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 59.Waheed AA, Freed EO. 2008. Peptide inhibitors of HIV-1 egress. ACS Chem. Biol. 3:745–747. 10.1021/cb800296j. [DOI] [PubMed] [Google Scholar]
- 60.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. 2014. Small molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J. Virol. 88:7294–7306. 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]