ABSTRACT
The influence of major histocompatibility complex class I (MHC-I) alleles on human immunodeficiency virus (HIV) diversity in humans has been well characterized at the population level. MHC-I alleles likely affect viral diversity in the simian immunodeficiency virus (SIV)-infected pig-tailed macaque (Macaca nemestrina) model, but this is poorly characterized. We studied the evolution of SIV in pig-tailed macaques with a range of MHC-I haplotypes. SIVmac251 genomes were amplified from the plasma of 44 pig-tailed macaques infected with SIVmac251 at 4 to 10 months after infection and characterized by Illumina deep sequencing. MHC-I typing was performed on cellular RNA using Roche/454 pyrosequencing. MHC-I haplotypes and viral sequence polymorphisms at both individual mutations and groups of mutations spanning 10-amino-acid segments were linked using in-house bioinformatics pipelines, since cytotoxic T lymphocyte (CTL) escape can occur at different amino acids within the same epitope in different animals. The approach successfully identified 6 known CTL escape mutations within 3 Mane-A1*084-restricted epitopes. The approach also identified over 70 new SIV polymorphisms linked to a variety of MHC-I haplotypes. Using functional CD8 T cell assays, we confirmed that one of these associations, a Mane-B028 haplotype-linked mutation in Nef, corresponded to a CTL epitope. We also identified mutations associated with the Mane-B017 haplotype that were previously described to be CTL epitopes restricted by Mamu-B*017:01 in rhesus macaques. This detailed study of pig-tailed macaque MHC-I genetics and SIV polymorphisms will enable a refined level of analysis for future vaccine design and strategies for treatment of HIV infection.
IMPORTANCE Cytotoxic T lymphocytes select for virus escape mutants of HIV and SIV, and this limits the effectiveness of vaccines and immunotherapies against these viruses. Patterns of immune escape variants are similar in HIV type 1-infected human subjects that share the same MHC-I genes, but this has not been studied for SIV infection of macaques. By studying SIV sequence diversity in 44 MHC-typed SIV-infected pigtail macaques, we defined over 70 sites within SIV where mutations were common in macaques sharing particular MHC-I genes. Further, pigtail macaques sharing nearly identical MHC-I genes with rhesus macaques responded to the same CTL epitope and forced immune escape. This allows many reagents developed to study rhesus macaques to also be used to study pigtail macaques. Overall, our study defines sites of immune escape in SIV in pigtailed macaques, and this enables a more refined level of analysis of future vaccine design and strategies for treatment of HIV infection.
INTRODUCTION
There is a need for a highly effective vaccine or immune-based therapies for human immunodeficiency virus (HIV) infection. The immune pressure on the virus by cytotoxic T lymphocytes (CTLs) can result in reduced virus levels (1) but usually leads to the emergence of escape mutations (2). Some of these mutations exact a fitness cost on the virus (3), which can result in a lowered viral load (VL) in the host (4). However, compensatory mutations that occur together with immune escape mutations are able to at least partially negate the fitness cost resulting from escape mutations (5, 6). Compensatory mutations can become fixed in viral sequences, since returning to the wild type requires multiple simultaneous changes (7). Although studies of HIV polymorphisms in humans have been illuminating and have led to the successful identification of important CTL epitopes (8 – 10), the infecting virus strain and timing of infection are not always known in humans, and this lack of knowledge complicates analyses. Simian immunodeficiency virus (SIV) infection of pig-tailed macaques (Macaca nemestrina) is a very useful model for the study of HIV pathogenesis. Recent studies suggest that SIV-infected pig-tailed macaques have a disease progression that is similar to if not more predictable than that in the more commonly used Indian rhesus macaque (Macaca mulatta) (11, 12).
The influence of major histocompatibility complex (MHC) class I (MHC-I) alleles on infection progression is not well characterized in the SIV-infected pig-tailed macaque model, and few CTL epitopes have been described. The most extensively studied CTL responses in pig-tailed macaques and their escape mutations are KP9 in Gag (13, 14) and KSA10 and KVA10 in Tat (15), all of which are restricted by the Mane-A1*084 allele. A compensatory mutation of the canonical monomorphic KP9 mutation K165R has also been recently identified (5). Immune escape at KP9 occurs early. However, the K165R escape mutation extracts a high fitness cost, and viruses which have escaped at the KP9 epitope rapidly revert to the wild type upon passage to naive Mane-A1*084-negative animals (16 – 18). Escape at KSA10 occurs more slowly than escape at KP9, while the escape kinetics of KVA10 are similar to those of KP9 (15). Escape mutations at the KVA10 and KSA10 Tat epitopes occur at multiple amino acid positions within the epitope, in contrast to the escape mutations at the KP9 Gag epitope. On the basis of the findings for 8 infected animals, it was reported that Mane-A1*084-positive animals maintain a lower SIV load, although this has not been rigorously tested with larger numbers of animals (13). Additional CTL epitopes in pig-tailed macaques have been described in the Env protein, but the restricting MHC-I alleles have not been identified, and the mutations did not appear to affect the replicative capacity of the virus adversely (19, 20).
Broadly reactive CTL responses against multiple SIV epitopes likely contribute toward the efficient control of viremia in macaques. Broader CTL responses, particularly to Gag, are associated with improved control of HIV-1 (21). Conversely, a vaccine directing CTL responses toward a smaller number of critical epitopes restricted by Mamu-B*008 in Indian rhesus macaques was able to control SIV infection efficiently (22).
There is a need to identify a broader range of SIV-specific CTL epitopes, their MHC restriction, and their role in controlling SIV infection in the pig-tailed macaque model. Incisive human studies have linked HIV-1 mutations with particular human leukocyte antigen (HLA) class I alleles, revealing important clues to CTL immunity, epitope discovery, and the control and evolution of HIV (8, 9, 23 – 25). Several studies have also linked hepatitis C virus (HCV) immune system-driven adaptations to HLA class I alleles, and elegant studies have estimated the false discovery rates of such linkage analyses (10, 26 – 28). Linking CTL escape mutations with pig-tailed macaque MHC-I haplotypes both will improve this animal model and could in turn facilitate the design of efficacious CTL vaccines that limit CTL escape. We conducted a large analysis of MHC-I typing and SIV deep sequencing in pig-tailed macaques. We amplified whole virus from plasma samples from 44 pig-tailed macaques challenged with SIVmac251 at approximately 15 weeks postinfection using Illumina deep sequencing. The same 44 macaques were MHC-I genotyped using Roche/454 pyrosequencing. The MHC typing and SIV sequencing data sets were analyzed using in-house bioinformatics pipelines to associate CTL escape mutations with specific MHC-I haplotypes. We identified a series of SIV polymorphisms linked to particular MHC-I haplotypes and validated that a subset of these polymorphisms corresponds to CTL escape mutations.
MATERIALS AND METHODS
Animals and SIV infection.
Forty-four SIVmac251-infected pig-tailed macaques from three separate trials were studied in total (Table 1). Eleven macaques received a Kunjin virus vector SIV or HIV vaccine that was ineffective in controlling SIV infection (29). Two macaques received an influenza virus that expressed KP9 as a vaccine that was also ineffective (30). Thirty-one macaques were infected with SIVmac251 and then received antiretroviral therapy (ART) (tenofovir and emtricitabine from weeks 3 to 10 after infection). Of these 31 macaques, 10 macaques remained unvaccinated, 10 macaques received a peptide-based SIV vaccine based on overlapping SIV Gag peptides, and another 11 macaques were given a peptide-based SIV vaccine based on overlapping peptides spanning all 9 SIV proteins (31). The vaccinations were administered from weeks 4 to 10 after SIVmac251 infection. All experiments were conducted in compliance with the ethics approval obtained from the University of Melbourne and the Commonwealth Scientific and Industrial Research Organization (CSIRO) Livestock Industries Animal Ethics Committees. Animals were sedated with 10 mg/kg of body weight of ketamine prior to withdrawing blood from the femoral vein into EDTA- and sodium heparin-coated Vacuette tubes. Macaques were euthanized before developing simian AIDS, as determined by their peripheral CD4+ T cell counts and plasma viral loads. Macaques were infected intrarectally or intravenously with the same stock of SIVmac251 (kindly provided by R. Pal and N. Miller through the NIH AIDS Research and Reference Reagent Repository), as shown in Table 1.
TABLE 1.
Details of SIVmac251-infected pig-tailed macaques used in this studya
| Study | No. of macaques | Vaccine | ART | Challengeb | Outcome of vaccine | Reference |
|---|---|---|---|---|---|---|
| 1 | 11 | Kunjin SIV | No | SIVmac251, intravenously | No effect | 29 |
| 2 | 31 | Peptide immunotherapy (n = 21) or control (n = 10) | Yesc | SIVmac251, intravenously | Immunotherapy effective | 31 |
| 3 | 2 | Influenza virus-SIV | No | SIVmac251, intrarectally | No effect | 30 |
A total of 44 macaques were studied.
The same stock of SIVmac251, supplied by R. Pal and N. Miller, was used for all infections.
The animals received ART from weeks 3 to 10 after infection.
Amplification of the whole SIV genome.
The whole viral RNA genome in plasma samples from SIVmac251-infected macaques and of the SIVmac251 challenge stock was amplified by reverse transcriptase PCR (RT-PCR) using specifically designed primers in four overlapping fragments for Illumina deep sequencing. Briefly, 140 μl of frozen plasma from whole blood collected in EDTA-coated Vacuette tubes was used to extract viral RNA using a QIAamp viral RNA minikit (Qiagen, USA). Viral RNA was reverse transcribed to DNA and amplified using a SuperScript III one-step RT-PCR kit with Platinum Taq high-fidelity DNA polymerase (Life Technologies, USA) with the following primer pairs (Sigma-Aldrich, Australia): 5′-AGATAGAGTGGGAGATGGG and 5′-CTCTAATTAACCTACAGAGATGTTTGG, 5′-AAAATTGAAGCAGTGGCCATTAT and 5′-CCTCTACTGTTCTGTTTAGCC, 5′-ATAGGGGATATGACTCCAGCAGAAAGATTA and 5′-AGTCCTATTATCCCTACCATGCCAGTAAAT, and 5′-CTTGCTTTTAAGTGTCTACGGGATCTATTG and 5′-GAATACAGAGCGAAATGCAGTG.
RT-PCR was performed in an Eppendorf Realplex Mastercycler (Eppendorf, Germany) under the following conditions: 50°C for 60 min; 94°C for 2 min; 2 cycles of 94°C for 15 s, 60°C for 1 min, and 68°C for 4 min; 2 cycles of 94°C for 15 s, 58°C for 1 min, and 68°C for 4 min; 41 cycles of 94°C for 15 s, 55°C for 1 min, and 68°C for 4 min; and 68°C for 10 min. PCR products were run on a 0.7% agarose gel (Promega, USA) alongside a 1-kb DNA ladder (New England BioLabs, USA), bands of the correct size were excised, and DNA was extracted using a QIAquick gel extraction kit (Qiagen, USA).
Illumina SIV sequencing.
Amplified plasma SIV DNA concentrations were measured using a Qubit double-stranded DNA high-sensitivity (HS) assay kit and a Qubit (version 2.0) fluorometer (Life Technologies, USA). Pools containing the complete SIVmac251 genome from each animal were created by combining 4 × 108 copies from each of the four PCR products. A NexteraXT DNA sample preparation kit (Illumina, USA) was used to prepare libraries for indexed paired-end sequencing, and these were sequenced with a MiSeq instrument (Illumina, USA). SIV deep sequencing data are available upon request.
MHC-I typing.
MHC-I genotyping was performed as previously described (32, 33). Briefly, cellular RNA was extracted from frozen macaque peripheral blood mononuclear cells (PBMCs) using an RNeasy minikit (Qiagen, USA). cDNA was synthesized using a SuperScript III first-strand synthesis system (Life Technologies, USA). Primary amplicons consisting of 568-bp fragments, including most of exon 2, all of exon 3, and part of exon 4, were generated by PCR using primers that bind highly conserved regions and universally amplify macaque MHC-I sequences. In addition, each primer contained a Roche/454 GS FLX titanium adaptor A or B and a distinct 10-bp multiplex identifier for each animal to facilitate pooling of amplicons for emulsion PCR and bidirectional pyrosequencing with a Roche/454 GS Junior instrument (Roche/454, USA). The resulting sequencing data were binned by multiplex identifier, and sequence reads from each animal were assembled into unidirectional contigs having 100% identity using a custom genotyping analysis work flow (LabKey, USA). The number of sequence reads comprising each unidirectional contig was enumerated, and the resulting consensus sequences were mapped against an in-house database containing known pig-tailed MHC-I allele sequences using the BLASTn program (33 – 35). Putative Mane-A and Mane-B haplotypes were inferred by identifying combinations of shared alleles between animals as described previously for rhesus macaques (35). MHC-I typing data for 5 out of the 44 macaques have been previously reported (33). The MHC-I typing data are available on request.
Identifying SNPs associated with Mane alleles.
The frequency of mutations in plasma virus was identified by comparison of the plasma virus sequence with a reference SIVmac251 sequence (GenBank accession number M19499). Sequence reads from Illumina sequencing were assembled using Geneious software (version 7.0.2; Biomatters, Auckland, New Zealand). We assembled the SIVmac251 sequence reads obtained from all 44 macaques against the stock reference sequence to call single nucleotide polymorphisms (SNPs). Using an in-house pattern-mining approach, we identified, in all animals, SIV sequence positions with nonsynonymous mutations present at a frequency of >5% compared to the reference sequence (Fig. 1). In order to identify if a mutation was associated with a particular Mane haplotype, we then compared the frequency of the mutation in animals with a particular Mane haplotype with the frequency in those without that haplotype, using Fisher's exact test (Fig. 1). Fisher's exact test then provided a P value for the significance of the difference in the frequency of mutations in Mane haplotype-positive animals versus Mane haplotype-negative animals. Spreadsheets evaluating the linkage between SIV polymorphisms and MHC-I alleles are available upon request.
FIG 1.
Flowchart of study used for identification of linkages between MHC-I haplotypes and SIV mutations in 44 SIVmac251-infected pig-tailed macaques.
A sliding window to identify mutated regions.
The method described above identifies only mutations where the same nonsynonymous mutation is present in multiple animals. For some escape mutations, animals may harbor different mutations within the same epitope. To identify such variable escape patterns, we also analyzed the frequency of mutations within a sliding window of 30 bp (in 3-bp increments), using in-house codes in Matlab and VB.NET (Fig. 1). This sliding window analysis is similar to previous work performed in linking mutations in HCV infection with HLA class I alleles in humans (10). We identified a sequence to be mutated (within a given window) if at least 1 nucleotide had a nonsynonymous mutation present at a >5% frequency. We then used Fisher's exact test to identify associations between mutated windows and Mane haplotypes and obtain a P value for each association. Spreadsheets evaluating the linkage between SIV polymorphisms across a 10-amino-acid window and MHC-I alleles are available upon request.
Permutation approach to evaluate the significance of associations.
The large number of sequence positions and Mane haplotypes means that there is a high risk of discovery of false associations, as is noted with similar linkage analyses (23, 28). In order to estimate the probability that any identified association was real, we sought to identify the probability of false associations being identified in our data. To do this, we utilized a permutation approach. That is, each animal has a set of Mane haplotypes associated with a set of viral sequences in the same animal. The question that we wish to ask is, how many random associations would we expect to see, given the Mane haplotype structure (i.e., the number of animals with the different Mane alleles) and the virus structure (number of sites mutated)? In order to keep the data structure of the Mane haplotypes and viral mutations, we need to permute all the Mane haplotypes of a given animal en bloc, as well as all the viral sequences from a given animal en bloc. Therefore, we permuted the order of the animals (and all of the associated Mane haplotypes), leaving the order of viral sequences intact. For each permutation, we had a new order of Mane haplotype sets randomly matched to viral sequence sets. We then again performed Fisher's exact test for the permuted population to identify the P values that we would obtain studying the associations between Mane alleles and viral mutations from this random matching of the Mane haplotype and virus sequences (Fig. 1). By performing this permutation 1,000 times, we observed the frequency with which associations of different strength (P value) would be identified in our data by chance. This was performed for both the SNP and sliding window analyses to obtain the expected frequency of associations of different strengths for each. By comparing, for a given P value, the observed number of associations in our data with the observed frequency in the permuted data sets, we identified the proportion of associations that we observed that were likely to have arisen by chance. By comparing, for a given threshold P value, the observed number of associations in our data with the observed frequency in the permuted data sets, we identified the proportion of associations that we observed that were likely to have arisen by chance (28). We note that in previous studies of MHC-viral mutation associations in HIV (8, 9, 36) and HCV (10, 26, 27), it was also necessary to account for phylogenetic relationships in the data (23). However, in our study, animals were infected with the same stock of virus. In addition, mutations were identified not by comparison to the sequences of other macaques but by comparison to this reference sequence. Therefore, we did not investigate the effects of phylogenetic linkage in our analysis.
ICS assays.
To validate that our approach can correctly identify CTL epitopes, we infected 3 additional pig-tailed macaques, one of which expressed the Mane-B028 haplotype, with SIVmac239, which was associated with a polymorphism in SIV Nef. We confirmed that infection had occurred (peak viral load, 108 to 109 copies/ml 2 weeks after infection) and obtained fresh blood 2 to 15 weeks after infection to analyze the responses to the 3 15-mer peptides (NIH AIDS Research and Reference Reagent Repository, USA) spanning the SIV Nef polymorphism by intracellular cytokine staining (ICS), as previously described (37). Briefly, 190 μl of whole blood was stimulated with specific 15-mer peptides at 1 μg/ml in the presence of anti-CD28 and anti-CD49d (BD, USA) and 10 μg/ml brefeldin A (Sigma, USA). Negative and positive controls were included in the assay with an equivalent concentration of dimethyl sulfoxide (DMSO; Sigma, USA) and 1 μg/ml staphylococcal enterotoxin B (Sigma, USA). Cells were incubated for 5.5 h at 37°C in 5% CO2. The cell surface was stained with CD3-Pacific Blue clone SP34-2 (BD, USA) and CD8-peridinin chlorophyll protein (PerCP) clone SK1 (BD, USA) for 30 min. Red blood cells were lysed using 1× BD fluorescence-activated cell sorting lysing solution, and cells were permeabilized using 1× BD permeabilizing solution 2 (BD, USA). Cells were incubated with anti-gamma interferon (IFN-γ)-allophycocyanin (APC) (BD, USA) for 45 min. A minimal epitope was mapped by obtaining individual 9- to 11-amino-acid peptides (Purar Chemicals, China) spanning the region of the overlapping 15-mer peptides and performing ICS as described above using dilutions of peptides from a 1-μg/ml stock. Once the minimal epitope was identified, whole blood was stimulated separately with dilutions of peptides from a 1-μg/ml stock of the wild-type and escape mutation 9-mer peptides (Purar Chemicals, China) as described above. Cells were washed and fixed with 1% paraformaldehyde (Sigma). Cells were acquired using a BD LSR II flow cytometer, and at least 2 × 105 lymphocytes were collected. Flow cytometry data were analyzed using FlowJo software (version 9.1; TreeStar, USA).
Tetramer staining for macaques positive for the Mane-B017 haplotype.
To identify that CD8 T cells specific for particular Mamu-B017 epitopes could be recovered from Mane-B017 macaques and identified using Mamu-B*017:01 tetramers, frozen PBMCs were thawed and washed in RPMI (Life Technologies, USA) supplemented with 10% fetal calf serum (DKSH, Australia) and expanded as previously described (38). Briefly, PBMCs were split into stimulators and responders at a 1:2 ratio. Stimulators were incubated with 15-mer peptides that span the desired CTL epitope region (NIH AIDS Research and Reference Reagent Repository, USA) at 1 μg/ml for 90 min in 5% CO2 at 37°C. Stimulators were washed two times and added to the responders. Cells were grown over 10 days at 37°C in 5% CO2. On day 3 and day 5, fresh medium containing interleukin-2 (IL-2) and interleukin-7 (IL-7) at 50 U/ml and 10 ng/ml, respectively (PeproTech, USA), was added. The same procedure was followed for control samples, whereby stimulators were pulsed with peptides restricted by a different Mane haplotype identified in our MHC-I and SNP correlation analysis. PBMCs were stained with the Live/Dead Fixable Aqua dead cell stain (Invitrogen, USA) for 30 min at room temperature and washed. Cells were stained with Env FW9-phycoerythrin and Nef IW9-APC using a titration-optimized volume of a Mamu-B*017:01 tetramer folded around the FW9 or IW9 epitope (kindly provided by N. A. Wilson Schlei, University of Miami [39]) for 60 min at room temperature. The cell surface was stained with CD3-Alexa Fluor 700 clone SP34-2, CD8-PerCP clone SK1, and CD20-fluorescein isothiocyanate (BD, USA) for 30 min. Cells were washed and fixed with 1% paraformaldehyde (Sigma, USA). Cells were acquired using a BD Canto II flow cytometer, and at least 2 × 105 lymphocytes were collected. Flow cytometry data were analyzed using FlowJo software (version 9.1; TreeStar, USA).
RESULTS
MHC-I typing of 44 pig-tailed macaques by pyrosequencing.
Multiple MHC-I alleles and/or haplotypes have been characterized in a limited number of pig-tailed macaques (33, 34, 40, 41). Macaques can express more than 20 MHC-I transcripts from a variable number of Mane-A and Mane-B loci that have undergone gene duplication, in contrast to their human counterparts (34, 40). Each macaque carries a pair of Mane-A and Mane-B haplotypes inherited from the parents. Abbreviated haplotype designations for Mane-A were based on the presence of a transcriptionally abundant Mane-A1 allele, while Mane-B haplotypes were assigned on the basis of a diagnostic major Mane-B transcript that is characteristic for a group of linked Mane-B alleles, as previously described for rhesus macaques (34, 42). We identified a diverse range of Mane-A and Mane-B haplotypes in our cohort of 44 SIVmac251-infected pig-tailed macaques, as shown in Table 2. The most common Mane-A and Mane-B haplotypes in this cohort were Mane-A084, Mane-A082, and Mane-B118, which represented 23%, 15%, and 15% of the MHC-I chromosomal regions, respectively.
TABLE 2.
MHC-I genotyping of the 44 pigtail macaques infected with SIVmac251
| Mane-A or -B haplotype | No. of haplotypes observed | Haplotype frequency (%) | Major alleles |
Minor alleles |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diagnostic | 2 | 3 | 4 | 5 | 1 | 2 | 3 | |||
| A084 | 20 | 22.7 | A1*084 | A3*13 | ||||||
| A082 | 13 | 14.8 | A1*082 | A3*13 | ||||||
| A052 | 6 | 6.8 | A1*052 | A3*13 | A5*30 | |||||
| A019 | 5 | 5.7 | A1*019 | A2*05 | A4*14 | |||||
| A010 | 4 | 4.5 | A1*010 | A3*13 | A4*14 | |||||
| A006 | 4 | 4.5 | A1*006 | A2*05 | ||||||
| A009 | 3 | 3.4 | A1*009 | A2*05 | ||||||
| A031 | 3 | 3.4 | A1*031 | A4*14 | ||||||
| A072 | 2 | 2.3 | A1*072 | A3*13 | ||||||
| A016 | 2 | 2.3 | A1*016 | A4*14 | ||||||
| A032 | 2 | 2.3 | A1*032 | A4*14 | ||||||
| A047 | 2 | 2.3 | A1*047 | A2*05 | A4*01 | |||||
| A053 | 2 | 2.3 | A1*053 | A2*05 | A4*14 | |||||
| A066 | 2 | 2.3 | A1*066 | A3*13 | ||||||
| A083 | 2 | 2.3 | A1*083 | A2*05 | A3*13 | |||||
| A114 | 2 | 2.3 | A1*114 | A2*05 | A3*13 | |||||
| A003 | 2 | 2.3 | A1*003 | A2*05 | ||||||
| A004 | 1 | 1.1 | A1*004 | A4*14 | ||||||
| A007 | 1 | 1.1 | A1*007 | A2*05 | ||||||
| A085 | 1 | 1.1 | A1*085 | A2*05 | A4*14 | |||||
| A018 | 1 | 1.1 | A1*018 | A2*05 | A4*14 | |||||
| A unknown | 8 | 9.1 | A1 novel | |||||||
| B118 | 13 | 14.8 | B*118 | B*122 | B*027 | B*030 | B*082 | |||
| B043 | 8 | 9.1 | B*043 | B*030 | B*045 | B*072 | ||||
| B015 | 8 | 9.1 | B*015 | B*068 | B*064 | B*088 | ||||
| B028 | 7 | 8.0 | B*028 | B*061 | B*124 | B*068 | B*021 | B*088 | B*079 | B*045 |
| B016 | 7 | 8.0 | B*016 | B*041 | B*089 | B*088 | ||||
| B120 | 6 | 6.8 | B*120 | B*107 | B*082 | B*078 | ||||
| B047 | 6 | 6.8 | B*047 | B*068 | B*112 | |||||
| B039 | 5 | 5.7 | B*039 | B*108 | B*088 | |||||
| B069 | 4 | 4.5 | B*069 | B*081 | B*107 | B*082 | B*079 | B*051 | ||
| B119 | 3 | 3.4 | B*119 | B*068 | ||||||
| B150 | 3 | 3.4 | B*150 | B*068 | B*057 | B*089 | B*072 | |||
| B104 | 3 | 3.4 | B*104 | B*144 | B*057 | B*046 | ||||
| B004 | 2 | 2.3 | B*004 | B*004 | ||||||
| B110 | 2 | 2.3 | B*110 | B*123 | B*082 | B*068 | B*072 | |||
| B017 | 2 | 2.3 | B*017 | B*060 | ||||||
| B056 | 2 | 2.3 | B*056 | B*017 | B*041 | B*082 | B*079 | |||
| B052 | 2 | 2.3 | B*052 | B*058 | ||||||
| B008 | 1 | 1.1 | B*008 | B*125 | B*082 | B*079 | B*072 | |||
| B019 | 1 | 1.1 | B*019 | B*014 | B*057 | B*116 | ||||
| B024 | 1 | 1.1 | B*024 | B*068 | B*089 | B*082 | ||||
| B099 | 1 | 1.1 | B*099 | B*058 | B*072 | B*054 | B*063 | |||
| B101 | 1 | 1.1 | B*101 | B*068 | B*060 | B*089 | ||||
Association between MHC-I and VL.
We first assessed the influence of Mane haplotypes with levels of chronic SIV viremia across our 44-macaque cohort. We calculated an average SIV plasma RNA level (viral load) for each animal during steady-state viremia from week 6 onwards. Of the 44 animals studied, 21 animals received ART and vaccination, 10 animals received ART only, and 13 animals received neither ART nor vaccination. To account for these variables, a two-way analysis of variance (ANOVA) was employed to study the link between viral load and ART, vaccination, and MHC-I haplotype (Table 3). In an analysis not corrected for the number of associations studied, of the individual MHC-I haplotypes, only Mane-A082 and Mane-B118 showed an association with modestly lower overall viral loads and Mane-B120 showed an association with modestly higher viral loads. However, the results after correction for multiple correlations showed no significant link between specific MHC-I haplotypes and a decrease or increase in viral load. Importantly, we did not find a significant association between Mane-A084 and lower viral loads in this study of 44 macaques (16 of which were Mane-A084 positive [Mane-A084+]), in contrast to the findings of a previous much smaller study (13). We also analyzed whether the Mane-A084+ macaques that mutate at the KP9 Gag CTL epitope have lower viral loads than those that remain wild type but did not detect significant differences. The 11 Mane-A084+ macaques that generated the K165R escape mutant had an overall mean VL of 4.535 log10 copies of RNA/ml plasma, whereas the 5 Mane-A084+ macaques that did not generate this mutant had a mean VL of 4.690 log10 copies of RNA/ml plasma (P = 0.740, Mann-Whitney test). However, we acknowledge that compensatory mutations may confound this analysis, and the use of larger numbers of animals would be helpful to study this more fully.
TABLE 3.
Influence of MHC-I genotype on SIV load
| Haplotype | No. of macaques |
VLa |
Uncorrected P valueb | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype positive | Haplotype negative | No ART, not vaccinated |
Early ART, not vaccinated |
Early ART, vaccinated |
|||||
| Mane positive | Mane negative | Mane positive | Mane negative | Mane positive | Mane negative | ||||
| Mane-A006 | 3 | 41 | 5.96 | 5.74 | NA | 4.84 | 3.31 | 4.43 | 0.56 |
| Mane-A082 | 13 | 31 | 5.96 | 5.62 | 5.48 | 4.57 | 4.7 | 4.3 | 0.04 |
| Mane-A084 | 16 | 28 | 5.61 | 5.83 | 4.55 | 5.04 | 4.25 | 4.48 | 0.22 |
| Mane-A114 | 2 | 42 | NA | 5.78 | 5.42 | 4.78 | 3.31 | 4.43 | 0.62 |
| Mane-B015 | 8 | 36 | 5.54 | 5.88 | 4.9 | 4.83 | 5.13 | 4.3 | 0.72 |
| Mane-B016 | 7 | 37 | 5.98 | 5.69 | 4.75 | 4.85 | 5.14 | 4.3 | 0.3 |
| Mane-B017 | 2 | 42 | 5.38 | 5.81 | NA | 4.84 | 4.7 | 4.36 | 0.94 |
| Mane-B028 | 7 | 37 | 5.39 | 5.85 | 4.28 | 4.98 | 4.18 | 4.41 | 0.12 |
| Mane-B043 | 7 | 37 | 5.67 | 5.82 | NA | 4.84 | 4.33 | 4.38 | 0.75 |
| Mane-B047 | 6 | 38 | 6.07 | 5.75 | 4.9 | 4.84 | 3.8 | 4.51 | 0.24 |
| Mane-B069 | 4 | 40 | 5.61 | 5.79 | 4.32 | 4.97 | 4.34 | 4.38 | 0.36 |
| Mane-B118 | 13 | 31 | 6.06 | 5.75 | 5.63 | 4.65 | 4.65 | 4.13 | 0.02 |
| Mane-B120 | 6 | 38 | 6.13 | 5.75 | 4.31 | 5.07 | 3.26 | 4.49 | 0.03 |
The VL (log10 number of copies of RNA/ml plasma) is the mean of all available VL measurements after week 6. NA, not applicable.
After correction for multiple comparisons, no MHC-I alleles were associated with a significant change in VL.
Validation of the method employed to associate pig-tailed macaque MHC-I haplotypes with SNPs in the SIV genome.
A previous study conducted in Mauritian cynomolgus macaques, together with a retrospective analysis of studies with Indian-origin rhesus macaques, demonstrated that transcriptionally abundant MHC-I alleles play a crucial role in presenting CTL epitopes in SIV infection (43). We developed an in-house bioinformatics pipeline to study associations between the four MHC-I haplotypes for each macaque across the nearly 10,000-bp-long SIV genome, as described in Materials and Methods and shown in Fig. 1. We first validated our approach by analyzing the data for previously characterized KP9, KSA10, and KVA10 CTL epitopes restricted by Mane-A1*084 (14, 15). We compared SIV sequences from animals with and without the Mane-A084 haplotype. The strongest association was with the previously identified KP9 epitope in SIV Gag (Table 4). In position 165 of Gag, we observed this mutation in 11 of 16 Mane-A084-positive animals and 0 of 28 Mane-A084 negative animals (P = 5.7 × 10−7). The second most significant set of mutations was also observed in a previously identified epitope, the KVA10 epitope in Tat, in which 9 and 7 of 16 Mane-A084-positive animals had a K-to-E mutation at position 1 and position 2, respectively, whereas 0 of 28 Mane A084-negative animals had these mutations (P = 1.6 × 10−5 and 3 × 10−4, respectively). We also identified 2 mutations within the KSA10 epitope of Tat (in positions 3 and 9 of the epitope) as being significantly linked with Mane-A084 (P = 1.1 × 10−3 and 4 × 10−3, respectively). Thus, our method identified the three known Mane-A1*084-restricted SIV CTL epitopes and their previously described escape mutations, confirming the ability of this approach to identify MHC-I-restricted CTL escape mutations.
TABLE 4.
Validation of the study approach to link pig-tailed macaque MHC-I haplotypes with CTL escape mutations by using previously defined CTL responses restricted by the Mane-A1*084 allele
| Protein | Epitope | Region of epitope within protein | No. of Mane-positive mutation-positive animals/total no. of Mane-positive animals | No. of Mane-negative mutation-positive animals/total no. of Mane-negative animals | P value (two-tailed) | Amino acid change | Escape motifa |
|---|---|---|---|---|---|---|---|
| Gag | KP9 | 164–172 | 11/16 | 0/28 | 5.7E−07 | K → R | KRFGAEVVP |
| Tat | KSA10 | 87–96 | 6/16 | 0/28 | 1.1E−03 | A → T | KKAKTNTSSA |
| KSA10 | 87–96 | 6/16 | 0/28 | 1.1E−03 | S → T or S → P | KKAKANTSTA or KKAKANTSPA | |
| KSA10 | 87–96 | 5/16 | 0/28 | 4.0E−03 | A → T | KKTKANTSSA | |
| Tat | KVA10 | 114–123 | 9/16 | 0/28 | 1.6E−05 | K → E | EKETVEKAVA |
| KVA10 | 114–123 | 7/16 | 0/28 | 3.0E−04 | K → E | KEETVEKAVA |
The mutated amino acids are in boldface and underlined.
Identification of novel MHC-I-linked mutations in SIV.
Using the approach described above, we searched for associations for the more common Mane haplotypes present in our cohort. The analysis of associations between four Mane-A and -B haplotypes for each macaque across the nearly 10,000 bp of the SIV genome results in a very large number of associations, many of which are random. Thus, the P values from our Fisher's exact test are not a precise indication of the probability that a mutation is genuine or random. To assess how often we might expect to see an association of a particular strength simply by chance, we can randomly sort the animal identities (and their Mane-A and -B haplotypes) with the viral mutation patterns. That is, we can permute the animal identities, leaving the order of viral sequences intact, so that, for example, the Mane haplotypes of animal 1 are a match with the viral sequences of animal 18, the Mane haplotypes of animal 2 are a match with the viral sequences of animal 6, and so on. Each time that we permute the animal identities, we can then perform the same analysis of associations between Mane haplotypes and mutations and observe the P values that arise from these random associations. By performing the permutation 1,000 times, we obtain the expected distribution of P values arising from chance associations (Fig. 1). Thus, for a given P value for the association, we observed a certain proportion of associations in the actual data set and another for the permuted data set. We were then able to identify threshold values below which we had a given level of confidence in the associations (Fig. 1). For example, we first identified a P value below which we estimated that 95% of observed associations were genuine. That is, at a P value cutoff of 1.0 × 10−3, there were 8 associations with lower P values in our data versus an average of 0.3 association in the permuted data (3 associations with P values of <1 × 10−3 out of 1,000 permutations). Similarly, we identified 2 thresholds of P values where either 84% of associations or 66% of associations were likely to be genuine (Fig. 1). The associations between SIV SNPs and MHC-I alleles are shown in Table 5.
TABLE 5.
Novel SIVmac251 SNPs linked to MHC-I haplotypes in pig-tailed macaques
| Haplotype | Protein | Nucleotide position | No. of Mane-positive mutation-positive animals/total no of Mane-positive animals | No. of Mane-negative mutation-positive animals/Total no. of Mane-animals | P valuea (two tailed) | Amino acid change(s) | Wild-type motifb |
|---|---|---|---|---|---|---|---|
| Mane-A*010 | Gag | 2179 | 2/4 | 0/40 | 0.0064 | P → S | EALKEALAPVPIPFAAAQQRG |
| Pol | 4405 | 2/4 | 0/40 | 0.0064 | N → S | CPTESESRLVNQIIEEMIKKS | |
| Pol | 4408 | 2/4 | 0/40 | 0.0064 | Q → R | PTESESRLVNQIIEEMIKKSE | |
| Env | 7483 | 2/4 | 0/40 | 0.0064 | D → E | NRTYIYWHGRDNRTIISLNKY | |
| Env | 7493 | 2/4 | 0/40 | 0.0064 | I → V | IYWHGRDNRTIISLNKYYNLT | |
| Env | 8019 | 2/4 | 0/40 | 0.0064 | T → I | VTSLIANIDWTDGNQTNITMS | |
| Mane-A*032 | Gag | 2179 | 2/2 | 0/42 | 0.0011 | P → S | EALKEALAPVPIPFAAAQQRG |
| Pol | 4405 | 2/2 | 0/42 | 0.0011 | N → S | CPTESESRLVNQIIEEMIKKS | |
| Pol | 4408 | 2/2 | 0/42 | 0.0011 | Q → R | PTESESRLVNQIIEEMIKKSE | |
| Env | 7483 | 2/2 | 0/42 | 0.0011 | D → E | NRTYIYWHGRDNRTIISLNKY | |
| Env | 7493 | 2/2 | 0/42 | 0.0011 | I → V | IYWHGRDNRTIISLNKYYNLT | |
| Vif | 5863 | 2/2 | 0/42 | 0.0064 | N → D | NPTWKQWRRDNRRGLRMAKQN | |
| Mane-A*053 | Nef | 9676 | 2/2 | 0/42 | 0.0011 | S → Y, S → F | RHYLMQPAQTSKWDDPWGEVL |
| Mane-A*082 | Env | 7001 | 5/13 | 0/31 | 0.0012 | T → A, T → S, T → P | LTKSSTTTAPTTTTAASKIDM |
| Env | 7033 | 8/13 | 5/31 | 0.0087 | M → I | TTTTAASKIDMVNETSSCITH | |
| Mane-A*114 | Gag | 2477 | 2/2 | 0/42 | 0.0011 | Q → R | PAVDLLKNYMQLGKQQREKQR |
| Mane-B004 | Vif | 5935 | 2/2 | 1/41 | 0.0032 | K → E | DKQRGGKPPTKGANFPGLAKV |
| Env | 6996 | 2/2 | 1/41 | 0.0032 | A → V | WGLTKSSTTTAPTTTTAASKI | |
| Env | 7017 | 2/2 | 2/42 | 0.0064 | A → G A → V | TTTAPTTTTAASKIDMVNETS | |
| Mane-B015 | Env | 9236 | 4/8 | 1/36 | 0.0024 | L → V | RIRQGLELTLL |
| Env | 7280 | 3/8 | 0/36 | 0.0042 | T → A | QESCDKHYWDTIRFRYCAPPG | |
| Mane-B016 | Gag | 1360 | 3/6 | 0/38 | 0.0015 | I → V | KVKHTEEAKQIVQRHLVVETG |
| Env | 6789 | 3/6 | 0/38 | 0.0015 | G → D | WGTTQCLPDNGDYSELALNVT | |
| Mane-B017 | Nef | 9568 | 2/2 | 0/42 | 0.0011 | I → T | DWQDYTSGPGIRYPKTFGWLW |
| Mane-B028 | Env | 7049 | 3/7 | 0/37 | 0.0027 | S → P | SKIDMVNETSSCITHDNCTGL |
| Env | 9105 | 4/7 | 3/37 | 0.0075 | Q → R | YGWSYFQEAVQAGWRSATETL | |
| Nef | 9105 | 4/7 | 3/37 | 0.0075 | K → E | MGGAISRRRSKPAGDLRQRLL | |
| Mane-B039 | Env | 7027 | 3/5 | 2/39 | 0.0070 | I → M | APTTTTAASKIDMVNETSSCI |
| Mane-B043 | Vif | 5917 | 3/8 | 0/36 | 0.0043 | G → S | KQNSRGDKQRGGKPPTKGANF |
| Nef | 9402 | 5/8 | 5/36 | 0.0091 | T → A | VPVMPRVPLRTMSYKLAIDMS | |
| Mane-B056 | Pol | 4618 | 2/2 | 1/42 | 0.0032 | R → K | KELVFKFGLPRIVARQIVDTC |
| Mane-B069 | Gag | 1880 | 2/4 | 0/40 | 0.0064 | R → K | WIQLGLQKCVRMYNPTNILDV |
| Tat | 8808 | 2/4 | 0/40 | 0.0064 | I → V, I → L | KPISNRTRHCQPE | |
| Env | 8804 | 2/4 | 0/40 | 0.0064 | T → A | FSSPPSYFQQTHIQQDPALPT | |
| Env | 8808 | 2/4 | 0/40 | 0.0064 | H → R, H → P | SSPPSYFQQTHIQQDPALPTR | |
| Mane-B104 | Tat | 6408 | 2/3 | 0/41 | 0.0032 | G → R | DATTPESANLGEEILSQLYRP |
| Env | 8019 | 2/3 | 0/41 | 0.0032 | T → I | VTSLIANIDWTDGNQTNITMS | |
| Mane-B118 | Env | 8981 | 4/13 | 0/31 | 0.0053 | G →A | FSNCRTLLSRAYQILQPILQR |
| Mane-B119 | Pol | 2538 | 2/3 | 0/41 | 0.0032 | D → N | RKQREALQGGDRGFAAPQFSL |
| Env | 9089 | 2/3 | 0/41 | 0.0032 | F → V | LTYLQYGWSYFQEAVQAGWRS | |
| Env | 9093 | 2/3 | 0/41 | 0.0032 | Q → R | TYLQYGWSYFQEAVQAGWRSA | |
| Nef | 9089 | 2/3 | 0/41 | 0.0032 | I → M | MGGAISRRRSKPAGD | |
| Nef | 9093 | 2/3 | 0/41 | 0.0032 | R → G | MGGAISRRRSKPAGDLR | |
| Tat | 6364 | 2/3 | 1/41 | 0.0094 | C → Y | SLESSNERSSCISEADATTPE | |
| Env | 7931 | 2/3 | 1/41 | 0.0094 | V → I | IRQIINTWHKVGKNVYLPPRE | |
| Mane-B150 | Vpr | 6055 | 2/3 | 0/41 | 0.0032 | M → I | YLCLIQKALFMHCKKGCRCLG |
P values are uncorrected (Fisher's exact test).
The mutated amino acids are in boldface.
Using this approach, we identified a total of 46 novel nonsynonymous mutations within SIV that were significantly associated with particular Mane class I A or B haplotypes at a level of at least 66% and likely to be genuine associations using our permutations analyses (P < 9.7 × 10−3; Table 5). An example of a polymorphism detected in Nef that was linked to the Mane-B028 haplotype is shown in Fig. 2. Within the 46 associations identified, 27 had P values sufficiently low (P < 3.1 × 10−3; Table 5) that they were 84% likely to be genuine associations using our permutation analyses. The associations identified were distributed across 18 Mane class I haplotypes and were in 7 SIV proteins (5 in Gag, 6 in Pol, 22 in Env, 6 in Nef, 3 in Vif, 3 in Tat, and 1 in Vpr).
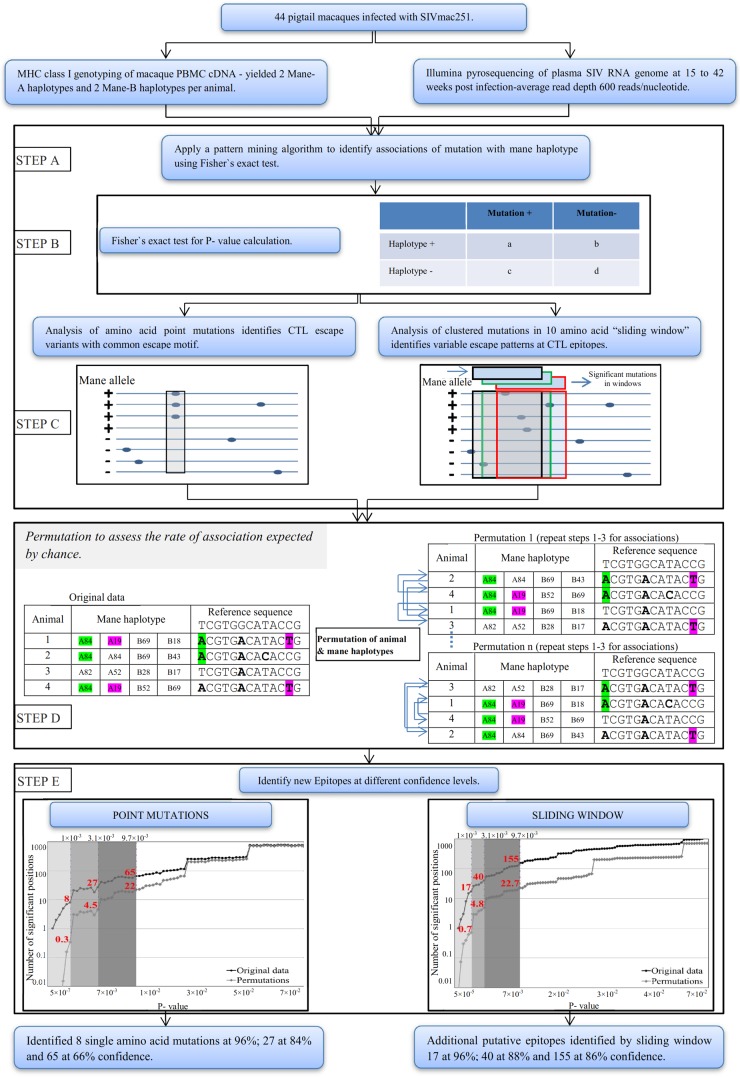
FIG 2.
Example of linkage analysis to identify mutations within the SIV proteome which are more common in macaques with specific MHC-I haplotypes. Shown are SIV sequences across Nef in all 44 macaques. Mutations at the shaded amino acid were observed in 4 of 7 Mane-B028+ macaques and 3 of the Mane-B028-negative macaques (P = 0.0075, Fisher's exact test).
Identification of mutated regions using a sliding window.
The identification of individual mutations that are associated with a particular MHC-I haplotype requires that escape occur in the same way in multiple animals. However, for some epitopes, escape occurs in different ways in different animals. In this case, we would see clusters of mutations within the epitope, but no single mutation may occur at a sufficient frequency to be significantly associated with a particular MHC-I haplotype. Indeed, we noted in the single nucleotide mutation analysis described above that some identified mutations linked to the same MHC-I haplotype were closely spaced, suggesting the likelihood that a single epitope spanned both associations. For example, the multiple Mane-A*010 associations in Pol and Env (Table 5) were <30 nucleotides apart. This was also observed in the validation exercise, where multiple mutations in the KVA10 and KSA10 epitopes were linked to the restricting MHC-I allele Mane-A1*084.
Thus, we developed a sliding window approach to link closely spaced mutations (<10 amino acids apart) with MHC-I haplotypes. This approach is similar to an approach previously used to link HCV mutations in humans with HLA class I alleles (10). This approach identifies all the closely spaced mutations linked to the same haplotype identified in the single nucleotide analyses presented in Table 5 but also identifies new sites where no single mutation is of sufficient frequency to be significantly associated with an MHC-I haplotype. The schematic of this approach is shown in Fig. 1, and a specific example of an association identified using this methodology is shown in Fig. 3. In the example shown in Fig. 3, 5 of 16 Mane-A1*084+ animals but none of 28 Mane-A1*084-negaitve animals have various mutations across a stretch of 5 amino acids in Gag.
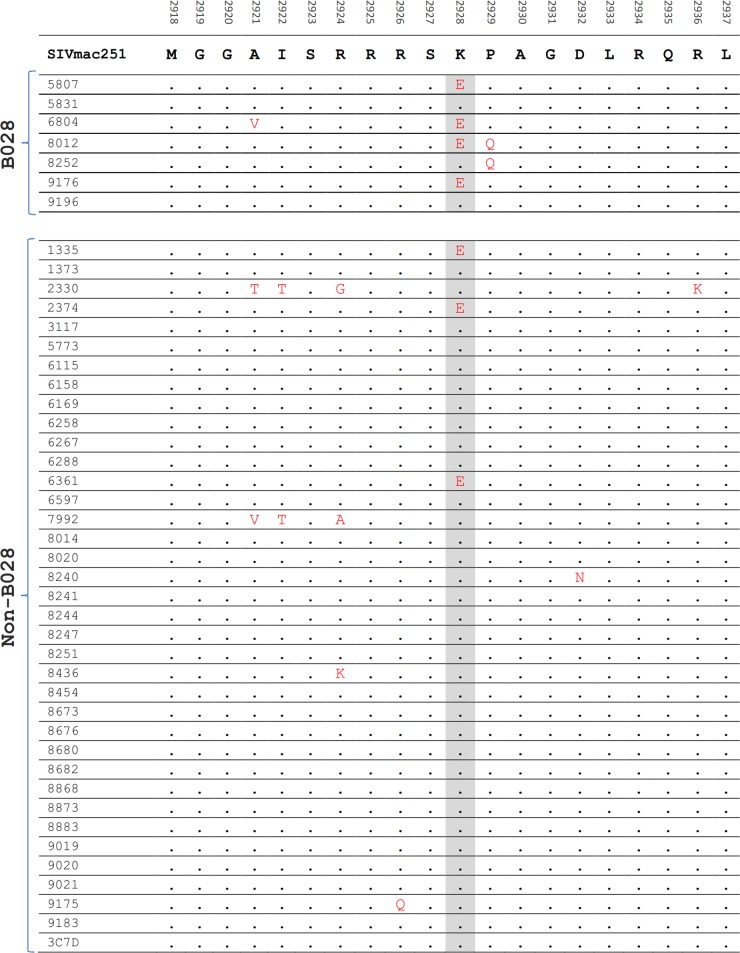
FIG 3.
Example of sliding window analysis to identify 10-amino-acid regions of the SIV proteome which undergo changes at different positions but which are more common in macaques with specific MHC-I haplotypes. Shown are SIV sequences across Gag in all 44 macaques. Five of the 16 Mane-A084+ macaques showed mutations, identified in the shaded 10-amino-acid window, that were not observed in any of the 28 Mane-A084-negative macaques.
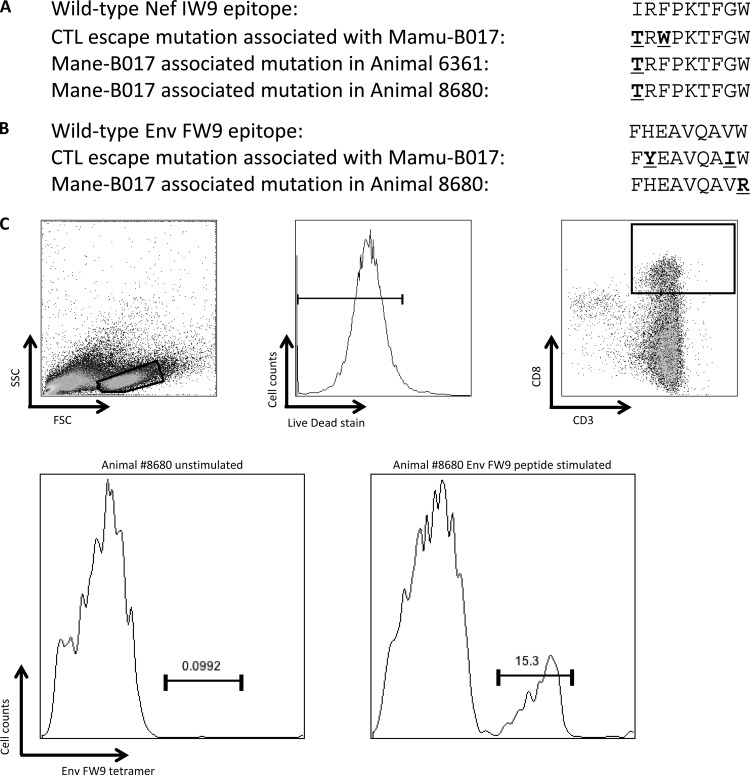
FIG 5.
Recognition of known Mamu-B017-restricted CTL epitopes identified in rhesus macaques by Mane-B017+ pig-tailed macaques. (A and B) The SIV sequences of 2 CTL epitopes previously identified in Mamu-B017+ rhesus macaques (Nef IW9 and Env FW9) were studied in Mane-B017+ pig-tailed macaques and compared to common mutations observed in Mamu-B017+ rhesus macaques. The mutations identified are underlined and in bold. (C) Identification of an Env FW9/Mane-B017 tetramer-specific CD8 T cell population in PBMCs from a Mane-B017+ pig-tailed macaque. PBMCs were unstimulated or stimulated for 10 days with the Env epitope, and gated CD3+ CD8+ lymphocytes were stained with an Env FW9/Mane-B017 tetramer.
This sliding window approach identified 142 windows where there was an association that we estimated had a >86% probability of being real and did not overlap the mutations observed in our previous analysis of individual sequence positions. This corresponds to a P value by Fisher's exact test of <3.1 × 10−3. Many of the windows overlapped each other in the same region (as would be expected around an epitope). Once we identified the unique sites covered, we found 32 sites within the SIV genome where mutations were linked to Mane-A or -B haplotypes that were not discovered when only individual SNPs were analyzed (Table 6). Four of the associations were in Gag, 9 were in Pol, 11 were in Env, 2 were in Tat, 4 were in Vif, and 1 each was in Nef and Vpx.
TABLE 6.
Identification of putative SIV-specific polymorphisms linked to pig-tailed macaque MHC-I haplotypes using a 10-amino-acid sliding window analysis
| Haplotype | Protein | Nucleotide region |
No. of Mane-positive mutation-positive animals/total no. of Mane-positive animals | No. of Mane-negative mutation-positive animals/total no. of Mane-negative animals | P value (two-tailed) | No. of windows positive for amino acid sequence | Amino acid sequence spanning positive sliding windows | |
|---|---|---|---|---|---|---|---|---|
| Start | End | |||||||
| Mane-A*006 | Gag | 2182 | 2226 | 2/3 | 1/41 | 0.0094 | 6 | IPFAAAQQRGPRKPI |
| Pol | 4047 | 4076 | 2/3 | 1/41 | 0.0094 | 1 | QWWTDYWQVT | |
| Env | 7538 | 7588 | 3/3 | 6/41 | 0.0063 | 8 | RPGNKTVLPVTIMSGLV | |
| Mane-A*019 | Pol | 3036 | 3095 | 4/5 | 2/39 | 0.0005 | 11 | AIKKKDKNKWRMLIDFRELN |
| Env | 9023 | 9091 | 4/5 | 2/39 | 0.0005 | 10 | ALQRIREVLRTELTYLSYF | |
| Mane-A*032 | Tat | 6393 | 6425 | 2/2 | 2/42 | 0.0063 | 2 | ESANLGEEILS |
| Mane-A*052 | Env | 6911 | 6958 | 3/5 | 2/39 | 0.0070 | 7 | KLSPLCITMRCNKSET |
| Mane-A*082 | Env | 7256 | 7309 | 4/13 | 0/31 | 0.0053 | 9 | SCDKHYWDTIRFRYCAPP |
| Mane-A1*084 | Gag | 2284 | 2313 | 5/16 | 0/28 | 0.0040 | 1 | RQGCWKCGKM |
| Env | 8795 | 8836 | 13/16 | 7/28 | 0.0005 | 5 | FQQTHIQQDPALPT | |
| Mane-A*114 | Gag | 2446 | 2475 | 2/2 | 1/42 | 0.0032 | 1 | PAVDLLKNYM |
| Pol | 4044 | 4073 | 2/2 | 2/42 | 0.0063 | 1 | EQWWTDYWQV | |
| Vif | 5701 | 5748 | 2/2 | 2/42 | 0.0063 | 7 | EVRRAIRGEQLLSCCK | |
| Mane-B017 | Pol | 3426 | 3455 | 2/2 | 2/42 | 0.0063 | 1 | DRTDLEHDRV |
| Mane-B039 | Tat | 8862 | 8900 | 3/5 | 1/39 | 0.0029 | 4 | EKAVATAPGLGR |
| Mane-B043 | Pol | 5277 | 5306 | 3/5 | 0/36 | 0.0042 | 1 | ILKVGTDIKV |
| Vif | 5533 | 5562 | 4/8 | 1/35 | 0.0023 | 1 | SHLEVQGYWH | |
| Vif | 5548 | 5589 | 4/8 | 2/36 | 0.0065 | 5 | QGYWHLTPERGWLS | |
| Env | 9137 | 9190 | 5/8 | 2/36 | 0.0009 | 8 | AGAWGDLWETLRRGRWI | |
| Mane-B047 | Env | 8837 | 8866 | 6/6 | 14/38 | 0.0055 | 1 | REGKEGDGGE |
| Nef | 9279 | 9314 | 5/6 | 9/38 | 0.0089 | 3 | PWRNPAEEREKL | |
| Mane-B052 | Pol | 2751 | 2780 | 2/2 | 2/42 | 0.0063 | 1 | VLGKRIKGTI |
| Pol | 3351 | 3413 | 2/2 | 1/42 | 0.0032 | 12 | VLEPFRKANPDVTLVQYMDDI | |
| Vif | 5869 | 5904 | 2/2 | 2/42 | 0.0063 | 3 | RGLRMAKQNSRG | |
| Mane-B069 | Gag | 2272 | 2304 | 2/4 | 0/40 | 0.0063 | 2 | RAPRRQGCWKC |
| Pol | 2568 | 2603 | 2/4 | 0/40 | 0.0063 | 3 | LWRRPVVTAHIE | |
| Env | 6815 | 6853 | 2/4 | 0/40 | 0.0063 | 4 | VTESFDAWENTVT | |
| Mane-B104 | Vpx | 6619 | 6657 | 3/3 | 7/41 | 0.0091 | 4 | ESAAYRHLAFKCL |
| Env | 7142 | 7189 | 2/3 | 1/41 | 0.0094 | 7 | KEYNETWYSTDLVCEQ | |
| Env | 8291 | 8341 | 2/3 | 1/41 | 0.0094 | 8 | QQQQLLDVVKRQQELLR | |
| Mane-B119 | Pol | 3822 | 3857 | 2/3 | 1/41 | 0.0094 | 3 | PLEATVIKSQDN |
| Env | 7907 | 7939 | 2/3 | 1/41 | 0.0094 | 2 | QIINTWHKVGK | |
Validation that a novel Mane-B028-associated polymorphism in Nef represents a CTL escape mutation.
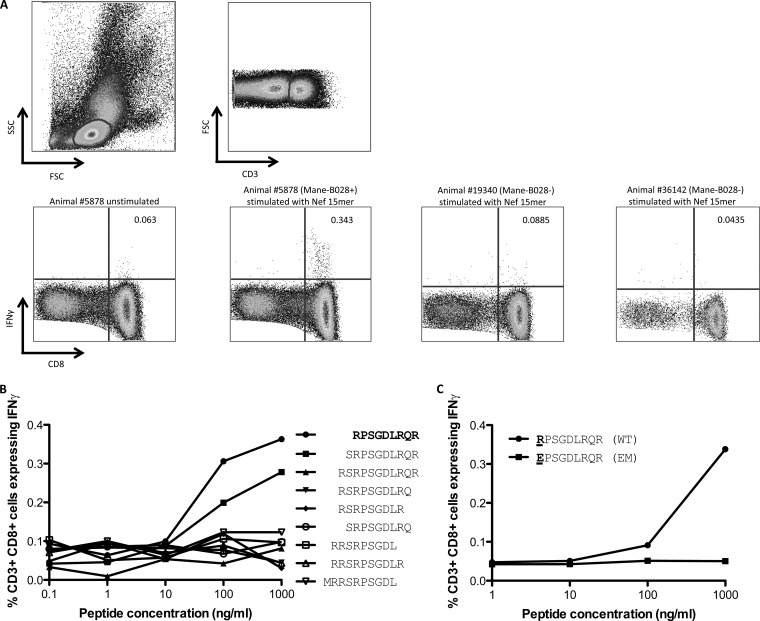
To validate that our approach successfully identified novel CTL escape mutations, we infected 3 additional pig-tailed macaques with the closely related strain SIVmac239. One macaque expressed the Mane-B028 haplotype that was linked to a polymorphism in Nef (shown in Fig. 2). We obtained blood samples to analyze CD8 T cell responses to specific 15-mer SIV peptides across the Nef polymorphism linked to Mane-B028 in the analyses described above. We found that ∼0.3% of CD8 T cells obtained 10 weeks after infection expressed IFN-γ in response to stimulation with the Nef peptides (Fig. 4A). This response was not identified in the other 2 Mane-B028-negative SIV-infected pig-tailed macaques studied. We subsequently obtained 9- to 11-mer peptides spanning this region and identified the minimal epitope to be a RE9 peptide (Fig. 4B). When the escape mutation identified in the linkage analyses was inserted into the minimal epitope (a mutation of R to E at the first position), a substantial loss of CD8 T cell recognition was observed (Fig. 4C). Thus, our approach successfully identified an escape mutation within a novel CD8+ CTL epitope within SIV in pig-tailed macaques.
FIG 4.
SIV Nef-specific CD8 T cell response predicted by linkage analysis. (A) Whole blood from 3 SIV-infected macaques was stimulated by 3 SIV Nef 15-mer peptides spanning the site of a putative Mane-B028-restricted CTL response identified by the MHC-I–SIV mutation linkage analysis. Gated CD3+ CD8+ lymphocytes were studied for IFN-γ expression, with the 1 Mane-B028-positive macaque showing a positive response and 2 Mane-B028-negative macaques showing no response. SSC, side scatter; FSC, forward scatter. (B) The minimal Nef-specific CTL epitope was mapped to a 9-mer peptide using serial dilutions of overlapping 9- to 11-mer peptides. (C) The putative CTL escape mutation (R to E at position 1) identified in the linkage analysis was evaluated for whether it abrogated CTL recognition.
Analysis of Mane-B017-associated polymorphisms and CTL responses.
Studies conducted in the Indian rhesus macaque model have demonstrated that Mamu-B017 is able to assist in the control of SIV replication in a subset of animals (39, 44). Furthermore, several CTL epitopes, which include epitopes in Env and Nef, have been shown to occur in association with the Mamu-B017 haplotype in rhesus macaques (39). The pig-tailed macaque Mane-B*017 allelic variants that were present in 2 of our SIV-infected animals differed from Mamu-B*017 by only single conservative amino acid substitutions in exons 2 and 4. Interestingly, although the cohort is very small, the 2 Mane-B017-positive pig-tailed macaques did not control SIVmac251 to low levels (Table 3).
We examined our sequencing data for the presence of mutations within known Mamu-B*017-restricted Env and Nef epitopes. We found that the Nef IW9 epitope (identified in Mamu-B017-positive [Mane-B017+] rhesus macaques) contained mutations within our Mane-B017+ pig-tailed macaques that were similar to those described in SIV-infected Mamu-B017+ rhesus macaques (Fig. 5A). We also found a mutation within the Env FW9 epitope in one Mane-B017+ pig-tailed macaque, although the particular mutation was different from that reported in Mamu-B017+ rhesus macaques (Fig. 5B). To validate that Mane-B017 presents the Nef IW9 epitope and Env FW9 epitope identified in Mamu-B017-positive rhesus macaques, we thawed frozen PBMCs from one of the Mane-B*017-positive macaques obtained 24 weeks after infection with SIVmac251 and stimulated the PBMCs with peptides containing the IW9 and FW9 epitopes. The cultures were stained with Mamu-B017 tetramers folded around the Nef IW9 and Env FW9 epitopes. We identified a tetramer-positive population for Env FW9 (Fig. 5C), although we did not identify such a population for Nef IW9 (not shown).
DISCUSSION
The important role that MHC-I alleles and CTL responses play in the progression of HIV infection has been well documented by linking HIV-1 polymorphisms with MHC-I alleles (24, 25, 45). Although a number of SIV-specific CTL epitopes have been described in most macaque models of SIV infection, a broad linking of MHC-I haplotypes and SIV mutations in macaques has not previously been performed, despite the advantage of knowing the time of infection and identity of the infecting strain in this animal model. In the study described in this report, we sequenced the plasma viral genome of 44 SIVmac251-infected pig-tailed macaques and determined their MHC-I haplotypes. The MHC-I haplotypes and the viral sequencing databases were analyzed using specifically designed in-house pipelines to identify an association between viral mutations and Mane-A and -B haplotypes. The methodology that we used was first validated using the well-described KP9, KSA10, and KVA10 escape mutations associated with Mane-A1*084. We then used this method to identify novel potential escape mutations, and we were able to identify 46 novel single nucleotide mutations throughout the SIVmac251 proteome linked to diverse Mane class I haplotypes with a reasonable likelihood of being genuine associations (we estimated >86% and >66% likelihoods in sliding window and point mutation analyses, respectively). Further, a novel sliding window methodology was used to identify 10-amino-acid regions of the SIVmac251 genome where closely spaced amino acids (potentially within the same CTL epitope) were mutated at different places in different animals. This analysis identified a further 32 potential MHC-restricted CTL epitopes undergoing escape. The novel potential epitopes identified will allow scientists to study in finer detail the CTL responses induced by vaccination or infection in this model and potentially better understand the principles of T cell control of SIV in this model.
The total of 78 sites in SIV undergoing mutational escape that we identified through this MHC-I linkage analysis likely represents an overestimate of the true number of CTL epitopes and their escape motifs at these sites, since (i) some of the observed associations may have occurred by chance, (ii) some mutations may reflect compensatory changes for CTL mutations elsewhere, and (iii) some mutations may reflect adaptation to high-avidity CD8 T cell responses rather than escape mutations (8, 46). We conducted a permutation analysis to estimate the likelihood of a false discovery rate, and we report only associations with greater than 86% and 66% likelihoods of being a true association by sliding window and point mutation analyses, respectively.
Interestingly, our analyses found a relatively large number of associations within the variable Env protein (33 of 78 associations). These likely reflect the higher levels of variability in Env than in other SIV proteins. Nonetheless, we also identified a total of 24 associations within the less variable Gag and Pol proteins. Given the associations between Gag CTL responses and control of both HIV and SIV (47), many of these new potential epitopes will be of interest to those studying CTL-based control of SIV in this model. As a proof of principle, our studies to date validated that our approach (i) correctly identifies immune escape mutations within 3 known pig-tailed macaque CTL epitopes restricted by Mane-A1*084, (ii) detected using functional T cell assays that a CTL epitope identified in our linkage and sequencing analyses was indeed directed to a Mane-B028-restricted Nef CTL epitope, and (iii) identified Mane-B017-linked mutations in epitopes previously defined in Mamu-B017+ rhesus macaques (39). However, more work remains to be done to validate a larger proportion of the putative CTL responses identified in this report, and we are currently infecting additional pig-tailed macaques to begin those studies. Identifying a wider suite of SIV-specific CTL epitopes and their escape patterns should assist with identification of which CTL responses or combinations of CTL responses (either by vaccination or by natural infection) are best able to assist with control of SIV. It is notable that our experiments validated that rhesus macaque MHC reagents, such as Mamu-B*017 tetramers, can detect T cell responses in pig-tailed macaques sharing very similar MHC-I alleles, which will allow some of the tools developed for rhesus macaque research to be used for the study of pig-tailed macaques.
Our use of a cohort of 44 SIV-infected pig-tailed macaques allowed us to ask if any particular MHC-I haplotypes were strongly associated with better control of SIV. Only weak VL associations with 3 Mane class I haplotypes were observed, and none were significant after adjusting for multiple comparisons. There are caveats in our analyses, since (i) there were only 44 animals studied, so we did not have a robust power to detect VL differences, and (ii) the 44 animals underwent a variety of treatments prior to and during SIV infection, although we attempted to control for these variables using a two-way ANOVA. We accept that there may well be MHC-I haplotypes that are associated with at least the modest control of SIV in pig-tailed macaques and that might be picked up with a much larger and more stringently selected cohort of animals. An important negative finding was that the presence of Mane-A084 was not associated with a significantly improved control of SIV, which contrasts with a preliminary report from our group studying a much smaller (n = 8) cohort (13), although the findings of the analysis described in this report are in agreement with those of another group who found no affect of Mane-A084 on plasma SIV load across 32 pig-tailed macaques (11 expressing Mane-A084) (48). We have developed a number of reagents for Mane-A1*084 (including MHC tetramers and genetic techniques to screen animals for this allele [14]) and have previously characterized the escape patterns at the known Mane-A1*084-restricted KP9, KVA10, and KSA10 CTL epitopes. Our analyses should allow scientists to proceed with studies using Mane-A084+ pig-tailed macaques without undue concern that these animals will control SIV to a much greater extent than Mane-A084-negative animals.
In summary, we employed a linkage analysis between MHC-I alleles and SIV sequences in SIV-infected pig-tailed macaques to identify potential CTL epitopes undergoing mutational escape. This work provides a basis to improve the pig-tailed macaque model of SIV infection and ultimately better understand the CTL-based control of SIV and HIV infection.
ACKNOWLEDGMENTS
This work was supported by Australian National Health and Medical Research Council (NHMRC) awards 628331 and 1025576. S.J.K. and M.P.D. are NHMRC Research Fellows.
We thank Sheilajen Alcantara, Thakshila Amarasena, Matt Scarlotta, and Georges Khoury for technical assistance.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.Harrer T, Harrer E, Kalams SA, Barbosa P, Trocha A, Johnson RP, Elbeik T, Feinberg MB, Buchbinder SP, Walker BD. 1996. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 156:2616–2623. [PubMed] [Google Scholar]
- 2.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. U. S. A. 94:1890–1895. 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez CS, Stratov I, De Rose R, Walsh K, Dale CJ, Smith MZ, Agy MB, Hu SL, Krebs K, Watkins DI, O'Connor DH, Davenport MP, Kent SJ. 2005. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J. Virol. 79:5721–5731. 10.1128/JVI.79.9.5721-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell KA, Rabi SA, Siliciano RF, Blankson JN. 2011. CD4+ T cells from elite suppressors are more susceptible to HIV-1 but produce fewer virions than cells from chronic progressors. Proc. Natl. Acad. Sci. U. S. A. 108:E689–E698. 10.1073/pnas.1108866108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reece JC, Alcantara S, Gooneratne S, Jegaskanda S, Amaresena T, Fernandez CS, Laurie K, Hurt A, O'Connor SL, Harris M, Petravic J, Martyushev A, Grimm A, Davenport MP, Stambas J, De Rose R, Kent SJ. 2013. Trivalent live attenuated influenza-simian immunodeficiency virus vaccines: efficacy and evolution of cytotoxic T lymphocyte escape in macaques. J. Virol. 87:4146–4160. 10.1128/JVI.02645-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedrich TC, Frye CA, Yant LJ, O'Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, Hughes AL, Wilson N, Watkins DI. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581–2585. 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Maarseveen NM, Wensing AM, de Jong D, Taconis M, Borleffs JC, Boucher CA, Nijhuis M. 2007. Persistence of HIV-1 variants with multiple protease inhibitor (PI)-resistance mutations in the absence of PI therapy can be explained by compensatory fixation. J. Infect. Dis. 195:399–409. 10.1086/510533. [DOI] [PubMed] [Google Scholar]
- 8.Almeida CA, Bronke C, Roberts SG, McKinnon E, Keane NM, Chopra A, Kadie C, Carlson J, Haas DW, Riddler SA, Haubrich R, Heckerman D, Mallal S, John M. 2011. Translation of HLA-HIV associations to the cellular level: HIV adapts to inflate CD8 T cell responses against Nef and HLA-adapted variant epitopes. J. Immunol. 187:2502–2513. 10.4049/jimmunol.1100691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida CA, Roberts SG, Laird R, McKinnon E, Ahmad I, Keane NM, Chopra A, Kadie C, Heckerman D, Mallal S, John M. 2010. Exploiting knowledge of immune selection in HIV-1 to detect HIV-specific CD8 T-cell responses. Vaccine 28:6052–6057. 10.1016/j.vaccine.2010.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merani S, Petrovic D, James I, Chopra A, Cooper D, Freitas E, Rauch A, di Iulio J, John M, Lucas M, Fitzmaurice K, McKiernan S, Norris S, Kelleher D, Klenerman P, Gaudieri S. 2011. Effect of immune pressure on hepatitis C virus evolution: insights from a single-source outbreak. Hepatology 53:396–405. 10.1002/hep.24076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batten CJ, De Rose R, Wilson KM, Agy MB, Chea S, Stratov I, Montefiori DC, Kent SJ. 2006. Comparative evaluation of simian, simian-human, and human immunodeficiency virus infections in the pigtail macaque (Macaca nemestrina) model. AIDS Res. Hum. Retroviruses 22:580–588. 10.1089/aid.2006.22.580. [DOI] [PubMed] [Google Scholar]
- 12.Klatt NR, Canary LA, Vanderford TH, Vinton CL, Engram JC, Dunham RM, Cronise HE, Swerczek JM, Lafont BA, Picker LJ, Silvestri G, Brenchley JM. 2012. Dynamics of simian immunodeficiency virus SIVmac239 infection in pigtail macaques. J. Virol. 86:1203–1213. 10.1128/JVI.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MZ, Dale CJ, De Rose R, Stratov I, Fernandez CS, Brooks AG, Weinfurter J, Krebs K, Riek C, Watkins DI, O'Connor DH, Kent SJ. 2005. Analysis of pigtail macaque major histocompatibility complex class I molecules presenting immunodominant simian immunodeficiency virus epitopes. J. Virol. 79:684–695. 10.1128/JVI.79.2.684-695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MZ, Fernandez CS, Chung A, Dale CJ, De Rose R, Lin J, Brooks AG, Krebs KC, Watkins DI, O'Connor DH, Davenport MP, Kent SJ. 2005. The pigtail macaque MHC class I allele Mane-A*10 presents an immundominant SIV Gag epitope: identification, tetramer development and implications of immune escape and reversion. J. Med. Primatol. 34:282–293. 10.1111/j.1600-0684.2005.00126.x. [DOI] [PubMed] [Google Scholar]
- 15.Mason RD, De Rose R, Kent SJ. 2009. Differential patterns of immune escape at Tat-specific cytotoxic T cell epitopes in pigtail macaques. Virology 388:315–323. 10.1016/j.virol.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Reece JC, Loh L, Alcantara S, Fernandez CS, Stambas J, Sexton A, De Rose R, Petravic J, Davenport MP, Kent SJ. 2010. Timing of immune escape linked to success or failure of vaccination. PLoS One 5:e12774. 10.1371/journal.pone.0012774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh L, Reece JC, Fernandez CS, Alcantara S, Center R, Howard J, Purcell DF, Balamurali M, Petravic J, Davenport MP, Kent SJ. 2009. Complexity of the inoculum determines the rate of reversion of SIV Gag CD8 T cell mutant virus and outcome of infection. PLoS Pathog. 5:e1000378. 10.1371/journal.ppat.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh L, Batten CJ, Petravic J, Davenport MP, Kent SJ. 2007. In vivo fitness costs of different Gag CD8 T-cell escape mutant simian-human immunodeficiency viruses for macaques. J. Virol. 81:5418–5422. 10.1128/JVI.02763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peut V, Kent SJ. 2007. Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen. J. Virol. 81:13125–13134. 10.1128/JVI.01408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peut V, Kent SJ. 2009. Substantial envelope-specific CD8 T-cell immunity fails to control SIV disease. Virology 384:21–27. 10.1016/j.virol.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Mothe B, Llano A, Ibarrondo J, Zamarreno J, Schiaulini M, Miranda C, Ruiz-Riol M, Berger CT, Herrero MJ, Palou E, Plana M, Rolland M, Khatri A, Heckerman D, Pereyra F, Walker BD, Weiner D, Paredes R, Clotet B, Felber BK, Pavlakis GN, Mullins JI, Brander C. 2012. CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS One 7:e29717. 10.1371/journal.pone.0029717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Capuano S, III, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M, Jr, Haase AT, Lifson JD, Allen TM, Watkins DI. 2012. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 491:129–133. 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson J, Kadie C, Mallal S, Heckerman D. 2007. Leveraging hierarchical population structure in discrete association studies. PLoS One 2:e591. 10.1371/journal.pone.0000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443. 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 25.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645. 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudieri S, Rauch A, Pfafferott K, Barnes E, Cheng W, McCaughan G, Shackel N, Jeffrey GP, Mollison L, Baker R, Furrer H, Gunthard HF, Freitas E, Humphreys I, Klenerman P, Mallal S, James I, Roberts S, Nolan D, Lucas M. 2009. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49:1069–1082. 10.1002/hep.22773. [DOI] [PubMed] [Google Scholar]
- 27.Rauch A, James I, Pfafferott K, Nolan D, Klenerman P, Cheng W, Mollison L, McCaughan G, Shackel N, Jeffrey GP, Baker R, Freitas E, Humphreys I, Furrer H, Gunthard HF, Hirschel B, Mallal S, John M, Lucas M, Barnes E, Gaudieri S. 2009. Divergent adaptation of hepatitis C virus genotypes 1 and 3 to human leukocyte antigen-restricted immune pressure. Hepatology 50:1017–1029. 10.1002/hep.23101. [DOI] [PubMed] [Google Scholar]
- 28.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent SJ, De Rose R, Mokhonov VV, Mokhonova EI, Fernandez CS, Alcantara S, Rollman E, Mason RD, Loh L, Peut V, Reece JC, Wang XJ, Wilson KM, Suhrbier A, Khromykh A. 2008. Evaluation of recombinant Kunjin replicon SIV vaccines for protective efficacy in macaques. Virology 374:528–534. 10.1016/j.virol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Sexton A, De Rose R, Reece JC, Alcantara S, Loh L, Moffat JM, Laurie K, Hurt A, Doherty PC, Turner SJ, Kent SJ, Stambas J. 2009. Evaluation of recombinant influenza virus-simian immunodeficiency virus vaccines in macaques. J. Virol. 83:7619–7628. 10.1128/JVI.00470-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Rose R, Fernandez CS, Smith MZ, Batten CJ, Alcantara S, Peut V, Rollman E, Loh L, Mason RD, Wilson K, Law MG, Handley AJ, Kent SJ. 2008. Control of viremia and prevention of AIDS following immunotherapy of SIV-infected macaques with peptide-pulsed blood. PLoS Pathog. 4:e1000055. 10.1371/journal.ppat.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, O'Connor DH. 2013. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) 3:1195–1201. 10.1534/g3.113.006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez CS, Reece JC, Saepuloh U, De Rose R, Ishkandriati D, O'Connor DH, Wiseman RW, Kent SJ. 2011. Screening and confirmatory testing of MHC class I alleles in pig-tailed macaques. Immunogenetics 63:511–521. 10.1007/s00251-011-0529-5. [DOI] [PubMed] [Google Scholar]
- 34.O'Leary CE, Wiseman RW, Karl JA, Bimber BN, Lank SM, Tuscher JJ, O'Connor DH. 2009. Identification of novel MHC class I sequences in pig-tailed macaques by amplicon pyrosequencing and full-length cDNA cloning and sequencing. Immunogenetics 61:689–701. 10.1007/s00251-009-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiseman RW, Karl JA, Bimber BN, O'Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O'Connor DH. 2009. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat. Med. 15:1322–1326. 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya T, Daniels M, Heckerman D, Foley B, Frahm N, Kadie C, Carlson J, Yusim K, McMahon B, Gaschen B, Mallal S, Mullins JI, Nickle DC, Herbeck J, Rousseau C, Learn GH, Miura T, Brander C, Walker B, Korber B. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586. 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 37.De Rose R, Batten CJ, Smith MZ, Fernandez CS, Peut V, Thomson S, Ramshaw IA, Coupar BE, Boyle DB, Venturi V, Davenport MP, Kent SJ. 2007. Comparative efficacy of subtype AE simian-human immunodeficiency virus priming and boosting vaccines in pigtail macaques. J. Virol. 81:292–300. 10.1128/JVI.01727-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jegaskanda S, Reece JC, De Rose R, Stambas J, Sullivan L, Brooks AG, Kent SJ, Sexton A. 2012. Comparison of influenza and SIV specific CD8 T cell responses in macaques. PLoS One 7:e32431. 10.1371/journal.pone.0032431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maness NJ, Yant LJ, Chung C, Loffredo JT, Friedrich TC, Piaskowski SM, Furlott J, May GE, Soma T, Leon EJ, Wilson NA, Piontkivska H, Hughes AL, Sidney J, Sette A, Watkins DI. 2008. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive simian immunodeficiency virus-infected rhesus macaques. J. Virol. 82:5245–5254. 10.1128/JVI.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lafont BA, Buckler-White A, Plishka R, Buckler C, Martin MA. 2003. Characterization of pig-tailed macaque classical MHC class I genes: implications for MHC evolution and antigen presentation in macaques. J. Immunol. 171:875–885. 10.4049/jimmunol.171.2.875. [DOI] [PubMed] [Google Scholar]
- 41.Pratt BF, O'Connor DH, Lafont BA, Mankowski JL, Fernandez CS, Triastuti R, Brooks AG, Kent SJ, Smith MZ. 2006. MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics 58:995–1001. 10.1007/s00251-006-0164-8. [DOI] [PubMed] [Google Scholar]
- 42.Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, Read DS, O'Connor DH. 2008. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics 60:37–46. 10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budde ML, Lhost JJ, Burwitz BJ, Becker EA, Burns CM, O'Connor SL, Karl JA, Wiseman RW, Bimber BN, Zhang GL, Hildebrand W, Brusic V, O'Connor DH. 2011. Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate SIV-specific CD8+ T cell responses. J. Virol. 85:3250–3261. 10.1128/JVI.02355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O'Connor DH, Carrington M, Watkins DI. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074–5077. 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJ. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775. 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 46.Keane NM, Roberts SG, Almeida CA, Krishnan T, Chopra A, Demaine E, Laird R, Tschochner M, Carlson JM, Mallal S, Heckerman D, James I, John M. 2012. High-avidity, high-IFNgamma-producing CD8 T-cell responses following immune selection during HIV-1 infection. Immunol. Cell Biol. 90:224–234. 10.1038/icb.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8(+) T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53. 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 48.Mankowski JL, Queen SE, Fernandez CS, Tarwater PM, Karper JM, Adams RJ, Kent SJ. 2008. Natural host genetic resistance to lentiviral CNS disease: a neuroprotective MHC class I allele in SIV-infected macaques. PLoS One 3:e3603. 10.1371/journal.pone.0003603. [DOI] [PMC free article] [PubMed] [Google Scholar]