ABSTRACT
Electron micrographs from the 1960s revealed the presence of an S-shaped tubular structure in the center of the vaccinia virion core. Recently, we showed that packaging of virus transcription enzymes is necessary for the formation of the tubular structure, suggesting that the structure is equivalent to a nucleocapsid. Based on this study and on what is known about nucleocapsids of other viruses, we hypothesized that in addition to transcription enzymes, the tubular structure also contains the viral DNA and a structural protein as a scaffold. The vaccinia virion structural protein L4 stands out as the best candidate for the role of a nucleocapsid structural protein because it is abundant, it is localized in the center of the virion core, and it binds DNA. In order to gain more insight into the structure and relevance of the nucleocapsid, we analyzed thermosensitive and inducible mutants in the L4R gene. Using a cryo-fixation method for electron microscopy (high-pressure freezing followed by freeze-substitution) to preserve labile structures like the nucleocapsid, we were able to demonstrate that in the absence of functional L4, mature particles with defective internal structures are produced under nonpermissive conditions. These particles do not contain a nucleocapsid. In addition, the core wall of these virions is abnormal. This suggests that the nucleocapsid interacts with the core wall and that the nucleocapsid structure might be more complex than originally assumed.
IMPORTANCE The vaccinia virus nucleocapsid has been neglected since the 1960s due to a lack of electron microscopy techniques to preserve this labile structure. With the advent of cryo-fixation techniques, like high-pressure freezing/freeze-substitution, we are now able to consistently preserve and visualize the nucleocapsid. Because vaccinia virus early transcription is coupled to the viral core structure, detailing the structure of the nucleocapsid is indispensable for determining the mechanisms of vaccinia virus core-directed transcription. The present study represents our second attempt to understand the structure and biological significance of the nucleocapsid. We demonstrate the importance of the protein L4 for the formation of the nucleocapsid and reveal in addition that the nucleocapsid and the core wall may be associated, suggesting a higher level of complexity of the nucleocapsid than predicted. In addition, we prove the utility of high-pressure freezing in preserving the vaccinia virus nucleocapsid.
INTRODUCTION
Vaccinia virus, the best-studied member of the family Poxviridae, is a large double-stranded DNA (dsDNA)-containing virus with a unique and complex virion structure. The vaccinia virion does not conform to the standard helical or icosahedral classifications but, instead, consists of a brick-shaped particle comprising an outer membrane that surrounds a biconcave core and two lateral bodies within each concavity of the core (1).
The vaccinia virus replicative cycle takes place entirely in the cell cytoplasm. Because of the cytoplasmic site of replication, vaccinia virus packages all of the enzymes necessary for early-stage viral transcription. The virus also encodes all of the enzymes required for DNA replication. Virion assembly occurs in cytoplasmic areas devoid of organelles, called viral factories. Viral membrane crescents are the first structures observed within factories during virion assembly. These membrane crescents grow and engulf an electron-dense viroplasm to form immature virions (IVs). IVs encapsidate viral DNA and evolve into mature virions (MVs) in a process that is coupled to cleavage of several membrane and core proteins (2, 3). Processing of these viral proteins is linked to a major structural rearrangement in the virion in which the spherical IV morphs into the classical brick-shaped virion. In addition to the change in the overall shape of the viral particle, the interior of the virion undergoes striking changes, with the reorganization of the viroplasm mass into a biconcave core with lateral bodies (4, 5).
Experiments from the early 1960s revealed a tubular structure inside the core (6, 7). This tubular structure has been largely overlooked for the last 50 years because of the difficulty in preserving it using conventional techniques for electron microscopy (EM). Recently, we demonstrated that formation of the tubular structure requires packaging of viral transcription enzymes into the virions (8). This observation prompted us to designate the tubular structure as a nucleocapsid. By analogy with other viruses, we hypothesized that the vaccinia virus nucleocapsid contains the viral DNA and a major structural protein. The virion structural protein L4 possesses several characteristics that suggest it as the best candidate for a nucleocapsid protein. L4 binds both RNA and DNA with high affinity, and it is highly abundant, being the sixth most abundant protein in the virions (9–11). Also, the absence of L4 from virions leads to formation of viral particles with fragile viral cores, from which the transcription enzymes are more easily extracted than from viral particles containing L4 (12). Interestingly, Sarov and Joklik (13) and Pedersen and colleagues (14), using biochemical and microscopy techniques, characterized intermediates of vaccinia virus that accumulate in the period immediately following virus entry and before the onset of virus DNA replication. These studies revealed that intermediates that accumulate after about 90 min of infection contained most of the proteins present in the mature virion core but lacked DNA, L4, and three other viral proteins (13, 14). The intermediates isolated by Sarov and Joklik also lacked DNA-dependent RNA polymerase activity, suggesting that these structures were no longer active for viral transcription. These two independent studies suggest that L4 and DNA are lost simultaneously during the initial steps of virus replication.
All poxviruses contain homologs to vaccinia virus L4, with amino acid identities ranging from a low of 43% (molluscum contagiosum) in a comparison of all mammalian poxviruses to 98% for the orthopoxviruses. L4 has no sequence homology with proteins outside the poxvirus family. L4 is synthesized late during infection and cleaved during maturation from IV to MV (12, 15, 16). Data showing that L4 stimulates the activity of the vaccinia virus I8 RNA helicase and that viral particles lacking L4 are inactive for core transcription even though they package all transcription factors suggest that L4 could participate in virus transcription (17, 18).
The main goal of this study was to understand in more detail the structure and function of the vaccinia virus nucleocapsid. For this, we used an approach that has been successful in revealing the basic biology of poxviruses, the study of conditionally lethal mutants. Poxvirus conditional lethal mutants comprise two main classes, inducible and thermosensitive. For some vaccinia virus genes, both thermosensitive and inducible mutants are available. In the majority of cases, the phenotypes of thermosensitive and inducible mutants are similar. However, in a few cases these two classes of mutants present blockage in different steps of replication (19–22). This difference in phenotypes reflects the mechanistic difference between the two classes of mutants: thermosensitive mutants often produce a nonfunctional protein which is nevertheless present under the nonpermissive condition, whereas the affected gene product is completely absent during infection with an inducible mutant. These observations demonstrate the importance of studying both types of mutants in order to obtain the most complete understanding of protein function.
In this study, we characterized a thermosensitive mutant previously mapped to the L4R gene, Ets85 (12, 23, 24). Importantly, because routine preparations for electron microscopy provide poor preservation of the nucleocapsid, we used high-pressure freezing followed by freeze-substitution to visualize the nucleocapsid. High-pressure freezing and freeze-substitution techniques ensure better preservation of fine ultrastructure than conventional fixation methods (26, 27). We then reinvestigated the morphogenesis of an inducible mutant in L4 that has been characterized previously (12).
The results with Ets85 show that during virus assembly under nonpermissive conditions, abnormal immature virions (IVs) are produced. Even though the immature virions are aberrant, they package all virion proteins tested as well as viral DNA. Some IVs are able to progress further in the maturation pathway to produce abnormal mature particles in the cytoplasm. Virtually identical abnormal mature virions also form when infection is performed with the inducible mutant in L4, vL4i. With both Ets85 and vL4i, aberrant mature virions appear to have a defect in core wall formation in addition to being devoid of a nucleocapsid, showing that the absence of L4 has a profound effect on the core structure that goes beyond the nucleocapsid structure.
MATERIALS AND METHODS
Cells and viruses.
Viruses were grown in BSC-40 cells, an African green monkey cell line. BSC-40 cells were cultured in Dulbecco's modified Eagle's medium (catalog number 12100-061; Life Technologies) containing 10% fetal bovine serum, 0.12 mg/ml penicillin (Sigma), 0.2 mg/ml streptomycin (Sigma), and 250 μg/ml amphotericin B (Fungizone; Sigma). Wild-type vaccinia virus strain WR, used as the control in this study, the temperature-sensitive mutant virus Ets85, the inducible mutant in the L4R gene (vL4i), and the inducible mutant in the E6R gene (vE6i) have been described previously (12, 21, 24, 28). (Ets85 was obtained indirectly from Marcia Ensinger. Upon her retirement Ensinger left her mutant collection with Hamish Young at Columbia University. Upon his retirement in 1995, Young provided us with the entire Ensinger collection. vL4i is our designation for the mutant vDW4, and was obtained from Geoffrey Smith.) The permissive temperature for Ets85 is 31°C. The nonpermissive temperature is 39.7°C. vL4i and vE6i were grown at 37°C in the presence (permissive condition) or absence (nonpermissive condition) of 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for vL4i and 50 μM IPTG for vE6i.
Plaque assay.
Ten-fold dilutions of the virus were used to infect BSC-40 cell monolayers in 60-mm dishes. After 1 h of adsorption, the inoculum was removed, and medium supplemented with 1% methylcellulose was added to the cells. Cells were incubated for 3 or 7 days at 31°C or 39.7°C and stained with crystal violet.
One-step growth.
BSC-40 cells were infected with WR or Ets85 at a multiplicity of infection (MOI) of 10. After a 30-min adsorption, the inoculum was removed, and medium was added to the cells. Infected cells were incubated at 31°C or 39.7°C and harvested at various times postinfection. Virus titer was determined by plaque assay at 31°C.
Western blot analysis.
BSC-40 cells in 35-mm dishes were infected at an MOI of 10 with WR or Ets85 and incubated at 31°C or 39.7°C. Infected cells were harvested at different times postinfection in sample buffer (50 mM Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 140 mM 2-mercaptoethanol, 0.1% bromophenol blue). Western blots were prepared and analyzed as described previously (29). The L4 antibody was supplied by Paula Traktman and used in a 1:50,000 dilution. Secondary anti-rabbit antibody was purchased from Santa Cruz Biotechnologies, Inc.
Protein pulse-labeling.
Cells were infected at an MOI of 10 with WR or Ets85 and incubated at 31°C or 39.7°C. For pulse-labeling, at each time point, cells were labeled with methionine-free culture medium containing 20 μCi/ml of [35S]methionine (PerkinElmer). After 30 min of incubation at 31°C or 39.7°C, the medium was removed, and the cells were harvested in sample buffer. Proteins were analyzed by SDS-PAGE, followed by autoradiography (29).
For pulse-chase experiments, cells were pulse-labeled at 8 h postinfection. After 15 min, medium was replaced with unlabeled, methionine-containing medium, and cells were incubated for various times at 31°C or 39.7°C. Cells were harvested in sample buffer and analyzed by SDS-PAGE and autoradiography.
Electron microscopy: conventional sample fixation for ultrastructure.
BSC-40 cells in 60-mm dishes were infected with either the WR or Ets85 vaccinia virus at an MOI of 10 and incubated at 31°C or 39.7°C for 24 h. Infected cells were fixed with 2% glutaraldehyde in cacodylate buffer (0.1 M sodium cacodylate, 2 mM MgCl2, 1 mM CaCl2, 43 mM NaCl, pH 7.2) and postfixed with 1% osmium tetroxide in water. After dehydration with increasing concentrations of ethanol, samples were embedded in Spurr's resin. Sections of 70- to 80-nm thickness were poststained with 2% uranyl acetate and lead citrate and visualized in a Hitachi H-7000 electron microscope (29). The electron microscopy was done with assistance of the University of Florida ICBR Electron Microscopy Core Laboratory.
Electron microscopy: high-pressure freezing/freeze-substitution.
BSC-40 cells in 100-mm dishes were infected at an MOI of 10 with WR, Ets85, or vL4i. For infections with Ets85, both mutant and control infections were incubated at 31°C or 39.7°C. For infections with vL4i, cells were incubated with drug-free medium or medium containing 200 μM IPTG at 37°C. At 24 h postinfection, cells were harvested and incubated with cacodylate buffer containing 4% paraformaldehyde and 1% glutaraldehyde for 30 min on ice. Fixed cells were pelleted by gentle centrifugation, washed twice with cacodylate buffer, and resuspended in 25 μl of cryoprotectant (20% [wt/vol] dextran [molecular weight, 39,000] prepared in serum-free medium). The dextran-protected cell suspension was loaded into a 6-mm aluminum type A planchette (Leica Microsystems, Buffalo Grove, IL) that was capped using the flat side of a 6-mm aluminum type B planchette. This assembly was loaded into a holder and immediately frozen using a high-pressure freezing system (HPM 100; Leica Microsystems, Buffalo Grove, IL). After freezing, planchettes containing cells were kept under liquid nitrogen and transferred to a vial containing 20% epoxy resin in acetone. The epoxy fixative was adapted from Matzko and Mueller (31). The vials containing the planchettes were transferred to the freeze-substitution unit (EM AFS2; Leica Microsystems, Buffalo Grove, IL). Freeze-substitution was performed using the following program: −90°C for 12 to 48 h, followed by slow warming from −90°C to −70°C for 16 h, from −70°C to −45°C for 12 h, from −45°C to −20°C for 2 h, and from −20°C to 4°C for 2 h. Cells were removed from the planchettes and washed three times with acetone, followed by 1 h of incubation with 1% tannic acid in acetone on ice. After three washes with 100% acetone, cells were incubated in 1% osmium tetroxide in acetone for 1 h on ice. Cells were washed three times with acetone at 4°C and then handled at room temperature for infiltration with increasing concentrations of Embed resin (30%, 50%, 70%, and 100%) using a Pelco BioWave laboratory microwave, ColdSpot platform, and EM Pro Vacuum chamber (Ted Pella, Redding, CA) set at a 20-Hg vacuum and 220 W for 3 min. Specimens were polymerized over a period of 2 days at 60°C. Ultrathin sections (70 to 80 nm) were poststained with 2% uranyl acetate and lead citrate and examined with an H-7000 transmission electron microscope (TEM; Hitachi High Technologies America, Inc., Schaumburg, IL) operated at 100 kV. Digital images were acquired with a Veleta camera and iTEM software (Olympus Soft-Imaging Solutions Corp., Lakewood, CO). Details of the development of these techniques for imaging poxvirus-infected cells will be published elsewhere (D. Jesus, unpublished data).
Immunoelectron microscopy.
BSC-40 cells in 60-mm dishes were infected at an MOI of 10 with WR or Ets85 (see Fig. 9 and 10) or with the inducible mutant in the E6R gene in the presence of IPTG (Fig. 1). (The experiments shown in Fig. 1 are control infections from a related project studying the role of E6 in virus assembly.) vE6i infections done in the presence of IPTG are equivalent to wild-type virus infections (21). After 24 h of infection at 31°C or 39.7°C, cells were washed with PHEM buffer (60 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) and fixed with a solution containing 0.6% glutaraldehyde, 4% paraformaldehyde, 5 mM CaCl2, and 0.5% sucrose in PHEM buffer for 1 h at 4°C. The monolayer was washed with 0.1 M glycine in PHEM buffer, and cells were harvested. Cell pellets were dehydrated in a series of ice-cold ethanol (30%, 50%, 70%, and 100%) and infiltrated with Lowicryl HM20 resin. Samples were polymerized with 100% Lowicryl at −20°C with UV light for 2 days. Ultrathin sections (70 to 80 nm) were collected on nickel grids and immunolabeled as described previously (32). The following antibodies were used: anti-L4 (at a dilution of 1:100) and anti-F17 (1:2,000) rabbit sera (supplied by Paula Traktman), anti-A3 (1:100) and anti-D13 (1:1,000) rabbit sera (supplied by Bernard Moss), anti-dsDNA (1:10) mouse serum (Santa Cruz Biotechnologies), and secondary antibodies anti-rabbit and anti-mouse (1:50) (BBI Solutions, Cardiff, United Kingdom).
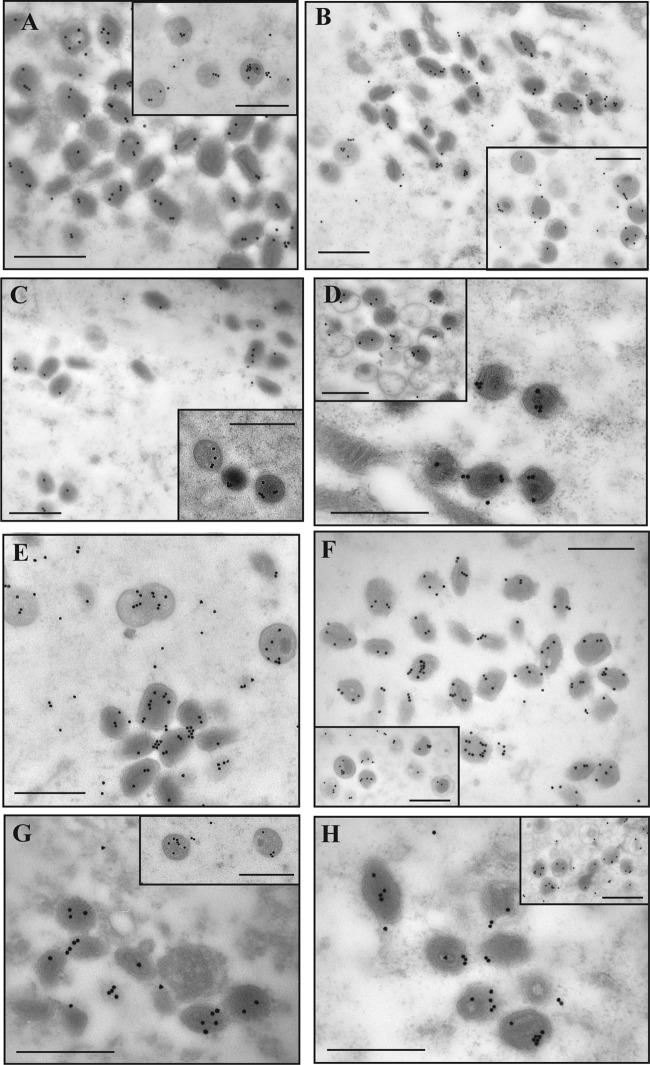
FIG 9.
Immunogold labeling of core proteins. Cells were infected with WR or Ets85 at an MOI of 10 and incubated at 31°C or 39.7°C. At 24 h postinfection, cells were processed for immunoelectron microscopy. Ultrathin sections were probed with antibodies against L4 (A to D) or A3 (E to H) and visualized in an electron microscope (Hitachi H-7000). (A and E) WR at 31°C. (B and F) WR at 40°C. (C and G) Ets85 at 31°C. (D and H) Ets85 at 39.7°C. The insets show the labeling of immature virions. Scale bar, 500 nm.
FIG 10.
Immunogold labeling of DNA. Cells were infected with WR or Ets85 at an MOI of 10 and incubated at 31°C or 39.7°C. At 24 h postinfection, cells were processed for immunoelectron microscopy. Sections of 70- to 90-nm thickness were probed with antibodies against double-strand DNA (5-nm gold particles) or against the viral protein D13 (20-nm gold) and visualized in an electron microscope (Hitachi H-7000). (A) WR at 31°C. (B) WR at 40°C. (C) Ets85 at 31°C. (D) Ets85 at 39.7°C. Scale bar, 200 nm.
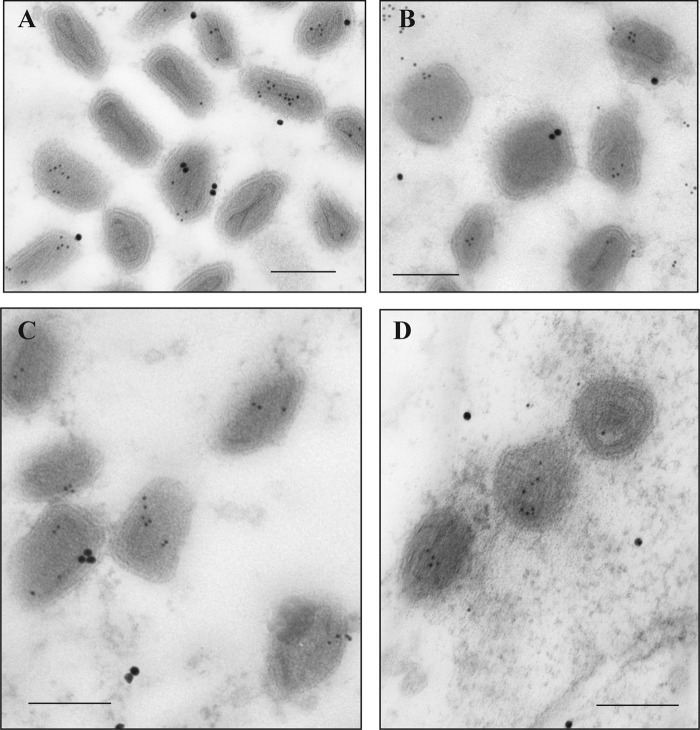
FIG 1.
Localization of vaccinia virus core proteins in the virions. Cells were infected at an MOI of 10 with vE6i in the presence of IPTG and incubated at 37°C. At 24 h postinfection, cells were processed for immunoelectron microscopy as described in Materials and Methods. Ultrathin sections were probed with antibodies against L4, A3, A4, and F17 (top) or only against L4 (bottom) and visualized in an electron microscope. Scale bar, 100 nm.
RESULTS
L4 protein is localized in the center of the virus core.
As described in the introduction, several characteristics of the L4 protein are consistent with a possible role as a nucleocapsid structural protein. Reasoning that a nucleocapsid protein should localize to the center of the virion, we compared the subvirion localization of L4 with several other major virion structural proteins using immunoelectron microscopy. Thin sections of virus-infected cells were probed with antibodies against the A3 (4b), A4 (p39), F17 (VP11), and L4 proteins. A3 and A4 are thought to be core wall proteins, and F17 is thought to be a lateral body protein (33–36). Figure 1 shows that antibodies against L4 decorate the center of the core, distinct from A3, A4, and F17, which have a more peripheral localization. The centralized localization of L4 reinforces our hypothesis that L4 is the structural protein of the vaccinia virus nucleocapsid.
Ets85 phenotype is due to an amino acid substitution in the L4R gene.
The Ets85 mutant has been mapped previously to the L4R gene; however, the mutation responsible for the thermosensitivity was not determined (24). In order to confirm the genotype of Ets85, we repeated the marker rescue, confirming that the mutant could be rescued only by the L4R gene (data not shown). To verify the precise mutation that renders Ets85 thermosensitive, we determined the complete nucleotide sequence of the L4R gene from Ets85. The sequencing revealed a missense mutation at nucleotide 412 (C412T) that led to a change of amino acid 138 from proline to serine (P138S). The sequence alignment represented in Fig. 2 shows that the proline-138 residue is highly conserved throughout the orthopoxviruses. In addition, secondary structure prediction using PredictProtein indicates that proline-138 is probably buried inside the protein structure and that a change from proline to serine has a high potential to affect the protein structure (25). Because proline and serine have different polarities (proline is nonpolar and serine is polar) as well as different structures, these differences probably account for the thermosensitivity of Ets85.
FIG 2.
Analysis of L4 sequence. The protein sequences were obtained from www.poxvirus.org and aligned using the software programs BioEdit and ClustalW Multiple. The gray box highlights residue 138 (proline) that is mutated in the mutant Ets85. Vaccinia virus (VACV) WR and variola, monkeypox, ectromelia, and cowpox viruses are representatives of the genus Orthopoxvirus; deerpox virus is a member of the genus Cervidpoxvirus; molluscum contagiosum virus is a member of the genus Molluscipoxvirus. Amino acid identity relative to the WR vaccinia virus is 98% for members of genus Orthopoxvirus, 63% for deerpox virus, and 43% for molluscum contagiosum. Asterisk (*), identical amino acid; colon (:), conserved substitutions; period (.), semiconserved substitutions.
Plaque phenotype and growth properties of Ets85.
To analyze the plaque phenotype of Ets85, we performed a plaque assay at permissive (31°C) and nonpermissive (39.7°C) temperatures. Under permissive conditions, Ets85 formed plaques that were similar in size to the wild-type plaques (Fig. 3A). However, under nonpermissive conditions, only the wild-type virus was able to form visible viral plaques after 7 days of infection.
FIG 3.
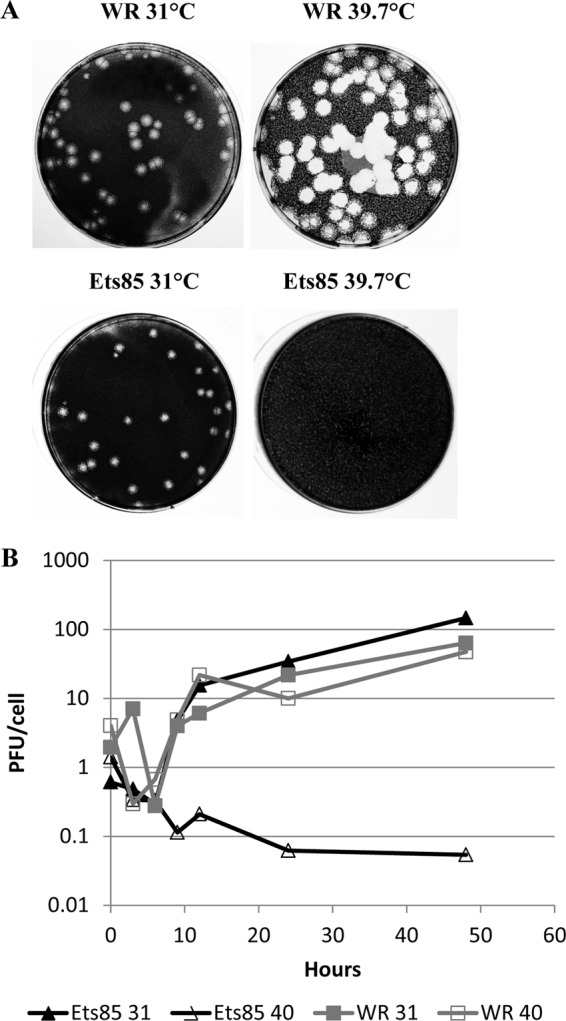
Characterization of the Ets85 mutant. (A) BSC-40 cells were infected with WR or Ets85 and incubated at 31°C or 39.7°C. After 7 days of infection, dishes were stained with crystal violet. (B) Cells were infected at an MOI of 10 with WR or Ets85 and incubated at the permissive or nonpermissive temperature. Samples were harvested at various times postinfection. Titers were determined by plaque assay at 31°C. The experiment was done three times. A representative experiment is shown.
The growth pattern of Ets85 was analyzed in a one-step growth experiment (Fig. 3B). We infected cells at an MOI of 10, and after incubation for various times postinfection at 31°C or 39.7°C, we harvested the infected cells and determined the virus yields by plaque assay. Figure 3B shows that wild-type virus was able to grow to normal titers irrespective of the temperature of incubation. In contrast, Ets85 was able to grow only when the infection proceeded at the low temperature. No infectious particles were produced at the high temperature after 48 h postinfection. Thus, the data presented in Fig. 3 corroborate the thermosensitivity of Ets85.
Accumulation of L4 protein during infection with Ets85.
In general, two fundamentally distinct mechanisms for thermosensitivity exist: a mutant protein may be rendered unstable and therefore be absent from the infection, or, alternatively, a mutation may cause a conformational change and loss of function without destabilizing the protein, in which case the mutant protein will persist during the infection. To discriminate between these alternatives, we analyzed the accumulation of L4 protein during infection. Cells were infected with WR or Ets85 at an MOI of 10 and incubated at permissive and nonpermissive temperatures, and samples taken at various times after infection were analyzed for the presence of the L4 protein by Western blotting (Fig. 4). During the WR infection, the L4 protein accumulated during the late phase of infection in similar quantities and with similar kinetics at both 31°C and 39.7°C. During an Ets85 infection, L4 protein accumulation was slightly reduced at both temperatures compared to the wild-type infection; however, the high temperature did not significantly destabilize L4. We can also infer from the data shown in Fig. 4 that the processing of the mutated L4 protein is slightly inhibited at 39.7°C compared to Ets85 at 31°C or to the wild type.
FIG 4.
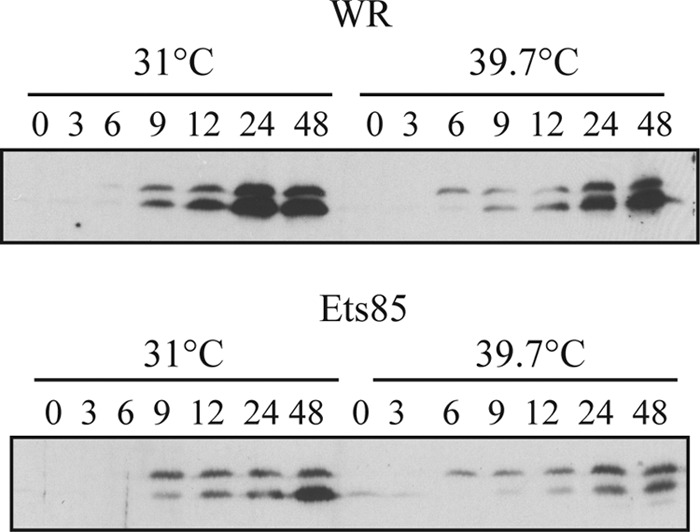
Accumulation of L4 during infection. Cells were infected with WR or Ets85 at an MOI of 10 and incubated at 31°C or 39.7°C. Samples were harvested at various times postinfection and analyzed by SDS-PAGE, followed by Western blotting using antibody against L4.
Protein synthesis and processing.
To analyze Ets85 gene expression throughout replication, we infected cells at an MOI of 10, pulse-labeled the cells with [35S]methionine at various times postinfection, and analyzed labeled proteins by SDS-PAGE and autoradiography. Figure 5 shows that for the wild-type infection, the pattern of protein synthesis was the same at both temperatures. Specifically, host protein synthesis apparent at 0 h postinfection was inhibited within 3 h, early viral protein synthesis was apparent at 3 h postinfection, and viral late protein synthesis commenced by 6 h postinfection and persisted through 24 h postinfection. The infection with Ets85 produced the same results as the wild-type infection at both temperatures, demonstrating that overall virus gene expression is not affected by the L4 mutation.
FIG 5.
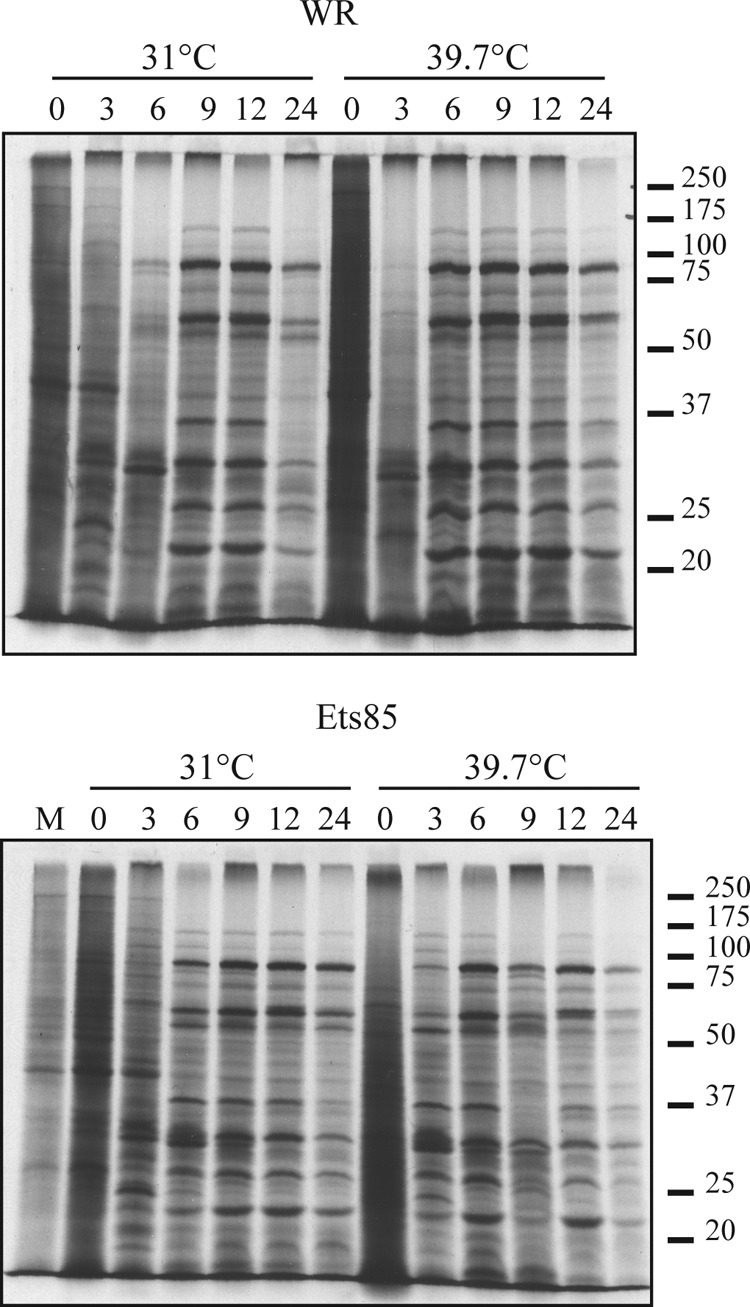
Viral protein synthesis during WR and Ets85 infection. Cells were infected at an MOI of 10 with WR or Ets85 and incubated at 31°C or 39.7°C. At various times postinfection, cells were pulse-labeled for 30 minutes with [35S]methionine. Samples were then harvested and analyzed by SDS-PAGE and autoradiography. Molecular masses in kilodaltons are indicated on the right. The numbers at the top indicate the time (in hours) postinfection.
Protein processing is a major step during vaccinia virus morphogenesis, and it is required for the transition from immature virions to mature, infectious virus. Because we observed a slight inhibition of L4 processing in infections done with Ets85 at a high temperature, we wanted to determine whether the processing of other proteins, specifically p4a/4a and p4b/4b, is also affected under these conditions. Cells infected at an MOI of 10 were incubated at 31°C or 39.7°C and pulse-labeled with [35S]methionine at 8 h postinfection. Some infections were harvested immediately after the pulse while others were incubated in the presence of excess unlabeled methionine for various times at the low or high temperature. Samples were analyzed by SDS-PAGE and autoradiography. Figure 6 shows that p4a and p4b were processed normally during both virus infections at both temperatures. These results were confirmed using Western blot assays with antibodies against p4a and p4b (data not shown). The quantification of the bands corresponding to p4a/4a and p4b/4b showed that the ratio of unprocessed to processed forms was the same under all conditions (data not shown). However, the quantification of the bands corresponding to unprocessed and processed forms of L4 showed 2- to 3-fold less processing in infections with the mutant at the nonpermissive temperature than with the wild type at both temperatures or than Ets85 at 31°C. The same result was obtained when the Western blot with anti-L4 (Fig. 4) was quantified. We conclude that L4 protein processing is slightly decreased in mutant infections done at the high temperature.
FIG 6.
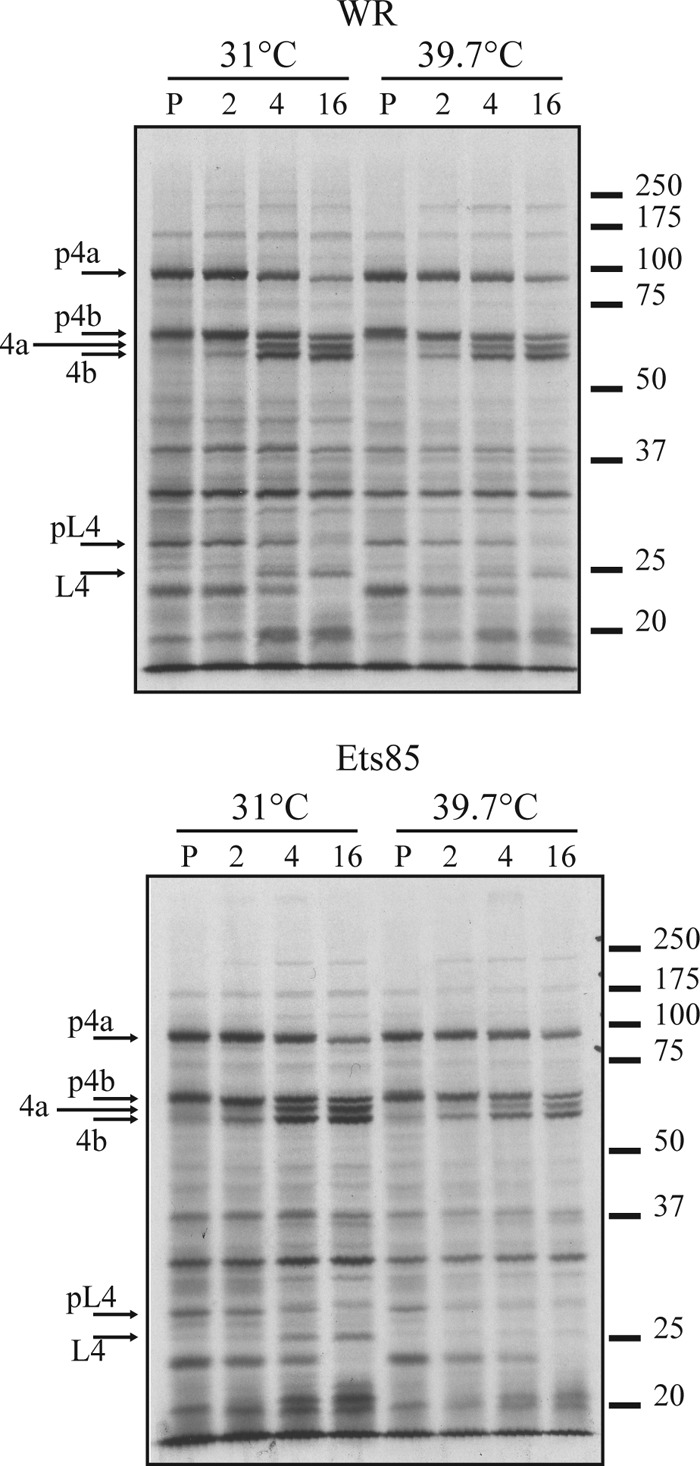
Analysis of protein processing during infection. Cells were infected at an MOI of 10 with WR or Ets85 and incubated at 31°C or 39.7°C. At 8 h postinfection, cells were pulse-labeled for 30 minutes with [35S]methionine. Some cells were harvested after the 30-min pulse (P), and some were incubated at 31°C or 39.7°C for 2, 4, or 16 h in medium lacking [35S]methionine. Samples were then harvested and analyzed by SDS-PAGE and autoradiography. Molecular masses in kilodaltons are indicated on the right.
Virus morphogenesis.
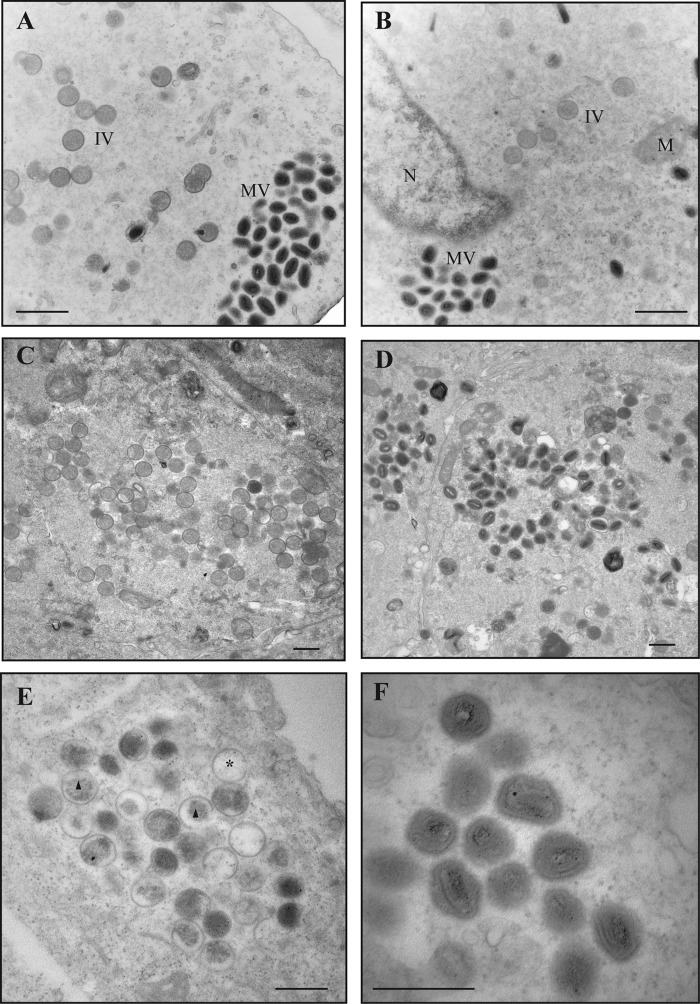
Because Ets85 produces a normal complement of late proteins under nonpermissive conditions, we hypothesize that the L4 mutation affects virus assembly or infectivity. To analyze virus assembly, we infected cells at an MOI of 10 with WR or Ets85, incubated the cells for 24 h at 31°C or 39.7°C, and prepared ultrathin sections for analysis by transmission electron microscopy using conventional fixation techniques, as described in Materials and Methods. Figure 7A, C, and D show that the wild-type virus produced normal immature and mature particles at both temperatures. The same normal phenotype was observed during Ets85 infections done at 31°C (Fig. 7B). However, in infections done with Ets85 at the high temperature, most of the particles seen in the cytoplasm were defective immature virions (IVs) (Fig. 7E). The viroplasm inside the IVs was not homogeneous as in wild-type IVs, and in many cases a gap was observed between the viroplasm and the IV membrane (Fig. 7E, triangle). Also, a high percentage of the IVs appeared empty (Fig. 7E, star). Some IVs were nevertheless able to develop into MVs. However, these MVs presented an aberrant structure (Fig. 7F). The defective mature particles contain a malformed core that is not dumbbell shaped and a core wall with the palisade layer not discernible.
FIG 7.
Viral morphogenesis: conventional fixation. Cells were infected with WR or Ets85 at an MOI of 10 and incubated at 31°C or 39.7°C. At 24 h postinfection, cells were fixed with 2% glutaraldehyde and 1% osmium tetroxide and processed for transmission electron microscopy as described in Materials and Methods. (A) WR at 31°C. (B) Ets85 at 31°C. (C and D) WR at 39.7°C. (E and F) Ets85 at 39.7°C. MV, mature virus particles; IV, immature virus particles; M, mitochondria; N, nucleus; asterisk, empty IV; triangle, IV with nonhomogeneous viroplasm. Scale bar, 500 nm.
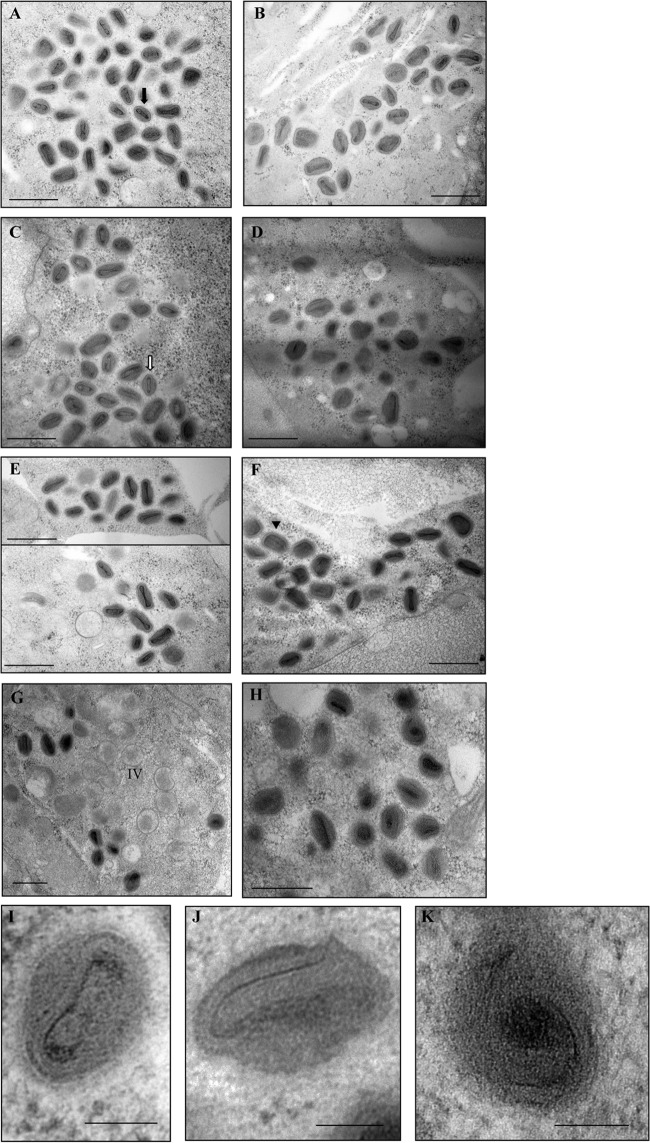
Our primary interest is in the role of L4 in the nucleocapsid structure. However, we cannot achieve good preservation of the nucleocapsid using conventional electron microscopy protocols for sample fixation and dehydration. In order to visualize the nucleocapsid in a consistent fashion, techniques that improve preservation of ultrastructure are necessary. Therefore, we turned to high-pressure freezing followed by freeze-substitution to analyze the nucleocapsid structure in the mutant particles. Furthermore, we were interested in analyzing whether the assembly of Ets85 would resemble the previously determined phenotype for the inducible mutant in L4R (vL4i), namely, the production of normal-looking mature particles (12). We infected cells at an MOI of 10 with WR, Ets85, or vL4i and incubated the cells under the permissive or nonpermissive conditions. At 24 h postinfection, we cryo-immobilized the cells using a high-pressure freezing system. The ice from frozen cells was replaced by a solution of 20% epoxy fixative in acetone during the freeze-substitution step. Following freeze-substitution, cells were poststained with tannic acid and osmium tetroxide in acetone as described in Materials and Methods. Subsequent steps were similar to the conventional protocol for electron microscopy. In all of the wild-type infections, we were able to visualize the nucleocapsid in several particles (Fig. 8A, B, and E). We observed the same phenotype in infections with the mutants Ets85 and vL4i under permissive conditions (Fig. 8C and F). However, both mutants produced mature particles with similar aberrant structures under nonpermissive conditions (Fig. 8D, G, and H). These particles have an electron-dense structure reminiscent of a core; however, the core wall palisade is not discernible, and the core wall is not shaped as a dumbbell surrounding the electron-dense part of the core. Instead, in some views the core wall seems to form a spiral around the electron-dense portion of the core (Fig. 8I, J, and K). More importantly, it was not possible to detect a normal nucleocapsid in any of the abnormal particles. Thus, the phenotype of the L4 mutants as visualized by electron microscopy is consistent with a role for L4 in the nucleocapsid structure. The data also suggest a relationship between the nucleocapsid and the core wall.
FIG 8.
Viral morphogenesis: high-pressure freezing/freeze-substitution. Cells were infected with WR, Ets85, or vL4i at an MOI of 10. For Ets85, cells were incubated at 31°C or 39.7°C. For vL4i, cells were incubated at 37°C in the presence or absence of 200 μM IPTG. At 24 h postinfection, cells were processed by high-pressure freezing and freeze-substitution as described in Materials and Methods. Ultrathin sections were visualized in an electron microscope. (A) WR at 31°C. (B) WR at 39.7°C. (C) Ets85 at 31°C. (D) Ets85 at 39.7°C. (E) WR at 37°C. (F) vL4i in the presence of IPTG. (G and H) vL4i in the absence of IPTG. (I) Higher magnification of a wild-type mature particle. (J and K) Higher magnification of mature particles produced in mutant infections under nonpermissive conditions. Arrows and arrowheads indicate mutually perpendicular sections of representative particles in which the nucleocapsid can be visualized. Filled arrow, transverse section (A); open arrow, sagittal section (C); arrowhead, coronal section (F). Scale bars, 500 nm (A to H) and 100 nm (I to K).
Protein and DNA content of mutant virus particles.
The abnormal structure of viral particles formed under nonpermissive conditions might be explained by the absence of L4 from the particles, by the presence of a mutated form of L4, or by the absence of other viral proteins. In order to discriminate among these alternatives, we analyzed the protein composition of both immature and mature wild-type and mutant particles. Attempts to purify viral particles produced during infections with Ets85 at the high temperature were unsuccessful. The apparent yield of viral particles for the mutant under nonpermissive conditions as judged by light scattering was approximately 10% of the yield obtained with wild-type infections at both temperatures or with mutant infections at the low temperature. The reasons for the low yields are unknown; however, given the previous observation that particles lacking L4 are relatively fragile (18), it seems most likely that the aberrant virions produced at the high temperature are also fragile to the extent that they cannot be efficiently purified. We therefore used immunoelectron microscopy to characterize the viral particles. We infected cells with WR or Ets85 at an MOI of 10. After 24 h of infection, cells were prepared for immunoelectron microscopy, and ultrathin sections were probed with antibodies against virion structural proteins (Fig. 9). We observed that both wild-type and mutant IVs and mature particles contain both L4 (Fig. 9A to D) and A3 (Fig. 9E to H). We tested additional virus core proteins (A10, A4, and F17), and all of them were present within particles formed at either temperature during both wild-type and Ets85 infections (data not shown). As noted previously, Ets85 infection at 39.7°C produces a large amount of immature virions that appear empty. Consistent with this appearance, the empty particles do not label with any of the antibodies tested (Fig. 9D and H and data not shown).
We hypothesize that the viral DNA is contained within the nucleocapsid and that L4 is the nucleocapsid scaffold protein. Because the mutated L4 protein causes a defect during assembly at a high temperature and L4 is able to bind DNA, we were interested to determine whether the defective particles contain viral DNA. For this, we used an antibody against double-stranded DNA in an immunogold labeling experiment. In this experiment, we also probed with anti-D13 antibody to ensure that the defective electron-dense particles were abnormal mature particles and not immature virions. D13 protein is present only during the immature stage, being released during virus maturation (37). Importantly, labeling with anti-DNA antibody showed a very specific pattern, with robust labeling of cell nuclei and viral factories (data not shown). This pattern was distinct from labeling observed with all other antibodies, giving us confidence in the specificity of the anti-DNA antibody used. Figure 10 shows that the defective particles formed at a high temperature contain viral DNA. Also, because we do not see significant labeling with D13, we conclude that these particles are abnormal mature particles. Immature particles labeled for D13 under all conditions (data not shown).
Taken together, these data imply that the defect observed with Ets85 at the nonpermissive temperature is due to the presence in the particles of a mutated form of L4.
DISCUSSION
The motivation for the present study was to gain insight into the structure of the vaccinia virus nucleocapsid. Determining the structure and biological significance of vaccinia virus nucleocapsid will help elucidate how viral proteins are organized in the virion and how viral early transcription occurs inside the virus core. By analogy with other virus nucleocapsids, we postulated that the vaccinia virus nucleocapsid contains a major structural protein. The best candidate for this role is the protein L4 because of its abundance, its localization in the center of the core, and its ability to bind DNA. In order to test the hypothesis that L4 is a structural protein of the vaccinia virus nucleocapsid, we analyzed two mutants in the L4R gene. Both mutants produce similar abnormal mature particles exhibiting a defective core wall and lacking the nucleocapsid. The lack of a nucleocapsid is consistent with a requirement for L4 in this structure. The unexpected observation that the core wall is malformed suggests an interaction between nucleocapsid and core wall and indicates a higher degree of complexity of the nucleocapsid than predicted.
The abnormality of the viral particles produced under nonpermissive conditions results from a loss of function of L4 and not from a complete absence of L4 or other viral proteins. The reasoning behind this conclusion is that the temperature-sensitive and the inducible mutant present equivalent phenotypes regardless of the total absence of L4 (in the case of vL4i infections) or the presence of a mutated form of L4 (in the case of Ets85 infections). Furthermore, all proteins tested as well as the viral DNA were present in the viral particles formed at high temperature. These data are in accordance with previously published results with vL4i showing that virions completely lacking L4 contain a normal complement of proteins (18).
The absence of a nucleocapsid, coupled with the abnormality of the core wall, suggests that these structures may be connected. Our previous work has shown that the absence of viral transcription enzymes from virions results in the lack of a nucleocapsid; however, the core wall is not affected (8). Therefore, disruptions of nucleocapsid structure do not necessarily impact the core wall, suggesting that the defect in the core wall observed in the present study is directly linked to the absence of L4 function (and not the nucleocapsid) and that L4 may be important for an association between the nucleocapsid and core wall. The major protein of the inner core wall is thought to be A3 (35; N. Moussatche, unpublished data); therefore, it would be interesting to determine if an interaction between L4 and A3 exists.
The defect in core structure probably explains the block in virus early transcription reported previously with the inducible mutant of L4 (18). Correspondingly, a thermosensitive mutant on the A3L gene (Cts8) presents a defect in core structure and a block in early transcription despite the fact that all enzymes are present and functional in the virus particles (30). In addition, mutants in the H1R (phosphatase), I8R (RNA helicase), and E8R (unknown function) genes produce mature virus particles that are indistinguishable from wild-type virions in structure and protein and DNA content but that are unable to perform core-directed early transcription (29, 38, 39). Thus, the present study reinforces the observation that changes in core structure are detrimental to viral early transcription even when these changes are subtle and not readily visualized by conventional electron microscopy. These facts, however, do not conflict with the notion that L4 might have a direct role in virus transcription. In fact, some evidence suggests that L4 might be important for early transcription by stimulating the virus RNA helicase activity or for transport of viral RNAs to accumulation areas (17, 40).
Our results are consistent with but do not definitively demonstrate that L4 is the structural protein of the nucleocapsid. However, the data prove that L4 is important for nucleocapsid formation and add the new perspective that the core wall and nucleocapsid may not be entirely separate structures as previously thought. Our data also strengthen the value of high-pressure freezing for studying vaccinia virus structure. Studies applying high-pressure freezing to mutants that are not able to transcribe early genes but produce normal-looking mature particles, as visualized by conventional electron microscopy, have the potential to offer a better understanding of the vaccinia virus nucleocapsid. Moreover, analysis of the nucleocapsid of mutants with a defective core wall could clarify the interaction between the core wall and nucleocapsid. These mutant studies combined with structural data acquired from cryo-electron microscopy, which permits examination of the fine structure of the virus core, will be vital for a comprehensive understanding of vaccinia virus nucleocapsid structure and function.
ACKNOWLEDGMENTS
We thank Karen Kelley and the University of Florida ICBR Electron Microscopy Core Laboratory for excellent advice and technical assistance and Nicole Kay, Travis Weber, and Agnieszka Boron for technical support.
This work was supported by NIH grant R01 AI055560 to R.C.
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1.Moss B. 2007. Poxviridae: the viruses and their replication, 2906–2945 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Dales S, Siminovitch L. 1961. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475–503. 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns J. 1960. The initiation of vaccinia infection. Virology 11:603–623. 10.1016/0042-6822(60)90103-3. [DOI] [PubMed] [Google Scholar]
- 4.Vanslyke JK, Lee P, Wilson EM, Hruby DE. 1993. Isolation and analysis of vaccinia virus previrions. Virus Genes 7:311–324. 10.1007/BF01703388. [DOI] [PubMed] [Google Scholar]
- 5.Ansarah-Sobrinho C, Moss B. 2004. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 78:6335–6343. 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters D, Mueller G. 1963. The fine structure of the DNA-containing core of vaccinia virus. Virology 21:267–269. [DOI] [PubMed] [Google Scholar]
- 7.Hyde JM, Peters D. 1971. The organization of nucleoprotein within fowlpox virus. J. Ultrastruct. Res. 35:626–641. 10.1016/S0022-5320(71)80015-1. [DOI] [PubMed] [Google Scholar]
- 8.McFadden BD, Moussatche N, Kelley K, Kang BH, Condit RC. 2012. Vaccinia virions deficient in transcription enzymes lack a nucleocapsid. Virology 434:50–58. 10.1016/j.virol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WP, Bauer WR. 1988. Purification and characterization of vaccinia virus structural protein VP8. Virology 167:578–584. 10.1016/0042-6822(88)90120-1. [DOI] [PubMed] [Google Scholar]
- 10.Bayliss CD, Smith GL. 1997. Vaccinia virion protein VP8, the 25 kDa product of the L4R gene, binds single-stranded DNA and RNA with similar affinity. Nucleic Acids Res. 25:3984–3990. 10.1093/nar/25.20.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung CS, Chen CH, Ho MY, Hwang CY, Liao CL, Chang W. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127–2140. 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcock D, Smith GL. 1994. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology 202:294–304. 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- 13.Sarov I, Joklik WK. 1972. Characterization of intermediates in the uncoating of vaccinia virus DNA. Virology 50:593–602. 10.1016/0042-6822(72)90410-2. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen K, Snijder EJ, Schleich S, Roos N, Griffiths G, Locker JK. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525–3536. 10.1128/JVI.74.8.3525-3536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B, Rosenblum EN. 1973. Letter: protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J. Mol. Biol. 81:267–269. 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 16.Yang WP, Kao SY, Bauer WR. 1988. Biosynthesis and post-translational cleavage of vaccinia virus structural protein VP8. Virology 167:585–590. 10.1016/0042-6822(88)90121-3. [DOI] [PubMed] [Google Scholar]
- 17.Bayliss CD, Wilcock D, Smith GL. 1996. Stimulation of vaccinia virion DNA helicase I8R, but not A18R, by a vaccinia core protein L4R, an ssDNA binding protein. J. Gen. Virol. 77:2827–2831. 10.1099/0022-1317-77-11-2827. [DOI] [PubMed] [Google Scholar]
- 18.Wilcock D, Smith GL. 1996. Vaccinia virions lacking core protein VP8 are deficient in early transcription. J. Virol. 70:934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szajner P, Jaffe H, Weisberg AS, Moss B. 2003. Vaccinia virus G7L protein Interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 77:3418–3429. 10.1128/JVI.77.6.3418-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercer J, Traktman P. 2005. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J. Virol. 79:7146–7161. 10.1128/JVI.79.11.7146-7161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd O, Turner PC, Moyer RW, Condit RC, Moussatche N. 2010. The E6 protein from vaccinia virus is required for the formation of immature virions. Virology 399:201–211. 10.1016/j.virol.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd O, Strahl AL, Rodeffer C, Condit RC, Moussatche N. 2010. Temperature-sensitive mutant in the vaccinia virus E6 protein produce virions that are transcriptionally inactive. Virology 399:221–230. 10.1016/j.virol.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ensinger MJ. 1982. Isolation and genetic characterization of temperature-sensitive mutants of vaccinia virus WR. J. Virol. 43:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ensinger MJ, Weir JP, Moss B. 1985. Fine structure marker rescue of temperature-sensitive mutations of vaccinia virus within a central conserved region of the genome. J. Virol. 56:1027–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B. 2014. PredictProtein–an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 42:W337–W343. 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monaghan P, Cook H, Hawes P, Simpson J, Tomley F. 2003. High-pressure freezing in the study of animal pathogens. J. Microsc. 212:62–70. 10.1046/j.1365-2818.2003.01245.x. [DOI] [PubMed] [Google Scholar]
- 27.McDonald KL, Auer M. 2006. High-pressure freezing, cellular tomography, and structural cell biology. Biotechniques 41:137–143. 10.2144/000112226. [DOI] [PubMed] [Google Scholar]
- 28.Condit RC, Motyczka A. 1981. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology 113:224–241. 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 29.Kato SE, Condit RC, Moussatche N. 2007. The vaccinia virus E8R gene product is required for formation of transcriptionally active virions. Virology 367:398–412. 10.1016/j.virol.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato SE, Strahl AL, Moussatche N, Condit RC. 2004. Temperature-sensitive mutants in the vaccinia virus 4b virion structural protein assemble malformed, transcriptionally inactive intracellular mature virions. Virology 330:127–146. 10.1016/j.virol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 31.Matsko N, Mueller M. 2004. AFM of biological material embedded in epoxy resin. J. Struct. Biol. 146:334–343. 10.1016/j.jsb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Castro AP, Carvalho TM, Moussatche N, Damaso CR. 2003. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 77:9052–9068. 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682–3695. 10.1128/JVI.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 35.Ichihashi Y, Oie M, Tsuruhara T. 1984. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J. Virol. 50:929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt FI, Bleck CK, Reh L, Novy K, Wollscheid B, Helenius A, Stahlberg H, Mercer J. 2013. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 4:464–476. 10.1016/j.celrep.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 37.Bisht H, Weisberg AS, Szajner P, Moss B. 2009. Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis. J. Virol. 83:9140–9150. 10.1128/JVI.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross CH, Shuman S. 1996. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J. Virol. 70:8549–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu K, Lemon B, Traktman P. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69:7823–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallardo M, Schleich S, Krijnse LJ. 2001. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs into distinct cytoplasmic structures. Mol. Biol. Cell 12:3875–3891. 10.1091/mbc.12.12.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]