Abstract
Stable isotope probing (SIP) of nucleic acids is a powerful tool for studying the functional traits of microbial populations within complex communities, but SIP involves a number of technical challenges. Many of the difficulties in DNA-SIP and RNA-SIP experiments can be effectively overcome with an efficient, sensitive method for quantitating the isotopic enrichment of nucleic acids. Here, we present a sensitive method for quantitating 13C enrichment of nucleic acids, requiring a few nanograms of sample, and we demonstrate its utility in typical DNA-SIP and RNA-SIP experiments. All five nucleobases (adenine, guanine, cytosine, thymine, and uracil) were separated and detected by using ultrahigh-performance liquid chromatography–tandem mass spectrometry. We detected all isotopic species in samples with as low as 1.5 atom% 13C above natural abundance, using 1-ng loadings. Quantitation was used to characterize the isotopic enrichment kinetics of cellulose- and lignin-based microcosm experiments and to optimize the recovery of enriched nucleic acids. Application of our method will minimize the quantity of expensive isotopically labeled substrates required and reduce the risk of failed experiments due to insufficient recovery of labeled nucleic acids for sequencing library preparation.
INTRODUCTION
The application of stable isotope probing (SIP) in molecular biology began over 15 years ago and has since become a powerful tool in microbiology, particularly in the study of complex communities in various environments. Since the last major review paper on SIP (1), and subsequent advances (2, 3), there continue to be innovative developments such as “stable isotope switching” (4). However, despite the promise of pairing DNA-SIP and RNA-SIP to high-throughput sequencing technology, research in this area has progressed slowly. This is owing, in part, to the risk of inconclusive results due to insufficient recovery of nucleic acids for downstream sequence analysis. Thus, SIP experiments can be risky, given the high cost of isotopically labeled substrates or the need to custom synthesize them and laborious experimental procedures. Another major challenge in performing a successful SIP experiment is optimizing the incubation time and substrate concentration to ensure detectable levels of enriched nucleic acids while minimizing dilution of the isotope via turnover of biomass. To avoid the risk of insufficient enrichment, researchers typically err on the side of excess substrate or incubation time. Here, we address these limitations by developing a technique for quantitating isotopic enrichment of nucleic acids using ultrahigh-performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS), requiring minimal consumption of sample. We demonstrate how one might use quantitation to optimize incubations with 13C-labeled lignin and cellulose substrates and maximize recovery of enriched nucleic acids for downstream analyses. Forest soil was chosen as a study system, since lignocellulose decomposition is a key process in that environment, and there is commercial potential for the valorization of lignocellulose.
There are a number of methods for determining the degree of enrichment of nucleic acids and the success of separation of heavy and light fractions of nucleic acids following density gradient centrifugation. For the latter, qualitative methods have been used to distinguish between 13C-labeled and control samples, such as visualizing the distribution of DNA among fractions with ethidium bromide (5) or comparing community composition in fractions via denaturing gradient gel electrophoresis (6). Quantitative PCR (or quantitative reverse transcriptase PCR) is also used to measure nucleic acids within a density gradient and has been the de facto method for measuring RNA, since lower quantities of starting material are added to the gradient (7). Until now, there have been only two quantitative methods which involve direct measurement of [13C]carbon in nucleic acids, but both require between 0.8 and 1.0 μg of nucleic acids and thus cannot be used for assessing enrichment postcentrifugation, where typical recovery is between 10 and 200 ng of nucleic acid. One method is based on elemental analysis using isotope-ratio mass spectrometry (8), while a more recent method uses liquid chromatography-mass spectrometry to measure 13C incorporation into thymine and is therefore unsuitable for the detection of RNA (9). We present a significant improvement of the latter methodology, based on ultrahigh-performance chromatography–tandem mass spectrometry (UHPLC-MS/MS), demonstrating the detection of all five nucleobases, with sufficient sensitivity for all stages of DNA-SIP and RNA-SIP and without substantial depletion of sample.
MATERIALS AND METHODS
DNA/RNA extraction, centrifugation, and fractionation.
Organic and mineral layer soils, originating from Ponderosa Pine plantations in California (38.91N, −120.66W), were incubated with 99 atom% 13C bacterial cellulose, produced from [13C]glucose by Gluconacetobacter xylinus grown on Yamanaka medium (10), or ring-labeled 60 atom% 13C dehydrogenatively polymerized lignin (DHP lignin), synthesized as previously described (11). Substrate was added at concentrations of 10% (wt/wt) dry weight of soil. Soil DNA extractions were performed using the FastDNA spin kit for soil (MP Biomedicals, USA). RNA was extracted from Gluconacetobacter cultures grown on either [12C]glucose or [13C]glucose using the RNA PowerSoil total RNA isolation kit (MoBio, California). RNA was treated with DNase (Turbo DNA-free; Life Technologies, Inc., Carlsbad, CA) and then washed twice with diethyl pyrocarbonate (DEPC)-treated water using Amicon Ultra 0.5-ml centrifugal filters (Millipore, Massachusetts) to remove residual DNA. An artificial 1:1 mixture of RNA was prepared by combining equal amounts of unlabeled and 13C-labeled RNA. Separation of enriched DNA was performed using cesium chloride according to the method outlined by Neufeld et al. (5), while RNA separation was performed with cesium trifluoroacetate according the method of Whiteley et al. (7), except for the following modifications: (i) we used a fixed angle rotor (Beckman-Coulter TLN100) run at 61,000 rpm (131,902 × g) for 52 h, and (ii) we used larger-volume tubes (Quick-Seal polyallomer tubes; 3.9 ml; 13 × 38 mm; Beckman Coulter, USA) with accordingly higher quantities of RNA (1 μg). During fractionation, a constant flow rate was maintained with a syringe pump (model R-E; Razel Scientific, USA). For DNA-SIP, a 60-ml syringe was used, and fractions were collected every 30 s, resulting in 20 fractions of ∼250 μl. The density was measured using refractometry and converted using the following formula: density = (refractive index × 10.927) − 13.593. The DNA in each fraction was suspended in 30 μl of pure water. For RNA-SIP, a 30-ml syringe was used, and fractions were collected every 37.5 s, resulting in 16 fractions of ∼200 μl. The density was measured by weighing samples with 0.1-mg precision. RNA from each fraction was suspended in 20 μl of DEPC-treated pure water.
UHPLC-MS/MS sample preparation.
For quantitation of recovered soil DNA, extracts were first assayed using a Quant-iT PicoGreen dsDNA assay kit (Life Technologies). Extracts were then diluted to ∼1 ng/μl, of which 5 μl was prepared for UHPLC-MS/MS analysis. For density gradient fractions, 5-μl portions of either DNA or RNA samples were used. Each 5-μl sample was mixed with 95 μl of 88% formic acid in 250-μl PCR tubes, followed by incubation at 70°C for 2 h to depolymerize the nucleic acids via hydrolysis. Samples were then completely dried with a Savant SpeedVac SC110A (Thermo Scientific, USA) with a custom-made acid trap (a 500-ml air-tight container packed with soda lime) to prevent damage to the equipment. The sample was then suspended in 40 μl of a UHPLC mobile phase, i.e., a 0.22-μm-pore-size-filtered solution consisting of 98% solvent A (water with 0.1% [vol/vol] formic acid) plus 2% solvent B (methanol with 0.1% [vol/vol] formic acid).
Quantitation was calibrated using DNA and RNA standards based on a dilution series of either lambda DNA (Quant-iT PicoGreen dsDNA assay kit; Life Technologies) or rRNA standard from Escherichia coli (Quant-iT RiboGreen RNA assay kit; Life Technologies). Quantitation was performed in triplicate and one batch of standards was used for all measurements in each experiment.
UHPLC-MS/MS instrumentation and protocol.
Standards were prepared for each nucleobase from the following sources: cytosine (>99%; Amresco, USA), thymine (>99%), guanine (>98%), and adenine (>99%) (all three from Sigma-Aldrich, USA), and uracil (>99%; Alfa Aesar, USA). The UHPLC-MS/MS system consisted of a 1290 Infinity binary pump, a 1290 Infinity sampler, a 1290 Infinity thermostat, and a 1290 Infinity thermostat column compartment (Agilent, Mississauga, Ontario, Canada) connected to an AB Sciex QTrap 5500 hybrid linear ion-trap triple quadrupole mass spectrometer equipped with a Turbo spray source (AB Sciex, Concord, Ontario, Canada). The mass spectrometer was operated in positive-ionization mode, and data were acquired and processed using Analyst software (v1.5.2; AB Sciex).
For the routine detection of adenine and guanine, chromatographic separation was performed by using a Waters Acquity UPLC BEH phenyl column (1.7 μm, 2.1 by 50 mm; Waters Corp., Milford, MA) maintained at 30°C with the autosampler tray temperature maintained at 10°C. The mobile phase was isocratic with solvent A (98%) plus solvent B (2%), as described above. The flow rate was 0.2 ml/min, the injection volume was 20 μl, and the run time was 2.0 min. The compounds eluted at a retention time of approximately 1.0 min, and the mobile phase flow was diverted to the waste before 0.7 min and after 1.5 min.
For the measurement of all five nucleobases, a Waters Acquity UPLC BEH phenyl column (1.7 μm, 2.1 by 150 mm) was used. The conditions were as described above, except that the mobile phase was 95% solvent A plus 5% solvent B, and the run time was 5.0 min, with no mobile-phase flow diversion.
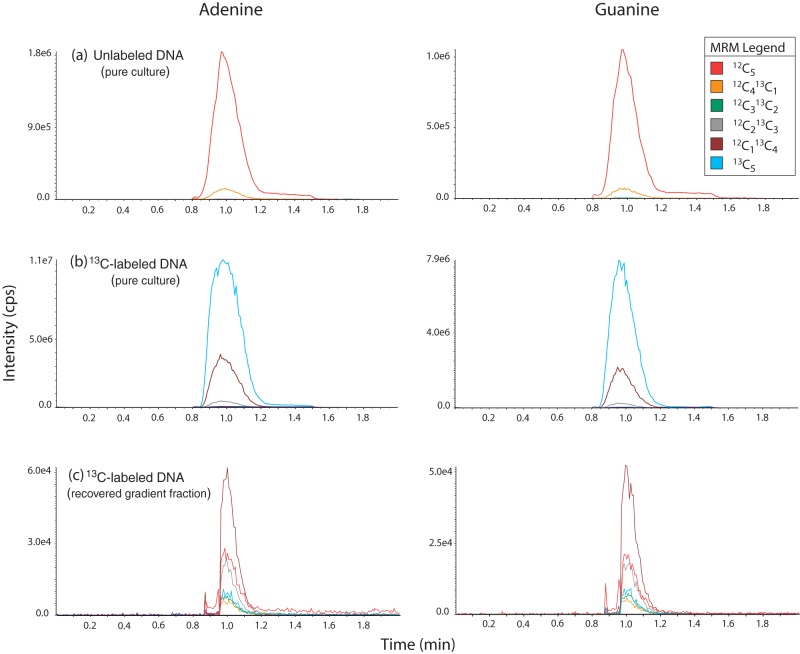
The 13C-enriched isotopic isomers were quantitated by using multiple reaction monitoring (MRM) mode. In MRM, a precursor ion is selected in the first quadrupole, which is fragmented in the collision cell (second quadrupole), and the product ions are detected in the third quadrupole. A MRM trace is generated, which consists of the signals of the precursor and product ions, and this trace can be plotted in separate channels, which allows the independent integration and quantitation of the analytes, even if their chromatographic peaks are not separated. MRM is a much improved technique over single quadrupole MS, because it is more selective and sensitive. The MRM traces with all carbons representing 12C species and the incremental substitution with 13C species were m/z 136.0/119.0 to m/z 141.0/124.0 for adenine and m/z 152.1/135.1 to m/z 157.1/140.1 for guanine (see Fig. 1).
FIG 1.
Examples of MRM spectra for adenine and guanine for three samples: unenriched DNA from a Gluconacetobacter sp. culture (a), 13C-enriched DNA from Gluconacetobacter sp. grown on pure [13C]glucose (b), and 13C-enriched DNA recovered from a high density fraction following the ultracentrifugation of DNA extracted from a soil incubation with 13C-cellulose (c). The differences in retention times between Fig. 1 and Fig. S2 in the supplemental material are due to the use of different chromatography columns and mobile phases, as described in Materials and Methods.
UHPLC-MS/MS data analysis.
As expected, unlabeled nucleic acids had measurable background natural abundance of 13C in MRMC12+1 and MRMC12+2. Based on the average natural abundance of 13C (1.11%), the theoretical probability of detecting a single 13C isotope in a five-carbon nucleobase is 5.9%. We observed average natural abundances of 5.9% ± 0.2% (standard error) from manufactured lambda DNA and 7.7% ± 0.2% with DNA extracted from unlabeled soil samples. We believe the discrepancy in natural abundance from environmental samples results from a combination of the following factors: (i) isotopic discrimination in nucleic acid synthesis, (ii) differences in the abundance of 13C in soil microbial biomass (although soil biomass is typically depleted in 13C carbon), and (iii) a technical artifact related to peak integration, where the less-abundant 13C-labeled species may have inflated peak areas due to widening of peaks near the detection limit of the instrument. In order to determine the amount of 13C above natural abundance in these MRM traces, we constructed a formula based on long-term averages of over 100 samples of unlabeled DNA from soil and culture samples (both were consistently comparable). The final formula used to calculate the 12C level was as follows: MRMC12 + MRMC12+1·0.077 + MRMC12+2·0.0048; the final formula used to calculate the 13C level was as follows: MRMC12+5 + MRMC12+4 + MRMC12+3 + MRMC12+2·0.9952 + MRMC12+1·0.923. It is advisable to always run unenriched standards in order to provide a robust baseline estimate for natural abundance.
DNA sequencing and analysis.
16S rRNA gene libraries were prepared from 12 soil microcosms fed [13C]cellulose and their corresponding 12C-labeled controls from both mineral and organic soil layers (n = 48). In the heavy fractions of 12C-labeled controls, sufficient quantities of DNA for PCR amplification were obtained only by including additional lighter fractions (1.724 to 1.727 g/ml, corresponding to fractions 8 and 9). Sequencing was performed as described previously (12) using 454 pyrosequencing technology (Genome Quebec, Montreal, Quebec, Canada). Libraries were quality trimmed, chimera checked, and processed with mothur, according to the standard operating procedure for 454 data (13). Principle coordinate analyses were performed in R (v3.1.0; R Development Core Team), using the bioconductor package phyloseq (14).
RESULTS
We detected uracil, adenine, thymine, guanine, and cytosine using either DNA or RNA samples with as low as 10-pg loadings to the column (see Fig. S1 in the supplemental material). Using MRM, we tracked the mass of each nucleobase with all carbons represented by 12C species and with incremental substitution of 13C to the most enriched species with mass corresponding to all 13C (Fig. 1). The typical sample requirement to accurately detect enrichment in the least-abundant isotopic species (i.e., complete 13C substitution) was 1.0 to 2.5 ng of nucleic acids. In theory, the detection of 13C depends on the degree of 13C enrichment in nucleic acids, so higher loadings may be required to discern lower levels of enrichment. In practice, we were able to detect all isotopic species in samples with as low as 1.5 atom% 13C above natural abundance using 1-ng loadings. For routine high-throughput applications, we measured only adenine and guanine, thereby shortening the run length from 5 to 2 min per sample.
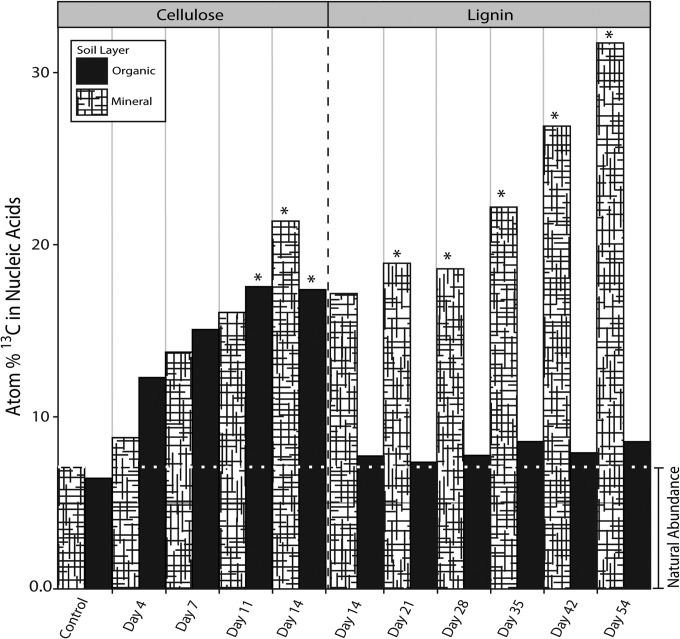
The UHPLC-MS/MS method facilitated optimization of DNA-SIP experiments where mineral and organic soil microcosms were incubated with 13C-labeled cellulose or lignin. We detected 13C enrichment of DNA from the first time point for each substrate (cellulose, day 4; lignin, day 14; Fig. 2), showing that both were readily metabolized by microbial biomass. However, adequate quantities of 13C-enriched DNA necessary for downstream analyses were only obtained at later time points, based on estimates of recovery after ultracentrifugation. Organic soil incubations with lignin did not produce sufficient 13C-enriched DNA for analysis. In mineral soil incubations, incorporation of 13C from lignin was comparable to cellulose. A follow-up experiment, using sterile mineral soil inoculated with a cell slurry from organic soil and incubated with [13C]lignin, resulted in substantial 13C enrichment of DNA.
FIG 2.
Quantitation of atom% 13C in total DNA during soil incubations with [13C]lignin or [13C]cellulose. *, samples that yielded adequate quantities of [13C]DNA (∼50 to 100 ng) for our downstream sequencing needs (i.e., preparation of 16S rRNA gene, ITS region, and metagenomic libraries). Note the difference in timescales along the x axis for the cellulose and lignin incubations.
The 13C content of soil DNA extracts (prior to ultracentrifugation) revealed further differences between soil layers in the incorporation of carbon from cellulose or lignin into DNA. Whereas the average total amount of DNA recovered from organic soils exceeded that from mineral soils, 29 versus 5 μg, respectively, the overall percentage of 13C was lower in organic soils, 3.5 versus 10.3 atom% 13C above natural abundance, respectively. Thus, the total amount of 13C incorporation was comparable, albeit slightly lower in mineral soils, 1 μg versus 0.6 μg, respectively. This information was put to practical use in order to recover maximal quantities of enriched DNA. In the case of organic layer samples, where the 13C was diluted by unenriched nucleic acids, each sample was run multiple times and corresponding density fractions were pooled, since one is limited by the amount of DNA one can add to the gradient. Quantification of 12C and 13C DNA in soil extracts was a consistent predictor of the amount of recoverable “heavy” DNA following ultracentrifugation, enabling the optimal processing of samples with variable levels of enrichment.
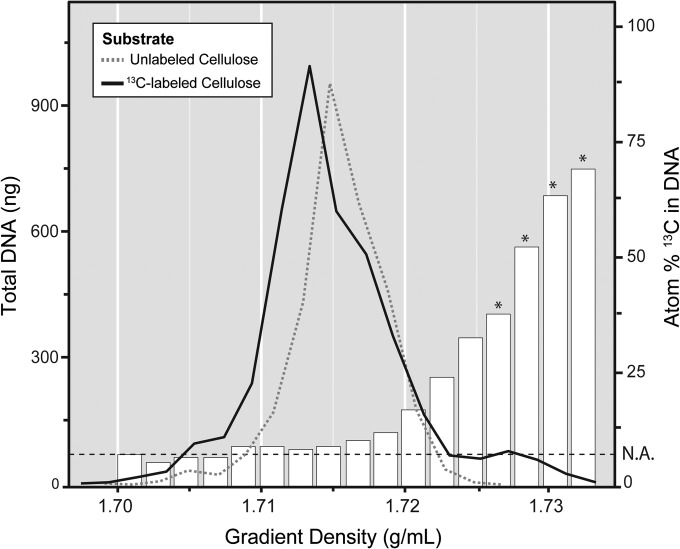
The UHPLC-MS/MS method was subsequently used to assess 13C enrichment in DNA recovered from density fractions after ultracentrifugation (Fig. 3). Only samples from cultures or soils incubated with 13C-labeled substrates had detectable DNA in high-density fractions (1.727 to 1.735 g/ml). Increasing atom% 13C of DNA corresponded with increasing gradient density. Following centrifugation, total recovered DNA amounted to ∼ 3 to 4 μg, and enriched fractions contained ∼ 50 to 200 ng (∼ 1 to 6%). Typically, fractions 4, 5, 6 and 7 were pooled, to yield a composite sample of ~50 atom% 13C for downstream analysis of 13C-enriched DNA.
FIG 3.
Quantitation of the distribution of unlabeled DNA and 13C-enriched DNA, derived from a soil microcosm incubated with [13C]cellulose, across density gradient fractions. The atom% 13C value, corresponding to the bar plots and right y axis, is based on an average enrichment of adenine and guanine. Heavy fractions (>1.736 g/ml) that had no detectable traces of DNA were not included. N.A., natural abundance, which corresponds to the average atom% 13C detected in nucleic acids not originating from an enrichment. *, fractions pooled for downstream analysis based on the requirement of having >50 atom% 13C in the pooled sample.
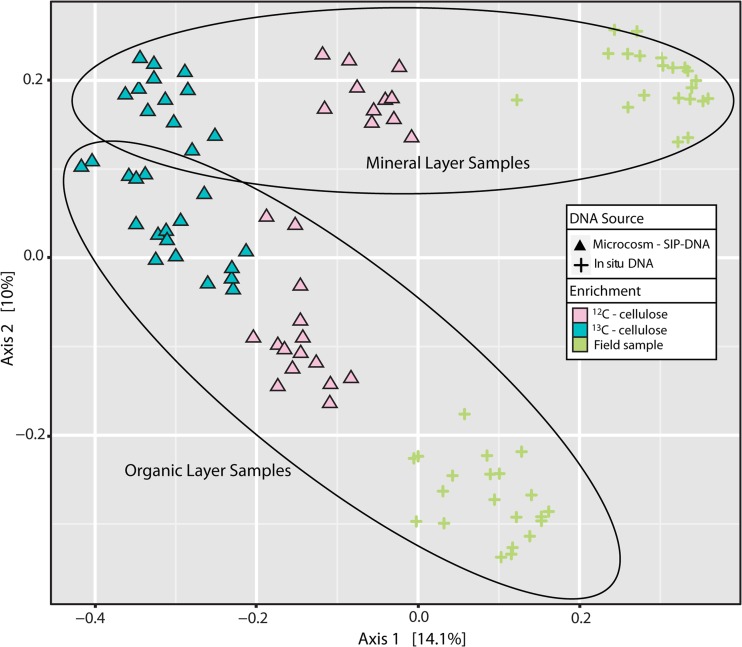
The successful separation and recovery of enriched nucleic acids was evident from the strong distinction between bacterial 16S rRNA gene libraries from soils incubated with 13C-labeled substrates versus unlabeled substrates (Fig. 4). The converging similarity of communities from the two soil layers is consistent with the enrichment of common cellulose-degrading populations from both layers.
FIG 4.
Principle coordinate analyses were conducted based on the Bray-Curtis dissimilarity of bacterial 16S rRNA gene pyrosequencing libraries from soil samples. Blue or pink symbols, respectively, correspond to DNA samples from high-density gradient fractions from incubations with labeled or unlabeled cellulose. Symbol shapes correspond to different sources of DNA template, namely, microcosm DNA (triangles) or unincubated soil DNA extract (cross-hairs [+]).
Finally, we tested the utility of the UHPLC-MS/MS method for RNA-SIP. A 1:1 mixture of RNA extracted from Gluconacetobacter sp. grown on 13C-labeled or unlabeled glucose was fractionated by density-gradient ultracentrifugation (see Fig. S2 in the supplemental material). The RNA-SIP data had higher variability than DNA-SIP data, likely due to the overall low quantities of RNA. However, increasing atom% 13C of RNA clearly corresponded to increasing gradient density. The total recovered RNA was ca. 75 to 100 ng, with enriched fractions containing ∼2.5 to 5 ng (2.5 to 6%).
DISCUSSION
Quantification of 13C-enriched nucleic acids by UHPLC-MS/MS greatly facilitated SIP experiments in several ways. Initially, the method allowed efficient analysis of a large number of samples from soil microcosm time course experiments to characterize the kinetics of 13C incorporation into DNA. These experiments further provided an estimate of the total recoverable 13C-enriched nucleic acids for the substrates and soil types used. Thus, we were able to optimize the incubation time in order to yield the necessary quantity of 13C-enriched nucleic acids for subsequent sequence analyses, while minimizing dilution of the isotope via biomass turnover.
In subsequent experiments, the UHPLC-MS/MS assay permitted quantification of 13C-enriched DNA in small subsamples of total DNA extracts prior to the time-consuming step of density gradient centrifugation, ensuring sufficient recovery of 13C-enriched DNA from the high-density fractions. The extracts were screened for adequate enrichment, and an optimal amount of DNA was then applied to the gradient. This was invaluable for processing different soil types, particularly where loadings of organic soil DNA had to be increased due to the dilution of enriched DNA by the higher overall DNA recovered. This facilitated the preparation of the largest sample size for any DNA-SIP experiment reported to date and exemplifies how quantification can assist in the consistent processing of a large number of samples.
By analysis of total DNA extracts, we were able to quantitatively compare the incorporation of carbon from substrates into biomass among experimental treatments. Interestingly, 13C incorporation from cellulose in organic versus mineral soil was comparable, despite mineral soils having lower overall biomass based on DNA recovery. This may indicate differing substrate accessibility but may also indicate a surprisingly high capacity for cellulose degradation by populations inhabiting the mineral layer. Surprisingly, 13C incorporation from lignin was far greater in mineral soil than in organic soil. The fact that an organic soil community transferred to sterile mineral soil achieved a high 13C incorporation rate suggests that the organic soil per se, rather than its community composition, was the factor limiting incorporation of 13C from lignin.
Finally, the UHPLC-MS/MS assay allowed us to measure 13C-enriched DNA in density gradient centrifugation fractions. The sensitivity of the assay required only small subsamples from the fractions, preserving most of the DNA for sequence analyses. We collected 20 gradient fractions per sample, more than typical experiments (∼12 to 16 fractions), and pooled fractions with only the highest proportion of [13C]DNA (see Fig. 2). As a result, we obtained 13C samples representing microbial populations very distinct from those in 12C controls, facilitating identification of populations that assimilated 13C. The sensitivity of our quantitation method was even more beneficial for RNA-SIP, where far smaller amounts of nucleic acids can be applied during gradient centrifugation, due to the formation of secondary structures (15). The UHPLC-MS/MS assay can replace quantitative reverse transcriptase PCR, which is the current standard method for assessing successful separation, providing a direct, and more relevant, measure of isotopic enrichment.
The successful recovery and quantitation of 13C-enriched RNA demonstrated the broad applicability of the method. The simple, fast, and direct method for assessing separation of 13C-enriched RNA facilitated adapting the previous protocol (7) for use with the equipment available to us. RNA was not as clearly density fractionated by centrifugation as DNA and demonstrated higher variability, since the concentrations were closer to the detection limit. Further, we observed a relatively high background signal across the entire gradient compared to that with DNA, which may be the result of residual DNA nucleobases from the DNase pretreatment. Such a high background level was previously reported in both DNA- and RNA-SIP (15), which is why unlabeled controls are typically fractionated and sequenced in parallel.
By demonstrating a rapid and direct assay for isotopically labeled nucleic acids, we hope to improve the quality of SIP data, build greater confidence in SIP experiments, and pave the way for new SIP approaches. The ongoing reduction in the quantity of nucleic acids required to prepare sequencing libraries increases the feasibility of SIP-metagenome and -metatranscriptome analyses and makes this quantitation method a helpful tool for tailoring experiments. More selective recovery of 13C-enriched nucleic acids will simplify the downstream process of resolving sequences that are differentially abundant between enriched samples and controls. Direct analysis of isotopically labeled nucleic acids will facilitate assessing isotope incorporation and transfer via cross-feeding in experimental systems, improving experimental design and the interpretation of results. The UHPLC-MS/MS instrumentation used for this analysis is becoming widely available at research institutions, and the supplemental material includes suggestions for adapting the protocol to alternative instrumentation.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a large-scale applied research project grant from Genome Canada and Genome BC. R.W. was supported by the Alexander Graham Bell Canada Graduate Scholarship (CGS) from the Natural Sciences and Engineering Research Council of Canada.
We thank Rahul Singh for assistance in preparing DHP lignin and Josh Neufeld for providing Gluconacetobacter xylinus.
Footnotes
Published ahead of print 12 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02223-14.
REFERENCES
- 1.Neufeld JD, Wagner M, Murrell JC. 2007. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 1:103–110. 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Murrell JC. 2010. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 18:157–163. 10.1016/j.tim.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Uhlik O, Leewis MC, Strejcek M, Musilova L, Mackova M, Leigh MB, Macek T. 2013. Stable isotope probing in the metagenomics era: a bridge towards improved bioremediation. Biotechnol. Adv. 31:154–165. 10.1016/j.biotechadv.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxfield PJ, Dildar N, Hornibrook ERC, Stott AW, Evershed RP. 2012. Stable isotope switching (SIS): a new stable isotope probing (SIP) approach to determine carbon flow in the soil food web and dynamics in organic matter pools. Rapid Commun. Mass Spectrom. 26:997–1004. 10.1002/rcm.6172. [DOI] [PubMed] [Google Scholar]
- 5.Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, Murrell JC. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860–866. 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A. 2013. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 7:2091–2104. 10.1038/ismej.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteley AS, Thomson B, Lueders T, Manefield M. 2007. RNA stable-isotope probing. Nat. Protoc. 2:838–844. 10.1038/nprot.2007.115. [DOI] [PubMed] [Google Scholar]
- 8.Rangel-Castro JI, Killham K, Ostle N, Nicol GW, Anderson IC, Scrimgeour CM, Ineson P, Meharg A, Prosser J. 2005. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ. Microbiol. 7:828–838. 10.1111/j.1462-2920.2005.00756.x. [DOI] [PubMed] [Google Scholar]
- 9.Auclair J, Lépine F, Villemur R. 2012. A liquid chromatography–mass spectrometry method to measure 13C-isotope enrichment for DNA stable-isotope probing. Can. J. Microbiol. 58:287–292. 10.1139/w11-133. [DOI] [PubMed] [Google Scholar]
- 10.Ruka DR, Simon GP, Dean KM. 2012. Altering the growth conditions of Gluconacetobacter xylinus to maximize the yield of bacterial cellulose. Carbohydr. Polym. 89:613–622. 10.1016/j.carbpol.2012.03.059. [DOI] [PubMed] [Google Scholar]
- 11.Gagnaire D, Robert D. 1977. A polymer model of lignin (D.H.P.) 13C selectively labelled at the benzylic positions: synthesis and NMR study. Die Makromolekulare Chemie 178:1477–1495. 10.1002/macp.1977.021780522. [DOI] [Google Scholar]
- 12.Hartmann M, Howes CG, Van Insberghe D, Yu H, Bachar D, Christen R, Nilsson RH, Hallam SJ, Mohn WW. 2012. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 6:2199–2218. 10.1038/ismej.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sah JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lueders T, Manefield M, Friedrich MW. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73–78. 10.1046/j.1462-2920.2003.00536.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.