ABSTRACT
The family Marseilleviridae consists of Acanthamoeba-infecting large DNA viruses with icosahedral particles ∼0.2 μm in diameter and genome sizes in the 346- to 380-kb range. Since the isolation of Marseillevirus from a cooling tower in Paris (France) in 2009, the family Marseilleviridae has expanded rapidly, with representatives from Europe and Africa. Five members have been fully sequenced that are distributed among 3 emerging Marseilleviridae lineages. One comprises Marseillevirus and Cannes 8 virus, another one includes Insectomime virus and Tunisvirus, and the third one corresponds to the more distant Lausannevirus. We now report the genomic characterization of Melbournevirus, the first representative of the Marseilleviridae isolated from a freshwater pond in Melbourne, Australia. Despite the large distance separating this sampling point from France, Melbournevirus is remarkably similar to Cannes 8 virus and Marseillevirus, with most orthologous genes exhibiting more than 98% identical nucleotide sequences. We took advantage of this optimal evolutionary distance to evaluate the selection pressure, expressed as the ratio of nonsynonymous to synonymous mutations for various categories of genes. This ratio was found to be less than 1 for all of them, including those shared solely by the closest Melbournevirus and Cannes 8 virus isolates and absent from Lausannevirus. This suggests that most of the 403 protein-coding genes composing the large Melbournevirus genome are under negative/purifying selection and must thus significantly contribute to virus fitness. This conclusion contrasts with the more common view that many of the genes of the usually more diverse large DNA viruses might be (almost) dispensable.
IMPORTANCE A pervasive view is that viruses are fast-evolving parasites and carry the smallest possible amount of genomic information required to highjack the host cell machinery and perform their replication. This notion, probably inherited from the study of RNA viruses, is being gradually undermined by the discovery of DNA viruses with increasingly large gene content. These viruses also encode a variety of DNA repair functions, presumably slowing down their evolution by preserving their genomes from random alterations. On the other hand, these viruses also encode a majority of proteins without cellular homologs, including many shared only between the closest members of the same family. One may thus question the actual contribution of these anonymous and/or quasi-orphan genes to virus fitness. Genomic comparisons of Marseilleviridae, including a new Marseillevirus isolated in Australia, demonstrate that most of their genes, irrespective of their functions and conservation across families, are evolving under negative selection.
INTRODUCTION
The existence of “giant” viruses, simply defined as those with particles large enough to be easily seen under a light microscope (i.e., >0.3 μm in diameter), was revealed by the discovery of Mimivirus in 2003 in an Acanthamoeba organism that was originally thought to be infected by an obligate intracellular bacterium (1). Further characterizations showed that the large Mimivirus icosahedral particle (0.7 μm in diameter) enclosed a DNA genome larger than a megabase and a number of genes comparable to that of many bacteria (2–5). As Mimivirus infects its host by mimicking the bacteria that constitute their normal food, it was then postulated that more giant viruses could be found using Acanthamoeba as bait to explore a variety of environments. This approach was very successful and led to the isolation of many relatives of Mimivirus, now constituting the rapidly expanding family Mimiviridae (6–8), which also includes viruses infecting marine phagocytic and mixotrophic unicellular protists (9, 10). In parallel, this search led to the discovery of 3 additional types of acanthamoeba-infecting viruses unrelated to the Mimiviridae: the two pandoraviruses (Pandoravirus dulcis and Pandoravirus salinus) (11), Pithovirus sibericum (12), and several members of the proposed family Marseilleviridae, whose founder is Marseillevirus (13). While the pandoraviruses and pithoviruses exhibit amphora-shaped particles of impressive dimensions (0.5 μm wide and >1 μm in length), members of the Marseilleviridae possess more typical icosahedral particles, 190 to 250 nm in diameter, putting them at the frontier between the previously largest known DNA viruses, such as chloroviruses (14), and the newly defined giant viruses. The genome sizes (347 to 383 kb) of Marseilleviridae are also comparable to those of other large microalgal viruses, such as the coccolithoviruses (∼410 kb) (15) and chloroviruses (∼350 kb) (16).
The genomes of 5 different members of the family Marseilleviridae have been fully sequenced, and many others have been partially characterized (17). The phylogenetic analysis of these viruses, which have been isolated in a variety of locations, suggested the existence of 3 subgroups (Fig. 1). Lineage A is centered on the prototype Marseillevirus and includes the 2 other fully sequenced viruses, Cannes 8 virus (18) and Senegalvirus (19), lineage B is represented by Lausannevirus (20) alone, and lineage C includes Tunisvirus (21) and Insectomime virus (22).
FIG 1.
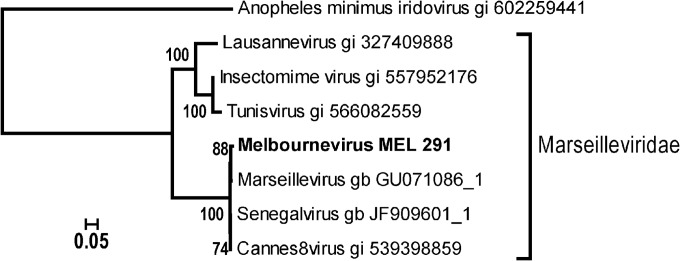
Clade structure of the Marseilleviridae. Neighbor-joining clustering was computed from 986 ungapped sites of a multiple alignment of 8 DNA polymerase catalytic-subunit amino acid sequences. The alignment and tree were computed using the Computational Biology Research Center (CBRC) server (http://www.cbrc.jp) using the Whelan and Goldman substitution model (estimated α = 1.9; 100 bootstrap resampling). The Anopheles minimus iridovirus ortholog is the best-matching homolog among viruses and is used as an outgroup. The known Marseilleviridae representatives appear to be distributed among 3 emerging subclades. The Marseillevirus subclade encompasses viruses exhibiting nearly identical genome sequences, although they were independently isolated from geographically diverse locations.
All viruses cited above belong to class I in the Baltimore classification (23), the class grouping all the viruses with a double-stranded DNA genome. This class includes the largest number of different families and the largest number of viral taxons for which at least one genome has been fully sequenced, close to a thousand (24). Despite sharing the same global scheme of genome expression, replication, and propagation, these class I viruses exhibit the widest distribution of genome sizes and number of genes among all “homogeneous” biological entities. A factor of 500 separates the gene content of the recently described Pandoravirus salinus (with a 2.77-Mbp genome encoding more than 2,500 proteins) (11) from that of the tiny polyomaviruses (with genome sizes of about 5 kb, encoding five proteins). This raises the question of the evolutionary processes able to generate and tolerate such a huge variation in genome complexity for biological entities exhibiting the same overall replication and particle-based propagation strategy. In other words, what is the need for a virus to possess thousand genes, if a handful is sufficient to ensure similar reproductive success? Moreover, within each virus family, the number of genes shared by all the members (the core genome) is often a small fraction of the total number of genes predicted in all individual genomes (<1/3 for the Marseilleviridae [21]). This flexibility in gene content also observed in other families of viruses infecting unicellular hosts (16, 25) might be taken as suggesting that many of these genes are simple genomic hitchhikers without much influence on virus fitness.
In this article, we report the complete genome analysis of the first representative of the Marseilleviridae isolated in Australia, which we named Melbournevirus. This is the first virus of this family isolated in our laboratory, while all previously described members of the Marseilleviridae have been isolated in Europe or Africa and characterized in two different laboratories (13, 20). Unexpectedly, Melbournevirus exhibits a genome sequence that is approximately 98% identical on average to that of Marseillevirus (isolated in Paris) and Cannes 8 virus (isolated on the French Riviera). We took advantage of this remarkable feature, rarely encountered for viruses independently isolated from remote locations, to compute the selection pressure exerted on genes displaying different conservation pattern within the Marseilleviridae. Contrary to the above hypothesis that many of the genes comprising their large and variable genomes might be dispensable, our results indicate that most of them seem to significantly contribute to the viral fitness, despite the variable conservation patterns they exhibit among the Marseilleviridae lineages or among other large DNA viruses.
MATERIALS AND METHODS
Virus isolation.
Melbournevirus was isolated from the same sample as Pandoravirus dulcis (11). This sample consisted of muddy fresh water collected in a pond near Melbourne, Australia (37°43′09.6″ S, 145°03′06.9″ E). After the mud and the water had been mixed, 30 ml was recovered from the bottle. Pure amphotericin B (Fungizone) (3 ml; 25-μg/ml final concentration) was added to the sample, which was vortexed and incubated overnight at 4°C on a stirring wheel. After filtration through a 20-μm sieve to remove microorganisms and debris, the filtrate was recovered and centrifuged at 500 × g for 5 min. A 100-μl portion of the supernatant was used to infect 1-ml cultures of our A. castellanii laboratory strain in a P12 plate at a confluence of 20,000 cells/cm2. To limit the contamination of the cultures by bacteria and fungi, these Acanthamoeba cells were initially adapted to increasing concentrations of amphotericin B up to 2.5 μg/ml. The cells were grown in PPYG medium [2% proteose peptone, 0.1% yeast extract, 2.5 mM KH2PO4, 2.5 mM Na2HPO4, 0.4 mM CaCl2, 4 mM MgSO4 · 7H2O, 50 μM Fe(NH4)2(SO4)2, 100 mM glucose (pH 6.5)]. All cultures were performed in the presence of amphotericin B (2.5 μg/ml), ampicillin (100 μg/ml), and penicillin-streptomycin (GIBCO) (100 μg/ml). As one of the P12 wells exhibited Acanthamoeba cell lysis (following rounding and loss of adherence) in the absence of visible Pandoravirus dulcis particles easily detectable by light microscopy, we suspected that another virus could be responsible for the cell death. The supernatant of the lysed culture was then recovered and used to infect A. castellanii in a T25 flask to amplify the virus. After serial passages exhibiting a steady increase in cell lysis, the medium was recovered and centrifuged 5 min at 500 × g to remove the cellular debris, and the supernatant was centrifuged 30 min at 6,000 × g. The pellet was resuspended in 500 μl phosphate-buffered saline (PBS) buffer complemented with one antiprotease tablet (Roche Diagnostics). This provided the starting material within which viral particles were initially visualized using electron microscopy.
Virus mass production and purification.
Acanthamoeba castellanii cultures in 50 T175 flasks were infected by 3 μl of the viral solution. The cultures were recovered after lysis completion and centrifuged 10 min at 500 × g to remove the cellular debris. The supernatant was then centrifuged for 1 h at 6,000 × g, and the pellet was resuspended in 60 ml PBS buffer. Viral pellets were washed twice in PBS, layered on a discontinuous sucrose gradient (10%, 15%, 20%, 25%, 30%, 35%, and 40% [wt/vol]), and centrifuged at 5,000 × g for 45 min. The virus particles produced a white disk around the 20% sucrose layer and a pearly pellet that was recovered, washed twice in PBS, and stored at 4°C or −80°C with 7.5% dimethyl sulfoxide (DMSO).
Electron microscopy.
Cell cultures of infected A. castellanii cells were fixed with glutaraldehyde 1% for 20 min at room temperature at various times postinfection. Cells were recovered and pelleted by centrifugation (20 min at 5,000 × g). The pellets were resuspended in PBS with 1% glutaraldehyde and incubated at 4°C for at least 1 h. Samples were washed twice in PBS and then coated in agarose before being embedded in Epon resin. Each pellet was mixed with low-melting-point 2% agarose and centrifuged to obtain small flanges of approximately 1 mm3 containing the sample coated with agarose. Using a standard method, the samples were embedded in Epon resin by being fixed overnight in 1% osmium tetroxide, dehydrated in increasing ethanol concentrations (50%, 70%, 90%, and 100% ethanol), and embedded in Epon-812. Ultrathin sections were poststained with 4% uranyl acetate and lead citrate and observed using a Zeiss EM 912 microscope operating at 100 kV.
Genome sequencing, assembly, and annotation.
Melbournevirus genomic DNA was recovered from approximately 2 × 109 particles using the Purelink genomic DNA kit (Invitrogen) following the manufacturer protocol for Gram-negative bacteria. Sequencing was performed under contract with GATC Biotech AG (Cologne, Germany) using 2 platforms: 454 GS FLX+ and Illumina HiSeq 2000. We first used Newbler 2.3 (454 Life Sciences) to assemble 454 GS FLX+ data (259,198 reads) into 135 contigs with an average length of 16,600 nucleotides (nt). The mapping of the Illumina data set (116,448,984 bp of HiSeq 2000 single-end reads) using Bowtie (26) resulted in a single contig of 369,360 nt mostly entirely colinear with the published genomes of Cannes 8 virus (374,041 nt, 484 predicted proteins) and Marseillevirus (368,454 nt, 428 predicted proteins). As these genomes are circular (or linear, circularly permutated, and terminally redundant), sequence numbering is arbitrary. For the sake of easier comparisons, we used the same numbering origin as in Cannes 8 virus. To facilitate the identification of orthologous genes, pairwise whole-genome alignments were generated between Melbournevirus and its closest relatives, Marseillevirus and Cannes 8 virus, using nucmer (27). Direct and inverted repeats were searched using Ugene (28). The GenemarkS web service (29) was used to ab initio predict protein-coding genes from the Melbournevirus genome sequence. These predictions were then cross-validated and annotated by comparing them with the published Marseillevirus (GenBank number NC_013756), Cannes 8 virus (KF261120), and Lausannevirus (NC_015326) predicted proteins. Orthologous protein-coding genes were determined using the reciprocal best BLASTP (30) hit criteria. The functional annotation of predicted proteins was verified by comparing them to the whole nonredundant protein database at NCBI (31).
Estimation of the selection pressure.
The selection pressure was determined from the analysis of carefully verified and visually inspected pairwise alignments of orthologous proteins, converted into codon alignments. The rate of nonsynonymous (dN) and synonymous (dS) substitutions and their ratio (ω [dN/dS]) were computed by PAML (32) version 4.6 using the options “model = 0” and “codonfreq = 2.” The following stringent criteria were imposed on coding regions to be included in the analysis: (i) the percentage of identical nucleotides had to be >90%, (ii) dS had to be between 0.0001 and 0.5, and (ii) the absolute number of synonymous substitutions had to be >5. These criteria were imposed on one hand to minimize the probability of double hit substitutions and on the other hand to have enough substituted positions to work with. Due to these criteria, 103 Melbournevirus-Cannes 8 virus gene pairs and 120 Melbournevirus-Marseillevirus gene pairs were retained for ω value calculations (Table 1). Most gene pairs not included in the analyses were excluded because of a lack of sufficient divergence. The rest corresponded to the few coding regions without detectable homologs between the pairs of viral genomes. The Mann-Whitney/Wilcoxon (MWW) nonparametric test was used to assess the statistical significance of the pairwise differences between the dN/dS values computed for various mutually exclusive categories genes.
TABLE 1.
Statistics of Melbournevirus genes used and not used in dN/dS computations
| Criterion | No. of genes in: |
|
|---|---|---|
| Cannes 8 virus | Marseillevirus | |
| Genes analyzed | 103 | 120 |
| No ortholog in virus | 9 | 46 |
| dS ≤ 0.0001 | 90 | 42 |
| dS > 0.5 | 17 | 35 |
| ≤5 synonymous substitutions | 184 | 160 |
| Total genes too similar | 274 | 202 |
| Total | 403 | 403 |
Nucleotide sequence accession number.
The genome sequence of Melbournevirus is available in GenBank (accession number KM275475).
RESULTS
Isolation of Melbournevirus.
Melbournevirus particles were present in the same muddy freshwater sample from which the giant virus Pandoravirus dulcis was isolated. As we focused on the characterization of virus-like particles large enough to be detected by light microscopy, Melbournevirus was overlooked until preliminary sequence data revealed the mixture of two viral genomes with markedly different characteristics: one exhibited a high G+C content (63.7%) and very little similarity with known viruses (11), while the other one had a lower G+C content (44.7%) and a strong similarity with genes from Marseilleviridae representatives. The two viruses were then pseudocloned by serial dilution and separately characterized. The spectacular features of Pandoravirus dulcis have been published elsewhere (11).
Melbournevirus replication cycle.
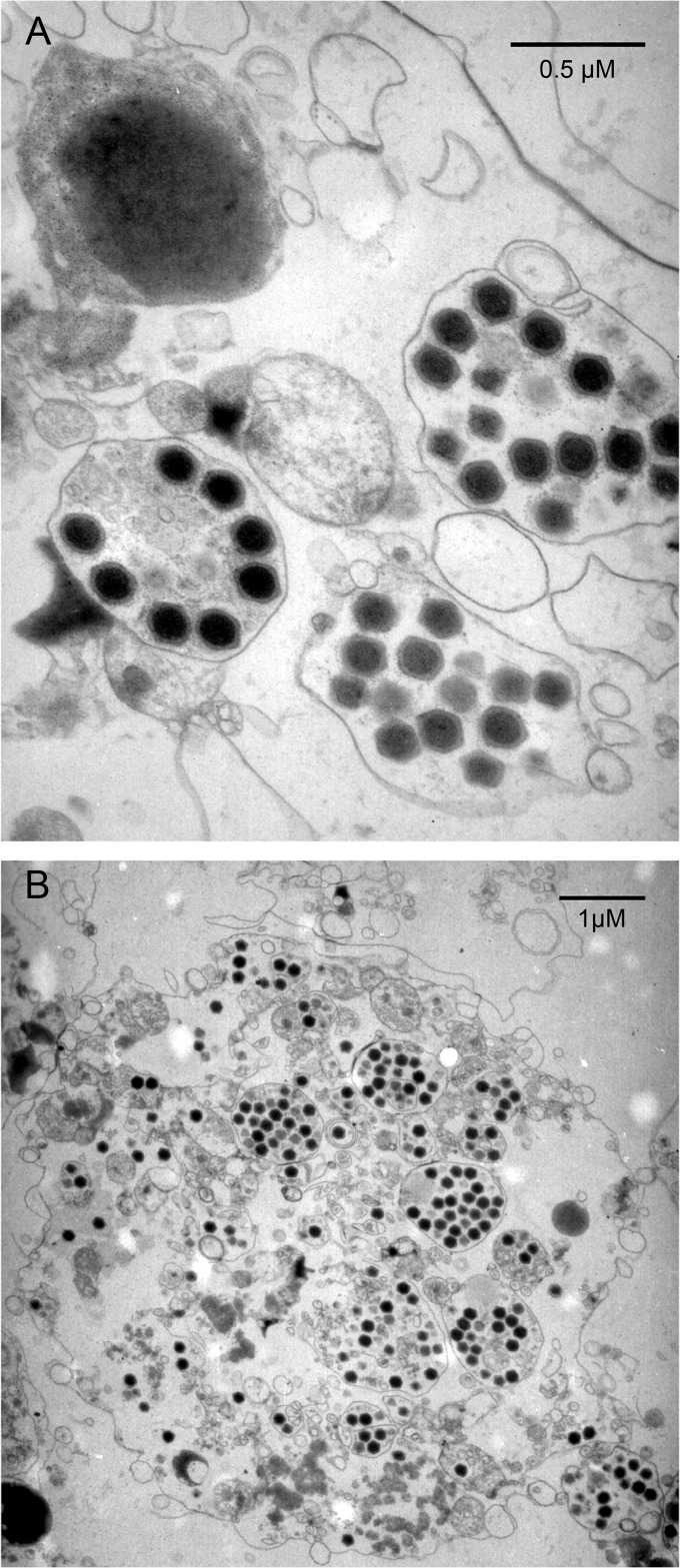
Observation of infected cells at various times postinfection with transmission electronic microscopy (TEM) revealed intracellular icosahedral particles 200 nm in diameter and devoid of surrounding fibrils (Fig. 2A). These particles are thus not visible under a light microscope. The Melbournevirus cycle lasts 12 h and is very similar to the previously described replicative cycle of Lausannevirus and Marseillevirus (17–22). The first stage of the infection corresponded to the internalization of icosahedral particles. The host nucleus appeared intact during the entire replicative cycle suggesting that, as for the other described marseilleviruses, the Melbournevirus replication cycle is entirely cytoplasmic. The new virions are assembled in the periphery of virion factories, first as empty-looking particles and later on as electron-dense, fully mature virions filled with DNA. The capsid's external layer appears translucent, in contrast to the denser internal compartment of the virions. A distinctive feature of the Melbournevirus replicative cycle is that the newly synthesized virions are ultimately gathered into intracytoplasmic vacuoles, suggesting an exocytosis-like mode of dissemination (Fig. 2). These virion-filled vacuoles may be encountered in the external medium after the complete lysis of the host cells.
FIG 2.
Electron microscopy images of ultrathin sections of Melbournevirus. (A) Enlarged view of mature Melbournevirus particles in intracytoplasmic vacuoles. (B) Overall view of an infected cell at a late stage of infection, when the cell is about to be lysed. The cell is filled by Melbournevirus mature particles, most of which are in vacuoles. The cytoplasm is disorganized, and the cell organelles are no longer recognizable.
Melbournevirus genome and gene content.
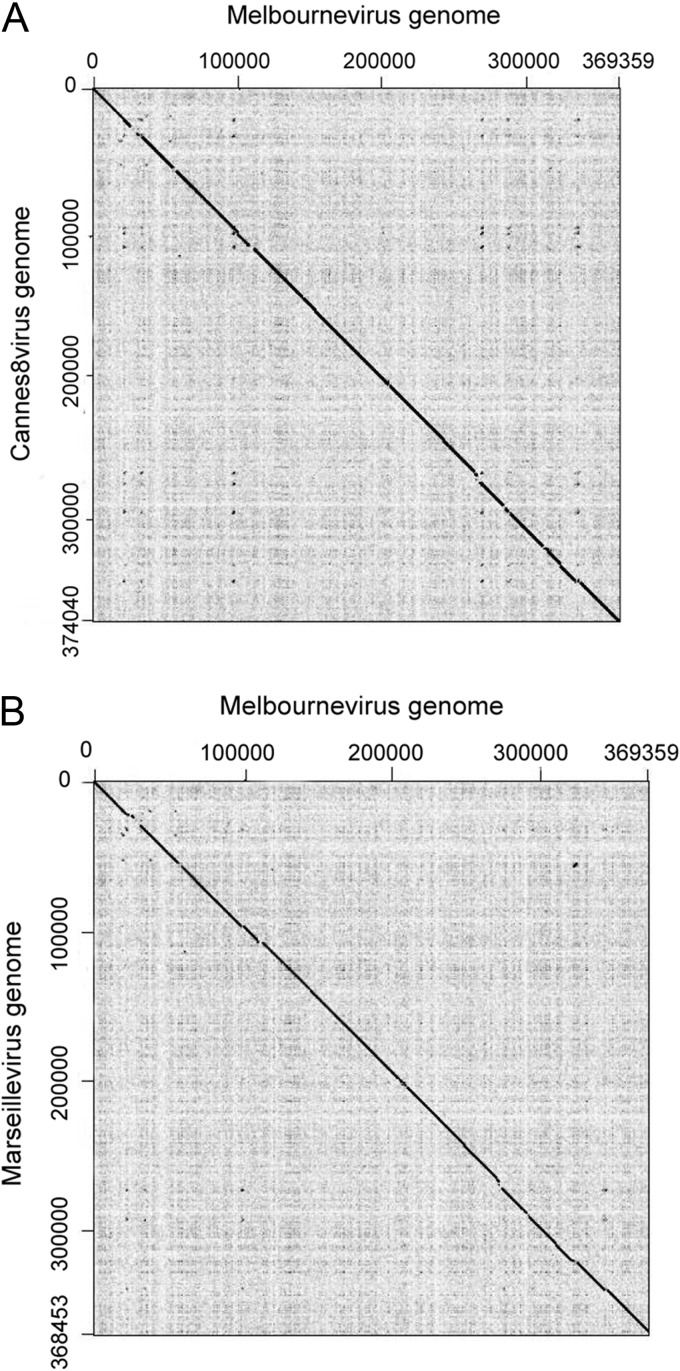
The very high coverage of the 454-flex and Illumina paired-end data sets and the paucity of repeated regions allowed the double-stranded DNA genome sequence of Melbournevirus to be readily assembled into a single contig of 369,360 bp with a G+C content of 44.7%. A total of 403 open reading frames (ORFs) were predicted to encode proteins ranging from 60 to 1,537 amino acids for an average length of 264 residues. Protein-coding regions occupy 86.7% of the genome and are separated by short intergenic regions of 122 nt on average. As for all Marseilleviridae family representatives that had been entirely sequenced, the Melbournevirus genome was assembled as a closed circle. However, it might be a linear DNA molecule that is circularly permutated and terminally redundant, as was demonstrated for some members of the Iridoviridae (33), the other family of large DNA viruses with which Marseilleviridae have the closest phylogenetic proximity (13, 20). The most unexpected genomic feature of Melbournevirus was its extremely close similarity to Cannes 8 virus and Marseillevirus. Orthologous ORFs (defined as reciprocal best matches) share 98.5% and 98% identical nucleotides on average between Melbournevirus versus Cannes 8 virus and Marseillevirus, respectively. Accordingly, in a comprehensive BLASTP search of the nonredundant (NR) NCBI database (31), 215 (53.3%) of Melbournevirus's predicted proteins exhibited a Cannes 8 virus homolog as a best match, 174 (43.2%) a Marseillevirus homolog as a best match, and 5 (1.2%) another Marseilleviridae representative, such as Tunisvirus (21) or Insectomime virus (22). Remarkably, only 9 (2.2%) of predicted Melbournevirus proteins had no homolog according to a BLASTP search. However, ORFs corresponding to 6 of these missing proteins (Mel_036, Mel_069, Mel_086, Mel_224, Mel_324, and Mel_401) are present and well conserved in both Cannes 8 virus and Marseillevirus genomes, in which they have not been annotated. Another one predicted as a Nudix hydrolase (Mel_248) appears to be truncated in Cannes 8 virus and Marseillevirus. Thus, only 2 (Mel_027 and Mel_125) of the 403 annotated Melbournevirus genes do not have a recognizable homolog in Cannes 8 virus or in any other microorganism (NR database) or environmental DNA sequence (24). These relatively short ORFs encoding 181 and 144 amino acids, respectively, may not correspond to actual proteins. Given its high level of similarity with Melbournevirus, the larger number (i.e., 484) of genes predicted in the Cannes 8 virus genome, which is only 4.6 kb longer (i.e., corresponding to 5 proteins on average), might seem inconsistent with the perfect colinearity exhibited by the Melbournevirus, Cannes 8 virus, and Marseillevirus genomes (Fig. 3). Upon closer inspection, the discrepancies are due to annotated Cannes 8 virus ORFs shorter than 60 amino acids (aa) or without an initial ATG or consist of adjacent ORFs merged into a single ortholog in Melbournevirus, such as CAN39-40 (merged in Melbournevirus as an ORF encoding D5-like helicase-primase), CAN179-180 (merged in Melbournevirus as an ORF encoding DNA topoisomerase II), and CAN319-320 (merged in a Melbournevirus as an ORF encoding an early transcription factor large subunit). Only 15 of the 81 “supplementary” Cannes 8 virus predicted ORFs are associated with functional attributes. The list of these discrepancies is available upon request.
FIG 3.
Colinearity of the Melbournevirus genome with that of Cannes 8 virus and Marseillevirus. (A) Comparison of the Melbournevirus and Cannes 8 virus nucleotide sequences. (B) Comparison of the Melbournevirus and Marseillevirus nucleotide sequences. Dotplots were generated with Gepard (40), using a word length of 10 and a window size of 0. Notice the scarcity of repeats in both genomes (i.e., absence of significant off-diagonal segments).
Estimation of the selection pressures on various Marseilleviridae gene types.
Synonymous substitutions are usually regarded as neutral, or at least as having a much smaller effect on fitness than nonsynonymous substitutions (i.e., those resulting in an amino acid change). The relative frequency of nonsynonymous versus synonymous changes, the so-called dN/dS ratio, computed from the comparison of orthologous genes, can thus be used to reveal the type of selection pressure acting on each individual gene. A low ratio (dN/dS ≪ 1) indicates purifying (resisting change) selection, whereas a high ratio (dN/dS > 1) indicates a pressure for diversification (positive selection). The calculation of dN/dS has thus become an accepted tool to identify genes encoding proteins whose changes are likely to be detrimental (i.e., under strong negative selection) or conversely, proteins engaged in a diversifying process, such as virulence factors which confront defense mechanisms of the host (34), or genes that are en route to becoming pseudogenes and disappearing.
The computation of the selection pressure would thus seem like a convenient approach to study the evolution process at work behind the significant variability in gene content and number between the various “species” of giant (Mimiviridae and Pandoraviridae) and large (Marseilleviridae and Chlorovirus) DNA viruses for which several full genome sequences are available. In other words, could we correlate the selection pressure associated with different genes with their predicted function or propensity to be shared by viruses from increasingly distant families? However, such analyses are feasible only if genomes with very similar sequences are used, for two reasons: the pairwise sequence alignment of the orthologous protein and associated coding regions has to be flawless, and the probability of multiple substitutions at a given position must be as small as possible. Until now, the various known members of large and giant DNA virus family were far too distant to make the use of such an approach reliable.
For the first time, the very close genomic sequences of the independently isolated viruses Melbournevirus, Marseillevirus, and Cannes 8 virus offered the possibility to compute the selection pressure associated with their genes and proteins in a reliable manner. With sequences differing by approximately 2% of nucleotides, pairwise alignments of protein and coding regions sequences become unambiguous, and the probability of a given position's having experienced multiple substitutions is negligible. Those are the two main requisites of the method. On the other hand, the small number of nucleotide changes that may pose a problem in achieving statistically significant results is compensated for by the availability of whole-gene contents and the negligible probability of sequence errors thanks to the high coverage and accuracy provided by today's high-throughput sequencing platforms. The fact that Melbournevirus was isolated in a different laboratory than Cannes 8 virus and Marseillevirus, as well as from a very distant location, suggests that it is not just a random variant from the European isolates that diverged recently. We interpret the overall low level of sequence divergence between Melbournevirus and the European isolates as suggesting a very slow evolutionary process, putatively due to a strong purifying selection applied to a large majority of its genes.
The three viruses mentioned above, as well as the more distant Lausannevirus, give us the first opportunity to study the microevolution within a family of large DNA viruses, the Marseilleviridae, and learn about the respective contributions of the different categories of proteins they encode to virus fitness. More specifically, we discuss the three following hypotheses. (i) None of their genes/proteins truly contribute to the virus fitness and the observed gene content is simply an instantaneous picture of a random (neutral) process of gene losses and gains. (ii) A few essential (core, conserved) genes significantly contribute to the virus fitness; the others are just (neutral) passengers or are undergoing diversification. (iii) Most of the genes/proteins do contribute to the virus fitness, and their presence is the result of an active purifying selection process. The results presented below argue in favor of the latter hypothesis.
If 2% of average nucleotide sequence divergence is convenient for the computation of selection pressure, accurately pinpointing the corresponding small number of substitutions requires perfect sequence alignments. To eliminate potential source of errors (in particular regarding the predicted position of the true N-terminal methionine), we thus discarded from the pairwise analyses all the coding regions not predicted to start and stop at the same position or exhibiting more than 10% divergence at the nucleotide sequence level. On the other hand, ORFs with fewer than 5 synonymous substitutions were also discarded for their potential lack of statistical/biological significance. These additional constraints decreased the number of ORFs available to 103 and 120 for pairwise comparison of Melbournevirus with Cannes 8 virus and Marseillevirus, respectively (Table 1).
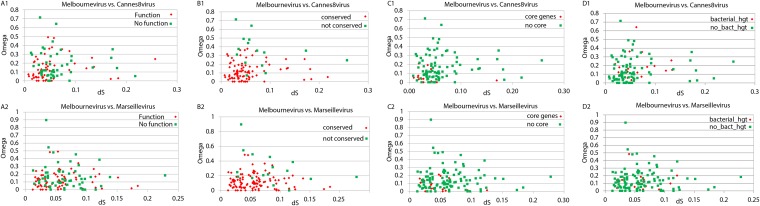
The first global message delivered by the computation of dN/dS ratios is that all Melbournevirus genes are evolving under purifying selection, suggesting that they all significantly contribute to the fitness of the virus. Using Cannes 8 virus or Marseillevirus as a reference, the computed values of ω are 0.16 ± 0.14 (median = 0.12) and 0.15 ± 0.14 (median = 0.12), respectively. These values are comparable to the dN/dS ratios of bacterial genes differing by 2% of nucleotides and assumed to be under stabilizing selection (35). Moreover, with the exception of 5 outliers (compared to Cannes 8 virus) or 6 outliers (compared to Marseillevirus), all ORFs' ω values are smaller than 0.4 (Fig. 4). This strong pressure to resist changes does not characterize only the 103 or 120 genes used to compute the value of ω, since 274 (68% of 403) and 202 (50.1% of 403) ORFs were eliminated from the dN/dS calculation, as they exhibited fewer than 5 synonymous substitutions compared to their orthologs in Cannes 8 virus or Marseillevirus, respectively. In contrast, Table 2 lists 32 ORFs that exhibit markedly lower sequence similarity levels or that have been lost or truncated in Cannes 8 virus or Marseillevirus. These ORFs might correspond to the few proteins whose functions have no significant impact on the virus fitness. Their amino acid sequences share 70% identical residues on average (median = 68.5%, lowest = 29%), when they do not vanish entirely. We noticed that many of these divergent proteins have paralogs, suggesting that their higher divergence rate could be due to redundancy. Nine of these ORFs encode endonucleases that may be the footprints of various mobile elements.
FIG 4.
Distribution of dN/dS values in function of dS for various categories of Melbournevirus protein-coding genes. The dN/dS values were computed from the alignments of Melbournevirus coding regions with their orthologous coding regions in Cannes 8 virus or Marseillevirus. (A) Protein of known or unknown functions; (B) proteins with or without homologs in Lausannevirus (C); core proteins versus regular proteins; (D) proteins of probable bacterial origin versus regular proteins.
TABLE 2.
Most divergent Melbournevirus ORFs
| Melbournevirus ORF (protein size [aa]) | % identical residuesa |
Predicted function and comment | ||
|---|---|---|---|---|
| Cannes 8 virus | Marseillevirus | Lausannevirus | ||
| 025 (384) | 71 | 99 | 76 | 6 paralogous remnants in Melbournevirus |
| 027 (181) | Absent | Absent | Absent | >40 paralogous remnants in Cannes 8 virus |
| 033 (118) | 99 | 38 | 30 | |
| 034 (495) | 39 | 27 | Absent | Helicase, 2 remnants in Cannes 8 virus |
| 035 (1,098) | 90; partial | 93; partial | 82 | Helicase, 2 remote paralogs in Melbournevirus |
| 036 (103) | 61; no annot. | 95; no annot. | Absent | 1 paralogs + 1 remnant in Cannes 8 virus |
| 037 (252) | 55 | 99 | 29 | F box containing, 6 paralogs in Cannes 8 virus and Melbournevirus |
| 038 (430) | 66 | 100 | 53 | Restriction endonuclease, 15 paralogs in Cannes 8 virus 10 paralogs in Melbournevirus |
| 054 (166) | 88 | 98 | 47 | |
| 058 (129) | 30; partial | 46; partial | 38; partial | 2 paralogs in Cannes 8 virus, 1 in Marseillevirus |
| 059 (398) | 38 | 38 | 28 | Zinc finger protease, 4 paralogs in Cannes 8 virus, 2 in Melbournevirus |
| 113 (172) | 99 | 26 | 24 | |
| 124 (1,083) | 99; partial | 83; partial | 99 | Helicase |
| 125 (144) | Absent | Absent | Absent | |
| 166 (155) | 99 | 31; partial | 22 | |
| 222 (242) | 91; partial | Absent | Absent | |
| 223 (398) | 50 | 51 | 38 | MutH/HJR-like endonuclease |
| 224 (134) | 88; no annot. | 87; no annot. | Absent | |
| 270 (537) | 89 | 87 | 45 | MutH/HJR family nuclease, 2 paralogs in Cannes 8 virus, 4 in Marseillevirus |
| 286 (458) | 29 | 24 | 25 | Zinc finger protein, 4 paralogs in Cannes 8 virus, 3 in Marseillevirus, 2 in Melbournevirus |
| 287 (124) | 30; partial | 32; partial | 28; partial | |
| 301 (106) | 100; partial | Absent | 67 | |
| 306 (193) | 65 | 55 | 59 | 2 paralogs in Melbournevirus, 1 in Cannes 8 virus |
| 307 (438) | 74 | 58; partial | 39; partial | MutH/HJR-like endonuclease, 1 paralog in Melbournevirus |
| 311 (434) | 59 | 100 | 53 | Restriction endonuclease, 10 paralogs in Melbournevirus and Cannes 8 virus, 8 paralogs in Marseillevirus |
| 312 (183) | 75 | 94; no annot. | 46 | |
| 336 (137) | 97 | 42 | Absent | |
| 345 (537) | 50 | 99 | 47 | HNH homing endonuclease |
| 346 (355) | 43 | 43 | 42 | Restriction endonuclease |
| 364 (504) | 61 | 96 | 53 | Restriction endonuclease |
| 365 (424) | 66 | 66 | 57 | Restriction endonuclease |
| 370 (104) | 100 | 30; partial | 26 | |
annot., annotation.
We then compared, in greater detail, the selection pressure corresponding to Melbournevirus ORFs distributed among several pairs of mutually exclusive categories. For instance, ORFs in which functional motifs are detected may be considered more likely to correspond to “actual” and useful proteins than ORFs that do not contain any recognizable motif and might simply be the result of bioinformatics overpredictions. In the absence of a real protein's being encoded, the computed dN/dS ratio should be close to 1 on average. Table 3 shows that this is not the case. If ORFs of unknown function exhibit a 23% increase in the computed dN/dS ratio over ORFs of known function, this value remains well below 0.2, and the difference is not statistically significant. If real, this slight difference might indicate that the set of ORFs without functional attributes might contain several wrongly predicted ORFs but that a large majority of them are real and contribute to the virus fitness to the same extent as the ORFs with a predicted function.
TABLE 3.
Selection pressure on various gene categories
| Virus and category | No. | Mean ω | SD | Mean SD | P |
|---|---|---|---|---|---|
| Cannes 8 virus | |||||
| Known function | 55 | 0.14 | 0.13 | 0.02 | >0.08 |
| Anonymous | 48 | 0.18 | 0.15 | 0.02 | |
| Conserved | 88 | 0.12 | 0.1 | 0.01 | <5 × 10−5 |
| Not conserved | 15 | 0.35 | 0.18 | 0.05 | |
| Core | 17 | 0.06 | 0.06 | 0.02 | <5 × 10−4 |
| Noncore | 86 | 0.18 | 0.14 | 0.02 | |
| Bacterial HGT | 18 | 0.19 | 0.15 | 0.03 | >0.11 |
| Nonbacterial HGT | 85 | 0.15 | 0.14 | 0.01 | |
| Marseillevirus | |||||
| Known function | 64 | 0.13 | 0.11 | 0.01 | >0.17 |
| Anonymous | 56 | 0.17 | 0.16 | 0.02 | |
| Conserved | 104 | 0.12 | 0.1 | 0.01 | <1 × 10−4 |
| Not conserved | 16 | 0.3 | 0.22 | 0.05 | |
| Core | 17 | 0.08 | 0.07 | 0.02 | <0.015 |
| Noncore | 103 | 0.16 | 0.14 | 0.01 | |
| Bacterial HGT | 16 | 0.13 | 0.11 | 0.03 | >0.65 |
| Nonbacterial HGT | 104 | 0.15 | 0.14 | 0.01 |
We then examined if the ω value computed for Melbournevirus genes in reference to their Cannes 8 virus and Marseillevirus orthologs was correlated to their presence (i.e., conservation) in the genome of the more distant Lausannevirus. Here, our underlying hypothesis was that genes encoding accessory functions are the ones preferentially lost during the course of the reductive evolution that characterizes all obligatory intracellular parasites. Table 3 shows that the Melbournevirus genes not conserved in Lausannevirus are indeed associated with a significantly larger dN/dS value (ω ≅ 0.3) than the conserved ones (ω ≅ 0.12) (MWW test; P < 0.0001). Thus, even if both categories of genes are under purifying selection (with a ω of ≪1) and contribute to the virus fitness, the one exhibiting less resistance to change will tend to be lost over time.
We then extended this analysis to Melbournevirus genes with (or without) orthologs beyond the family Marseilleviridae but also in more remote representatives of the nucleocytoplasmic large DNA virus (such as the Poxviridae, the Mimiviridae, or the Iridoviridae). Identity between these conserved proteins (the so-called “core” proteins) (36) is sometimes difficult to assess due to their large divergence in sequence. To eliminate any tendency toward subjectivity, we carried out the comparison between the core and noncore Melbournevirus proteins on the basis of the identification previously performed for Marseillevirus (13). Out of the 31 genes identified as encoding core proteins, 14 had to be excluded from the dN/dS calculation because they were too similar to their Melbournevirus orthologs (or had <5 substitutions). Omega values were thus computed for the remaining 17 core genes. As expected from the results already obtained for the genes conserved in Lausannevirus, the Melbournevirus genes classified as encoding core proteins exhibit on average an even lower dN/dS ratio (ω ≅ 0.07) than the genes encoding proteins solely conserved within the family Marseilleviridae (Table 3). However, 10 genes (including the DNA polymerase-catalytic subunits or the DNA-directed RNA polymerase large subunit) exhibited ω values which were in the 0.2-to-0.1 range and were thus comparable to the one computed for many regular genes not associated with a remarkable function or a wide conservation pattern.
Selection pressure on Marseilleviridae genes likely acquired from bacteria.
The genome of Marseillevirus was initially noted to encompass a larger than usual repertoire of genes of putatively cellular origins, including 58 genes (>10% of the whole predicted gene content) most likely acquired by horizontal gene transfer (HGT) from bacteria or phages (13). Hosts feeding on bacteria such as Acanthamoeba, already documented to serve as a DNA “melting pot” between intracellular bacteria (37), might thus also facilitate HGT between prokaryotes and eukaryotes by the intermediary of large DNA viruses (13). Identifying the subset of potentially horizontally transferred genes is usually less problematic when they originate from bacteria (rather than from an unidentified eukaryote), as the discrepant phylogenetic and sequence similarity signal they exhibit is usually stronger. We thus chose to focus on their analysis.
Out of the 58 bacterial/phage HGT events proposed by Boyer et al. (13), 6 do not have homologs in the Melbournevirus genome (Mar34, Mar44, Mar66, Mar318, Mar387a, and Mar387b). Among the 52 remaining HGT candidates, 6 and 10 were too divergent (<90% identical nucleotides) from their Cannes 8 virus and Marseillevirus homologs, respectively, to be incorporated in our analysis. Thus, a total of 12 (21%) or 16 (28%) exhibited a larger-than-average divergence from their orthologs in Cannes 8 virus and Marseillevirus, respectively. On the other hand, respectively, 28 (48%) and 26 (45%) of these bacterial HGT-derived genes were too close (fewer than 5 synonymous substitutions) from their Cannes 8 virus and Marseillevirus orthologs to be taken into account in our selection pressure computations. These statistics already suggested that the genes putatively acquired by HGT from bacteria or phages did not exhibit a general trend of evolving much faster than regular genes. This was confirmed by the results of the computation of dN/dS for the 18 and 16 Melbournevirus genes with orthologs in Cannes 8 virus and Marseillevirus, respectively, falling into the suitable similarity bracket. As shown in Table 3, all of them are under negative selection with a distribution of ω values not significantly different from that of the rest of the genes. This somewhat unexpected result suggests that these genes, if truly laterally transferred from bacteria, immediately had a beneficial effect on the virus fitness without first undergoing a period of accelerated evolution to accompany the transition from a prokaryote to a eukaryotic cellular environment. As this is probably true of the orthologous gene pairs that are too similar to allow the computation of dN/dS, only approximately one quarter of the genes putatively transferred from bacteria or phage exhibited an accelerated evolution that may eventually lead to their disappearance from the recipient Marseilleviridae genome.
DISCUSSION
As indicated by their names, Marseillevirus, Cannes 8 virus, Tunisvirus, and Lausannevirus, all the known representatives of the rapidly expanding family Marseilleviridae, were isolated in Western Europe or North Africa. We now report the first member of this family isolated from a freshwater pond in Melbourne, Australia. In accord with the naming scheme for previous isolates, we called this virus Melbournevirus.
The complete genome sequencing of Melbournevirus revealed that it was nearly identical to the previously described Marseillevirus (isolated from a cooling tower in Paris, France) and Cannes 8 virus (isolated from a cooling tower in Cannes, a coastal city on the French Riviera). Fortunately, the possibility of a cross-contamination could be rejected for two reasons: first, our laboratory never isolated or handled any member of the Marseilleviridae before processing the Australian sample, and second, Melbournevirus was coisolated with Pandoravirus dulcis, and no Pandoravirus had been isolated from a European sample. We can thus confidently conclude that Melbournevirus truly originated from Australia. This then raises the question of how viruses that have been separated long enough to reach locations 15,000 kilometers away and across the Pacific Ocean could have kept their genome sequence more than 98% identical on average. This finding is even more puzzling given that other members of the family Marseilleviridae, such as Tunisvirus and Lausannevirus, exhibit a much lower percentage of nucleotide sequence identity (<60%) while still being able to infect the same acanthamoeba host with the same apparent efficiency (20, 21). Extreme sequence conservation between large DNA viruses isolated from very distant locations has been reported previously but was limited to individual genes (38). The extreme genomic stability of some of the Marseilleviridae members might be due to the presence of three histone-like proteins (20). However, Tunisvirus and Lausannevirus also encode these histone-like proteins. One possible explanation was that genes responsible for the extreme stability of the genome of the Marseillevirus/Melbournevirus subclade were lost, giving rise to the more rapidly evolving Lausannevirus/Tunisvirus subclade. We investigated this possibility by specifically looking at genes present in Melbournevirus, Marseillevirus, and Cannes 8 virus that are absent from the other Marseilleviridae. We identified 26 of them, unfortunately including 22 without predicted function. Further scrutiny of these anonymous sequences using 3D fold recognition tools (39) did not provide any additional clue to their function. Among the 4 genes associated with a predicted function, only Mel016, encoding a DNA-adenine-methyltransferase domain (DAM), was found to have a possible relationship with the fidelity of viral genome replication. Proteins encoding a DAM have been implicated in methylation-directed DNA mismatch repair, replication, recombination, and restriction/modification systems. Homologs of this protein are found in a number of large eukaryotic DNA viruses (such as chloroviruses and some Mimiviridae), as well as phages. In any case, the evolutionary advantage conferred to a virus by a genome replication with exquisitely high fidelity is not obvious, while viruses are usually pictured as fast-evolving, fast-adapting microorganisms. Alternatively, the extreme genome similarity of the independently and remotely isolated Melbournevirus, Cannes 8 virus, and Marseillevirus might be due to a fast spreading of these viruses over long distances through a mechanism that remains to be elucidated.
The availability of the highly similar whole-genome sequences of the three independent isolates Melbournevirus, Cannes 8 virus, and Marseillevirus, as well as that of the more distant Lausannevirus, allowed the measurement of the selection pressures at work on many genes during their microevolution with an unprecedented accuracy for large DNA viruses. As the gene content of different large and giant DNA viruses infecting the same Acanthamoeba host varies enormously across different families, such as the Marseilleviridae, the Mimiviridae, the pandoraviruses, and the pithoviruses, and between members of the same family, one could have expected that a large proportion of the proteins encoded in each individual genome would appear quite dispensable, thus corresponding to neutral or near-neutral (ω ≅ 1) dN/dS values. To our surprise, our computation supported the hypothesis that, with the exception of 30 rapidly evolving genes/proteins (7.5% of the total), a large majority of genes/proteins are associated with a ω value of <0.3, for an overall average of 0.16. Furthermore, this low value must be taken as an upper estimate, due to the well-documented effect of incomplete fixation (i.e., elimination) of slightly deleterious nonsynonymous substitutions in highly similar (98% identity) variants (35). To the 103 and 120 Melbournevirus genes for which purifying selection (ω ≪ 1) was documented in reference to Cannes 8 virus and Marseillevirus, respectively, 274 and 202 were found to be too similar to their orthologs to allow reliable dN/dS calculations, although this lack of divergence is also compatible with a strong purifying selection process. Altogether, this corresponds to up to 377 (93.5% of 403) of Melbournevirus genes that appear to contribute to its fitness.
Although the previous result indicates that most genes play a nonnegligible role in the Melbournevirus life cycle, significantly higher purifying selection was associated with genes conserved among distant Marseilleviridae representatives, reaching a maximal resistance to change for genes encoding core proteins conserved in different families of large DNA viruses beyond the Marseilleviridae. This result indicates that the ω values computed from the 3 very close sequences of the Melbournevirus, Cannes 8 virus, and Marseillevirus are indeed meaningful and correlate with the presence/absence of protein-coding genes in increasingly divergent large DNA viruses. This pattern is consistent with a stochastic evolutionary process of genome reduction during which genes exhibiting the least resistance to change are also the ones whose loss has the highest probability of being tolerated and fixed.
A large majority (approximately 75%) of the genes likely to have been acquired by HGT from bacteria or phage were not found to be subject to positive selection (i.e., accelerated evolution). This result argues against the hypothesis that a large number of viral proteins without cellular homologs could have originated from horizontally transferred genes from which all detectable phylogenetic signals would have been subsequently erased. Elucidating the source of these “virus-only” and/or so-called “ORFan” (i.e., suspected genes without known relatives) proteins thus remains a central challenge to the understanding of the origin and evolution of large and giant DNA viruses.
Finally, no significant difference was found in the selection pressure applied to genes of unknown function (i.e., “anonymous ORFs”) compared to the one computed for the genes associated with functional attributes. This result indicates that the numerous proteins of unknown function that often constitute the majority of ORFs predicted in large and giant virus genomes evolve as real proteins positively contributing to virus fitness. We noticed that many anonymous genes were associated with dN/dS values as low (ω < 0.2) as those associated with core genes. (Fig. 4A and C). This result suggests that the corresponding functions, albeit unknown, have a significant impact on the virus fitness may be through the fine-tuning of virus/host interactions. This finding justifies the idea that more efforts should be invested in elucidating the biological and cellular functions of the large proportion of anonymous viral genes, as it may lead to basic discoveries in metabolism and cell biology as well as to innovative biomedical and pharmaceutical applications.
ACKNOWLEDGMENTS
We thank Victoria Schmidt for her assistance in Melbournevirus isolation, Jean-Paul Chauvin, Fabrice Richard, and Aïcha Aouane for their expert assistance on the IBDML imagery facility, and Artemis Kosta from the IMM. We thank Michael Ciancanelli for reading the manuscript and Patricia Bonin for her help. We thank Jean-Laurent Casanova for his generous hospitality at Rockefeller University.
This work was partially supported by CNRS and Aix Marseille University and the Region Provence Alpes-Côte-d'Azur. G.D. is the recipient of a Ph.D. fellowship from Aix-Marseille University.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Legendre M, Audic S, Poirot O, Hingamp P, Seltzer V, Byrne D, Lartigue A, Lescot M, Bernadac A, Poulain J, Abergel C, Claverie JM. 2010. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 20:664–674. 10.1101/gr.102582.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legendre M, Santini S, Rico A, Abergel C, Claverie JM. 2011. Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 8:99. 10.1186/1743-422X-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claverie JM, Abergel C. 2013. Open questions about giant viruses. Adv. Virus Res. 85:25–56. 10.1016/B978-0-12-408116-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 6.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. U. S. A. 108:17486–17491. 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boughalmi M, Saadi H, Pagnier I, Colson P, Fournous G, Raoult D, La Scola B. 2013. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 15:2000–2007. 10.1111/1462-2920.12068. [DOI] [PubMed] [Google Scholar]
- 8.Pagnier I, Reteno DG, Saadi H, Boughalmi M, Gaia M, Slimani M, Ngounga T, Bekliz M, Colson P, Raoult D, La Scola B. 2013. A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba. Intervirology 56:354–363. 10.1159/000354556. [DOI] [PubMed] [Google Scholar]
- 9.Fischer MG, Allen MJ, Wilson WH, Suttle CA. 2010. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. U. S. A. 107:19508–19513. 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Barbe V, Wommack KE, Noordeloos AA, Brussaard CP, Claverie JM. 2013. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 110:10800–10805. 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 12.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Couté Y, Rivkina E, Abergel C, Claverie JM. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. U. S. A. 111:4274–4279. 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U. S. A. 106:21848–21853. 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Etten JL. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37:153–195. 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 15.Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. 2005. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309:1090–1092. 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 16.Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, Van Etten JL, Blanc G. 2013. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14:158. 10.1186/1471-2164-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colson P, Pagnier I, Yoosuf N, Fournous G, La Scola B, Raoult D. 2013. “Marseilleviridae,” a new family of giant viruses infecting amoebae. Arch. Virol. 158:915–920. 10.1007/s00705-012-1537-y. [DOI] [PubMed] [Google Scholar]
- 18.Aherfi S, Pagnier I, Fournous G, Raoult D, La Scola B, Colson P. 2013. Complete genome sequence of Cannes 8 virus, a new member of the proposed family “Marseilleviridae.” Virus Genes 47:550–555. 10.1007/s11262-013-0965-4. [DOI] [PubMed] [Google Scholar]
- 19.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18:1185–1193. [DOI] [PubMed] [Google Scholar]
- 20.Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G. 2011. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 13:1454–1466. 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 21.Aherfi S, Boughalmi M, Pagnier I, Fournous G, La Scola B, Raoult D, Colson P. 2014. Complete genome sequence of Tunisvirus, a new member of the proposed family Marseilleviridae. Arch. Virol. 10.1007/s00705-014-2023-5. [DOI] [PubMed] [Google Scholar]
- 22.Boughalmi M, Pagnier I, Aherfi S, Colson P, Raoult D, La Scola B. 2013. First isolation of a Marseillevirus in the Diptera Syrphidae Eristalis tenax. Intervirology 56:386–394. 10.1159/000354560. [DOI] [PubMed] [Google Scholar]
- 23.Baltimore D. 1971. Expression of animal virus genomes. Bacteriol. Rev. 35:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Krasnov S, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Karsch-Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. 2012. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 40:D13–D25. 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer M, Azza S, Barrassi L, Klose T, Campocasso A, Pagnier I, Fournous G, Borg A, Robert C, Zhang X, Desnues C, Henrissat B, Rossmann MG, La Scola B, Raoult D. 2011. Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. U. S. A. 108:10296–10301. 10.1073/pnas.1101118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okonechnikov K, Golosova O, Fursov M, the UGENE Team 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 29.Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451–W454. 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. 2011. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 39:D38–D51. 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 33.Jakob NJ, Müller K, Bahr U, Darai G. 2001. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology 286:182–196. 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- 34.Smith NH, Maynard Smith J, Spratt BG. 1995. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol. Biol. Evol. 12:363–370. [DOI] [PubMed] [Google Scholar]
- 35.Rocha EPC, Maynard Smith J, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil E. 2006. Comparisons of dN/dS are time-dependent for closely related bacterial genomes. J. Theor. Biol. 239:226–235. 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 36.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734. 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:e76. 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Adams B, Sun L, Burbank DE, Van Etten JL. 2001. Intron conservation in the DNA polymerase gene encoded by Chlorella viruses. Virology 285:313–321. 10.1006/viro.2001.0935. [DOI] [PubMed] [Google Scholar]
- 39.Shi J, Blundell TL, Mizuguchi K. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243–257. 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 40.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]