Abstract
Streptococcus gordonii is a commensal species of human oral flora. It initiates dental biofilm formation and provides binding sites for later colonizers to attach to and generate mature biofilm. Smoking is the second highest risk factor for periodontal disease, and cigarette smoke extract has been reported to facilitate Porphyromonas gingivalis-S. gordonii dual-species biofilm formation. Our hypothesis is that nicotine, one of the most important and active components of tobacco, stimulates S. gordonii multiplication and aggregation. In the present study, S. gordonii planktonic cell growth (kinetic absorbance and CFU), biofilm formation (crystal violet stain and confocal laser scanning microscopy [CLSM]), aggregation with/without sucrose, and 11 genes that encode binding proteins or regulators of gene expression were investigated. Results demonstrated planktonic cell growth was stimulated by 1 to 4 mg/ml nicotine treatment. Biofilm formation was increased at 0.5 to 4 mg/ml nicotine. CLSM indicated bacterial cell mass was increased by 2 and 4 mg/ml nicotine, but biofilm extracellular polysaccharide was not significantly affected by nicotine. Cell aggregation was upregulated by 4, 8, and 16 mg/ml nicotine with sucrose and by 16 mg/ml nicotine without sucrose. Quantitative reverse transcriptase PCR indicated S. gordonii abpA, scaA, ccpA, and srtA were upregulated in planktonic cells by 2 mg/ml nicotine. In conclusion, nicotine stimulates S. gordonii planktonic cell growth, biofilm formation, aggregation, and gene expression of binding proteins. Those effects may promote later pathogen attachment to tooth surfaces, the accumulation of tooth calculus, and the development of periodontal disease in cigarette smokers.
INTRODUCTION
Smoking is strongly associated with periodontal disease. Smokers have more supra- and subgingival calculus than nonsmokers (1, 2), and smoking is ranked after diabetes mellitus as the second highest risk factor for periodontal diseases (3). Generally, smokers usually have more calculus deposit than nonsmokers, and the calculus from smokers is stiffer and more tightly attached to teeth than that from nonsmokers. Dental calculus is a form of hardened oral biofilm. Its major components are fossilized bacteria and calcium phosphate salts. Acquired pellicle attaching to a tooth surface is the initial step for dental biofilm formation, followed by bacterial cell attachment to the acquired pellicle. After that, calcium phosphate salts bind to the bacterial layer through electrostatic attraction (4). Subgingival calculus is an irritant for local chronic inflammation which continuously destroys periodontal tissues (3).
Streptococcus gordonii, a Gram-positive facultative anaerobe, is a commensal species of human oral flora. It plays a central role in initiating dental biofilm formation and providing binding sites for later colonizers, such as Porphyromonas gingivalis, to attach to and generate mature biofilm. Its DNA has been detected in dental calculus (5). Cigarette smoke extract stimulates P. gingivalis-S. gordonii biofilm formation, and it potentially contributes to periodontal disease development (6). Higher counts of S. gordonii also are associated with periodontal inflammation (7).
The specific aim of this study was to investigate the effect of nicotine, an alkaloid in tobacco, on S. gordonii planktonic cell growth, biofilm formation, aggregation, and binding-related gene expression. Our null hypothesis was that nicotine has no effect on S. gordonii.
MATERIALS AND METHODS
Bacterial strain and growth media.
S. gordonii (ATCC 35105) was used in the present study. S. gordonii was stored at −80°C before use. To initiate the bacterial culture, S. gordonii was streaked on anaerobic blood agar plates (bioMerieux, Inc., Durham, NC, USA), followed by overnight growth in BBL trypticase soy broth (TSB; Becton, Dickinson, and Company, Sparks, MD, USA). Unless stated otherwise, the growing atmosphere was 5% CO2 at 37°C. Nicotine (Sigma-Aldrich, St. Louis, MO, USA) and sucrose (Fisher Scientific, Fair Lawn, NJ, USA) were used. The optical densities (OD) were measured by a spectrophotometer (Spectramax 190; Molecular Devices, Sunnyvale, CA, USA).
Planktonic cell growth.
For kinetic planktonic cell growth, overnight-grown S. gordonii (5 × 107 CFU/ml; optical density at 595 nm [OD595] of 0.45) cells were diluted 1:100 in TSB with 0, 0.25, 0.5, 1, 2, and 4 mg/ml nicotine. Triplicate samples were grown in sterile 96-well flat-bottom microtiter plates (Fisher Scientific, Newark, DE, USA) for 12, 24, and 48 h. After incubation, planktonic cells were carefully transferred to another microtiter plate to eliminate the reading error generated from biofilm formed on the bottom of the original microtiter plates. The planktonic culture turbidity was read at OD595 by a spectrophotometer.
To detect the CFU of S. gordonii with nicotine treatment, overnight-grown S. gordonii cells were diluted 1:100 in TSB with 0, 0.25, 0.5, 1, 2, and 4 mg/ml nicotine and grown in sterile 96-well flat-bottom microtiter plates for 24 h. After incubation, planktonic cells were diluted in saline (0.9% NaCl) at an appropriate dilution (1:10,000 in the present study) and spiral plated on tryptic soy agar (TSA; Difco, Detroit, MI, USA) plates by a Spiral System automatic spiral plater (Spiral Biotech, Inc., Bethesda, MD, USA). The plates were incubated for 48 h, and colonies were counted by a ProtoCOL automated colony counter (Synbiosis, Inc., Frederick, MD, USA). The log10 of CFU/ml was calculated.
Biofilm formation.
Overnight-grown S. gordonii cells were diluted 1:100 in TSB plus 1% sucrose (TSBS) with 0, 0.25, 0.5, 1, 2, and 4 mg/ml nicotine. Triplicate samples were grown in a sterile 96-well flat-bottom microtiter plate for 24 h. Biofilm formation was determined by a crystal violet assay described previously (8). Briefly, biofilm was fixed with formaldehyde, stained with crystal violet, and washed twice with saline to remove unbound crystal violet. Bound crystal violet was extracted by 2-propanol. The extracted crystal violet in 2-propanol was further diluted 1:5 with 2-propanol, and its absorbance was read at OD490 by a spectrophotometer.
Aggregation assay.
The aggregation assay was modified from Xu et al. as described previously (9). Briefly, late-log-phase S. gordonii cells (OD595 of 0.4) were harvested by centrifugation, washed twice with sterile phosphate-buffered saline (PBS), and diluted in PBS to an OD595 of 0.85. One hundred fifty μl of the bacterial suspension was mixed with 150 μl of 0, 4, 8, 16, and 32 mg/ml nicotine in PBS or in PBS with 2% sucrose (PBS+S) in microtiter plates, yielding final nicotine concentrations of 0 to 16 mg/ml with or without 1% sucrose. The plates were incubated for 2 h at 37°C. One hundred twenty μl of undisturbed supernatants of the mixture was carefully transferred to another microtiter plate, and the absorbance was read at OD595. The aggregation formula was calculated as aggregation = (1 − [ODexp − ODblk′]/[ODctl − ODblk]) × 100, where ODexp was the nicotine-treated group value, ODctl was the control (no-nicotine) group value, and ODblk′ and ODblk were the blank without bacteria suspension of nicotine-treated and control groups, respectively.
Confocal laser scanning microscope (CLSM).
The confocal microscope assay was modified from Xiao and Koo as described previously (10). Overnight-grown S. gordonii cells were diluted 1:100 in 0, 1, 2, and 4 mg/ml nicotine in TSBS with 0.1 μM Alexa Fluor 647 fluorescently labeled dextran conjugate (Molecular Probes Inc., Eugene, OR, USA). This dextran was used as a glucosyltransferase (Gtf) primer by bacteria in extracellular polysaccharide (EPS) synthesis (10). Cells were grown in four-well Lab-Tek chamber slides (Thermo Fisher Scientific, Rochester, NY) for 24 h. Biofilm was washed twice with distilled H2O (dH2O), incubated with 0.1 μM SYTO 9 green nucleic acid stain (Molecular Probes Inc., Eugene, OR, USA) for 20 min in the dark at room temperature, and washed again with dH2O. After slides were air dried, biofilm was covered by ProLong Gold antifade reagent (Molecular Probes Inc.). Green fluorescence for bacteria and red fluorescence for EPS were captured by an Olympus FV1000-MPE confocal/multiphoton microscope (Olympus Corp., USA) with Olympus FV10-ASW software (Olympus Corp., USA) in the Division of Nephrology, Indiana Center for Biological Microscopy, Indiana University. Biofilm mass and coverage of biofilm were calculated based on the principles described earlier (11) by MATLAB R2012a (The MathWorks, Inc.) codes (see S1 in the supplemental material). Biofilm thickness was recorded by the microscope and software.
qRT-PCR.
The quantitative reverse transcriptase-PCR (qRT-PCR) assay was modified from Xu et al. as described previously (9). Briefly, overnight-grown S. gordonii cells were diluted 1:100 in TSBS with 0 or 2 mg/ml nicotine for 8 h. Planktonic cells were harvested, washed three times by PBS, stabilized by 1 ml of RNAprotect bacterial reagent (Qiagen, MD, USA), and suspended in 100 μl lysis buffer (30 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 15 mg/ml lysozyme) with 10 μl mutanolysin (Sigma-Aldrich, MO, USA) and 15 μl proteinase K (Qiagen). Samples were incubated for 90 min at 37°C with agitation and sonicated for 10 s with 5 cycles (52% amplitude; Model 500 sonic dismembrator; Fisher Scientific). Bacterial RNA was extracted using an RNeasy minikit (Qiagen). The quality and quantity of the RNA samples were determined by a NanoDrop 2000 (Thermo Fisher Scientific Inc., USA). RNA samples were incubated at 37°C for 30 min with DNase I and appropriate buffer (New England BioLabs Inc., Ipswich, MA) to remove genomic DNA. The reaction was stopped by adding 0.5 mM EDTA and incubating at 75°C for 10 min. Twenty μg RNA was used to synthesize cDNA by a high-capacity cDNA reverse transcription kit with random primers (Applied Biosystems, Life Technologies Corp., CA, USA), and the manufacturer's protocol was followed. Two μg of cDNA samples and appropriate primers (0.375 μM) (Table 1) were processed with Fast SYBR green master mix (Applied Biosystems). The qRT-PCR amplification was performed on an ABI Prism 7000 sequence detection system (Applied Biosystems) with default program settings. The original RNA samples and distilled H2O were loaded with Faster SYBR green master mix to serve as an operative system control. The value of 2−ΔCT instead of 2−ΔΔCT of each sample was recorded (where CT represents the qRT-PCR amplifying cycle number once the threshold is reached), and the value for 16S rRNA data equals 1. Since 16S rRNA has the highest expression, all of the other gene results were less than 1. Using 2−ΔCT instead of 2−ΔΔCT makes between-gene comparisons available.
TABLE 1.
Primers used in this study
| Primer | Sequence | Description | Source or reference |
|---|---|---|---|
| Sg 16S-F | 5′-AAGCAACGCGAAGAACCTTA-3′ | rRNA-16S rRNA | Zheng et al. (27) |
| Sg 16S-R | 5′-GTCTCGCTAGAGTGCCCAAC-3′ | ||
| apbA-F | 5′-TGATGCAGTTGAAGGTGGAA-3′ | Amylase-binding protein AbpA | |
| abpA-R | 5′-TAGCTGCACCAACACGTTTC-3′ | ||
| abpB-F | 5′-CAAAAACTCCGGAAAAACCA-3′ | Amylase-binding protein AbpB | |
| abpB-R | 5′-GGAGCTTGACTCGGTTCTTG-3′ | ||
| scaA-F | 5′-CACCGAAGAAGAAGGCACTC-3′ | Metal ABC transporter substrate-binding lipoprotein | |
| scaA-R | 5′-TGTCTCCATCTTCGCCTTTT-3′ | ||
| gtfG-F | 5′-CCAAGACTTTGCAACCCGTG-3′ | Glucosyltransferase G | |
| gtfG-R | 5′-GACACTGTGGAGAGCACGAA-3′ | ||
| hsa-F | 5′-CAGAGCTGCAAATCCAAACA-3′ | Streptococcal hemagglutinin | |
| hsa-R | 5′-GCCGAGATACTTGCGCTTAC-3′ | ||
| cshA-F | 5′-CAGACGATGCAACCCCTATT-3′ | Surface-associated protein CshA | |
| cshA-R | 5′-TAACGGTCAAGGTCACCACA-3′ | ||
| cshB-F | 5′-CGTTGTTCAGCAAGGATCAA-3′ | Surface-associated protein CshB | |
| cshB-R | 5′-GCCGTTCTGTTGTCCAGTAG-3′ | ||
| srtA-F | 5′-CCATCAGCTGCATCGTCAAC-3′ | Sortase A | This study |
| srtA-R | 5′-TGGGAAAAGGCAACTACGCT-3′ | ||
| CcpA-F | 5′-GGGGTTTCTATGGCGACAGT-3′ | Catabolite control protein A | |
| CcpA-R | 5′-GCTAGCCAAACCACGAGCTA-3′ | ||
| sspA-F | 5′-GGTTGTCCAGCCAGCCTTAT-3′ | Streptococcal surface protein A | |
| sspA-R | 5′-CTGCCTTAATTTGGGCCTGC-3′ | ||
| sspB-F | 5′-TACCAAGCTAAGCTAGCGGC-3′ | Streptococcal surface protein B | |
| sspB-R | 5′-TTACTCTCCGCTTCTGCGAG-3′ |
Statistical analysis.
Each experiment was independently repeated at least three times. Two-way analysis of variance (ANOVA) and the Hole-Sidak multiple-comparison test were used for kinetic planktonic cell growth analysis by SigmaPlot software (version 12.0; Systat Software Inc., San Jose, CA). One-way ANOVA and post hoc Tukey's multiple-comparison test were used for planktonic cell growth (CFU/ml), biofilm formation, aggregation, and CLSM data statistical analysis by SPSS software (version 20.0; SPSS, Chicago, IL, USA). Student t test was used for qRT-PCR data analysis. P < 0.05 was considered significantly different.
RESULTS
Nicotine stimulates planktonic cell growth, biofilm formation, and sucrose-dependent aggregation.
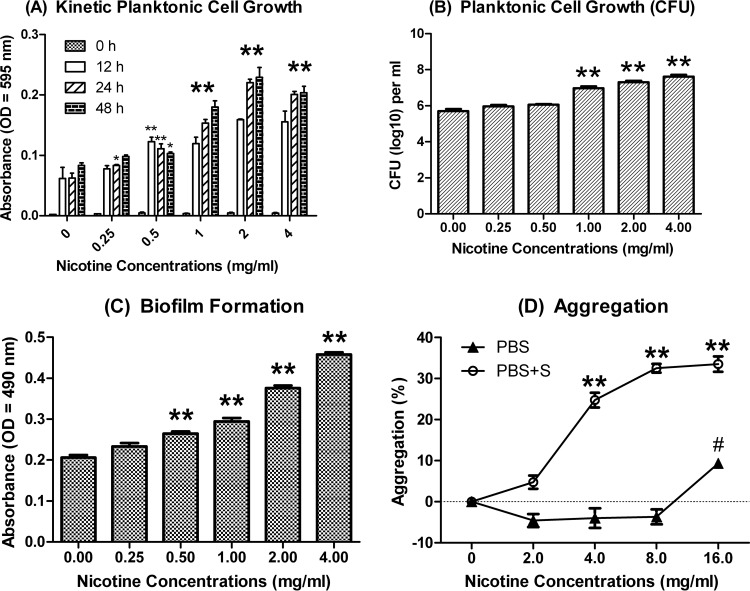
Nicotine significantly stimulates S. gordonii kinetic planktonic cell growth at 0.5, 1, 2, and 4 mg/ml through the three different time points (12, 24, and 48 h) (Fig. 1A). The mean CFU/ml values at 24 h were (5.33 ± 2.21) × 105, (9.85 ± 3.61) × 106, (2.10 ± 0.74) × 107, and (4.40 ± 1.97) × 107 for 0, 1, 2, and 4 mg/ml nicotine-treated groups, respectively (Fig. 1B). Nicotine significantly increased S. gordonii biofilm formation at 0.5, 1, 2, and 4 mg/ml (P ≤ 0.01). The mean crystal violet assay results at 24 h were 0.21 ± 0.01, 0.26 ± 0.01, 0.29 ± 0.01, 0.38 ± 0.01, and 0.45 ± 0.01 relative light units (RLU) for 0, 0.5, 1, 2, and 4 mg/ml nicotine-treated groups, respectively (Fig. 1C). Nicotine significantly promoted S. gordonii sucrose-independent aggregation at 16 mg/ml and sucrose-dependent aggregation at 4, 8, and 16 mg/ml. With sucrose, nicotine promoted aggregation by 24.76, 32.51, and 33.53% at 4, 8, and 16 mg/ml, respectively (Fig. 1D). Therefore, the effect of nicotine on S. gordonii aggregation is stronger in the presence of sucrose than without it.
FIG 1.
S. gordonii planktonic cell growth, biofilm formation, and aggregation with nicotine. (A) Kinetic planktonic cell growth (absorbance). S. gordonii cells grown overnight were treated with 0 to 4 mg/ml nicotine for 0, 12, 24, and 48 h in TSB. Planktonic cell cultures were transferred to another microtiter plate, and the OD595 was read. Results indicated S. gordonii planktonic cell growth was increased at 0.5, 1, 2, and 4 mg/ml nicotine at all time points. Oversized asterisks demonstrate that all of the subgroups were significantly different from their nontreated controls, while smaller asterisks demonstrate the result was significantly different from that for its nontreated controls at the same time point. *, P < 0.05; **, P < 0.01. (B) Planktonic cell growth (CFU). S. gordonii cells grown overnight were treated with 0 to 4 mg/ml nicotine for 24 h in TSB. Planktonic cells were diluted and spiral plated on TSA agar plates. Results indicated S. gordonii planktonic cell growth was increased in a nicotine concentration-dependent manner. **, P < 0.01. (C) Biofilm formation. S. gordonii cells grown overnight were treated with 0 to 4 mg/ml nicotine for 24 h in TSB plus 1% sucrose (TSBS). The volume of biofilm was determined by crystal violet assay. Results indicated S. gordonii biofilm formation was increased in a nicotine concentration-dependent manner. **, P < 0.01. (D) Aggregation. S. gordonii cells in exponential phase were washed, diluted to an OD595 of 0.85, and suspended in PBS with 0, 2, 4, 8, and 16 mg/ml nicotine with or without 1% sucrose. After 2 h of incubation, the OD595 of the supernatant was measured. With sucrose, cell aggregation was detected in the 4 to 16 mg/ml nicotine groups. Without sucrose, aggregation was detected only in the 16 mg/ml nicotine group. In the sucrose group, P < 0.01 (**). In the no-sucrose group, P < 0.05 (#).
CLSM.
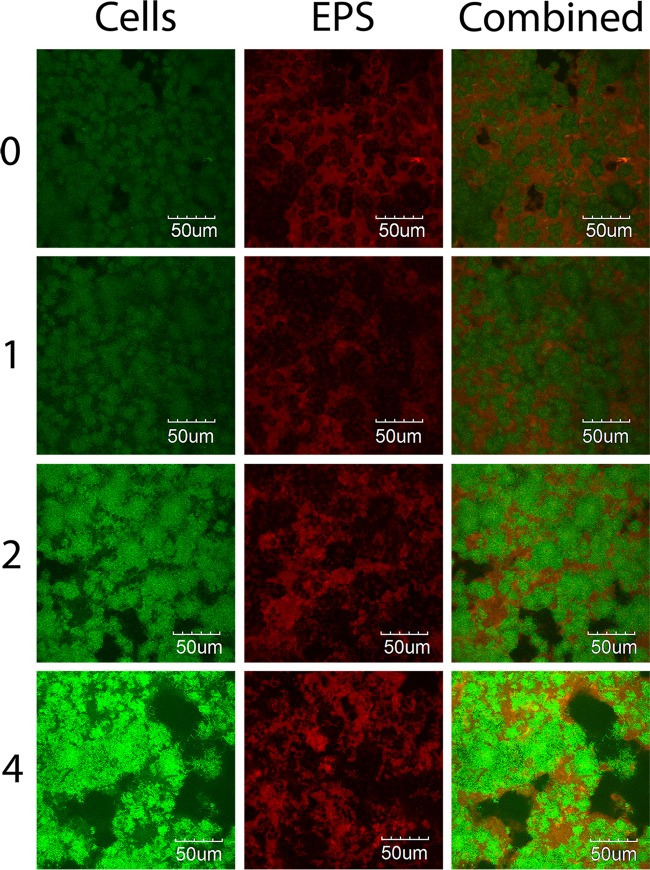
The major components of S. gordonii biofilm are bacterial cells and EPS. The crystal violet assay cannot differentiate one from the other. Therefore, a CLSM study was conducted to investigate how those two components are affected by nicotine. From the images, both bacterial cells and EPS were increased in a nicotine-dependent manner. At higher nicotine concentrations, bacterial cells were more aggregated, leaving more blank areas outside thicker biofilm stacks (Fig. 2). The green fluorescence intensity, which represents bacterial cell volume, was significantly increased at nicotine concentrations of 2 and 4 mg/ml (Table 2). The green fluorescence coverage, which represents the area of biofilm surface coverage, was increased by nicotine, but the increase was not statistically significant. The volume/coverage ratio of green fluorescence, which reflects the cell numbers in one unit area and partially demonstrates cell aggregation, was increased with nicotine treatment. The EPS volume was increased by nicotine, but it was not significant. The EPS coverage was not changed, while the EPS volume/coverage ratio was slightly, but not significantly, increased (Table 2). The biofilm thickness indicated an increased trend from 25.0 to 32.5 μm by nicotine, but the difference was not statistically significant (Table 2).
FIG 2.
S. gordonii bacterial cell multiplication and EPS synthesis with nicotine. S. gordonii cells grown overnight were treated with 0 to 4 mg/ml nicotine for 24 h in TSBS with 0.1 μM Alexa Fluor 647 dextran-conjugated red fluorescence. The biofilm was washed and further incubated with 0.1 μM SYTO 9 nucleic acid green fluorescence. Bacterial cells were labeled green, and EPS was labeled red. Results indicate that bacterial cell multiplication and EPS synthesis were stimulated by nicotine.
TABLE 2.
Biofilm volume and coverage of S. gordonii bacterial cells and EPS treated with nicotine
| Nicotine (mg/ml) | Bacteria |
EPS |
Thickness (μm) | ||||
|---|---|---|---|---|---|---|---|
| Volumea (μm2/μm) | Coverage (%) | V/C ratiob | Volume (μm2/μm) | Coverage (%) | V/C ratio | ||
| 0 | 45.4 ± 6.1 | 71.1 ± 17.7 | 63.87 | 45.9 ± 5.9 | 62.8 ± 11.8 | 73.14 | 25.0 ± 2.1 |
| 1 | 55.1 ± 19.5 | 72.1 ± 21.2 | 76.42 | 50.9 ± 9.9 | 67.4 ± 8.5 | 75.59 | 29.5 ± 2.5 |
| 2 | 96.0 ± 10.7* | 90.5 ± 4.2 | 106.17 | 47.8 ± 2.0 | 60.8 ± 4.0 | 78.60 | 31.5 ± 3.5 |
| 4 | 107.3 ± 30.6* | 97.2 ± 3.3 | 110.42 | 57.8 ± 6.8 | 65.1 ± 3.9 | 88.74 | 32.5 ± 7.1 |
Asterisks indicate significant difference (P ≤ 0.05) from the value for the no-nicotine control sample.
V/C ratio is the volume/coverage ratio, which reflects the cell numbers in one unit area.
qRT-PCR.
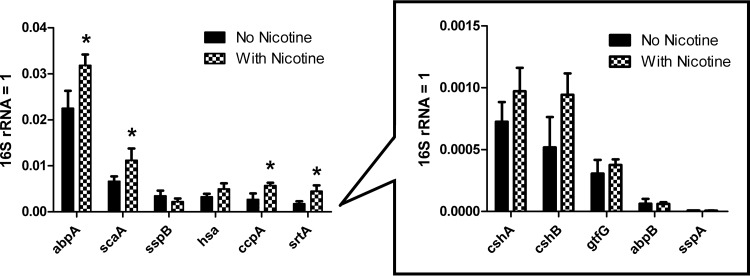
The amounts of RNA obtained from control- and nicotine-treated samples were 917 to 1,212 and 1,558 to 1,810 ng/μl, respectively. The 260/280 ratio (the ratio of absorbances at wavelengths of 260 and 280 nm) was above 2.0 in every sample. The levels of expression of planktonic cell genes abpA, scaA, ccpA, and srtA were significantly upregulated by nicotine treatment (Fig. 3). Although there were only 4 out of 11 that provided significant differences, those 4 genes accounted for the majority of the total gene expression. For better fold change comprehension, the traditional 2−ΔΔCT value was calculated (see S2 in the supplemental material). The S. gordonii binding-related genes generally were upregulated by nicotine because 10 of those 11 genes exhibited an increase, although only four demonstrated significant increases. The expression of sspB was slightly decreased but not significantly so. Since the same amount of RNA was used for reverse transcription and the raw 16S rRNA CT value was slightly higher in the control (7.08) than in the nicotine-treated group (6.38), the 16S rRNA gene was not downregulated by nicotine.
FIG 3.
S. gordonii gene expression with nicotine. Overnight S. gordonii cells were treated with 0 and 2 mg/ml nicotine for 8 h in TSBS, and planktonic cells were harvested for qRT-PCR. The value for 2−ΔCT instead of 2−ΔΔCT of each sample was recorded. The 16S rRNA data were standardized to 1, and all other gene data were less than 1. Using 2−ΔCT instead of 2−ΔΔCT makes between-gene comparisons available. The gene expression levels of the no-nicotine controls were ranked from highest to lowest (left to right). Because the gene expression was very low in cshA, cshB, gtfG, abpB, and sspA, their results were placed in another figure with a different scale than the original one. Results indicated nicotine significantly upregulated abpA, scaA, ccpA, and srtA expression. Although there were only 4 out of 11 genes that indicated significant changes, those four genes accounted for the majority of the total gene expression. *, P < 0.05.
DISCUSSION
The nicotine concentration in human saliva of smokers varies from 0 to 2.27 mg/ml (12–15). It is affected by the inhalation characteristics of the smokers (noninhalers, slight inhalers, medium inhalers, and deep inhalers), saliva properties (unstimulated or stimulated), and cigarette consumption (13, 15). The nicotine concentrations used in the present study were mostly in the 0 to 2.27 mg/ml range, except the aggregation assay.
S. gordonii is critical in initiating dental plaque and providing subsequent binding sites for P. gingivalis. Without S. gordonii, P. gingivalis can barely establish biofilm under a low challenging shear force. However, with S. gordonii, P. gingivalis successfully establishes biofilm under the same shear force (16). P. gingivalis fimbriae, S. gordonii autoinducer 2, and streptococcal surface protein B (SspB) have been reported to be involved in the S. gordonii-P. gingivalis interaction (17–19). Among them, P. gingivalis major fimbrial antigen (FimA) instead of the minor fimbrial antigen (Mfa1) was demonstrated to be responsible for the P. gingivalis-S. gordonii interaction with cigarette smoke extract treatment (6).
The doubling time of S. gordonii (60 min) is shorter than that of many other oral bacterial species, such as Streptococcus mutans (73 min) and Streptococcus sobrinus (64 min) (20). Faster multiplication is one of the mechanisms bacteria use to compete with other species; bacteria gain more growing space and nutrients by multiplying faster. In the present study, nicotine was demonstrated to stimulate S. gordonii biofilm formation and planktonic growth, which means nicotine is a preferred environmental factor for S. gordonii. Nicotine has a similar stimulation effect on S. mutans biofilm formation, but it has no effect or even reduces S. mutans planktonic cell growth (8). One bacterial nicotinic acetylcholine receptor (nAChR) homologue, which is a prokaryotic proton-gated ion channel, has been found in several bacterial species (21). Previous study in our laboratory has found significant homology of the S. mutans genome with the β2 subunit gene of nAChRs, and β2 subunit gene activity was enhanced at 1 mg/ml nicotine (A. Jouravlev, K. DelaCruz, R. L. Gregory, unpublished data). Nicotine may regulate S. gordonii multiplication-related downstream targets of this signaling pathway.
From the present study, we established that nicotine promotes the formation of S. gordonii biofilm. From the CLSM experiments, both bacterial cell numbers and EPS synthesis within biofilm were elevated, although the latter was not statistically significant. Biofilm accumulation has two major sources of gaining cell numbers, cells multiplying inside biofilm and cells attaching to biofilm from the environment. The former source can be explained by the same rationale, as it multiplies in the planktonic form. The latter can be explained by two reasons: since there are more planktonic cells present in the environment, there are more cells available for binding, and the surface-binding proteins could be upregulated by nicotine treatment, resulting in better binding ability. This will be discussed below. In this regard, nicotine stimulates S. mutans bacterial biofilm formation (8) and facilitates S. mutans to compete with Streptococcus sanguinis, a commensal species in the dental flora (22).
The major components of extracellular polymeric components are EPS, proteins, extracellular DNA (eDNA), and lipids. Among these, EPS and proteins account for 75 to 89% of extracellular polymeric components (23). eDNA promotes biofilm formation and stability (24, 25). Glucosyltransferase G (GtfG) of S. gordonii utilizes monosaccharides catabolized from sucrose and synthesizes water-insoluble and water-soluble glucans (26). Nicotine also has been reported to stimulate EPS synthesis of S. mutans biofilm (22). Since S. gordonii and S. mutans belong to the same genus, they may share similar nicotine-related receptors and pathways that lead to increased EPS synthesis.
Our data demonstrate that the effect of nicotine on S. gordonii aggregation is stronger in the presence of sucrose. S. gordonii expresses at least 11 surface-binding-related proteins (27–30). Our preliminary reverse transcription-PCR indicated that the gene expression of those binding proteins was much higher in planktonic cells than in biofilm cells (data not shown). That means bacteria synthesize more binding proteins when they are in the planktonic state and searching for a surface to attach to. That is the reason we focused on planktonic cell gene expression rather than biofilm cells. Amylase is one of the most abundant salivary enzymes. It metabolizes starch to monosaccharide and also serves as a receptor for bacterial binding to the tooth surface (31). S. gordonii binds to amylase via a 20-kDa amylase-binding protein A (abp-A) and 42-kDa abp-B (28, 29). Generally, abp-A is specific to the S. gordonii species (32), and the abp-A mutant S. gordonii strain has difficulty generating biofilm (31). Unlike abp-A, the abp-B mutant strain retains the ability to bind to soluble amylase (29). abp-A, instead of abp-B, is the major protein for S. gordonii tooth surface binding (33), and the present study confirmed that the expression of abp-A was much higher than that of abp-B (Fig. 3). abp-A and abp-B are mediated by carbon catabolite protein A (CcpA) and are indirectly related to sucrose metabolism (27). CcpA has central control of S. gordonii EPS production and biofilm formation. The ability to generate exopolysaccharide from the CcpA mutant was severely impaired, and the CcpA mutant grows slower than the wild type (27). In the present study, CcpA expression was significantly upregulated by nicotine, and this observation may explain the increased cell growth. GtfG, which synthesizes water-soluble and -insoluble glucans, also regulates S. gordonii adhesion (26). gtfG expression was relatively low in S. gordonii and not significantly upregulated by nicotine treatment. Sortase A (SrtA) is an LPXTG motif-regulated S. gordonii cell wall anchoring transpeptidase, and it specifically cleaves LPXTG sequences and transfers polypeptides to peptidoglycan (34). An SrtA mutant of S. gordonii demonstrated less biofilm and impaired binding ability (35). Previous studies in our laboratory demonstrated that sortase-defective S. mutans forms less biofilm than the wild type under the same nicotine treatment (33). The promotion effect of nicotine on S. gordonii srtA expression partially explains the increased biofilm formation in nicotine-treated groups. Streptococcal surface protein A (SspA) and SspB encode antigen I/II salivary adhesins (36). SspA mediates S. gordonii salivary agglutinin aggregation (37). SspB mediates S. gordonii aggregation with P. gingivalis or Candida albicans (6, 38). Streptococcal hemagglutinin (Hsa) is required for S. gordonii sialic acid binding (39). Other proteins, including surface-associated proteins CshA and CshB and metal ABC transporter substrate-binding lipoprotein (ScaA), also are related to S. gordonii aggregation with C. albicans or attachment to fimbriae (19, 36). ScaA is a fibronectin-binding protein that regulates S. gordonii cell coaggregation (27). The increased scaA activity of S. gordonii by nicotine contributed to the increased cell aggregation observed in the present study. Further experiments are needed to ascertain the complete mechanism of how the genes mentioned above are involved in S. gordonii biofilm formation and aggregation regulated by nicotine. Future studies also will focus on the effect of nicotine on other S. gordonii strains and on how nicotine regulates the S. gordonii quorum-sensing system.
In conclusion, nicotine stimulates S. gordonii planktonic cell growth, biofilm formation, aggregation, and gene expression of binding proteins. These effects may promote later pathogen attachment to tooth surfaces, the accumulation of tooth calculus, and the development of periodontal disease in cigarette smokers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Malgorzata M. Kamocha (Gosia) for CLSM operation guidance, and we thank Junjie Zhang for qRT-PCR technology support.
This work was partially funded by the Indiana Branch of the American Society of Microbiology (IBASM) Student Research Grant Fund, the Indiana University Purdue University Indianapolis Tobacco Cessation and Biobehavioral Group, and the Indiana University School of Dentistry Ph.D. Student Research Fund.
Footnotes
Published ahead of print 12 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02395-14.
REFERENCES
- 1.Bergstrom J. 1999. Tobacco smoking and supragingival dental calculus. J. Clin. Periodontol. 26:541–547. 10.1034/j.1600-051X.1999.260808.x. [DOI] [PubMed] [Google Scholar]
- 2.Bergstrom J. 2005. Tobacco smoking and subgingival dental calculus. J. Clin. Periodontol. 32:81–88. 10.1111/j.1600-051X.2004.00638.x. [DOI] [PubMed] [Google Scholar]
- 3.Genco RJ. 1996. Current view of risk factors for periodontal diseases. J. Periodontol. 67:1041–1049. [DOI] [PubMed] [Google Scholar]
- 4.White DJ. 1997. Dental calculus: recent insights into occurrence, formation, prevention, removal and oral health effects of supragingival and subgingival deposits. Eur. J. Oral Sci. 105:508–522. 10.1111/j.1600-0722.1997.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 5.Fuente CDL, Flores S, Moraga M. 2012. DNA from human ancient bacteria: a novel source of genetic evidence from archaeological dental calculus. Archaeometry 55:767–778. 10.1111/j.1475-4754.2012.00707.x. [DOI] [Google Scholar]
- 6.Bagaitkar J, Daep CA, Patel CK, Renaud DE, Demuth DR, Scott DA. 2011. Tobacco smoke augments Porphyromonas gingivalis-Streptococcus gordonii biofilm formation. PLoS One 6:e27386. 10.1371/journal.pone.0027386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Socransky SS, Haffajee AD, Smith C, Duff GW. 2000. Microbiological parameters associated with IL-1 gene polymorphisms in periodontitis patients. J. Clin. Periodontol. 27:810–818. 10.1034/j.1600-051x.2000.027011810.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang R, Li M, Gregory RL. 2012. Effect of nicotine on growth and metabolism of Streptococcus mutans. Eur. J. Oral Sci. 120:319–325. 10.1111/j.1600-0722.2012.00971.x. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Zhou XD, Wu CD. 2012. Tea catechin epigallocatechin gallate inhibits Streptococcus mutans biofilm formation by suppressing gtf genes. Arch. Oral Biol. 57:678–683. 10.1016/j.archoralbio.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J, Koo H. 2010. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J. Appl. Microbiol. 108:2103–2113. 10.1111/j.1365-2672.2009.04616.x. [DOI] [PubMed] [Google Scholar]
- 11.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. [DOI] [PubMed] [Google Scholar]
- 12.Dhar P. 2004. Measuring tobacco smoke exposure: quantifying nicotine/cotinine concentration in biological samples by colorimetry, chromatography and immunoassay methods. J. Pharm. Biomed. Anal. 35:155–168. 10.1016/j.jpba.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 13.Feyerabend C, Higenbottam T, Russell MA. 1982. Nicotine concentrations in urine and saliva of smokers and non-smokers. Br. Med. J. 284:1002–1004. 10.1136/bmj.284.6321.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann D, Adams JD. 1981. Carcinogenic tobacco-specific N-nitrosamines in snuff and in the saliva of snuff dippers. Cancer Res. 41:4305–4308. [PubMed] [Google Scholar]
- 15.Robson N, Bond AJ, Wolff K. 2010. Salivary nicotine and cotinine concentrations in unstimulated and stimulated saliva. Afr. J. Pharm. Pharmacol. 4:61–65. [Google Scholar]
- 16.Cook GS, Costerton JW, Lamont RJ. 1998. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J. Periodontal. Res. 33:323–327. 10.1111/j.1600-0765.1998.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 17.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627–1636. [DOI] [PubMed] [Google Scholar]
- 18.McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274–284. 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander PE, Andersen RN, Ganeshkumar N. 1994. Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect. Immun. 62:4469–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooshima T, Osaka Y, Sasaki H, Osawa K, Yasuda H, Matsumura M, Sobue S, Matsumoto M. 2000. Caries inhibitory activity of cacao bean husk extract in in-vitro and animal experiments. Arch. Oral Biol. 45:639–645. 10.1016/S0003-9969(00)00042-X. [DOI] [PubMed] [Google Scholar]
- 21.Bocquet N, Prado de Carvalho L, Cartaud J, Neyton J, Le Poupon C, Taly A, Grutter T, Changeux JP, Corringer PJ. 2007. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445:116–119. 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Huang R, Zhou X, Zhang K, Zheng X, Gregory RL. 2013. Effect of nicotine on dual-species biofilms of Streptococcus mutans and Streptococcus sanguinis. FEMS Microbiol. Lett. 28:1574–6968. 10.1111/1574-6968.12317. [DOI] [PubMed] [Google Scholar]
- 23.Tsuneda S, Aikawa H, Hayashi H, Yuasa A, Hirata A. 2003. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol. Lett. 223:287–292. 10.1016/S0378-1097(03)00399-9. [DOI] [PubMed] [Google Scholar]
- 24.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76:3405–3408. 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilain S, Pretorius JM, Theron J, Brozel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 75:2861–2868. 10.1128/AEM.01317-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vickerman MM, Sulavik MC, Nowak JD, Gardner NM, Jones GW, Clewell DB. 1997. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene, gtfG. DNA Seq. 7:83–95. [DOI] [PubMed] [Google Scholar]
- 27.Zheng L, Chen Z, Itzek A, Herzberg MC, Kreth J. 2012. CcpA regulates biofilm formation and competence in Streptococcus gordonii. Mol. Oral Microbiol. 27:83–94. 10.1111/j.2041-1014.2011.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers JD, Haase EM, Brown AE, Douglas CW, Gwynn JP, Scannapieco FA. 1998. Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology 144:1223–1233. 10.1099/00221287-144-5-1223. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Tanzer JM, Scannapieco FA. 2002. Identification and analysis of the amylase-binding protein B (AbpB) and gene (abpB) from Streptococcus gordonii. FEMS Microbiol. Lett. 212:151–157. 10.1111/j.1574-6968.2002.tb11259.x. [DOI] [PubMed] [Google Scholar]
- 30.Aguirre A, Levine MJ, Cohen RE, Tabak LA. 1987. Immunochemical quantitation of alpha-amylase and secretory IgA in parotid saliva from people of various ages. Arch. Oral Biol. 32:297–301. 10.1016/0003-9969(87)90024-0. [DOI] [PubMed] [Google Scholar]
- 31.Rogers JD, Palmer RJ, Jr, Kolenbrander PE, Scannapieco FA. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046–7056. 10.1128/IAI.69.11.7046-7056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown AE, Rogers JD, Haase EM, Zelasko PM, Scannapieco FA. 1999. Prevalence of the amylase-binding protein A gene (abpA) in oral streptococci. J. Clin. Microbiol. 37:4081–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li MY, Huang RJ, Zhou XD, Gregory RL. 2013. Role of sortase in Streptococcus mutans under the effect of nicotine. Int. J. Oral Sci. 5:206–211. 10.1038/ijos.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson GK, Mitchell TJ. 2004. The biology of Gram-positive sortase enzymes. Trends Microbiol. 12:89–95. 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Nobbs AH, Vajna RM, Johnson JR, Zhang Y, Erlandsen SL, Oli MW, Kreth J, Brady LJ, Herzberg MC. 2007. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 153:4088–4097. 10.1099/mic.0.2007/007252-0. [DOI] [PubMed] [Google Scholar]
- 36.Holmes AR, McNab R, Jenkinson HF. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkinson HF, Terry SD, McNab R, Tannock GW. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78:4644–4652. 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi Y, Konishi K, Cisar JO, Yoshikawa M. 2002. Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70:1209–1218. 10.1128/IAI.70.3.1209-1218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.