Abstract
Microbial communities that deconstruct plant biomass have broad relevance in biofuel production and global carbon cycling. Biomass pretreatments reduce plant biomass recalcitrance for increased efficiency of enzymatic hydrolysis. We exploited these chemical pretreatments to study how thermophilic bacterial consortia adapt to deconstruct switchgrass (SG) biomass of various compositions. Microbial communities were adapted to untreated, ammonium fiber expansion (AFEX)-pretreated, and ionic-liquid (IL)-pretreated SG under aerobic, thermophilic conditions using green waste compost as the inoculum to study biomass deconstruction by microbial consortia. After microbial cultivation, gravimetric analysis of the residual biomass demonstrated that both AFEX and IL pretreatment enhanced the deconstruction of the SG biomass approximately 2-fold. Two-dimensional nuclear magnetic resonance (2D-NMR) experiments and acetyl bromide-reactive-lignin analysis indicated that polysaccharide hydrolysis was the dominant process occurring during microbial biomass deconstruction, and lignin remaining in the residual biomass was largely unmodified. Small-subunit (SSU) rRNA gene amplicon libraries revealed that although the dominant taxa across these chemical pretreatments were consistently represented by members of the Firmicutes, the Bacteroidetes, and Deinococcus-Thermus, the abundance of selected operational taxonomic units (OTUs) varied, suggesting adaptations to the different substrates. Combining the observations of differences in the community structure and the chemical and physical structure of the biomass, we hypothesize specific roles for individual community members in biomass deconstruction.
INTRODUCTION
Complex microbial communities, typically consisting of bacteria, archaea, fungi, and protists, depolymerize plant biomass across natural environments (1). These communities have important roles in the cycling of carbon in terrestrial ecosystems and as sources of enzymes to hydrolyze plant polysaccharides to sugars for conversion to biofuels (2). It has been reported that the microbial community structure corresponds with the complexity of the cell wall structures in plants (3), making it challenging to understand the structure and dynamics of natural biomass-deconstructing microbial communities. Investigations of simplified communities that deconstruct biomass with predictable plant cell wall structures are vital, as they can provide insights into how microbes deconstruct plant biomass that can be translated to more complex systems (4–8).
Chemical pretreatment of plant biomass provides a well-established method to generate differential biomass substrates that have predictable physiochemical properties (9). Pretreatment reorients the physical structure of the plant cell wall and reduces the recalcitrance of the polysaccharides, cellulose, and hemicellulose (10). Depending on the pretreatment, some plant polymers, primarily hemicellulose and lignin, are depolymerized, and the resulting substrate is enriched in cellulose (11). Two prominent chemical pretreatments of biomass are ammonia fiber expansion (AFEX) pretreatment, in which gaseous ammonia disrupts the plant cell wall while maintaining the composition of the biomass (10, 12, 13), and ionic-liquid (IL) pretreatment, in which molten organic salts are used to achieve partial dissolution of biomass, yielding a cellulose-enriched substrate in which the polysaccharides are much more accessible than in the native biomass. Both pretreatments substantially increase the rate and extent of enzymatic polysaccharide hydrolysis compared to untreated biomass. AFEX- and IL-pretreated samples are useful in comparative studies because the native biomass structure is extensively disrupted in IL pretreatment but is less perturbed during AFEX pretreatment.
Here, we describe parallel enrichments of compost-derived consortia grown under aerobic conditions on untreated and pretreated (AFEX-pretreated and IL-pretreated) switchgrass. These parallel enrichments demonstrated that community structure was influenced by the physiochemical properties of the biomass substrate and that polysaccharides in the pretreated substrates were preferably depolymerized over lignin.
MATERIALS AND METHODS
Sample collection and enrichment of thermophilic consortia.
Compost samples were purchased from a municipal green waste compositing facility, the Newby Island Sanitary Landfill (NIC) in Milpitas, CA, USA (37°27′15.52″N, 121°55′17.35″W). The green waste consisted of yard trimmings and discarded food waste from an end-stage compost pile. Samples were transported to the lab at room temperature and inoculated within 24 h. The ionic-liquid (IL)-pretreated switchgrass was prepared by heating switchgrass with 1-ethyl-3-methylimidazolium acetate as previously described and washed with water to remove the ionic liquid (11). Ammonium fiber expansion (AFEX)-pretreated switchgrass was prepared as previously described and washed with water after pretreatment to remove readily solubilized compounds (14). Untreated switchgrass was extracted with water and ethanol to remove soluble compounds (6). The adaptation of thermophilic consortia to purified biomass substrates was described previously (5). Briefly, biomass substrates (0.5%, wt/vol) were the sole supplemented carbon and energy source in 50 ml of M9 medium augmented with vitamins and buffered with 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) at a final pH of 6.5 (6). Approximately 0.5 g of the NIC compost material was inoculated into the initial enrichments. Three biological replicates for each carbon source were incubated in parallel at 60°C under aerobic conditions at 200 rpm. The enrichments were serially passed through five sets of liquid cultures (10% [vol/vol] inoculum). Total DNA recovered from the cultures after 2 weeks of growth ranged from ca. 100 to 300 μg of DNA per 50 ml culture. DNA was not recovered from the control samples without substrate after the first passage, as previously reported (5). Additional control experiments were performed in duplicate with each biomass substrate in the absence of inoculation. No microbial growth was evident in these uninoculated controls.
Measurement of glycoside hydrolase activity.
At the end of each serial passage, glycoside hydrolase (GH) activity was measured using the 3,5-dinitrosalicyclic acid assay with 50 mM MES at a pH 6.0 as described previously (5). Standard curves ranging from a 0 to 5 mM concentration of either xylose (xylanase assay) or glucose (cellulase assay) were run with every assay to ensure that the insoluble substrates did not interfere with the accuracy of the assay. The R2 value for the standard curves ranged from 0.955 to 0.997.
Small-subunit (SSU) rRNA amplicon pyrosequencing.
Total DNA was extracted from 0.5-ml aliquots from each replicate across the treatments using a MoBio PowerSoil DNA extraction kit (MoBio, Carlsbad, CA). SSU rRNA amplicon pyrosequencing of each DNA sample targeting the 16S rRNA gene using 926F (5′-AAACTYAAAKGAATTGACGG-3′) and 1392R (5′-ACGGGCGGTGTGTRC-3′) primers from the enrichments were performed at the Department of Energy Joint Genome Institute (Walnut Creek, CA) following a previously established protocol (15). Sequence tags were trimmed and analyzed using the pyroclust method at a sequence similarity of 97% (OTU97) in the PyroTagger Program (http://pyrotagger.jgi-psf.org) with a 200-bp sequence length and accuracy of 10% for low-quality bases (7). Singletons and low-quality reads along with putative chimeras were removed from the final data set. Taxonomic identification of the PyroTagger-defined OTUs was determined with the RDP classifier (http://rdp.cme.msu.edu/), and the closest validated isolate to each sequence was determined by a BLAST search against the collection of 16S rRNA genes for bacteria and archaea found in GenBank.
Diversity statistics were generated using the mothur software package, normalized to the smallest pyrosequencing library (n = 1,934) (16). The PyroTagger cluster classification output file was modified to a shared file, which was used as the input for mothur. Bacterial richness, which is a measure of the number of different species, was estimated using Chao and the abundance coverage estimator (ACE) at the operational taxonomic unit (OTU) of 0.03, which correlates to a sequence similarity of 97% (here referred to as OTU97). Bacterial diversity, which is a combined measure of the number of different species along with the relative abundance of those species, was estimated using the Shannon and Simpson indices of diversity (transformed using the equation −lnD) at OTU97. Beta diversity (as measured using the Bray-Curtis dissimilarity index), analysis of variance, and indicator species analysis were performed with the R program (http://www.r-project.org/).
Biomass analysis.
The biomass content of each switchgrass substrate before cultivation was determined using a modification of a published protocol (12) (see Table 4). Approximately 200 mg of pretreated (IL and AFEX) and unpretreated switchgrass samples were treated with 2 ml of 72% H2SO4 in duplicate (with stir bars) and incubated at 30°C (120 to 200 rpm) for 60 min. Following the incubation, 56 ml of distilled water was added to each sample after incubation and autoclaved at 121°C for 60 min. Subsequent filtration through porcelain crucibles yielded filtrates for quantitative analysis. Each filtrate was analyzed for glucose, xylose, and cellobiose using the Agilent high-performance liquid chromatography (HPLC) (1100 series) system equipped with a 1200 series refractive index (RI) detector as previously described (17). The retained acid-insoluble fractions were dried at 105°C and burnt to ash in a muffled furnace at 575°C for 6 h to determine the acid-insoluble lignin content. The ash content was determined by burning approximately 50 mg of each biomass in duplicate in a muffled furnace at 575°C for 6 h.
TABLE 4.
Compositional measurements of switchgrass substrates
| Sample | Average biomass composition (g/g)a |
||
|---|---|---|---|
| Glucanb,c | Xylanb | Acid-insoluble lignin (Klason lignin)d | |
| Untreated switchgrass | 0.421 | 0.259 | 0.172 |
| AFEX-pretreated switchgrass | 0.338 | 0.242 | 0.182 |
| IL-pretreated switchgrass | 0.548 | 0.239 | 0.116 |
Biomass composition measurements were performed in duplicate.
Glucan and xylan measurements varied ≤2% between replicates.
Average cellobiose composition was ∼0.007 g/g (1.6% of total glucan).
Soluble lignin content was not determined. Acid-insoluble lignin measurements varied ≤5% between replicates.
Residual biomass after microbial cultivation was insufficient to perform complete compositional analysis, so the samples were compared using gravimetric acetyl bromide-reactive lignin (ABL) and acid-precipitable polymeric lignin (APPL) analyses. Residual biomass analysis was performed on duplicates in passage 4 (replicate 1 and replicate 3) and reported as the average for two biological replicates. The residual biomass was collected by centrifugation at 10,000 × g for 10 min and filtered through Miracloth (EMD Millipore, Billerica, MA, USA). The retained biomass samples were washed several times with distilled water to remove microbial cells, then lyophilized, and weighed. The amount of APPL released into the medium during enrichment was determined by sequentially filtering 22 ml of each culture supernatant through 0.45- and 0.22-μm filters (18). The filtered supernatants were acidified to pH 1.0 to 2.0 with ca. 150 μl of 10 N HCl and kept at 4°C to precipitate APPL. The APPL samples were precipitated, washed, lyophilized, and weighed. Gravimetric analysis and APPL release were also measured in the uninoculated control experiments. The ABL content of each residual biomass was determined as previously described and is reported as a relative increase in the residual biomass compared to the substrate before cultivation (19).
Two-dimensional (2D) 13C-1H heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectroscopy.
Plant cell wall samples from passage 4 replicates were extracted with water-ethanol and ball milled as previously described (20, 21). Gels were formed using dimethyl sulfoxide d6 (DMSO-d6) and 1-ethyl-3-methylimidazolium acetate as a cosolvent (22, 23) and homogenized by sonication in a Branson 2510 tabletop cleaner (Branson Ultrasonic Corporation, Danbury, CT). The temperature of the bath was closely monitored and maintained below 55°C. The homogeneous solutions were transferred to NMR tubes. HSQC spectra were acquired at 25°C using a Bruker Avance-600 MHz instrument equipped with a 5-mm inverse-gradient 1H/13C cryoprobe using a q_hsqcetgp pulse program (ns = 200, ds = 16, number of increments = 256, d1 = 1.0 s) (24). Chemical shifts were referenced normalized to the central DMSO peak (δC/δH, 39.5/2.5 ppm). Assignment of the HSQC spectra has been described elsewhere (20, 25). A semiquantitative analysis of the volume integrals of the HSQC correlation peaks was performed using Bruker's Topspin 3.1 (Windows) processing software. A Gaussian apodization in F2 (LB = −0.50; GB = 0.001) and squared cosine bell in F1 (line broadening [LB] = −0.10; Gaussian line broadening [GB] = 0.001) were applied prior to 2D Fourier transformation.
Data access.
Amplicon pyrosequencing data are available on MG-RAST under the project Newby Island Enrichments (http://metagenomics.anl.gov/linkin.cgi?project=7437).
RESULTS
Community structure of thermophilic switchgrass-deconstructing consortia.
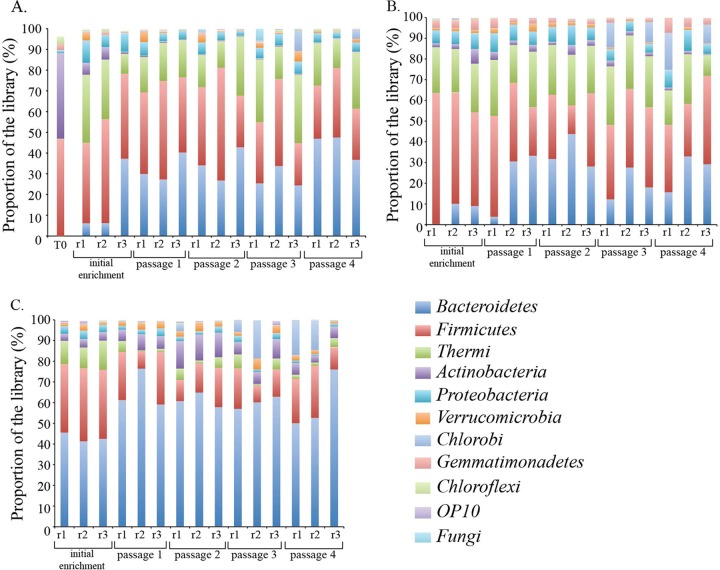
Triplicate thermophilic consortia adapted to grow on untreated, AFEX-pretreated, and IL-pretreated switchgrass were enriched in parallel through multiple passages at 60°C using green-waste compost. Small subunit rRNA gene amplicon pyrosequencing was used to characterize the bacterial diversity, richness, composition, and development of these consortia (Table 1; Fig. 1). The starting microbial community in the Newby Island Compost (NIC) was a diverse microbial community (Shannon = 3.69; Simpson = 2.95), composed primarily of Firmicutes and Actinobacteria with a fungal component (Fig. 1) (5). Microbial communities distinct from the starting inoculum were reproducibly (n = 3) enriched on AFEX-treated, IL-treated, and untreated switchgrass that were dominated by bacteria. The relative proportion of the bacterial phyla differed across these carbon sources based on the SSU amplicon libraries. For example, Bacteroidetes OTUs were typically more prevalent in IL-pretreated switchgrass (ca. 40 to 75%), than untreated switchgrass and AFEX-pretreated switchgrass (typically less than 40%). AFEX-treated and untreated switchgrass had a higher proportion of Firmicutes (Fig. 1).
TABLE 1.
Summary of species richness and diversity estimatesa
| Switchgrass | Time of sampling | Diversity index |
Richness index |
||
|---|---|---|---|---|---|
| Shannon | Simpson | ACE | Chao | ||
| None (NIC) | T0 | 3.69 | 2.95 | 411 | 315 |
| Untreated | Initial enrichment | 2.39 ± 0.13 | 1.76 ± 0.02 | 261 ± 87 | 161 ± 43 |
| Passage 1 | 2.08 ± 0.18 | 1.68 ± 0.21 | 104 ± 22 | 87 ± 15 | |
| Passage 2 | 1.94 ± 0.22 | 1.43 ± 0.14 | 189 ± 60 | 125 ± 32 | |
| Passage 3 | 2.20 ± 0.15 | 1.77 ± 0.21 | 143 ± 37 | 99 ± 11 | |
| Passage 4 | 1.91 ± 0.06 | 1.49 ± 0.03 | 109 ± 52 | 68 ± 9 | |
| P | <0.022 | >0.05 | <0.0324 | <0.0105 | |
| AFEX pretreated | Initial enrichment | 2.44 ± 0.23 | 1.84 ± 0.2 | 217 ± 113 | 156 ± 73 |
| Passage 1 | 2.21 ± 0.17 | 1.63 ± 0.12 | 167 ± 28 | 87 ± 15 | |
| Passage 2 | 2.34 ± 0.25 | 1.80 ± 0.38 | 132 ± 27 | 125 ± 32 | |
| Passage 3 | 2.27 ± 0.16 | 1.75 ± 0.19 | 186 ± 127 | 99 ± 11 | |
| Passage 4 | 2.30 ± 0.17 | 1.85 ± 0.25 | 147 ± 30 | 68 ± 9 | |
| P | >0.679 | >0.812 | >0.715 | >0.387 | |
| IL pretreated | Initial enrichment | 1.99 ± 0.06 | 1.41 ± 0.09 | 216 ± 95 | 111 ± 23 |
| Passage 1 | 1.48 ± 0.22 | 0.81 ± 0.25 | 102 ± 5 | 91 ± 19 | |
| Passage 2 | 1.77 ± 0.09 | 0.98 ± 0.08 | 179 ± 87 | 117 ± 47 | |
| Passage 3 | 1.77 ± 0.13 | 0.98 ± 0.08 | 123 ± 65 | 100 ± 42 | |
| Passage 4 | 1.49 ± 0.25 | 0.95 ± 0.35 | 82 ± 16 | 64 ± 9 | |
| P | <0.0194 | >0.13 | >0.142 | >0.34 | |
The P values from an analysis of variance are listed.
FIG 1.
Plot of the relative abundance of dominant phyla (≥3%) based on SSU rRNA amplicon pyrosequencing for the untreated (A)-, AFEX-pretreated (B)-, and IL-pretreated (C)-switchgrass-adapted consortia for the initial enrichment and passages 1 through 4. Biological replicates are depicted for each passage. Newby Island compost time zero (T0) is also depicted. A cutoff of 3% or more abundance was chosen to highlight the most abundant organisms present in the community.
Compared to those in the starting community, the richness and diversity decreased in the initial enrichment (IE) and subsequent passages (P1 to P4) for untreated SG, AFEX-pretreated switchgrass, and IL-pretreated switchgrass (Table 1). Statistically significant differences were noted primarily in untreated SG, based on the Shannon diversity index (IE ≠ P4, P < 0.02; IE ≠ P2, P < 0.04), the abundance coverage estimator (ACE) (IE ≠ P1, P < 0.04; IE ≠ P4, P < 0.03), and the Chao richness estimate (IE ≠ P1, P < 0.03; IE ≠ P4, P < 0.009). The Shannon diversity was also different for IL-pretreated switchgrass (IE ≠ P1, P < 0.03; IE ≠ P4, P < 0.03) (Table 1).
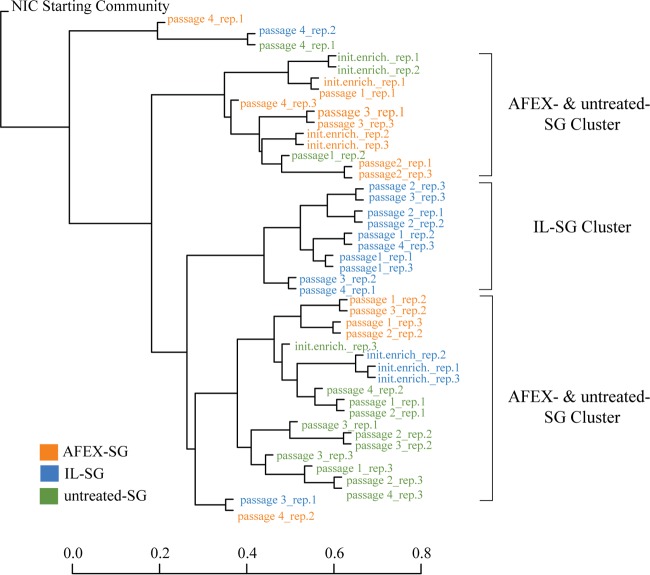
The similarity of the SSU amplicon libraries generated across the passages for the adapted thermophilic consortia was assessed using the Bray-Curtis dissimilarity index and visualized using agglomerative hierarchical clustering (Fig. 2) (26). All adapted thermophilic consortia were distinct from the starting community (Fig. 2). Even though the triplicates did not always cluster together, the IL-pretreated switchgrass-adapted communities across the passages (IE and P1 to P4) tended to cluster together and were distinct from the AFEX-treated- and untreated-switchgrass communities, whereas the AFEX-treated- and untreated-switchgrass-adapted communities tended to cluster across two groups. IL-pretreated-switchgrass-adapted consortia appear to be more evenly distributed than untreated-switchgrass- and AFEX-pretreated-switchgrass-adapted consortia.
FIG 2.
Agglomerative hierarchical cluster dendrograms of the untreated (green)-, IL-pretreated (blue)-, and AFEX-pretreated (orange)-switchgrass-enriched thermophilic consortia across passages and triplicates based on the Bray-Curtis dissimilarity index. The scale bar indicates the similarity of the communities.
OTU analysis.
OTUs associated with the phyla Bacteroidetes, Firmicutes, and Deinococcus-Thermus primarily populated the SSU amplicon libraries (Table 2). Indicator species analysis identified OTUs that were significantly associated with AFEX-pretreated switchgrass (12 species)- and untreated switchgrass (2 species)-adapted consortia, respectively, at passage 4. The indicator species of the AFEX-pretreated switchgrass treatment were members of the phyla Firmicutes (OTU97_24, OTU97_25, and OTU97_33, OTU97_47 and OTU97_84), Actinobacteria (OTU97_49), Gemmatimonadetes (OTU97_15 and OTU97_30), Bacteroidetes (OTU97_194), Chloroflexi (OTU97_74), and Gammaproteobacteria (OTU97_43 and OTU97_16). The indicator species of the untreated switchgrass treatment were members of the phyla Chloroflexi (OTU97_78) and Bacteroidetes (OTU97_854).
TABLE 2.
Relative proportion of the phylotypes (OTU97, >1%) for passage 4 samplesa
| Taxon | Phylotype | Proportion in: |
BLAST hit (% identity) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Untreated switchgrass |
AFEX-pretreated switchgrass |
IL-pretreated switchgrass |
|||||||||
| Rep. 1 | Rep. 2 | Rep. 3 | Rep. 1 | Rep. 2 | Rep. 3 | Rep. 1 | Rep. 2 | Rep. 3 | |||
| Bacteroidetes | OTU97_1 | 40.1 | 40.8 | 32.6 | 0.78 | 29.4 | 3.6 | 49.9 | 52.3 | 75.8 | Crenotalea thermophila STH-1-Y1 (99) |
| OTU97_9 | 6.67 | 6.48 | 3.91 | 14.56 | 3.18 | 25.11 | 0 | 0 | 0 | Rhodothermus marinus SG0.5JP17-172 (100) | |
| Firmicutes | OTU97_3 | 17.2 | 22.0 | 19.5 | 14.6 | 18.12 | 29.2 | 13.5 | 15.8 | 0.93 | Thermobacillus composti KWC4 (97) |
| OTU97_4 | 3.46 | 6.67 | 3.81 | 9.1 | 0.88 | 9.15 | 6.61 | 7.41 | 4.36 | Paenibacillus kobensis NBRC 15730 (96) | |
| OTU97_41 | 0.52 | 0.79 | 0.37 | 2.25 | 0.35 | 1.14 | 0.28 | 0.82 | 0.34 | Thermobacillus composti KWC4 (100) | |
| OTU97_47 | 0.05 | 0.03 | 0.01 | 0.96 | 0.73 | 0.51 | 0.02 | 0.1 | 0.13 | Geobacillus stearothermophilus CICC 10392 (100) | |
| Deinococcus-Thermus | OTU97_2 | 12.4 | 9.42 | 0.9 | 16.67 | 23.6 | 9.89 | 1.76 | 1.06 | 1.09 | Truepera radiovictrix RQ-24 (93) |
| OTU97_7 | 7.91 | 4.54 | 26.1 | 0 | 0 | 0.01 | 0.19 | 0.48 | 3.35 | Thermus thermophilus SG0.5JP17-16 (100) | |
| Other | OTU97_25 | 3.57 | 3.47 | 0.75 | 2.16 | 2.52 | 0.11 | 0.44 | 0.14 | 3.96 | Geobacter daltonii FRC-32 (95)b |
| OTU97_12 | 1.03 | 0.45 | 1.7 | 0.18 | 1.33 | 0.41 | 2.43 | 1.83 | 1.17 | Alterococcus agarolyticus ADT3 (91) | |
| OTU97_14 | 0 | 0 | 4.36 | 17.7 | 0 | 8.96 | 16.8 | 14.3 | 0 | Chlorobaculum tepidum TLS (87) | |
| OTU97_17 | 0.05 | 0.1 | 0.09 | 0.09 | 0.02 | 0.02 | 1.87 | 0.77 | 1.26 | Thermobispora bispora DSM 43833 (100) | |
| OTU97_18 | 3.46 | 2.43 | 3.02 | 0.96 | 1.53 | 0.51 | 0.3 | 0.14 | 0.61 | Plasticicumulans lactativorans YD (94) | |
| OTU97_27 | 0.16 | 0.01 | 0.14 | 0.46 | 0.82 | 0.27 | 0.3 | 0.1 | 1.92 | Thermobispora bispora DSM 43833 (95) | |
| OTU97_31 | 0 | 0.1 | 0.04 | 6.71 | 4.08 | 2.94 | 0 | 0 | 0 | Thermaerobacter composti Ni80 (95) | |
| OTU97_37 | 0 | 0 | 0 | 5.65 | 4.35 | 3.16 | 0.33 | 0.24 | 0 | Lysobacter oryzae YC6269 (99) | |
| OTU97_53 | 0.1 | 0.11 | 0.07 | 0.41 | 0.9 | 0.41 | 0.33 | 0.24 | 0.2 | Enhydrobacter aerosaccus PAGU 1624 (97) | |
| OTU97_63 | 0 | 0.06 | 0 | 0.73 | 0.6 | 0.31 | 0.03 | 0.05 | 0.09 | Thiobacter subterraneus C55 16S (98) | |
| OTU97_73 | 0.1 | 0.03 | 0.01 | 0.18 | 0.25 | 0.13 | 0.08 | 0.05 | 0.11 | Rhodomicrobium udaipurense JA643 (96) | |
| OTU97_79 | 0.05 | 0.02 | 0 | 0 | 0 | 0.11 | 0 | 0 | 0 | Planifilum yunnanense LA5 (100) | |
Phylum description was assigned by the Ribosomal Database Project Classifier, release 10, and the OTU identification was assigned by GenBank. Rep., replicate.
Uncultivated clones with 99% identity to OTU_25 are affiliated with the Myxococcales.
There were also shifts in the relative abundances of more prevalent OTUs across these treatments. OTU97_1, representing a novel Bacteroidetes strain previously isolated from this compost material by enrichment on microcrystalline cellulose (strain NYFB), primarily dominated the untreated switchgrass and IL-pretreated switchgrass thermophilic consortia but not the AFEX-pretreated switchgrass thermophilic consortia. The other abundant Bacteriodetes OTU was 100% identical to Rhodothermus marinus, a population previously observed in switchgrass enrichments (6). OTU97_3, a member of the Firmicutes most closely related to clone JP2339, a sequence from previous compost enrichments on switchgrass, was prevalent across all treatments (6). OTU97_2, a novel phylotype that was affiliated with the Trueperaceae, a family in the Deinococcus-Thermus phylum, was primarily prevalent in the AFEX-pretreated switchgrass thermophilic consortia, whereas another phylotype affiliated with the Deinococcus-Thermus phylum, OTU97_7, was primarily present in untreated switchgrass and was 100% identical to Thermus thermophilus.
Glycoside hydrolase activities of the AFEX-treated-, IL-treated-, and untreated-switchgrass-adapted consortia.
The adapted consortia were assayed for cellulase and xylanase activities. Across all the AFEX-treated-, IL-treated-, and untreated-switchgrass-adapted passages, cellulase activity was low (0.003 to 0.03 U/min/ml). In contrast, the AFEX-treated-, IL-treated-, and untreated-switchgrass-adapted passages had high levels of xylanase activity (0.32 to 5.11 U/ml) (Table 3). Statistically significant differences were noted in passages 1, 2, and 4 for endoglucanase (P1, SG ≠ AFEX, P < 0.03; P1 SG ≠ IL, P < 0.01; P2 SG ≠ AFEX, P < 0.002; P2 SG ≠ IL, P < 0.002; P4 SG ≠ IL, P < 0.04) and passage 1 for xylanase (P1 IL ≠ AFEX, P < 0.0001; P1 SG ≠ IL, P < 0.0001).
TABLE 3.
Average xylanase and cellulase activities measured in whole-cell fractions across the five passages for the various substrates at 65°C and pH 6.0a
| Sample | Xylanase activity (U/ml/min) |
Cellulase activity (U/ml/min) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IE | Passage 1 | Passage 2 | Passage 3 | Passage 4 | IE | Passage 1 | Passage 2 | Passage 3 | Passage 4 | |
| Untreated switchgrass | 0.86 ± 0.40 | 1.1 ± 0.3 | 1.1 ± 0.1 | 0.48 ± 0.43 | 0.67 ± 0.51 | 0.007 ± 0.0006 | 0.004 ± 0.0006 | 0.032 ± 0.001 | 0.010 ± 0.001 | 0.003 ± 0.0005 |
| AFEX-pretreated switchgrass | 0.76 ± 0.17 | 1.4 ± 0.2 | 1.2 ± 0.1 | 0.45 ± 0.57 | 0.32 ± 0.36 | 0.010 ± 0.004 | 0.010 ± 0.003 | 0.011 ± 0.002 | 0.009 ± 0.003 | 0.010 ± 0.002 |
| IL-pretreated switchgrass | 0.93 ± 0.17 | 5.1 ± 0.4 | 1.1 ± 0.1 | 0.55 ± 0.78 | 0.43 ± 0.53 | 0.011 ± 0.002 | 0.012 ± 0.003 | 0.011 ± 0.007 | 0.008 ± 0.002 | 0.012 ± 0.002 |
| Pb | >0.749 | <0.0001 | >0.951 | >0.978 | >0.678 | >0.282 | <0.0105 | <0.001 | >0.597 | <0.039 |
Values are means ± standard deviations for 3 biological replicates. Activities for the control samples (without supplemented substrates) were typically below the detection limit: activities were detected only in the IE (xylanase, 0.11 ± 0.08; cellulase, 0.007 ± 0.002).
P values from an analysis of variance.
Biomass composition and soluble lignin content after enrichment on switchgrass substrates.
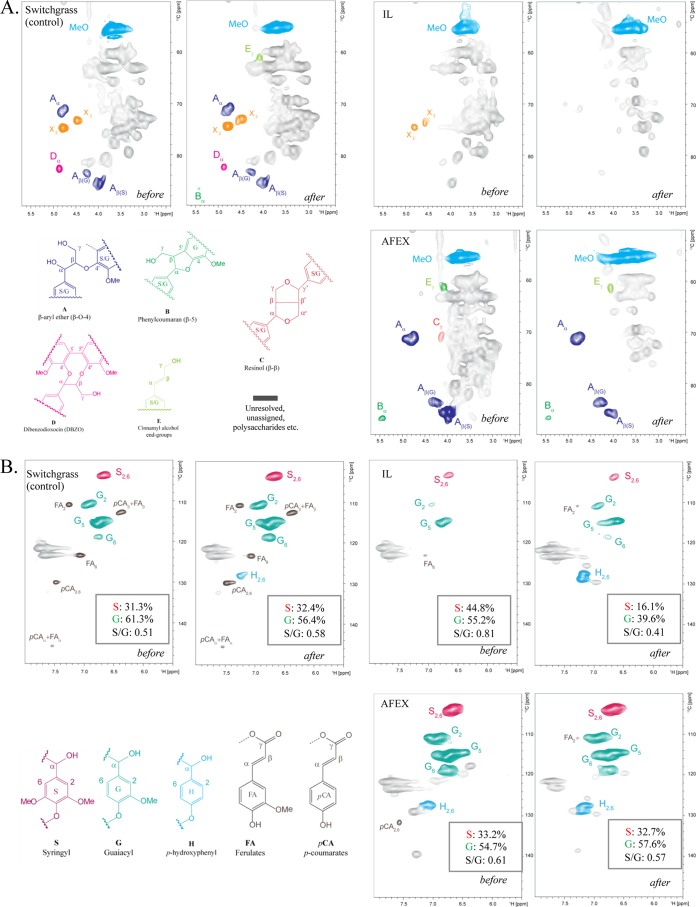
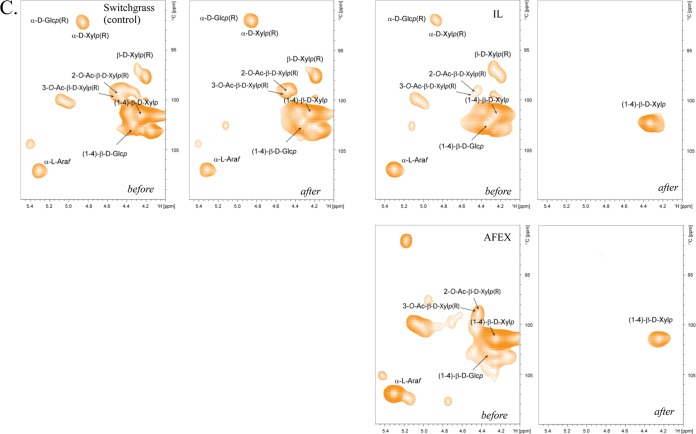
The starting biomass composition of ionic-liquid-pretreated switchgrass had higher glucan content (1.3-fold higher than in untreated SG and 1.6-fold higher than in AFEX-pretreated SG) and a reduced lignin content (ca. 1.7-fold less for untreated and AFEX-pretreated SG) (Table 4). The residual biomass after microbial deconstruction was analyzed to determine the effect of microbial metabolism on these biomass substrates. Gravimetric analysis of residual biomass demonstrated that ca. 80% of the biomass was liberated from the pretreated switchgrass (IL- and AFEX-pretreated switchgrass), while only ca. 40% of the biomass was liberated from untreated switchgrass (Table 5). Two-dimensional NMR spectroscopy (2D-NMR) documented changes of aliphatic (lignin side chain units) (Fig. 3A), aromatic (Fig. 3B), and polysaccharide anomeric (Fig. 3C) regions before and after microbial deconstruction of the supplemented substrates. Aliphatic regions of untreated switchgrass and IL-pretreated switchgrass prior to microbial deconstruction (Fig. 3A) exhibited two distinct peaks of 2-O-Ac-β-d-Xylp(R) (X′2) and 3-O-Ac-β-d-Xylp(R) (X′3). However, X′2 and X′3 were not observed in AFEX-pretreated switchgrass, suggesting deacetylation of hemicelluloses upon AFEX pretreatment. The aromatic region (Fig. 3B) of IL-pretreated switchgrass before microbial deconstruction showed weaker aromatic signals than those of untreated switchgrass and AFEX-pretreated switchgrass, corroborating the lower lignin content in IL-pretreated switchgrass (Table 4). The NMR resonances present in the aromatic regions of the spectra of the residual biomass of the untreated switchgrass, AFEX-pretreated switchgrass, and IL-pretreated switchgrass enrichments were slightly more distinct, but virtually no changes in the bond linkages were observed. The 2D-NMR results demonstrating that lignin was largely unmodified were complemented by relative measurements of residual lignin in biomass, which showed substantial increases in the pretreated biomass samples (50% for AFEX-pretreated switchgrass; 131% for IL-pretreated switchgrass) but only a small increase in the untreated switchgrass (Table 5). Evidence for lignin solubilization, especially in the untreated switchgrass and AFEX-pretreated switchgrass cultivations, was obtained by the observation of acid-precipitable polymeric lignin (APPL), a solubilized form of lignin that precipitates from the enrichment supernatant upon acidification (18, 27). Examination of the anomeric region (Fig. 3C) demonstrated that while the untreated switchgrass did not show any alteration in the signals for major polysaccharides linkages (β-1,4 glucan and β-1,4 xylan), the AFEX-pretreated switchgrass showed complete disappearance of the β-1,4 glucan linkages while β-1,4 xylan linkages remained after the 2-week incubation. Hemicellulose side chains, including acetyl esters and α-l-arabinofuranosides, were hydrolyzed in the AFEX-pretreated-switchgrass and IL-pretreated-switchgrass samples.
TABLE 5.
Gravimetric analysis of residual biomass of switchgrass substrates upon microbial decompositiona
| Sample | Mass loss (%)b | APPL (μg/ml)c | Increase in ABL (%)d |
|---|---|---|---|
| Untreated switchgrass | 40 | 250 | 10 ± 1 |
| AFEX-pretreated switchgrass | 83 | 210 | 50 ± 2 |
| IL-pretreated switchgrass | 82 | 59 | 131 ± 15 |
Values are means for biological duplicates.
Mass loss values varied ≤5% between biological replicates. Mass loss in duplicate control experiments that were not inoculated was ∼2 to 5%.
Recovered acid-precipitable polymeric lignin (APPL) varied ≤8% between biological replicates. An average of 73 μg/ml of APPL was recovered from untreated switchgrass controls; no APPL was detected in AFEX- and IL-pretreated SG controls.
Acetyl bromide-reactive lignin (ABL) was measured in triplicate for each biological replicate.
FIG 3.
Partial 13C-1H (HSQC) NMR spectra (aliphatic, aromatic, and anomeric regions) of untreated-, IL-pretreated, and AFEX-pretreated switchgrass before and after cultivation. (A) Analysis of the lignin side chain region (aliphatic region) of the HSQC spectra; 2-O-Ac-β-d-Xylp(R) (X′2) and 3-O-Ac-β-d-Xylp(R) (X′3) are derived by polysaccharides. (B) Partial short-range 13C-1H (HSQC) spectra (aromatic region). Lignin monomer ratios are provided. (C) Polysaccharide anomeric regions of 2D 13C-1H (HSQC) spectra. α-d-Glcp, α-d-glucopyranoside; β-d-Glcp, β-d-glucopyranoside; β-d-Xylp, β-d-xylopyranoside; α-l-Araf, α-l-arabinofuranoside; 2-O-Ac-β-d-Xylp, acetylated β-d-Xylp; 3-O-Ac-β-d-Xylp, acetylated β-d-Xylp; R, reducing end; NR, nonreducing end.
DISCUSSION
Our triplicate enrichments on untreated and pretreated switchgrass demonstrated substrate-specific development of these consortia. Despite the observed differences of these microbial communities as measured by diversity indices, common OTUs were observed as the most abundant populations in the consortia. These abundant OTUs are very similar to those in previous studies of aerobic thermophilic consortia adapted to grow on biomass, confirming that community adaptation is a reproducible process (6, 28).
The AFEX-pretreated switchgrass has increased physiochemical complexity compared to IL-pretreated switchgrass (13). Furthermore, the isolation of APPL from the AFEX-pretreated switchgrass culture supernatant indicates that some lignin deconstruction is required to access the polysaccharides, which is consistent with structural studies of AFEX-pretreated switchgrass (10). The microbial community adapted to grow on AFEX-pretreated switchgrass was more diverse and rich than the IL-pretreated switchgrass, consisting of members of the Bacteroidetes, Firmicutes, and Deinococcus-Thermus in similar proportions (Fig. 1).
The 2D-NMR for both IL-pretreated switchgrass and AFEX-pretreated switchgrass suggested complete digestion of cellulose in both substrates and the presence of residual xylan. These results were surprising, since enzymatic assays of the supernatant recovered much higher levels of xylanase activity than cellulase activity. Therefore, it is possible that cellulase activity is underestimated based on measurement of the supernatants using model cellulase substrates and that such estimates do not necessarily reflect the cellulose hydrolysis activity in the consortia (29).
The untreated switchgrass exhibited a markedly different pattern of deconstruction, yet the community structure was similar to the AFEX-pretreated-switchgrass communities (Fig. 1). Except for minor alterations, the residual biomass had NMR resonances identical to those of the starting material, and the relative ratio of lignin as assayed by the acetyl bromide method was virtually unchanged. These observations are reminiscent of the anaerobic growth of hyperthermophile Caldicellulosiruptor bescii on untreated switchgrass, in which no change was observed in the ratio of polysaccharides to lignin between the starting material and the residual biomass after cultivation (30). The comparison of residual biomass after microbial growth between pretreated and untreated switchgrass suggests that the consortia may have fundamentally different mechanisms of growth on untreated switchgrass, despite the similarity in community composition.
Across all treatments, the lignin in the residual biomass was largely unaltered based on 2D-NMR studies. This suggests that lignin remodeling is not an important component of biomass deconstruction by these microbial communities. This observation is in contrast to 2D-NMR studies of the brown rot fungus Postia placenta, which showed extensive remodeling of the lignin component of aspen wood during biomass deconstruction (31).
Residual biomass analyzed upon microbial deconstruction demonstrated a number of remarkable features. Ionic-liquid pretreatment provided a substrate enriched in cellulose, providing polysaccharides with more accessibility for deconstruction (12), compared to untreated switchgrass. The simplicity of this substrate is reflected in the distinct adapted microbial community, which was primarily composed of members of the Bacteroidetes (Fig. 1). Taking these data together, we hypothesize that given the minimal solubilization of the lignin during growth on IL-pretreated switchgrass, the microbial communities along with their associated glycoside hydrolases had direct access to polysaccharides and therefore did not need to break bonds in the lignin to access the polysaccharides.
The patterns of abundant OTUs in the enrichments on switchgrass substrates provide further insights into the roles of these populations in biomass deconstruction. The dominant OTU in many of the enrichments, which is represented by the Bacteriodetes strain NYFB, was identical to the dominant OTU observed in cultures adapted from the same compost inoculum to grow on purified fractions of cellulose and hemicellulose (5). Strain NYFB can degrade mono-, di- and polysaccharides but was unable to grow on insoluble polysaccharides. This observation, along with the predominance of members of the Firmicutes at early time points during the 2-week cultivations on cellulose, led to the proposal of a successional structure for these biomass-deconstructing microbial communities. This structure appears to be conserved in the communities growing on the untreated and pretreated switchgrass substrates. In the adapted enrichments that were not dominated by strain NYFB (e.g., AFEX-pretreated switchgrass passage 4), an OTU representing Rhodothermus marinus (OTU97_9) (Table 2) was abundant. In previous enrichments from compost with switchgrass and cellulose as the sole carbon source, R. marinus was observed as the dominant Bacteriodetes population (6, 7). R. marinus is a thermophilic Bacteriodetes species that produces high levels of cellulases and hemicellulases when grown on biomass substrates and may occupy a functional niche in the adapted cultures similar to that of strain NYFB (32).
In the enrichments across all three switchgrass substrates, a single Firmicutes OTU predominated (OTU97_3) (Table 2), which was previously identified in compost-derived enrichments on switchgrass, indicating that this OTU may represent a population specifically adapted to deconstruct complex plant biomass substrates (6). Interestingly, this Firmicutes OTU is not abundant (<0.5% of SSU amplicon reads) in enrichments with microcrystalline cellulose and wheat arabinoxylan (hemicellulose) (5). A population closely related to Thermus thermophilus, which is consistently observed in these thermophilic enrichment on biomass substrates, is present in the untreated switchgrass and IL-pretreated switchgrass cultures but is noticeably absent in the AFEX-pretreated switchgrass cultures.
In contrast, a population affiliated with the Trueperaceae, in the Deinococcus-Thermus phylum, was observed at high relative abundance in the AFEX-pretreated switchgrass cultivations (OTU97_2, average of 16.73% for three AFEX-pretreated switchgrass passage 4 replicates). This population was also observed at high abundance in compost-derived enrichments on a purified hemicellulose fraction, wheat arabinoxylan, while T. thermophilus was absent (5). These results imply that the presence of hemicellulose, which is intact and readily accessible in AFEX-pretreated switchgrass, may select for this Trueperaceae population. There is currently only one described species in this family, Truepera radiovictrix, which is thermophilic and tolerant to very high levels of radiation (33, 34). Therefore, natural samples where thermophilic biomass deconstruction occurs may be fruitful sources for new isolates related to Truepera radiovictrix.
Our study revealed that the thermophilic consortia adapted to grow on switchgrass, a complex lignocellulosic substrate, preferentially hydrolyzed polysaccharides compared to lignin deconstruction. This preference is evident in the similarity of the microbial community composition to that of adapted communities grown on purified fractions of cellulose and hemicellulose (5) and in the increase in lignin content in the residual biomass of the pretreated switchgrass substrates. To identify thermophilic bacterial populations and pathways that primarily deconstruct and metabolize lignin, adaptions need to be performed on lignin-enriched biomass substrates that have minimal polysaccharide content.
ACKNOWLEDGMENTS
This work was performed as part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. Pyrotag sequencing was conducted by the Joint Genome Institute, which is supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231.
We thank Susannah Tringe, Tijana Glavina Del Rio, and Stephanie Malfatti of the Joint Genome Institute for their assistance in obtaining and processing pyrotag sequencing data. Bruce Dale (Michigan State University; Great Lakes Bioenergy Center) is acknowledged for providing AFEX-pretreated switchgrass. John Gladden (Sandia National Laboratories) is acknowledged for assistance in preparing ionic-liquid-pretreated switchgrass. We thank Ken Vogel at the U.S. Department of Agriculture, Lincoln, NE, for supplying the switchgrass cultivars.
Footnotes
Published ahead of print 26 September 2014
REFERENCES
- 1.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo SJ, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 2.Tilman D, Socolow R, Foley JA, Hill J, Larson E, Lynd L, Pacala S, Reilly J, Searchinger T, Somerville C, Williams R. 2009. Beneficial biofuels-the food, energy, and environment trilemma. Science 325:270–271. 10.1126/science.1177970. [DOI] [PubMed] [Google Scholar]
- 3.McCann MC, Carpita NC. 2008. Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 11:314–320. 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 4.D'haeseleer P, Gladden JM, Allgaier M, Chain PSG, Tringe SG, Malfatti SA, Aldrich JT, Nicora CD, Robinson EW, Pasa-Tolic L, Hugenholtz P, Simmons BA, Singer SW. 2013. Proteogenomic analysis of a thermophilic bacterial consortium adapted to deconstruct switchgrass. PLoS One 8:e68465. 10.1371/journal.pone.0068465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichorst SA, Varanasi P, Stavila V, Zemla M, Auer M, Singh S, Simmons BA, Singer SW. 2013. Community dynamics of cellulose-adapted thermophilic bacterial consortia. Environ. Microbiol. 15:2573–2587. 10.1111/1462-2920.12159. [DOI] [PubMed] [Google Scholar]
- 6.Gladden JM, Allgaier M, Miller CS, Hazen TC, VanderGheynst JS, Hugenholtz P, Simmons BA, Singer SW. 2011. Glycoside hydrolase activities of thermophilic bacterial consortia adapted to switchgrass. Appl. Environ. Microbiol. 77:5804–5812. 10.1128/AEM.00032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gladden JM, Eichorst SA, Hazen TC, Simmons BA, Singer SW. 2012. Substrate perturbation alters the glycoside hydrolase activities and community composition of switchgrass-adapted bacterial consortia. Biotechnol. Bioeng. 109:1140–1145. 10.1002/bit.24388. [DOI] [PubMed] [Google Scholar]
- 8.Park JI, Steen EJ, Burd H, Evans SS, Redding-Johnson AM, Batth T, Benke PI, D'Haeseleer P, Sun N, Sale KL, Keasling JD, Lee TS, Petzold CJ, Mukhopadhyay A, Singer SW, Simmons BA, Gladden JM. 2012. A thermophilic ionic liquid-tolerant cellulase cocktail for the production of cellulosic biofuels. PLoS One 7:e37010. 10.1371/journal.pone.0037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar P, Barrett DM, Delwiche MJ, Stroeve P. 2009. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind. Eng. Chem. Res. 48:3713–3729. 10.1021/ie801542g. [DOI] [Google Scholar]
- 10.Chundawat SPS, Vismeh R, Sharma LN, Humpula JF, Sousa LD, Chambliss CK, Jones AD, Balan V, Dale BE. 2010. Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Biores. Technol. 101:8429–8438. 10.1016/j.biortech.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Dibble DC, Li CL, Sun L, George A, Cheng ARL, Cetinkol OP, Benke P, Holmes BM, Singh S, Simmons BA. 2011. A facile method for the recovery of ionic liquid and lignin from biomass pretreatment. Green Chem. 13:3255–3264. 10.1039/c1gc15111h. [DOI] [Google Scholar]
- 12.Li C, Knierim B, Manisseri C, Arora R, Scheller HV, Auer M, Vogel KP, Simmons BA, Singh S. 2010. Comparison of dilute acid and ionic liquid pretreatment of switchgrass: biomass recalcitrance, delignification and enzymatic saccharification. Biores. Technol. 101:4900–4906. 10.1016/j.biortech.2009.10.066. [DOI] [PubMed] [Google Scholar]
- 13.Li CL, Cheng G, Balan V, Kent MS, Ong M, Chundawat SPS, Sousa LD, Melnichenko YB, Dale BE, Simmons BA, Singh S. 2011. Influence of physico-chemical changes on enzymatic digestibility of ionic liquid and AFEX pretreated corn stover. Biores. Technol. 102:6928–6936. 10.1016/j.biortech.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Bals B, Rogers C, Jin M, Balan V, Dale B. 2010. Evaluation of ammonia fibre expansion (AFEX) pretreatment for enzymatic hydrolysis of switchgrass harvested in different seasons and locations. Biotechnol. Biofuels 3:1. 10.1186/1754-6834-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelbrektson A, Kunin V, Wrighton KC, Zvenigorodsky N, Chen F, Ochman H, Hugenholtz P. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647. 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 16.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshua CJ, Dahl R, Benke PI, Keasling JD. 2011. Absence of diauxie during simultaneous utilization of glucose and xylose by Sulfolobus acidocaldarius. J. Bacteriol. 193:1293–1301. 10.1128/JB.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford DL, Pometto AL, Crawford RL. 1983. Lignin degradation by Streptomyces viridosporus: isolation and characterization of a new polymeric lignin degradation intermediate. Appl. Environ. Microbiol. 45:898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima RS, Kerley MS. 2011. Use of lignin extracted from different plant sources as standards in the spectrophotometric acetyl bromide lignin method. J. Agric. Food Chem. 59:3505–3509. 10.1021/jf104826n. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, Ralph J. 2010. Solution-state 2D NMR of ball-milled plant cell wall gels in DMSO-d6/pyridine-d5. Org. Biomol. Chem. 8:576–591. 10.1039/b916070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansfield SD, Kim H, Lu F, Ralph J. 2012. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 7:1579–1589. 10.1038/nprot.2012.064. [DOI] [PubMed] [Google Scholar]
- 22.Cheng K, Sorek H, Zimmermann H, Wemmer DE, Pauly M. 2013. Solution-state 2D NMR spectroscopy of plant cell walls enabled by a dimethylsulfoxide-d6/1-ethyl-3-methylimidazolium acetate solvent. Anal. Chem. 85:3213–3221. 10.1021/ac303529v. [DOI] [PubMed] [Google Scholar]
- 23.Sathitsuksanoh N, Holtman KM, Yelle DJ, Morgan T, Stavila V, Pelton J, Blanch H, Simmons BA, George A. 2014. Lignin fate and characterization during ionic liquid biomass pretreatment for renewable chemicals and fuels production. Green Chem. 16:1236–1247. 10.1039/C3GC42295J. [DOI] [Google Scholar]
- 24.Heikkinen S, Toikka MM, Karhunen PT, Kilpeläinen IA. 2003. Quantitative 2D HSQC (Q.-HSQC) via suppression of J-dependence of polarization transfer in NMR spectroscopy: application to wood lignin. J. Am. Chem. Soc. 125:4362–4367. 10.1021/ja029035k. [DOI] [PubMed] [Google Scholar]
- 25.Yelle DJ, Ralph J, Frihart CR. 2008. Characterization of nonderivatized plant cell walls using high-resolution solution-state NMR spectroscopy. Magn. Reson. Chem. 46:508–517. 10.1002/mrc.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sul WJ, Cole JR, Jesus EDC, Wang Q, Farris RJ, Fish JA, Tiedje JM. 2011. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc. Natl. Acad. Sci. U. S. A. 108:14637–14642. 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown ME, Walker MC, Nakashige TG, Iavarone AT, Chang MCY. 2011. Discovery and characterization of heme enzymes from unsequenced bacteria: application to microbial lignin degradation. J. Am. Chem. Soc. 133:18006–18009. 10.1021/ja203972q. [DOI] [PubMed] [Google Scholar]
- 28.Plecha S, Hall D, Tiquia-Arashiro SM. 2013. Screening for novel bacteria from the bioenergy feedstock switchgrass (Panicum virgatum L.). Environ. Technol. 34:1895–1904. 10.1080/09593330.2013.818701. [DOI] [PubMed] [Google Scholar]
- 29.McClendon SD, Baath T, Petzold CJ, Adams PD, Simmons BA, Singer SW. 2012. Thermoascus aurantiacus is a promising source of enzymes for biomass deconstruction under thermophilic conditions. Biotechnol. Biofuels 5:54. 10.1186/1754-6834-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kataeva I, Foston MB, Yang SJ, Pattathil S, Biswal AK, Poole FL, Basen M, Rhaesa AM, Thomas TP, Azadi P, Olman V, Saffold TD, Mohler KE, Lewis DL, Doeppke C, Zeng YN, Tschaplinski TJ, York WS, Davis M, Mohnen D, Xu Y, Ragauskas AJ, Ding SY, Kelly RM, Hahn MG, Adams MWW. 2013. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ. Sci. 6:2186–2195. 10.1039/c3ee40932e. [DOI] [Google Scholar]
- 31.Yelle DJ, Wei DS, Ralph J, Hammel KE. 2011. Multidimensional NMR analysis reveals truncated lignin structures in wood decayed by the brown rot basidiomycete Postia placenta. Environ. Microbiol. 13:1091–1100. 10.1111/j.1462-2920.2010.02417.x. [DOI] [PubMed] [Google Scholar]
- 32.Bjornsdottir SH, Blondal T, Hreggvidsson GO, Eggertsson G, Petursdottir S, Hjorleifsdottir S, Thorbjarnardottir SH, Kristjansson JK. 2006. Rhodothermus marinus: physiology and molecular biology. Extremophiles 10:1–16. 10.1007/s00792-005-0466-z. [DOI] [PubMed] [Google Scholar]
- 33.Albuquerque L, Simoes C, Nobre MF, Pino NM, Battista JR, Silva MT, Rainey FA, da Costa MS. 2005. Truepera radiovictrix gen. nov., sp nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiology Lett. 247:161–169. 10.1016/j.femsle.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Ivanova N, Rohde C, Munk C, Nolan M, Lucas S, Del Rio TG, Tice H, Deshpande S, Cheng JF, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Brambilla E, Rohde M, Goker M, Tindall BJ, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lapidus A. 2011. Complete genome sequence of Truepera radiovictrix type strain (RQ-24(T)). Stand. Genomic Sci. 4:91–99. 10.4056/sigs.1563919. [DOI] [PMC free article] [PubMed] [Google Scholar]