ABSTRACT
Although effective hepatitis C virus (HCV) antivirals are on the horizon, a global prophylactic vaccine for HCV remains elusive. The diversity of the virus is a major concern for vaccine development; there are 7 major genotypes of HCV found globally. Therefore, a successful vaccine will need to protect against HCV infection by all genotypes. Despite the diversity, many monoclonal antibodies (MAbs) with broadly cross-neutralizing activity have been described, suggesting the presence of conserved epitopes that can be targeted to prevent infection. Similarly, a vaccine comprising recombinant envelope glycoproteins (rE1E2) derived from the genotype 1a HCV-1 strain has been shown to be capable of eliciting cross-neutralizing antibodies in guinea pigs, chimpanzees, and healthy human volunteers. In order to investigate the basis for this cross-neutralization, epitope mapping of anti-E1E2 antibodies present within antisera from goats and humans immunized with HCV-1 rE1E2 was conducted through peptide mapping and competition studies with a panel of cross-neutralizing MAbs targeting various epitopes within E1E2. The immunized-goat antiserum was shown to compete with the binding of all MAbs tested (AP33, HC33.4, HC84.26, 1:7, AR3B, AR4A, AR5A, IGH526, and A4). Antisera showed the best competition against HC84.26 and AR3B and the weakest competition against AR4A. Furthermore, antisera from five immunized human vaccinees were shown to compete with five preselected MAbs (AP33, AR3B, AR4A, AR5A, and IGH526). These data show that immunization with HCV-1 rE1E2 elicits antibodies targeting multiple cross-neutralizing epitopes. Our results further support the use of such a vaccine antigen to induce cross-genotype neutralization.
IMPORTANCE An effective prophylactic vaccine for HCV is needed for optimal control of the disease burden. The high diversity of HCV has posed a challenge for developing vaccines that elicit neutralizing antibodies for protection against infection. Despite this, we have previously shown that a vaccine comprising recombinant envelope glycoproteins derived from a single genotype 1a strain was capable of eliciting a cross-neutralizing antibody response in human volunteers. Here, we have used competition binding assays and peptide binding assays to show that antibodies present in the antisera from vaccinated goats and humans bind epitopes overlapping with those of a variety of well-characterized cross-neutralizing monoclonal antibodies. This provides a mechanism for the cross-neutralizing human antisera: antibodies present in the antisera bind to conserved regions associated with cross-neutralization. Importantly, this work provides further support for a vaccine comprising recombinant envelope glycoproteins, perhaps in a formulation with a vaccine component eliciting strong anti-HCV CD4+ and CD8+ T cell responses.
INTRODUCTION
Hepatitis C virus (HCV), the causative agent of hepatitis C, poses a global health problem, with an estimated ∼150 million people infected worldwide and up to 3 to 4 million new infections per year (1). Nearly complete cures are on the horizon with the advent of directly acting antivirals (DAAs). However, given the high cost of DAAs, they are unlikely to completely reduce the global health burden. A prophylactic vaccine for HCV remains a crucial requirement for the eventual control of HCV (2, 3). One of the major obstacles to HCV vaccine development is the considerable genetic diversity of the virus; there are 7 major genotypes found around the world, with differences up to 30 to 40% at the primary nucleotide sequence level (4). Many factors contribute to the diversity of HCV: an error-prone RNA-dependent RNA polymerase that lacks proofreading activity, resulting in a mutation rate of 2.5 × 10−5 mutations per nucleotide per genome replication, the long-lived nature of infected cells, the presence of multiple replication complexes in an infected cell, and the low turnover rate of these replication complexes (5). As a result of this diversity, HCV exists as a quasispecies in an infected individual, which has been suggested to facilitate immune evasion via mutations. One of the domains identified as important in immune evasion is the N-terminal hypervariable region I of E2 (reviewed in reference 6).
There have been studies clearly proving the importance of virus-specific cell-mediated immunity in control of infection, and furthermore, chimpanzees were shown to eradicate infection despite poor anti-HCV antibody responses (7 – 10). However, there is evidence that neutralizing antibodies targeting the envelope glycoproteins (E1 and E2) on the surface of the virion play a role in spontaneous clearance of infection by preventing cell entry. Reports have shown that in two instances of single-source outbreaks, an early robust neutralizing antibody response was closely associated with clearance of the infection, while a late neutralizing antibody response was seen in patients who proceeded to develop a chronic infection (11, 12). A more recent study has shown that an early broadly neutralizing antibody response against a panel of genotype 1 viruses in vitro was predictive of clearance in a cohort of prospectively monitored young injection drug users (13). There has been much work done in studying cross-neutralizing patient sera and antisera from animals immunized with E1E2, with a particular focus on cross-neutralizing monoclonal antibodies (MAbs) isolated from the patients and animals (reviewed in reference 14). This has resulted in an extensive collection of MAbs with well-characterized epitopes and neutralizing activities. Both patient sera and MAbs have been shown to prevent chronic infection in vivo using the chimeric human liver SCID/uPa mouse model and chimpanzees (15 – 18). Taking the data together, while cell-mediated immunity is protective, a prompt cross-neutralizing antibody response likely contributes to protection, as well. Given the vast diversity of the virus, much of which is located within E1E2, an optimal global prophylactic vaccine may need to elicit antibodies capable of neutralizing the entry of highly variable strains.
A vaccine based on the recombinant envelope glycoproteins (rE1E2) from a single genotype 1a strain (HCV-1) protected chimpanzees from chronic infection following homologous and heterologous genotype 1a (gt1a) viral challenge (reviewed in reference 19). Antisera from the immunized chimpanzees were shown to exhibit in vitro cross-neutralizing activity (20). A phase I clinical trial was conducted in human volunteers with a similar antigen (21). Antisera from selected vaccinated individuals were similarly capable of neutralizing chimeric cell culture-derived viruses (HCVcc) expressing the structural proteins of strains representing all 7 major HCV genotypes in vitro (22). We hypothesized that there are antibodies present in the antisera that recognize conserved structural determinants on the surfaces of virions despite the vast heterogeneity of the virus, enabling cross-neutralization.
In this study, we performed peptide mapping and competition enzyme-linked immunosorbent assays (ELISAs) with well-described cross-neutralizing antibodies to explore the antigenic regions of E1E2 antibodies present in neutralizing antisera. This was used to investigate the basis of cross-genotype neutralization in vitro using rE1E2 antisera.
MATERIALS AND METHODS
Cell cultures and antibodies.
Chinese hamster ovary (CHO) cells stably expressing E1E2 from the genotype 1a H77c strain (GenBank accession number AF009606) were propagated in Iscove's modified Dulbecco's medium (IMDM) (Thermo) containing 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 0.1 mM/0.016 mM sodium hypoxanthine/thymidine (HT supplement; Gibco), 0.002 mM methotrexate, 100 units/ml penicillin, and 100 μg/ml streptomycin (PenStrep; Invitrogen). Huh-7.5 cells were propagated in Dulbecco's modified Eagle medium (DMEM) (Gibco) containing 10% heat-inactivated FBS (Omega Scientific), 0.1 mM nonessential amino acids (NEAA) (Invitrogen), and penicillin and streptomycin (PenStrep; Invitrogen). The MAbs mouse anti-cluster of differentiation 81 (CD81) clone JS-81 (BD Biosciences), mouse isotype control IgG1 (R&D Systems), mouse anti-NS5A (9E10), anti-HCV MAbs (IGH526, A4, AP33, HC33.4, 1:7, AR3B, AR4A, and AR5A), and human anti-HIV antibody B6 have been described previously (15, 16, 23 – 31).
HC33.4 and HC84.26 were provided by Steven K. H. Foung; B6, AR3B, AR4A, AR5A, and IGH526 were provided by Mansun Law; AP33 was provided by Arvind Patel; 1:7 was provided by Mats Persson; A4 was provided by Jean Dubuisson; and 9E10 was provided by Charlie Rice and Tim Tellinghuisen.
Antiserum samples from immunized goats and human volunteers.
Two goats (G757 and G714) were immunized five times with 10 μg of rE1E2 derived from the genotype 1a HCV-1 strain (Novartis; GenBank accession number M62321.1). For the first three immunizations, the vaccine was formulated with the Addavax adjuvant (Invivogen), a squalene-based oil-in-water emulsion similar to MF59, followed by one immunization formulated with complete Freund's adjuvant (CFA) and one immunization formulated with incomplete Freund's adjuvant (IFA) (Rockland Immunochemicals). The three antiserum samples examined in this study were collected in blood draws performed prior to the immunizations (Pre), after the 3 rounds of immunizations using the Addavax adjuvant (Post-Addavax), and after the last two immunizations with CFA-IFA (Post-Freund's). Goat immunizations were performed by Rockland Immunochemicals under approved IACUC protocols.
Human antisera were obtained from an NIH-funded clinical trial (DMID 01-012) in which the safety and immunogenicity of a vaccine constituting a recombinant E1E2 immunogen derived from a genotype 1a isolate (HCV-1) formulated with the MF59C.1 adjuvant was tested (21). The trial was approved by the Saint Louis University Institutional Review Board (IRB number 15719). Two samples were used for this study: prior to immunization (Pre) and 2 weeks post-3rd immunization (Post).
Antisera from goats and humans were heat-inactivated at 56°C for 30 min prior to all assays in order to inactivate complement.
ELISA. (i) Binding to E1E2 glycoproteins.
Microtiter plates (Corning) were coated with Galanthus nivalis lectin antigen (GNA) (20 μg/ml; Sigma), and the plates were blocked with blocking buffer (phosphate-buffered saline [PBS], 1% casein, 0.5% Tween 20). One hundred micrograms of lysates from CHO cells stably expressing H77c E1E2 were incubated in each well. Antisera from goats and humans or MAbs were then added to test binding. For titration experiments, MAbs were added in 6-fold dilutions starting at 36 μg/ml. Alkaline phosphatase (AP)-conjugated goat anti-human, goat anti-mouse, or rabbit anti-goat secondary antibody (1:10,000; JacksonImmuno) was incubated in the wells, and p-nitrophenyl phosphate (Sigma) was used to develop the signal. Absorbance was read at 405 nm in an Enspire 2300 Multilabel Plate Reader (Perkin-Elmer).
(ii) Competition ELISA.
GNA-captured H77c E1E2 was used as the target antigen. Goat or human antisera in different dilutions were added for 1 h prior to removal and adding MAb (at a concentration normally resulting in 70% maximal binding) for 1 h. Binding of the MAb was measured with AP-conjugated anti-human or anti-mouse secondary antibodies (1:10,000; JacksonImmuno). When human antisera were used to block binding of human MAbs, the MAbs used were biotinylated using the EZ-Link Micro NHS-PEG4-Biotinylation Kit (Thermo), and binding was detected using AP-conjugated streptavidin (1:4,000; Sigma). Fifty percent inhibitory concentrations (IC50) were calculated with Prism 5 (GraphPad software).
Binding to linear synthetic peptides.
N-terminal biotinylated peptides (GLBiochem, Shanghai, China) were diluted in acetonitrile and water to 1 mg/ml. Peptides were diluted to 10 μg/ml in peptide blocking buffer (PBS, 1% casein, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA) and added to NeutrAvidin-coated microtiter plates (Thermo). Antisera from vaccinated goats and human MAbs were diluted in blocking buffer and added to the plates. The same alkaline phosphatase-conjugated secondary antibodies described above were diluted to 1:10,000 in a 1:1 ratio of peptide blocking buffer and Odyssey blocking buffer (Li-Cor Biosciences). The peptides used were amino acids (aa) 409 to 423 of the HCV-1 (gt1a) strain (QNVQLINTNGSWHLN), aa 433 to 447 of the HCV-1 (gt1a) strain (LNTGWLAGLFYHHKF), and an irrelevant peptide (GYIRGLFPNVLRE) derived from a rice protein (Oryza sativa japonica). For peptide competition assays, different dilutions of goat antisera were added to the peptides for 1 h prior to removal and adding the MAb for 1 h. The binding of the MAb was then measured as described above.
HCVcc production and neutralization.
Cell culture-derived chimeric HCVcc were produced using a previously described protocol (32, 33). Briefly, 6 × 106 Huh7.5 cells were mixed with 5 μg of HCV RNA in a 2-mm-gap electroporation cuvette. Five pulses of 860 V (99 μs; 1.1-s intervals) were delivered with an electroporator (ElectroSquare Porator ECM 830; BTX). The cells electroporated with HCV RNA were incubated in DMEM-10% FBS-0.1 mM NEAA, and supernatant was collected to produce virus stocks at 3 and 5 days postelectroporation. The virus titer was calculated as the 50% tissue culture infectious dose (TCID50) as described previously (32).
Neutralization was evaluated using the HCVcc in vitro model. Briefly, dilutions of antisera from immunized goats and humans were preincubated with 300 TCID50 of HCVcc for 1 h at 37°C. They were then added to 96-well plates seeded with Huh-7.5 cell monolayers (104 cells/well) 24 h previously. The medium was removed 12 h postinfection and replaced with fresh medium. The cells were fixed with 2% paraformaldehyde 48 h postinfection. Mouse anti-NS5A antibody (9E10) or mouse isotype control IgG1 (R&D Systems) was added at 1 μg/ml as a primary antibody, and an Alexa Fluor 647-conjugated goat anti-mouse secondary antibody was added (1:400; Molecular Probes). Cells that stained positive for the presence of NS5A were counted using the fluorescence-activated cell sorter (FACS) LSRFortessa (BD Biosciences). The results were analyzed with FlowJo (TreeStar).
RESULTS
Recombinant E1E2 heterodimer elicits cross-neutralizing antibodies in goats.
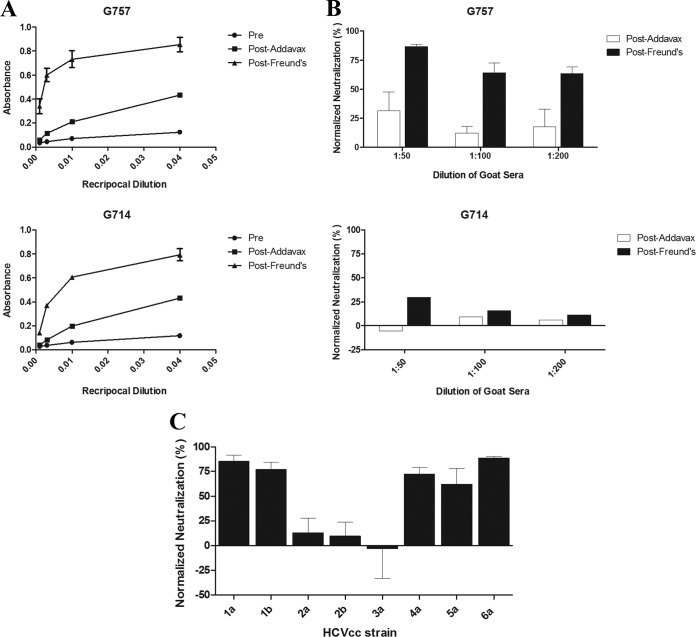
Two goats (G757 and G714) were immunized with rE1E2, the same genotype 1a (HCV-1) immunogen previously utilized in a phase I clinical trial (21). The first three vaccinations were formulated with the adjuvant Addavax, followed by two sequential immunizations using CFA-IFA. The vaccine was immunogenic: Post-Addavax and Post-Freund's antisera exhibited binding to E1E2 derived from the genotype 1a H77c sequence, as measured by ELISA (Fig. 1A). Goat antisera also neutralized infection by the chimeric HCVcc H77c/JFH-1 (H77c represents a heterologous subgenotype 1a isolate). Of the two goats, antisera from G757 exhibited higher neutralizing activity than antisera from G714 (Fig. 1B). The immunogenicity was improved after boosting with the CFA-IFA formulation; there was an increase in E1E2 binding and neutralization potency after the boosts with CFA-IFA for both goats. G757 was studied for neutralizing activity against the same panel of HCVcc strains (representing all the major global genotypes) that was previously used for the vaccinated human volunteers. The antiserum was found to have broadly cross-neutralizing activity, and notably, the neutralizing profile was comparable with human vaccinee antisera (Fig. 1C) (22). In the subsequent characterizations of the immune response, we focused on antiserum from G757 due to its greater neutralizing activity.
FIG 1.
Antisera from two goats (G757 and G714) immunized with recombinant E1E2 (HCV-1) were tested for E1E2 binding (A) and activity to prevent HCV infection (B and C). The goat antisera tested were collected prior to immunization (Pre), following the immunizations formulated with Addavax (Post-Addavax), and after the two sequential immunizations with complete and incomplete Freund's adjuvant (Post-Freund's). (A) GNA was used to capture E1E2 in lysates from CHO cells stably expressing recombinant E1E2 (strain H77c). The goat antisera were diluted in blocking buffer in 4-fold serial dilutions (1:25, 1:100, 1:400, and 1:1,600) and added to ELISA microtiter wells containing E1E2. Antibody binding was detected with an AP-conjugated anti-goat secondary antibody. The error bars represent standard deviations between triplicates. (B and C) Neutralization of HCVcc expressing the Core-NS2 regions of various strains by immunized-goat antisera. HCVcc were preincubated with diluted goat antisera and used to infect Huh-7.5 cells. The cells were fixed 48 h postinfection, and infected cells were detected by flow cytometry (see Materials and Methods). The results are shown as percent neutralizing activity by postvaccination antisera normalized to activity by prevaccination antisera. The error bars represent standard deviations between at least two independent experiments. (B) Neutralization of chimeric H77c HCVcc (genotype 1a) by diluted Post-Addavax and Post-Freund's antisera (1:50, 1:100, and 1:200) from both goats. (C) Neutralization of chimeric HCVcc expressing the Core-NS2 regions of the H77c (1a), J4 (1b), J6 (2a), J8 (2b), S52 (3a), ED43 (4a), SA13 (5a), and HK6a (6a) strains by Post-Freund's antiserum from G757 at a 1:50 dilution.
Goat antiserum prevents the binding of cross-neutralizing monoclonal antibodies to E1E2.
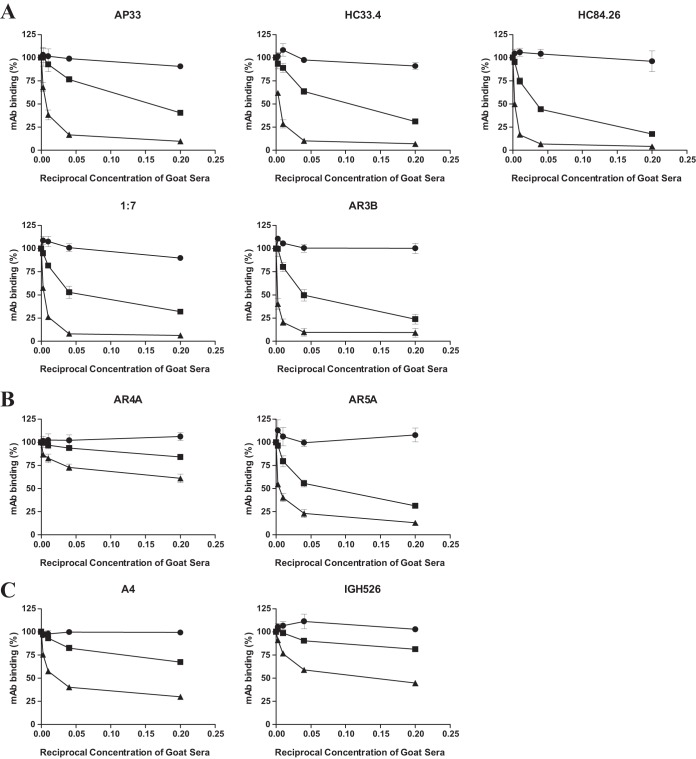
Table 1 shows the cross-neutralizing antibodies examined in this study, and Fig. 2 provides a model describing the epitopes on E1E2. Five of the antibodies (AP33, HC33.4, HC84.26, 1:7, and AR3B) are capable of binding soluble E2. These MAbs target the disparate regions of E2 that form the CD81 receptor binding site (CD81bs) to block the interaction between E2 and CD81 (15, 23 – 28). Two other antibodies (AR4A and AR5A) recognize unique epitopes outside the CD81bs and bind to the native E1E2 heterodimer and do not bind denatured E1E2 or either E1 or E2 alone (16). A4 and IGH526 recognize epitopes in E1 alone. A4 is a nonneutralizing MAb, and IGH526 is capable of cross-neutralizing various genotypes of HCV (29 – 31). Competition ELISAs were used to determine if vaccine-elicited antibodies targeted similar epitopes. First, we performed titration experiments with MAbs in E1E2-binding ELISAs. We elected to use the concentrations of the MAbs resulting in 70% maximal binding in the competition studies (data not shown). We then tested the effects that various dilutions of antisera from the immunized goat had on binding of the MAbs. Both G757 Post-Addavax and Post-Freund's antisera were capable of partially inhibiting the binding of all MAbs to H77c E1E2, while G757 Pre demonstrated minimal nonspecific inhibition (Fig. 3A to C). The dilution of antiserum capable of inhibiting the binding of the MAbs by 50% (IC50) was calculated (Table 2). Both Post-Addavax and Post-Freund's antisera were able to inhibit the binding of AP33, HC33.4, HC84.26, 1:7, AR3B, and AR5A by more than 50% at the lowest dilution of antiserum tested (1:5). While substantial competition was observed with IGH526, A4, and AR4A, it was noticeably less; either Post-Addavax (IGH526 and A4) or both postimmunization bleeds (AR4A) were incapable of competing with the binding of these MAbs by at least 50% at a dilution of 1:5 (Table 2). Our results showed that goat antisera competed with the binding of MAbs targeting E1, E2, and the E1E2 heterodimer. While the anti-E1 and anti-E2 MAbs varied in their dependence on the native E1E2 conformation for binding, both the anti-E1E2 MAbs were conformation sensitive (Table 1 and Fig. 2). Although E1 is usually considered less immunogenic, we found that the goat antisera competed with the binding of two anti-E1 antibodies (Fig. 3C).
TABLE 1.
Monoclonal antibodies used in competition ELISAs
| MAb | Protein targeted | Conformation dependence | Species | Critical binding residuesa | Interaction targeted | Neutralization potencyb | Reference |
|---|---|---|---|---|---|---|---|
| AP33 | E2 | No | Mouse | 413, 415, 418, 420 | CD81 | 2a (HCVcc); 1a, 1b, 2a, 3a, 4a, 5a, 6a (HCVpp) | 23 – 25 |
| HC33.4 | E2 | No | Human | 413, 418, 420, 421 | CD81 | 1a, 2a, 3a, 4a, 5a, 6a (HCVcc) | 28 |
| HC84.26 | E2 | Yes | Human | 429, 441, 442, 446, 616 | CD81 | 1a, 2a, 3a, 4a, 5a, 6a (HCVcc) | 27 |
| 1:7 | E2 | Yes | Human | 415, 417, 484, 491, 523, 525, 526, 527, 529, 530, 533, 535, 540 | CD81 | 2a (HCVcc); 1a, 1b, 2a, 3a, 4a, 5a, 6a (HCVpp) | 26 |
| AR3B | E2 | Yes | Human | 412, 416, 418, 423, 424, 523, 525, 530, 535, 540 | CD81 | 2a (HCVcc); 1a, 1b, 2a, 2b, 4, 5, 6 (HCVpp) | 15 |
| AR4A | E1E2 | Yes | Human | 201, 204, 205, 206, 487, 657, 658, 692, 698 | Unknown | 1a, 1b, 2a, 3a, 4a, 5a, 6a (HCVcc); 1a, 1b, 2a, 2b, 2i, 2x, 3a, 4, 5, 6 (HCVpp) | 16 |
| AR5A | E1E2 | Yes | Human | 201, 204, 205, 206, 639, 657, 658, 665, 692 | Unknown | 1a, 2a, 4a, 5a, 6a (HCVcc); 1a, 1b, 4, 5, 6 (HCVpp) | 16 |
| IGH526 | E1 | Untested | Human | 313–327 | Unknown | 1a, 1b, 2a, 4a, 5a, 6a (HCVpp) | 31 |
| A4 | E1 | No | Mouse | 197–207 | None | NA | 29, 30 |
FIG 2.
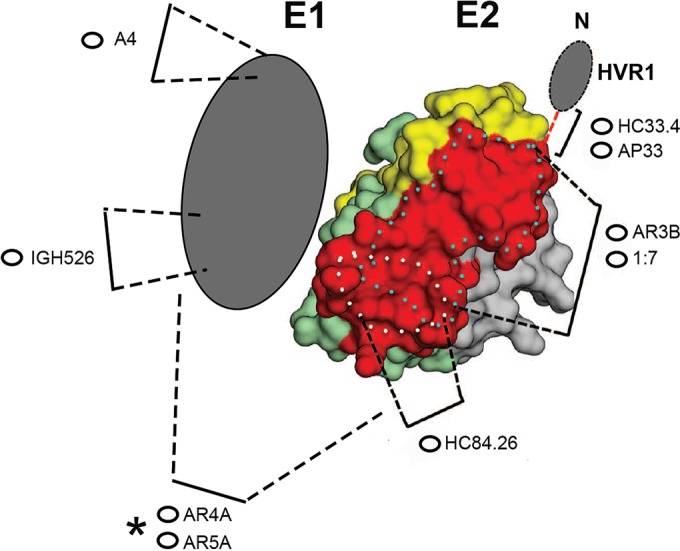
Model of E1E2 showing the locations of epitopes targeted by the MAbs used in the study. The model of the E2 core (reprinted from Science [55] with permission of the publisher) has been modified to include representations of the epitopes of the MAbs used in this study. Hypervariable region I (HVR1) is indicated at the N-terminal end of the E2 protein. With the exception of A4, which is nonneutralizing, all of the MAbs have cross-neutralizing activity (Table 1). A4 and IGH526 bind to different epitopes in E1. The MAbs directly targeting epitopes in E2 (AP33, HC33.4, HC84.26, 1:7, and AR3B) block the interaction between E2 and CD81. The epitopes targeted by these MAbs all contain residues that facilitate or modulate the E2-CD81 interaction. The asterisk highlights the fact that AR4A and AR5A bind conformation-dependent epitopes on the native E1E2 heterodimer.
FIG 3.
Competition studies with G757 goat antisera (●, Pre; ■, Post-Addavax; ▲, Post-Freund's) and a panel of MAbs. Microtiter wells containing GNA-captured E1E2 of H77c were first incubated with diluted antiserum samples from G757 (1:5, 1:25, 1:100, and 1:400) for 1 hour, followed by incubation with the indicated MAb. Monoclonal antibodies were added at a concentration resulting in 70% maximal binding to E1E2, as determined in prior titration experiments. Binding of the MAbs was detected with species-specific AP-conjugated secondary antibodies. The percentage of MAbs binding in the presence of goat antisera normalized to binding in the absence of any antiserum was calculated. The data are reported as the mean values of at least 2 experiments performed in triplicate. The error bars represent the standard deviations of all replicates. The eight MAbs used in competition ELISAs are grouped based on their reactivities: antibodies that bind E2 (A), antibodies that require E1E2 for binding (B), and antibodies that target E1 (C).
TABLE 2.
Goat G757 antiserum IC50
| MAb | G757 antiserum IC50a |
|
|---|---|---|
| Post-Addavax | Post-Freund's | |
| AP33 | 1:7.4 | 1:145 |
| HC33.4 | 1:12.4 | 1:252 |
| HC84.26 | 1:30 | <1:400 |
| 1:7 | 1:18.0 | 1:296 |
| AR3B | 1:22.7 | <1:400 |
| AR4A | >1:5 | >1:5 |
| AR5A | 1:17.2 | 1:244 |
| IGH526 | >1:5 | 1:8.75 |
| A4 | >1:5 | 1:44.3 |
IC50 were calculated with Prism 5 (GraphPad Software). “>1:5” indicates the immunized goat antiserum sample was incapable of inhibiting binding of a MAb by at least 50% at the lowest dilution (1:5).
Antisera from the other rE1E2-vaccinated goat (G714) showed a similar pattern of competition with the binding of some of the MAbs in the panel, but competition was less efficacious than with G757 antisera, since the vaccine was generally less reactive in G714 (Fig. 1A and B and data not shown).
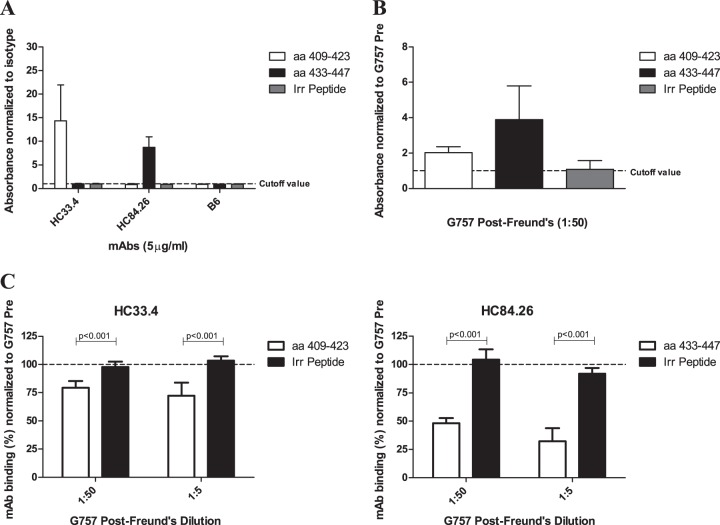
The MAbs HC33.4 and HC84.26 bind to linear synthetic peptides, so we explored whether goat antiserum exhibits similar properties (27, 28). Consistent with the literature, HC33.4 and HC84.26 bound linear peptides spanning aa 409 to 423 and aa 433 to 447 from the E2 protein, respectively. While immunization elicited antibodies in G757 capable of binding both peptides, the reactivity was higher for peptide aa 433 to 447 (Fig. 4A and B). Consistently, we observed better competition with G757 antisera against HC84.26 than HC33.4 (Fig. 3A and Table 2). To explore the specificity of the competition with binding of MAbs to E1E2 further, we studied the ability of goat antisera to compete with the binding of HC33.4 and HC84.26 to their respective peptides. We found G757 antisera could compete with the binding of these MAbs to their respective peptides but did not affect nonspecific binding to an irrelevant peptide (Fig. 4C). This indicates G757 antiserum competes with the binding of HC33.4 and HC84.26 by directly blocking interaction with their target epitopes.
FIG 4.
Binding of immunized-goat antisera and MAbs to linear-synthetic biotinylated peptides by ELISA. (A and B) Human MAbs (HC33.4, HC84.26, and human IgG1 isotype control B6) at 5 μg/ml (A) or immunized-goat antisera from G757 diluted 1:50 (B) were incubated with biotinylated peptides precaptured in NeutrAvidin-coated microtiter plate wells. Two peptides from E2 (aa 409 to 423 and aa 433 to 447) and an irrelevant (Irr) biotinylated peptide of similar length were used (see Materials and Methods for the sequences). Binding was detected with species-specific AP-conjugated secondary antibodies. The results are presented as fold binding signal to peptide over the cutoff value. The cutoff value was determined by the mean binding to the same peptide plus 3 standard deviations by either G757 Pre (A) or human IgG1 isotype control MAb B6 (B). (C) The ability of goat sera to compete with MAb binding to peptide was determined. Diluted goat antisera were added to wells containing peptide for 1 hour, followed by incubation of the MAb at 5 μg/ml. The MAb and the target peptide are indicated. Binding of the MAbs was detected, and the percent MAb binding in the presence of G757 antisera was normalized to binding in the presence of G757 Pre. The error bars show standard deviations of all replicates from at least two independent experiments done in triplicate. Statistical evaluation comparing the competition with the binding of MAbs to target peptide or irrelevant peptide was performed using a two-tailed unpaired t test, and the P values are shown.
E1E2 vaccine induces antibodies recognizing epitopes associated with broad neutralization in humans.
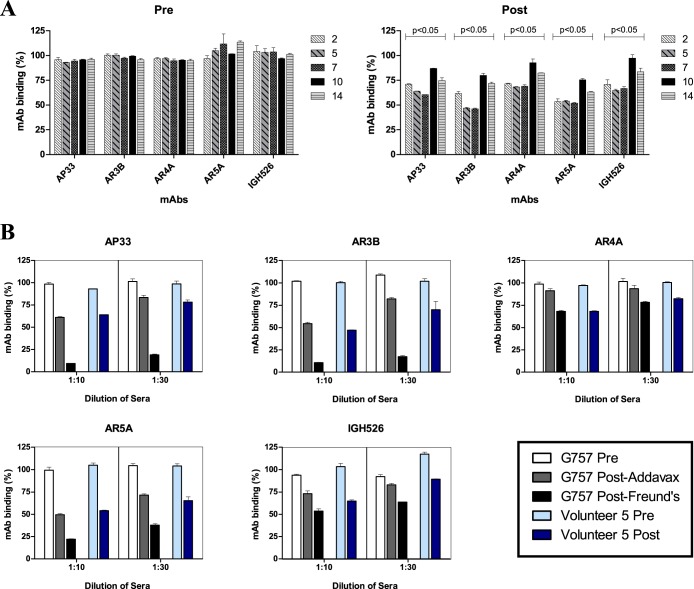
We extended our findings to the vaccinated human volunteers and investigated if antibodies targeting the epitopes of cross-neutralizing MAbs were elicited. Competition assays were conducted with antisera from five of the volunteers vaccinated in the clinical phase I trial in the 100-μg-dose group. The antiserum samples from these volunteers (2, 5, 7, 10, and 14) identified as Pre and Post were the same samples previously studied for in vitro neutralizing activity. Post samples from two of the volunteers (5 and 7) were found to have broad cross-neutralizing activity against chimeric HCVcc expressing the structural proteins from strains from all 7 HCV genotypes (22). Due to the paucity of vaccinee antisera, we tested competition with just five MAbs (AP33, AR3B, AR4A, AR5A, and IGH526). Since all the MAbs with the exception of AP33 were human derived, the MAbs were biotinylated to enable detection in competition ELISAs. Biotinylation did not affect binding of MAbs to E1E2 or competition by goat antisera (data not shown). Prevaccination antisera from all the volunteers exhibited no effect on binding of all tested MAbs compared to a no-serum control. Antibodies recognizing epitopes of these MAbs were present in the vaccinees; similar patterns of competition with binding of the MAbs were exhibited by postvaccination antisera from all the volunteers, albeit with varying degrees of competition (Fig. 5A). Postvaccination antiserum from volunteer 5 blocked binding of the MAbs to an extent comparable with postvaccination goat antiserum from G757. We also observed a dose-dependent effect, as competition was enhanced at 1:10 compared to 1:30 (Fig. 5B).
FIG 5.
Competition studies with human volunteers' antisera and a panel of neutralizing MAbs. Antiserum samples collected prior to immunization (Pre) and 2 weeks post-3rd immunization (Post) from five volunteers (2, 5, 7, 10, and 14) were tested. Microtiter wells containing GNA-captured E1E2 of H77c were first incubated with diluted volunteers' antisera or G757 for 1 hour, followed by incubation with the indicated MAb. Each MAb (AP33, AR3B, AR4A, AR5A, and IGH526) was added at a concentration resulting in 70% maximal binding. AR3B, AR4A, AR5A, and IGH526 were biotinylated to enable specific detection. Binding was detected with AP-conjugated NeutrAvidin or AP-conjugated anti-mouse secondary antibody. The results are shown as the percent binding of MAbs in the presence of human or goat antisera normalized to binding in the absence of antisera. (A) Competition assay results from antiserum samples from five volunteers at a dilution of 1:10. The statistical significance of the competition between Pre and Post antiserum samples for each of the MAbs was evaluated by a two-tailed paired t test. (B) Results comparing immunized-goat antisera and matched antiserum samples from one of the volunteers (5) at dilutions of 1:10 and 1:30. Due to the paucity of human vaccinee antisera, experiments were performed only once in duplicate with a 1:10 dilution and twice in duplicate with a 1:30 dilution. The error bars show standard deviations between replicates.
DISCUSSION
Previously, antisera from human volunteers immunized with genotype 1a rE1E2 (HCV-1) were found to have cross-genotype neutralizing activity (22). In this study, we characterized the cross-neutralizing epitope repertoire within these antisera. We found evidence of antibodies present in the antisera of vaccinated goats and humans targeting numerous epitopes associated with broad cross-neutralization.
HCV has a complex entry process involving numerous entry coreceptors and cofactors, including heparan sulfate proteoglycans (HSPG) (34), scavenger receptor BI (SR-BI) (35), CD81 (36), the cholesterol absorption receptor Niemann-Pick C1-like 1 (NPC1L1) (37), and the tight-junction proteins claudin 1 (CLDN1) (38) and occludin (OCLN) (39). Addtionally, the receptor tyrosine kinases epidermal growth factor receptor (EGFR) and ephrin type A receptor 2 (EphA2) have been identified as important factors that regulate the CLDN1-CD81 interactions necessary for entry (40). Transferrin receptor 1 has also been identified as an entry factor (41). Both soluble E2 and the E1E2 heterodimer have been shown to directly bind to CD81 and SR-BI, and a direct interaction between the E1E2 heterodimer and CLDN1 has also recently been described (35, 36, 42 – 47). Interaction with HSPG has been shown to be facilitated by both E1 and E2 (34, 48, 49). In addition, virion-associated lipid components (apolipoprotein E, apolipoprotein B, apolipoprotein C-1, and cholesterol) have a role in facilitating and modulating interactions with HSPG, low-density lipoprotein receptor (LDLR), SR-BI, and NPC1L1 (37, 50 – 53).
Many monoclonal antibodies with broad cross-neutralizing activity have been isolated and characterized (reviewed in reference 14). The majority of these antibodies recognize overlapping regions on the E2 protein: E2 aa 412 to 424, 433 to 447, 523 to 540, and 613 to 616 contain residues critical for binding to CD81 (27, 54). The CD81bs on E2 is a conserved conformational structure that interacts with CD81 during entry, and many cross-neutralizing antibodies bind various residues in the CD81bs. The interactions of E2 with CD81and these antibodies are described in two recent papers characterizing the crystal structure of E2 (55, 56). All the anti-E2 MAbs used in this study (AR3B, HC33.4, AP33, HC84.26, and 1:7) bind to various domains of the CD81bs to inhibit the E2-CD81 interaction (Table 1 and Fig. 2). G757 antisera competed with the binding of all of these MAbs similarly, indicating that the immunogen elicited antibodies covering the broad CD81bs on E2 (Fig. 3A). Similar results were obtained using human volunteer antisera to compete with the binding of AP33 and AR3B (Fig. 5A and B). This is supported by previous findings showing that the human vaccinees' antisera inhibited the E2-CD81 interaction (21).
AR4A and AR5A are two recently identified MAbs targeting epitopes outside the CD81bs and do not inhibit interaction with CD81 (16). These MAbs require the native E1E2 heterodimer for binding and do not bind E1 alone, E2 alone, or denatured E1E2. The MAbs were isolated through an exhaustive-panning strategy of an antibody antigen-binding fragment (Fab) phage display library generated from a patient chronically infected with HCV using the E1E2 heterodimer protein preblocked with previously isolated anti-E2 MAbs to obtain unique and rare MAbs. They are broadly cross-neutralizing, but the exact binding modes are currently unknown. AR4A and AR5A can bind E1E2 simultaneously without cross-competition. However, alanine-scanning mutagenesis studies revealed that the two MAbs share a dependency to bind E1E2 on several residues in the N terminus of E1 and the C terminus of E2. D698 and D639 of E2 were critical for the binding of AR4A and AR5A, respectively. Interestingly, D698 is within the highly conserved membrane-proximal external region (MPER), which is important for E1E2 heterodimerization and membrane fusion (57). There is still much to be discovered about how the epitopes of these MAbs are formed by E1E2 heterodimerization. While G757 and human antisera competed with the binding of AR4A and AR5A, there was a distinctly reduced competition against AR4A (Fig. 3B and 5A and B). Although recognition of AR5A requires native E1E2, binding to E1E2 was shown to be effectively competed by CBH-7, an anti-E2 MAb that inhibits the E2-CD81 interaction (16, 58, 59). As such, it is possible that the antibodies present in vaccine antisera compete with the binding of AR5A in a fashion similar to that of CBH-7 and prevent E2-CD81 interaction. The reduced competition against AR4A binding is intriguing. There may be differences in the recognition of the AR4A epitope between species, as human antisera exhibited higher competition against AR4A binding than G757 Post-Addavax antiserum, while competition with the other MAbs was similar (Fig. 5A). It may also be possible that the epitope for AR4A is somewhat inaccessible in the recombinant E1E2 structure compared to the native HCV virion, rendering it less immunogenic. It will be of future interest to compare the relative competition of AR4A by both vaccine antisera and infected patient sera.
There are very few anti-E1 antibodies available, limiting the study of such antibodies in vaccine antisera. Nevertheless, our studies revealed that G757 antisera competed with the binding of two anti-E1 antibodies (A4 and IGH526), indicating that an immune response was elicited against the E1 component of the E1E2 vaccine (Fig. 3C). Similar findings were observed using human antisera to compete with the binding of IGH526 (Fig. 5A and B). IGH526 has cross-neutralizing activity and binds a linear peptide spanning aa 313 to 327 from the E1 protein (31). Competition with IGH526 by goat antisera was reduced in comparison with the other MAbs. Future work will compare the relative competition afforded by both vaccine antisera and patient sera to see if it reflects an altered structure of recombinant E1E2 compared with that of virions circulating in infected humans. More attention has been paid to E2 as a neutralizing antibody target, and anti-E2 MAbs have been used to demonstrate protection in both chimeric human liver mice and chimpanzees (15 – 17). However, anti-E1 antibodies have also been shown to confer protection. Recombinant E1 protein derived from a genotype 1b strain elicited neutralizing antibodies in two chimpanzees and protected them from heterologous genotype 1b viral challenge (60). With only one neutralizing epitope exclusively in E1 characterized so far (aa 313 to 327), there is likely much more to learn about neutralizing epitopes on E1 and their roles in protection.
We have shown that vaccine antiserum binds directly to linear peptides targeted by cross-neutralizing antibodies HC33.4 and HC84.26 and competes for the binding of numerous other discrete cross-neutralizing antibodies targeting conformational epitopes. Our data suggest that the HCV-1 rE1E2 immunogen is capable of eliciting antibodies that bind conserved residues overlapping the epitopes of many broadly cross-neutralizing antibodies targeting the E2-CD81 interaction (represented by AP33, HC33.4, HC84.26, 1:7, and AR3B) and other regions (represented by IGH526, AR4A, and AR5A). These antibodies are likely responsible for the cross-neutralizing activity exhibited by the antisera. This study supports and confirms earlier studies showing that this vaccine induced antibodies in human volunteers capable of inhibiting the E2-CD81 interaction and neutralization of HCVcc representing strains from all 7 major global genotypes, even though rE1E2 was derived from a single strain (21, 22). Future work will focus on characterizing the nature of cross-neutralizing monoclonal antibodies elicited by the vaccine and on elucidating the mechanisms of neutralization by rE1E2 antisera in the sequential process of viral entry.
Early studies performed both in vitro and in vivo suggested that antibody-mediated neutralization was isolate specific (61, 62). Later findings implicated hypervariable region I in E2 as a major determinant for both isolate-specific neutralizing antibodies and progression to chronic infection (63 – 65). These studies posed a challenge to the goal of a prophylactic vaccine based on eliciting neutralizing antibodies. However, our previous study has shown that E1E2 derived from a single strain can induce antibodies capable of cross-neutralizing diverse HCV strains, and similarly, the current work shows that these antibodies are capable of competing with the binding of broadly cross-neutralizing monoclonal antibodies (22). While the most impressive cross-neutralization and competition activity was obtained after vaccination with the CFA-IFA adjuvant system, the human volunteers' antisera and both the Post-Addavax and Post-Freund's bleeds from G757 showed similar trends in both the cross-neutralization and competition profiles. Importantly, this work indicates that the HCV-1 rE1E2 immunogen is capable of eliciting antibodies targeting multiple conserved regions on E1E2 that confer cross-neutralizing activity. This work provides further support for the clinical development of rE1E2 as a vaccine antigen. In fact, this is the only HCV vaccine candidate for which statistically significant preclinical efficacy exists (19). More powerful adjuvants are also being investigated in order to further increase immune responses. The addition of rE1E2 to a vaccine component eliciting broad anti-HCV CD4+ and CD8+ T cell responses could elicit broadly protective responses from both the humoral and cell-mediated arms of the immune system and offer enhanced protection.
ACKNOWLEDGMENTS
We are grateful to Janelle Johnson, Heather Gordon, and Darci Loewen-Dobler for technical help. We are also grateful to Andrea Holme (Faculty of Medicine and Dentistry Flow Cytometry Core). We thank Steven K. H. Foung for providing HC33.4 and HC84.26; Mansun Law for providing B6, AR3B, AR4A, AR5A, and IGH526; Arvind Patel for providing AP33; Mats Persson for providing 1:7; Jean Dubuisson for providing A4; Charlie Rice and Tim Tellinghuisen for providing 9E10 and other HCV-related reagents; and Judith M. Gottwein and Jens Bukh for providing reagents for HCVcc production. We thank Rajen Koshy (NIH) for facilitating access to the vaccinee samples. Recombinant E1E2 used to immunize the goats was kindly provided by Novartis.
J.A.J.-X.W. is supported by the National CIHR Research Training Program in Hepatitis C and the Canadian Liver Foundation. M.H. is a recipient of the Canada Excellence in Research Chairs Program.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.WHO. 2013. Hepatitis C. WHO fact sheet 164. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs164/en/index.html. [Google Scholar]
- 2.Liang TJ. 2013. Current progress in development of hepatitis C virus vaccines. Nat. Med. 19:869–878. 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas DL. 2013. Global control of hepatitis C: where challenge meets opportunity. Nat. Med. 19:850–858. 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakano T, Lau GMG, Lau GML, Sugiyama M, Mizokami M. 2012. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int. 32:339–345. 10.1111/j.1478-3231.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- 5.Ribeiro RM, Li H, Wang S, Stoddard MB, Learn GH, Korber BT, Bhattacharya T, Guedj J, Parrish EH, Hahn BH, Shaw GM, Perelson AS. 2012. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog. 8:e1002881. 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farci P. 2011. New insights into the HCV quasispecies and compartmentalization. Semin. Liver Dis. 31:356–374. 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 7.Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA, Walker CM. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 197:1645–1655. 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302:659–662. 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 9.Cooper S, Erickson AL, Adams EJ, Kansopon J, Weiner AJ, Chien DY, Houghton M, Parham P, Walker CM. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439–449. 10.1016/S1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 10.Logvinoff C, Major ME, Oldach D, Heyward S, Talal A, Balfe P, Feinstone SM, Alter H, Rice CM, McKeating JA. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 101:10149–10154. 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, Patel AH, Meisel H, Baumert J, Viazov S, Rispeter K, Blum HE, Roggendorf M, Baumert TF. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 104:6025–6030. 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavillette D, Morice Y, Germanidis G, Donot P, Soulier A, Pagkalos E, Sakellariou G, Intrator L, Bartosch B, Pawlotsky JM, Cosset FL. 2005. Human serum facilitates hepatitis C virus infection, and neutralizing responses inversely correlate with viral replication kinetics at the acute phase of hepatitis C virus infection. J. Virol. 79:6023–6034. 10.1128/JVI.79.10.6023-6034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, Cox AL, Ray SC. 2014. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology 59:2140–2151. 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sautto G, Tarr AW, Mancini N, Clementi M. 2013. Structural and antigenic definition of hepatitis C virus E2 glycoprotein epitopes targeted by monoclonal antibodies. Clin. Dev. Immunol. 2013:450963. 10.1155/2013/450963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14:25–27. 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 16.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 109:6205–6210. 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin TJ, Broering TJ, Leav BA, Blair BM, Rowley KJ, Boucher EN, Wang Y, Cheslock PS, Knauber M, Olsen DB, Ludmerer SW, Szabo G, Finberg RW, Purcell RH, Lanford RE, Ambrosino DM, Molrine DC, Babcock GJ. 2012. Human monoclonal antibody HCV1 effectively prevents and treats HCV infection in chimpanzees. PLoS Pathog. 8:e1002895. 10.1371/journal.ppat.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meuleman P, Bukh J, Verhoye L, Farhoudi A, Vanwolleghem T, Wang RY, Desombere I, Alter H, Purcell RH, Leroux-Roels G. 2011. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology 53:755–762. 10.1002/hep.24171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houghton M. 2011. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol. Rev. 239:99–108. 10.1111/j.1600-065X.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 20.Meunier JC, Gottwein JM, Houghton M, Russell RS, Emerson SU, Bukh J, Purcell RH. 2011. Vaccine-induced cross-genotype reactive neutralizing antibodies against hepatitis C virus. J. Infect. Dis. 204:1186–1190. 10.1093/infdis/jir511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frey SE, Houghton M, Coates S, Abrignani S, Chien D, Rosa D, Pileri P, Ray R, Di Bisceglie AM, Rinella P, Hill H, Wolff MC, Schultze V, Han JH, Scharschmidt B, Belshe RB. 2010. Safety and immunogenicity of HCV E1E2 vaccine adjuvanted with MF59 administered to healthy adults. Vaccine 28:6367–6373. 10.1016/j.vaccine.2010.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, Belshe RB, Wakita T, Bukh J, Jones CT, Rice CM, Abrignani S, Tyrrell DL, Houghton M. 2013. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One 8:e59776. 10.1371/journal.pone.0059776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owsianka A, Tarr AW, Juttla VS, Lavillette D, Bartosch B, Cosset FL, Ball JK, Patel AH. 2005. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79:11095–11104. 10.1128/JVI.79.17.11095-11104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potter JA, Owsianka AM, Jeffery N, Matthews DJ, Keck ZY, Lau P, Foung SKH, Taylor GL, Patel AH. 2012. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J. Virol. 86:12923–12932. 10.1128/JVI.02052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarr AW, Owsianka AM, Timms JM, McClure CP, Brown RJP, Hickling TP, Pietschmann T, Bartenschlager R, Patel AH, Ball JK. 2006. Characterization of the hepatitis C virus E2 epitope defined by the broadly neutralizing monoclonal antibody AP33. Hepatology 43:592–601. 10.1002/hep.21088. [DOI] [PubMed] [Google Scholar]
- 26.Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MAA. 2007. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 104:16269–16274. 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keck Z-Y, Xia J, Wang Y, Wang W, Krey T, Prentoe J, Carlsen T, Li AY-J, Patel AH, Lemon SM, Bukh J, Rey FA, Foung SKH. 2012. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 8:e1002653. 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keck Z, Wang W, Wang Y, Lau P, Carlsen THR, Prentoe J, Xia J, Patel AH, Bukh J, Foung SKH. 2013. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J. Virol. 87:37–51. 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helle F, Vieyres G, Elkrief L, Popescu CI, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J. Virol. 84:11905–11915. 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meunier JC, Russell RS, Goossens V, Priem S, Walter H, Depla E, Union A, Faulk KN, Bukh J, Emerson SU, Purcell RH. 2008. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J. Virol. 82:966–973. 10.1128/JVI.01872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 33.Gottwein JM, Scheel TKH, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377. 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 34.Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003–41012. 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 35.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017–5025. 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 37.Sainz B, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 18:281–285. 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 39.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset F-L, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoël M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 17:589–595. 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin DN, Uprichard SL. 2013. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc. Natl. Acad. Sci. U. S. A. 110:10777–10782. 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset F-L, Lavillette D. 2014. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 59:776–788. 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 43.Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, Abrignani S, Grandi G. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824–4830. 10.1128/JVI.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higginbottom A, Quinn ER, Kuo C, Flint M, Wilson EB, Nicosia A, Monk PN, McKeating J, Levy S. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642–3649. 10.1128/JVI.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummer HE, Wilson KA, Poumbourios P. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143–11147. 10.1128/JVI.76.21.11143-11147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk PN, Higginbottom A, Levy S, McKeating J. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, Fremont DH, Diamond MS. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J. Virol. 85:7005–7019. 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi F, Yamada S, Taguwa S, Kataoka C, Naito S, Hama Y, Tani H, Matsuura Y, Sugahara K. 2012. Specific interaction of the envelope glycoproteins E1 and E2 with liver heparan sulfate involved in the tissue tropismatic infection by hepatitis C virus. Glycoconj. J. 29:211–220. 10.1007/s10719-012-9388-z. [DOI] [PubMed] [Google Scholar]
- 49.Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579–10590. 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. 2012. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J. Virol. 86:7256–7267. 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agnello V, Ábel G, Elfahal M, Knight GB, Zhang Q-X. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. U. S. A. 96:12766–12771. 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Germi R, Crance J-M, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RWH, Zarski J-P, Drouet E. 2002. Cellular glycosaminoglycans and low density lipoprotein receptor are involved in hepatitis C virus adsorption. J. Med. Virol. 68:206–215. 10.1002/jmv.10196. [DOI] [PubMed] [Google Scholar]
- 53.Dao Thi VL, Granier C, Zeisel MB, Guerin M, Mancip J, Granio O, Penin F, Lavillette D, Bartenschlager R, Baumert TF, Cosset FL, Dreux M. 2012. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 287:31242–31257. 10.1074/jbc.M112.365924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Owsianka AM, Timms JM, Tarr AW, Brown RJP, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80:8695–8704. 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rychlowska M, Owsianka AM, Foung SKH, Dubuisson J, Bienkowska-Szewczyk K, Patel AH. 2011. Comprehensive linker-scanning mutagenesis of the hepatitis C virus E1 and E2 envelope glycoproteins reveals new structure-function relationships. J. Gen. Virol. 92:2249–2261. 10.1099/vir.0.034314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hadlock KG, Lanford RE, Perkins S, Rowe J, Yang Q, Levy S, Pileri P, Abrignani S, Foung S. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407–10416. 10.1128/JVI.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owsianka AM, Tarr AW, Keck ZY, Li TK, Witteveldt J, Adair R, Foung SKH, Ball JK, Patel AH. 2008. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J. Gen. Virol. 89:653–659. 10.1099/vir.0.83386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verstrepen BE, Depla E, Rollier CS, Mares G, Drexhage JAR, Priem S, Verschoor EJ, Koopman G, Granier C, Dreux M, Cosset FL, Maertens G, Heeney JL. 2011. Clearance of genotype 1b hepatitis C virus in chimpanzees in the presence of vaccine-induced E1-neutralizing antibodies. J. Infect. Dis. 204:837–844. 10.1093/infdis/jir423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farci P, Alter H, Wong D, Miller Roger H, Govindarajan S, Engle RE, Shapiro M, Purcell R. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. U. S. A. 91:7792–7796. 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimizu YK, Hijikata M, Iwamoto A, Alter H, Purcell R, Yoshikura H. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 68:1494–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344. 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 64.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu YK, Shapiro M, Alter H, Purcell R. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. U. S. A. 93:15394–15399. 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimizu YK, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R, Yoshikura H. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223:409–412. 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 66.Kuiken C, Simmonds P. 2009. Nomenclature and numbering of the hepatitis C virus. Methods Mol. Biol. 510:33–53. 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]