ABSTRACT
Many viruses utilize cell adhesion molecules of the immunoglobulin superfamily as receptors. In particular, viruses of different classes exploit nectins. The large DNA viruses, herpes simplex and pseudorabies viruses, use ubiquitous nectins 1 and 2. The negative-strand RNA virus measles virus (MeV) uses tissue-specific nectin-4, and the positive-strand RNA virus poliovirus uses nectin-like 5 (necl-5), also known as poliovirus receptor. These viruses contact the BC, C′C″, and FG loops on the upper tip of their receptor's most membrane-distal domain. This location corresponds to the newly defined canonical adhesive interface of nectins, but how viruses utilize this interface has remained unclear. Here we show that the same key residues in the BC and FG loops of nectin-4 govern binding to the MeV attachment protein hemagglutinin (H) and cell entry, nectin-4 homodimerization, and heterodimerization with nectin-1. On the other hand, residues in the C′C″ loop necessary for homo- and heterotypic interactions are dispensable for MeV-induced fusion and cell entry. Remarkably, the C′C″ loop governs dissociation of the nectin-4 and H ectodomains. We provide formal proof that H can interfere with the formation of stable nectin-1/nectin-4 heterodimers. Finally, while developing an alternative model to study MeV spread, we observed that polarized primary pig airway epithelial sheets cannot be infected. We show that a single amino acid variant in the BC loop of pig nectin-4 fully accounts for restricted MeV entry. Thus, the three loops forming the adhesive interface of nectin-4 have different roles in supporting MeV H association and dissociation and MeV-induced fusion.
IMPORTANCE Different viruses utilize nectins as receptors. Nectins are immunoglobulin superfamily glycoproteins that mediate cell-cell adhesion in vertebrate tissues. They interact through an adhesive interface located at the top of their membrane-distal domain. How viruses utilize the three loops forming this interface has remained unclear. We demonstrate that while nectin-nectin interactions require residues in all three loops, the association of nectin-4 with the measles virus hemagglutinin requires only the BC and FG loops. However, we discovered that residues in the C′C″ loop modulate the dissociation of nectin-4 from the viral hemagglutinin. Analogous mechanisms may support cell entry of other viruses that utilize nectins or other cell adhesion molecules of the immunoglobulin superfamily as receptors.
INTRODUCTION
Measles virus (MeV) is a highly contagious and immunosuppressive virus that still causes more than 120,000 deaths in developing countries each year. Recently, the virus has been responsible for small epidemics throughout North America and Europe, where vaccination coverage has waned (1, 2). After host entry and dissemination in lymphatic cells through the receptor signaling lymphocytic activation molecule (SLAM/CD150) (3, 4), rapid spread in respiratory epithelia depends on the adherens junction protein nectin-4 (5, 6). Vaccine strain MeV enters cells also through the ubiquitous membrane cofactor protein (MCP/CD46) (7, 8). The tetrameric MeV attachment protein hemagglutinin (H) binds these receptors and mediates virus entry in concert with the fusion protein trimers (3, 9–12). Structural analyses of H in complex with either receptor have identified overlapping surfaces buried after complex formation (13–15). Functional studies have further defined the individual residues governing the receptor interactions, revealing remarkable differences (16).
Nectins are type I transmembrane proteins of the immunoglobulin (Ig) superfamily that initiate cell-cell adhesion in vertebrate tissues (17, 18). Four family members, nectin-1 through nectin-4 (also known as poliovirus receptor-like proteins 1 to 4 or PVRL1-4), are well conserved among vertebrate species (19). Nectins mediate calcium-independent cell-cell adhesion in many tissues, including epithelia. They are localized to the subapical region of lateral membranes in columnar epithelial cells and are one of the main constituents of the adherens junction (20). Nectins are characterized by an ectodomain composed of one membrane-distal Ig variable-like domain (V), two Ig constant-like domains, a transmembrane segment, and a cytosolic tail (17). The V domain has the characteristic Ig fold, defined by two opposing antiparallel β sheets connected in a unique manner (21, 22). The core of the Ig fold is formed by four β-strands (B, C, E, and F) augmented with additional β-strands (A, C′, C″, D, and G). Recent functional and structural analyses have identified an adhesive interface involved in the homophilic interaction of nectins, mapping to the B, C, C′, C″, F, and G strands of the V domain (20, 23, 24).
Remarkably, viruses of different classes exploit nectins for cell entry. Large DNA viruses like herpes simplex virus (HSV) and pseudorabies virus use ubiquitous nectins 1 and 2 (25–27), and the small RNA virus poliovirus uses nectin-like 5 (necl-5), also known as PVR (poliovirus receptor) (28). Recently, the cocrystal structure of the herpes simplex virus attachment protein gD in complex with nectin-1 became available (29, 30). Interestingly, most of the nectin-1 residues involved in gD binding (Fig. 1, blue residues) are part of the adhesive interface (Fig. 1, residues in boxes) (29). In addition, early functional data (31, 32) in conjunction with recent structural analyses of nectins 1 to 4 and necl-5 homodimers (20) suggest that targeting of the adhesive interface could be the basis for the exploitation of nectins as viral receptors.
FIG 1.

Sequences and interactions of the Ig V domain of nectins and necl-5. The protein sequences of human nectin-4 (Q96NY8), nectin-1 (Q15223), nectin-2 (Q92692), nectin-3 (Q9NQS3), and necl-5 (P15151) from the UniProt database (http://www.uniprot.org/) were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Residues in boxes define the canonical nectin adhesive interface (20). Colored residues may interact with the viral attachment proteins of MeV (nectin-4, green), HSV (nectin-1 and nectin-2, blue and orange, respectively), or poliovirus (necl-5, pink) (29, 32). The β-strands A, B, C, C′, C″, D, E, F, and G (20) are indicated with arrows above the nectin-4 sequence.
Taking advantage of the available structure of the nectin-4 homodimer (20) and the MeV H-nectin-4 complex (15), we assessed how MeV H interacts with the nectin-4 adhesive interface. We show that the three loops forming this interface have different roles in supporting MeV association and dissociation and MeV-induced membrane fusion and cell entry. We discovered that a single residue variant in the nectin-4 BC loop restricts MeV entry in pig airway epithelial (PAE) cells. We also address the question of the heterophilic nectin interactions. All nectins can form heterodimers, and certain heterophilic interactions are much stronger than the homophilic interactions. For example, nectin-4/nectin-1 interactions are at least 1,000 times more stable than the nectin-4 homodimers (20, 33, 34). We show that MeV H can compete for the formation of nectin-4/nectin-1 trans-heterodimers.
MATERIALS AND METHODS
Cells.
Vero, VerohSLAM (kindly provided by Y. Yanagi), VerohN4 (kindly provided by C. Richardson), 293, and Cos-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; HyClone, South Logan, UT) supplemented with 10% fetal bovine serum (FBS). G418 at a concentration of 0.5 mg/ml was added to the medium of VerohSLAM and VerohN4 cells. Chinese hamster ovary (CHO) cells were maintained in RPMI medium (Corning, Manassas, VA) supplemented with 10% FBS and 0.5 mg/ml of nonessential amino acids. CHO-N1 and CHO-N4 cells were grown in RPMI medium complemented with 0.5 mg/ml of G418. Baby hamster kidney cells expressing stably the T7 polymerase (BHK-T7) were maintained in DMEM supplemented with 10% FBS and 0.1 mg/ml G418. All the cell lines were kept at 37°C with 5% CO2.
Viruses and infections.
To generate MV323LucII, the firefly luciferase open reading frame (ORF) was transferred into an infectious MV (i.e., recombinant MeV) genome in an additional transcription unit upstream of N, using the MluI and AatII restriction sites (35, 36). MV323GFP expressing green fluorescent protein (GFP) has already been described (37). Recombinant viruses were rescued as described previously (38). To prepare virus stocks, VerohSLAM cells were infected at a multiplicity of infection (MOI) of 0.01 and then incubated at 37°C for 36 to 48 h. To harvest virus, cells were scraped into Opti-MEM I reduced-serum medium (Life Technologies, Grand Island, NY) and freeze-thawed twice. Titers were determined by 50% tissue culture infective dose (TCID50) titration on VerohSLAM cells. For infections, cells were inoculated at the indicated MOIs for 1 h at 37°C in Opti-MEM I reduced-serum medium, washed, and incubated in 5% FBS medium at 37°C with 5% CO2 for the indicated times. For infections of N4-expressing cells in the presence of soluble N4 ectodomains, cells were infected with MV323LucII at an MOI of 0.2 in the presence of 10 μg/ml of the corresponding recombinant soluble nectin-4 ectodomain. Luciferase expression was determined at 16 h postinfection by using the Steady-Glo luciferase activity system (Promega Corporation, Madison, WI) and a Topcount NXT luminometer (Packard Instrument Company, Meriden, CT).
Plasmids and mutagenesis.
A codon-optimized ORF of human nectin-4 tagged by a carboxy-terminal (C-ter) FLAG epitope was cloned into a pCG vector. The plasmid coding for a C-ter FLAG-tagged nectin-1 was obtained by subcloning of the nectin-1 ORF from the pLX1.12 plasmid (39). Plasmids pCFR1 and pCFR4, encoding the full-length soluble ectodomain of nectin-1 or nectin-4 fused to the human IgG1 Fc fragment, were described elsewhere (34). The H and F open reading frames of vaccine MeV were cloned into a pCG vector (16). To clone the pig nectin-4 (pN4), RNA was extracted from cultures of pig airway epithelial cells using the RNeasy kit (Qiagen, Valencia, CA). A reverse transcription step using the Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) followed by a PCR using specific primers (sense, 5′-ATGCCTCTGTCCCTGGGAGCC-3′; antisense, 5′-TCAGACCAGGTGCCCCCGC-3′; antisense FLAG, 5′-TTATCACTTGTCGTCATCGTCTTTGTAATCGACCAGGTGCCCCCGC-3′) allowed amplification of the pN4 cDNA and addition of the FLAG-epitope sequence. The pN4 cDNA encoding a C-ter FLAG-tagged protein was cloned in the pCG vector. For all these plasmids, desired mutations were introduced by using the QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) protocol. Introduction of the mutations was confirmed by sequencing.
Entry assay.
To measure viral entry through mutated receptors, Vero cells were transfected with the pCG vectors expressing the indicated N4 mutants by using Lipofectamine 2000 (Life Technologies, Grand Island, NY). Forty-eight hours posttransfection, cells were infected with MV323LucII at an MOI of 0.2. Luciferase expression was determined at 16 h postinfection by using the Steady-Glo luciferase activity system (Promega Corporation, Madison, WI) and a Topcount NXT luminometer (Packard Instrument Company, Meriden, CT). In parallel, cell surface and total expression of the recombinant N4 were analyzed by flow cytometry and immunoblotting at 48 h posttransfection. For flow cytometry, cells were detached using Versene (Life Technologies, Grand Island, NY) and then incubated in fluorescence-activated cell sorter (FACS) buffer (1× phosphate-buffered saline [PBS], 2% FBS, 0.1% NaN3) with primary antibodies for 1 h at 4°C, followed by an incubation of 45 min at 4°C with phycoerythrin (PE)-conjugated secondary antibodies. The cells were fixed in 2% paraformaldehyde and read by FACSCalibur (BD Biosciences, San Jose, CA). The results were analyzed by FlowJo software (Tree Star Inc., Ashland, OR). For immunoblotting, cells were lysed in coimmunoprecipitation (co-IP) buffer (10 mM HEPES [pH 7.4], 50 mM Na-Pyro-PO4, 50 mM Na-F, 50 mM NaCl2, 5 mM EDTA, 5 mM EGTA, 100 μM sodium vanadate, 1% Triton X-100) 48 h posttransfection. The protein samples were resolved on 4 to 15% gradient SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes (PVDF; Millipore, Billerica, MA), and then probed with horseradish peroxidase (HRP)-conjugated monoclonal antibodies FLAG-M2-HRP and β-actin-HRP (Sigma-Aldrich, St. Louis, MO). Membranes were visualized using Supersignal West Dura chemiluminescent substrate (Thermo Scientific, Rockford, IL) and autoradiography (Denville Scientific, Metuchen, NJ).
Production and purification of recombinant soluble proteins.
Cos-7 cells were transfected with pCFR1 and pCFR4 encoding the full-length nectin-1 and nectin-4 ectodomains (wild type or mutants) using X-tremeGENE 9 DNA transfection reagent (Roche). Supernatants were harvested at days 5 and 7 posttransfection, from which proteins were purified through their Fc tag using protein A chromatography (Pierce). Purification was monitored by Coomassie Easy Blue (Life Technologies) SDS-PAGE and immunoblotting using human IgG Fc HRP-conjugated antibodies (Bethyl Laboratories, Montgomery, TX). Protein concentrations were determined by enzyme-linked immunosorbent assay (ELISA) and spectrophotometry. The soluble H protein ectodomain was expressed and purified as described previously (11).
Surface plasmon resonance.
The receptor binding kinetics of the mutant nectin-4 ectodomains to the MeV H protein were determined essentially as described previously (11). Briefly, the interactions of the soluble nectin-4 ectodomains with the soluble H protein ectodomain were monitored by using a BIAcore T200 instrument (GE Healthcare, Pittsburgh, PA). HBS-EP+ (GE Healthcare, Pittsburgh, PA) buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.05% [vol/vol] surfactant P20) was used for all protein dilutions and as the running buffer. The FLAG-tagged H protein was captured onto the CM5 sensor chip surface by an immobilized anti-FLAG antibody (Sigma, St. Louis, MO). To monitor receptor binding, different concentrations of soluble receptors (0.05 μM to 0.8 μM) were injected over the captured MeV H proteins as indicated. The soluble receptors were injected over the CM5 sensor chip surfaces either exhibiting or lacking captured H protein. The surfaces lacking captured H served as control surfaces for nonspecific receptor binding. The antibody surface was regenerated by injection of 50 mM citric acid (pH 3.0) for 60 s followed by 10 mM glycine (pH 3.0) for 60 s, with little change in the activity of the monoclonal antibody surface monitored. The curve-fitting function of Biacore T200 Evaluation software (version 1.0) was used to fit rate equations derived from the 1:1 Langmuir binding model to the experimental data. The equilibrium dissociation constant (KD) was determined from the kinetic rate constants kon (association) and koff (dissociation).
Coimmunoprecipitation.
Cells were transfected with plasmids encoding the corresponding FLAG-tagged nectin-4 (wild type or mutant, as indicated) or nectin-1. Forty-eight hours posttransfection, cells were lysed in co-IP buffer and cleared lysates were incubated with an anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO). Immunoprecipitates were then incubated with the corresponding soluble nectin ectodomains. Coimmunoprecipitates were eluted and analyzed by SDS-PAGE followed by immunoblotting using monoclonal antibodies FLAG-M2-HRP (Sigma-Aldrich, St. Louis, MO) and human IgG Fc HRP-conjugated antibodies (Bethyl).
Competition binding assay.
CHO-N1 cells expressing human nectin-1 were incubated with 1 μg of soluble nectin-4 ectodomain in the presence of 10 μg/μl of an antibody directed to the V domain of nectin-1 (R1.302), or an antibody directed to the V domain of nectin-4 (N4.61), or an antibody directed to a C domain of nectin-4 (N4.40). Additional competition experiments included a soluble H ectodomain (sH) or 10 μg/μl of an sH in the presence of 100 μg/μl of H-neutralizing antibody 20H6 (40). Antibodies R1.302, N4.61, and N4.40 were described previously (33). The binding of the soluble nectin-4 ectodomain was then assessed by flow cytometry using anti-human Fc goat PE antibodies (Abcam, Cambridge, MA).
Pig airway epithelial cells.
Primary cultures of pig airway epithelial cells were prepared from trachea and bronchi by enzymatic dispersion and seeded onto collagen-coated, semipermeable membranes with a 0.4-μm pore size (Millicell-HA; surface area, 0.6 cm2; Millipore Corporation, Billerica, MA) as previously described for human airway epithelia (41). Only well-differentiated cultures (>3 weeks old; resistance, >1,000 Ω × cm2) were used. Transepithelial resistance was measured with a volt-ohm meter (World Precision Instruments, Sarasota, FL). Values were corrected for the blank filter resistance and further standardized against baseline readings and uninfected cultures. Neither corrected nor raw numbers resulted in a statistically significant variation from measurements in uninfected epithelia as determined by analysis of variance (ANOVA). For transductions, adenovirus type 5 (Ad5) vectors expressing mCherry or the human nectin-4 were diluted in sterile PBS to an MOI of 50, and 100 μl of the solution was applied for 4 h to the basolateral surface by inverting the culture insert. For infections, MeV preparations were diluted in sterile PBS to an MOI of 0.1 (as determined on Vero-SLAM cells), and 100 μl of the solution was applied to the basolateral surface.
RESULTS
Cross-linking the nectin-4 adhesive interface prevents infection.
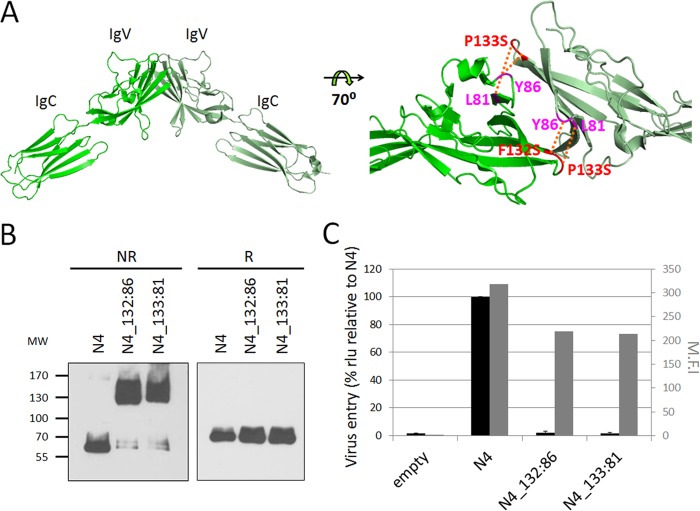
While it was long thought that cis and trans nectin-nectin interactions are governed by different V domain surfaces (19, 24), recent structural analyses strongly suggest that all these interactions depend on one single adhesive interface (20). We reasoned that if MeV uses the dimerization interface, cross-linking it by covalent bridges should prevent MeV entry. To test this hypothesis, we introduced disulfide bonds at the homodimer interface. We identified suitable locations for introduction of cysteine residues on a model of the nectin-4 homodimer based on the nectin-1 homodimer structure (23). Paired residues F132 and Y86 as well as P133 and L81 were chosen for cysteine substitution because they face each other across the antiparallel dimer interface and are located at an appropriate distance. Figure 2A shows the crystal structure of the nectin-4 homodimer (20) and the location of the disulfide bridges that may prevent access to the adhesive interface.
FIG 2.
Blocking access to the nectin-4 adhesive interface prevents infection. (A) Nectin-4 homodimer (PDB 4FRW) with cross-linking disulfide bridges. Structural coordinates were processed using PyMOL. The left panel shows the two membrane-distal domains of the dimer. The right panel shows the dimer interface with the residues mutated to Cys and the expected disulfide bonds (red dotted lines). (B) Immunoblot analysis of the dimerization capacity and protein expression levels of the nectin-4 cysteine substitution mutants. Protein extracts from transfected cells were separated by 4 to 15% SDS-PAGE under nonreducing (NR) or reducing (R) conditions. MW, molecular weight (in thousands). (C) Entry of MeV through the nectin-4 cysteine substitution mutants. Forty-eight hours posttransfection, cells were infected with MV323LucII at an MOI of 0.2. At 16 h postinfection, cells were analyzed for luciferase expression levels. Averages and standard deviations relative to the standard nectin-4 (N4) are indicated (black bars). rlu, relative luciferase units. Cell surface expression of the different nectin-4 constructs is also shown as mean fluorescence intensity (M.F.I., gray bars).
Figure 2B (left panel, nonreducing conditions [NR]) documents that while a FLAG-tagged human nectin-4 (N4) migrates as monomer, both N4_132:86 and N4_133:81 mutant proteins migrate almost exclusively as dimers. Thus, these proteins did fold correctly and the SH groups of the new cysteine residues formed disulfide bonds stabilizing the dimers. To assess whether these stabilized dimers are functional MeV receptors, we transiently expressed nectin-4 mutants in Vero cells, which do not express endogenous nectin-4, and then infected these cells with a luciferase-expressing wild-type MeV (MV323-LucII) (Fig. 2C). While standard nectin-4 supported MeV entry and production of luciferase, only background levels of luciferase were detected in the N4_132:86- and N4_133:81-expressing cells. We verified that N4_132:86 and N4_133:81 proteins are expressed at the cell surface (about 69% and 67% of nectin-4, respectively) (Fig. 2C, gray bars). Thus, MeV needs to access the nectin adhesive interface to enter cells.
The nectin-4 adhesive interface loops BC and FG are critical for MeV entry.
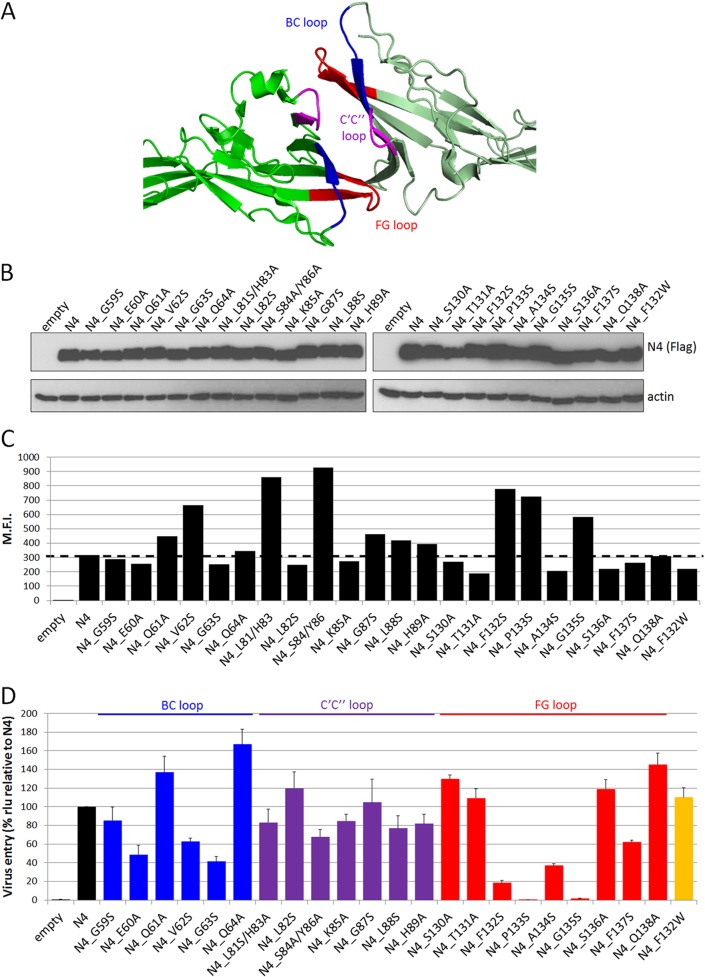
To assess whether all three loops forming the nectin-4 adhesive interface are functionally important for MeV entry, we performed an alanine/serine scan. Charged polar residues in the BC, C′C″, and FG loops (Fig. 3A; blue, purple, and red, respectively) were mutated to alanine, and uncharged apolar residues were replaced by serine. Total protein expression and cell surface protein expression were measured for all the mutants (Fig. 3B and C). Nectin-4 or its mutants were expressed in Vero cells for 48 h before infection with MV323-LucII. Luciferase levels were measured at 16 h postinfection to assess cell entry.
FIG 3.
Residues of the nectin-4 adhesive interface supporting MeV entry. (A) Nectin-4 homodimer (PDB 4GJT) showing the adhesive interface and the localization of the BC (blue), C′C″ (purple), and FG (red) loops. The molecules are oriented as in Fig. 2A, right panel. (B) Immunoblots of cell extracts of cells expressing the mutant proteins harvested 48 h posttransfection. Top panels, nectin-4 expression; bottom panels, actin expression. (C) Cell surface expression of the nectin-4 mutant proteins analyzed 48 h posttransfection by flow cytometry. M.F.I., mean fluorescence intensity. The dashed line indicates standard nectin-4 expression levels. (D) MeV entry efficiency through the nectin-4 mutants. Forty-eight hours posttransfection, cells were infected with MV323LucII at an MOI of 0.2. At 16 h postinfection, cells were analyzed for luciferase expression levels. Averages and standard deviations relative to the standard nectin-4 (N4) are indicated. rlu, relative luciferase units.
Figure 3D documents that mutations of the FG loop had the strongest effect on MeV entry: mutations of the highly conserved 131-TFP-X-G-135 motif had strong effects, with substitutions P133S and G135S abrogating entry and substitution F132S reducing entry by more than 80%. Importantly, the conservative mutation F132W had no impact on entry efficiency (Fig. 3D, orange bar). Substitutions A134S and F137S reduced entry by 60% and 40%, respectively. Other mutations of the FG loop slightly increased MeV entry. In the BC loop, mutations of residues E60, V62, and G63 reduced entry by 40 to 60% while substitutions of residues Q61 and Q64 enhanced entry. Mutations of C′C″ loop residues had little to no effect on MeV entry. Accordingly, mutations of the MeV H residues facing the C′C″ loop of nectin-4 (PDB 4GJT) did not affect the ability of H to support fusion of cells expressing nectin-4 (data not shown). Thus, the integrity of the BC and FG loops in the nectin-4 adhesive interface is required for nectin-4-dependent entry of MeV.
The nectin-4 BC and FG loops support MeV H binding.
Having observed differential effects of mutations in the BC, C′C″, and FG loops of nectin-4 on entry of viral particles, we sought to determine the role of each loop in supporting binding of the viral attachment protein. To this purpose, we engineered soluble recombinant nectin-4 ectodomains with individual loops mutated to either alanine or serine residues. Based on the interactions observed in the nectin-4/H cocrystal, we generated as previously described (33) three mutant proteins, namely, 61-QVGQ-64 (BC loop, QVGQ→ASSA), 83-HSKY-86 (C′C″ loop, HSKY→AAAA), and 132-FPAG-135 (FG loop, FPAG→SSSS) (Fig. 4A). These soluble ectodomains were added to the medium of nectin-4-expressing CHO cells with MV323-LucII (Fig. 4B). As expected, the soluble N4-Fc ectodomain blocked infection by more than 95%. The soluble 83-HSKY-86 mutant also blocked infection, suggesting that the C′C″ loop residues are not critical for MeV binding. On the other hand, both soluble 61-QVGQ-64 and 132-FPAG-135 mutant ectodomains failed to block infection, implying the importance of the BC and FG loops for MeV H binding.
FIG 4.
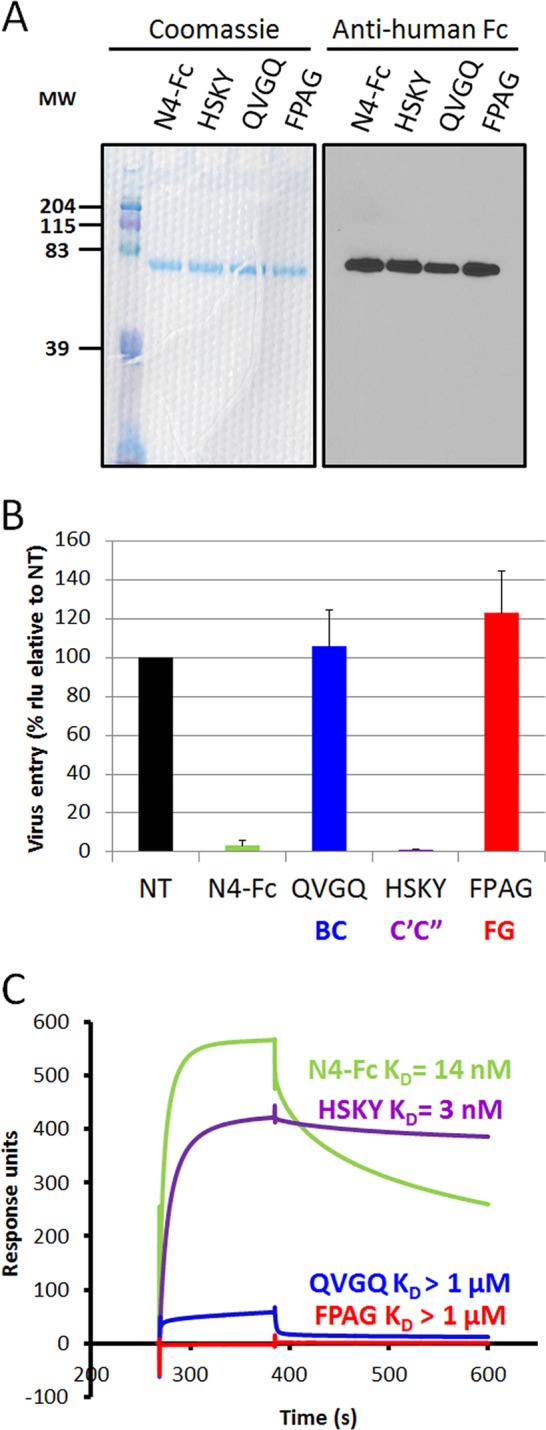
The BC and FG loops of nectin-4 are required for infection and for binding of the MeV attachment protein. (A) The soluble ectodomain of wild-type nectin-4 (N4-Fc), a mutant of the BC loop (61-QVGQ-64→61-ASSA-64), a mutant of the C′C″ loop (83-HSKY-86→83-AAAA-86), and a mutant of the FG loop (132-FPAG-135→132-SSSS-135) were expressed in Cos-7 cells. Eluted soluble nectin-4 ectodomains were separated by SDS-PAGE and visualized by Coomassie blue staining (left panel) or immunoblotting (right panel). MW, molecular weight (in thousands). (B) CHO cells expressing nectin-4 were infected with MV323LucII at an MOI of 0.2 in the absence (NT, not treated) or presence of soluble nectin-4 ectodomain (N4-Fc, green), or soluble mutants of nectin-4 with substitutions in the BC (QVGQ, blue), C′C″ (HSKY, purple) or FG loop (FPAG, red). At 16 h postinfection, cells were analyzed for luciferase expression levels. (C) Kinetics of binding of soluble H to the corresponding soluble nectin-4 ectodomains, as determined by surface plasmon resonance. KD, equilibrium dissociation constant. Color coding is as described for panel B.
We further determined the binding kinetics and affinities of the mutant soluble nectin-4 ectodomains for MeV H using surface plasmon resonance. As expected, the equilibrium dissociation constant (KD) of N4-Fc for MeV H was about 14 nM (Fig. 4C). In contrast, the ectodomains with mutations in the BC and FG loops, 61-QVGQ-64 and 132-FPAG-135, respectively, had on rates (kon) reduced 38- and 67-fold, and their affinities were in the micromolar range (Fig. 4C and Table 1). In contrast, the ectodomain with mutations in the C′C″ loop (83-HSKY-86) had a 2-fold-reduced kon. Interestingly, the off rate (koff) was also reduced, resulting in a 4- to 5-fold-higher affinity than standard nectin-4.
TABLE 1.
Binding kinetics of soluble H ectodomains to soluble nectin-4 ectodomains
| Ligand | kon (105 M−1 s−1) | koff (10−3 s−1) | KD (koff/kon) |
|---|---|---|---|
| N4-Fc | 2.71 | 3.83 | 14 nM |
| QVGQ | 0.07 | 136 | >1 μM |
| HSKY | 1.21 | 0.37 | 3 nM |
| FPAG | 0.04 | 19.3 | >1 μM |
One variant residue in the BC loop of pig nectin-4 disallows MeV entry.
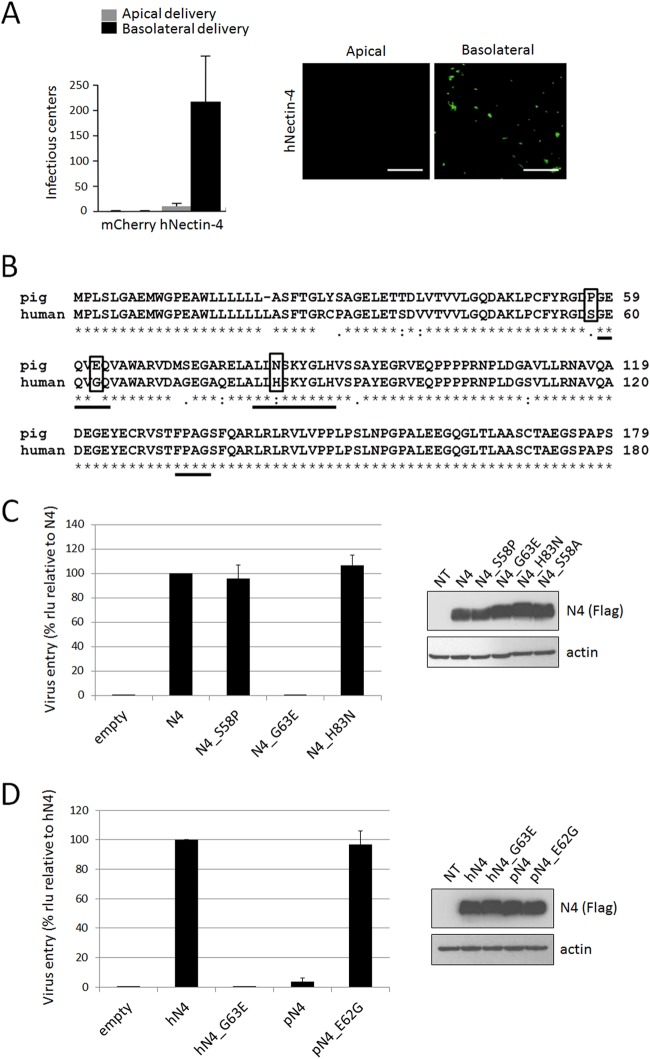
While developing an alternative model to study MeV spread in epithelia, we observed that polarized primary pig airway epithelia cannot be infected with MeV (Fig. 5A, left panel, mCherry) despite efficient pig nectin-4 transcription (data not shown). Expression of human nectin-4 through an adenovirus vector rendered these epithelial sheets susceptible to basolateral MeV infection (Fig. 5A, left panel [hNectin-4] and right panels). These observations prompted us to compare the protein sequences of human and pig nectin-4.
FIG 5.
A single amino acid variant in pig nectin-4 accounts for defective MeV entry in PAE cells. (A) PAE cells were transduced with a control Ad5 vector expressing mCherry (mCherry) or a vector expressing human nectin-4 (hNectin-4) and then infected apically or basolaterally with a GFP-expressing MeV. At 72 h postinfection, infectious centers were counted. The left panel shows the number of infectious centers per well (averages and standard deviations). The right panel shows GFP-positive infectious centers following apical or basolateral infection of PAE transduced with Ad5-nectin-4. Scale bars, 100 μm. (B) Sequence alignment of the IgV domains of pig and human nectin-4. Key residues in the BC, C′C″, and FG loops are underlined. The variant amino acids tested functionally are boxed. (C and D) MeV entry in cells expressing recombinant nectin-4 proteins. Forty-eight hours posttransfection, cells were infected with MV323LucII at an MOI of 0.2. At 16 h postinfection, cells were analyzed for luciferase expression levels. Right panels show immunoblots documenting expression of different recombinant nectin-4 proteins.
In the alignment (Fig. 5B), we noted two differences mapping in the BC loop (positions 58 and 63) and one mapping in the C′C″ loop (position 83). We replaced the corresponding residues in human nectin-4 by the pig counterpart (S58P, G63E, and H83N). Residue serine 58 was also replaced by an alanine (S58A), as this mutant was not in our previous screen. All these mutants were expressed at levels similar to that of wild-type nectin-4 (Fig. 5C, right panel). When tested for their ability to support entry and replication of MV323-LucII in Vero cells, all the mutants except G63E were susceptible (Fig. 5C, left panel).
To assess whether G63E is the sole difference accounting for MeV restrictions in PAE cells, we cloned the pig nectin-4 cDNA and assessed its ability to support MeV infection in human cells not expressing endogenous nectin-4 (Fig. 5D). As expected, the standard pig nectin-4 did not support entry of MeV in transfected cells. However, substitution of glutamic acid with glycine at position 62 (N4_E62G), which is found in the human homologue, reestablished MeV receptor activity to pig nectin-4. Altogether, these results highlight the importance of the BC loop for the H/nectin-4 interaction.
MeV H prevents nectin-4/nectin-1 heterodimer formation.
Since heterophilic interactions of nectin-4 with nectin-1 are much stronger than both types of homophilic interactions (33), nectin-4 molecules are likely to be present on the cell surface as nectin-4/nectin-1 heterodimers. Therefore, we assessed whether MeV H can prevent the nectin-4 to nectin-1 interaction. We developed a flow cytometry-based competition assay. Figure 6 demonstrates that binding of the nectin-4 ectodomain (N4-Fc) to CHO cells expressing nectin-1 (CHO-N1) is abrogated in the presence of antibodies against the V domain of nectin-1 (R1.302) or against the V domain of nectin-4 (N4.61). In contrast, an antibody against one of the C domains (N4.40) did not prevent N4-Fc binding to the CHO-N1 cells. In addition, neither N4.61 nor N4.40 cross-reacted with CHO-N1 cells (data not shown). In the presence of a soluble MeV H (sH), N4-Fc binding to CHO-N1 cells was reduced by more than 65%. However, sH did not prevent the N4-Fc interaction in the presence of the anti-H antibody 20H6 (40). Therefore, the MeV attachment protein can prevent nectin-4/nectin-1 heterodimer formation.
FIG 6.
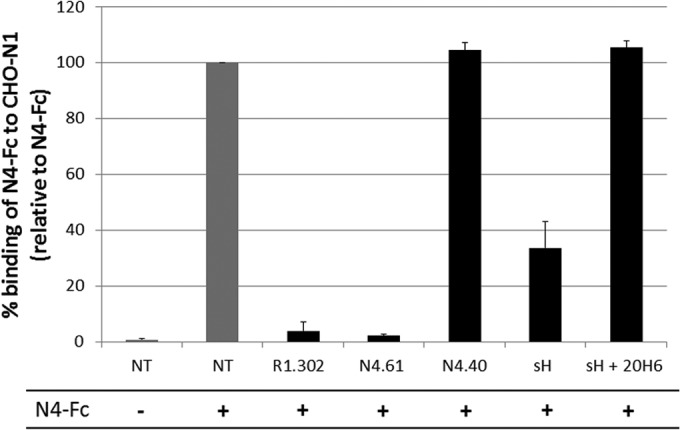
MeV H prevents nectin-4/nectin-1 heterodimer formation. Binding of soluble nectin-4 (N4-Fc) to CHO-N1 cells expressing nectin-1 in the absence (NT, not treated) or presence of anti-IgV nectin-1 antibodies (R1.302), anti-IgV nectin-4 antibodies (N4.61), anti-IgC nectin-4 antibodies (N4.40), soluble hemagglutinin ectodomain (sH), or sH- and H-neutralizing antibodies (sH + 20H6). The negative control (first column) consisted of nontreated cells not incubated with N4-Fc. Binding was assessed by flow cytometry using an anti-human IgG1 Fc antibody directed to the Fc fragment of human IgG1 fused to the soluble nectin-4 ectodomain (N4-Fc).
Nectin-4 uses a single interface to interact with itself and with nectin-1.
Structural analyses of all four nectins and the necl-5 homodimers identified a canonical adhesive interface (20). It was also suggested that the heterodimer's stability may be controlled by a few charged residues (20), but no heterodimer structure has been obtained yet. While our observation that MeV H can prevent the nectin-4 trans-interaction with nectin-1 is consistent with a model in which the adhesive interface also governs heterophilic dimer interactions, formal proof is missing.
To characterize directly the nectin-4/nectin-1 dimer interactions, we developed a coimmunoprecipitation assay based on FLAG-tagged nectin-4 proteins (Fig. 7). We first assessed whether the disulfide bridge-stabilized recombinant N4_132:86 and N4_133:81 homodimers can interact with nectin-1. As expected by the single-interface model, the stabilized homodimer did not coimmunoprecipitate soluble nectin-1 while the wild-type control nectin-4 (N4) did (Fig. 7, left panel). We then used the whole set of alanine/serine mutants of the nectin-4 adhesive interface to identify those residues involved in nectin-1 binding. Mutations of residues V62 and Q64 in the BC loop interfered with the ability to coimmunoprecipitate nectin-1 (Fig. 7, center panels, left half). In the C′C″ loop, mutants L81S/H83A, NS84A/Y86A, G87S, and H89A lost their ability to interact with nectin-1 (Fig. 7, center panels, right half). The FG loop mutants T131A, F132S, P133S, G135S, and F137S also lost binding to nectin-1 (Fig. 7, right panel). Similarly, the soluble recombinant nectin-4 ectodomains 61-QVGQ-64, 83-HSKY-86, and 132-FPAG-135 did not coimmunoprecipitate with a FLAG-tagged nectin-1 (data not shown). Altogether, these results demonstrate that nectin-4 uses the same key residues in the BC and FG loops to bind H and to make heterotypic and homotypic interactions. For the nectin interactions, unlike for H binding, nectin-4 also requires the participation of amino acids in the C′C″ loop.
FIG 7.
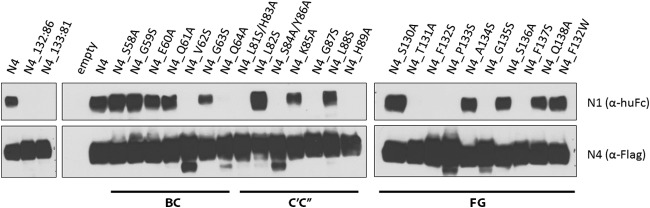
Residues involved in nectin-4/nectin-1 heterodimerization. Immunoblot analysis of soluble nectin-1 binding to nectin-4 proteins with mutations in the adhesive interface. At 48 h posttransfection, nectin-4 proteins were immunoprecipitated through their C-terminal FLAG tag. The immunoprecipitates were incubated with soluble nectin-1 ectodomain. Eluates were separated by 4 to 15% SDS-PAGE. Proteins were detected using antibodies to the human Fc fragment of IgG1 (huFc) for soluble nectin-1 or the FLAG epitope for the FLAG-tagged nectin-4.
DISCUSSION
Viruses depend on cell surface molecules to enter cells and begin their replicative cycle. Many viruses utilize cell adhesion molecules of the Ig superfamily as receptors (42). In particular, HIV uses CD4 (43), adenovirus uses the coxsackie virus and adenovirus receptor (44), reovirus uses the junctional adhesion molecule A (45), and coronaviruses use members of the carcinoembryonic antigen glycoprotein family (46, 47). MeV uses two cell adhesion molecules of the Ig superfamily as receptors, SLAM and nectin-4 (3–5). Other viruses also exploit nectin-like proteins: herpes simplex viruses use nectins 1 and 2 (25–27), and poliovirus uses necl-5 (28). Since all these viruses belong to different families, these viral attachment protein-Ig superfamily receptor interactions may have evolved independently. It has been proposed that early viruses may have shaped the evolution of the primordial Ig fold prior to the existence of the vertebrate immune system (42).
Viruses contact these receptors through the upper “tip” of their most membrane-distal domain (42), which includes the BC, C′C″, and FG loops. This location corresponds to the newly defined canonical adhesive interface of nectins (20). Here we characterize how MeV appropriates this interface of nectin-4, preventing dimerization. Based on proximity in the cocrystal structure, it was suggested that contacts of MeV H with all three loops are necessary for efficient virus entry (15). We show here that only the contacts with the C β-sheet and the FG loop are critical for MeV H binding and cell entry.
Figure 8A illustrates how the nectin-4 FG loop (red backbone) protrudes into an H-protein hydrophobic groove (orange surface). The BC loop (blue backbone) covers the groove and helps stabilize the complex. In agreement with the costructure (15), our functional screen identifies E60, V62, and G63 as the main residues supporting contacts in the BC loop: mutations of the 61-QVGQ-64 stretch abrogated binding of nectin-4 to MeV H. The nature of the amino acid at position 63 in human nectin-4 is particularly important. While substitution G63S reduces nectin-4 receptor activity, substitution G63E completely abrogates it. In addition, this residue can determine species specificity: pig nectin-4 presents a glutamic acid instead of a glycine at the corresponding position and cannot function as a receptor for MeV. We show here that pig nectin-4 with a glycine at the critical position has full MeV receptor activity. The longer side chain and the negative charge of glutamic acid are likely to induce a steric clash preventing complex stabilization.
FIG 8.
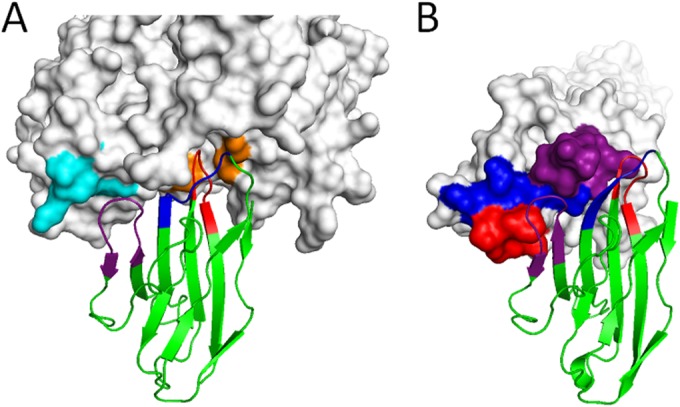
Nectin-4 binding to the MeV hemagglutinin or to itself. (A) Nectin-4 (backbone representation) in complex with the MeV hemagglutinin (surface representation [PDB 4GJT]). The nectin-4 FG loop (red) protrudes in the MeV H hydrophobic groove (orange). The BC loop (dark blue) crosses over the FG loop in the hydrophobic pocket. The C′C″ loop (purple) contacts H residues in the β4 blade (light blue). (B) Structure of the nectin-4 homodimeric adhesive interface (4FRW). Color coding is as described for panel A. Note that the C′C″ loop contacts the BC and FG loops of the opposite monomer.
Residues in the highly conserved 131-TFP-X-G-135 motif of the nectin-4 FG loop are also critical for binding MeV H. These residues protrude deep in the groove and participate in hydrophobic interactions with residues L482, Y483, Y541, and Y543 on Mev H (16). Indeed, Zhang et al. demonstrated that a 132-SSSS-135 mutant of the FG loop does not bind the surface of MeV H-expressing cells (15). We show here that the same mutant does not bind MeV H in a surface plasmon resonance assay. Recently, Delpeut et al. showed that the attachment protein of canine distemper virus binds the V domain of canine nectin-4 through the TFP-X-G motif (48). Remarkably, the same motif in the FG loop of nectin-1 is also targeted by HSV (29). Moreover, poliovirus targets the corresponding motif on necl-5 (32). As the phenylalanine in this motif is critical for nectin dimerization and the establishment of the adherens junction (20), all viruses using nectins target the same key motif in the adhesive interface for cell entry.
We show here formally that the conserved F132 in the nectin-4 FG loop is critical not only for homodimer formation (20) but also for heterodimer formation. Our studies also document that all three loops forming the adhesive surface are functionally important for nectin-nectin heterophilic interactions. Indeed, the structure of the nectin-4 homodimer shows that the symmetric interface is stabilized mainly by interactions of the BC and FG loops (Fig. 8B, blue and red backbone) with the C′C″ loop of the facing nectin-4 monomer (Fig. 8B, purple surface). Although integrity of the BC loop is required for the interaction, the contacts between the FG and C′C″ loops are the strongest, with residue F132 fitting in a hydrophobic pocket formed by H83, G87, Y86, and H89 of the C′C″ loop (20). Strikingly, the recently published structure of necl-5 in complex with the TIGIT Ig superfamily protein documents an analogous binding mode (49).
All these observations highlight the relevance of the interactions of viral attachment proteins with the tip of the V domain of cell adhesion molecules of the Ig superfamily. Interactions between soluble forms of viral attachment proteins and their receptors can be much stronger than homophilic interactions between receptor subunits: HSV gD can interfere with the binding of nectin-3 and nectin-4 to nectin-1 (33). We present here the first detailed analysis of how a viral attachment protein prevents heterodimerization of its Ig superfamily receptor. Displacement of receptor dimers forming in trans between infected and noninfected cells is of central importance for MeV spread: since this virus remains strongly cell associated (50–52), H protein expressed on infected cells must disrupt nectin-4/nectin-1 trans-cellular dimers to allow spread.
In conclusion, the C′C″ loop is the principal discriminator of the functional interactions of the nectin-4 adhesive interface with its viral and cellular partners. This has implications for antiviral strategies. We also discovered that different contact surfaces documented in the H-nectin-4 cocrystals are critical for different phases of the MeV-elicited membrane fusion process: while the BC and FG loops govern binding, the C′C″ loop must support a successive phase. It will be interesting to assess whether the same adhesive interface loops similarly govern the association and the dissociation of attachment proteins of other viruses.
ACKNOWLEDGMENTS
We thank Patricia Devaux, Marie Frenzke, Swapna Apte-Sengupta, Christian Pfaller, Tanner Miest, and Vladimir Gainullin for helpful discussions.
This work was supported by U.S. National Institutes of Health grants R01 AI063476 to Roberto Cattaneo and R01 HL-105821 to Patrick L. Sinn.
Mathieu Mateo is a Merck fellow of the Life Sciences Research Foundation.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.Griffin DE. 2013. Measles virus, p 1042–1069. Fields virology, 6th ed, vol 1 Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Kupferschmidt K. 2012. Public health. Europe's embarrassing problem. Science 336:406–407. 10.1126/science.336.6080.406. [DOI] [PubMed] [Google Scholar]
- 3.Mateo M, Navaratnarajah CK, Cattaneo R. 2014. Structural basis of efficient contagion: measles variations on a theme by parainfluenza viruses. Curr. Opin. Virol. 5:16–23. 10.1016/j.coviro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. 10.1038/35022579. [DOI] [PubMed] [Google Scholar]
- 5.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Jr, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. 10.1038/nature10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD. 2011. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7:e1002240. 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorig RE, Marcil A, Chopra A, Richardson CD. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295–305. [DOI] [PubMed] [Google Scholar]
- 8.Naniche D, Varior-Krishnan G, Cervoni F, Wild TF, Rossi B, Rabourdin-Combe C, Gerlier D. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535–19540. 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. 2011. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 18:128–134. 10.1038/nsmb.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navaratnarajah CK, Vongpunsawad S, Oezguen N, Stehle T, Braun W, Hashiguchi T, Maenaka K, Yanagi Y, Cattaneo R. 2008. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J. Biol. Chem. 283:11763–11771. 10.1074/jbc.M800896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4(4):e00413-13. 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18:135–141. 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 14.Santiago C, Celma ML, Stehle T, Casasnovas JM. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17:124–129. 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Lu G, Qi J, Li Y, He Y, Xu X, Shi J, Zhang CW, Yan J, Gao GF. 2013. Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4. Nat. Struct. Mol. Biol. 20:67–72. 10.1038/nsmb.2432. [DOI] [PubMed] [Google Scholar]
- 16.Mateo M, Navaratnarajah CK, Syed S, Cattaneo R. 2013. The measles virus hemagglutinin beta-propeller head beta4-beta5 hydrophobic groove governs functional interactions with nectin-4 and CD46 but not those with the signaling lymphocytic activation molecule. J. Virol. 87:9208–9216. 10.1128/JVI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takai Y, Ikeda W, Ogita H, Rikitake Y. 2008. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu. Rev. Cell Dev. Biol. 24:309–342. 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- 18.Takai Y, Nakanishi H. 2003. Nectin and afadin: novel organizers of intercellular junctions. J. Cell Sci. 116:17–27. 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- 19.Rikitake Y, Mandai K, Takai Y. 2012. The role of nectins in different types of cell-cell adhesion. J. Cell Sci. 125:3713–3722. 10.1242/jcs.099572. [DOI] [PubMed] [Google Scholar]
- 20.Harrison OJ, Vendome J, Brasch J, Jin X, Hong S, Katsamba PS, Ahlsen G, Troyanovsky RB, Troyanovsky SM, Honig B, Shapiro L. 2012. Nectin ectodomain structures reveal a canonical adhesive interface. Nat. Struct. Mol. Biol. 19:906–915. 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bork P, Holm L, Sander C. 1994. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242:309–320. [DOI] [PubMed] [Google Scholar]
- 22.Harpaz Y, Chothia C. 1994. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238:528–539. [DOI] [PubMed] [Google Scholar]
- 23.Narita H, Yamamoto Y, Suzuki M, Miyazaki N, Yoshida A, Kawai K, Iwasaki K, Nakagawa A, Takai Y, Sakisaka T. 2011. Crystal structure of the cis-dimer of nectin-1: implications for the architecture of cell-cell junctions. J. Biol. Chem. 286:12659–12669. 10.1074/jbc.M110.197368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samanta D, Ramagopal UA, Rubinstein R, Vigdorovich V, Nathenson SG, Almo SC. 2012. Structure of Nectin-2 reveals determinants of homophilic and heterophilic interactions that control cell-cell adhesion. Proc. Natl. Acad. Sci. U. S. A. 109:14836–14840. 10.1073/pnas.1212912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. [DOI] [PubMed] [Google Scholar]
- 27.Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH, Eisenberg RJ. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendelsohn CL, Wimmer E, Racaniello VR. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855–865. [DOI] [PubMed] [Google Scholar]
- 29.Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 7:e1002277. 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang N, Yan J, Lu G, Guo Z, Fan Z, Wang J, Shi Y, Qi J, Gao GF. 2011. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat. Commun. 2:577. 10.1038/ncomms1571. [DOI] [PubMed] [Google Scholar]
- 31.Struyf F, Martinez WM, Spear PG. 2002. Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions. J. Virol. 76:12940–12950. 10.1128/JVI.76.24.12940-12950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. U. S. A. 105:18284–18289. 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabre S, Reymond N, Cocchi F, Menotti L, Dubreuil P, Campadelli-Fiume G, Lopez M. 2002. Prominent role of the Ig-like V domain in trans-interactions of nectins. Nectin3 and nectin4 bind to the predicted C-C'-C”-D beta-strands of the nectin1 V domain. J. Biol. Chem. 277:27006–27013. 10.1074/jbc.M203228200. [DOI] [PubMed] [Google Scholar]
- 34.Reymond N, Fabre S, Lecocq E, Adelaide J, Dubreuil P, Lopez M. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276:43205–43215. 10.1074/jbc.M103810200. [DOI] [PubMed] [Google Scholar]
- 35.Msaouel P, Iankov ID, Allen C, Morris JC, von Messling V, Cattaneo R, Koutsilieris M, Russell SJ, Galanis E. 2009. Engineered measles virus as a novel oncolytic therapy against prostate cancer. Prostate 69:82–91. 10.1002/pros.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda M, Takeuchi K, Miyajima N, Kobune F, Ami Y, Nagata N, Suzaki Y, Nagai Y, Tashiro M. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643–6647. 10.1128/JVI.74.14.6643-6647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard VH, Sinn PL, Hodge G, Miest T, Devaux P, Oezguen N, Braun W, McCray PB, Jr, McChesney MB, Cattaneo R. 2008. Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118:2448–2458. 10.1172/JCI35454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iankov ID, Penheiter AR, Griesmann GE, Carlson SK, Federspiel MJ, Galanis E. 2013. Neutralization capacity of measles virus H protein specific IgG determines the balance between antibody-enhanced infectivity and protection in microglial cells. Virus Res. 172:15–23. 10.1016/j.virusres.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, Welsh MJ. 2002. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol. Biol. 188:115–137. 10.1385/1-59259-185-X:115. [DOI] [PubMed] [Google Scholar]
- 42.Dermody TS, Kirchner E, Guglielmi KM, Stehle T. 2009. Immunoglobulin superfamily virus receptors and the evolution of adaptive immunity. PLoS Pathog. 5:e1000481. 10.1371/journal.ppat.1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. [DOI] [PubMed] [Google Scholar]
- 44.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323. [DOI] [PubMed] [Google Scholar]
- 45.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. 10.1016/S0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 46.Dveksler GS, Dieffenbach CW, Cardellichio CB, McCuaig K, Pensiero MN, Jiang GS, Beauchemin N, Holmes KV. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams RK, Jiang GS, Holmes KV. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 88:5533–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delpeut S, Noyce RS, Richardson CD. 2014. The V domain of dog PVRL4 (nectin-4) mediates canine distemper virus entry and virus cell-to-cell spread. Virology 454–455:109–117. 10.1016/j.virol.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 49.Stengel KF, Harden-Bowles K, Yu X, Rouge L, Yin J, Comps-Agrar L, Wiesmann C, Bazan JF, Eaton DL, Grogan JL. 2012. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc. Natl. Acad. Sci. U. S. A. 109:5399–5404. 10.1073/pnas.1120606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohn W, Rutter G, Hohenberg H, Mannweiler K. 1983. Inhibition of measles virus budding by phenothiazines. Virology 130:44–55. [DOI] [PubMed] [Google Scholar]
- 51.Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rentier B, Hooghe-Peters EL, Dubois-Dalcq M. 1978. Electron microscopic study of measles virus infection: cell fusion and hemadsorption. J. Virol. 28:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]