Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a Shiga-toxigenic pathogen capable of inducing severe forms of enteritis (e.g., hemorrhagic colitis) and extraintestinal sequelae (e.g., hemolytic-uremic syndrome). The molecular basis of colonization of human and animal hosts by EHEC is not yet completely understood, and an improved understanding of EHEC mucosal adherence may lead to the development of interventions that could disrupt host colonization. FdeC, also referred to by its IHE3034 locus tag ECOK1_0290, is an intimin-like protein that was recently shown to contribute to kidney colonization in a mouse urinary tract infection model. The expression of FdeC is tightly regulated in vitro, and FdeC shows promise as a vaccine candidate against extraintestinal E. coli strains. In this study, we characterized the prevalence, regulation, and function of fdeC in EHEC. We showed that the fdeC gene is conserved in both O157 and non-O157 EHEC and encodes a protein that is expressed at the cell surface and promotes biofilm formation under continuous-flow conditions in a recombinant E. coli strain background. We also identified culture conditions under which FdeC is expressed and showed that minor alterations of these conditions, such as changes in temperature, can significantly alter the level of FdeC expression. Additionally, we demonstrated that the transcription of the fdeC gene is repressed by the global regulator H-NS. Taken together, our data suggest a role for FdeC in EHEC when it grows at temperatures above 37°C, a condition relevant to its specialized niche at the rectoanal junctions of cattle.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) is a Shiga-toxigenic pathogen capable of inducing severe forms of enteritis, such as hemorrhagic colitis (HC), and extraintestinal sequelae, such as hemolytic-uremic syndrome (1). Ruminant livestock are the principal hosts of Shiga-toxigenic E. coli (STEC) (2, 3), including EHEC, which is often carried at the herd and individual-animal levels as commensal enteric flora (4). Food-borne transmission of STEC through contamination of primary produce is a significant factor in the epidemiology of EHEC infections, which are prevalent worldwide. E. coli of the O157 serotype is considered the archetypal EHEC, although a small number of non-O157 serotypes are more commonly associated with severe disease than other non-O157 serotypes. In the United States, the Food and Drug Administration recently designated six non-O157 serotypes (O26, O45, O103, O111, O121, O145) as food adulterants, highlighting these as having enhanced public health significance, and increasing the stringency of STEC food safety assurance regulation and testing requirements at a global level (5).
The molecular mechanisms associated with the colonization of human and animal hosts by EHEC are not yet completely understood. Colonization is generally considered to involve three steps: (i) initial attachment, (ii) translocation of bacterial components to mammalian cells via a type III secretion system (T3SS), and (iii) intimate attachment, characterized by the formation of attaching and effacing lesions and mediated by the bacterial effector intimin (6). Research has focused on the locus of enterocyte effacement (LEE), which encodes a T3SS produced by strains of EHEC and closely related enteropathogenic E. coli (EPEC), as well as a range of translocated effector molecules. It is likely that other mediators of colonization are yet to be identified, particularly for non-O157 EHEC, and particularly with respect to the colonization of bovine hosts. An improved understanding of EHEC mucosal adherence may lead to the development of interventions that could inhibit host colonization. This could benefit public health either by disrupting the colonization of humans to prevent the acquisition of such strains leading to disease or by reducing the colonization of livestock and thus decreasing the transmission of EHEC to humans via decreases in food contamination. An intimin-based vaccine is already available for the reduction of the frequency of colonization of cattle with O157 strains; however, no such control measure is available for non-O157 EHEC (7).
A recently described E. coli adhesin, FdeC (GenBank accession no. ADE88959), contributes to E. coli colonization of the upper urinary tract in a mouse infection model (8). Immunization with FdeC, also known by its IHE3034 locus tag ECOK1_0290, is also protective in mouse models of urinary tract infection (UTI) and sepsis (8, 9), demonstrating that FdeC represents a potential vaccine antigen against uropathogenic E. coli (UPEC)-mediated UTI. While FdeC expression is undetectable under normal laboratory growth conditions, the protein is expressed during coculture with mammalian cells and in vivo (8). FdeC shares structural similarity with intimin, containing an N-terminal β-barrel and a C-terminal extracellular domain, and belongs to the type Ve group of autotransporter proteins (10). FdeC was identified in genome sequences from a range of different E. coli pathotypes; to date, however, it has been studied only for its potential role in uropathogenesis.
In this study, we have described conditions under which FdeC is expressed by EHEC and have demonstrated that the level of FdeC expression can be manipulated by minor alteration of these conditions. We have also shown that FdeC expression is repressed by the global regulator H-NS (11) and that FdeC mediates biofilm formation following overexpression in a recombinant strain under continuous-flow conditions. Taken together, our data suggest a potential role for FdeC in the colonization of the terminal rectums of cattle.
MATERIALS AND METHODS
Strains and growth conditions.
N39 (also known as EC673) is an eae- and stx1-positive O26:H11 strain of E. coli originally isolated from the feces of an Australian calf. EDL933 is a well-characterized O157:H7 EHEC outbreak isolate (12). E. coli MS427 (K-12 strain MG1655Δflu [i.e., lacking antigen 43 {Ag43}]) and OS56 (green fluorescent protein [GFP]-tagged MG1655Δflu) have been described previously (13–15). Plasmid pHNS has been described previously (16). Cells were grown at 37°C on solid or in liquid Luria-Bertani (LB) medium supplemented with the appropriate antibiotics (100 μg/ml ampicillin, 50 μg/ml kanamycin) unless otherwise stated. Where necessary, gene expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
DNA manipulations and genetic techniques.
Genomic DNA was extracted from overnight cultures by using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer's procedure for Gram-negative bacteria. Plasmid DNA was isolated by using the QIAprep Spin Miniprep kit (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions. Restriction digestion, ligation, and T4 polymerase treatment followed the manufacturer's recommendations (New England BioLabs [NEB], Ipswich, MA, USA). PCRs for which high fidelity was required were performed by using the Expand High Fidelity polymerase system (Roche, Basel, Switzerland) according to the manufacturer's recommendations. Taq DNA polymerase (New England BioLabs) was used for screening PCRs. DNA sequencing was performed by the Australian Equine Genetics Research Centre.
Construction of plasmids.
The fdeC gene was amplified by PCR from N39 with primers designed using the available genome sequence of O26:H11 strain 11368 (primers 3018 [5′-GGCGAGCTCCATCAGGAAATTATCTCAATGTCA] and 3019 [5′-GGCAAGCTTAAATTATTTCATCGCCTCCTCGTC]). The PCR product was digested with SacI (forward primer) and HindIII (reverse primer) and was ligated to SacI-HindIII-digested plasmid pSU2718 (17) to generate plasmid pFdeC. In this construct, the transcription of fdeC was under the control of the IPTG-inducible lac promoter.
Antiserum production and immunoblotting.
A six-histidine-tagged, 97-kDa truncated form of FdeC was constructed. Primers 2871 (5′-TACTTCCAATCCAATGCGCTTGACGGTCAGAGCCGTAT) and 2830 (5′-TTATCCACTTCCAATGTTAGTTCATCGCCTCCTCGCCCT) were used to amplify the portion of the gene coding for the predicted extracellular component of the protein (amino acids 467 to 1417), which was then inserted into the pMCSG7 vector by ligation-independent cloning and was maintained in E. coli DH5α (18). This plasmid was then transferred to E. coli BL21 for expression of the recombinant protein by induction with 1 mM IPTG and purification by Ni-nitrilotriacetic acid (NTA) Superflow columns (Qiagen) under native conditions (according to the manufacturer's instructions). Protein purity was assessed by SDS-PAGE analysis as described previously (19). A polyclonal anti-FdeC serum was raised in rabbits by the Institute of Medical and Veterinary Sciences (South Australia). For immunoblotting, cell lysates were subjected to SDS-PAGE and were transferred to polyvinylidene difluoride (PVDF) microporous membrane filters as described previously (19). Culture supernatants were prepared by filtering (pore size, 0.22 μm) to remove intact bacterial cells and concentrating 100× by size exclusion centrifugal filtration. A serum raised against the extracellular portion of FdeC was used as the primary serum, and the secondary antibody was alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Sigma, Castle Hill, NSW, Australia). 5-Bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium (NBT) was used as the substrate in the detection process (Sigma).
Construction of fdeC and hns mutants.
The fdeC gene was deleted from E. coli N39 by using a modification of the λ Red recombinase gene replacement system (20). The kanamycin resistance cassette was amplified from pKD4 using primers 747 (5′-GTGTAGGCTGGAGCTGCTTC) and 748 (5′-CATATGAATATCCTCCTTAGTTC). Approximately 500-bp regions flanking the fdeC gene were amplified using primers 2925 (5′-AGTGTTGCGATGGGTAGTGC) and 2926 (5′-GAAGCAGCTCCAGCCTACACGCAGCGGGTCAGAACTGAAT) for the upstream region and primers 2927 (5′-GAACTAAGGAGGATATTCATATGTGACCTATGGCGGGTACGAA) and 2928 (5′-TTTGCCGGTACGGTATGCAA) for the downstream region. The kanamycin resistance cassette was inserted between these flanking sequences using three-way PCR, and the construct was amplified sufficiently to allow the transformation of N39 with approximately 1 μg of DNA. Mutants were selected by growth on kanamycin and were confirmed by DNA sequencing using primers 3025 (5′-CCAGCCAGTTCCTTTCGATGCCT) and 3026 (5′-CCGTGTATACCGCCACCGTGA). The mutant strain was referred to as N39fdeC. A modification of the method described above was used to inactivate the hns gene in N39, by amplifying the kanamycin resistance cassette and 500-bp flanking regions directly from EDL933hns (strain MS3832) (21) using primers 2361 (5′-TTGTATGAAGATTGCAAC) and 2364 (5′-TTTATCACAGGATAACCT). The replacement of the hns gene with the kanamycin resistance cassette was confirmed by DNA sequencing with primers 1935 (5′-TTGCCTGCTGTTCATGTTCC) and 2449 (5′-GCTGGTTACTCACTCATC). This strain was referred to as N39hns. To construct a mutant strain lacking both fdeC and hns, the kanamycin resistance cassette was first removed from N39fdeC by using pCP20 as described previously (20), and hns was inactivated in this strain by a method identical to that described above.
Construction of an fdeC::lacZ reporter in N39.
The lacIZ genes were replaced with the chloramphenicol resistance cassette from pKD3 in N39 by using the method described above, with primers 3286 (5′-TTTGCCCGGAACAAGACCGC), 3287 (5′-GAAGCAGCTCCAGCCTACACTGCGACATCGTATAGCGTTACTGG), 3288 (5′-GAACTAAGGAGGATATTCATATGTAACCGGGCAGGCCATGTCT), and 3289 (5′-ACAACGCCCAGCCAACACAG). The deletion was confirmed by DNA sequencing using primers 3314 (5′-GGCCTGCATCGCACCACTGT) and 3315 (5′-CCCGCGTACCCTGTTCACCG). This strain was referred to as N39lacIZ and was used as a negative control in the β-galactosidase assays. Similarly, the fdeC gene was then replaced with the lacZ-zeo cassette amplified from strain UGB1969 (a gift from J. M. Ghigo) with primers 3290 (5′-ATGACCATGATTACGGATTCACTG) and 3291 (5′-TCAGTCCTGCTCCTCGGCCACGAA). Flanking regions were amplified using primers 3292 (5′-TCGTTCATGAGAAGCATAACGTA), 3293 (5′-CAGTGAATCCGTAATCATGGTCATTGAGATAATTTCCTGATGAAC), 3294 (5′-TTCGTGGCCGAGGAGCAGGACTGATTTTAAGAATATGAAAGTAACATTCT), and 3295 (5′-AAGACGGATATCTATTTCGTCTC). The lacZ fusion and correct replacement of the fdeC gene were confirmed by DNA sequencing using the same primers that were used to confirm the fdeC mutant in combination with lacZ-zeo cassette-specific screening primers 3312 (5′-TGGTGCCGGACAACACCCTG) and 3313 (5′-GCGGGCCTCTTCGCTATTACGC). The chloramphenicol resistance cassette was removed using pCP20 in order to facilitate further mutagenesis using this antibiotic selection marker, and this reporter strain was referred to as N39fdeC::lacZ.
Transposon mutagenesis.
Random transposon mutants were generated in N39fdeC::lacZ using the chloramphenicol cassette from pKD3 (20) with mini-Tn5 ends introduced by amplifying the cassette using primers 2279 (5′-CTGTCTCTTATACACATCTCACGTCTTGAGCGATTGTGTAGG) and 2280 (5′-CTGTCTCTTATACACATCTGACATGGGAATTAGCCATGGTCC) from the NotI-digested plasmid. The mini-Tn5::Cm PCR product was phosphorylated with T4 polynucleotide kinase (NEB) for 90 min at 37°C, followed by inactivation for 20 min at 60°C. The phosphorylated DNA was concentrated to at least 400 ng/μl, and transposomes were assembled by incubation with transposase (Epicentre, Madison, WI, USA) at 37°C for 1 h. Electrocompetent N39fdeC::lacZ was transformed with the mini-Tn5::Cm transposomes, plated onto LB medium containing chloramphenicol and 5-bromo-4-chloro-3-indoly-β-d-galactopyranoside (X-gal), and incubated at 37°C overnight (16 to 18 h). Blue colonies were selected for further analysis, and the plates were transferred to 43°C for a further overnight incubation. Those colonies that did not develop a blue color upon incubation at the higher temperature were also selected for further analysis. The locations of the transposon insertions were determined using inverse PCR. Genomic DNA was digested with Taqα1 and was religated with T4 DNA ligase (both from NEB), and recircularized DNA containing the transposon sequence was amplified using primers 2240 (5′-GCCGATCAACGTCTCATTTT) and 4027 (5′-AATAAGATCACTACCGGGCG). DNA sequencing using primer 2240 was used to ascertain the sequence immediately flanking the insertion, and the genomic location was determined by using a BLASTn search of the O26:H11 strain 11368 genome sequence.
β-Galactosidase assay.
Reporter strain lacZ activity was assayed using a β-galactosidase assay with ortho-nitrophenyl-β-galactoside (ONPG) as the substrate (22). The optical density of the bacterial cultures was measured by diluting 75 μl of culture into 900 μl of LB medium and reading at 600 nm in a 1-cm cuvette. A matching 75-μl aliquot of each culture was also lysed with 50 μl chloroform, 25 μl 0.1% SDS, and 675 μl Z-buffer (60 mM Na2HPO4·2H2O, 40 mM NaH2PO4·2H2O, 50 mM β-mercaptoethanol, 10 mM KCl, 1 mM MgSO4·7H2O [pH 7.0]). Lysed bacteria were equilibrated to 28°C, and 150 μl of ONPG substrate (4 mg/ml) was added to each sample. Reactions were stopped by the addition of 375 μl 1 M Na2CO3, and the time (T) was recorded. Color development was measured by the optical density at 420 nm (OD420) in 1-cm cuvettes. Miller units were calculated using the following formula: (750 × OD420)/(T × V × OD600), where V is the volume in milliliters (0.075).
Rapid amplification of 5′ cDNA ends (5′-RACE).
The transcription start site of fdeC was determined by using the 5′ RACE System for Rapid Amplification of cDNA Ends, version 2.0 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Two gene-specific primers were used for this assay: GSP1 (primer 4083 [5′-AGGAAATACAGGAGTGGC]) and GSP2 (primer 4084 [5′-AACTTTATTACCGGAATGGCCAC]).
DNA curvature prediction and DNA PAGE analysis.
The fdeC promoter region was analyzed in silico using bend.it, a program that enables the prediction of a curvature propensity plot calculated with DNase I-based parameters (23). The curvature is calculated as a vector sum of dinucleotide geometries (roll, tilt, and twist angles) and is expressed as degrees per helical turn (10.5°/helical turn = 1°/bp). Experimentally tested curved motifs produce curvature values of 5 to 25°/helical turn, whereas straight motifs give values below 5°/helical turn. The 250-bp fdeC promoter region was PCR amplified using primers 4062 (5′-AGGTATCTATTTAATGACTTGCAC) and 4063 (5′-TGAGATAATTTCCTGATCAACG), and its curvature was experimentally tested by comparing its electrophoretic mobility with that of a 100-bp DNA ladder (NEB) on a 0.5× Tris-borate-EDTA (TBE)–7.5% PAGE gel at 4°C.
Electromobility shift assay.
Electromobility shift assays were performed essentially as described previously (24). H-NS protein was prepared by amplifying hns from N39 using primers 4059 (5′-TACTTCCAATCCAATGCAATGAGCGAAGCACTTAAAATTCTG) and 4060 (5′-TTATCCACTTCCAATGTTATTGCTTGATCAGGAAATCGTC), cloned into the pMCSG7 vector using a ligation-independent method, and maintained in E. coli DH5α. This plasmid was then transferred to E. coli BL21 for expression of the recombinant protein by induction with 1 mM IPTG and purification by Ni-NTA Superflow columns (Qiagen) under native conditions (according to the manufacturer's instructions). Protein purity was assessed by SDS-PAGE analysis as described previously (19). The fdeC promoter region was amplified using primers 4062 (5′-AGGTATCTATTTAATGACTTGCAC) and 4063 (5′-TGAGATAATTTCCTGATGAACG). A DNA mixture comprising the PCR-amplified fdeC promoter region and TaqI-SspI-digested pBR322 at an equimolar ratio was incubated at room temperature for 15 min with 4 μM native purified H-NS protein in 30 μl of reaction mixture containing H-NS binding buffer (40 mM HEPES [pH 8], 60 mM potassium glutamate, 8 mM magnesium aspartate, 5 mM dithiothreitol, 10% glycerol, 0.1% octylphenoxypolyethoxyethanol, 0.1 mg/ml bovine serum albumin [BSA]). DNA fragments and DNA-protein complexes were resolved by gel electrophoresis (0.5 × TBE, 3% Resolution Plus Agarose [catalog no. 200-0040; Quantum Scientific]; 50 V at 4°C) and were visualized after staining with ethidium bromide.
Epithelial cell and ECM adherence assays.
The ability of E. coli strains to adhere to HeLa, HEp-2, or primary bovine rectal epithelial cells was tested as described previously (25, 26). Briefly, confluent cell monolayers were infected with E. coli strains at a multiplicity of infection (MOI) of 1:10 and were incubated for 1 h. Nonadherent bacteria were removed by 5 washes with phosphate-buffered saline (PBS), and cell lysates were serially diluted and were plated onto LB agar plates for the enumeration of adherent bacteria. Adherence to extracellular matrix (ECM) proteins was determined as described previously (16, 21, 27).
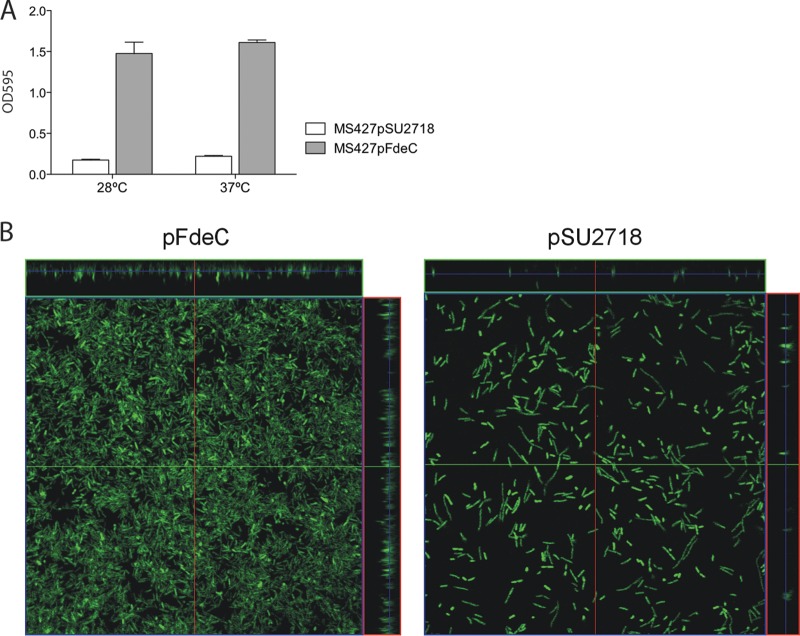
Analysis of biofilm formation.
Biofilm formation on polystyrene surfaces was assessed in 96-well microtiter plates (Iwaki) essentially as described previously (13). Briefly, the pFdeC and pSU2718 plasmids were transferred to the Ag43-negative strain MS427 to create MS427(pFdeC) and MS427(pSU2718). These were grown for 24 h in M9 glucose minimal medium (containing 1 mM IPTG for the induction of gene expression and 30 μg/ml chloramphenicol) at 28°C or 37°C, washed to remove unbound cells, and stained with crystal violet. Quantification of bound cells was assessed by adding acetone-ethanol (20:80) to dissolve the crystal violet, and the optical density was measured at 595 nm. To analyze biofilm formation under continuous-flow conditions, pFdeC or pSU2718 (vector control) was transferred into E. coli OS56, which is identical to MS427 except that it constitutively expresses GFP to allow analysis by confocal microscopy (12, 13). Flow cells were inoculated with cultures standardized by the optical density at 600 nm after overnight growth in M9 glucose minimal medium. OS56(pFdeC) and OS56(pSU2718) biofilms were grown in M9 minimal medium at 28°C on Rinzl plastic coverslips (ProSciTech, Kirwan, QLD, Australia) in the presence of chloramphenicol and IPTG for the maintenance of plasmids and the induction of gene expression as described previously (16, 28, 29). Biofilm development was monitored by confocal scanning laser microscopy at 24 h or 36 h after inoculation. For analysis of the flow cell biofilms, 14 z-stacks were collected for each strain and were analyzed using the COMSTAT software program (30). COMSTAT data were analyzed using the nonparametric Kruskal-Wallis test within the Minitab (version 14) software package. P values of <0.05 were considered significant.
Microscopy and image analysis.
Polyclonal FdeC-specific rabbit antiserum was used for immunofluorescence microscopy (100× objective) essentially as described previously (28, 31). Immunofluorescence microscopy and image acquisition were performed on a scanning confocal laser microscope (Axioplan 2; Zeiss) equipped with detectors and filters for monitoring fluorescein isothiocyanate (FITC) fluorescence. Microscopic observations of biofilms and image acquisition were performed on a scanning confocal laser microscope (LSM 510 META; Zeiss, Jena, Thuringia, Germany) equipped with detectors and filters for monitoring GFP. Vertical cross sections through the biofilms were visualized using the Zeiss LSM Image Examiner. Images were further processed for display by using Photoshop software (Adobe, Mountain View, CA, USA).
RESULTS
Genomic context of fdeC.
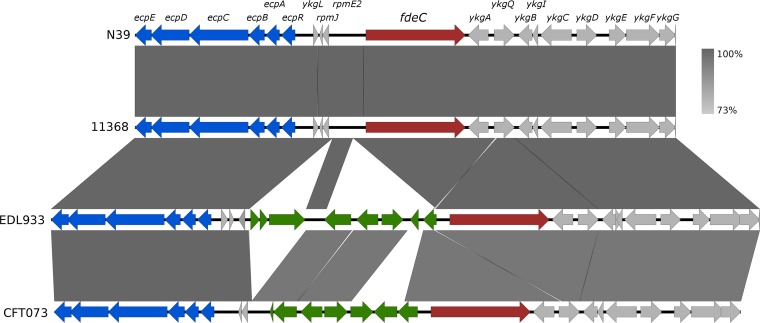
The genomic context of the fdeC gene in non-O157:H7 and O157:H7 EHEC strains was examined using complete and draft genome sequences available in the NCBI database and our own collection. All non-O157:H7 EHEC strains possessed a conserved fdeC sequence organization that is represented by the O26:H11 strains N39 and 11368 (Fig. 1). In contrast, all O157 EHEC strains, as represented by the archetypal strain EDL933, possess two additional DNA segments in the region upstream of the fdeC gene and downstream of the adjacent ecp locus. The sequence between fdeC and the ecp locus in non-O157:H7 and O157:H7 EHEC strains differs from that described previously for the extraintestinal pathogenic E. coli (ExPEC) strains CFT073 and IHE3034 (8). Despite these differences, the 550-bp region upstream of the fdeC coding sequence was conserved in all strains, even in E. coli K-12, where fdeC is truncated to 875 nucleotides (GenBank accession no. U00096.3; range, nucleotides 314357 to 315232) (32). We hypothesized that this 550-bp region contained the regulatory elements that control the transcription of fdeC, and we demonstrated this in the series of experiments described below.
FIG 1.
Genomic context of fdeC. The genomic context of fdeC (red) is shown in the O26:H11 STEC strains (N39 and 11368) and other pathogenic E. coli strains (O157:H7 STEC strain EDL933 and UPEC strain CFT073). Variable regions (green) between fdeC and the mat (or ecp) operon (blue) are shown. The genomic comparison was performed using Easyfig (54). Gray shading indicates high nucleotide identity between sequences (95 to 100%).
The fdeC promoter is active at high temperatures.
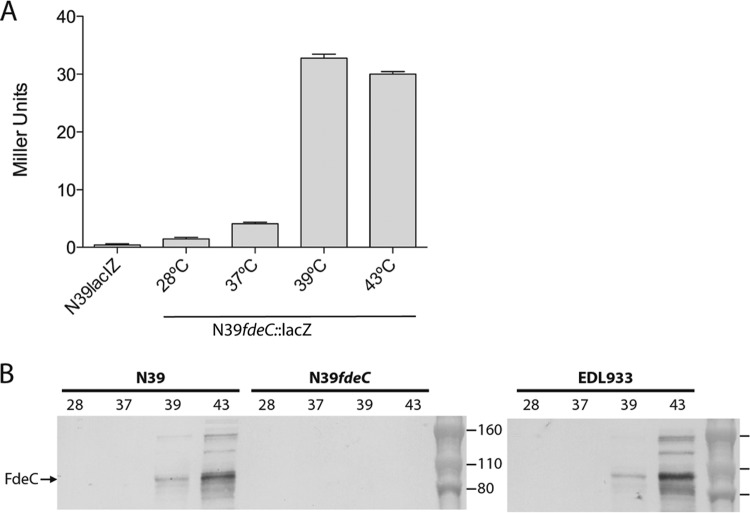
In order to investigate the regulation of fdeC, a reporter strain was constructed using the O26:H11 strain N39. The lacIZ genes of N39 were initially inactivated to generate strain N39lacIZ, and this strain was subsequently modified by inserting the lacZ gene as a chromosomally located transcriptional fusion to the fdeC promoter to generate strain N39lacIZ fdeC::lacZ-zeo. When grown on X-gal plates, all N39lacIZ fdeC::lacZ-zeo colonies were white (indicating no transcription). No color heterogeneity was observed among the N39lacIZ fdeC::lacZ-zeo colonies at either 37°C or 43°C, indicating that the transcription of fdeC is not phase variable. The N39lacIZ fdeC::lacZ-zeo reporter strain was grown under various laboratory culture conditions, including different NaCl concentrations, pHs, and acetate concentrations, without any difference in β-galactosidase activity (data not shown). However, when cells were grown at or above 39°C, the level of β-galactosidase activity increased significantly over that at 37°C and below (Fig. 2A). This indicated that the fdeC promoter is active at temperatures of ≥39°C but is repressed at lower temperatures.
FIG 2.
Temperature-dependent expression of FdeC. (A) β-Galactosidase activities (in Miller units) of N39lacIZ (control) and N39fdeC::lacIZ grown at various temperatures. The fdeC promoter was active only at 39°C and 43°C. (B) Western blot analysis of STEC strains N39, N39fdeC, and EDL933 grown at 28°C, 37°C, 39°C, and 43°C. As in the reporter assay, FdeC expression was detected only at or above 39°C.
FdeC is expressed by wild-type EHEC strains at high temperatures.
Based on the transcriptional data presented above, we used an FdeC-specific antibody to examine the expression of FdeC in wild-type strains. Western blot analysis of whole-cell lysates prepared from N39 revealed strong expression of FdeC at 43°C and a lower level of expression at 39°C, but no expression at 28°C or 37°C (Fig. 2B). An fdeC mutant of N39 did not produce FdeC at any of these temperatures, confirming the specificity of the antibody. The same experiment was also performed with the O157:H7 strain EDL933 and the UPEC strain CFT073, and the same temperature-dependent expression of FdeC was observed (Fig. 2B and data not shown). Taken together, the data demonstrate that FdeC expression is temperature regulated and is stimulated at or above 39°C. Furthermore, the temperature regulation of fdeC in N39, EDL933, and CFT073 is identical, suggesting that the genetic basis of this transcriptional control lies within the conserved 550-bp DNA region upstream of the fdeC open reading frame.
Promoter organization of fdeC.
The transcription start site for fdeC was determined using 5′-RACE; it was located at a position 184 bp upstream of the fdeC ATG translation start site (Fig. 3). The transcription start site is preceded by a strong −10 promoter consensus sequence (5′-TATAAT-3′). A weak −35 promoter consensus sequence (5′-CTAAAT-3′) was identified with a 19-bp spacer region. In line with the temperature regulation observed for fdeC transcription, bioinformatic analysis revealed the presence of a putative H-NS nucleation site (5′-TGGATATATT-3′) (33, 34) downstream of the fdeC −10 promoter sequence. The presence of this nucleation site suggested that H-NS may be responsible for the strong repression of fdeC at temperatures below 37°C. Indeed, genes repressed by H-NS are often regulated in response to changes in temperature; thus, this prediction is consistent with our data.
FIG 3.
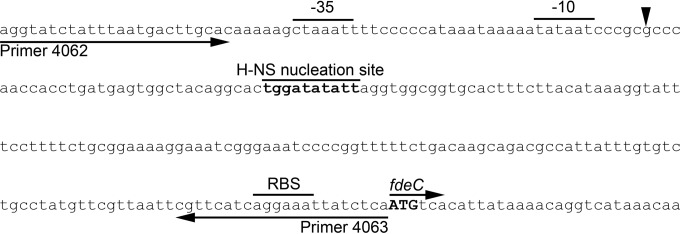
Promoter organization of the fdeC gene. Shown is the structural organization of the fdeC promoter, indicating the −10 and −35 elements, transcription start site (arrowhead), ribosome binding site (RBS), and ATG start codon (boldface). Also indicated is a putative H-NS nucleation site (boldface). The primer binding sites for the region amplified for use in the electromobility shift assay are indicated by arrows below the sequence.
Transcription from the fdeC promoter is repressed by H-NS.
To test if the transcription of fdeC is repressed by H-NS, we mutated the hns gene in the N39lacIZ fdeC::lacZ-zeo reporter strain. The β-galactosidase activity of the N39lacIZ fdeC::lacZ-zeo hns mutant was significantly higher than that of the parent strain N39lacIZ fdeC::lacZ-zeo following growth at 37°C (Fig. 4A). Complementation of the N39lacIZ fdeC::lacZ-zeo hns mutant with an hns-containing plasmid (pHNS) restored repression, while a vector control strain possessed β-galactosidase activity at a level similar to that of strain N39lacIZ fdeC::lacZ-zeo hns (Fig. 4A). An independent transposon mutagenesis approach was also utilized to identify regulatory genes that alter β-galactosidase activity in N39fdeC::lacZ when grown at 37°C. A library of approximately 20,000 mutants was constructed and screened by color development on agar containing X-gal. The transposon insertion site of those colonies that differed from the parent strain in blue color development was determined by inverse PCR. In this experiment, only the hns gene was identified, further supporting the observation that fdeC is repressed by H-NS at 37°C.
FIG 4.
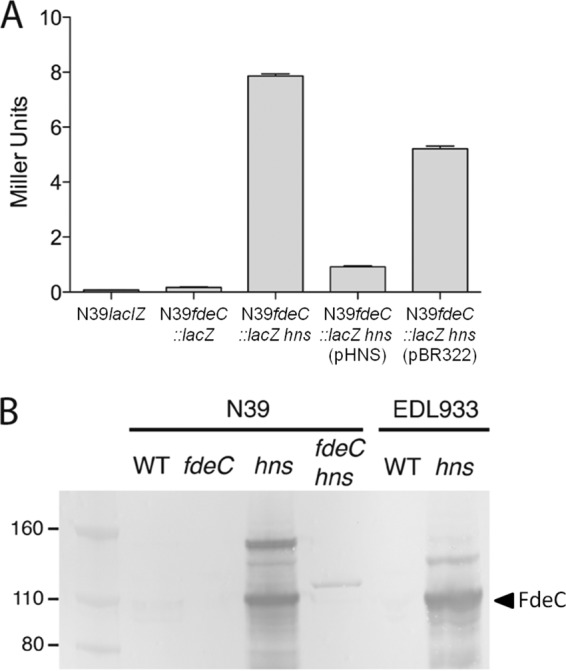
Repression of fdeC transcription by H-NS. (A) β-Galactosidase activities (in Miller units) of N39lacIZ (control), N39fdeC::lacIZ, N39fdeC::lacIZ hns, N39fdeC::lacIZ hns(pHNS), and N39fdeC::lacIZ hns(pBR322). Mutation of the hns gene (N39fdeC::lacIZ hns) resulted in an increase in β-galactosidase activity, which could be reduced by complementation with hns on a plasmid [N39fdeC::lacIZ hns(pHNS)]. (B) Western blot analysis of FdeC performed using whole-cell lysates prepared from wild-type (WT) N39, N39fdeC, N39hns, N39fdeC hns, WT EDL933, and EDL933hns. The band representing the predicted FdeC extracellular domain (∼100 kDa) is indicated. The higher-molecular-weight cross-reacting band likely represents the full-length FdeC protein.
FdeC expression is regulated by H-NS.
To directly examine the effect of H-NS on FdeC expression, the hns gene was mutated in N39 and EDL933 to generate strains N39hns and EDL933hns. Western blot analysis of whole-cell lysates prepared from N39hns and EDL933hns grown at 37°C revealed strong expression of FdeC. In contrast, no expression of FdeC was observed in either parent strain (Fig. 4B). These results demonstrate that fdeC is repressed by H-NS in EHEC at 37°C during growth in LB broth.
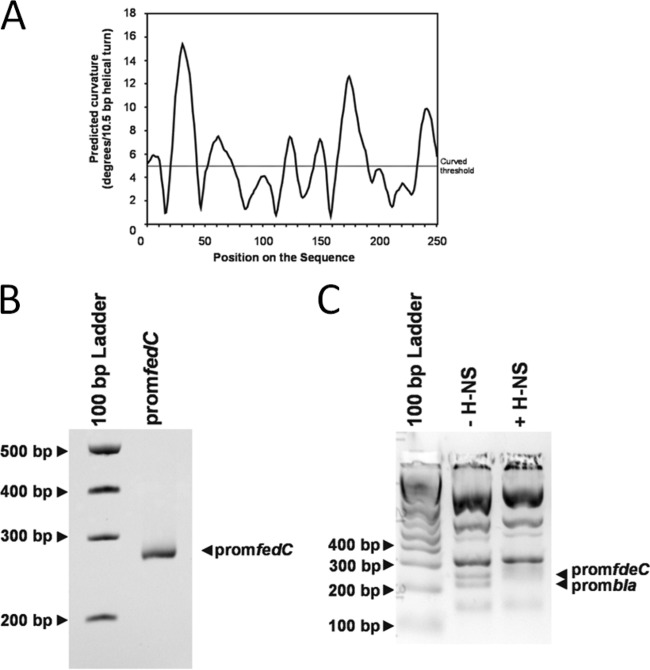
H-NS binds to the promoter region of fdeC.
H-NS binds to intrinsically curved regions of DNA. An in silico-generated curvature propensity plot calculated with DNase I-based parameters suggested that the promoter region of fdeC may assume a curved conformation (Fig. 5A). To test this curvature, we amplified the 250-bp PCR product containing the predicted fdeC promoter region and examined it by polyacrylamide gel electrophoresis at 4°C as described previously (35, 36). In line with our prediction, the 250-bp fdeC promoter region displayed retarded gel electrophoretic mobility relative to that of noncurved DNA standards (Fig. 5B). In order to investigate whether H-NS influences fdeC transcription by direct binding to the fdeC promoter, we performed electrophoretic mobility shift assays as described previously (24, 27). A 250-bp PCR product containing the fdeC promoter region was generated and was mixed with TaqI-SspI-digested pBR322 DNA (which contains the bla promoter, which has been shown previously to be bound by H-NS [37]). The DNA was incubated with or without purified H-NS protein and was subsequently visualized by gel electrophoresis. The 250-bp fdeC promoter region and the fragment containing the bla promoter were both retarded in mobility by the addition of 4 μM H-NS (Fig. 5C). In contrast, the pBR322 fragments not containing the bla promoter did not bind H-NS at this concentration.
FIG 5.
H-NS binds to the promoter region of fdeC. (A) In silico curvature propensity plot showing predicted regions of curved DNA in the 250-bp PCR fragment containing the fdeC promoter. (B) Electrophoretic mobility of the 250-bp PCR fragment containing the fdeC promoter. The migration of this band is slightly retarded in the gel, suggesting that the DNA is curved. (C) Electrophoretic band shift of the 250-bp PCR fragment containing the fdeC promoter and the bla promoter from TaqI-SspI-digested pBR322 DNA in the absence or presence of 4 μM H-NS. The migration of pBR322 fragments not containing the bla promoter was not altered by H-NS. The image depicts a gel representative of three independent experiments.
fdeC gene expression is suppressed by acetate and low pH.
The fdeC gene is located proximally to the mat (or ecp) fimbria-encoding genes, which in addition to regulation by temperature and H-NS, are also regulated by pH and acetate (38). To examine if these factors also influence fdeC transcription, we examined the reporter activity of N39fdeC::lacZ in response to altered pHs and acetate concentrations when grown at the permissible temperature of 43°C. Reporter activity was decreased at lower pHs and increased at higher pHs, while increasing the acetate concentration decreased reporter activity (Fig. 6). No difference in the growth rate was observed under these conditions (data not shown). Taken together, our data suggest that FdeC is expressed optimally at pH 8.0 and at low acetate concentrations.
FIG 6.
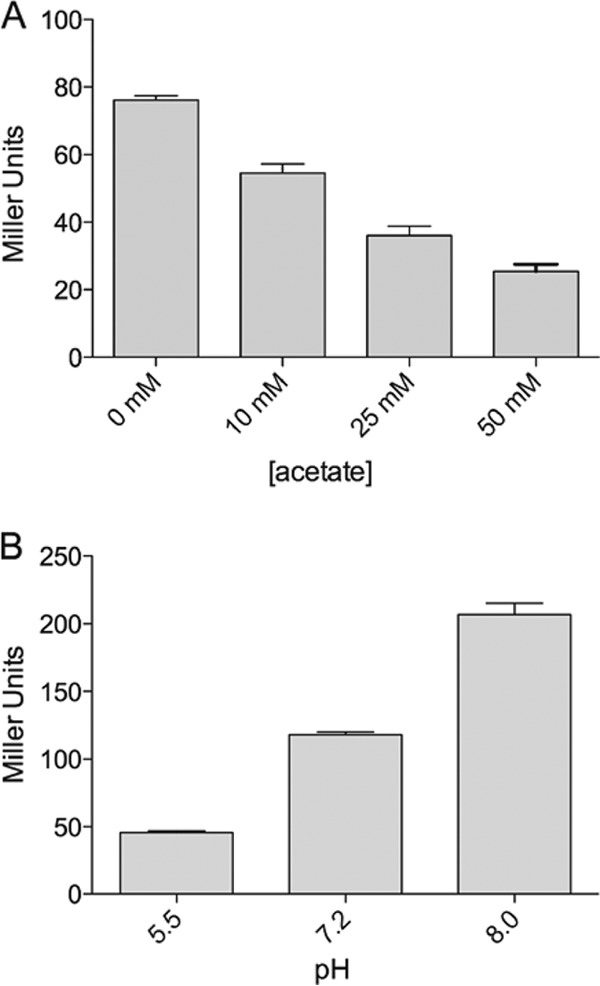
Influence of pH and acetate on fdeC promoter activity. Shown are the β-galactosidase activities (in Miller units) of N39fdeC::lacIZ grown in the presence of increasing concentrations of acetate (A) or at increasing pHs (B). The fdeC promoter was most active in the absence of acetate and at pH 8.0.
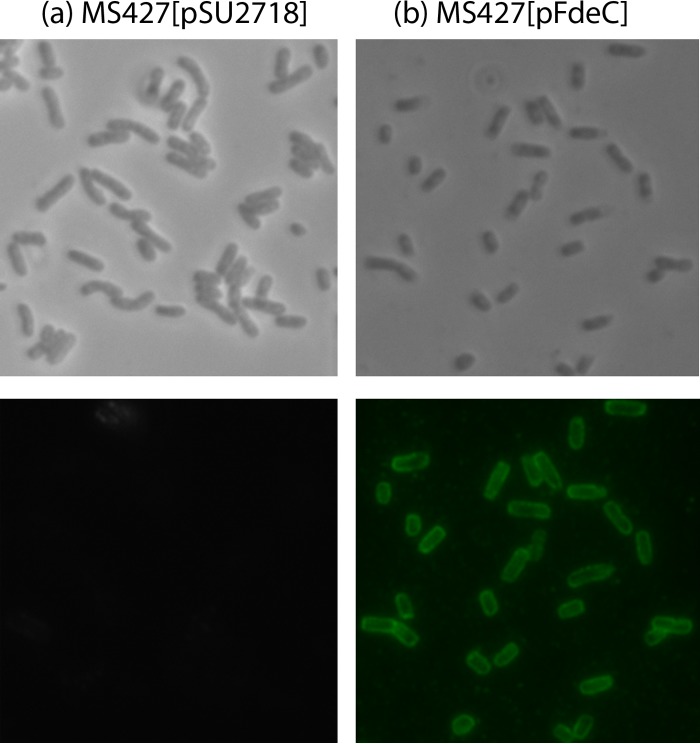
FdeC is located at the cell surface.
To examine the cellular location of FdeC, the full-length fdeC gene from STEC strain N39 was cloned into the pSU2718 expression vector to generate plasmid pFdeC, allowing inducible expression of the recombinant FdeC from the lac promoter (17). This plasmid was transferred to the E. coli K-12 flu deletion strain MS427. This strain does not contain an intact fdeC gene and is unable to mediate the classical cell aggregation and biofilm phenotypes associated with Ag43 expression (15). FdeC protein expression in MS427(pFdeC) was confirmed by Western blotting (data not shown), and surface exposure of FdeC was confirmed by immunofluorescence microscopy (Fig. 7). Thus, the EHEC fdeC gene is functional and encodes a protein located at the cell surface.
FIG 7.
FdeC is localized at the cell surface. Cells of the E. coli vector control strain MS427(pSU2718) (a) and the complemented strain MS427(pFdeC) (b) were observed by phase-contrast (top) or immunofluorescence (bottom) microscopy using an FdeC-specific antiserum. Strains were grown in the presence of 1 mM IPTG, fixed, and incubated with anti-FdeC serum, followed by incubation with goat anti-rabbit IgG coupled to Alexa Fluor 488. A positive reaction indicating surface localization of FdeC was detected for MS427(pFdeC).
FdeC mediates biofilm formation.
To assess the functional properties of FdeC, we tested its ability to mediate binding to the extracellular matrix (ECM) protein collagen and to epithelial cells. In our assays, MS427(pFdeC) did not promote significant adherence to collagen or to HeLa, HEp-2, or primary bovine rectal epithelial cells (data not shown). Next, we assessed the ability of MS427(pFdeC) to form a biofilm in a static nontreated polystyrene microtiter plate model. MS427(pFdeC) was able to form a strong biofilm at both 28°C and 37°C, in contrast to the vector control strain (Fig. 8A). Finally, biofilm formation mediated by FdeC was assessed under dynamic conditions using the continuous-flow chamber model (13, 31). In this assay, OS56(pFdeC) produced significantly more biofilm growth at both 24 h and 36 h than the negative control, OS56(pSU2718), after induction with IPTG (Fig. 8B). Comparative analysis of the biofilms formed by these two strains after 36 h revealed significant differences in the level of biovolume (P < 0.001), substratum coverage (P < 0.001), surface area-to-biovolume ratio (P < 0.001), mean thickness (P = 0.001), and biofilm roughness (P = 0.001). These results demonstrate that FdeC promotes biofilm formation under continuous-flow growth conditions in an overexpressing strain.
FIG 8.
FdeC mediates biofilm formation. (A) Biofilm formation in polystyrene microtiter plates by the vector control strain MS427(pSU2718) and the complemented strain MS427(pFdeC) at 28°C and 37°C. Results are averages of OD595 readings from at least 4 independent experiments ± standard errors of the means. The expression of FdeC resulted in strong biofilm formation at both temperatures. (B) Dynamic flow chamber assay examining biofilm formation by E. coli OS56 containing pFdeC (left) or the control plasmid pSU2718 (right). Biofilm development was monitored by confocal scanning laser microscopy after 36 h. The images are representative horizontal sections collected within each biofilm and vertical sections (on the right and above each larger panel, representing the yz and xz planes, respectively). FdeC expression resulted in strong biofilm formation.
DISCUSSION
Bacterial adherence to mammalian cells is a complex process involving a wide range of different adhesins, with the complement of surface proteins and their regulation dramatically influencing the outcome of infection. The adherence of EHEC is attributed mainly to the intimin/Tir system and other LEE-associated adhesins; however, some non-O157 EHEC strains are able to cause disease although they lack the LEE (39). This suggests that other adhesins are able to compensate for the lack of intimin (for example, the long polar fimbriae [40]), although no proteins characterized to date appear to completely explain this phenomenon. The majority of E. coli genomes include a gene annotated as eaeH, which was first described in a comparison of an enterotoxigenic E. coli (ETEC) genome (strain H10407) with a K-12 laboratory strain (41). This gene is truncated in K-12 strains and was computationally predicted to encode an adhesin/invasin in genome-wide studies, based on its predicted structural similarity with intimin and the Yersinia invasin. This gene is highly prevalent and well conserved across all pathotypes, and the protein encoded has a structure similar to that of intimin, with an N-terminal β-barrel domain and a C-terminal extracellular domain. This family of proteins with an inverted autotransporter structure has recently been designated type Ve and has been shown to be secreted in a manner similar to that of traditional autotransporters (10). This intimin-like protein was recently characterized in UPEC and was named FdeC (8). FdeC was shown to influence the adherence of UPEC to bladder epithelial cells and collagen I, III, V, and VI and to enhance virulence in a mouse model of UTI. Here we have examined the regulation and function of fdeC in EHEC.
The previous study with UPEC reported that FdeC is not expressed under laboratory conditions but is induced when UPEC comes into contact with mammalian cells and that it can influence the progression of infection in a mouse UTI model (8). Our initial aim was therefore to investigate the mechanism of repression of fdeC and to determine the conditions under which this protein is produced by EHEC. We therefore constructed a lacZ reporter strain that could be used to analyze fdeC promoter activity. After testing various in vitro growth conditions, we detected a striking increase in promoter activity during growth at or above 39°C over that at 37°C, and we confirmed that FdeC was expressed by EHEC following growth at ≥39°C.
EHEC strains are zoonotic pathogens that colonize the terminal rectums of cattle and other ruminants, where the normal temperature is approximately 39°C (42). It has also been demonstrated that the rectal temperature of cattle can rise to around 41°C when cattle are exposed to high ambient temperatures (43). This suggests that EHEC may possess specific adaptations to growth at temperatures greater than 37°C. Indeed, heat treatment of O157 EHEC at 48°C has been shown to increase the expression of the virulence-associated genes eae, hcpA, iha, lpfA, and toxB (44). Interestingly, we could replicate the same pattern of thermoregulation not only in the O157:H7 strain but also in the UPEC strain CFT073, indicating that the mechanism of regulation is likely conserved.
Analysis of the genomic context of fdeC in a range of E. coli pathotypes indicated that there is a conserved 550-bp region immediately upstream of fdeC that we hypothesized to be involved in its regulation. The transcription start site of fdeC was determined by 5′-RACE to be 184 bp upstream of the predicted ATG start codon, and we therefore chose to analyze this sequence in more detail. Bioinformatic analysis detected a putative H-NS nucleation site (33, 34). Additionally, analysis of a random transposon mutant library revealed that a mutation in the hns gene led to upregulation of fdeC expression.
H-NS is a global regulator involved in the silencing of a number of E. coli genes (45, 46). Many E. coli H-NS-regulated genes are also thermoregulated; these include the enteroinvasive E. coli (EIEC) gene virF (47), the ETEC gene encoding heat-labile enterotoxin (LT) (48), the O157:H7 ehx operon (49), and the EPEC AIDA-I gene and its associated glycosyltransferase (50). We therefore sought to determine whether fdeC is repressed by H-NS. Targeted deletion of hns in the reporter strain, along with complementation with plasmid-encoded H-NS (pHNS), allowed us to confirm that repression of fdeC promoter activity at 37°C is relieved in the absence of H-NS. The construction of hns mutants in the EHEC strains N39 and EDL933 allowed us to confirm this result at the protein level by Western blotting. Analysis of a CFT073hns mutant also revealed conservation of this regulation in UPEC. A gel shift assay was used to demonstrate direct binding of H-NS proximally to the fdeC promoter. Together, these findings strongly suggest that fdeC is directly repressed by H-NS, most likely via its binding to the predicted H-NS nucleation site. However, this alone does not entirely explain the mechanism of fdeC regulation.
A predicted RNA thermometer was identified in the 5′ untranslated region (5′ UTR) of the fdeC transcript. RNA thermometers are regulatory elements that form secondary structures at normal (or lower) temperatures that occlude the ribosome binding site and thereby prevent translation. A rise in temperature destabilizes the RNA secondary structure and allows translation to proceed (51, 52). It would seem likely that in addition to the repression of fdeC transcription by H-NS, the translation of fdeC is also directly regulated by temperature via an RNA thermometer. We hypothesize that in the presence of H-NS, low-level transcription may be possible; however, at low temperatures, translation is prevented by the RNA secondary structure. If the temperature increases, then translation is allowed to occur, and a low level of protein production is possible. The absence of H-NS results in much higher levels of transcription; however, a temperature effect on translation should still occur. Although such a temperature effect was not clear by Western blotting due to the very high concentration of protein, it was evident in our β-galactosidase assays using the reporter strains (data not shown). In the absence of H-NS, FdeC can be produced at a lower temperature, indicating that repression of translation by the RNA thermometer is not absolute and that very high levels of transcript can overcome this regulatory element.
In the ExPEC strain IHE3034, MatA regulates the mat (ecp) operon by forming a feedback loop that destabilizes H-NS binding, allowing expression at low temperatures, acidic pHs, and high acetate concentrations (53). Since the very highly conserved mat (or ecp) operon is located near fdeC (Fig. 1) and is also regulated by H-NS and temperature, we sought to determine whether pH and acetate concentration would also influence fdeC regulation. Analysis of fdeC promoter activity under various growth conditions indicated that fdeC expression is influenced by pH and acetate concentration inversely to their influence on the mat (or ecp) operon, just as with altered temperature. That is, assuming that the mat (or ecp) operon is regulated similarly in EHEC and in ExPEC, while mat (or ecp) is upregulated by low temperatures, low pHs, and high acetate concentrations, fdeC is upregulated by high temperatures, high pHs, and the absence of acetate. Whether or not this apparent inverse relationship is indicative of a linked regulatory mechanism remains to be determined.
FdeC functions as an adhesin in E. coli IHE3034 through its ability to mediate adherence to bladder epithelial cells and collagen (8). We were unable to replicate these results using FdeC cloned from our O26:H11 EHEC strain; our recombinant E. coli MS427(pFdeC) strain failed to adhere to HeLa, Hep2, or primary bovine rectal epithelial cells, and to a range of ECM proteins, including collagen. While this may suggest that the cellular adherence function of FdeC is specific to uroepithelial cells, Nesta et al. (8) used a truncated recombinant protein for investigating adherence to ECM proteins, while we employed an FdeC overexpression strain, which may account for the discrepancy between the two studies. It is possible that in our system the predicted extracellular domain of FdeC that was demonstrated by Nesta et al. to mediate attachment (8) either is cleaved from the surface of the recombinant strain, is not folded correctly when overexpressed, or is occluded by other surface proteins. Further study of the function of FdeC in different pathotypes is therefore required. The previous study also demonstrated a role for FdeC in cell aggregation and adherence to glass (8). In this study, we examined the role of FdeC in biofilm formation and demonstrated that its expression in a recombinant K-12 background led to greatly enhanced biofilm formation both in a static plate-based assay and in a continuous-flow model.
In summary, we have characterized the regulation of the intimin-like protein FdeC in EHEC and have identified H-NS as a repressor of fdeC transcription. We also show that the expression of FdeC is exquisitely controlled by temperature. FdeC mediates biofilm formation following overexpression in a recombinant strain background, and its expression at ≥39°C suggests that it may have a role in EHEC when the organism resides in the terminal rectums of cattle, where such temperatures are not abnormal. Future studies will seek to test this hypothesis.
ACKNOWLEDGMENTS
This work was supported by grants from the Queensland Government Smart Futures National and International Research Alliances Program (2008004328) and the Australian Research Council (DP1097032). M.A.S. is supported by an ARC Future Fellowship (FT100100662).
We thank researchers from the CSIRO Animal, Food and Health Sciences Division for the provision of STEC isolates.
Footnotes
Published ahead of print 19 September 2014
REFERENCES
- 1.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295:405–418. 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45–E62. 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 4.Matthews L, Low JC, Gally DL, Pearce MC, Mellor DJ, Heesterbeek JA, Chase-Topping M, Naylor SW, Shaw DJ, Reid SW, Gunn GJ, Woolhouse ME. 2006. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl. Acad. Sci. U. S. A. 103:547–552. 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith JL, Fratamico PM, Gunther NW., IV 2014. Shiga toxin-producing Escherichia coli. Adv. Appl. Microbiol. 86:145–197. 10.1016/B978-0-12-800262-9.00003-2. [DOI] [PubMed] [Google Scholar]
- 6.Bardiau M, Szalo M, Mainil JG. 2010. Initial adherence of EPEC, EHEC and VTEC to host cells. Vet. Res. 41:57. 10.1051/vetres/2010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potter AA, Klashinsky S, Li Y, Frey E, Townsend H, Rogan D, Erickson G, Hinkley S, Klopfenstein T, Moxley RA, Smith DR, Finlay BB. 2004. Decreased shedding of Escherichia coli O157:H7 by cattle following vaccination with type III secreted proteins. Vaccine 22:362–369. 10.1016/j.vaccine.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3(2):e00010-12. 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077. 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oberhettinger P, Schütz M, Leo JC, Heinz N, Berger J, Autenrieth IB, Linke D. 2012. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS One 7:e47069. 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atlung T, Ingmer H. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7–17. 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 12.Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 13.Kjaergaard K, Schembri MA, Ramos C, Molin S, Klemm P. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695–702. 10.1046/j.1462-2920.2000.00152.x. [DOI] [PubMed] [Google Scholar]
- 14.Sherlock O, Schembri MA, Reisner A, Klemm P. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058–8065. 10.1128/JB.186.23.8058-8065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933–946. 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 16.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect. Immun. 80:321–332. 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez E, Bartolomé B, de la Cruz F. 1988. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19 plasmids. Gene 68:159–162. 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. 2006. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr. Purif. 47:446–454. 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulett GC, Webb RI, Schembri MA. 2006. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 152:2101–2110. 10.1099/mic.0.28607-0. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo J-M, Schembri MA. 2012. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl. Environ. Microbiol. 78:2179–2189. 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria, vol 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Vlahovicek K, Kajan L, Pongor S. 2003. DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res. 31:3686–3687. 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beloin C, Dorman CJ. 2003. An extended role for the nucleoid structuring protein H-NS in the virulence gene regulatory cascade of Shigella flexneri. Mol. Microbiol. 47:825–838. 10.1046/j.1365-2958.2003.03347.x. [DOI] [PubMed] [Google Scholar]
- 25.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10:589–604. 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 26.Mabbett AN, Ulett GC, Watts RE, Tree JJ, Totsika M, Ong CL, Wood JM, Monaghan W, Looke DF, Nimmo GR, Svanborg C, Schembri MA. 2009. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int. J. Med. Microbiol. 299:53–63. 10.1016/j.ijmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo JM, Schembri MA. 2012. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. J. Bacteriol. 194:5769–5782. 10.1128/JB.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 190:4147–4161. 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 75:3233–3244. 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407. [DOI] [PubMed] [Google Scholar]
- 31.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669. 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 33.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. 2007. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 14:441–448. 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 34.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A. 2007. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 35:6330–6337. 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ussery DW, Higgins CF, Bolshoy A. 1999. Environmental influences on DNA curvature. J. Biomol. Struct. Dyn. 16:811–823. 10.1080/07391102.1999.10508294. [DOI] [PubMed] [Google Scholar]
- 36.Yamada H, Muramatsu S, Mizuno T. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 108:420–425. [DOI] [PubMed] [Google Scholar]
- 37.Zuber F, Kotlarz D, Rimsky S, Buc H. 1994. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol. Microbiol. 12:231–240. 10.1111/j.1365-2958.1994.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 38.Lehti TA, Bauchart P, Kukkonen M, Dobrindt U, Korhonen TK, Westerlund-Wikstrom B. 2013. Phylogenetic group-associated differences in regulation of the common colonization factor Mat fimbria in Escherichia coli. Mol. Microbiol. 87:1200–1222. 10.1111/mmi.12161. [DOI] [PubMed] [Google Scholar]
- 39.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, Robins-Browne RM, Paton JC, Whittam TS, Paton AW, Hartland EL. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372–380. 10.3201/eid1503.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galli L, Torres AG, Rivas M. 2010. Identification of the long polar fimbriae gene variants in the locus of enterocyte effacement-negative Shiga toxin-producing Escherichia coli strains isolated from humans and cattle in Argentina. FEMS Microbiol. Lett. 308:123–129. 10.1111/j.1574-6968.2010.01996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Savarino SJ, Venkatesan MM. 2006. Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152:1041–1054. 10.1099/mic.0.28648-0. [DOI] [PubMed] [Google Scholar]
- 42.Reid ED, Fried K, Velasco JM, Dahl GE. 2012. Correlation of rectal temperature and peripheral temperature from implantable radio-frequency microchips in Holstein steers challenged with lipopolysaccharide under thermoneutral and high ambient temperatures. J. Anim. Sci. 90:4788–4794. 10.2527/jas.2011-4705. [DOI] [PubMed] [Google Scholar]
- 43.Gaughan JB, Bonner S, Loxton I, Mader TL, Lisle A, Lawrence R. 2010. Effect of shade on body temperature and performance of feedlot steers. J. Anim. Sci. 88:4056–4067. 10.2527/jas.2010-2987. [DOI] [PubMed] [Google Scholar]
- 44.Slanec T, Schmidt H. 2011. Specific expression of adherence-related genes in Escherichia coli O157:H7 strain EDL933 after heat treatment in ground beef. J. Food. Prot. 74:1434–1440. 10.4315/0362-028X.JFP-11-018. [DOI] [PubMed] [Google Scholar]
- 45.Dorman CJ. 2013. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem. Soc. Trans. 41:542–547. 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- 46.Fang FC, Rimsky S. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113–120. 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colonna B, Casalino M, Fradiani PA, Zagaglia C, Naitza S, Leoni L, Prosseda G, Coppo A, Ghelardini P, Nicoletti M. 1995. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 177:4703–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trachman JD, Yasmin M. 2004. Thermo-osmoregulation of heat-labile enterotoxin expression by Escherichia coli. Curr. Microbiol. 49:353–360. 10.1007/s00284-004-4282-y. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Granat A, Stewart V, Gillespie JR. 2008. RpoS, H-NS, and DsrA influence EHEC hemolysin operon (ehxCABD) transcription in Escherichia coli O157:H7 strain EDL933. FEMS Microbiol. Lett. 285:257–262. 10.1111/j.1574-6968.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- 50.Benz I, van Alen T, Bolte J, Wormann ME, Schmidt MA. 2010. Modulation of transcription and characterization of the promoter organization of the autotransporter adhesin heptosyltransferase and the autotransporter adhesin AIDA-I. Microbiology 156:1155–1166. 10.1099/mic.0.032292-0. [DOI] [PubMed] [Google Scholar]
- 51.Chowdhury S, Maris C, Allain FH, Narberhaus F. 2006. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25:2487–2497. 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narberhaus F, Waldminghaus T, Chowdhury S. 2006. RNA thermometers. FEMS Microbiol. Rev. 30:3–16. 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- 53.Lehti TA, Heikkinen J, Korhonen TK, Westerlund-Wikström B. 2012. The response regulator RcsB activates expression of Mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 194:3475–3485. 10.1128/JB.06596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]