Abstract
Bacillus coagulans 2-6 is an excellent producer of optically pure l-lactic acid. However, little is known about the mechanism of synthesis of the highly optically pure l-lactic acid produced by this strain. Three enzymes responsible for lactic acid production—NAD-dependent l-lactate dehydrogenase (l-nLDH; encoded by ldhL), NAD-dependent d-lactate dehydrogenase (d-nLDH; encoded by ldhD), and glycolate oxidase (GOX)—were systematically investigated in order to study the relationship between these enzymes and the optical purity of lactic acid. Lactobacillus delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer) and Lactobacillus plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer) were also examined in this study as comparative strains, in addition to B. coagulans. The specific activities of key enzymes for lactic acid production in the three strains were characterized in vivo and in vitro, and the levels of transcription of the ldhL, ldhD, and GOX genes during fermentation were also analyzed. The catalytic activities of l-nLDH and d-nLDH were different in l-, d-, and dl-lactic acid producers. Only l-nLDH activity was detected in B. coagulans 2-6 under native conditions, and the level of transcription of ldhL in B. coagulans 2-6 was much higher than that of ldhD or the GOX gene at all growth phases. However, for the two Lactobacillus strains used in this study, ldhD transcription levels were higher than those of ldhL. The high catalytic efficiency of l-nLDH toward pyruvate and the high transcription ratios of ldhL to ldhD and ldhL to the GOX gene provide the key explanations for the high optical purity of l-lactic acid produced by B. coagulans 2-6.
INTRODUCTION
Lactic acid is among the top 30 building block chemicals of biomass, and it has versatile applications in the food, pharmaceutical, cosmetic, and chemical industries. The use of lactic acid in the synthesis of polylactic acid (PLA) has been consistently on the rise (1). PLA is a renewable and biodegradable material and is regarded as an environmentally friendly alternative to traditional plastics derived from fossil fuels (2). Since only the optically pure l- and d-lactic acid monomers can be used as precursors of PLA, the production of optically pure lactic acid is a prerequisite for polymer synthesis (1, 3, 4).
Microbial fermentation is an efficient approach for producing lactic acid with high optical purity. Many microorganisms, such as fungi, Lactobacillus species, Bacillus coagulans, and various genetically modified strains, can produce lactic acid (1, 2, 5, 6). The bacterial genera that form lactic acid are often called lactic acid bacteria (LAB). The fermentative metabolism of LAB is characterized by the glycolytic breakdown of carbohydrates, and a late step in this pathway is distinguished by the conversion of pyruvate into lactic acid, a reaction that oxidizes the NADH formed during glycolysis, thus maintaining cellular redox balance (7). The enzymes responsible for the conversion of pyruvate to lactic acid and back are lactate dehydrogenases (LDHs), which fall into two broad classes: NAD-dependent lactate dehydrogenases (nLDHs) and NAD-independent lactate dehydrogenases (iLDHs). nLDHs are cytoplasmic proteins that catalyze the conversion of pyruvate to lactic acid in a reversible or irreversible manner. iLDHs are membrane bound, and they catalyze the oxidation of lactic acid to pyruvate (8). Members of these two classes exist in a stereospecific (l- or d-specific) form.
Almost all LAB have LDHs, but the optical purities of lactic acids produced by various LAB are markedly different (1). NAD-dependent l-lactate dehydrogenases (l-nLDHs; EC 1.1.1.27) are responsible for the synthesis of l-lactic acid, whereas d-lactic acid can be produced in either of two ways: through NADH-dependent reduction of pyruvate by NAD-dependent d-lactate dehydrogenases (d-nLDHs; EC 1.1.1.28) or through racemization of l-lactic acid by an l-lactic-acid-inducible lactate racemase (9, 10). l-nLDHs differ across genera and species and even among strains of the same species; the same is true for d-nLDHs (11). Sequence alignment has shown that l-nLDHs and d-nLDHs, encoded by ldhL and ldhD, respectively, belong to two distinct families, the NAD-dependent l- and d-2-hydroxyacid dehydrogenases, respectively (12). Lactate racemase (EC 5.1.2.1) can be found only in strains that form dl-lactic acid, such as Staphylococcus ureae, Lactobacillus curvatus, Lactobacillus paracasei, Lactobacillus plantarum WCFS1, Lactobacillus sakei, and Clostridium butylicum (1, 11). Although l-nLDHs and d-nLDHs are the key synthetic enzymes for l-lactic acid and d-lactic acid, respectively, studies have shown that in L. plantarum, an organism producing a mixture of 50% l-lactic acid and 50% d-lactic acid, mutation of both l- and d-nLDH-encoding genes does not result in a complete lack of lactic acid production (10). The same situation has also been found for other strains, such as Lactobacillus fermentum (13) and Lactobacillus casei BL23 (14). The different activities of l- and d-nLDHs contribute to differences in the ratio of the two isomers (1, 15). Because the optical purities of lactic acids differ, and few studies focusing on the relationship between the enzymes and the optical purity of lactic acid have been reported, a study of the crucial factors affecting the optical purity of lactic acid is necessary.
Thermophilic Bacillus species have several traits, such as the capacity to withstand relatively low pHs, high temperatures, and various other harsh conditions, which could be beneficial in industrial strains and could improve the commercial competitiveness of lactic acid production (16). B. coagulans can metabolize an extensive range of sugars, such as pentoses and hexoses in lignocellulosic biomass (16, 17). In contrast to the most frequently used lactic acid producers, such as Lactococcus lactis and Lactobacillus rhamnosus, B. coagulans can grow optimally at 50 to 55°C, a temperature range that is expected to minimize contamination in industrial-scale fermentations under nonsterile fermentation conditions. Therefore, in recent years, there has been an interest in studies on the optical purity of l-lactic acid produced by this species (17–19). However, the mechanism of production of highly optically pure l-lactic acid by this species has never been demonstrated.
In this study, a producer of l-lactic acid with high (>99%) optical purity, B. coagulans 2-6, was chosen as a representative strain. Three enzymes responsible for lactic acid production—l-nLDH, d-nLDH, and glycolate oxidase (GOX; EC 1.1.3.15)—were annotated from the whole-genome sequence of B. coagulans 2-6 (20). To systematically investigate the relationship between these enzymes and the optical purity of lactic acid, Lactobacillus delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer) and Lactobacillus plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer) were selected as comparative strains for study.
MATERIALS AND METHODS
Bacteria and culture conditions.
B. coagulans 2-6 was isolated by our laboratory and was used in this study. It is a homofermentative producer of l-lactic acid with an optical purity of 99% (20, 21). L. delbrueckii subsp. bulgaricus DSM 20081 (producing d-lactic acid at an optical purity of 98%) and L. plantarum subsp. plantarum DSM 20174 (1:1 ratio of l-lactic acid to d-lactic acid) were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). These three strains were chosen because they produce different types of lactic acid (l-, d-, and dl-lactic acid, respectively). The strains were maintained on de Man, Rogosa, and Sharpe (MRS) agar slants. The pH was adjusted to 6.2 to 6.5. Unless otherwise stated, the incubation temperatures for B. coagulans 2-6, L. delbrueckii subsp. bulgaricus DSM 20081, and L. plantarum subsp. plantarum DSM 20174 were 50°C, 37°C, and 30°C, respectively. Fully grown slants were stored at 4°C.
The seed culture was prepared as follows: a loop of cells from the fully grown slant was inoculated into 50 ml of MRS broth in 100-ml Erlenmeyer flasks and was incubated for 12 h at an appropriate temperature without agitation.
Cloning and overexpression of the enzymes responsible for lactic acid production.
Both the ldhL and ldhD genes were shown to be distributed in the three strains. According to the whole-genome sequence of B. coagulans 2-6 (GenBank accession number CP002472), the GOX gene was also present. No lactate racemase genes or homologs were found in any of the three strains (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). Genes of interest were amplified by PCR using Pfu DNA polymerase (Thermo Fisher Scientific) from the genomic DNAs (gDNAs) of the respective organisms. Seven DNA fragments were cloned into the expression vector (pETDuet-1, with the T7 promoter). The resultant recombinant plasmids were as follows: pETDuet-1-2-6ldhL, pETDuet-1-2-6ldhD, pETDuet-1-2-6go, pETDuet-1-20081ldhL, pETDuet-1-20081ldhD, pETDuet-1-20174ldhL, and pETDuet-1-2017ldhD. The recombinant plasmids containing the ldhL, ldhD, and GOX genes were transformed into competent Escherichia coli BL21(DE3) cells. For the expression of the recombinant proteins, transformed cells were first grown to an A600 of 0.6 at 37°C, then induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside, and grown for an additional 5 h at 23°C.
Purification of the enzymes.
The E. coli cultures were pelleted by centrifugation (10,540 × g, 30 min). Cells were resuspended in binding buffer (20 mM sodium phosphate, 500 mM sodium chloride, and 10 mM imidazole [pH 7.0]) and were then disrupted by sonication for 10 min to obtain crude enzyme solutions. After the cell debris was removed by centrifugation (13,000 × g, 30 min, 4°C), the supernatant was used to obtain purified nLDHs and GOX using a fast protein liquid chromatography (FPLC) system with nickel column chromatography. Proteins were resolved by SDS-PAGE using a 4-to-16% Bis-tris gel (Invitrogen) and were detected by Coomassie blue staining. The protein content was determined with a Bradford protein assay kit (Bio-Rad). Absorbance at 595 nm was measured and was compared to a standard curve prepared with bovine serum albumin. This purification was performed at 4 to 10°C, and the buffer used was potassium phosphate (pH 7.0) unless otherwise stated.
In vitro enzymatic activity assays.
Enzyme activity assays were performed in transparent 96-well plates. Briefly, the reaction mixture (total volume, 200 μl) containing 100 mM sodium phosphate (pH 6.5), 200 μM NADH, and 0.1 mg/ml enzyme was preincubated at 45°C for 10 min. To start the reaction, sodium pyruvate was added to a final concentration of 20 mM, and NADH oxidation was monitored at 340 nm in a 96-well plate reader. Assay conditions for the nLDHs originating from L. delbrueckii subsp. bulgaricus DSM 20081 and L. plantarum subsp. plantarum DSM 20174 were similar to those described for B. coagulans 2-6, except that the reaction temperature was 35°C for strain DSM 20081 and 30°C for strain DSM 20174.
To measure the reverse enzymatic activities of nLDHs from lactic acid to pyruvate, the reaction mixture (total volume, 200 μl) contained 100 mM sodium phosphate (pH 6.5), 15 mM NAD, and 0.1 mg/ml enzyme. Different concentrations of l- or d-lactic acid were added to the reaction mixture to start the reaction, and the NAD reduction was monitored at 340 nm at the respective optimal temperature.
The assay of the GOX enzyme from B. coagulans 2-6 was conducted under conditions described previously (22). The reaction mixture (total volume, 200 μl) contained 200 mM Tris-HCl (pH 7.5), 100 μM dichlorophenolindophenol, and 0.1 mg/ml enzyme. The reaction was initiated with the addition of 20 mM sodium lactate, and the change in absorbance at 578 nm was determined in a 96-well plate reader at 30°C.
The kinetic parameters of purified enzymes were determined by varying the concentration of one substrate over a wide range while maintaining the other at near-saturating levels. The reaction mixture (total volume, 200 μl) contained 100 mM sodium phosphate (pH 6.5), 200 μM NADH, and 0.1 mg/ml enzyme. To start the reaction, different concentrations of pyruvate were added to the reaction mixture, and NADH oxidation was monitored at 340 nm in a 96-well plate reader.
One unit of nLDH activity was defined as the amount of enzyme required to reduce 1 μmol NAD per min. One unit of GOX activity was defined as the amount of enzyme that caused an initial change in absorbance of 0.001/min at 578 nm (22).
In vivo enzymatic activity assays.
To assay the enzymatic activities in vivo, whole-cell extracts were used as described previously (21), with some modifications. Fermentations were performed in 500-ml Erlenmeyer flasks containing 200 ml of MRS broth. The cultures were inoculated for 24 h at an appropriate temperature without agitation. Exponentially growing cells were harvested by centrifugation (10,540 × g, 30 min, 4°C) and were washed with 0.85% (wt/vol) physiological saline. Cell pellets were subsequently suspended in 100 mM potassium phosphate buffer (pH 7.0) and were disrupted by sonication in an ice bath. After centrifugation at 12,000 × g for 10 min, the supernatants were used as the crude cell extracts. The reaction mixture, containing 100 mM sodium phosphate (pH 6.5), 200 μM NADH, 20 mM pyruvate, and 0.1 mg/ml crude cell extract, was preincubated at 37°C for 10 min. After incubation, the concentrations of l-lactic acid and d-lactic acid were analyzed by high-performance liquid chromatography (HPLC).
In vivo enzymatic activities were also confirmed by active staining of nLDHs after native PAGE according to a previous report (21) with some modifications. Crude enzymes of the three representative strains were concentrated by ultrafiltration and were separated by native PAGE on native polyacrylamide gradient (4-to-20%) gels. After electrophoresis, gels were cut into sections for active staining. The gel section with the molecular weight marker was soaked in Coomassie brilliant blue R-250. The other parts were soaked with 100 mM Tris-HCl buffer (pH 8.0) containing 0.1 mM phenazine methosulfate, 0.1 mM nitrotetrazolium blue chloride, 2 mM NAD, and 100 mM d-lactate, l-lactate, or dl-lactate, depending on the enzyme substrate preference. The staining mixture and gel sections were shaken gently until clear blue bands appeared.
Quantitative RT-PCR.
Quantitative real-time (RT)-PCR was employed to determine the transcription levels of the genes encoding key enzymes in different fermentation periods. Initially, B. coagulans 2-6, L. delbrueckii subsp. bulgaricus DSM 20081, and L. plantarum subsp. plantarum DSM 20174 were inoculated in MRS broth at 50°C, 37°C, and 30°C, respectively, to measure the growth curves of the strains and the time courses of optical purity values during fermentation for the determination of sample points. Cells of the three representative strains were harvested by centrifugation (5,000 × g, 10 min, 4°C) for RNA isolation by using an E.Z.N.A. bacterial RNA kit (Omega). The total RNA concentration was determined from the absorbance at 260 nm (NanoVue spectrophotometer; GE). By using appropriate gene-specific primers (Table 1), cDNA copies were synthesized with a FastQuant RT kit (with gDNase) (Tiangen, China) and were amplified with SYBR Premix Ex Taq (TaKaRa, China). The threshold cycles (CT) for each PCR with different concentrations of cDNA were determined and were compared with that for a standard DNA (the 16S rRNA gene) that was also analyzed at the same time (23, 24). From these results, a ratio of the concentration of gene-specific mRNA to total mRNA in the sample was calculated. The results reported are the averages of results from at least three experiments with variations of <15%.
TABLE 1.
Specific primers for quantitative real-time PCR analyses
| Strain | Target gene | Sequence (5′ → 3′) |
|---|---|---|
| B. coagulans 2-6 | ldhL | GTGGTTGGAACGGGTGCAGTTGGTAC |
| GTCTCCGCAACTCTTTAAGCGCCCAC | ||
| ldhD | CCTGAAACGGCAATGAAAGC | |
| CCCGTCTCCATAAAGGATACAAG | ||
| GOX gene | AAAATCTGACGGTGACGGTTGAGC | |
| CAACCCGCTCCATTTCGTCC | ||
| 16S rRNA | GCATGGAGGAAAAAGGAA | |
| TAAAACTCTGTTGCCGGG | ||
| L. delbrueckii subsp. bulgaricus DSM 20081 | ldhL | TGACAGAAGCAGCCTTAGATG |
| GCCACGCCATAGTAGGTTG | ||
| ldhD | AAGATGAGCCTGCGTAAC | |
| TTCGTCCATAGCCTTGTC | ||
| 16S rRNA | TCAGTGCCGCAGCAAACG | |
| CGGGACTTAACCCAACATCTCA | ||
| L. plantarum subsp. plantarum DSM 20174 | ldhL | ATCCTCGTTCCGTTGATG |
| AAGTTGATGATGTCGTAAGC | ||
| ldhD | TGGTGTTATCGGTACTGGTC | |
| TGTGGTAGTTATCCTTCAATGC | ||
| 16S rRNA | CAGCCTACAATCCGAACTGAGAA | |
| TCGTGTCGTGAGATGTTGGGTT |
Analytical methods.
The optical purities of l-lactic acid and d-lactic acid were analyzed using an HPLC (Agilent 1260 series; Hewlett-Packard, USA) equipped with a chiral column (MCI Gel CRS10W; Japan) (1). The mobile phase was 2 mM CuSO4 at a flow rate of 0.5 ml/min (25°C), with UV detection at 254 nm. The optical purity of l-lactic acid was defined as [l-lactic acid]/([l-lactic acid] + [d-lactic acid]) × 100%. The optical purity of d-lactic acid was defined as [d-lactic acid]/([l-lactic acid] + [d-lactic acid]) × 100%. Glucose and l-lactic acid concentrations were measured by an SBA-40C biosensor analyzer (Institute of Biology, Shandong Academy of Sciences, China). For the growth curves, A600 was measured by a 7200 visible spectrophotometer (Unico, Shanghai, China).
RESULTS
Characterization of specific activities of l- and d-nLDHs and GOX in vivo and in vitro.
After the purified heterologously expressed His-tagged l- and d-nLDHs and GOX had been obtained, the kinetic parameters of the purified enzymes were studied (Table 2). For B. coagulans 2-6, both l-nLDH and d-nLDH were found to have catalytic activities. The catalytic efficiencies of recombinant nLDHs were calculated as kcat/Km. The catalytic efficiency of l-nLDH toward pyruvate (5.8 × 103 M−1 s−1) was higher than that of d-nLDH (1.9 × 103 M−1 s−1). Furthermore, the Km of l-nLDH (3.8 mM) was lower than that of d-nLDH (5.9 mM), indicating that l-nLDH had a higher affinity for pyruvate than d-nLDH. Although the GOX genes were found in the B. coagulans 2-6 genome, heterologously expressed GOX exhibited no detectable activity. Furthermore, our study showed that B. coagulans 2-6 nLDHs could convert pyruvate to lactic acid in a reversible manner. They could convert lactic acid to pyruvate when strains were grown in a medium containing l- or d-lactic acid (Table 3). For L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer), l-nLDH activity was not detected. Furthermore, the d-nLDH of L. delbrueckii subsp. bulgaricus DSM 20081 exhibited the highest catalytic efficiency among the three d-nLDHs. For L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), both l-nLDH and d-nLDH activities were detected, and the d- and l-nLDHs showed similar catalytic efficiencies with pyruvate as the substrate.
TABLE 2.
Kinetic parameters of purified His-tagged l- and d-nLDHs and GOXs from the three representative strains
| Strain and enzymea | Km (mM) | Vmax (U/mg) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Bacillus coagulans 2-6 | ||||
| d-nLDH | 5.9 ± 0.2 | 18.7 ± 2.8 | 11.3 ± 1.7 | (1.9 ± 0.2) × 103 |
| l-nLDH | 3.8 ± 0.3 | 38.5 ± 1.0 | 22.1 ± 0.6 | (5.8 ± 0.2) × 103 |
| GOX | NDb | ND | ND | ND |
| Lactobacillus delbrueckii subsp. bulgaricus DSM 20081 | ||||
| d-nLDH | 0.3 ± 0.1 | 384.6 ± 2.6 | 235.5 ± 1.6 | (7.9 ± 0.5) × 106 |
| l-nLDH | ND | ND | ND | ND |
| Lactobacillus plantarum subsp. plantarum DSM 20174 | ||||
| d-nLDH | 2.7 ± 0.3 | 149.6 ± 1.5 | 91.3 ± 0.9 | (3.4 ± 0.0) × 104 |
| l-nLDH | 1.24 ± 0.1 | 32.3 ± 0.7 | 19.0 ± 0.4 | (1.5 ± 0.0) × 104 |
d-nLDH, NAD-dependent d-lactate dehydrogenase; l-nLDH, NAD-dependent l-lactate dehydrogenase; GOX, glycolate oxidase.
ND, not detected.
TABLE 3.
Kinetic parameters of purified heterologously expressed His-tagged l- and d-nLDHs of B. coagulans 2-6 for the conversion of lactic acid to pyruvate
| nLDH | Km (mM) | Vmax (U/mg) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| l-nLDH | 4.96 ± 2.37 | 0.10 ± 0.15 | 0.06 ± 0.08 | 0.012 ± 0.003 |
| d-nLDH | 9.54 ± 1.12 | 0.12 ± 0.38 | 0.07 ± 0.09 | 0.008 ± 0.080 |
To further address whether nLDHs showed different catalytic activities in vivo, the enzymatic activities of l-nLDHs and d-nLDHs in the three strains were determined using whole-cell extracts of the strains. Exponentially growing cells were collected, and the reduced products, l-lactic acid and d-lactic acid, were measured in order to evaluate enzymatic activity. As shown in Table 4, in contrast to the results obtained in vitro, B. coagulans 2-6 d-nLDH showed no enzymatic activity. High activity was observed for B. coagulans 2-6 l-nLDH. In L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer), the specific activity of d-nLDH (8.30 μmol min−1 mg−1) was much higher than that of l-nLDH (0.17 μmol min−1 mg−1). In L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), the specific activities of l- and d-nLDHs were similar.
TABLE 4.
Specific activities of l- and d-nLDHs from the three strains in exponential phase
| Strain | Sp act (μmol min−1 mg−1) |
|
|---|---|---|
| l-nLDH | d-nLDH | |
| Bacillus coagulans 2-6 | 11.35 ± 1.17 | NDa |
| Lactobacillus delbrueckii subsp. bulgaricus DSM 20081 | 0.17 ± 0.02 | 8.30 ± 1.08 |
| Lactobacillus plantarum subsp. plantarum DSM 20174 | 4.72 ± 0.38 | 2.51 ± 0.45 |
ND, not detected.
The in vivo enzymatic activities were also confirmed by active staining of nLDHs after native PAGE (Fig. 1). d-nLDH activities were detected in L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer) and L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer) but not in B. coagulans 2-6. Both d- and l-nLDH activities were detected in L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), while only d-nLDH activities were detected in L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer). Furthermore, GOX has been reported to catalyze the oxidation of lactic acid to pyruvate (11). For B. coagulans 2-6, only one fragment was observed when gels were soaked with l-lactate and dl-lactate, which means that only l-nLDH activities were detected in B. coagulans 2-6. GOX activities were undetectable by active staining after native PAGE analysis.
FIG 1.
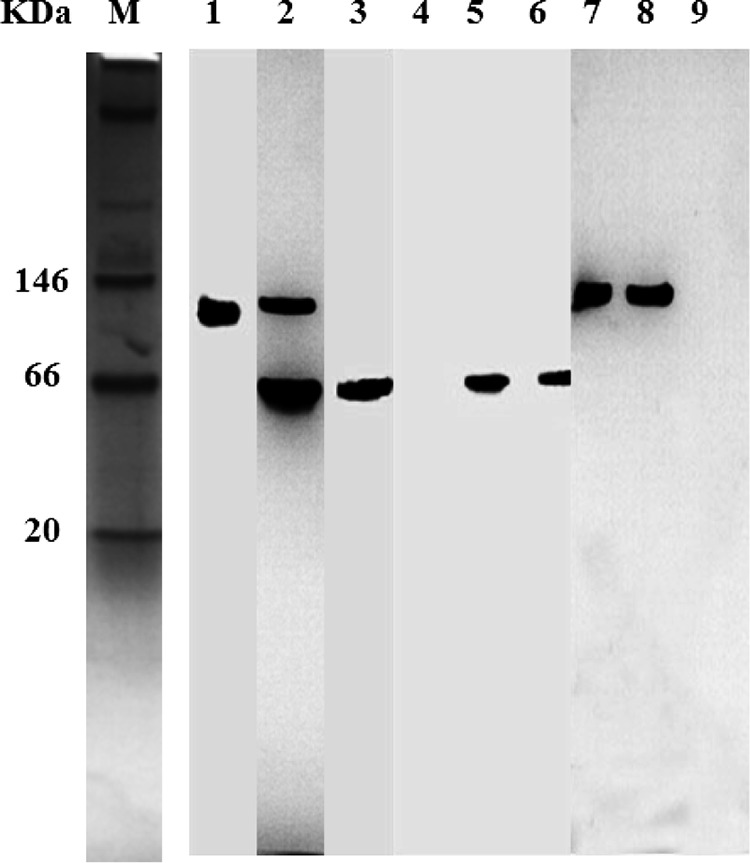
Active staining of nLDHs after native PAGE. Extracts of B. coagulans 2-6 (lanes 1, 4, and 7), L. plantarum subsp. plantarum DSM 20174 (lanes 2, 5, and 8), and L. delbrueckii subsp. bulgaricus DSM 20081 (lanes 3, 6, and 9) cells were used for native PAGE. dl-Lactate (lanes 1, 2, and 3), d-lactate (lanes 4, 5, and 6), and l-lactate (lanes 7, 8, and 9) were used as substrates for active staining. M, molecular size marker.
Changes in optical purity during fermentation.
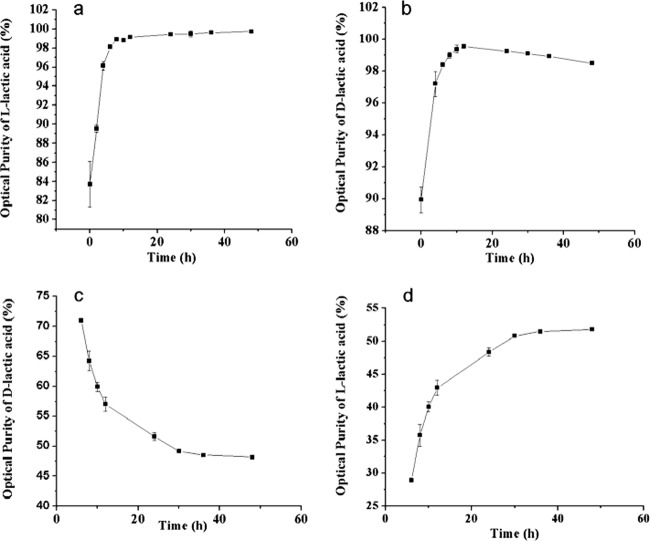
The optical purities of lactic acids produced by the three representative strains differed during fermentation. For B. coagulans 2-6, the optical purity of l-lactic acid was initially 83.7%. The optical purity dramatically increased to 98.1% within 6 h and then reached 99.8% at 48 h (Fig. 2a). For L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer), the upward trend in the optical purity of d-lactic acid was similar to that of l-lactic acid produced by B. coagulans 2-6. The optical purity of d-lactic acid increased from 89.9% at 0 h to 98.9% at 48 h (Fig. 2b). For L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), the optical purity of l-lactic acid increased from 29.0% at 6 h to 51.8% at 48 h (Fig. 2d), and the optical purity of d-lactic acid decreased from 71.6% at 6 h to 48.2% at 48 h (Fig. 2c). The ratio of the optical purity of l-lactic acid to that of d-lactic acid was approximately 1:1 at the end of the experiment.
FIG 2.
Changes in the optical purity of lactic acid during the fermentation process. Shown are the time courses of the optical purity values of l-lactic acid produced by B. coagulans 2-6 (a), d-lactic acid produced by L. delbrueckii subsp. bulgaricus DSM 20081 (b), d-lactic acid produced by L. plantarum subsp. plantarum DSM 20174 (c), and l-lactic acid produced by L. plantarum subsp. plantarum DSM 20174 (d).
Changes in transcription levels of ldhL, ldhD, and the GOX gene during fermentation.
To examine the transcription levels of the genes encoding nLDHs and GOX, RT-PCR assays were performed. According to the growth curves of the representative strains (data not shown) and the time courses of optical purity values, cells of the three representative strains at the different phases (exponential phase, stationary phase, and decline phase) were collected in order to measure the transcription levels.
The 2−ΔΔCT relative quantification method was used to determine the mRNA levels, and 16S rRNA was used as the internal reference. For B. coagulans 2-6, ldhL transcription levels were higher than those of ldhD and the GOX gene in different growth phases (Fig. 3a and b). The transcription ratio of ldhL to ldhD increased from 130-fold in the exponential phase to 216-fold in the decline phase. Although GOX genes were found in the genome of B. coagulans 2-6, no GOX enzymatic activity was detectable in vivo or in vitro. Thus, as expected, the GOX gene transcription levels were also rather low, and the transcription ratio of ldhL to the GOX gene was 1,323-fold in the exponential phase. For L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer), ldhD transcription levels were higher than those of ldhL; transcript ratios of ldhD to ldhL ranged from 13-fold to 22-fold in different growth phases (Fig. 3c). For L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), ldhD transcription levels were slightly higher than those of ldhL; transcript ratios of ldhD to ldhL ranged from 3-fold to 12-fold in different growth phases (Fig. 3d). The transcription levels of key enzymes encoding genes in the three representative strains were high in the exponential phase, decreased gradually from the exponential phase to the stationary phase, and then reached their minimums in the decline phase (see Fig. S1 in the supplemental material).
FIG 3.
Determination of the relative transcription levels of ldhD, ldhL, and the GOX gene by RT-PCR analyses. (a and b) Transcript ratios of ldhL to ldhD (a) and ldhL to the GOX gene (b) at different growth phases in B. coagulans 2-6. (c and d) Transcript ratios of ldhD to ldhL at different growth phases in L. delbrueckii subsp. bulgaricus DSM 20081 (c) and L. plantarum subsp. plantarum DSM 20174 (d). Error bars represent the standard deviations of the means for three independent experiments.
DISCUSSION
Lactobacillus species are representative lactic acid producers, and they are known to produce high yields of lactic acid (25). However, a limitation of lactic acid production is that the optical purity does not satisfy the demands for polymer-grade PLA (21). Efforts have been made to inactivate the nLDHs responsible for l- or d-lactic acid formation in Lactobacillus species (9, 10). Studies have shown that in the absence of one active nLDH, pyruvate can be metabolized by several alternative pathways. Thus, this method does not dramatically increase the optical purity of lactic acid (7). B. coagulans 2-6 has the advantages of a high fermentation temperature, high productivity, high yield, and a product with high optical purity (21). The 99.8% optical purity of l-lactic acid produced by B. coagulans 2-6 signifies that it is a good producer of polymer-grade l-lactic acid. In addition, B. coagulans 2-6 grows optimally at 50 to 55°C, which is also expected to minimize the contamination caused by mesophilic dl-lactic acid producers in industrial-scale fermentation. Nonsterilized batch fermentations for l-lactic acid production have been performed using B. coagulans 2-6, and studies have shown that the optical purity of l-lactic acid was higher than 99%, without contamination during the open operations (21). In recent years, there has been interest in lactic acid production by B. coagulans. Although optically pure lactates are synthesized from pyruvate by the catalysis of chirally specific d- or l-LDHs (26), the mechanism of production of highly optically pure l-lactic acid by B. coagulans has not been demonstrated.
l-nLDH (encoded by ldhL), d-nLDH (encoded by ldhD), and GOX (encoded by the GOX gene) were annotated from the whole-genome sequence of B. coagulans 2-6 (20). In fact, both ldhL and ldhD are widely distributed in almost all sequenced or reported LAB (1). As a predominant redox product of catabolism, NADH plays an important role in >700 biochemical reactions, a number of which are synthetically practical enzymatic reactions (27). The physiological role of nLDHs in bacteria is to regenerate NAD+ in a balanced way during fermentation. During this process, NADH, which is generated during the conversion of hexoses to pyruvate, is used as a cofactor. This reaction is an important step in the metabolism and energy conversion of living cells, because it allows the reoxidation of NADH, which is necessary for glycolysis. nLDHs play a complex role in B. coagulans. They catalyze the transformation of pyruvate to lactic acid and NADH oxidation. Furthermore, glucose consumption and growth rates have been found to be impaired in a Bacillus subtilis strain defective in nLDHs (16).
In this study, the catalytic efficiencies of nLDHs from B. coagulans 2-6 were investigated. In vitro, both l-nLDH and d-nLDH activities were detected, and the catalytic efficiency of l-nLDH toward pyruvate was 3-fold higher than that of d-nLDH. The Km of l-nLDH for pyruvate, 3.8 mM, in comparison to the Km of d-nLDH, 5.9 mM, suggests that d-nLDH has a lower affinity for pyruvate in B. coagulans 2-6 (Table 2). Although both l-nLDH and d-nLDH catalytic activities were detected in vitro, no d-nLDH activity was detectable in B. coagulans 2-6 by in vivo analysis and active-staining studies. The low or undetectable d-nLDH activity under native conditions may explain the high optical purity of the l-lactic acid produced by B. coagulans 2-6.
Furthermore, the catalytic activities of l-nLDH and d-nLDH from L. delbrueckii subsp. bulgaricus DSM 20081 and L. plantarum subsp. plantarum DSM 20174 were studied in vivo and in vitro for comparison. For L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer), the catalytic activity of d-nLDH (kcat/Km) was the highest among the three strains and was also higher than the catalytic activity of d-nLDH reported previously (1). Moreover, only d-nLDH activity was detected by active staining. For L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer), both l-nLDH and d-nLDH activities were detected in vivo and in vitro, with a ratio of approximately 1:1. Our study confirmed that l-nLDH and d-nLDH are mainly responsible for primary l- and d-lactic acid production in LAB.
According to previous reports on other L. plantarum strains, the catalytic activity of nLDH changes during the exponential phase (28). For B. coagulans 2-6, the optical purity of l-lactic acid increased gradually during the exponential phase until it reached its maximum in the stationary phase (Fig. 2). For the other two representative strains (L. delbrueckii subsp. bulgaricus DSM 20081 and L. plantarum subsp. plantarum DSM 20174), the optical purity of lactic acid also differed at different growth phases. The transcription of genes encoding key enzymes was analyzed to determine the relationship between changes in optical purity and gene transcription levels. B. coagulans 2-6 ldhL, ldhD, and GOX gene transcription levels decreased gradually from the exponential phase to the stationary phase, and these levels reached their minimums in the decline phase (see Fig. S1 in the supplemental material). The level of gene transcription was high in the exponential phase, which correlates with the marked change in the optical purity of l-lactic acid when the cells were in the exponential phase of growth. Subsequently, gene transcription decreased gradually, and accordingly, the optical purities were constant. However, the relative transcription levels of ldhL in B. coagulans 2-6 were much higher than those of ldhD and the GOX gene in different growth phases, and the transcription ratio of ldhL to ldhD increased from the logarithmic phase to the decline phase (Fig. 3). Previous reports on various Lactobacillus strains showed that the transcription levels of ldhD were higher than those of ldhL in representative LAB, and there are no other obvious distinctions between ldhL and ldhD transcript levels among various Lactobacillus strains (1). In this study, we comparatively investigated the transcription of ldhL and ldhD in L. delbrueckii subsp. bulgaricus DSM 20081 (a d-lactic acid producer) and L. plantarum subsp. plantarum DSM 20174 (a dl-lactic acid producer). ldhD transcription levels were higher than those of ldhL in both strains (Fig. 3), findings similar to those of the previous report (1). d-Lactic acid is important for the biosynthesis of the L. plantarum cell wall, and d-lactic acid is incorporated at the last position of the peptidoglycan precursor instead of the usual d-alanine. This feature has also been observed in other Lactobacillus strains, such as L. casei (15). Therefore, d-lactic acid is required for the growth of Lactobacillus strains. Even for l-lactic acid producers (such as L. casei, with a 92% enantiomeric excess of l-lactic acid), ldhD transcription levels were higher than those of ldhL (1).
In contrast to the situation in Lactobacillus strains, ldhL transcription in B. coagulans 2-6 was much higher than ldhD transcription in all growth phases, which may explain the high optical purity of the l-lactic acid produced. Although d-nLDH activities were not detected under the native condition, the transcription of d-nLDH-encoding genes was detected by RT-PCR analysis. Western blot analyses were also conducted to detect the expression of d-nLDH using whole-cell extracts of Bacillus coagulans 2-6. Although d-nLDH could not be detected by native PAGE, expression of d-nLDH could be detected by Western blotting (see Fig. S2 in the supplemental material). Since Western blotting, which can detect less than 1 pg of protein per band, has higher sensitivity than native PAGE (about 0.5 μg of protein per band), it was thought that the d-nLDH activity of B. coagulans 2-6 was too low to be detected by native PAGE, and its contribution to the optical purity of lactic acid appeared to be minimal in vivo. Meanwhile, the presence of a very low level of ldhD transcription also explains why B. coagulans 2-6 produced very small amounts of d-lactic acid.
GOX belongs to the flavoprotein oxidase class of enzymes, a large family of enzymes including dehydrogenases and reductases that utilize nucleotide cofactors for oxidation-reduction reactions (29). GOX and iLDHs are members of the α-hydroxyacid-oxidizing flavoprotein group, and their main function is to oxidize lactate to pyruvate by a flavin-dependent mechanism (28). All iLDHs convert lactic acid to pyruvate, and no evidence of a reverse reaction has been found (11). Although the genes encoding GOX were annotated in the B. coagulans 2-6 genome, the heterologously expressed enzyme showed no detectable activity in vitro (Table 2). Furthermore, active-staining studies also showed that only l-nLDH activities were detected in B. coagulans 2-6 (Fig. 1), which means that GOX activities were too low to be detected in vivo using native PAGE. However, GOX expression could be detected by Western blot analysis (see Fig. S3 in the supplemental material), which means that the observed transcripts of the gene encoding GOX were not silenced. Notably, ldhL transcription levels were 209-fold to 1,323-fold higher than those of the GOX gene in vivo (Fig. 3). Lactic acid is a weak acid (pKa, 3.86) that is uncharged and small enough to permeate the lipid membranes of cells (30). B. coagulans 2-6 cells likely encounter this acidic condition, especially in the decline phase of growth. Under these conditions, GOX could catalyze the oxidation of l-lactic acid to pyruvate in vivo and could allow strains to grow well in a medium containing l-lactic acid (31). Indeed, the transcription ratios of ldhL to the GOX gene decreased from 1,323-fold in the exponential phase to 209-fold in the decline phase, indicating that GOX might play a growing role in B. coagulans 2-6. However, the contribution of GOX to the optical purity of lactic acid appeared to be minimal, if any, in B. coagulans 2-6.
In conclusion, the catalytic activities of l-nLDH and d-nLDH differed in l-, d-, and dl-lactic acid producers. Although l-nLDH (encoded by ldhL), d-nLDH (encoded by ldhD), and GOX (encoded by the GOX gene) were annotated from the complete genome sequence of B. coagulans 2-6, only l-nLDH activity was detected in vivo. d-nLDH and GOX contributed weakly to l-lactic acid production in B. coagulans 2-6. The high catalytic efficiency of l-nLDH toward pyruvate and the high transcription ratios of ldhL to ldhD and the GOX gene explain the mechanism of production of highly optically pure l-lactic acid by B. coagulans 2-6. Studies revealing the mechanism of synthesis of highly optically pure l-lactic acid will provide direction for further strain improvements.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (31270108), the Chinese National Programs for High Technology Research & Development (2011AA02A202), and the Key Deployment Project of the Chinese Academy of Sciences (KSZD-EW-Z-016-3).
Footnotes
Published ahead of print 12 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01864-14.
REFERENCES
- 1.Zheng Z, Sheng B, Ma C, Zhang H, Gao C, Su F, Xu P. 2012. Relative catalytic efficiencies of ldhL- and ldhD-encoded products is crucial for optical purity of lactic acid produced by Lactobacillus strains. Appl. Environ. Microbiol. 78:3480–3483. 10.1128/AEM.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma K, Maeda T, You H, Shirai Y. 2014. Open fermentative production of l-lactic acid with high optical purity by thermophilic Bacillus coagulans using excess sludge as nutrient. Bioresour. Technol. 151:28–35. 10.1016/j.biortech.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Jiang X, Xue YF, Wang AY, Wang LM, Zhang GM, Zeng QT, Yu B, Ma YH. 2013. Efficient production of polymer-grade l-lactate by an alkaliphilic Exiguobacterium sp. strain under nonsterile open fermentation conditions. Bioresour. Technol. 143:665–668. 10.1016/j.biortech.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Wang LM, Ju JS, Yu B, Ma YH. 2013. Efficient production of polymer-grade d-lactate by Sporolactobacillus laevolacticus DSM442 with agricultural waste cottonseed as the sole nitrogen source. Bioresour. Technol. 142:186–191. 10.1016/j.biortech.2013.04.124. [DOI] [PubMed] [Google Scholar]
- 5.Ye L, Zhou X, Hudari MSB, Li Z, Wu JC. 2013. Highly efficient production of l-lactic acid from xylose by newly isolated Bacillus coagulans C106. Bioresour. Technol. 132:38–44. 10.1016/j.biortech.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A. 2010. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl. Microbiol. Biotechnol. 85:413–423. 10.1007/s00253-009-2280-5. [DOI] [PubMed] [Google Scholar]
- 7.Viana R, Yebra MJ, Galán JL, Monedero V, Pérez-Martínez G. 2005. Pleiotropic effects of lactate dehydrogenase inactivation in Lactobacillus casei. Res. Microbiol. 156:641–649. 10.1016/j.resmic.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Sharkey MA, Maher MA, Guyonvarch A, Engel PC. 2011. Kinetic characterization of recombinant Corynebacterium glutamicum NAD+-dependent LDH over-expressed in E. coli and its rescue of an lldD− phenotype in C. glutamicum: the issue of reversibility re-examined. Arch. Microbiol. 193:731–740. 10.1007/s00203-011-0711-z. [DOI] [PubMed] [Google Scholar]
- 9.Ferain T, Garmyn D, Bernard N, Hols P, Delcour J. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferain T, Hobbs JN, Richardson J, Bernard N, Garmyn D, Hols P, Allen NE, Delcour J. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178:5431–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garvie EI. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razeto A, Kochhar S, Hottinger H, Dauter M, Wilson KS, Lamzin VS. 2002. Domain closure, substrate specificity and catalysis of d-lactate dehydrogenase from Lactobacillus bulgaricus. J. Mol. Biol. 318:109–119. 10.1016/S0022-2836(02)00086-4. [DOI] [PubMed] [Google Scholar]
- 13.Aarnikunnas J, von Weymarn N, Ronnholm K, Leisola M, Palva A. 2003. Metabolic engineering of Lactobacillus fermentum for production of mannitol and pure l-lactic acid or pyruvate. Biotechnol. Bioeng. 82:653–663. 10.1002/bit.10615. [DOI] [PubMed] [Google Scholar]
- 14.Rico J, Yebra MJ, Pérez-Martínez G, Deutscher J, Monedero V. 2008. Analysis of ldh genes in Lactobacillus casei BL23: role on lactic acid production. J. Ind. Microbiol. Biotechnol. 35:579–586. 10.1007/s10295-008-0319-8. [DOI] [PubMed] [Google Scholar]
- 15.Goffin P, Deghorain M, Mainardi JL, Tytgat I, Champomier-Vergès MC, Kleerebezem M, Hols P. 2005. Lactate racemization as rescue pathway for supplying d-lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum. J. Bacteriol. 187:6750–6761. 10.1128/JB.187.19.6750-6761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero S, Merino E, Bolívar F, Gosset G, Martinez A. 2007. Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl. Environ. Microbiol. 73:5190–5198. 10.1128/AEM.00625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LM, Zhao B, Liu B, Yu B, Ma CQ, Yang CY, Su F, Hua DL, Li QG, Ma YH, Xu P. 2010. Efficient production of l-lactic acid from corncob molasses, a waste by-product in xylitol production, by a newly isolated xylose utilizing Bacillus sp. strain. Bioresour. Technol. 101:7908–7915. 10.1016/j.biortech.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 18.Wang LM, Xue ZW, Zhao B, Yu B, Xu P, Ma YH. 2013. Jerusalem artichoke powder: a useful material in producing high-optical-purity l-lactate using an efficient sugar-utilizing thermophilic Bacillus coagulans strain. Bioresour. Technol. 130:174–180. 10.1016/j.biortech.2012.11.144. [DOI] [PubMed] [Google Scholar]
- 19.Peng LL, Wang LM, Che CC, Yang G, Yu B, Ma YH. 2013. Bacillus sp. strain P38: an efficient producer of l-lactate from cellulosic hydrolysate, with high tolerance for 2-furfural. Bioresour. Technol. 149:169–176. 10.1016/j.biortech.2013.09.047. [DOI] [PubMed] [Google Scholar]
- 20.Su F, Yu B, Sun J, Ou HY, Zhao B, Wang L, Qin J, Tang H, Tao F, Jarek M, Scharfe M, Ma C, Ma Y, Xu P. 2011. Genome sequence of the thermophilic strain Bacillus coagulans 2-6, an efficient producer of high-optical-purity l-lactic acid. J. Bacteriol. 193:4563–4564. 10.1128/JB.05378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, Zhao B, Wang X, Wang L, Yu B, Ma Y, Ma C, Tang H, Sun J, Xu P. 2009. Non-sterilized fermentative production of polymer-grade l-lactic acid by a newly isolated thermophilic strain Bacillus sp. 2-6. PLoS One 4:e4359. 10.1371/journal.pone.0004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doelle HW. 1971. Nicotinamide adenine dinucleotide-dependent and nicotinamide adenine dinucleotide-independent lactate dehydrogenases in homofermentative and heterofermentative lactic acid bacteria. J. Bacteriol. 108:1284–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Y, Rhee MS, Ingram LO, Shanmugam KT. 2011. Physiological and fermentation properties of Bacillus coagulans and a mutant lacking fermentative lactate dehydrogenase activity. J. Ind. Microbiol. Biotechnol. 38:441–450. 10.1007/s10295-010-0788-4. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Ingram LO, Shanmugam KT. 2008. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol. 190:3851–3858. 10.1128/JB.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding S, Tan T. 2006. l-Lactic acid production by Lactobacillus casei fermentation using different fed-batch feeding strategies. Process Biochem. 41:1451–1454. 10.1016/j.procbio.2006.01.014. [DOI] [Google Scholar]
- 26.Gu SA, Jun C, Joo JC, Kim S, Lee SH, Kim YH. 2014. Higher thermostability of l-lactate dehydrogenases is a key factor in decreasing the optical purity of d-lactic acid produced from Lactobacillus coryniformis. Enzyme Microb. Technol. 58–59:29–35. 10.1016/j.enzmictec.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Weckbecker A, Gröger H, Hummel W. 2010. Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Adv. Biochem. Eng. Biotechnol. 120:195–242. 10.1007/10_2009_55. [DOI] [PubMed] [Google Scholar]
- 28.Goffin P, Lorquet F, Kleerebezem M, Hols P. 2004. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J. Bacteriol. 186:6661–6666. 10.1128/JB.186.19.6661-6666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindqvist Y, Brändén CI. 1985. Structure of glycolate oxidase from spinach. Proc. Natl. Acad. Sci. U. S. A. 82:6855–6859. 10.1073/pnas.82.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philp A, Macdonald AL, Watt PW. 2005. Lactate—a signal coordinating cell and systemic function. J. Exp. Biol. 208:4561–4575. 10.1242/jeb.01961. [DOI] [PubMed] [Google Scholar]
- 31.Gao C, Jiang T, Dou P, Ma C, Li L, Kong J, Xu P. 2012. NAD-independent l-lactate dehydrogenase is required for l-lactate utilization in Pseudomonas stutzeri SDM. PLoS One 7:e36519. 10.1371/journal.pone.0036519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.