ABSTRACT
H6N6 viruses are commonly isolated from domestic ducks, and avian-to-swine transmissions of H6N6 viruses have been detected in China. Whether subsequent adaptation of H6N6 viruses in mammals would increase their pathogenicity toward humans is not known. To address this, we generated a mouse-adapted (MA) swine influenza H6N6 virus (A/swine/Guangdong/K6/2010 [GDK6-MA]) which exhibited greater virulence than the wild-type virus (GDK6). Amino acid substitutions in PB2 (E627K), PA (I38M), and hemagglutinin ([HA] L111F, H156N, and S263R) occurred in GDK6-MA. HA with the H156N mutation [HA(H156N)] resulted in enlarged plaque sizes on MDCK cells and enhanced early-stage viral replication in mammalian cells. PA(I38M) raised polymerase activity in vitro but did not change virus replication in either mammalian cells or mice. These single substitutions had only limited effects on virulence; however, a combination of HA(H156N S263R) with PA(I38M) in the GDK6 backbone led to a significantly more virulent variant. This suggests that these substitutions can compensate for the lack of PB2(627K) and modulate virulence, revealing a new determinant of pathogenicity for H6N6 viruses in mice, which might also pose a threat to human health.
IMPORTANCE Avian H6N6 influenza viruses are enzootic in domestic ducks and have been detected in swine in China. Infections of mammals by H6N6 viruses raise the possibility of viral adaptation and increasing pathogenicity in the new hosts. To examine the molecular mechanisms of adaptation, a mouse-adapted avian-origin swine influenza H6N6 virus (GDK6-MA), which had higher virulence than its parental virus, was generated. Specific mutations were found in PB2 (E627K), PA (I38M), and HA (L111F, H156N, and S263R) and were assessed for their virulence in mice. The combination of HA(H156N S263R) and PA(I38M) compensated for the lack of PB2(627K) and showed increased pathogenicity in mice, revealing a novel mechanism that can affect the virulence of influenza viruses. H6N6 viruses should be monitored in the field for more virulent forms that could threaten human health.
INTRODUCTION
Influenza A viruses are important pathogens that infect both avian and mammalian species although these viruses are believed to have originated from avian influenza viruses (1, 2). Sixteen hemagglutinin (HA) and 9 neuraminidase (NA) subtypes of influenza A viruses have been isolated from birds (1, 3). Predicting which of the subtypes of avian influenza viruses might be able to cross the species barrier and infect humans has proved difficult. This was demonstrated in 2013 by the unexpected emergence in humans of a low-pathogenic avian influenza (LPAI) H7N9 virus in China, which has caused over 400 confirmed human infections, with a fatality rate of about 30%, according to the WHO (http://www.who.int/influenza/human_animal_interface/influenza_h7n9/Risk_Assessment/en/).
The H6 subtype of influenza viruses has the potential to cross the species barrier. It was first isolated from a turkey in 1965 and has been detected worldwide since then (4–7). Influenza virus surveillance indicated that H6 viruses had become enzootic in the domestic ducks of southern China, mostly as H6N6 viruses (8). Approximately one-third of H6 viruses isolated from live poultry markets in southern China could bind to human-type receptors, many could replicate in mice, and some could transmit efficiently among guinea pigs (9). Serological surveys conducted in China and America indicated that H6 influenza viruses might have previously infected swine and humans (10, 11), and one-third of human volunteers inoculated with H6N1 or H6N2 viruses showed mild clinical symptoms with virus shedding (12). H6N1 viruses caused significant morbidity and mortality in mice without prior adaptation (13), and an H6N5 virus was lethal to mice and could be transmitted between ferrets by direct contact (14).
In 2011, an H6N6 virus, A/swine/Guangdong/K6/2010 (GDK6), was isolated from a swine farm in southern China, and 3.4% of the pigs in the swine farms of this region were seropositive for H6 viruses (15). The GDK6 virus belongs to the group II lineage of H6 viruses, which are the predominant H6 viruses in domestic ducks of southern China (8). Subsequently, another H6N6 virus was isolated from swine in eastern China (16), and the first known natural case of human infection with an avian H6N1 influenza virus was reported in Taiwan in 2013 (17).
Adaptation is considered a primary impetus in evolution, and the process of natural selection in influenza A viruses appears to be mimicked by experimental adaptation in mice (18). Although some avian influenza viruses can replicate efficiently in mice without prior adaptation, they are mostly avirulent, and adaptation of a virus in mice can lead to the emergence of mutations causing higher pathogenicity (18–21). However, there is still limited knowledge of the molecular basis for the virulence of H6 viruses in mammals (20, 22).
Here, we focus on the molecular basis of pathogenicity in a mouse-adapted (MA) variant of the GDK6 (GDK6-MA) H6N6 virus. Using reverse genetics to generate reassortants of the wild type and a mouse-adapted virus, we identified the genes and mutations responsible for its higher pathogenicity in mice. A novel combination of substitutions in HA (H156N and S263R) and PA (I38M) led to the enhanced virulence of this H6N6 influenza A virus in mice.
MATERIALS AND METHODS
Ethics statement.
All animal studies were conducted under the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China. The animal experiments in our study were approved by the animal experimental ethics committee of the South China Agricultural University (approval number 2013-07).
Cells and viruses.
Adenocarcinomic human alveolar basal epithelial cells (A549), human embryonic kidney cells (293T), and Madin-Darby canine kidney (MDCK) cells were obtained from the Shanghai Cell Bank, Type Culture Collection Committee, Chinese Academy of Sciences, China. Cells were propagated in growth medium containing Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel) and 1% Glutamax (Gibco, USA) at 37°C and 5% CO2 until they reached ∼80% confluence.
A/swine/Guangdong/K6/2010 (H6N6) (GDK6) was isolated as described previously (15). Virus stocks were propagated in the allantoic cavities of 10-day-old specific-pathogen-free (SPF) embryonated hens' eggs (Merial, Beijing, China) at 37°C for 48 h. Allantoic fluid containing virus was harvested, aliquoted, and frozen at −80°C until used in experiments. Virus titers were measured by plaque assays in MDCK cells, and results were expressed as PFU counts (23). All experiments with live viruses were performed in an enhanced animal biosafety level 3 (ABSL-3+) facility at the South China Agricultural University, Guangzhou, China.
Adaptation of the GDK6 virus in mice.
A mouse-adapted variant was derived from a series of sequential lung-to-lung passages in mice. Five 6-week-old female BALB/c mice (Guangdong Medical Laboratory Animal Center, China) were lightly anesthetized with dry ice, and 50 μl of allantoic fluid containing 106 PFU of wild-type GDK6 virus was inoculated intranasally (i.n.). At 3 days postinoculation (p.i.), three inoculated mice were euthanized, and the lungs were harvested and homogenized in 2 ml of sterile cold phosphate-buffered saline (PBS). The homogenate was centrifuged at 6,000 × g for 5 min at 4°C and filtered through a 0.22-μm-pore-size cellulose acetate filter (Millipore, USA). Fifty microliters of the centrifuged homogenate was used as the inoculum for the next passage. After 12 passages, the virus in the lung homogenate was cloned three times by plaque purification in MDCK cells, and the cloned virus, designated GDK6-MA, was inoculated into 10-day-old SPF embryonated hens' eggs and held for 48 h at 37°C to prepare a virus stock.
Molecular cloning and sequencing of the viral genes.
Viral RNA was extracted from aliquots containing virus using a NucleoSpin RNA Virus kit (Macherey-Nagel, Germany). As described previously, with minor modification in restriction enzyme cutting sites (24), specific primers for each gene segment were used to perform reverse transcription of viral RNA and subsequent PCR. Amplified reverse transcription-PCR (RT-PCR) products were purified using a Wizard SV Gel and PCR Clean-Up System (Promega, USA). To sequence the viruses, the PCR products were cloned into the pJET 1.2 blunt-end cloning vector (Thermo Scientific, USA). At least three clones per gene were sequenced (performed by Shanghai Life Technologies Biotechnology Co., Ltd.).
Molecular modeling.
I-TASSER (25, 26) was used to predict a structural model of the HA of GDK6 based on its amino acid sequence (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The structural interpretation of the PA N-terminal domain utilized the structure of the PA of A/Victoria/3/1975 (PDB identification number 2W69). PyMol, version 1.6.0.0 (www.pymol.org), was used for visualization.
Construction of plasmids and virus rescue.
All reassortant viruses and the GDK6 and GDK6-MA viruses were generated by plasmid-based reverse genetics with the gene segments of GDK6 and GDK6-MA viruses cloned into the plasmid vector pHW2000 and amplified (27). Mutations in the HA gene were introduced by PCR-based site-directed mutagenesis with primer pairs containing point mutations and confirmed by sequencing.
A monolayer of 293T cells with approximately 90% confluence in six-well plates was transfected with a mix of PB2, PB1, PA, HA, NP, NA, M, and NS plasmids (wild type or mutated; 0.25 μg of plasmid) using Lipofectamine LTX reagent (Invitrogen, USA) according to the manufacturer's instructions. Specifically, 0.25 μg of each plasmid was mixed and incubated with 6 μl of Lipofectamine LTX and 2 μl of Plus Reagent in 300 μl of Opti-MEM (Gibco, USA) for 15 min at room temperature and then added to the cells. After incubation for 1 h at 37°C with 5% CO2, 1 ml of Opti-MEM containing 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich, USA) and 0.2% bovine serum albumin (BSA; Sigma-Aldrich, USA) was added to each well. The supernatant of the 293T culture was harvested after a 24-h incubation, and 300 μl of supernatant was inoculated into 10-day-old SPF hens' eggs and held for 48 h at 37°C to prepare virus stock. The parental and recombinant viruses were plaque purified and confirmed by a hemagglutination inhibition (HI) test and sequencing. The virus fluid was stored at −80°C until used.
Mouse pathogenicity experiments.
Groups of eight 6-week-old female BALB/c mice (Guangdong Medical Laboratory Animal Center, China) were lightly anesthetized with dry ice and i.n. inoculated with 50 μl of allantoic fluid containing 106.4 PFU of virus or 50 μl of sterile PBS (mock group). At 3 days p.i., four mice were euthanized, three for virus titration of the organs and one for histology and immunohistochemistry. Virus was titrated in lungs, brains, kidneys, thymuses, spleens, and livers by plaque assays in MDCK cells. The remaining four mice in each group were monitored for weight loss and clinical signs for 12 days. The percent changes in body weights were calculated relative to the weight on day 0. Mice losing more than 25% of their body weight were humanely euthanized.
To exclude the possibility that unwanted mutations might have arisen in the PB2 segment within the mice during the single round of infection, the lungs of dead mice infected with viruses carrying residue 627E in PB2 [PB2(627E)] were collected, and the PB2 genes of these viruses were confirmed by sequencing. To determine the 50% mouse lethal dose (MLD50), groups of four 6-week-old female BALB/c mice were intranasally inoculated with 50 μl of 10-fold serial dilutions containing 103.4 to 106.4 PFU of virus in sterile PBS and observed for signs of morbidity over 12 days. The MLD50 value was calculated by the method of Reed and Muench and expressed as log10PFU.
For histopathological examinations, the lungs collected from mice at 3 days p.i. were immersed into 10% phosphate-buffered formalin, embedded in paraffin, and then cut into 5-μm-thick sections and stained with hematoxylin and eosin (H&E). The organs were examined microscopically. Additional sections were processed for immunohistological staining with rabbit polyclonal antibody against GDK6 virus (prepared in our laboratory). Goat anti-rabbit IgG-horseradish peroxidase (HRP; SunShineBio, China) directed against the primary antibody was used as a secondary antibody. Specific antigen-antibody reactions were visualized by means of 3,3′-diaminobenzidine tetrahydrochloride.
Growth dynamics of viruses.
The parental and recombinant viruses were inoculated into MDCK cell monolayers (multiplicity of infection [MOI] of 0.01 PFU) or A549 cell monolayers (MOI of 0.1 PFU) with DMEM containing 0.5% BSA and 1 μg/ml TPCK-treated trypsin and incubated at 37°C with 5% CO2. Cell supernatants were harvested every 12 h until 60 h postinoculation (hpi).
Single-replication-cycle experiments were performed to confirm the early-stage viral replication. The parental and recombinant viruses were inoculated into MDCK cell monolayers or A549 cell monolayers (MOI of 5 PFU) with DMEM containing 0.5% BSA and 1 μg/ml TPCK-treated trypsin and incubated at 37°C with 5% CO2. Cell supernatants were harvested at 6 hpi.
Virus in supernatants was titrated for infectivity in MDCK cells by plaque assays.
Minigenome assay for polymerase activity.
293T cells were transfected with 0.25 μg of pHW2000-PB2 (wild type or mutated), pHW2000-PB1, pHW2000-PA (wild type or mutated), pHW2000-NP, the luciferase reporter plasmid pPolI-Luci-T, and an internal control plasmid expressing Renilla luciferase (Promega, USA) using Lipofectamine LTX reagent (Invitrogen, USA) as recommended by the manufacturer. After 48 h of transfection, cell extracts were harvested and lysed with a Dual-Luciferase Reporter Assay System (Promega, USA), and luciferase activity was measured using a Modulus Single Tube reader (Turner Biosystems, USA).
RESULTS
Adaptation of an H6N6 influenza virus in mice.
To adapt an H6N6 influenza virus to a mammalian host, the GDK6 strain was serially passaged in mouse lungs, with intranasal inoculations of 106.4 PFU of virus per mouse. Survival of infected animals was assessed after each passage. While the wild-type GDK6 virus was almost avirulent in mice, after five passages one mouse showed a continuous trend of weight loss and died at 8 days p.i. After eight passages, all inoculated mice died within 5 days p.i. (data not shown), indicating that the virus populations had acquired mutations profoundly affecting virulence. After 12 passages, the virus present in the lung homogenate was plaque purified three times in MDCK cells and designated GDK6-MA.
The mouse-adapted H6N6 virus exhibited enhanced virulence in mice.
Two groups of BALB/c mice were infected with 106.4 PFU of either GDK6-MA or GDK6 virus. Mice inoculated with GDK6 had a slight weight loss and recovered by day 6 p.i. In contrast, more serious clinical signs of disease, including decreased activity, huddling, and ruffled fur, were observed in the group inoculated with GDK6-MA, and these mice lost weight progressively, with all dying before day 6 p.i. (Fig. 1A and B).
FIG 1.
Virulence of GDK6 and GDK6-MA viruses in mice. Six-week-old female BALB/c mice (n = 4 per group) were infected with either 106.4 PFU of the indicated viruses or mock infected with 50 μl of PBS. (A) Body weight changes over a 12-day period are plotted as a percentage of body weight at day 0 p.i. (data are expressed as means ± standard deviations). (B) Survival plot of mice in each infection group. dpi, days p.i.
Sequence determinants of the increased virulence.
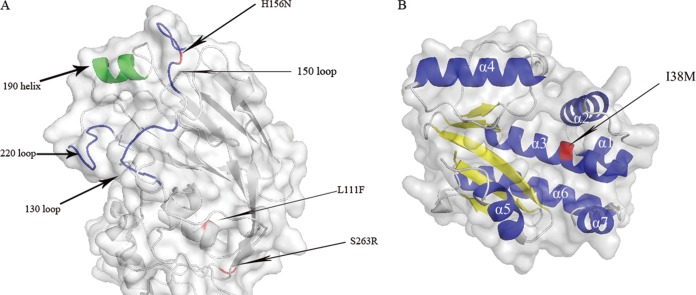
The complete genomes of the wild-type and mouse-adapted viruses were sequenced and compared to available influenza A virus sequences. Five amino acid substitutions were identified in the PB2 (E627K), PA (I38M), and HA (L111F, H156N, and S263R; H3 numbering) proteins (Table 1) of GDK6-MA. All HA substitutions were located in the globular head region of the HA1 domain (Fig. 2A). The substitution HA(H156N) results in a potential N-linked glycosylation site, which has not been observed in other H6 viruses and lies in the 150 loop (positions 153 to 160) proximal, but not immediately adjacent, to the 190 helix. Both of these regions flank the receptor-binding site (RBS) (28). Two sites within the RBS of GDK6 differ from those in the majority of H6 viruses, A138S and G228S. Apart from GDK6, only A/duck/Fujian/2606/2006 (H6N6) had S at both of these positions, while 25 other avian viruses and the human-isolated H6 virus had HA(228S). The three substitutions relative to GDK6 in the HA of GDK6-MA occur in some avian H6 viruses, but none carry all simultaneously.
TABLE 1.
Molecular characteristics of GDK6 and GDK6-MA viruses
| Virus protein | Position or feature | Presence of feature or amino acid(s)a |
|||
|---|---|---|---|---|---|
| GDK6 | GDK6-MA | Avian H6 | Mammalian H6 | ||
| HA (RBS)b | 138 | S | S | A (1,134/1,449), S (312/1,449), V (3/1,449) | A (2/3), S (1/3) |
| 190 | E | E | E (1,382/1,449), V (30/1,449), A (23/1,449), L (14/1,449) | E (3/3) | |
| 194 | L | L | L (1,449/1,449) | L (3/3) | |
| 225 | G | G | G (1,421/1,449), S (28/1,449) | G (3/3) | |
| 226 | Q | Q | Q (1,426/1,449), L (3/1,449) | Q (3/3) | |
| 228 | S | S | G (1,423/1,449), S (26/1,449) | S (2/3), G (1/3) | |
| Glycosylation at 150 loopf | No | Yes | No (1,449/1,449) | No (3/3) | |
| HA(HA1 domain)c | 111 | L | F | L (1,309/1,449), F (140/1,449) | L (3/3) |
| 156 | H | N | T (892/1,449), H (207/1,449), I (145/1,449), P (135/1,449), N (8/1,449), X (62/1,449)d | H (3/3) | |
| 263 | S | R | N (658/1,449), S (564/1,449), R (108/1,449), D (56/1,449), X (63/1,449)d | ||
| NA (N6) | Stalk deletion | Yes | Yes | Yes (6/226), No (220/226) | Yes (1/2), No (1/2) |
| PAg | 38 | I | M | I (931/931) | I (3/3) |
| PB2 | 627 | E | K | E (814/815), K (1/815)e | E (3/3) |
Values in parentheses represent the number of viruses with the amino acid or feature/total number of viruses. H6 sequences were obtained from the NCBI Influenza Viruses Resource and the Global Initiative on Sharing All Influenza Data (GISAID).
Comparison of conserved amino acids in the RBS between avian and mammalian H6 viruses.
Comparison of mutations identified in HA of GDKA-MA with wild-type avian and mammalian H6 viruses.
X represents amino acids which were seldom found in this position.
A/laughing gull/NJ/276/1989 (H6N8).
150 loop, positions 153 to 160.
PA variants were also found in other subtypes. In avian viruses A/mallard/QC/2323-25/2006 (H5N2), A/American green winged teal/Mississippi/11OS255/2011 (H7N7), and A/mallard/Berlin/453-4/1980 (mixed), variants were I (1,792/1,795) and M (3/1,795). In mammalian viruses A/swine/Iowa/13C020/2013 (H3N2), A/Malaysia/10807/1996 (H3N2), A/Netherlands/42/2006 (H3N2), and A/Boston/134/2009 (H1N1), variants I (1,892/1,896) and M (4/1,896) were present.
FIG 2.
Structural locations of mutations found in the mouse-adapted virus. The H6 HA and PA N-terminal domain models are illustrated using PyMol; mutated residues are shown in red. (A) Structure of the globular head of the HA1 portion of H6 HA. Receptor binding regions (130 loop, 150 loop, 220 loop, and 190 helix) are indicated on the model. (B) Structure of the PA N-terminal domain with α-helixes in blue and β-strands in yellow.
PA(I38M) is located on helix α3 of the N-terminal subunit (positions 1 to 256) of PA (Fig. 2B). PA(38M) does not occur in other H6 viruses, but is found in four mammalian and three avian influenza viruses. Of the available H6 virus PB2 sequences, only A/laughing gull/NJ/276/1989 (H6N8) had residue 627K. The NA of GDK6 had an 11-amino-acid deletion in the stalk region at positions 59 to 69, which was found in six avian H6N6 viruses and reported previously (8).
The HA and PA genes were dominant in the higher virulence of GDK6-MA.
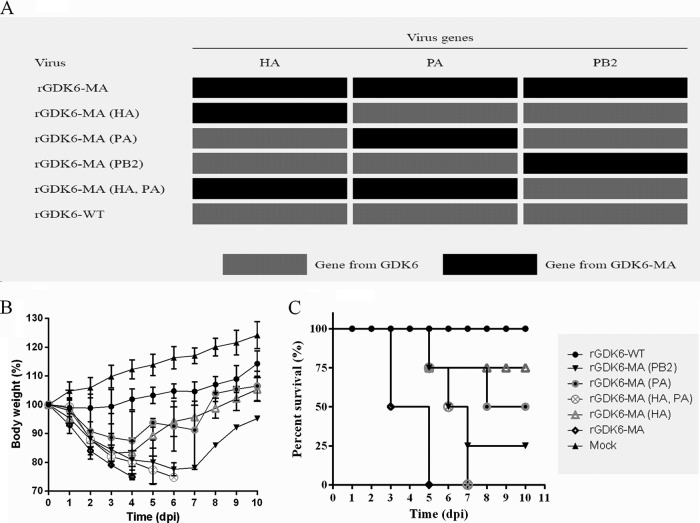
Recombinant viruses containing one or more of the GDK6-MA-associated segments in a GDK6 background were generated to determine the genetic basis for the higher virulence of GDK6-MA (Fig. 3A). The recombinant GDK6 (rGDK6) virus containing the HA of GDK6-MA was named rGDK6-MA (HA), and the other recombinants were named similarly. MLD50 values and viral titers in organs were assessed to evaluate the pathogenicity of these recombinants. The rGDK6 and rGDK6-MA viruses showed pathogenicity levels similar to their wild-type forms. As previous studies demonstrated (28–30), the PB2(E627K) substitution enhances pathogenicity in mammals, and this was observed in the rGDK6-MA (PB2) virus (Table 2; Fig. 3B and C). When either the GDK6-MA HA or PA segment replaced the GDK6 segment, 1 of 4 or 2 of 4, respectively, of the mice inoculated with 106.4 PFU of virus died (Fig. 3B and C). However, the log10MLD50 values for both viruses were greater than 6.4 (Table 2). The double substitution of HA and PA from rGDK6-MA caused all 4 mice inoculated with 106.4 PFU of virus to die. This virus had a log10MLD50 value of 4.39, which was close to that of rGDK6-MA, indicating that an interaction between the HA and PA genes affects virulence (Table 2; Fig. 3B and C).
FIG 3.
Virulence of the rescued viruses in mice. (A) Recombinant viruses were generated by introducing segments from GDK6-MA into the GDK6 backbone. The PB1, NP, NA, M, and NS segments were not replaced. (B) Body weight changes of mice (n = 4) infected with 106.4 PFU of the indicated virus over a 10-day period, plotted as a percentage of body weight at day 0 p.i. (data are expressed as means ± standard deviations). (C) Survival plot of groups of mice (n = 4) infected with 106.4 PFU of the indicated virus. WT, wild type.
TABLE 2.
Virulence and replication in mice of the recombinant virusesa
| Virus | MLD50 (log10PFU) | Lung titer (log10PFU)b |
|---|---|---|
| rGDK6-MA | 3.89 | 4.78 ± 0.16c |
| rGDK6-MA (HA) | >6.40 | 3.30 ± 0.44 |
| rGDK6-MA (PA) | >6.40 | 3.56 ± 0.55 |
| rGDK6-MA (PB2) | 4.73 | 3.53 ± 0.19 |
| rGDK6-MA (HA, PA) | 4.39 | 4.17 ± 0.22 |
| rGDK6 | >6.40 | 3.69 ± 0.10 |
Groups of 8 BALB/c mice were inoculated intranasally with 106.4 PFU of H6N6 influenza virus. Three mice from each group were euthanized at 3 days p.i. for virus titration.
Values are means ± standard deviations (n = 3). Virus was not detected in any organs except the lungs.
ignificantly different from titer of rGDK6 virus (P < 0.01, one-way ANOVA).
In all infected mice, viruses were detected only in the lungs. The viral titer of rGDK6-MA was significantly higher than that of rGDK6 (Table 2) (P < 0.01, one-way analysis of variance [ANOVA]), while the other viruses replicated in lungs to titers similar to the rGDK6 titer (Table 2) (P > 0.05, one-way ANOVA). Although the virus titers in lungs in each group of infected mice were similar (Table 2), viral antigen-positive alveolar epithelial cells in the lungs of mice inoculated with rGDK6-MA (PA), rGDK6-MA (HA, PA), or rGDK6-MA were more common than those in the lungs of mice infected with rGDK6 or rGDK6-MA (HA) (Fig. 4C, F, I, L, and O).
FIG 4.
Pathological findings in infected mice. Sections of lungs of mice infected with 106.4 PFU of rGDK6 (A to C), rGDK6-MA (HA) (D to F), rGDK6-MA (PA) (G to I), rGDK6-MA (HA, PA) (J to L), or rGDK6-MA (M to O) at 3 days p.i. are shown. Lung sections are hematoxylin and eosin (H&E) stained or immunostained. H&E-stained sections are at a magnification of ×100 (A, D, G, J, and M), with the boxed areas at a magnification of ×400 (B, E, H, K, and N). Immunostained sections are shown at a magnification of ×400 (C, F, I, L, and O) with viral antigen-positive cells marked by brown pigment.
Histopathology of the lungs of inoculated mice was conducted at 3 days p.i. GDK6-inoculated mice appeared almost normal (Fig. 4A and B). Mice inoculated with rGDK6-MA (HA) showed inflammatory cell infiltration and moderate thickening of the alveolar walls (Fig. 4D and E). Hemorrhage, moderate bronchopneumonitis, and moderate thickening of the alveolar walls and interstitium were observed in the lungs of mice infected with rGDK6-MA (PA) (Fig. 4G and H). Lungs of mice inoculated with either the rGDK6-MA or rGDK6-MA (HA, PA) strain showed more serious lung tissue damage, with hemorrhage, lymphocyte infiltration, severe peribronchiolar inflammation, and localized interstitial pneumonia (Fig. 4J, K, M, and N). The histopathology and immunohistochemistry tests suggest that the substitutions in the HA and PA proteins play critical roles in inflammation in the lungs, and the mutant PA accelerated viral replication in alveolar epithelial cells, as seen at 3 days p.i.
Combination of HA(H156N S263R) and PA(I38M) significantly increases virulence.
Recombinant GDK6 viruses with PA(I38M) and different point mutations in the HA protein were generated to assess the roles of the HA substitutions in virulence (MLD50) and viral replication in the lungs (Table 3). Of the six recombinants, only rGDK6 expressing both HA(H156N S263R) and PA(38M) [rGDK6-HA(156N 263R)-PA(38M)] had a virulence level similar to that of rGDK6-MA (HA, PA) (Table 3). However, viral titers in the lungs of all groups were similar to the rGDK6 titer (P > 0.05, one-way ANOVA). This suggests that the combination of HA(H156N S263R) and PA(I38M) is responsible for the higher virulence of the mouse-adapted H6N6 virus.
TABLE 3.
Virulence and replication in mice of the viruses with point mutationsa
| Virus | MLD50 (log10PFU) | Lung titer (log10PFU)b |
|---|---|---|
| rGDK6-HA(111F)-PA(38M) | >6.40 | 3.00 ± 0.41 |
| rGDK6-HA(156N)-PA(38M) | 5.90 | 4.12 ± 0.67 |
| rGDK6-HA(263R)-PA(38M) | >6.40 | 3.47 ± 0.52 |
| rGDK6-HA(111F 156N)-PA(38M) | >6.40 | 3.99 ± 0.55 |
| rGDK6-HA(111F 263R)-PA(38M) | 6.16 | 3.79 ± 0.22 |
| rGDK6-HA(156N 263R)-PA(38M) | 4.06 | 3.45 ± 0.17 |
| rGDK6-MA (HA, PA)c | 4.39 | 3.87 ± 0.26 |
| rGDK6c | >6.40 | 3.52 ± 0.29 |
Groups of 8 BALB/c mice were inoculated intranasally with 106.4 PFU of H6N6 influenza virus. Three mice from each group were euthanized at 3 days p.i. for virus titration.
Values are means ± standard deviations.
The titers for these viruses differs marginally from those in Table 2 as this is a separate replication.
To ensure that compensatory mammalian adaptation mutations in the PB2 protein at positions 627, 701, or 590 to 591 had not arisen during these experiments, viruses were isolated from mice, and the PB2 genes were sequenced, revealing no mutations (data not shown).
The HA(H156N) mutation increases GDK6 viral replication in mammalian cells.
The growth properties of the parental and mutated viruses were studied in MDCK and A549 cells. The plaque assays in MDCK cells revealed that recombinant viruses containing the HA(H156N) substitution formed larger plaques than other viruses and that other substitutions, including PB2(627K), did not influence plaque size (Fig. 5A), suggesting that, at least in MDCK cells, HA(H156N) increases the cytopathic effect of GDK6 variants. Multiple replication cycle growth curves in MDCK and A549 cell monolayers showed that all viruses reached a maximum at 36 hpi. The rGDK6-MA (PA) virus and the parental rGDK6 virus had similar replication kinetics in both MDCK and A549 cells (P > 0.05, two-way ANOVA), suggesting that the PA(I38M) mutation did not change growth properties in vitro (Fig. 5B and C). In contrast, viruses containing HA(H156N) had significantly higher titers at 12 and 24 hpi than rGDK6 and the other viruses in the two mammalian cells, but had similar replication rates at later time points. Single-replication-cycle experiments in MDCK and A549 cells also revealed that viruses containing HA(H156N) replicated more efficiently than rGDK6 at 6 hpi (Fig. 5D and E). Thus, HA(H156N) appeared to increase early-stage virus replication in vitro.
FIG 5.
Growth properties of the parental and mutated viruses. (A) Plaques formed by the indicated viruses in MDCK cells 48 h after inoculation. (B and C) Growth curves after inoculation of each virus at MOIs of 0.01 and 0.1 into MDCK and A549 cells, respectively. Each point on the curve is the mean ± standard deviation from four independent experiments. Titers of viruses that are significantly different from those of rGDK6 are marked as follows: *, P < 0.01; **, P < 0.001; ***, P < 0.0001. Statistical analysis was performed by two-way ANOVA. (D and E) Single-replication-cycle experiments after inoculation of each virus at MOIs of 5 and 10 into MDCK and A549 cells, respectively. Each column indicates the mean ± standard deviation from three independent experiments at 6 hpi. Titers of viruses that are significantly different from those of rGDK6 are marked as described for panels B and C. Statistical analysis was performed by one-way ANOVA.
PA(I38M) and PB2(E627K) upregulate transcription activity of RNP complex.
The transcription activity of reconstituted RNP complexes containing various substitutions was measured in 293T cells by a luciferase minigenome assay. A significant upregulation of activity was observed with PB2(627K) (Fig. 6). Although not statistically significant (P = 0.3; one-way ANOVA test), the transcription activity of PB2(627E)-PA(38M) rose to 2.6 times that of PB2(627E)-PA(38I), while the transcription activity of PB2(627K)-PA(38M) was 1.5 times higher than that of PB2(627K)-PA(38I) and statistically significantly different (P < 0.001; one-way ANOVA test). This suggests that Met-38 in PA and Lys-627 in PB2 increase the transcription activity of the RNP complex.
FIG 6.
Viral RNA transcription activity by minigenome assay. 293T cells were transfected with the plasmids PHW2000-PB2, PHW2000-PB1, PHW2000-PA, PHW2000-NP, the luciferase reporter plasmid pPolI-Luci-T, and an internal control plasmid expressing Renilla luciferase. The PB2 and PA plasmids were wild type or contained the mutations indicated. Luciferase activity was measured at 48 h after transfection. Values shown are the means ± standard deviations of six independent experiments. (**, P < 0.001; one-way ANOVA).
DISCUSSION
The swine H6N6 viruses from China were the first known H6 subtype viruses to infect mammals (15, 16). The HA and NA of the virus isolated from Guangdong (GDK6) differed from those in the majority of H6 viruses. Substitutions A138S and G228S at highly conserved positions in the receptor binding region of GD6K HA would be expected to favor binding of α2,6-sialic acids (SAs) (31–33). The 11-amino-acid deletion (positions 59 to 69) in the NA stalk region should affect NA activity (34). These factors would alter the balance between the HA and NA (35, 36) and may have facilitated the infection by GDK6 in swine.
As seen in other swine or avian H6N6 viruses (9, 16), the GDK6 virus was almost avirulent in mice although it replicated efficiently in their lungs. Whether and, if so, how it could adapt to become pathogenic in mice and potentially other mammals was not known. After multiple serial passages of GDK6 virus in mouse lungs, a virulent mouse-adapted virus GDK6-MA was generated. This virus differed from GDK6 by 5 amino acid substitutions in three gene segments (PB2 E627K, PA I38M, and HA L111F, H156N, and S263R) and replicated to higher titers than GDK6 in the lungs of infected mice.
Consistent with earlier studies (29), a recombinant GDK6 virus containing PB2(E627K), rGDK6-MA (PB2), had enhanced pathogenicity and replication in mice. Polymerase activity increased when PB2(627K) was present. While recombinant GDK6 viruses containing either the mouse-adapted HA or PA segment alone were not as virulent, the virus containing both segments had similar virulence to rGDK6-MA. Further investigation of the HA substitutions in the mouse-adapted virus showed that PA(I38M) and HA(H156N S263R) were responsible for the increased virulence in mice.
Methionine at position 38 of PA is rare in influenza A viruses, with no H6 and only seven other viruses having this residue. The N-terminal domain of PA (positions 1 to 256) is involved in protein stability, endonuclease activity, cap binding, promoter binding, and induction of the nuclear accumulation of PB1 (37–39). The PA(I38M) mutation increased transcription activity by the viral polymerase in 293T cells in vitro but did not influence the replication kinetics in MDCK or A549 cells.
The HA H156N and S263R substitutions that contribute to virulence are located on the globular head of HA1. H156N results in a potential N-linked glycosylation site not seen in other H6 viruses. Glycosylation in this region of HA can reduce receptor avidity (40) or reduce dependence on the NA for release from the receptor (41). This may be beneficial for HA-NA balance in the presence of a stalk deletion in the NA (41), which occurs in the NA of GDK6. H156N in HA, or the potentially additional N-linked glycoside, significantly enhanced the cytopathic effect on MDCK cells and increased early-stage viral replication in MDCK cells, probably reflecting a contribution to the HA-NA balance of the virus in mammalian systems. HA(S263R) did not enhance viral replication either in vivo or in vitro, and the mechanism by which it contributes to the enhanced virulence of rGDK6-MA (HA, PA) is unknown.
The viral titers in the lungs of mice infected with the rGDK6-MA (HA, PA) or rGDK6-MA (PB2) were similar to the titer in rGDK6-infected mice at 3 days p.i. This suggests that the increased virulence of the mouse-adapted virus did not directly result from the viral titer in the lungs, as has been reported previously (20, 29, 42).
The adaptation and pathogenicity of an influenza virus to a new host are clearly polygenic effects, as demonstrated here and in other studies (22, 42–44). Here, we found that while the individual mouse-adapted HA or PA segment only moderately enhanced the virulence of the GDK6 virus, the combination of these segments led to a virulent virus. We have identified key amino acids in the PA and HA proteins that significantly enhance viral pathogenicity in mice and can compensate in mammals for the lack of 627K in PB2. Our study suggests that multiple strategies can be utilized by influenza viruses for efficient adaptation in mammals, and it has identified a novel virulence determinant for mammalian influenza viruses.
ACKNOWLEDGMENTS
We acknowledge scientific support from George Fu Gao, Institute of Microbiology, Chinese Academy of Sciences, and the colleagues of Nanchang Center for Disease Control and Prevention. We thank Yi Guan, University of Hong Kong, for his helpful critical review and revision of the manuscript.
This work was supported by the National Key Basic Research Program (Project 973) of China (grant no. 2011CB504700-G), the Science and Technology Projects of Guangdong province (2012B020306005), the International Science and Technology Cooperation Program (2010DFB33920), and the Modern Agricultural Industry Technology System (CARS-36).
The funding organizations had no role in the study design, data collection and analysis, ownership of the materials, or preparation of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 1 October 2014
REFERENCES
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long LP, Cai Z, Zhu X, Liao M, Wan XF. 2010. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect. Genet. Evol. 10:1286–1288. 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822. 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinde H, Read DH, Daft BM, Hammarlund M, Moore J, Uzal F, Mukai J, Woolcock P. 2003. The occurrence of avian influenza A subtype H6N2 in commercial layer flocks in Southern California (2000-02): clinicopathologic findings. Avian Dis. 47:1214–1218. 10.1637/0005-2086-47.s3.1214. [DOI] [PubMed] [Google Scholar]
- 5.Webby RJ, Woolcock PR, Krauss SL, Walker DB, Chin PS, Shortridge KF, Webster RG. 2003. Multiple genotypes of nonpathogenic H6N2 influenza viruses isolated from chickens in California. Avian Dis. 47:905–910. 10.1637/0005-2086-47.s3.905. [DOI] [PubMed] [Google Scholar]
- 6.Rimondi A, Xu K, Craig MI, Shao H, Ferreyra H, Rago MV, Romano M, Uhart M, Sutton T, Ferrero A, Perez DR, Pereda A. 2011. Phylogenetic analysis of H6 influenza viruses isolated from rosy-billed pochards (Netta peposaca) in Argentina reveal the presence of different HA gene clusters. J. Virol. 85:13354–13362. 10.1128/JVI.05946-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang K, Zhu H, Fan X, Wang J, Cheung CL, Duan L, Hong W, Liu Y, Li L, Smith DK, Chen H, Webster RG, Webby RJ, Peiris M, Guan Y. 2012. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 86:6075–6083. 10.1128/JVI.06389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Deng G, Shi J, Luo W, Zhang G, Zhang Q, Liu L, Jiang Y, Li C, Sriwilaijaroen N, Hiramatsu H, Suzuki Y, Kawaoka Y, Chen H. 2014. H6 influenza viruses pose a potential threat to human health. J. Virol. 88:3953–3964. 10.1128/JVI.03292-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers KP, Setterquist SF, Capuano AW, Gray GC. 2007. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin. Infect. Dis. 45:4–9. 10.1086/518579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su S, Qi W, Chen J, Zhu W, Huang Z, Xie J, Zhang G. 2013. Seroepidemiological evidence of avian influenza A virus transmission to pigs in southern China. J. Clin. Microbiol. 51:601–602. 10.1128/JCM.02625-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beare AS, Webster RG. 1991. Replication of avian influenza viruses in humans. Arch. Virol. 119:37–42. 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 13.Gillim-Ross L, Santos C, Chen Z, Aspelund A, Yang CF, Ye D, Jin H, Kemble G, Subbarao K. 2008. Avian influenza H6 viruses productively infect and cause illness in mice and ferrets. J. Virol. 82:10854–10863. 10.1128/JVI.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam JH, Kim EH, Song D, Choi YK, Kim JK, Poo H. 2011. Emergence of mammalian species-infectious and -pathogenic avian influenza H6N5 virus with no evidence of adaptation. J. Virol. 85:13271–13277. 10.1128/JVI.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Kong W, Qi W, Long L-P, Cao Z, Huang L, Qi H, Cao N, Wang W, Zhao F, Ning Z, Liao M, Wan X-F. 2011. Identification of an H6N6 swine influenza virus in southern China. Infect. Genet. Evol. 11:1174–1177. 10.1016/j.meegid.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, Chen C, Huang J, Wang Y, Peng D, Liu X. 2013. Characterisation of one H6N6 influenza virus isolated from swine in China. Res. Vet. Sci. 95:434–436. 10.1016/j.rvsc.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Wei S-H, Yang J-R, Wu H-S, Chang M-C, Lin J-S, Lin C-Y, Liu Y-L, Lo Y-C, Yang C-H, Chuang J-H, Lin M-C, Chung W-C, Liao C-H, Lee M-S, Huang W-T, Chen P-J, Liu M-T, Chang F-Y. 2013. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir. Med. 1:771–778. 10.1016/S2213-2600(13)70221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown EG, Liu H, Kit LC, Baird S, Nesrallah M. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc. Natl. Acad. Sci. U. S. A. 98:6883–6888. 10.1073/pnas.111165798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84:8607–8616. 10.1128/JVI.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin PS, Peiris M, Shortridge KF, Webster RG. 2000. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? J. Virol. 74:6309–6315. 10.1128/JVI.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Sun Y, Xu Q, Tan Y, Pu J, Yang H, Brown EG, Liu J. 2012. Mouse-adapted H9N2 influenza A virus PB2 protein M147L and E627K mutations are critical for high virulence. PLoS One 7:e40752. 10.1371/journal.pone.0040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Y, Wang H, Chen Q, Zhang H, Zhang T, Chen J, Xu B, Sun B, Chen Z. 2013. Characterization of low-pathogenic H6N6 avian influenza viruses in central China. Arch. Virol. 158:367–377. 10.1007/s00705-012-1496-3. [DOI] [PubMed] [Google Scholar]
- 23.Matrosovich M, Matrosovich T, Garten W, Klenk HD. 2006. New low-viscosity overlay medium for viral plaque assays. Virol. J. 3:63. 10.1186/1743-422X-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289. 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113. 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinya K, Watanabe S, Ito T, Kasai N, Kawaoka Y. 2007. Adaptation of an H7N7 equine influenza A virus in mice. J. Gen. Virol. 88:547–553. 10.1099/vir.0.82411-0. [DOI] [PubMed] [Google Scholar]
- 29.Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842. 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 30.Tian J, Qi W, Li X, He J, Jiao P, Zhang C, Liu GQ, Liao M. 2012. A single E627K mutation in the PB2 protein of H9N2 avian influenza virus increases virulence by inducing higher glucocorticoids (GCs) level. PLoS One 7:e38233. 10.1371/journal.pone.0038233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234. 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 32.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. 2007. Receptor binding specificity of recent human H3N2 influenza viruses. Virol. J. 4:42. 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Z, Zhou H, Kim L, Jin H. 2012. The receptor binding specificity of the live attenuated influenza H2 and H6 vaccine viruses contributes to vaccine immunogenicity and protection in ferrets. J. Virol. 86:2780–2786. 10.1128/JVI.06219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castrucci MR, Kawaoka Y. 1993. Biologic importance of neuraminidase stalk length in influenza A virus. J. Virol. 67:759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J. Virol. 74:6015–6020. 10.1128/JVI.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J. Virol. 86:9221–9232. 10.1128/JVI.00697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara K, Schmidt FI, Crow M, Brownlee GG. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J. Virol. 80:7789–7798. 10.1128/JVI.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fodor E, Smith M. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 78:9144–9153. 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 40.Das SR, Hensley SE, David A, Schmidt L, Gibbs JS, Puigbo P, Ince WL, Bennink JR, Yewdell JW. 2011. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. U. S. A. 108:E1417–E1422. 10.1073/pnas.1108754108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baigent SJ, McCauley JW. 2001. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 79:177–185. 10.1016/S0168-1702(01)00272-6. [DOI] [PubMed] [Google Scholar]
- 42.Sakabe S, Ozawa M, Takano R, Iwastuki-Horimoto K, Kawaoka Y. 2011. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res. 158:124–129. 10.1016/j.virusres.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Qiao C, Marjuki H, Bawa B, Ma J, Guillossou S, Webby RJ, Richt JA, Ma W. 2012. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J. Virol. 86:1233–1237. 10.1128/JVI.05699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imai H, Shinya K, Takano R, Kiso M, Muramoto Y, Sakabe S, Murakami S, Ito M, Yamada S, Le MT, Nidom CA, Sakai-Tagawa Y, Takahashi K, Omori Y, Noda T, Shimojima M, Kakugawa S, Goto H, Iwatsuki-Horimoto K, Horimoto T, Kawaoka Y. 2010. The HA and NS genes of human H5N1 influenza A virus contribute to high virulence in ferrets. PLoS Pathog. 6:e1001106. 10.1371/journal.ppat.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]