ABSTRACT
The RNA-dependent RNA polymerase (RdRp) of influenza A virus is a heterotrimeric complex composed of the PB1, PB2, and PA subunits. The interplay between host factors and the three subunits of the RdRp is critical to enable viral RNA synthesis to occur in the nuclei of infected cells. In this study, we newly identified host factor DnaJA1, a member of the type I DnaJ/Hsp40 family, acting as a positive regulator for influenza virus replication. We found that DnaJA1 associates with the bPB2 and PA subunits and enhances viral RNA synthesis both in vivo and in vitro. Moreover, DnaJA1 could be translocated from cytoplasm into the nucleus upon influenza virus infection. The translocation of DnaJA1 is specifically accompanied by PB1-PA nuclear import. Interestingly, we observed that the effect of DnaJA1 on viral RNA synthesis is mainly dependent on its C-terminal substrate-binding domain and not on its typical J domain, while the J domain normally mediates the Hsp70-DnaJ interaction required for regulating Hsp70 ATPase activity. Therefore, we propose that DnaJA1 is co-opted by the influenza A virus to enter the nucleus and to enhance its RNA polymerase activity in an Hsp70 cochaperone-independent manner.
IMPORTANCE The interplay between host factors and influenza virus RNA polymerase plays a critical role in determining virus pathogenicity and host adaptation. In this study, we newly identified a host protein, DnaJA1/Hsp40, that is co-opted by influenza A virus RNA polymerase to enhance its viral RNA synthesis in the nuclei of infected cells. We found that DnaJA1 associates with both PB2 and PA subunits and translocates into the nucleus along with the nuclear import of the PB1-PA dimer during influenza virus replication. Interestingly, the effect of DnaJA1 is mainly dependent on its C-terminal substrate-binding domain and not on its typical J domain, which is required for its Hsp70 cochaperone function. To our knowledge, this is the first report on a member of the Hsp40s that is specifically involved in regulating influenza virus RNA polymerase. Targeting the interactions between polymerase subunits and DnaJA1 may provide a novel strategy to develop antiviral drugs.
INTRODUCTION
Influenza A virus belongs to the Orthomyxoviridae family of RNA viruses. The segmented, negative-sense genome and the utilization of the cell nucleus as the viral RNA synthesis site are key features for the family (1). The viral ribonucleoprotein complex (vRNP) of influenza A virus is the minimal functional unit for viral RNA transcription (vRNA→mRNA) and replication (vRNA→cRNA→vRNA) to occur in the nuclei of infected cells. The vRNP consists of an RNA-dependent RNA polymerase complex (RdRp), a viral RNA, and multiple copies of nucleoprotein (NP). The RNA polymerase is responsible for the synthesis of the three viral RNAs species (vRNA, mRNA, and cRNA), which play critical roles in determining virus pathogenicity and host adaptation (2, 3). The RdRp is a heterotrimeric complex composed of polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA) (1, 4). PB1 subunit contains the conserved motif characteristics of RNA-dependent RNA polymerases (5). PB2 is responsible for cap binding (6, 7), while PA contains an endonuclease domain that cleaves the capped primer from host pre-mRNAs to initiate viral mRNA synthesis (8, 9). During influenza virus replication, the three subunits are synthesized in the cytoplasm of infected cells individually. They must be transported into the nucleus and assembled into a trimeric complex before further assembly with NP and viral RNAs into vRNP complexes (10). An assembly model has proposed that PB1 and PA associate in the cytoplasm and are transported into the nucleus as a dimer, while PB2 enters the nucleus on its own and assembles with PB1-PA dimer in the nucleus to form the 3P complexes (11, 12).
Since the RNA polymerase is one of the pathogenicity and host range determinants of influenza A viruses and is an attractive target for antiviral development, great efforts have been made to specifically identify host factors that could interact with the polymerase subunits and regulate viral RNA synthesis. Over 120 host factors have been identified to be potential interacting partners of the viral RNA polymerase (13–15), and a number of them have been studied in detail for how they are involved in modulating viral RNA synthesis (reviewed in references 16 and 17).
Among those host factors, heat shock proteins (Hsps, e.g., Hsp90 and Hsp70) have been identified to be major host factors involved in regulating the viral RNA synthesis. Hsps normally act as molecular chaperones to facilitate protein folding, trafficking, prevention of aggregation, and degradation by proteolysis in cells (18–21). It has been reported that during influenza virus replication, Hsp90 is able to stimulate viral RNA polymerase activity and is involved in the trimeric polymerase complex assembly and nuclear import of the virus polymerase subunits by binding with PB2 monomers or PB2/PB1 heterodimers (22, 23). Hsp90 inhibitors, such as geldanamycin or its derivative 17-AAG, have been shown to be able to inhibit viral growth by affecting viral RNA polymerase assembly (24). In contrast, the roles of Hsp70 in regulating viral RNP polymerase activity are diverse. It has been shown that Hsp70 is related to thermal inhibition of the nuclear export of the RNP complex (25). On the other hand, it could disrupt the binding of viral polymerase with viral RNA by interacting with PB1 and PB2 of RNP (26). More recently, it has been demonstrated that Hsp70 could modulate viral RNA polymerase differentially at different phases of heat shock response, which appears to be a consequence of directional movement of Hsp70 between cytoplasmic and nuclear compartments (27).
In this study, we newly identified a heat shock protein, DnaJA1 (also designated Hdj2), a member of the type I DnaJ/Hsp40 family, as a positive regulator for influenza A virus replication. The proteins within type I DnaJ family are very diverse at the primary sequence level, but they all contain four typical domains: a highly conserved N-terminal J domain (28), followed by a Gly/Phe-rich region (G/F-rich domain), four repeats of the CxxCxGxG type zinc finger, and a less well-conserved C-terminal substrate-binding domain (SBD) (28, 29). DnaJ/Hsp40 and its homologous proteins normally act as Hsp70 cochaperones through their typical J domain to recruit specific substrates to Hsp70 and regulate Hsp70 ATPase activity (30). DnaJ/Hsp40s have been reported to be involved in regulating a wide range of viral infections by various mechanisms (reviewed in reference 31). DnaJA1 was identified as an interacting partner of NS5 (the largest viral protein exhibiting RNA-dependent RNA polymerase activity) of Japanese encephalitis virus (JEV) to facilitate viral replication (32). In addition, through a high-throughput approach based on random inactivation of cellular genes, DnaJA1 was also identified to be involved in regulating human immunodeficiency virus type 1 (HIV-1) replication (33). However, the mechanisms by which DnaJA1 regulates the replication of these viruses are still unknown.
In this study, we found that DnaJA1 interacted with both the PB2 and PA subunits of influenza A virus RNA polymerase and enhanced the viral RNA polymerase activity both in vivo and in vitro. We also observed that DnaJA1 was translocated into the nucleus along with the nuclear import of PA-PB1 dimer. Interestingly, we found that the stimulatory effects of DnaJA1 on viral RNA polymerase activities were mainly dependent on its C-terminal substrate-binding domain and independent of its highly conserved J domain. Therefore, we propose that DnaJA1 is specifically used by influenza virus to enhance its RNA polymerase activity in an Hsp70 cochaperone-independent manner.
MATERIALS AND METHODS
Viruses and cells.
Human lung carcinoma cell line A549, human embryonic kidney cell line 293T, human cervical cancer cell line HeLa, and the Madin-Darby canine kidney (MDCK) cell line were purchased from the American Type Culture Collection (ATCC; Manassas, VA). These cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA) supplemented with penicillin (100 U/ml), streptomycin (10 mg/ml), and 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA) at 37°C in a humidified incubator with 5% CO2 in air. Influenza A/WSN/33 virus was propagated in MDCK cells with 0.5% FBS-DMEM with 0.5 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Invitrogen, Carlsbad, CA). Vesicular stomatitis virus (VSV) was generously provided by Zhuo Zhou (Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College).
Plasmids and antibodies.
The full-length DnaJA1 coding sequence was amplified from a cDNA library of HeLa cells by PCR and cloned into pCMV6-entry vector at NheI/XhoI sites and into pcDNA3.1 vector at XhoI/BamHI sites for expressing DnaJA1-Flag and DnaJA1-myc/His, respectively. The DnaJA1 truncation mutants were generated by amplifying DnaJA1 fragments from DnaJA1-Flag-expressing plasmid and then cloning them into the pCMV6-entry vector at NheI/XhoI sites. The RNP reconstitution system of influenza virus A/HongKong/1/1968 (H3N2) was described before (34). The tandem affinity purification (TAP)-tagged or nontagged pcDNA plasmids encoding PB1, PB2, PA, and NP and the two viral RNA-expressing plasmids pPOLI-NA-RT and pPOLI-HA-RT were derived from influenza A/WSN/33 virus (34). The mouse anti-DnaJA1 antibody and the mouse anti-M1 antibody were purchased from Thermo Fisher (San Jose, CA), the mouse anti-influenza NP antibody (MAB8251) was from Millipore (Temecula, CA), the rabbit antiactin antibody was from Cell Signaling Technology (Danvers, MA), the rabbit anti-TAP (sc-25768) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), the mouse anti-Flag antibody was from Sigma-Aldrich (St. Louis, MO), and the rabbit anti-PB1 antibody and the rabbit anti-PB2 antibody were from Abmart Inc. (Shanghai, China). The rabbit anti-PA antibody was kindly provided by Ervin Fodor (Oxford University, United Kingdom).
RNA interference (RNAi).
DnaJA1 small interfering RNA (siRNA) (5′-GAUAUCAAGUGUGUACUAA-3′) and control siRNA (5′-UCACUGCGCUCGAUGCAGU-3′) were synthesized by Genepharma (Shanghai, China). The siRNA transfection reagent Hiperfect was purchased from Qiagen (Hilden, Germany). A549 cells were transfected with 50 nM concentrations of the siRNAs indicated below for 48 h, followed by virus infection or transfection. The knockdown efficiency was examined by Western blotting.
Growth curve analysis.
DnaJA1 knockdown or control A549 cells were infected with either wild-type influenza A/WSN/1933 (WSN) virus or VSV at a multiplicity of infection (MOI) of 0.001. At 0, 12, 24, 36, 48, 60, and 72 h postinfection (h.p.i.), supernatants were collected. The virus titers were determined by plaque assay on MDCK cells.
MUNANA fluorescence assay.
293T cells transfected with empty vector or DnaJA1-Flag, or its truncation mutant-expressing plasmids individually, were infected with WSN virus (MOI = 0. 1) at 24 h posttransfection (h.p.t.). Supernatants were then collected at 24 h.p.i. The virus titers were determined by quantifying the neuraminidase activity (NA) using a MUNANA (4-methylumbelliferyl-N-acetyl-neuraminicacid) assay. Briefly, 40 μl of cell supernatant was added into a 96-well black flat-bottom plate and mixed with 60 μl of MUNANA stock solution (20 μM; Sigma-Aldrich) at room temperature for 1 h. The amount of fluorescent product released by MUNANA hydrolysis was measured in SpectraMax M5 microplate reader (Molecular Devices, USA) with excitation and emission wavelengths of 355 nm and 460 nm.
Western blotting.
Cells were collected and lysed with CytoBuster (Merck Biosciences, Bad Soden, Germany) for 25 min at room temperature, and then the debris were removed by centrifugation at 12,000 rpm for 15 min. The lysates were separated by SDS-PAGE and transferred to nitrocellulose membranes (NC) and immunoblotted with indicated antibodies and IRDye secondary antibodies (LI-COR Biosciences, Lincoln, NE). The protein expression levels were determined with the Odyssey infrared imaging system (LI-COR Biosciences). The relative protein expression level was analyzed using the integrated software of the Odyssey system.
Sample preparation for mass spectrometry analysis.
293T cells which were 70% confluent in 15-cm dishes were transfected with 15 μg of pcDNA-PB2-TAP or pcDNA-TAP-NP with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for 40 h, and cells were harvested and lysed with 900 μl of lysis buffer (25% glycerol, 50 mM Tris-HCl [pH 8.0], 0.5% NP-40, 200 mM NaCl, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1% protease inhibitors). After centrifugation at 12,000 rpm for 15 min, the cell lysates were diluted in 5 ml of 150 mM NaCl solution and incubated with 100 μl of IgG Sepharose for 2 h at 4°C. The beads were then washed three times with 5 ml of binding buffer (50 mM Tris-HCl [pH 8.0], 1% glycerol, 0.2% NP-40, 150 mM NaCl, 1 mM PMSF). Finally, the bound proteins were released by cleavage with the Tobacco etch virus (TEV) protease (Invitrogen, Carlsbad, CA) for 4 h at 16°C. The purified proteins were separated by 8% SDS-PAGE, and the bands were visualized by silver staining.
LC-MS/MS analysis.
Samples were digested with trypsin using the filter-aided sample preparation (FASP) method (35). Peptides obtained by FASP were desalted using an Oasis HLB-1cc cartridge (Waters, Milford, MA). All peptide fractions were concentrated with a SpeedVac centrifuge (Savant, Farmingdale, NY) and solubilized in 0.1% formic acid. The resulting peptides were analyzed on an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Germany) coupled with a nano-flow liquid chromatography (LC) system (nanoACQUITY ultraperformance liquid chromatograph [UPLC]; Waters). The detailed experimental procedure for the peptide analysis is available on request. Tandem mass spectrometry (MS/MS) spectra were searched with SEQUEST search algorithm against a Swiss-Prot database (537505 sequence entries) using the following settings. (i) Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine was chosen as variable modification; (ii) a maximum of two missed cleavages was allowed; and (iii) MS and MS/MS tolerances of 10 ppm and 0.6 Da, respectively, were used. After the database search, the following filter criteria were applied to all results: high confidence corresponding to a false-discovery rate (FDR) of <1% and at least two unique peptide identifications per protein.
Primer extension analysis.
The total RNAs from either infected or transfected cells were extracted by TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Levels of neuraminidase (NA) segment-specific mRNA, cRNA, and vRNA were determined by primer extension assay as described previously (36, 37). The primers used in this study were 5′-TGGACTAGTGGGAGCATCAT-3′ (to detect vRNA) and 5′-TCCAGTATGGTTTTGATTTCCG-3′ (to detect mRNA and cRNA). The 32P-labeled primers were incubated with RNA sample and reverse transcriptase (Super Reverse Transcriptase III; Invitrogen) for 1.5 h at 50°C. The reaction was terminated by incubation for 5 min at 95°C. The transcription products were resolved on 6% PAGE gels containing 7 M urea and visualized by autoradiography. Quantification of the RNA bands was performed by phosphor image analysis with Typhoon Trio Plus (GE Healthcare, Waukesha, WI) and ImageQuant TL software (GE Healthcare).
Indirect immunofluorescence assay.
HeLa or A549 cells were infected with WSN viruses, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in 0.5 ml of phosphate-buffered saline (PBS) for 20 min, and blocked with bovine serum albumin–0.5% Tween 20–PBS (BSA–PBS-T) for 2 h at room temperature. Cells were then incubated with the primary antibodies (either anti-DnaJA1 antibody and anti-PA antibody or anti-DnaJA1 antibody and anti-PB2 antibody, respectively) for 2 h and further incubated with secondary antibodies Alexa Fluor 594 goat anti-mouse IgG and Alexa Fluor 488 goat anti-rabbit IgG (Zhongshanjinqiao Biotech Co., Beijing, China) for 1 h at room temperature, and cell nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI) reagent (Sigma-Aldrich). After 3 washes with 0.5 ml of PBS-T, cells were examined with a Leica TCS SP5 laser scanning confocal microscope (Leica, Cambridge, United Kingdom).
Coimmunoprecipitation assay.
293T cells in 3.5-cm dishes were transfected with DnaJA1-Flag and influenza viral polymerase subunits and NP expression plasmids. After 40 h of transfection, cells were harvested and lysed in 200 μl of RIPA buffer (50 mM Tris-HCl [pH 8.0], 1% NP-40, 150 mM NaCl, 0.25% sodium deoxycholate, and 1 mM EDTA). The lysates were either incubated for 2 h at 4°C with anti-Flag antibody and protein A/G agarose (Santa Cruz) for detecting Flag-tagged proteins or incubated with IgG Sepharose (GE Healthcare) for detecting TAP-tagged proteins. The beads were washed with 1 ml of washing buffer 6 times, followed by Western blotting. To detect endogenous DnaJA1 proteins, 293T cells were left uninfected or infected with influenza virus A/WSN/33 (MOI = 0.5). At 8 h postinfection, the cells were collected, washed with PBS, and lysed in 0.5 ml of RIPA buffer. The lysates were incubated with anti-PB2 or anti-PA antibody and protein A/G agarose beads for 4 h at 4°C. Beads were washed 6 times with 1 ml of washing buffer, and the samples were analyzed by Western blotting.
GST pulldown assay.
Glutathione S-transferase (GST)-fused DnaJA1 or GST proteins expressed from Escherichia coli cells were bound onto glutathione-Sepharose beads (GE Healthcare) and then incubated with lysates from 293T cells transfected with pcDNA-PB1, pcDNA-PB2, and pcDNA-PA at 4°C for 16 h. The beads were washed 3 times with PBS, and the samples were analyzed by Western blotting.
Recombinant protein preparations for in vitro transcription assay.
293T cells were transfected with pcDNA-PA-His, pcDNA-PB1, and pcDNA-PB2 for generating recombinant viral RNA polymerase and with pcDNA-DnaJA1-His for generating DnaJA1 protein. In addition, the recombinant RNA polymerase was also prepared in parallel in either DnaJA1 knockdown or control knockdown 293T cells. At 40 h posttransfection, cells were collected and lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 25% glycerol, 0.5% NP-40, 200 mM NaCl). The lysates were then incubated with a Ni-nitrilotriacetic acid column for 2 h and eluted with an elution buffer (50 mM Tris-HCl [pH 8.0], 5% glycerol, 200 mM NaCl, 100 mM imidazole). The purified recombinant protein samples were stored in 40% glycerol at −20°C.
In vitro ApG-primed transcription assay.
In vitro ApG-primed transcription was carried out at 30°C for 1 h in a total reaction volume of 3 μl containing 1 μl of purified recombinant proteins (see above), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ApG, 1 mM ATP, 0.5 mM CTP, 0.05 mM [α-32P]GTP (3,000 Ci/mmol; Amersham), 2 U/μl of RNase inhibitor, 0.5 μM 3′-end vRNA (5′-GGCCUGCUUUUGCU-3′), and 0.5 μM 5′-end vRNA (5′-AGUAGAAACAAGGCC-3′) (Dharmacon). The RNA transcription products were analyzed on 18% PAGE gels containing 7 M urea and visualized by autoradiography. The quantification was performed with Typhoon Trio Plus (GE Healthcare).
Statistical analysis.
A two-tailed Student t test was used to determine statistical significance. A P value of <0.05 was considered to be significant.
RESULTS
Identification of DnaJA1 to be a positive regulator for influenza virus replication.
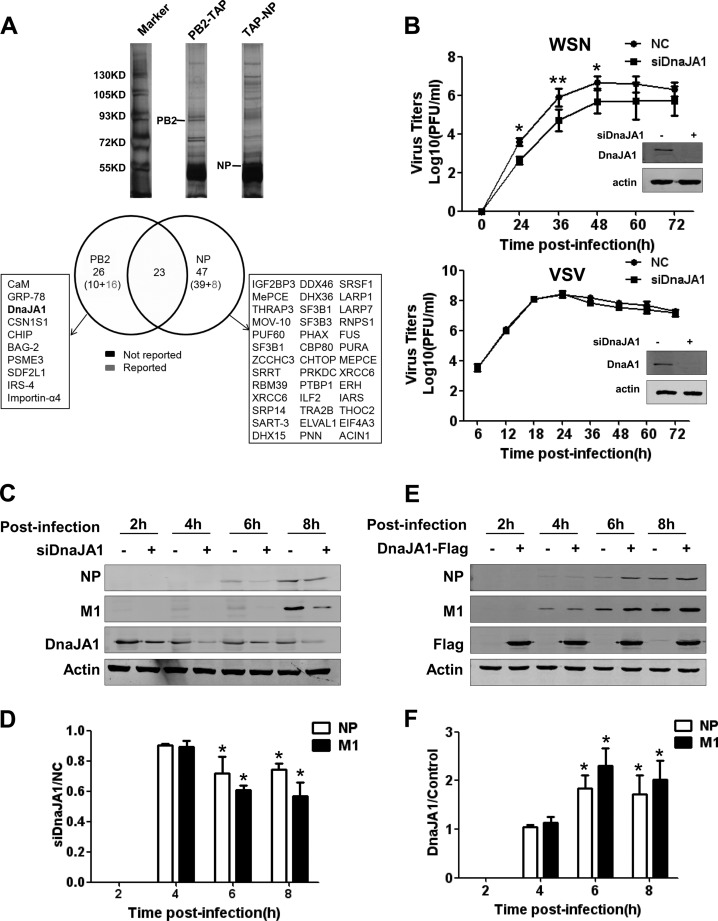
In an attempt to search for more host factors that could be involved in regulating viral RNA synthesis, we performed pulldown experiments with the polymerase subunits and NP as baits, followed by mass spectrometry analysis (14, 38) (see Materials and Methods). In a comparison of pulldown proteins between PB2-TAP or TAP-NP samples (Fig. 1A, top), 26 preliminary proteins in total were identified as PB2-TAP sample-specific proteins, and 10 of them have not been reported before. In contrast, 47 preliminary proteins in total were identified as TAP-NP sample-specific proteins, and 39 of them have not been reported before (Fig. 1A, bottom). In addition, there were 23 proteins to be present in both PB2-TAP and TAP-NP samples. Either they might interact with both PB2 and NP or they were pulled down by the TAP tag nonspecifically. Surely, these preliminary proteins await further systematic studies.
FIG 1.
Identification of DnaJA1 as a positive regulator for influenza virus replication. (A) DnaJA1 copurifies with the PB2 subunit. 293T cells were transfected with pcDNA-PB2-TAP or pcDNA-TAP-NP. At 40 h posttransfection, cells were lysed and subjected to IgG Sepharose purification. The purified proteins were released with TEV protease. A small of amount of the purified proteins were resolved on an 8% PAGE gel and visualized by silver staining. The rest were subjected to nano-LC-MS/MS analysis. There were 23 proteins which overlapped in both samples and are not listed individually. Preliminary proteins that were identified as either PB2-TAP sample-specific proteins or TAP-NP sample-specific proteins are shown in the boxes. (B) DnaJA1 acts as positive regulator specifically for influenza virus replication. A549 cells treated with DnaJA1-specific siRNA or control siRNA were infected with either influenza WSN virus or VSV at an MOI of 0.001. Every 12 h postinfection, the viral titers in the supernatant were determined by plaque assay on MDCK cells. The values are means of results from three independent experiments. Representative Western blot results for examining the knockdown efficiency of DnaJA1 are shown. (C) Knockdown of DnaJA1 reduces influenza virus protein expression. A549 cells treated with DnaJA1-specific siRNA or control siRNA were infected with influenza A/WSN/33 virus (MOI = 1). At the indicated times points postinfection, cells were harvested and lysed for Western blot analysis with NP- and M1-specific antibodies. (D) Statistical analysis of NP and M1 levels in panel C. Values for NP and M1 were standardized to the actin level and normalized to the levels of NP and M1 in cells treated with control siRNA (means ± SEMs of results from three independent experiments). (E) Overexpression of DnaJA1 increases influenza virus protein expression. 293T cells were transfected with pCMV-DnaJA1-Flag or pCMV6 empty vector for 24 h, followed by infection with WSN virus (MOI = 1). At the indicated times postinfection, cells were harvested and lysed for Western blot analysis with NP- and M1-specific antibodies. (F) Statistical analysis of NP and M1 levels in panel E. Values for NP and M1 were standardized to the actin level and normalized to levels of NP and M1 in cells transfected with control vector (means ± SEMs of the results from three independent experiments). *, P < 0.05; **, P < 0.01.
Among these host factors, we noticed that DnaJA1 was also previously identified in our recent cellular transcription profiling-based RNA interference (RNAi) screen (34). We observed a significant decrease in NP level in the cells treated with DnaJA1 siRNA in comparison with that in control cells. Therefore, we decided to examine this host factor further. To further confirm whether DnaJA1 affects influenza virus replication, we examined virus growth rate in A549 cells which were treated with DnaJA1 siRNA or control siRNA, followed by infection of WSN virus at an MOI of 0.001. We found that the virus titer decreased about 1 log in the DnaJA1 knockdown cells at various time points postinfection compared to that in the control cells (Fig. 1B, top). In order to examine whether the effect of the DnaJA1 is specific to influenza viruses, we also infected the DnaJA1 knockdown and control A549 cells with vesicular stomatitis virus (VSV) and examined its growth rate. The results showed that knockdown of DnaJA1 did not affect VSV growth rate (Fig. 1B, bottom).
Furthermore, we examined the effects of DnaJA1 knockdown on influenza virus protein expression in a single-cycle replication. Western blot analyses of lysates from DnaJA1 knockdown A549 cells infected with WSN virus at an MOI of 1 showed that the levels of viral NP and M1 proteins were lower in DnaJA1 knockdown cells than in control cells from 4 h.p.i. onward (Fig. 1C and D). In line with this, we observed that the levels of NP and M1 were increased when 293T cells were overexpressed with DnaJA1 protein at various time points postinfection (Fig. 1E and F). Taken together, our findings confirmed that DnaJA1 is co-opted by the influenza virus to act as a positive regulator for influenza virus replication.
DnaJA1 interacts with both the PB2 and PA subunits of the viral RNA polymerase.
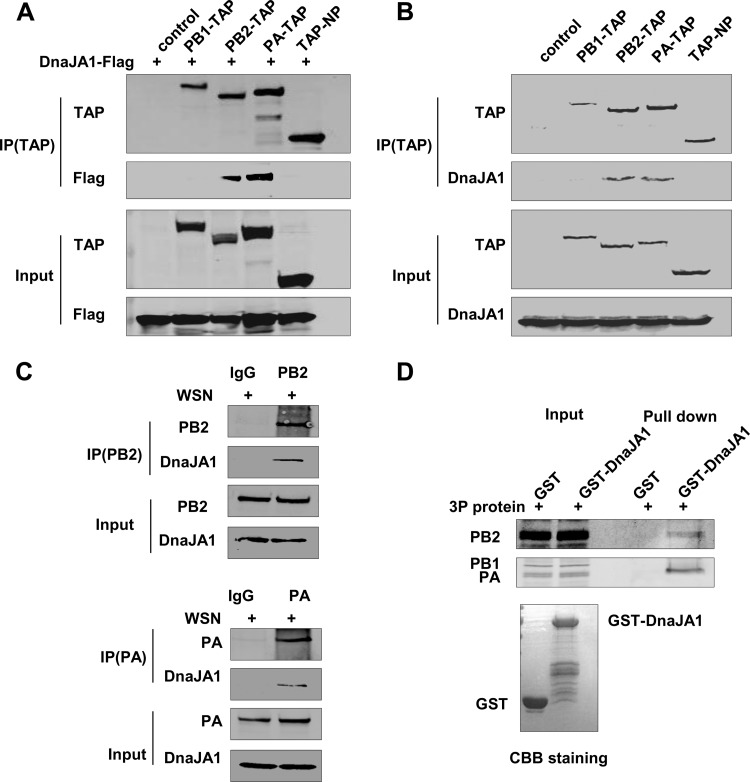
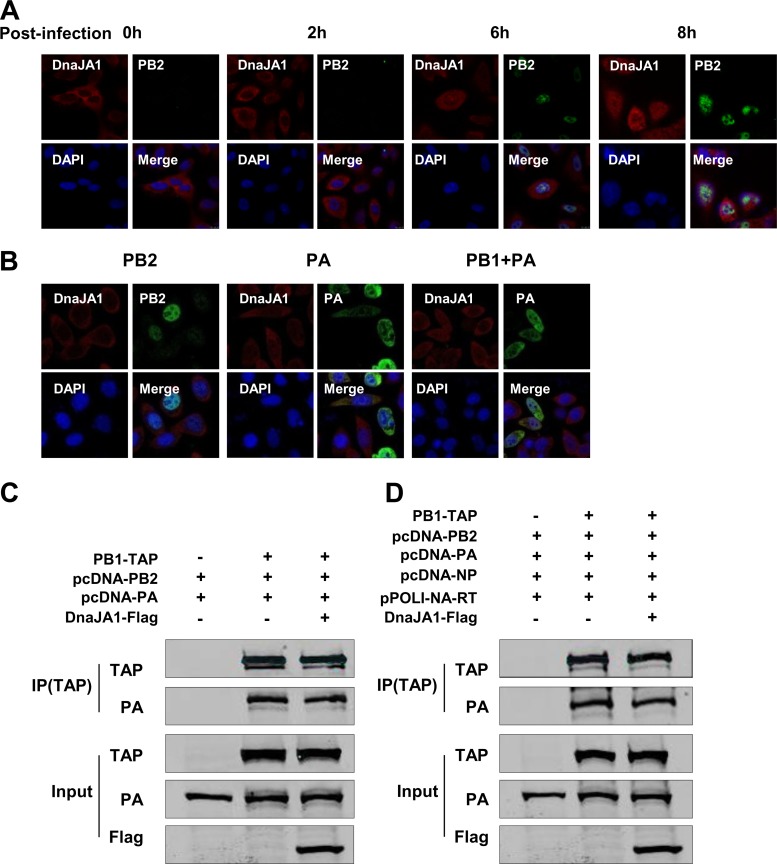
As described above, DnaJA1 was identified as an interacting partner of PB2 in our pulldown assay. To further confirm this interaction and to explore other potential interactions that may occur between DnaJA1 and other RNP components, we transfected DnaJA1-Flag-expressing plasmid into 293T cells, along with PB1-TAP-, PB2-TAP-, PA-TAP-, or TAP-NP-expressing plasmid individually. Cell lysates were then purified with IgG Sepharose and analyzed with an anti-Flag antibody. As shown in Fig. 2A, Flag-tagged DnaJA1 could be specifically associated with PB2 and PA but not with PB1 or NP. We also detected cellular DnaJA1 that could be specifically pulled down by PB2-TAP or PA-TAP (Fig. 2B). We further examined the associations between cellular DnaJA1 and PB2 or PA by coimmunoprecipitation assays with infected 293T cells. The results showed that endogenous DnaJA1 could specifically associate with PB2 and PA in WSN virus-infected cells (Fig. 2C). To further confirm the interaction between DnaJA1 and PB2 or PA, an in vitro GST pulldown assay was carried out. We used partially purified GST-DnaJA1 or GST proteins from E. coli cells and recombinant 3P complex from 293T cells for this assay. As shown in Fig. 2D, GST-tagged DnaJA1 could specifically pull down PB2 and PA but not PB1 of the 3P complex in vitro.
FIG 2.
DnaJA1 associates with PB2 and PA subunits. (A) Flag-tagged DnaJA1 copurifies with both PB2 and PA. The control vector, TAP-tagged WSN PA, PB1, PB2, or NP expression plasmids were individually transfected into 293T cells along with Flag-tagged DnaJA1. At 40 h posttransfection, cells were harvested and lysed, followed by IgG Sepharose purification and Western blot analyses. IP, immunoprecipitation. (B) Endogenous DnaJA1 interacts with PB2 and PA. The control vector, TAP-tagged WSN PA, PB1, PB2, and NP expression constructs were individually transfected into 293T cells. At 40 h posttransfection, cells were harvested and lysed, followed by IgG Sepharose purification and Western blot analyses with an anti-DnaJA1 antibody. (C) Endogenous DnaJA1 interacts with PB2 and PA in virus-infected cells. 293T cells were infected with influenza WSN virus (MO1 = 0.5). Cells were lysed at 16 h.p.i. and subjected to immunoprecipitation with either normal IgG or anti-PB2/anti-PA antibodies, followed by Western blot analyses with an anti-DnaJA1 antibody. (D) Exogenous DnaJA1 interacts with PB2 and PA. 293T cells were transfected with the pcDNA-PB1, pcDNA-PB2. and pcDNA-PA for 40 h. Cells were lysed and subjected to GST pulldown assay with the recombinant GST or GST-DnaJA1 prepared from E. coli cells. The pulldown samples were analyzed by Western blotting with anti-PB2 or anti-PA antibody. Coomassie brilliant blue (CBB) staining of purified protein is shown at the bottom.
The C-terminal substrate-binding domain of DnaJA1 is responsible for upregulating influenza virus replication.
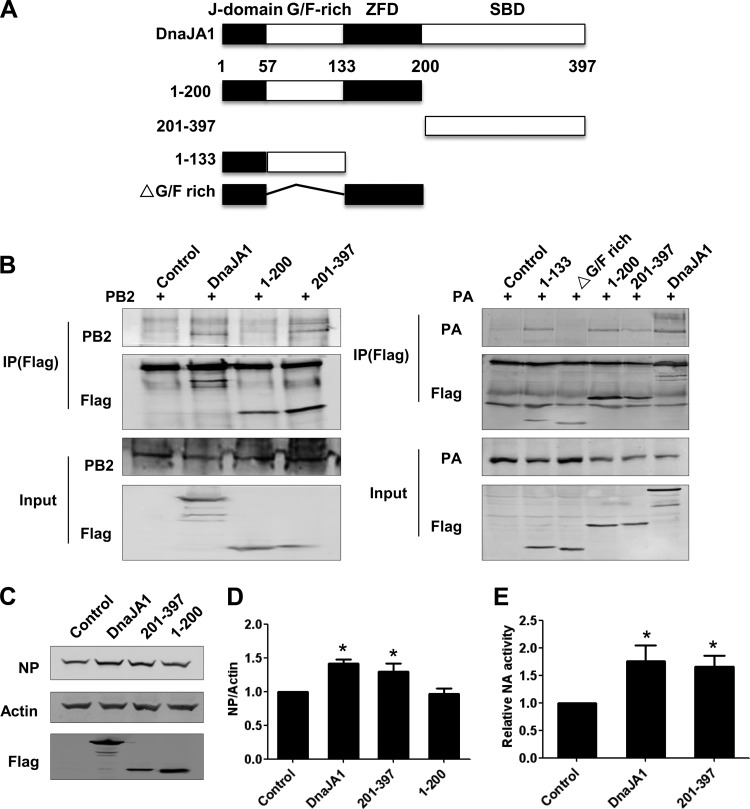
We next mapped the domain(s) of DnaJA1 that is responsible for the interactions between DnaJA1 and PB2 or PA. The four domains of DnaJA1, organized from the N terminus to the C terminus, are the highly conserved signature J domain, Gly/Phe-rich (G/F-rich) region, zinc finger domain (ZFD), and substrate-binding domain (SBD) (Fig. 3A). We first generated two DnaJA1 truncation mutants: the N-terminal DnaJA1(1–200) mutant and the C-terminal DnaJA1(201–397) mutant. We further split the N-terminal DnaJA1(1–200) truncation mutant into the DnaJA1(1–133) and ΔG/P-rich mutants (Fig. 3A). The associations of PB2 or PA with these truncation mutants were then examined by coimmunoprecipitation. Figure 3B showed that PB2 could specifically associate with the DnaJA1(201–397) mutant but not the DnaJA1(1–200) mutant, while PA could interact with both the Gly/Phe-rich region and the DnaJA1(201–397) mutant.
FIG 3.
The C-terminal substrate-binding domain of DnaJA1 is responsible for upregulating influenza virus replication. (A) Schematic diagram of Flag-tagged DnaJA1 truncation mutants. (B) Mapping of the PB2 or PA binding domain(s) of DnaJA1. 293T cells were transfected with the full-length DnaJA1-expressing or the two truncation mutant-expressing plasmids, respectively, together with pcDNA-PB2 or pcDNA-PA individually for 40 h. Cells were lysed and immunoprecipitated with an anti-Flag antibody. The co-IP samples and lysates were then analyzed by Western blotting with an anti-Flag antibody. (C) The C-terminal substrate-binding domain of DnaJA1 alone enhances influenza virus protein expression. 293T cells were transfected with either empty vector or plasmids expressing DnaJA1-Flag or its truncation mutants individually, followed by infection with WSN virus (MOI = 0.1). At 16 h.p.i., cell extracts were prepared and analyzed by Western blotting with anti-NP antibody. (D) Quantitative analysis of viral protein expression levels in panel C. Values of NP were standardized to the actin level and normalized to levels of NP in cells transfected with control vector. The results are presented as means ± SEMs of results from three independent experiments. (E) DnaJA1 promotes influenza virus replication. 293T cells were transfected with either empty vector or plasmids expressing DnaJA1-Flag or its truncation mutants individually, followed by infection with WSN virus (MOI = 0.1). At 24 h.p.i., the virus titers were determined by MUNANA assay. *, P < 0.05.
Subsequently, in order to test which domain of DnaJA1 plays a role in facilitating influenza virus replication, we transfected 293T cells with the two DnaJA1 truncation mutant-expressing plasmids individually, followed by WSN virus infection. We found that the DnaJA1(201–397) mutant, in addition to full-length DnaJA1, could significantly increase the level of NP upon virus infection, whereas the DnaJA1(1–200) mutant could not (Fig. 3C and D). To examine whether the overexpression of DnaJA1(201–397) could increase virus titers, a standard MUNANA assay based on quantifying NA activity was performed. Figure 3E showed that full-length DnaJA1 and DnaJA1(201–397) increased NA activity significantly. Taken together, these data indicated that the C-terminal substrate-binding domain is mainly responsible for upregulating influenza virus replication.
DnaJA1 enhances viral RNA synthesis in an RNP reconstitution system.
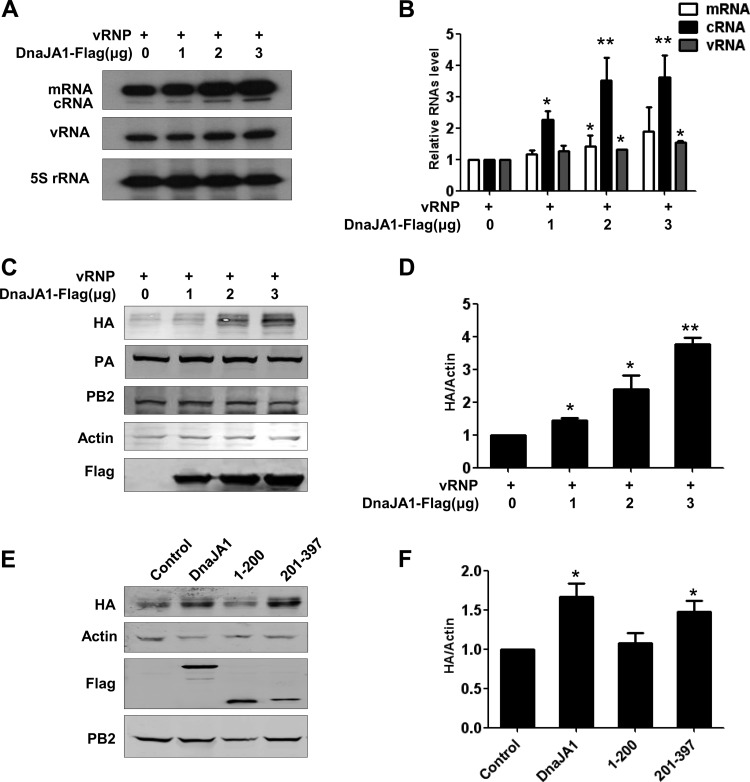
Since DnaJA1 was found to interact with both the PB2 and PA subunits specifically, it can be deduced that DnaJA1 may play a role in viral RNA synthesis machinery. To confirm this, we examined the effects of DnaJA1 on viral RNA synthesis in an RNP reconstitution system derived from influenza A/HongKong/1/1968 (H3N2) (34). 293T cells were transfected with the five RNP reconstitution plasmids (pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-NA-RT), together with increasing amounts of the DnaJA1 expression plasmid. We found that the levels of all three viral RNA species (vRNA, mRNA, and cRNA) were increased in a DnaJA1 dose-dependent manner (Fig. 4A and B). In the RNP reconstitution system, PB2 and PA are constantly produced by polymerase II (Pol II)-driven expression plasmids, while NA vRNA was first generated from a Pol I-driven RNA expression plasmid which was then amplified by the viral RNA polymerase. Since the NA vRNA level produced from the pPOLI plasmid was very low, the observed NA vRNA levels mainly reflected the activities of the viral RNA polymerase. Due to the observations that DnaJA1 could affect the protein expression levels of some cellular proteins (39–41), we asked whether the expression levels PB2 and PA could be affected by DnaJA1 overexpression. To test this, we performed a Western blot analysis to examine protein expression levels of the RNP reconstitution system, in which we replaced the pPOLI-NA-RT plasmid with the pPOLI-HA-RT for convenience of detection. Figure 4 shows that the levels of both PB2 and PA were comparable in all samples but that the expression levels of HA were increased in a DnaJA1 dose-dependent manner (Fig. 4C and D). These results suggest that DnaJA1 could upregulate viral RNA synthesis but not affect PB2 and PA expression.
FIG 4.
DnaJA1 enhances viral RNA synthesis in vivo. (A) DnaJA1 overexpression enhances viral RNA synthesis in a dose-dependent manner in an influenza virus RNP reconstitution system. 293T cells were transfected with influenza A/Hong Kong/1/1968 (H3N2) virus (HK68) RNP reconstitution plasmids (pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-NA-RT), along with increasing doses of the DnaJA1 expressing plasmid. Total RNAs were extracted and the levels of three viral RNA species were analyzed by primer extension analysis. (B) Statistical analysis of viral RNAs in panel A. Values of viral RNAs were standardized to the 5S rRNA level and normalized to levels of viral RNAs in cells transfected with control vector. Data represent the means ± SEMs of results from three independent experiments. (C) Overexpression of DnaJA1 does not affect PB2 and PA expression in the RNP reconstitution system. 293T cells were transfected as for panel A, except that the pPOLI-NA-RT plasmid was replaced with pPOLI-HA-RT plasmid. Cells were harvested after 24 h, and lysates were analyzed by Western blotting. (D) Statistical analysis of HA levels in panel C. Values of HA were standardized to the actin level and normalized to levels of HA in cells transfected with control vector (means ± SEMs of results from three independent experiments). (E) The C-terminal substrate-binding domain of DnaJA1 stimulates viral protein expression in the RNP reconstitution system. 293T cells were transfected as for panel C, except that the full-length DnaJA1-expressing plasmid was replaced with DnaJA1 truncation mutant-expressing plasmids individually. Cell extracts were prepared and analyzed by Western blotting. (F) Statistical analysis of HA levels in panel C. The data are presented as means ± SEMs of results from three independent experiments. *, P < 0.05; **, P < 0.01.
We next were interested in examining whether the C-terminal domain of DnaJA1 is able to stimulate viral RNA synthesis in the RNP reconstitution system. To this end, we transfected the two DnaJA1 truncation mutants individually, along with the RNP reconstitution plasmids, and examined the HA protein expression level with Western blot analysis. Interestingly, we found that the DnaJA1(201–379) mutant, containing the C-terminal substrate-binding domain alone, could stimulate HA protein expression to a level comparable to that stimulated by full-length DnaJA1, while the DnaJA1(1–200) mutant could not (Fig. 4E and F). These results strongly suggested that DnaJA1 stimulates viral RNA synthesis mainly dependent on its C-terminal substrate-binding domain and independent of its typical J domain.
DnaJA1 translocates into nucleus upon influenza virus infection but does not affect viral RNP assembly.
To study the mechanism(s) by which DnaJA1 regulates influenza A virus RNA synthesis, we first studied the localization of DnaJA1 upon influenza virus infection. A549 cells were infected with influenza virus at a high multiplicity (MOI = 3), and the subcellular localization of DnaJA1 was examined by indirect immunofluorescence assay at various time points postinfection. At the early stages of virus infection, DnaJA1 was present predominately in cytoplasm. However, at later stages, when the viral proteins were synthesized at a significant level as shown by the level of PB2, we observed that DnaJA1 was distributed all over the cells (Fig. 5A). This result supports the notion that DnaJA1 is specifically recruited into the nucleus during influenza virus replication.
FIG 5.
DnaJA1 is recruited into the nucleus upon influenza virus infection. (A) DnaJA1 translocates into the nucleus during influenza virus replication. A549 cells were infected with WSN virus at an MOI of 3. At the indicated times postinfection, cells were stained for nuclei (blue), PB2 (green), and DnaJA1 (red) by indirect immunofluorescence assay. (B) DnaJA1 translocates into the nucleus along with PB1-PA dimer. HeLa cells were transfected with either pcDNA-PB2 or pcDNA-PA or with a combination of pcDNA-PB1 and pcDNA-PA plasmids for 48 h. Cells were stained for nuclei (blue), PB2 or PA (green), and DnaJA1 (red). (C) DnaJA1 does not affect viral RNA polymerase assembly. 293T cells were transfected with the indicated plasmids. Cell lysates were purified with IgG Sepharose, and the lysates and products were analyzed by Western blot analysis. (D) DnaJA1 does not affect viral RNP assembly. 293T cells were transfected with the indicated plasmids. Cell lysates were purified with IgG Sepharose, and the lysates and products were analyzed by Western blot analysis.
According to the RdRp assembly model, PB1 associates with PA in the cytoplasm and is transported into the nucleus as a PB1-PA dimer, and then it assembles with individually transported PB2 into a trimeric complex (11, 22). Together with our observation in this study that DnaJA1 could interact with PB2 and PA, we further examined DnaJA1 nuclear translocation in HeLa cells expressingPB2 or PA alone or with both PB1 and PA. We found that the localization of DnaJA1 did not change in the cells when PB2 or PA was expressed alone. In contrast, when PB1 was coexpressed with PA, translocation of DnaJA1 into the nucleus was apparent, suggesting that DnaJA1 is recruited into the nucleus along with the PB1-PA dimer upon influenza virus infection (Fig. 5B).
Since both Hsp90 and Hsp70 could affect vRNP assembly by affecting nuclear transport of the polymerase subunits (11, 22), we then tested whether DnaJA1 affected the 3P or RNP assembly (Fig. 5C and D). We found that neither the 3P nor the vRNP assembly efficiency was associated with DnaJA1 overexpression in 293T cells. We concluded that DnaJA1 enters the nucleus upon influenza virus infection, but it does not affect the viral RNP complex assembly.
DnaJA1 enhances viral RNA polymerase activity in vitro.
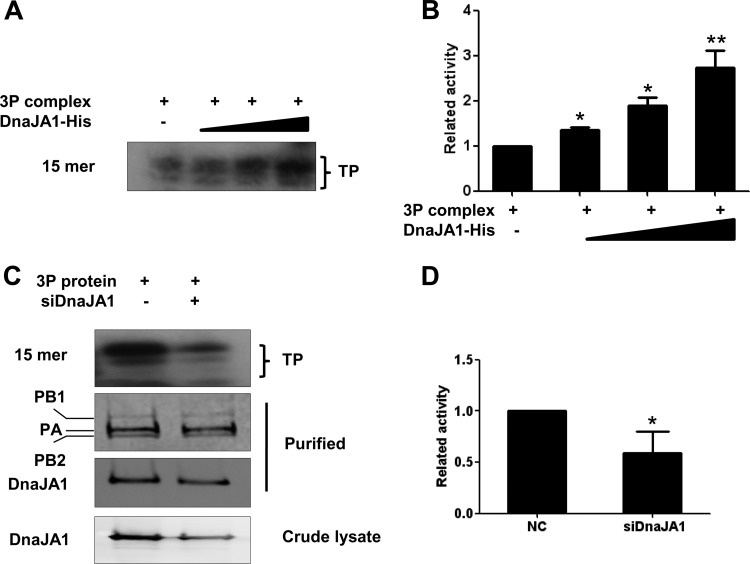
It is known that viral RNA synthesis efficiency is dependent on both RNP assembly efficiencies and RdRp activities. We were interested in examining whether DnaJA1 could affect viral RNA polymerase activities. To test this, we used an in vitro ApG-primed transcription assay that was developed to specifically examine the transcriptional activity of influenza viral RNA polymerase in vitro (42). We first performed the in vitro ApG-primed transcription assay with purified recombinant viral RNA polymerase (3P complex) and with increasing amounts of purified recombinant DnaJA1 protein. It can be seen that the recombinant DnaJA1 could stimulate transcriptional activity of the viral RNA polymerase in vitro in a dose-dependent manner (Fig. 6A and B). To further confirm this result, we also performed the in vitro ApG-primed transcription assay with the 3P complexes purified from DnaJA1 knockdown cells and from control cells. As shown in Fig. 6C, equal amounts of recombinant 3P complexes were purified, but more DnaJA1 was copurified with the 3P complex in the control cells than that in the DnaJA1 knockdown cells. As expected, we found that significantly higher levels of RNA transcripts were synthesized in the ApG-primed transcription assay with the polymerase prepared from the control cells than in the DnaJA1 knockdown cells (Fig. 6C and D). Therefore, our in vitro data suggested that DnaJA1 is used by influenza virus to specifically enhance its viral polymerase activity.
FIG 6.
DnaJA1 enhances influenza virus polymerase activity in vitro. (A) DnaJA1 stimulates the transcriptional activity of viral RNA polymerase activity in vitro. The ApG-primed transcription was conducted in the absence or presence of increasing amounts (50 ng, 150 ng, and 250 ng) of recombinant DnaJA1 protein. The transcription product (TP) is indicated. (B) Statistical analysis of TP levels in panel A. The values were normalized to levels of RNAs in the reaction without adding DnaJA1 protein. The data represent the means ± SEMs of results from three independent experiments. (C) The recombinant polymerase prepared in DnaJA1 knockdown cells is less active than the polymerase prepared in control cells. The polymerase complex from 293T cells treated with DnaJA1-specific siRNA and control siRNA was used for the ApG-primed transcription. (D) Statistical analysis of TP levels in panel C. The values were normalized to levels of RNAs with control siRNA. The data represent the means ± SEMs of results from three independent experiments, *, P < 0.05; **, P < 0.01.
DISCUSSION
The RNA polymerase of influenza virus is the most complex polymerase among the RNA-dependent RNA polymerases of RNA viruses because of its nature of being a heterotrimeric complex and its utilization of the nucleus as the site for viral RNA synthesis. As a result, the interplay between influenza virus RNA polymerase and host factors is much more complicated than those of other RNA viruses. In this study, we identified a host factor, DnaJA1/Hsp40, as an interacting partner of PB2 and PA subunits of the viral RNA polymerase through combined copurification and mass spectrometry methods. We found that DnaJA1 is specifically recruited into nucleus along with the PB1-PA nuclear import to enhance viral RNA polymerase activity. Previously, members of the Hsp40 family have been reported to be involved in regulating influenza virus replication through modulating protein kinase R (PKR) activity. The type III Hsp40 p58IPK, a cellular regulator of PKR, has been reported to regulate influenza virus mRNA translation and infection through a PKR-mediated mechanism (43). DnaJB1/Hsp40 was reported to be involved in activating the PKR pathway through its interaction with the viral M2 protein (44). On the other hand, DnaJB11/Hsp40 was identified as an interacting partner of NP and the DnaJB11-NP interaction was implicated in inhibition of PKR activity upon virus infection (45). To our knowledge, this is the first report that an Hsp40 protein, DnaJA1, is specifically involved in association with the influenza viral RNA polymerase subunits and is able to enhance viral RNA polymerase activity.
DnaJA1 is a typical member of type I Hsp40 proteins which contain four domains. The J domain is the most important domain for the Hsp70 cochaperone function of Hsp40 (30, 31, 46). Hsp40 proteins normally interact with their client proteins through their substrate-binding region and deliver the client protein to Hsp70 proteins through its J domain for subsequent protein folding (47, 48). In this study, we found that both the PB2 and PA subunits of influenza virus RNA polymerase are able to associate with the C-terminal substrate-binding domain of DnaJA1, while PA also associates with the G/F-rich domain of DnaJA1. In terms of the biological function of these associations, we found that the role of DnaJA1 in stimulating influenza viral RNA polymerase activity is mainly dependent on its C-terminal substrate-binding domain and completely independent of its J domain. It has been previously reported that the C-terminal substrate-binding fragments of other Hsp40 proteins, such as DnaJB6b and DnaJB8 in Homo sapiens and Ydj1 in yeast, could inhibit protein aggregation and that the antiaggregation activities of these Hsp40 proteins are independent of their J domains (49, 50). Considering the fact that the influenza viral RNA polymerase might form oligomers intracellularly (51, 52), together with our observation that the presence of DnaJA1 could increase the transcriptional activity of the RNA polymerase in vitro, we speculate that DnaJA1 is likely to play a role in preventing influenza virus RNA polymerase oligomerization by binding to the polymerase through its C-terminal substrate-binding domain. However, we should point out that our evidence here could not exclude possibilities that DnaJA1 may affect the structure and/or stability of the viral polymerase complex. Further identification of the DnaJA1 interaction sites on PB2 and PA and examination of the role of the DnaJA1 in polymerase activity are important to elucidate the mechanism.
In addition to DnaJA1 identified here, there were other host factors that have been reported to be able to stimulate influenza virus RNA polymerase activities in vitro. Momose et al. reported that the Hsp90 was able to stimulate in vitro virus RNA synthesis by stabilizing the viral polymerase activity (23). Although they found that Hsp90 interacted with the PB2 subunit through both the N-terminal chaperone domain and the middle region, the virus RNA synthesis stimulatory activity of Hsp90 was only dependent on an acidic domain of the middle region (23). In addition, a host factor, Ebp1, was reported to interact with the catalytic region of PB1 subunit and thus interfere with in vitro RNA synthesis activity of influenza virus RNA polymerase (3P complex) (53). Therefore, our findings in this study further extend our knowledge on host factors that are specifically involved in stimulating viral RNA polymerase activities.
In summary, we have newly identified a member of Hsp40 family, DnaJA1, that acts as a positive regulator for influenza virus replication. Our detailed biochemical analysis revealed that DnaJA1 could specifically associate with the influenza virus RNA polymerase PB2 and PA subunits and enter the nucleus along with the nuclear import of PB1-PA dimer to specifically enhance viral RNA polymerase activities. These results not only increase our knowledge on host factors involved in regulating influenza virus RNA polymerase but also provide a novel basis for development of drugs against influenza virus.
ACKNOWLEDGMENTS
This work was supported by grants from Chinese Science and Technology Key Projects (2013ZX10004601) and the National Natural Science Foundation of China (31471329 and 81271829).
Footnotes
Published ahead of print 24 September 2014
REFERENCES
- 1.Palese P, Shaw ML. 2013. Orthomyxoviridae: the viruses and their replication, p 1151–1185 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889–893. 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 3.Mänz B, Schwemmle M, Brunotte L. 2013. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 87:7200–7209. 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fodor E, Brownlee GG. 2002. Influenza virus replication, p 1–29 In Potter CW. (ed), Influenza. Elsevier Science, New York, NY. [Google Scholar]
- 5.Biswas SK, Nayak DP. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fechter P, Brownlee GG. 2005. Recognition of mRNA cap structures by viral and cellular proteins. J. Gen. Virol. 86:1239–1249. 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
- 7.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500–506. 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 8.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 9.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA (N) reveals an endonuclease active site. Nature 458:909–913. 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 10.Palese P, Shaw ML. 2013. Orthomyxoviridae, p 1151–1185 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 11.Deng T, Sharps J, Fodor E, Brownlee GG. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 79:8669–8674. 10.1128/JVI.79.13.8669-8674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor E, Smith M. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144–9153. 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tafforeau L, Chantier T, Pradezynski F, Pellet J, Mangeot PE, Vidalain PO, Andre P, Rabourdin-Combe C, Lotteau V. 2011. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J. Virol. 85:13010–13018. 10.1128/JVI.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088. 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 15.Bradel-Tretheway BG, Mattiacio JL, Krasnoselsky A, Stevenson C, Purdy D, Dewhurst S, Katze MG. 2011. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J. Virol. 85:8569–8581. 10.1128/JVI.00496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fodor E. 2013. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 57:113–122. 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 17.Nagata K, Kawaguchi A, Naito T. 2008. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18:247–260. 10.1002/rmv.575. [DOI] [PubMed] [Google Scholar]
- 18.Gething MJ, Sambrook J. 1992. Protein folding in the cell. Nature 355:33–45. 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 19.Young JC, Agashe VR, Siegers K, Hartl FU. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5:781–791. 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 20.Bukau B, Weissman J, Horwich A. 2006. Molecular chaperones and protein quality control. Cell 125:443–451. 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Hartl FU. 1996. Molecular chaperones in cellular protein folding. Nature 381:571–579. 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 22.Naito T, Momose F, Kawaguchi A, Nagata K. 2007. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 81:1339–1349. 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 277:45306–45314. 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 24.Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. 2008. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology 377:431–439. 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 25.Hirayama E, Atagi H, Hiraki A, Kim J. 2004. Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J. Virol. 78:1263–1270. 10.1128/JVI.78.3.1263-1270.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Zhang J, Tong X, Liu W, Ye X. 2011. Heat shock protein 70 inhibits the activity of influenza A virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo. PLoS One 6:e16546. 10.1371/journal.pone.0016546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manzoor R, Kuroda K, Yoshida R, Tsuda Y, Fujikura D, Miyamoto H, Kajihara M, Kida H, Takada A. 2014. Heat shock protein 70 modulates influenza A virus polymerase activity. J. Biol. Chem. 289:7599–7614. 10.1074/jbc.M113.507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheetham ME, Caplan AJ. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28–36. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cyr DM, Langer T, Douglas MG. 1994. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19:176–181. 10.1016/0968-0004(94)90281-X. [DOI] [PubMed] [Google Scholar]
- 30.Qiu XB, Shao YM, Miao S, Wang L. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63:2560–2570. 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knox C, Luke GA, Blatch GL, Pesce ER. 2011. Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res. 160:15–24. 10.1016/j.virusres.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Wang RY, Huang YR, Chong KM, Hung CY, Ke ZL, Chang RY. 2011. DnaJ homolog Hdj2 facilitates Japanese encephalitis virus replication. Virol. J. 8:471. 10.1186/1743-422X-8-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dziuba N, Ferguson MR, O'Brien WA, Sanchez A, Prussia AJ, McDonald NJ, Friedrich BM, Li G, Shaw MW, Sheng J, Hodge TW, Rubin DH, Murray JL. 2012. Identification of cellular proteins required for replication of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 28:1329–1339. 10.1089/aid.2011.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, Cao M, Guo Y, Zhao L, Wang J, Jia X, Li J, Wang C, Gabriel G, Xue Q, Yi Y, Cui S, Jin Q, Deng T. 2014. Fragile X mental retardation protein stimulates ribonucleoprotein assembly of influenza A virus. Nat. Commun. 5:3259. 10.1038/ncomms4259. [DOI] [PubMed] [Google Scholar]
- 35.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat. Methods 6:359–362. 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 36.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989–9001. 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vreede FT, Jung TE, Brownlee GG. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568–9572. 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. 2006. Role of Ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 80:11911–11919. 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker VE, Wong MJ, Atanasiu R, Hantouche C, Young JC, Shrier A. 2010. Hsp40 chaperones promote degradation of the HERG potassium channel. J. Biol. Chem. 285:3319–3329. 10.1074/jbc.M109.024000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abisambra JF, Jinwal UK, Suntharalingam A, Arulselvam K, Brady S, Cockman M, Jin Y, Zhang B, Dickey CA. 2012. DnaJA1 antagonizes constitutive Hsp70-mediated stabilization of tau. J. Mol. Biol. 421:653–661. 10.1016/j.jmb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orthwein A, Zahn A, Methot SP, Godin D, Conticello SG, Terada K, Di Noia JM. 2012. Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1. EMBO J. 31:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brownlee GG, Sharps JL. 2002. The RNA polymerase of influenza A virus is stabilized by interaction with its viral RNA promoter. J. Virol. 76:7103–7113. 10.1128/JVI.76.14.7103-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, Katze MG. 2007. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 81:2221–2230. 10.1128/JVI.02151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan Z, Liu D, Mi S, Zhang J, Ye Q, Wang M, Gao GF, Yan J. 2010. Interaction of Hsp40 with influenza virus M2 protein: implications for PKR signaling pathway. Protein Cell 1:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma K, Tripathi S, Ranjan P, Kumar P, Garten R, Deyde V, Katz JM, Cox NJ, Lal RB, Sambhara S, Lal SK. 2011. Influenza A virus nucleoprotein exploits Hsp40 to inhibit PKR activation. PLoS One 6:e20215. 10.1371/journal.pone.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Qian X, Sha B. 2009. Heat shock protein 40: structural studies and their functional implications. Protein Pept. Lett. 16:606–612. 10.2174/092986609788490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Qian X, Sha B. 2003. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11:1475–1483. 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858. 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 49.Hageman J, Rujano MA, van Waarde MA, Kakkar V, Dirks RP, Govorukhina N, Oosterveld-Hut HM, Lubsen NH, Kampinga HH. 2010. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell 37:355–369. 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Lu Z, Cyr DM. 1998. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273:5970–5978. 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 51.Jorba N, Area E, Ortin J. 2008. Oligomerization of the influenza virus polymerase complex in vivo. J. Gen. Virol. 89:520–524. 10.1099/vir.0.83387-0. [DOI] [PubMed] [Google Scholar]
- 52.Resa-Infante P, Recuero-Checa MA, Zamarreno N, Llorca O, Ortin J. 2010. Structural and functional characterization of an influenza virus RNA polymerase-genomic RNA complex. J. Virol. 84:10477–10487. 10.1128/JVI.01115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda A, Okamoto T, Ishihama A. 2007. Host factor Ebp1: selective inhibitor of influenza virus transcriptase. Genes Cells 12:133–142. 10.1111/j.1365-2443.2007.01047.x. [DOI] [PubMed] [Google Scholar]