ABSTRACT
Double-stranded RNA (dsRNA) is an important molecular pattern associated with viral infection and is detected by various extra- and intracellular recognition molecules. Poxviruses have evolved to avoid producing dsRNA early in infection but generate significant amounts of dsRNA late in infection due to convergent transcription of late genes. Protein kinase R (PKR) is activated by dsRNA and triggers major cellular defenses against viral infection, including protein synthesis shutdown, apoptosis, and type I interferon (IFN-I) production. The poxviral E3 protein binds and sequesters viral dsRNA and is a major antagonist of the PKR pathway. We found that the highly replication-restricted modified vaccinia virus Ankara (MVA) engineered to produce excess amounts of dsRNA early in infection showed enhanced induction of IFN-β in murine and human cells in the presence of an intact E3L gene. IFN-β induction required a minimum overlap length of 300 bp between early complementary transcripts and was strongly PKR dependent. Excess early dsRNA produced by MVA activated PKR early but transiently in murine cells and induced enhanced systemic levels of IFN-α, IFN-γ, and other cytokines and chemokines in mice in a largely PKR-dependent manner. Replication-competent chorioallantois vaccinia virus Ankara (CVA) generating excess early dsRNA also enhanced IFN-I production and was apathogenic in mice even at very high doses but showed no in vitro host range defect. Thus, genetically adjuvanting MVA and CVA to generate excess early dsRNA is an effective method to enhance innate immune stimulation by orthopoxvirus vectors and to attenuate replicating vaccinia virus in vivo.
IMPORTANCE Efficient cellular sensing of pathogen-specific components, including double-stranded RNA (dsRNA), is an important prerequisite of an effective antiviral immune response. The prototype poxvirus vaccinia virus (VACV) and its derivative modified vaccinia virus Ankara (MVA) produce dsRNA as a by-product of viral transcription. We found that inhibition of cellular dsRNA recognition established by the virus-encoded proteins E3 and K3 can be overcome by directing viral overexpression of dsRNA early in infection without compromising replication of MVA in permissive cells. Early dsRNA induced transient activation of the cellular dsRNA sensor protein kinase R (PKR), resulting in enhanced production of interferons and cytokines in cells and mice. Enhancing the capacity of MVA to activate the innate immune system is an important approach to further improve the immunogenicity of this promising vaccine vector.
INTRODUCTION
Nucleic acids are an important class of pathogen-associated molecular patterns (PAMPs) sensed by the innate immune system to detect the presence of various pathogens and particularly of viruses. Pathogen-specific or aberrantly localized nucleic acids are recognized by members of various families of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), RIG-like receptors (RLRs) RIG-I and MDA5, NOD-like receptors (NLRs), AIM2-like receptors (ALRs), DExD/H-box helicases, and others like IFIT1 and nucleic acid-activated enzymes (1–3). Endosomal TLRs monitor the presence of nucleic acids from the extracellular compartment, such as double-stranded RNA (dsRNA) (TLR3), single-stranded RNA (ssRNA) (TLR7/8), rRNA (TLR13), and CpG motif-containing DNA (TLR9), whereas the other PRRs sense nucleic acids localized in the cytoplasm. Stimulation of PRRs leads to cellular activation and to the secretion of type I interferon (IFN-I) as well as other cytokines and chemokines. Induction of IFN-I is a critical and important cellular response upon detection of viral infection. IFN-α and IFN-β, as the key IFN-I, induce an antiviral state, restricting viral replication, and significantly contribute to the activation and coordination of the adaptive immune response. They increase major histocompatibility complex (MHC) class I and II expression, cross-presentation by CD8+ dendritic cells (DCs), and antibody secretion as well as immunoglobulin isotype switching by B cells (1, 4, 5). IFN-I has been shown to promote expansion of modified vaccinia virus Ankara (MVA)-induced CD8 T cells (6). Deletion of the viral C6L-encoded inhibitor of IFN-β induction from MVA enhanced vector and transgene-specific CD8 T cell responses quantitatively and qualitatively (7).
dsRNA is an important signature of virus infection and thus a prototypic viral PAMP. Host cells and organisms have devised a variety of recognition receptors for dsRNA. While TLR3 is expressed mainly in immune cells and can sense extracellular and endosomal dsRNA, most other dsRNA sensors, like the RIG-like receptors RIG-I and MDA-5 and the dsRNA-activated enzymes protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase (2′-5′-OAS), are localized in the cytoplasm and are ubiquitously expressed in all cell types (8, 9). RIG-I and MDA-5 have recently gained much interest as important cytosolic dsRNA sensors for induction of IFN-I in response to infection with RNA and DNA viruses (10, 11). In contrast, for orthopoxviruses like vaccinia virus (VACV) and also for herpes simplex virus 1, PKR appears to represent a critical nucleic acid sensor (12, 13), although these viruses have a dsDNA genome. PKR is present in small amounts in the cell cytoplasm, and its kinase activity is triggered upon binding to dsRNA via the N-terminal dsRNA binding domain. The best-characterized substrate of PKR is the α subunit of eukaryotic initiation factor 2 (eIF2α). Phosphorylation of eIF2α leads to a shutdown in translation, thereby restricting viral replication (9, 14). Other consequences of PKR activation include apoptosis and mitogen-activated protein kinase activation (15, 16). In addition, a role of PKR in the induction of IFN-I is increasingly recognized (17–19).
Poxviruses produce large amounts of dsRNA, because the termination of transcription of poxviral intermediate and late genes is not tightly regulated, leading to viral mRNA with long 3′ untranslated regions (3′ UTRs) of heterogeneous lengths (20, 21). When such intermediate or late transcripts originate from two convergently transcribed genes, overlapping complementary mRNAs form dsRNA (22). Late viral mRNA can also anneal with mRNA from an adjacent, convergently transcribed early gene (22). Thus, generation of dsRNA is largely confined to the late phase of the poxviral replication cycle (23, 24). Early in infection, only minor amounts of viral dsRNA are detectable (22, 25, 26). Transcription of early genes is actively terminated by the poxviral early transcription machinery shortly after the stop codon within 20 to 50 nucleotides downstream of the sequence motif TTTTTNT (27). Early genes transcribed toward each other typically contain multiple transcription termination signals, suggesting that tight control of termination of early convergent RNA transcription is advantageous for the evolutionary fitness of poxviruses (28). Very likely, this represents a mechanism minimizing the generation of dsRNA early in infection and hence contributes to the viral countermeasures against triggering of the cellular dsRNA recognition systems.
Poxviruses encode at least two proteins, E3 and K3, to inhibit PKR and counteract its dsRNA-triggered antiviral effects, highlighting the importance of PKR in poxvirus biology. E3 binds and sequesters dsRNA, thereby preventing activation of both PKR and 2′-5′-OAS (29, 30). In addition, E3 inhibits PKR by direct binding (31, 32). VACV mutants lacking the E3L gene (VACV-ΔE3L) showed higher sensitivity to IFN-I treatment than the wild type (33), had a restricted host range, and induced apoptosis in many cell types (34, 35). The VACV K3 protein is a viral eIF2α-like pseudosubstrate of PKR blocking eIF2α phosphorylation (36, 37).
MVA is a highly replication restricted VACV derived by over 570 passages in chicken embryo cells from its replication-competent ancestor chorioallantois vaccinia virus Ankara (CVA) (38). Numerous MVA recombinants have been shown to efficiently induce immune responses in animals and humans against MVA's own viral proteins as well as against the heterologous antigens (39, 40). The large body of preclinical and clinical evidence demonstrating that MVA is a safe and immunogenic vaccine has paved the way for the recent approvals of the proprietary MVA-BN (Bavarian Nordic) (41) as a smallpox vaccine in the European Union and Canada under the trade names IMVANEX and IMVAMUNE, respectively. Here, we describe that recombinant production of excess amounts of dsRNA early in MVA and CVA infection increased the induction of IFN-α/β and other cytokines both in murine and human cells and in mice. In the case of CVA, forced early dsRNA overproduction resulted in an almost complete loss of virulence in mice, which was mediated mainly via IFN-I. Increased induction of IFN-I in vitro and in vivo by MVA expressing excess early dsRNA was largely dependent on PKR. Hence, tight control of dsRNA formation and prevention of PKR activation early in infection is crucial for VACV to restrict IFN-I induction and to ensure replicative fitness within the infected host. Engineering a favorable innate immune activation profile by genetic adjuvantation has the potential to further improve the antigenic properties of MVA for its application as a recombinant vaccine vector platform.
MATERIALS AND METHODS
Cells and viruses.
The BALB/3T3 clone A31, BALB/3T12-3, and HeLa cell lines were obtained from ATCC or the European Collection of Cell Cultures. HaCaT cells were obtained from N. E. Fusenig, German Cancer Research Center (DKFZ), Heidelberg, Germany. IFN-I receptor-deficient (IFNAR−/−) and IPS-1−/− mouse embryo fibroblasts (MEFs) were prepared from 15-day-old C57BL/6-IFNAR−/− and 14-day-old C57BL/6-IPS-1−/− embryos by standard procedures, and PKR−/− MEFs and corresponding PKR-sufficient control MEFs (42) (wild-type [wt] MEFs) were kindly provided by J. Pavlovic, Institute for Medical Virology, University of Zürich, Switzerland. All cell lines were cultivated in Dulbecco's modified Eagle medium (DMEM; Gibco/Invitrogen, Darmstadt, Germany) supplemented with 10% fetal calf serum (FCS; Pan Biotech, Aidenbach, Germany). Primary chicken embryo fibroblast (CEF) cells were prepared from 11-day-old embryonated chicken eggs and cultured in VP-SFM (Gibco) for virus stock production or DMEM supplemented with 10% FCS for replication analysis. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-dependent dendritic cells (GM-DCs) were generated from freshly prepared murine bone marrow by cultivation with recombinant murine GM-CSF (tebu-bio, Offenbach, Germany) as previously described (43).
CVA and MVA used in this study were derived from bacterial artificial chromosome (BAC) clones constructed from MVA-BN (Bavarian Nordic) and wt CVA and have been characterized previously (44). Wild-type MVA (MVA) and MVA mutants were propagated on secondary CEF or DF-1 cells and titrated on CEF cells using the 50% tissue culture infective dose (TCID50) method as described previously (45). Wild-type CVA (CVA) and CVA mutants were propagated on Vero cells and titrated by the TCID50 method on CV-1 cells. Shope fibroma virus was obtained from the ATCC (VR-364) and was propagated and titrated on rabbit cornea SIRC cells. Sendai virus strain Cantell was obtained from Charles River Laboratories at 2,000 hemagglutinin (HA) units/ml. All viruses used in animal experiments were purified twice through a 36% sucrose cushion.
Mice.
BALB/cJ mice were purchased from Harlan Winkelmann (Germany), C57BL/6 mice were purchased from Janvier Labs (Le Genest Saint Isle, France), and B6129SF2/J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). C57BL/6–129/Sv PKR−/− mice (42) were kindly provided by J. Pavlovic and, like C57BL/6-IFNAR−/− mice lacking a functional IFN-I receptor, were obtained from the animal facility of the University of Zürich. C57BL/6-IPS−/− mice (46) were kindly provided by J. Tschopp, University of Lausanne, Switzerland. Animals were bred and maintained in the animal facility of Bavarian Nordic GmbH in accordance with the institutional guidelines and German regulations for Animal Experimentation.
BAC recombineering and reactivation of infectious virus.
Construction of the CVA- and MVA-BAC clones has been described previously (35). Briefly, the inserted BAC cassette contains mini-F plasmid sequences derived from pMBO131 plasmid (38) for maintenance in Escherichia coli and a neomycin-phosphotransferase II (NPTII)-enhanced green fluorescent protein (EGFP) marker cassette. Transcription of the complete neo-EGFP cassette is driven by a poxviral synthetic early/late promoter (9), and translation of the EGFP cistron from the bicistronic mRNA is achieved by an encephalomyocarditis virus internal ribosomal entry site (IRES) preceding the EGFP open reading frame (ORF). The BAC cassette was inserted between the MVA and CVA orthologues, respectively, of the I3L and I4L genes. The CVA-BAC clone was modified by allelic exchange mutagenesis in E. coli strain DH10B utilizing the λ Red system for homologous recombination. MVA-BAC clones were also modified by allelic exchange mutagenesis using the counterselectable rpsL/neo cassette (44, 47) and E. coli strains DH10B and MDS42 as described. In the latter, all known insertion sequence (IS) elements have been deleted (48). The presence of the introduced mutations and the genetic integrity of neighboring DNA regions were confirmed by sequencing the respective regions containing the mutation sites. Absence of IS elements from all BAC clones propagated in E. coli DH10B was confirmed using IS element-specific PCR as described previously (44, 49). For generation of MVA recombinants expressing sense and antisense EGFP mRNA and all corresponding controls, the neo-IRES-EGFP cassette in the BAC backbone of these constructs was replaced by a bacterial tetracycline resistance cassette. The MVA-EGFP recombinant contained a bacterial kanamycin resistance cassette (NPTI) downstream of the EGFP ORF under the control of the strong early/late pHyb promoter (50). MVA-ΔE3L was obtained by replacing nucleotides 42697 to 43269 (ORF MVA050L) with an NPTI kanamycin resistance cassette by homologous recombination in E. coli.
The complete coding region of CVA mutants CVA-dsneo-ΔB15 and CVA-ΔB15 encompassing CVA ORFs 001 to 229 was sequenced by Eurofins MWG Operon (Ebersberg, Germany) using a pooled set of overlapping 5-kbp PCR fragments and next-generation sequencing with GS FLX Titanium series chemistry. Sequence ambiguities were reanalyzed by classical Sanger sequencing of short PCR fragments covering the ambiguous sites.
For reactivation of infectious virus, 106 BHK-21 cells were transfected with 3 μg of BAC DNA using Fugene HD (Promega, Mannheim, Germany) and 60 min later infected with Shope fibroma virus to provide the required helper functions. Reactivated virus was isolated, and helper virus was removed as previously described (44).
Real-time RT-qPCR.
RNA was isolated from 5 × 105 to 10 × 105 infected cells per sample using the RNeasy Plus minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, including a cell lysate homogenization step using QIAshredder columns (Qiagen) and a DNA removal step using gDNA eliminator columns (Qiagen). RNA was eluted from the RNeasy spin columns with 50 μl RNase-free water. The remaining viral and cellular DNA in the sample was digested with Turbo DNase (Ambion/Life Technologies, Darmstadt, Germany). The samples were incubated with the DNase for 30 min at 37°C, and DNase was subsequently inactivated by adding 0.5 M EDTA (Sigma-Aldrich, Hamburg, Germany) to a final concentration of 7.5 mM and heating to 75°C for 10 min. Reverse transcription of 3 μl of isolated RNA was performed using the Omniscript RT kit (Qiagen) in the presence of an RNase inhibitor (Sigma-Aldrich) at a final concentration of 10 U/μl for 90 min at 37°C using random nonamer primers (Sigma-Aldrich). A total of 1 μl of reverse transcriptase (RT) reaction mixture was added to 20 μl of total PCR mixture for quantitative real-time PCR (qPCR) based on the TaqMan gene expression master mix (Life Technologies, Darmstadt, Germany) according to the manufacturer's instructions. Quantitative PCR of cDNA was performed using the TaqMan gene expression assay IFNB1 (Hs01077958_s1) for detection of human interferon beta 1 transcripts and TaqMan gene expression assay IFNB1 (Mm00439552_s1) for detection of murine interferon beta 1 transcripts. Custom TaqMan gene expression assays were used for detection of rpsL-containing transcripts (forward primer, ACTTGGAACGAGCCTGCTTAC; reverse primer, GTTCGTTACCACACCGTACGT; probe, 6-carboxyfluorescein [FAM]-CTGCTCCGGCGTTAAA-MGBNFQ), of viral late F17R gene transcripts (forward primer, CGTTTATGAGGACGGACATGCTA; reverse primer, AAAAGTCTAGAAGCTACATTATCGCGATT; probe, FAM-CCGCGAACATATTTTG-MGBNFQ), and of viral early C7L gene transcripts (forward primer, TTTAGATTCATTATACGCCCAGATTGGT; reverse primer, TCACCGCATAGTTGTTTGCAAATAC; probe, FAM-ACCTCGTCGATTTCC). Total EGFP RNA (sense and antisense) generated by the set of recombinants expressing a nested set of antisense EGFP RNAs of different lengths (see Fig. 3A) was quantified using a custom RT-qPCR assay (AJ6RN3J, Life Technologies) targeting a sequence located at the 5′ end of the antisense RNA within the first 100 bp, which is common to all EGFP sense and antisense RNAs except the 50-bp antisense transcript of MVA-dsEGFP-50. Murine and human input cDNA was normalized using TaqMan gene expression assay 18S (Hs99999901_s1) detecting eukaryotic 18S rRNA transcripts. All real-time qPCR assays were performed in duplicate using an Applied Biosystems 7500 Fast real-time PCR system and the following protocol: initial incubation at 50°C for 2 min (uracil DNA glycosylase activation to remove carryover amplicon contamination), 10-min incubation at 95°C, 40 cycles of denaturation for 15 s at 95°C, and annealing/extension for 1 min at 60°C. No-template controls and no-RT controls were always included and were always negative. Transfection of high-molecular-weight poly(I·C) (Invivogen, Toulouse, France) served as the positive control for IFN-β induction. Fold induction of IFN-β was calculated relative to that of mock-infected cells. Threshold cycle (CT) values were arbitrarily set to 36 if the CT value was “undetermined” or >36. For calculation reasons, “undetermined” samples, e.g., from mock-infected cells, were assigned a fold induction value of 1.
FIG 3.
Threshold of dsRNA length for increased IFN-β induction. (A) Schematic representation of MVA recombinants generating excess early dsRNA of decreasing lengths from truncated antisense EGFP and full-length sense EGFP transcripts. (B) MEFs in 6-well plates were mock infected or infected with crude stocks of the indicated MVA recombinants expressing EGFP dsRNAs of decreasing lengths in duplicate at an MOI of 10. Fold induction of murine IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample with 18S rRNA as the endogenous control using total RNA isolated from cells at 7 h p.i. As a positive control, 2 μg of poly(I·C) per well was transfected using Fugene HD. (C) IFN-β in supernatants of MEFs at 14 h p.i. infected in parallel to the MEF cultures shown in panel B was quantified by ELISA.
Demonstration of EGFP dsRNA formation.
To demonstrate the formation of dsRNA from sense and antisense transcripts of the EGFP gene in MVA-dsEGFP-720(o)-infected cells, we isolated and combined total RNA from two wells of a 6-well plate per virus of AraC-treated (40 μg/ml) BALB/3T3-A31 cells using the TRIzol method. DNase-treated total RNA samples were digested with single-strand-specific RNases A and T1 (Ambion) or with RNase A/T1 plus dsRNA-specific RNase V1 (Ambion) in a total volume of 20 μl for 1 h at 37°C. The digested RNA samples and the untreated control sample (undigested) were purified using the RNA Clean & Concentrator kit (Zymo Research, Freiburg, Germany) and denatured at 95°C for 3 min. RT-qPCR was performed as described above by employing a commercially available EGFP TaqMan assay (Mr04329676_mr; Life Technologies). The mean of the fold induction of EGFP RNA over mock from the duplicate qPCRs of undigested RNA samples was calculated and set to 100%. The percentage of the remaining EGFP RNA after RNase A/T1 and A/T1/V1 digests was calculated by employing the fold induction values of the respective EGFP RNA samples.
Viral replication analysis.
For analysis of multicycle virus replication, confluent monolayers in 6-well culture plates were infected at a multiplicity of infection (MOI) of 0.05 in 500 μl of DMEM without FCS. After 60 min of adsorption at 37°C, cells were washed once with DMEM and were further incubated in DMEM-2% FCS. Cells and supernatant were harvested at the indicated time points, freeze-thawed, and sonicated. For the 0-h time point, cells were kept on ice after the 60-min adsorption period, washed with ice-cold DMEM, supplemented with 2 ml ice-cold DMEM-2% FCS, and immediately frozen. MVA titers were determined on CEF cells, and CVA titers were determined on CV-1 cells using the TCID50 method as described previously (45).
Immunoblot analysis of protein phosphorylation status and steady-state levels.
BALB/3T3-A31 and MEFs from PKR-deficient mice and from the corresponding wild-type controls were seeded on the day before infection in 12-well tissue culture plates. Infection, cell lysate preparation, immunoblotting, immunodetection, and stripping were performed as previously described (47). For detection of protein bands by chemiluminescence, two different substrate reagents were used: SuperSignal West Pico (Thermo Scientific, Bonn, Germany) as the standard reagent and the ECL Advance Western blotting detection kit (GE Healthcare, Munich, Germany) for high-sensitivity detection of phospho-IRF3 (p-IRF3) in PKR−/− MEF lysates. Antibodies with the following specificities were used: phospho-eIF2α (p-Ser51, dilution 1:2,000, catalog no. 9721), p-IRF3 (p-Ser396, 1:1,000, catalog no. 4947S), both from Cell Signaling, Danvers, USA, and anti-mouse-β-tubulin (clone SAP.4G5, 1:20,000; Sigma-Aldrich).
Mouse infection experiments.
Mice were anesthetized by ketamine-xylazine injection prior to intranasal infection with 2 × 106, 1 × 107, and 5 × 107 TCID50 of CVA and CVA mutants diluted in phosphate-buffered saline (PBS) to a final volume of 50 μl per mouse. Animals were weighed and inspected daily for 2 weeks, and the signs of illness were scored on an arbitrary scale from 0 to 4 as previously described (44). For analysis of systemic cytokine levels, mice were injected intravenously (i.v.) with 200 μl of the respective virus dilutions and bled 6 h later by the tail vein. All animal experiments have been approved according to the regulations of the German animal welfare law by the responsible authority, the Government of Upper Bavaria (Regierung von Oberbayern), and were carried out in accordance with the regulations set forth by the responsible authority and with the guidelines for animal experiments of Bavarian Nordic GmbH.
Systemic cytokine level analysis by cytometric bead assay.
Cytokine concentrations in mouse sera drawn 6 h after i.v. infection were determined by the bead-based FlowCytomix assay for the indicated mouse cytokines (eBioscience, Frankfurt, Germany) according to the manufacturer's instructions. The statistical significance for differences between treatment groups was analyzed using the nonparametric Mann-Whitney U test. The overall significance level (0.05) was Bonferroni corrected by dividing by the number of groups (e.g., 11 cytokines and chemokines tested in B6129SF2/J mice), i.e., the overall Bonferroni corrected level was set to 0.05/11 = 0.00455 for comparison between the B6129SF/2 treatment groups.
ELISA.
IFN-β protein levels secreted by infected cell lines were always determined at 14 to 16 h p.i., when maximal amounts of IFN-β had accumulated in supernatants of MVA-infected cells. Enzyme-linked immunosorbent assay (ELISA) kits for murine IFN-β (VeriKine mouse interferon beta) and human IFN-β (VeriKine-HS human interferon beta) were purchased from PBL Assay Science (Piscataway, NJ, USA). Murine IFN-α in supernatants of GM-DCs was determined using commercially available ELISA reagents as previously described (43).
RESULTS
VACV generating excess dsRNA induces increased IFN-β responses in cultured cells.
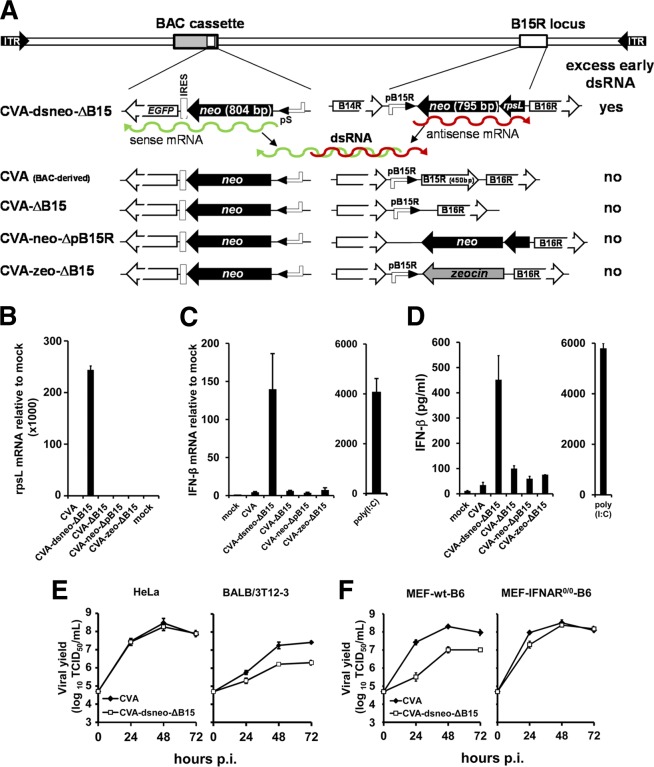
We generated a CVA mutant (CVA-dsneo-ΔB15) expressing partial cRNA transcripts from two separate insertions of the neomycin (neo) resistance gene (Fig. 1A). The neo sense transcript was derived from the neo-IRES-EGFP selection marker contained in the BAC cassette of our CVA constructs required for propagation of the CVA-BAC clone in E. coli (44). Transcription of the neo-IRES-EGFP mRNA is driven by the well-characterized early/late poxvirus pS promoter (51). The second neo transcript was generated from the neo open reading frame (ORF) in the neo/rpsL-positive/negative selection marker at the B15R locus, replacing the B15R ORF downstream of the B15R promoter (Fig. 1A). Since the neo/rpsL cassette is inserted in reverse orientation to the B15R promoter, a neo antisense transcript is generated which does not encode a protein. We confirmed the presence of the neo/rpsL transcript by RT-qPCR targeting the rpsL portion of this RNA (Fig. 1B). CVA-dsneo-ΔB15 infection of murine embryo fibroblasts (MEFs) resulted in increased levels of IFN-β transcripts as determined by RT-qPCR (Fig. 1C). In contrast, CVA-ΔB15 also lacking the B15R ORF but, without the additional neo/rpsL cassette (Fig. 1A), induced very low IFN-β gene expression comparable to that of wild-type CVA (Fig. 1C), demonstrating that the loss of the B15R gene per se was not responsible for increased IFN-β gene expression. When the B15R promoter (pB15R) directing transcription of antisense neo mRNA was deleted from CVA-dsneo-ΔB15, the resulting CVA-neo-ΔpB15R mutant failed to express detectable neo/rpsL transcripts (Fig. 1B), and IFN-β mRNA levels induced by this mutant were similar to the background levels induced by CVA (Fig. 1C). Thus, transcription of the neo/rpsL cassette inserted in antisense orientation was required for enhanced IFN-β mRNA induction in MEFs. A CVA mutant containing a zeo ORF in place of the neo/rpsL cassette in reverse orientation to the B15R promoter (CVA-zeo-ΔB15; Fig. 1A) also did not increase IFN-β mRNA expression compared to that of CVA (Fig. 1C). In line with the IFN-β mRNA data, increased levels of IFN-β protein were observed only in supernatants of CVA-dsneo-ΔB15-infected MEFs (Fig. 1D) or BALB/3T3-A31 cells (data not shown).
FIG 1.
A CVA mutant expressing excess early dsRNA induces increased expression of IFN-β. (A) Schematic representation of a CVA mutant expressing sense and antisense mRNA (indicated by green and red wiggly lines) from two neo inserts and of control and reference constructs. Promoters driving transcription of the neo inserts are indicated, as well as flanking ORFs at the B15R locus. (B) Mouse embryo fibroblasts (MEFs) in 6-well plates were mock infected or infected with crude stocks of the indicated CVA recombinants at an MOI of 10 in duplicate. The relative amount of neo-rpsL antisense transcripts (rpsL mRNA) compared to that of the mock was determined by duplicate RT-qPCR per sample using total RNA isolated from cells at 6 h p.i. and custom TaqMan primers for the rpsL portion of the neo-rpsL antisense transcript. An 18S rRNA-specific probe served as the endogenous control in RT-qPCR analysis. (C) MEFs in 6-well plates were mock infected or infected with crude stocks of the indicated MVA recombinants in duplicate at an MOI of 10. Fold induction of IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample using total RNA isolated from cells at 6 h p.i. Poly(I·C) was transfected using Fugene HD at 2 μg/well. Where error bars are not visible, the standard error was negligible. (D) IFN-β amounts in supernatants of MEF cultures infected in parallel to those shown in panel C were determined at 14 h p.i. by ELISA. Human HeLa and murine BALB/3T12-3 cells (E) and MEFs from C57BL/6 mice (MEF-wt-B6) and from IFNAR−/− C57BL/6 mice (“MEF-IFNAR0/0-B6”) in 6-well plates (F) were infected in triplicate at an MOI of 0.025 of the indicated viruses. Cells and supernatants were harvested at the indicated time points, and infectious viral titers in lysates were determined by standard titration assays on CEF cells using the TCID50 method. Total viral output at the indicated times is plotted, and each data point represents results from single titrations of three independent wells.
CVA-dsneo-ΔB15 replication was indistinguishable from that of CVA in HeLa cells but was moderately impaired in murine BALB/3T12-3 fibroblasts (Fig. 1E). A moderate replication impairment of CVA-dsneo-ΔB15 was also observed in MEFs (Fig. 1F) but was largely absent in MEFs from mice lacking a functional IFN-I receptor (Fig. 1F).
To exclude unwanted mutations in CVA-dsneo-ΔB15 as the cause of its enhanced IFN-β stimulatory capacity and altered replication behavior, we determined the nucleotide sequence of the complete coding regions of CVA-dsneo-ΔB15 (202,615 nucleotides) and CVA-ΔB15 (201,296 nucleotides). Both viruses contained only the deliberately introduced mutations. These findings collectively indicate that dsRNA formed by annealed sense and antisense neo transcripts was the trigger for increased induction of IFN-β by CVA-dsneo-ΔB15.
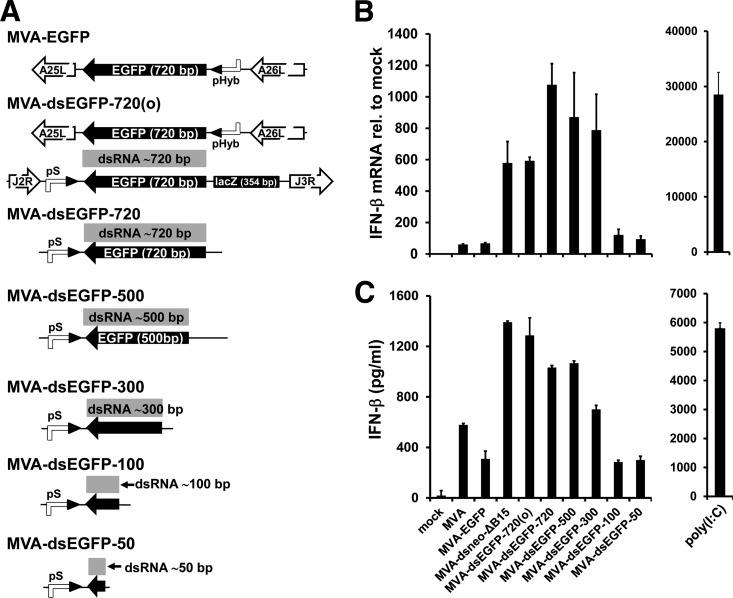
MVAs producing excess early dsRNA.
To further confirm the IFN-I stimulatory effect of early dsRNA generation during vaccinia virus infection, we constructed two pairs of recombinant MVA vectors expressing two complementary transcripts of either neo or the EGFP gene. This was achieved by inserting two copies of the neo or the EGFP ORF at distant sites in the MVA genome, each under the control of a poxviral early/late promoter (Fig. 2A). The BAC-derived wild-type MVA already harbored one copy of the neo/EGFP cassette within the BAC backbone insert (Fig. 2A). The BAC insert, including the neo/EGFP cassette, does not alter the properties of BAC-derived MVA and is thus considered to be equivalent to wild-type MVA (44). MVA-dsneo-ΔB15 was engineered to contain a second neo ORF as part of the neo/rpsL cassette at the B15R locus in reverse orientation relative to the endogenous B15R promoter (Fig. 2A), thus reproducing the constellation of neo inserts in the CVA-dsneo-ΔB15 mutant described above. MVA-dsneo-ΔB15 induced strongly increased expression of IFN-β mRNA and enhanced secretion of IFN-β protein into culture supernatants, whereas the MVA-ΔB15 mutant and MVA stimulated IFN-β to very similar extents (Fig. 2C and D). Excision of the complete loxP-flanked BAC insert from MVA-dsneo-ΔB15 by site-specific Cre recombination, resulting in the MVA-neo-ΔB15/ΔBAC mutant, abrogated the increased induction of IFN-β mRNA and protein (Fig. 2C and D). In conjunction with the results of the CVA mutants described above, we conclude that complementary neo transcripts forming dsRNA represented the stimulus for the observed increase in IFN-β induction.
FIG 2.
MVA recombinants expressing excess early dsRNA from neo or EGFP transgenes induce increased IFN-β expression. (A) Schematic representation of the two types of MVA recombinants generating excess early dsRNA either from two neo inserts (top) or from two EGFP inserts (bottom), each with the corresponding control and reference constructs. IGR, intergenic region. (B) Total RNA from murine BALB/3T3-A31 cells infected with the indicated viruses (MOI 10) or mock infected for 6 h was digested with RNase A/T1 (ssRNase digest) or RNase A/T1/V1 (ss+dsRNase digest) or not digested, and duplicate RT-qPCR quantification of total EGFP transcript (both sense and antisense) was performed as described in Materials and Methods. The mean of the fold induction values of EGFP or C7L transcripts over mock in undigested samples was set to 100%, and the mean percentage of the remaining qPCR signals after the indicated RNase digests was calculated for EGFP and C7L transcripts. Shown is one out two independent experiments. Where error bars are not visible, the standard error was negligible. (C) MEFs in 6-well plates were mock infected or infected with crude stocks of the indicated MVA recombinants at an MOI of 10 in duplicate. Fold induction of IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample using total RNA isolated from cells at 6 h p.i. using a commercially available TaqMan assay (Life Technologies) for the murine IFN-β gene. Poly(I·C) was transfected using Fugene HD at 2 μg/well. 18S rRNA served as the endogenous control in all RT-qPCR analyses. Where error bars are not visible, the standard error was negligible. (D) IFN-β amounts in supernatants of MEF cultures infected in parallel to those shown in panel C were determined at 14 h p.i. by ELISA. (E) Murine A31 cells were either preincubated with 40 μg/ml of AraC for 1 h or left untreated and infected in duplicate at an MOI of 10 with the indicated MVAs with either 40 μg/ml AraC throughout infection or without AraC. Cells were harvested at 6 h p.i. for isolation of total RNA. Messenger RNAs for murine IFN-β and the late F17R VACV gene were quantified by qRT-PCR analysis as described above.
For the second set of early dsRNA-generating MVA mutants, two inserts of the EGFP gene generating sense and antisense EGFP transcripts were integrated at distant sites into the genome of MVA (Fig. 2A). In this set of MVA mutants, the neo/EGFP cassette in the BAC backbone had been replaced by a tetracycline selection cassette. The basic MVA-dsEGFP-720(o) construct was designed to include an extended noncomplementary 3′ overhang at the antisense strand derived from a 354-bp-long coding sequence of the bacterial β-galactosidase (lacZ) gene. In contrast to the neo-based constructs, no autologous MVA gene was deleted in the MVA-EGFP constructs. In addition, the previously described strong early/late pS promoter (51) was utilized instead of the endogenous B15R promoter for antisense mRNA expression. Transcription of antisense EGFP-lacZ mRNA was verified by RT-qPCR targeting the lacZ portion of this mRNA (data not shown). Replication of MVA-dsEGFP-720(o) as determined by multicycle replication analysis and comparison of viral yields was only marginally decreased, and MVA-dsEGFP-720(o) expressed levels of EGFP similar to those of MVA-EGFP in MRC-5 cells both at 5 and 25 h p.i. (data not shown).
To demonstrate that sense and antisense EGFP transcripts indeed formed dsRNA, we conducted differential RNase digests of total RNA from MVA-dsEGFP-720(o)-infected cells with single-stranded RNA (ssRNA)- and dsRNA-specific RNases prior to RT-qPCR quantification of the remaining EGFP RNA. To prove that EGFP dsRNA is already increased early in infection before the onset of intermediate and late transcription, we isolated total RNA from infected cells treated with AraC. The applied method is very similar to a previously described protocol (26). While >99% of EGFP RNA from MVA-EGFP-infected cells could be degraded with the ssRNA-specific RNases A and T1, 28% of the EGFP RNA from MVA-dsEGFP-720(o)-infected cells remained resistant to ssRNase treatment (Fig. 2B). This indicates that a significant proportion of the complementary EGFP RNAs in MVA-dsEGFP-720(o)-infected cells had formed dsRNA, rendering this RNA resistant to RNase A/T1 treatment. The EGFP RNA signal in samples from MVA-dsEGFP-720(o)- and MVA-EGFP-infected cells was reduced to levels close to the detection limit when the RNA remaining after the ssRNase digest was further incubated with dsRNA-specific RNase V1 (Fig. 2B). This demonstrates that the vast majority of the 28% of ssRNase-resistant EGFP RNA molecules from MVA-dsEGFP-720(o)-infected cells were indeed EGFP dsRNA. As a control for the ssRNase digest, we analyzed degradation of the early viral C7L transcript by RT-qPCR (Fig. 2B, bottom) by using the same digested RNA samples as those used for the EGFP RNA analysis (Fig. 2B). More than 99% of C7L RNA from both MVA-EGFP- and MVA-dsEGFP-720(o)-infected cells was sensitive to ssRNase treatment (Fig. 2B), indicating that the ssRNase digest of the RNA from MVA-dsEGFP-720(o)-infected cells was efficient and that only trace amounts of C7L dsRNA were present, as expected. Thus, MVA-dsEGFP-720(o) is indeed able to generate significant amounts of EGFP dsRNA in infected cells.
MVAs producing excess early dsRNA induce increased expression of IFN-β.
MVA-dsEGFP-720(o) induced increased levels of IFN-β mRNA and protein in MEFs compared to those of MVA-EGFP expressing only a sense EGFP mRNA (Fig. 2C and D). IFN-β gene expression and protein secretion in MEF cells induced by the reference construct MVA-EGFP expressing only the sense EGFP transcript was not detectably different from that induced by MVA (Fig. 2C and D). Therefore, MVA-EGFP was used as the sole reference virus in all further experiments with EGFP-based MVA dsRNA recombinants.
MVA-dsEGFP-720(o) enhanced IFN-β gene induction in MEFs as described above, although MEFs did not support late viral gene expression by MVA (data not shown), suggesting that early transcription is sufficient for enhanced IFN-β induction. AraC treatment of MVA-infected murine A31 cells, which efficiently support late gene expression by MVA, as demonstrated by the presence of large amounts of transcripts from the late F17R gene in untreated cells at 6 h p.i., did not abrogate increased IFN-β induction by MVA-dsEGFP-720(o) (Fig. 2E). In contrast, AraC treatment drastically decreased the amount of F17R transcripts, demonstrating that it was able to effectively block postreplicative gene expression in A31 cells (Fig. 2E). Notably, IFN-β gene expression induced by MVA-dsEGFP-720(o) as well as by the MVA-EGFP reference virus was increased by the AraC treatment per se (Fig. 2E). The reason for this effect is presently unclear. In conclusion, excess production of viral dsRNA in the early phase of MVA infection is sufficient to achieve the IFN-β enhancing effect.
Length requirements for early dsRNA to stimulate IFN-β gene expression.
The length of the EGFP-ORF fragment used for generation of EGFP antisense transcripts was progressively shortened from the 720 bp of the wt EGFP ORF to 50 bp of the 3′ end of the ORF (Fig. 3A). All constructs except MVA-dsEGFP-720(o) lack the lacZ-derived noncomplementary sequence attached to the 3′ end of the antisense EGFP transcript (Fig. 3A). In wt MEFs, the group of MVA recombinants expressing complementary EGFP transcripts with overlaps between 720 and 300 bp induced increased levels of IFN-β mRNA and protein (Fig. 3B and C). The β-galactosidase (β-Gal)-derived 3′ overhang did not significantly alter the IFN-β enhancing effect in MEFs (Fig. 3B and C). Enhancement of IFN-β gene expression was largely undetectable, with MVA-dsEGFP constructs containing only 100 or 50 bases of EGFP sequence directing antisense expression (MVA-dsEGFP-100 and MVA-dsEGFP-50; Fig. 3B and C). As an approximation of the EGFP dsRNA amount, we quantified the total amount of sense and antisense EGFP RNA in cells infected with the various mutants (except MVA-dsEGFP-50) by RT-qPCR. We found that the amounts of total EGFP RNA generated by MVA-dsEGFP-720(o) and MVA-dsEGFP-100 were indistinguishable, indicating that early dsRNA of 100-bp length was too short to stimulate enhanced IFN-β expression (data not shown). For the group of recombinants expressing antisense RNAs between 720 and 300 bp of length, we could not exclude a correlation of the differences in IFN-β induction (Fig. 3B and C) with the total EGFP RNA amount (data not shown) rather than with the length of the EGFP dsRNA. Thus, the threshold length of MVA-expressed early dsRNA for inducing a detectable increase in IFN-I induction lies between 100 and 300 bp.
MVA-dsEGFP mutants enhance IFN-β production also in human cells.
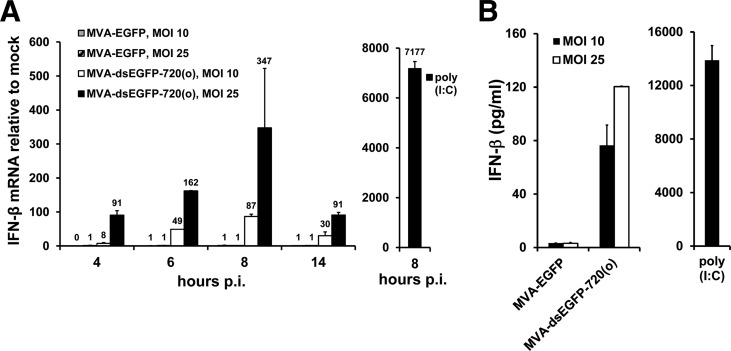
MVA-dsEGFP-720(o) induced significantly enhanced IFN-β mRNA expression in cultures of the interferon-competent human diploid lung fibroblast cell line MRC-5 with a peak around 8 h p.i., whereas basal IFN-β gene induction by MVA-EGFP was low and occasionally even undetectable in human MRC-5 cells (data not shown and Fig. 4A). Increasing the multiplicity of infection (MOI) further enhanced IFN-β mRNA induction (Fig. 4A). IFN-β protein levels in the supernatants of MRC-5 cells were also enhanced by MVA-dsEGFP-720(o) in a dose-dependent manner (Fig. 4B). Of note, the basal levels of IFN-β induction by MVA appeared to be much lower in human MRC-5 cells than in MEFs (compare Fig. 4 with Fig. 2C and D and Fig. 3B and C). MRC-5 cells infected with MVA constructs generating progressively shortened early EGFP-dsRNA showed a pattern of decreased IFN-β gene induction similar to that obtained with murine cells (data not shown). Thus, increased IFN-β induction by excess dsRNA generated early during MVA infection was not restricted to murine cells but was at least as prominent in human cells.
FIG 4.
MVA generating excess early dsRNA induces increased IFN-β expression in human cells. (A) Human MRC-5 cells in 6-well plates were mock infected or infected with crude stocks of the indicated MVA recombinants in duplicate at MOIs of 10 and 25. Fold induction of IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample with 18S rRNA as the endogenous control using total RNA isolated from cells after the indicated time points p.i. As a positive control, 2 μg of poly(I·C) per well was transfected using Fugene HD. Data labels were included in panel A to identify bars of low or zero height. (B) IFN-β in supernatants of MRC-5 cells at 14 h p.i. infected in parallel to the MRC-5 cultures shown in panel A was quantified by ELISA.
MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) activate PKR.
PKR is constitutively synthesized in mammalian cells at moderate levels as an inactive kinase that is activated by binding to dsRNA. The best-characterized substrate of activated PKR is the translation initiation factor eIF2α, which is phosphorylated by PKR at serine residue 51. Ser-51 phosphorylation of eIF2α in cells infected with MVA mutants generating excess early dsRNA was analyzed as an indicator of PKR activation. MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) activated PKR as early as 1 h after infection of A31 cells, whereas MVA wt did not detectably activate PKR throughout infection (Fig. 5). The amount of phospho-eIF2α (p-eIF2α) in cells infected with both neo- and EGFP-based early dsRNA producer mutants of MVA increased further until 4 h p.i. (Fig. 5). At 6 h p.i., the amounts of p-eIF2α in MVA-dsneo-ΔB15- and MVA-dsEGFP-720(o)-infected cells had declined and were only weakly if at all detectable at 8 h p.i. (Fig. 5). In contrast, an MVA mutant lacking the ORF encoding the vaccinia virus E3 protein (MVA-ΔE3L) showed a late onset of eIF2α phosphorylation (Fig. 5). The p-eIF2α signal induced by MVA-ΔE3L became detectable earliest after 3 h p.i. and then remained high for the rest of the observation period up to 8 h p.i. (Fig. 5) and beyond (data not shown). Thus, the early kinetics of eIF2α phosphorylation observed in MVA-dsEGFP-720(o)-infected cells argue for a very early but transient activation of PKR, which is most likely caused by the use of early or immediate early promoters driving transcription of both sense and antisense neo and EGFP transcripts.
FIG 5.

The MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) mutants induce PKR activation. Murine A31 cells were infected at day 1 after seeding with crude stocks of the indicated viruses at an MOI of 10. Phosphorylation of eIF2α (p-eIF2α) in cell lysates prepared after the indicated times of infection was analyzed by immunoblotting using an antibody against the phospho-epitope Ser-51 of eIF2α (“p-eIF2α” panels). Loading was monitored by stripping the blots and reprobing with an antibody against murine β-tubulin.
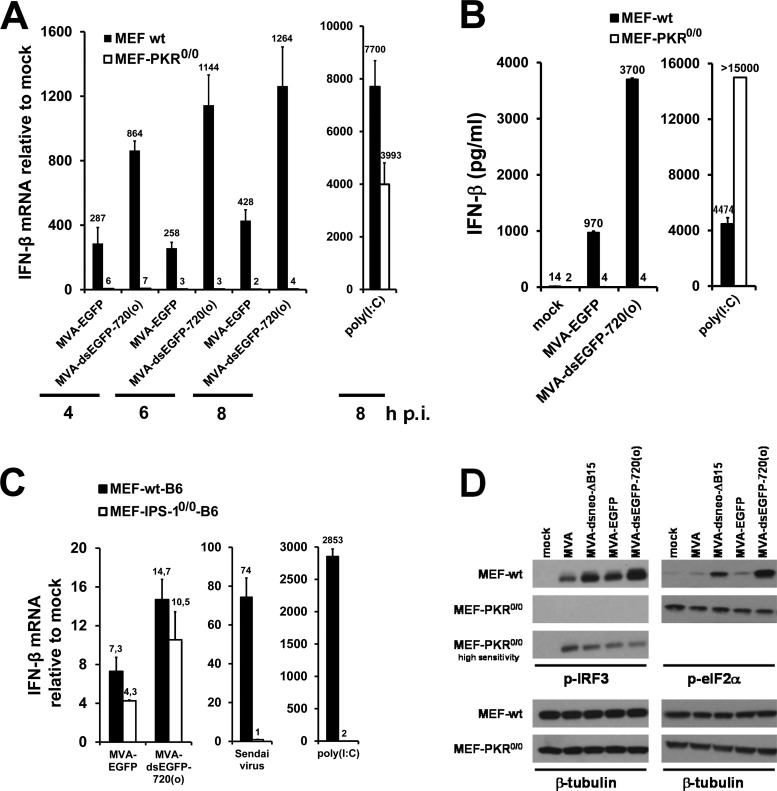
PKR but not IPS-1 is essential for increased IFN-β expression by MEFs.
To further examine the role of PKR in the enhanced IFN-β induction by MVA mutants expressing excess early dsRNA, we analyzed IFN-β induction in MEFs lacking a functional PKR. MVA-dsEGFP-720(o) did not induce enhanced IFN-β gene expression in PKR-deficient MEFs at any time point analyzed, while compared to MVA-EGFP it enhanced IFN-β mRNA synthesis in the corresponding wt MEFs, as expected (Fig. 6A). Secretion of IFN-β protein by PKR−/− MEFs infected with MVA-dsEGFP-720(o) and MVA-EGFP was undetectable (Fig. 6B), indicating that enhanced IFN-β production by MVA-dsEGFP-720(o) was dependent on functional PKR. Similar results were obtained for MVA-dsneo-ΔB15-infected PKR−/− MEFs (data not shown). PKR−/− MEFs transfected with the dsRNA mimic poly(I·C) secreted large amounts of IFN-β, demonstrating that the PKR−/− MEFs used here were fully competent to generate IFN-β mRNA and secrete IFN-β and that poly(I·C)-mediated IFN-β induction is not PKR dependent (Fig. 6A and B). MVA-EGFP induced lower levels of IFN-β in PKR−/− MEFs than in wt MEFs, suggesting a general role of this dsRNA sensor in induction of the IFN-I in MEFs by MVA (Fig. 6A and B). In summary, PKR appeared to be a critical cellular sensor involved in the enhanced induction of IFN-β by MVA-dsEGFP-720(o) and appeared to be involved in IFN-β gene induction by MVA in MEFs in general.
FIG 6.
Role of PKR in enhanced IFN-β induction by MVAs producing excess early dsRNA. (A) PKR−/− MEFs and the corresponding wt MEFs were mock infected or infected with the indicated MVAs in duplicate at an MOI of 10 for the indicated times. As a positive control, 2 μg of poly(I·C) was transfected and incubated for 8 h. Fold induction of IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample using total RNA isolated from cells at 5 h p.i. (B) IFN-β from supernatants of parallel infections of wt and PKR−/− MEFs shown in panel A at 14 h p.i. was analyzed by ELISA. Where error bars are not visible, the standard error was negligible. (C) MEFs from wt and IPS-1−/− mice were infected in duplicate with the indicated viruses at an MOI of 10. Fold induction of IFN-β mRNA over mock was determined by duplicate RT-qPCR per sample using total RNA isolated from cells at 6 h p.i. Sendai virus infection and transfection of 2 μg poly(I·C)/well served as controls. (D) Wild-type and PKR−/− MEFs were infected with MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) and the respective control viruses at an MOI of 10 for 4 h. Immunoblots of cell lysates from wt and PKR−/− MEFs were incubated with a monoclonal antibody against p-IRF3, and blots were first developed using a standard chemiluminescence substrate and applying identical exposure times. The blot membrane with samples from PKR−/− MEFs was then reincubated with a high-sensitivity chemiluminescence substrate and exposed to X-ray film for longer times than in the panels above to enhance the signal. Equal loading was monitored by stripping the blots and reprobing with an antibody against murine β-tubulin using standard ECL substrate (lower panels).
To evaluate a possible role of the IPS-1 signaling adaptor molecule and thus of the RIG-like dsRNA receptors in PKR-mediated enhanced IFN-I induction, we analyzed MEFs from IPS-1-deficient (IPS-1−/−) mice and the corresponding C57BL/6 MEFs for IFN-β gene induction by MVA-dsEGFP-720(o). MEFs from IPS-1−/− mice showed enhanced IFN-β gene expression upon infection by MVA-dsEGFP-720(o) comparable to that observed in wt MEFs, while IFN-β induction by poly(I·C) was almost completely dependent on IPS-1, as expected (Fig. 6C). In addition, IFN-β induction by Sendai virus, a negative-strand RNA virus known to depend on RIG-I and IPS-1 for IFN-I induction, was completely abolished in the IPS-1−/− MEFs (Fig. 6C), confirming that the IPS-1 pathway was indeed nonfunctional in these cells.
MVA-dsEGFP-720(o) drives PKR-dependent phosphorylation of IRF3.
IRF3 is a central transcription factor for generation of IFN-β mRNA, and its phosphorylated form is an essential component of the so-called enhanceosome, which also contains transcription factors NF-κB and ATF-2/c-jun and which binds to the enhancer element in the 5′ nontranslated region of the IFN-β gene driving its transcription (52). Clearly, MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) induced a stronger IRF3 phosphorylation than their corresponding reference constructs MVA-ΔB15 and MVA-EGFP (Fig. 6D). Thus, increased levels of IFN-β mRNA in cells infected with MVA-dsneo-ΔB15 and MVA-dsEGFP-720(o) most likely resulted at least in part from increased transcription of the IFN-β gene. Whether enhanced IFN-β mRNA stabilization, which was recently reported to be mediated by PKR (53), contributed to the increased IFN-β mRNA levels remains unknown at present. Increased IRF3 phosphorylation by MVAs overproducing early dsRNA coincided with increased eIF2α phosphorylation (Fig. 6D). In contrast to wild-type MEFs, neither increased eIF2α phosphorylation nor increased IRF3 phosphorylation was detectable in PKR−/− MEFs infected with MVA-dsneo-ΔB15 or MVA-dsEGFP-720(o) (Fig. 6D), indicating that PKR is a major regulator not only of eIF2α but also of IRF3 phosphorylation in the context of MVA infection. The enhanced basal levels of eIF2α phosphorylation in PKR−/− MEFs independent of treatment were observed in all experiments (Fig. 6D). The reasons for the latter effect are presently unknown. The levels of phosphorylated IRF3 were generally very low in MVA-infected PKR-deficient MEFs and could be detected only by long exposure of immunoblots developed with a high-sensitivity luminescence substrate (Fig. 6D), suggesting that at least in MEFs, PKR has an important role in MVA-induced IRF3 activation in general.
MVA-dsEGFP-720(o) induces an enhanced innate immune response in mice.
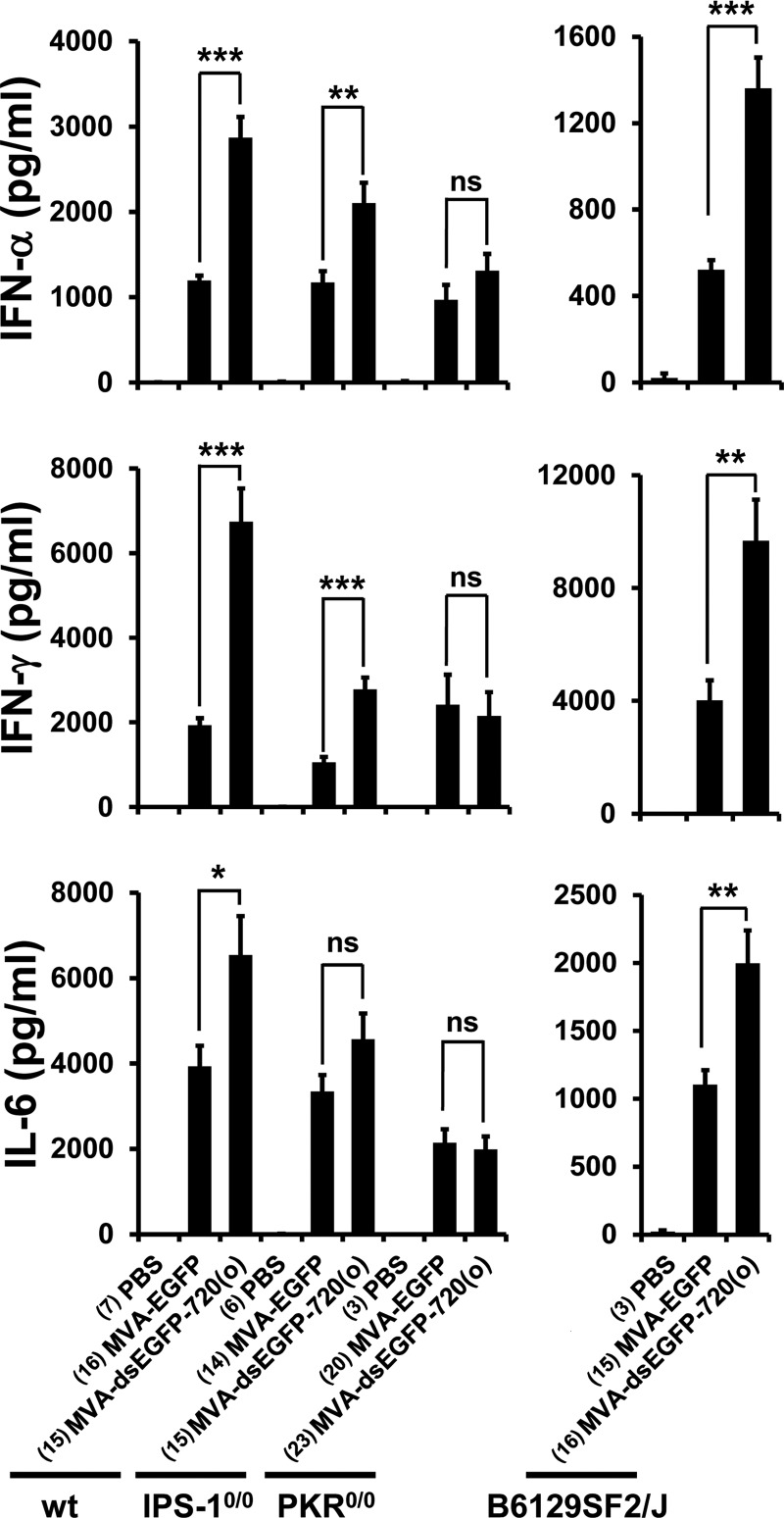
Levels of systemic cytokines in response to MVA-dsEGFP-720(o) were analyzed at 6 h after intravenous infection of C57BL/6 and B6129SF2/J mice, which served as control strains for IPS-1−/− mice and for the mixed genetic background of PKR−/− mice, respectively. IFN-α levels in the blood of MVA-dsEGFP-720(o)-infected mice were significantly higher than those after MVA-EGFP infection of C57BL/6 and B6129SF2/J mice (P < 0.0001 for all comparisons by Mann-Whitney U test) (Fig. 7). Systemic IFN-γ was also significantly increased in MVA-dsEGFP-720(o)-infected C57BL/6 and B6129SF2/J mice at 6 h p.i. compared to that in MVA-EGFP-infected mice (P < 0.0001 and P < 0.0047, respectively) (Fig. 7). Except for IFN-γ levels in B6129SF2/J mice, the differences in IFN-α and IFN-γ induction were still statistically significant after applying the very conservative Bonferroni correction (Bonferroni corrected alpha = 0.0021 for C57BL/6, IPS-1−/−, and PKR−/− mice and 0.0045 for B6129SF2/J mice). In addition, the inflammatory cytokine interleukin 6 (IL-6) was always elevated after MVA-dsEGFP-720(o) infection above levels induced by MVA in both mouse strains (P = 0.03 and P = 0.0047) (Fig. 7), although the increase did not reach statistical significance when the Bonferroni correction was applied. In contrast, in MVA-dsEGFP-720(o)-infected PKR−/− mice, none of the above-described cytokines showed a statistically significant difference in systemic levels at 6 h p.i. compared to MVA-EGFP infection (Fig. 7), demonstrating that PKR was a critical factor in increased induction of IFN-α, IFN-γ, and IL-6 by MVA-dsEGFP-720(o) also in vivo. It should be noted that there is still a slight increase of IFN-α levels in PKR−/− mice infected with MVA-dsEGFP-720(o), which is statistically nonsignificant (Fig. 7) but nevertheless suggests that factors other than PKR also play a role. Interestingly, PKR was not critical for the basal MVA-induced innate immune response in mice after intravenous application (Fig. 7), in contrast to the findings with MEFs (Fig. 6).
FIG 7.
MVA-dsEGFP-720(o) induces enhanced IFN and cytokine responses in vivo. Wild-type C57BL/6 (wt), IPS-1−/−, PKR−/−, and B6129SF2/J female mice were infected intravenously at a dose of 108 TCID50/mouse. Animals were bled 6 h after infection, and serum was analyzed for the concentration of selected murine cytokines using a bead-based flow cytometric assay. Data are averages from four (wt, IPS-1−/−, PKR−/−) or two (B6129SF2/J) independent experiments. Numbers of mice per group are indicated in parentheses. P values were calculated using Mann-Whitney U test, comparing mice infected with the MVA-EGFP and MVA-dsEGFP-720(o) strains. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant (P > 0.05).
IPS-1−/− mice are unable to relay signals from the dsRNA sensors RIG-I and MDA5 to transcription factors promoting IFN-I gene expression. The increase in IFN-γ levels appeared unchanged in IPS-1−/− mice, while the increase in systemic levels of IFN-α and IL-6 induced by MVA-dsEGFP-720(o) compared to MVA-EGFP was less pronounced than that in wt mice but was still statistically significant for IFN-α and IFN-γ (P < 0.0023 and P <0.0001, respectively) (Fig. 7). The statistical significance of the difference in IFN-α levels did not withstand Bonferroni correction. In conclusion, the IPS-1 pathway appears to contribute to the MVA-dsEGFP-720(o)-induced effect of enhanced cytokine production in vivo, but the PKR pathway is more critical for this effect.
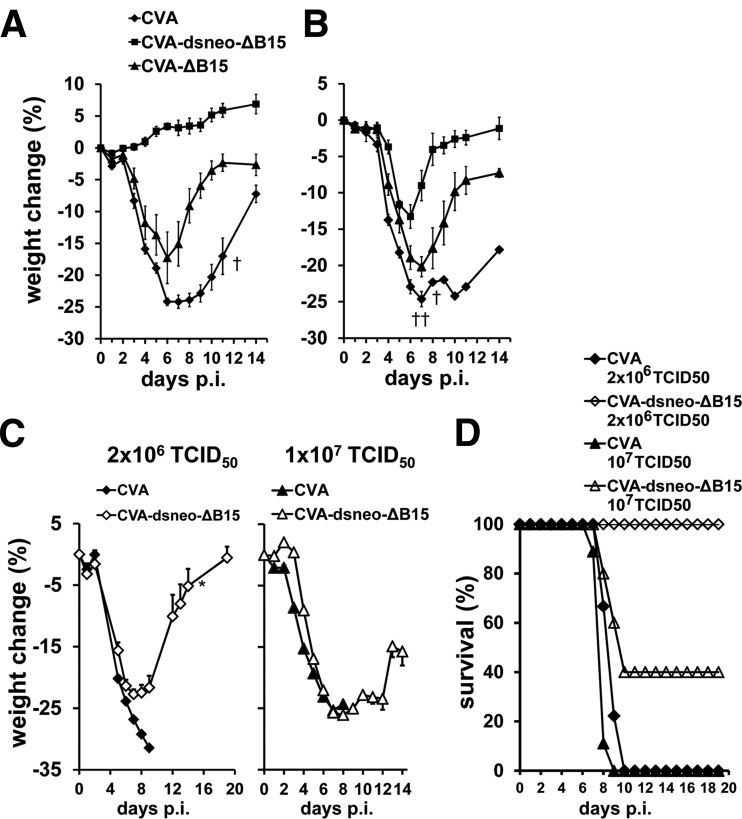
A CVA mutant producing early dsRNA is highly attenuated.
The virulence of a CVA mutant generating early dsRNA was assessed by disease monitoring following intranasal infection of C57BL/6 mice. CVA caused severe weight loss at a dose of 5 × 107 TCID50 in C57BL/6 mice, with some mice succumbing to disease (Fig. 8A). In contrast, CVA-dsneo-ΔB15 neither showed weight loss nor any signs of disease upon intranasal infection even at an inoculation dose as high as 5 × 107 TCID50 and was thus highly attenuated (Fig. 8A). Very similar results were obtained by intranasal infection of BALB/c mice (data not shown). The deletion of the B15R gene apparently had a detectable but minor role in the strong attenuation of CVA-dsneo-ΔB15, since CVA-ΔB15, lacking B15 as well as the neo antisense transcription at the B15 locus, showed only moderate attenuation (Fig. 8A). PKR-deficient mice infected intranasally with CVA-dsneo-ΔB15 showed clear signs of disease, indicating that PKR was crucial for the attenuation of this virus (Fig. 8B). Notably, CVA-dsneo-ΔB15 did not show the degree of virulence of CVA-ΔB15 in PKR-deficient mice, indicating that additional host factors contribute to attenuation of CVA-dsneo-ΔB15 in mice (Fig. 8B). CVA-ΔB15 also showed a certain degree of attenuation in PKR−/− mice, indicating that the effect of B15 absence is not PKR dependent.
FIG 8.
Groups of 5 to 9 wt mice (A), PKR−/− mice (B), and IFNAR−/− mice (C, D) were intranasally infected with a dose of 5 × 107 TCID50/mouse (wt and PKR−/−) or 2 × 106 and 1 × 107 TCID50/mouse (IFNAR−/−) of purified stocks of wild-type CVA, CVA-dsneo-ΔB15, and CVA-ΔB15 strains. Animals were inspected and weighed daily. Indicated is the average percentage of weight change (±standard error of the mean [SEM]) compared to body weight determined at day 0 (A to C) or the survival rate (D).
To assess the role of the IFN-I system in CVA-dsneo-ΔB15 attenuation, IFNAR−/− mice lacking a functional IFN-I receptor were intranasally infected. CVA-dsneo-ΔB15 inoculated at a dose of 2 × 106 TCID50 induced significant weight loss (Fig. 8C), but all CVA-dsneo-ΔB15-infected IFNAR−/− mice survived (Fig. 8D), while wt CVA-infected IFNAR−/− mice succumbed to infection (Fig. 8C and D). At an intermediate dose of 1 × 107 of CVA-dsneo-ΔB15, a subset of IFNAR−/− mice succumbed to infection, and weight loss was very prominent (Fig. 8C and D). A dose of 5 × 107 TCID50 of CVA-dsneo-ΔB15 as well as of CVA was uniformly lethal in IFNAR−/− mice (data not shown). CVA was also uniformly lethal for IFNAR−/− mice at the lower doses of 2 × 106 TCID50 and 1 × 107 TCID50 (Fig. 8D). These results demonstrate that the observed attenuation of CVA-dsneo-ΔB15 is strongly dependent on a functional IFN-I system, since CVA-dsneo-ΔB15 showed a very similar virulence to wild-type CVA at the high doses of 1 × 107 and 5 × 107 TCID50 in IFNAR−/− mice. The survival of CVA-dsneo-ΔB15-infected IFNAR−/− mice but not of CVA-infected IFNAR−/− mice at doses of 2 × 106 and 107 TCID50 suggests that host factors other than IFN-I are also important for CVA-dsneo-ΔB15 attenuation. In summary, PKR is partly responsible for the strong attenuation of the CVA-dsneo-ΔB15 in vivo, and attenuation almost completely depends on the IFN-I system. IPS-1 did not play a detectable role in attenuation, since the virulence of CVA-dsneo-ΔB15 in IPS-1-deficient mice was indistinguishable from virulence in wt mice (data not shown). This argues for a similar mode of action of early dsRNA-producing MVA and CVA mutants in vitro and in vivo, involving mainly PKR-mediated IFN-I induction.
DISCUSSION
VACV, the type species of the family Poxviridae, blocks the cellular recognition of viral dsRNA by PRRs and the ensuing immune activation by two major mechanisms timed during the viral replication cycle: (i) restricting the abundance of early overlapping complementary RNAs and (ii) expression of inhibitors of dsRNA recognition, primarily viral proteins E3 and K3, to counteract detection mainly of late dsRNA. We reasoned that overproduction of dsRNA from a single additional pair of complementary transcripts would not be able to override viral E3- and K3-mediated blockade of late dsRNA recognition, since levels and timing of expression of E3 and K3 are most probably well adjusted to counteract the effects of the large quantities of late dsRNA, which is much more abundant than early dsRNA in infected cells (22, 25, 26, 54). In contrast, early expression of additional dsRNA might override the low levels of E3 and K3 at this time point as well as bypass the viral mechanism that limits the length of early convergent transcripts to prevent overlap and consequently formation of activating quantities of dsRNA. Thus, we speculated that early during the VACV replication cycle, there might exist a window of opportunity for expressing excess dsRNA to induce an increased innate immune activation.
There are a number of reports indicating that dsRNA-triggered innate immune activation is avoided by maintaining a well-adapted balance between the levels of viral dsRNA and the expression levels of viral dsRNA recognition inhibitors. One example is the temperature-sensitive VACV mutant ts23 that harbors a mutation in the viral negative transcription elongation factor A18. This affected the transcription of all intermediate and late genes and led to the generation of mRNAs with much longer than normal 3′ untranslated regions at the nonpermissive temperature, leading to strongly increased quantities of postreplicative dsRNA (55, 56). The resulting phenotype of this mutant was a blockade of viral late gene expression (56) closely resembling that of E3L gene deletion mutants of VACV strain Copenhagen (57) and of MVA (58) in HeLa cells. This in turn indicates that in VACV mutant ts23, the abundance of late dsRNA and the quantity of viral dsRNA recognition inhibitors were out of balance. Another example illustrating the importance of this balance was an evolution experiment in cell culture using an E3L gene-deleted VACV. Mutants emerging in the course of cell culture passages of this VACV-ΔE3L were able to compensate the lack of the E3L gene by amplification of the K3L gene leading to enhanced K3 protein levels. The enhanced K3 levels were apparently able to achieve wild-type like inhibition of PKR and to restore VACV-ΔE3L replication (59). Our finding of transient eIF2α phosphorylation suggests that EGFP dsRNA generation directed by the late elements of the pS and the pHyb promoter in MVA-dsEGFP-720(o) did not activate PKR, implying that E3 and K3 expression levels at later times in infection were sufficient to compensate the additional late dsRNA generated from a single pair of transcription units.
Inevitably, early transcription and consequently also dsRNA formation from early mRNAs precede the translation of potential viral dsRNA recognition inhibitors from such early mRNAs. This suggests that at least one reason for the evolution of a viral mechanism that tightly restricts the length of early mRNAs was to avoid generation of dsRNA before sufficient amounts of viral dsRNA recognition inhibitors are produced in infected cells. Early transcription is terminated upon recognition of a TTTTTNT sequence motif in the coding strand of most early genes within approximately 20 to 50 nucleotides downstream of this signal and thus closely downstream of the translational stop codon (27). Notably, in instances where two adjacent early genes are transcribed toward each other, there are mostly two or three instead of only one early transcription termination motif found per ORF (28). Indeed, no early dsRNA derived from one such site of potential early transcript overlap could be detected in rabbit RK-13 cells infected with the VACV WR strain (26). Interestingly, early dsRNA from this site produced during infection with a K1L gene-deleted VACV WR was able to activate PKR, implying a role for K1 in preventing early dsRNA generation (26). Even if short early transcript overlaps of between 50 to 100 nucleotides were produced, these would most likely not be efficiently recognized by cellular receptors, as indicated by the fact that our mutants producing short EGFP dsRNA did not induce enhanced IFN-β. In conclusion, previous observations as well as our data indicate that there is a fine-tuned balance between the abundance of dsRNA of critical length produced during the different phases of VACV infection and the amount of poxviral inhibitors of dsRNA recognition. Using recombinant overexpression of early dsRNA, we were able to skew this balance in cells infected with our early dsRNA-generating MVA and CVA mutants and to achieve enhanced innate immune activation.
The characteristics of the poxviral replication cycle in infected cells apparently place PKR in a central position as a critical cell-intrinsic pattern recognition molecule alerting the host cell to an ongoing poxviral infection. This view is supported by the observation that multiple orthopoxvirus immunomodulators, including E3, K3, K1, and C7, inhibit the activation of PKR (26, 29, 37, 60, 61). The MVA mutants generating excess early dsRNA allowed us to further dissect the pathways by which PKR mediates IFN-I production in VACV-infected cells. It is well known that PKR triggers the activation of the NF-κB pathway, resulting in secretion of proinflammatory cytokines and chemokines (9). PKR-mediated NF-κB activation has also been postulated to be involved in enhanced expression of the IFN-β gene (62, 63), because NF-κB is a constitutive part of the enhanceosome triggering IFN-β gene expression (52). NF-κB activation by PKR was proposed to result at least in part from PKR-mediated phosphorylation of eIF2α, leading to translational control of the negative NF-κB regulator IκB (63). Wild-type MVA infection generally triggered IκB degradation in MEFs (our unpublished observation), indicative of NF-κB activation, as expected from a number of previously published reports (64, 65). However, IκB degradation induced by the MVA mutants expressing excess early dsRNA was indistinguishable from that induced by wild-type MVA (data not shown), indicating largely similar NF-κB activating capacities. Thus, enhanced NF-κB activation was unlikely to be responsible for the enhanced PKR-mediated transcription of the IFN-β gene reported here, which was rather mediated by the observed PKR-dependent increase in phosphorylated and thus activated IRF3.
In human HeLa cells infected with VACV, PKR-mediated IRF3 phosphorylation was reported to be dependent on RLRs and IPS-1 (66). In addition, a role for the dsRNA sensor MDA5 in MVA-induced IFN-β secretion by a human macrophage-like cell line has previously been reported (67). In contrast to these findings, our in vitro and in vivo data using IPS-1−/− MEFs and mice (Fig. 6 and 7) as well as IPS-1−/− GM-DCs (data not shown) argue against an important role of IPS-1 both for the effect of enhanced IFN-I induction by MVAs overproducing dsRNA early in infection and for basal wild-type MVA-induced IFN-I levels. The reason for this discrepancy is presently unclear. In our study, primary murine cells as well as mice were analyzed, while the above-cited studies were conducted with human cell lines. Possibly, primary murine cells do not use RLRs to induce IFN-I in the context of MVA infection. In mice, there was a moderate effect of IPS-1 deficiency on enhanced induction of IFN-α and on basal induction of IFN-γ, but IPS-1 was clearly neither essential for cytokine induction by MVA nor for enhanced induction of cytokines by MVA-dsEGFP-720(o). Depending on the cell type or species, activated PKR might not only enhance signals relayed via IPS-1 from the RIG-like dsRNA receptors, like in HeLa cells (66), but might also directly activate the IRF3 pathway. This could occur via interaction of PKR with TRAF3 (68), a signaling adaptor protein which mediates IFN-I production in a number of pathways downstream of PRRs.
The unexpected increase in IFN-β induction by AraC treatment of MVA-infected cells has been observed not only in A31 cells (see Fig. 2E) but also in MEFs and human MRC-5 cells (unpublished observation). It has previously been noted that AraC treatment can enhance the expression levels of early genes at later times both in MVA and VACV WR infection (69, 70). This effect might be caused by accumulation of early mRNA over the course of the artificially prolonged early phase, thus also increasing formation of dsRNA from complementary early mRNAs. This might in turn cause increased IFN-I induction, especially in the case of MVA that has lost a number of genes encoding antagonists of IFN-I-related pathways (e.g., B19R, N1L, C10L, A52R, VACV WR C16L). In particular, MVA does not express a functional K1 protein, which has been shown to counteract early dsRNA generation (25, 26). Alternatively, truncated viral genomic DNA might accumulate under AraC treatment and act as an additional PAMP, increasing the activation of cellular DNA recognition receptors. Such shorter, irregular genomic DNA molecules are thought to be generated by premature termination of viral genomic DNA synthesis due to AraC incorporation into newly synthesized DNA strands. In addition, viral factories are not formed under AraC treatment, which might expose these genomic DNA fragments to recognition by cellular receptors.
The reasons for the largely unaffected replication of CVA-dsneo-ΔB15 in HeLa cells and MVA-dsEGFP-720(o) in CEFs are unknown. Using MVA-dsEGFP-720(o)-infected murine cells, we observed a transient eIF2α phosphorylation, indicating transient rather than sustained activation of PKR, which is most likely also the case in CVA-dsneo-ΔB15-infected cells. Early transient PKR activation was apparently sufficient to upregulate IFN-β expression (see Results), but not apoptosis, and only marginally affected steady-state levels of MVA-expressed EGFP (unpublished data). Thus, potential eIF2α-mediated effects of PKR activation appeared to be minor. Very similar reasons might account for the largely unaffected replication of MVA-dsEGFP-720(o) in CEFs. In addition, the antiviral effect of enhanced IFN-I induction by CVA-dsneo-ΔB15 is likely negligible in human cells, since it has been reported that VACV replication in HeLa as well as in other human cells is relatively resistant to IFN-I treatment (71, 72). In contrast, the susceptibility of VACV replication to IFN-I treatment was much higher in murine cells than in human cells (71), possibly explaining our finding of reduced replication of CVA-dsneo-ΔB15 in IFN-I-competent, but not in IFN-I-deficient murine cells (see Fig. 1F). In CEF cells, neither MVA nor MVA-dsEGFP-720(o) induced detectable IFN-β mRNA (unpublished data). In addition, MVA could apparently afford the inactivation of the K1L gene during CEF cell adaptation, which encodes a protein with early dsRNA inhibitory function (25), suggesting that early transient PKR activation might not be critical for MVA replication in CEFs.
The attenuation in mice of the VACV mutant CVA-dsneo-ΔB15 expressing excess early dsRNA was remarkably strong and is mediated in large part by IFN-I, as shown by the greatly enhanced virulence of CVA-dsneo-ΔB15 in IFNAR−/− mice. The degree of attenuation in wt mice is similar to that observed for the E3L gene-deleted VACV WR strain (73, 74). The fact that all or some IFNAR−/− mice from treatment groups inoculated with doses of 2 × 106 and 1 × 107 TCID50 of CVA-dsneo-ΔB15 survived, whereas all IFNAR−/− mice infected with wild-type CVA at these doses died, suggested the contribution of IFN-I-independent factors to attenuation. CVA-ΔB15 not producing excess early dsRNA was far less attenuated than CVA-dsneo-ΔB15. This indicated that (i) neo-dsRNA generation contributed essentially to attenuation and that (ii) the lack of the B15R gene might have contributed to attenuation of CVA-dsneo-ΔB15. The B15R gene product is an inhibitor of NF-κB activation in VACV-infected cells (75). Therefore, an antiviral effect resulting from enhanced NF-κB activation due to the lack of B15 might have caused the moderate attenuation of CVA-ΔB15 in wt mice, as well as the remaining moderate attenuation of CVA-dsneo-ΔB15 in IFNAR−/− mice. The attenuation of CVA-ΔB15 is partly contradictory to previous observations with a VACV WR B15R deletion mutant. This mutant was not attenuated in BALB/c mice when inoculated via the intranasal route but showed attenuation in an intradermal lesion model (76). This discrepancy might have been caused by genetic differences between the two VACV strains. In contrast to the VACV WR strain, CVA lacks functional versions of several known (A39R, A55R) and potential (M1L, C9L) virulence factors (45) and is not mouse adapted, providing a rationale for potentially stronger effects of deletions of innate immune inhibitors on CVA virulence.
We have described here that deliberately increased dsRNA production early in the infection cycle of both replicating VACV and replication-restricted MVA was able to override viral countermeasures against PRR activation. The activation of PKR as an important cellular sensor of viral dsRNA resulted in enhanced innate immune activation, which is a desirable property to increase the immunogenicity of MVA-based vectors. The transient nature of PKR activation prevents induction of apoptosis, which is thought to require sustained PKR activation (9). The upper limit of innate immune activation achievable with MVA by this approach should be further explored, as well as alternative strategies to produce excess early dsRNA, such as using autologous viral sequences as the templates for sense and antisense mRNA production or deleting transcriptional termination signals from convergently transcribed early genes. Such studies will provide the basis for optimally exploiting the strategy of alerting the innate immune system in a temporally controlled manner for the development of MVA vectors with enhanced immunogenicity.
ACKNOWLEDGMENTS
We are employees of Bavarian Nordic GmbH, which funded the study, except for Mark Suter, who is a consultant to Bavarian Nordic. Paul Chaplin is also a shareholder of Bavarian Nordic.
We thank Jutta Kramer and Johannes Poddobrjanski from the Vaccine Generation Department of Bavarian Nordic for producing purified viral stocks, the Preclinical Department for animal husbandry, and Niels H. Wulff for help with statistical analysis of cytometric bead array data. We are grateful to Jovan Pavlovic, Institute for Medical Virology, University of Zürich, Switzerland, and Jürg Tschopp, University of Lausanne, Switzerland, for generously providing PKR-deficient mice and mouse embryo fibroblasts and for providing IPS-1-deficient mice, respectively.
Footnotes
Published ahead of print 8 October 2014
REFERENCES
- 1.Desmet CJ, Ishii KJ. 2012. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 12:479–491. 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki A. 2012. A virological view of innate immune recognition. Annu. Rev. Microbiol. 66:177–196. 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pichlmair A, Lassnig C, Eberle CA, Górna MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL, Rülicke T, Weber F, Colinge J, Müller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12:624–630. 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 4.Haller O, Kochs G, Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130. 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki A, Medzhitov R. 2010. Regulation of adaptive immunity by the innate immune system. Science 327:291–295. 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenz T, Waibler Z, Hofmann J, Hamdorf M, Lantermann M, Reizis B, Tovey MG, Aichele P, Sutter G, Kalinke U. 2010. Concomitant type I IFN receptor-triggering of T cells and of DC is required to promote maximal modified vaccinia virus Ankara-induced T-cell expansion. Eur. J. Immunol. 40:2769–2777. 10.1002/eji.201040453. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244. 10.1371/journal.pone.0024244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti A, Jha BK, Silverman RH. 2011. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 31:49–57. 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060. 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243:91–98. 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 11.Rehwinkel J, Reis e Sousa C. 2010. RIGorous detection: exposing virus through RNA sensing. Science 327:284–286. 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 12.Langland JO, Jacobs BL. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141. 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 13.Malmgaard L, Melchjorsen J, Bowie AG, Mogensen SC, Paludan SR. 2004. Viral activation of macrophages through TLR-dependent and -independent pathways. J. Immunol. 173:6890–6898. 10.4049/jimmunol.173.11.6890. [DOI] [PubMed] [Google Scholar]
- 14.Williams BR. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112–6120. 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 15.Gil J, Esteban M. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5:107–114. 10.1023/A:1009664109241. [DOI] [PubMed] [Google Scholar]
- 16.Goh KC, deVeer MJ, Williams BR. 2000. The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J. 19:4292–4297. 10.1093/emboj/19.16.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barry G, Breakwell L, Fragkoudis R, Attarzadeh-Yazdi G, Rodriguez-Andres J, Kohl A, Fazakerley JK. 2009. PKR acts early in infection to suppress Semliki Forest virus production and strongly enhances the type I interferon response. J. Gen. Virol. 90:1382–1391. 10.1099/vir.0.007336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilfoy FD, Mason PW. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 81:11148–11158. 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludwig H, Mages J, Staib C, Lehmann MH, Lang R, Sutter G. 2005. Role of viral factor E3L in modified vaccinia virus Ankara infection of human HeLa cells: regulation of the virus life cycle and identification of differentially expressed host genes. J. Virol. 79:2584–2596. 10.1128/JVI.79.4.2584-2596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper JA, Wittek R, Moss B. 1981. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J. Virol. 39:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahr A, Roberts BE. 1984. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J. Virol. 49:510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boone RF, Parr RP, Moss B. 1979. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J. Virol. 30:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colby C, Duesberg PH. 1969. Double-stranded RNA in vaccinia virus infected cells. Nature 222:940–944. 10.1038/222940a0. [DOI] [PubMed] [Google Scholar]
- 24.Moss B. 2013. Poxviridae, p 2129–2159 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Lynch HE, Ray CA, Oie KL, Pollara JJ, Petty IT, Sadler AJ, Williams BR, Pickup DJ. 2009. Modified vaccinia virus Ankara can activate NF-kappaB transcription factors through a double-stranded RNA-activated protein kinase (PKR)-dependent pathway during the early phase of virus replication. Virology 391:177–186. 10.1016/j.virol.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willis KL, Langland JO, Shisler JL. 2011. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-kappaB activation, when the K1 protein is absent or mutated. J. Biol. Chem. 286:7765–7778. 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuen L, Moss B. 1987. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc. Natl. Acad. Sci. U. S. A. 84:6417–6421. 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith GL, Symons JA, Alcami A. 1998. Poxviruses: interfering with interferon. Sem. Virol. 8:409–418. 10.1006/smvy.1997.0145. [DOI] [Google Scholar]
- 29.Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 89:4825–4829. 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas C, Gil J, Melkova Z, Esteban M, Diaz-Guerra M. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406–414. 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 31.Romano PR, Zhang F, Tan SL, Garcia-Barrio MT, Katze MG, Dever TE, Hinnebusch AG. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304–7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp TV, Moonan F, Romashko A, Joshi B, Barber GN, Jagus R. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302–315. 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 33.Beattie E, Paoletti E, Tartaglia J. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L- and E3L- mutant viruses. Virology 210:254–263. 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- 34.Beattie E, Kauffman EB, Martinez H, Perkus ME, Jacobs BL, Paoletti E, Tartaglia J. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89–94. 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- 35.Lee SB, Esteban M. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491–496. 10.1006/viro.1994.1151. [DOI] [PubMed] [Google Scholar]
- 36.Beattie E, Tartaglia J, Paoletti E. 1991. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183:419–422. 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 37.Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:1943–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayr A, Stickl H, Müller HK, Danner K, Singer H. 1978. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism (author's transl). Zentralbl. Bakteriol. B 167:375–390 (In German.) [PubMed] [Google Scholar]
- 39.Gomez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. 2011. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr. Gene Ther. 11:189–217. 10.2174/156652311795684731. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy JS, Greenberg RN. 2009. IMVAMUNE: modified vaccinia Ankara strain as an attenuated smallpox vaccine. Expert Rev. Vaccines 8:13–24. 10.1586/14760584.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suter M, Meisinger-Henschel C, Tzatzaris M, Hülsemann V, Lukassen S, Wulff NH, Hausmann J, Howley P, Chaplin P. 2009. Modified vaccinia Ankara strains with identical coding sequences actually represent complex mixtures of viruses that determine the biological properties of each strain. Vaccine 27:7442–7450. 10.1016/j.vaccine.2009.05.095. [DOI] [PubMed] [Google Scholar]
- 42.Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams BR, Aguet M, Weissmann C. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hochrein H, Schlatter B, O'Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, Suter M, Wagner H. 2004. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc. Natl. Acad. Sci. U. S. A. 101:11416–11421. 10.1073/pnas.0403555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meisinger-Henschel C, Späth M, Lukassen S, Wolferstätter M, Kachelriess H, Baur K, Dirmeier U, Wagner M, Chaplin P, Suter M, Hausmann J. 2010. Introduction of the six major genomic deletions of modified vaccinia virus Ankara (MVA) into the parental vaccinia virus is not sufficient to reproduce an MVA-like phenotype in cell culture and in mice. J. Virol. 84:9907–9919. 10.1128/JVI.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, Hausmann J. 2007. Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J. Gen. Virol. 88:3249–3259. 10.1099/vir.0.83156-0. [DOI] [PubMed] [Google Scholar]
- 46.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 47.Schweneker M, Lukassen S, Späth M, Wolferstätter M, Babel E, Brinkmann K, Wielert U, Chaplin P, Suter M, Hausmann J. 2012. The vaccinia virus O1 protein is required for sustained activation of extracellular signal-regulated kinase 1/2 and promotes viral virulence. J. Virol. 86:2323–2336. 10.1128/JVI.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posfai G, Plunkett G, III, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046. 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 49.Cottingham M. 2012. Genetic manipulation of poxviruses using bacterial artificial chromosome recombineering, p 37–57 In Isaacs S. (ed), Vaccinia virus and poxvirology: methods and protocols, vol 890 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 50.Baur K, Brinkmann K, Schweneker M, Pätzold J, Meisinger-Henschel C, Hermann J, Steigerwald R, Chaplin P, Suter M, Hausmann J. 2010. Immediate-early expression of a recombinant antigen by modified vaccinia virus Ankara breaks the immunodominance of strong vector-specific B8R antigen in acute and memory CD8 T-cell responses. J. Virol. 84:8743–8752. 10.1128/JVI.00604-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarti S, Sisler JR, Moss B. 1997. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques 23:1094–1097. [DOI] [PubMed] [Google Scholar]
- 52.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609–620. 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 53.Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. 2010. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe 7:354–361. 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colby C, Jurale C, Kates JR. 1971. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J. Virol. 7:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bayliss CD, Condit RC. 1993. Temperature-sensitive mutants in the vaccinia virus A18R gene increase double-stranded RNA synthesis as a result of aberrant viral transcription. Virology 194:254–262. 10.1006/viro.1993.1256. [DOI] [PubMed] [Google Scholar]
- 56.Xiang Y, Simpson DA, Spiegel J, Zhou A, Silverman RH, Condit RC. 1998. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J. Virol. 72:7012–7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arsenio J, Deschambault Y, Cao J. 2008. Antagonizing activity of vaccinia virus E3L against human interferons in Huh7 cells. Virology 377:124–132. 10.1016/j.virol.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 58.Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. 2006. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J. Gen. Virol. 87:1145–1155. 10.1099/vir.0.81623-0. [DOI] [PubMed] [Google Scholar]
- 59.Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150:831–841. 10.1016/j.cell.2012.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Backes S, Sperling KM, Zwilling J, Gasteiger G, Ludwig H, Kremmer E, Schwantes A, Staib C, Sutter G. 2010. Viral host-range factor C7 or K1 is essential for modified vaccinia virus Ankara late gene expression in human and murine cells, irrespective of their capacity to inhibit protein kinase R-mediated phosphorylation of eukaryotic translation initiation factor 2alpha. J. Gen. Virol. 91:470–482. 10.1099/vir.0.015347-0. [DOI] [PubMed] [Google Scholar]
- 61.Meng X, Chao J, Xiang Y. 2008. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372:372–383. 10.1016/j.virol.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McAllister CS, Samuel CE. 2009. The RNA-activated protein kinase enhances the induction of interferon-beta and apoptosis mediated by cytoplasmic RNA sensors. J. Biol. Chem. 284:1644–1651. 10.1074/jbc.M807888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAllister CS, Taghavi N, Samuel CE. 2012. Protein kinase PKR amplification of interferon beta induction occurs through initiation factor eIF-2alpha-mediated translational control. J. Biol. Chem. 287:36384–36392. 10.1074/jbc.M112.390039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oie KL, Pickup DJ. 2001. Cowpox virus and other members of the orthopoxvirus genus interfere with the regulation of NF-kappaB activation. Virology 288:175–187. 10.1006/viro.2001.1090. [DOI] [PubMed] [Google Scholar]
- 65.Shisler JL, Jin XL. 2004. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J. Virol. 78:3553–3560. 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang P, Samuel CE. 2008. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J. Biol. Chem. 283:34580–34587. 10.1074/jbc.M807029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaloye J, Roger T, Steiner-Tardivel QG, Le RD, Knaup RM, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, Pantaleo G, Esteban M, Calandra T. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480. 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208–211. 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 69.Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc. Natl. Acad. Sci. U. S. A. 105:2140–2145. 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nemeckova S, Hainz P, Otahal P, Gabriel P, Sroller V, Kutinova L. 2001. Early gene expression of vaccinia virus strains replicating (Praha) and non-replicating (modified vaccinia virus strain Ankara, MVA) in mammalian cells. Acta Virol. 45:243–247. [PubMed] [Google Scholar]
- 71.Stewart WE, Scott WD, Sulkin SE. 1969. Relative sensitivities of viruses to different species of interferon. J. Virol. 4:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thacore HR. 1976. Rescue of vesicular stomatitis virus from homologous and heterologous interferon-induced resistance in human cell cultures by poxviruses. Infect. Immun. 14:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandt TA, Jacobs BL. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850–856. 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice AD, Turner PC, Embury JE, Moldawer LL, Baker HV, Moyer RW. 2011. Roles of vaccinia virus genes E3L and K3L and host genes PKR and RNase L during intratracheal infection of C57BL/6 mice. J. Virol. 85:550–567. 10.1128/JVI.00254-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. 2008. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 4:e22. 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen RA, Jacobs N, Smith GL. 2006. Vaccinia virus strain Western Reserve protein B14 is an intracellular virulence factor. J. Gen. Virol. 87:1451–1458. 10.1099/vir.0.81736-0. [DOI] [PubMed] [Google Scholar]