Abstract
Bdellovibrio bacteriovorus is a Gram-negative predator of other Gram-negative bacteria. Interestingly, Bdellovibrio bacteriovorus 109J cells grown in coculture with Escherichia coli ML-35 prey develop into a spatially organized two-dimensional film when located on a nutrient-rich surface. From deposition of 10 μl of a routine cleared coculture of B. bacteriovorus and E. coli cells, the cells multiply into a macroscopic community and segregate into an inner, yellow circular region and an outer, off-white region. Fluorescence in situ hybridization and atomic force microscopy measurements confirm that the mature film is spatially organized into two morphologically distinct Bdellovibrio populations, with primarily small, vibroid cells in the center and a complex mixture of pleomorphic cells in the outer radii. The interior region cell population exhibits the hunting phenotype while the outer region cell subpopulation does not. Crowding and high nutrient availability with limited prey appear to favor diversification of the B. bacteriovorus population into two distinct, thriving subpopulations and may be beneficial to the persistence of B. bacteriovorus in biofilms.
INTRODUCTION
The predatory Gram-negative deltaproteobacterium Bdellovibrio bacteriovorus uses its single polar flagellum to swim rapidly through liquid medium and collide with potential prey cells. Upon recognition of a Gram-negative prey cell, B. bacteriovorus burrows into the prey's periplasm, forming a bdelloplast. The periplasmic bdellovibrio consumes its prey while elongating, septating, and then lysing the remains of the bdelloplast, allowing several new bdellovibrios to continue as hunters (1–4). This process typically requires 3 to 4 h and takes place in a spherical bdelloplast, but under certain conditions, such as altered nutrient regimes (5) and downregulation of genes affecting motility (6), multiple bdelloplast morphologies and delayed bdelloplast lysis (5–8) have been documented. A review of much of the literature for B. bacteriovorus from its discovery in the early 1960s through 2008 will efficiently introduce the reader to this fascinating bacterium (3, 9).
Although wild-type Bdellovibrio bacteriovorus is generally considered to be a predator, it can be isolated in what have historically been called host-independent (HI) forms that grow on medium in the absence of prey. Several investigators isolated HI variants, observing that both mutational events and environmental conditions such as crowding, nutrient availability, and signaling appeared to contribute to the observed lifestyle phenotype (5, 10–15). In 1992, Cotter and Thomashow linked mutational events in the hit locus, specifically of the Bd0108 gene, to generation of the HI phenotype (16). Sequencing of the Bdellovibrio genome in 2004 revealed that genes of the hit locus are involved in motility (17). Though this location in the genome is clearly tied to the predation phenotype, subsequent groups of investigators have isolated HI variants that are wild type in sequence at the hit locus, (18–20) indicating that mutations at other loci and/or gene regulation leading to altered phenotypes plays a critical role in the balance between predatory (host-dependent, or HD) and HI forms (21) or variants.
Many bacteria establish biofilm communities at surfaces and interfaces during part of their natural life cycle, living a sessile existence in a crowded environment (22–25). Although for many years investigations of B. bacteriovorus concentrated on its predatory nature in planktonic environments, significant recent attention has been directed to predation in the more diverse and complex bacterial cell environments characteristic of biofilms (1, 21, 26–28). In previous studies, our collaboration has explored interactions between wild-type Bdellovibrio predators and their prey at interfaces in model biofilm communities. When trapped atop a nutrient-poor flat surface with Escherichia coli prey cells, bdellovibrios multiply robustly and quickly devour their prey (29). In model E. coli biofilms submerged in liquid, B. bacteriovorus hunts vigorously, decimating or eradicating its prey in a single-component biofilm (1). However, B. bacteriovorus's effectiveness as a predator is inversely proportional to the nutrient concentration of the medium, leading us to wonder about the relative fitness for switching between predatory and HI Bdellovibrio or its variants in crowded, nutrient-rich environments. At the opposite extreme, we found that populations of purified Bdellovibrio predators began to grow robustly in an HI form in a nutrient-rich, interfacial surface environment lacking significant numbers of prey cells (30). Moreover, a strain of B. bacteriovorus isolated from the River Tiber, a river with high inputs of sewage and other sources of bacterial nutrients, was found to have genetically identical predatory and host-independent variants coexisting in the same ecological niche (31). These findings led us to consider the balance among populations of HI variants and HD Bdellovibrio and the role that nutrient availability, prey abundance, crowding, motility, and other factors play in that balance among these lifestyle forms.
In this investigation we explore an intermediate case in which a population of wild-type B. bacteriovorus predators with a small fraction of surviving E. coli prey is grown on a rich nutrient surface. In the resulting communities, HD bdellovibrios coexist with bdelloplasts, pleomorphic bdellovibrios, and E. coli; remarkably, the cells spatially segregate on a macroscopic scale, forming two distinct regions with different colors and predation phenotypes. We explore the genotypic and phenotypic variability within this community and consider its relevance to B. bacteriovorus lifestyle variation in biofilms and at surfaces in general. If we consider a biofilm to be a single entity with complex sensing, regulation, and communication (32), this investigation provides a new impetus for understanding B. bacteriovorus diversity and its role within bacterial communities in the environment.
MATERIALS AND METHODS
Bacteria and strains.
B. bacteriovorus 109J was purchased from ATCC (ATCC 43826) as a cleared prey lysate or coculture with the prey E. coli ML-35. We isolated and produced freezer stocks of E. coli ML-35 from the Bdellovibrio coculture. Our environmental isolate of B. bacteriovorus was collected from direct isolation and enrichment of Bdellovibrio cells following the detailed protocols of Ruby (2). In brief, several 50-ml samples of partially treated sewage water were obtained from a treatment plant in La Cañada Flintridge, CA. Small volumes of sewage water were combined with E. coli ML-35 and grown up in liquid culture with rapid shaking at 29°C. Cultures were checked daily by phase-contrast microscopy for the presence of Bdellovibrio. When bdellovibrios developed and showed vitality, the enriched cultures were centrifuged for 5 min at 8,000 × g and filtered through either a 0.45- or 0.8-μm-pore-size filter to remove organisms larger than Bdellovibrio. Serial dilutions of filtered cultures were plated onto lawns of E. coli ML-35 prey bacteria on yeast extract-peptone-sulfate-cysteine (YPSC) double-layered agar plates and incubated for 4 to 7 days until plaques formed. An agar plug containing a single plaque was collected and suspended in HEPES-metals (HM) buffer; then, the process of serial dilutions, growth on double-layer plates, and growth of plaques was repeated twice.
Media, buffers, and agar plates.
Luria broth (LB) was made with the following reagents in 1 liter of deionized water: 10 g of tryptone, 5 g of yeast extract, and 5 g of sodium chloride. For LB agar plates, 15 g of agar was also added. HEPES metals (HM) buffer is a nutrient-poor medium made from 6 g of HEPES free acid (CAS 7365-45-9) added to 1 liter of deionized water. The pH of this solution was adjusted to 7.6 using 1 M NaOH(aq) and 1 M HCl(aq). After the solution was autoclaved, previously autoclaved stock solutions of CaCl2 and MgCl2 were added to a final concentration of 0.25 g/liter and 0.60 g/liter, respectively. Yeast extract-peptone-sulfate-cysteine (YPSC) top and bottom agar plates were used together to form PFU of E. coli and B. bacteriovorus. These plates were also utilized for the predation phenotype assay, but in this case only E. coli was added to the top agar. The reagents were mixed together as follows: 0.125 g of CaSO4, 0.25 g of sodium acetate, 0.5 g of peptone, and 0.5 g of yeast extract were added to 1 liter of deionized water with 6 g of agar for the top layer or 10 g of agar for the bottom layer.
Preparation of cultures.
For the liquid cultures, cocultures (also termed prey lysates), and cultures for preparing PFU and single-colony growth on agar, we followed the following protocols based on methods from previous publications (1, 2, 9).
(i) HD B. bacteriovorus prey lysate derived from PFU.
For samples of HD B. bacteriovorus hunters where we wished to know that all the B. bacteriovorus cells were generated from a recent single ancestor, serial dilutions of HD B. bacteriovorus prey lysate were mixed with E. coli, added to YPSC top agar, and poured onto YPSC bottom agar. These plates were incubated at room temperature until clonal plaques similar to those used in the isolation of phage (9) appeared (3 to 5 days).
PFU were used to inoculate fresh E. coli cultures as follows. E. coli liquid cultures were grown to saturation and isolated by gentle centrifugation. The cells were washed and concentrated by resuspension and recentrifugation. The final suspended pellet was added to 7 ml of HM buffer in a 50-ml capped tube. Next, a mature and isolated single PFU of B. bacteriovorus was removed using a sterilized metal spatula and added to the 50-ml tube. The capped tube was then incubated at 29°C with shaking for 2 to 5 days until the liquid culture changed from murky to a comparatively clear suspension. Clearing is indicative of B. bacteriovorus consuming the prey cells; each cleared lysate contained approximately 107 to 108 B. bacteriovorus cells and about 105 E. coli cells per ml.
(ii) HD B. bacteriovorus non-PFU prey lysates.
HD B. bacteriovorus prey lysates were also generated in HM buffer with E. coli ML-35 augmented with a recently cleared prey lysate, clearings of B. bacteriovorus prey lysate atop an E. coli lawn in YPSC medium (but not formally PFU), or freezer stock of B. bacteriovorus prey lysate. Such cocultures are robust but are likely to contain more genetic variability in the bdellovibrio population than a plaque. All other parameters for generating a cleared prey lysate were the same as described above.
(iii) Preparation and growth of organized films.
Once the prey lysate cleared, this Bdellovibrio-prey coculture was deposited for biofilm growth onto a sterile filter paper on a medium plate. Four sterile 0.22-μm-pore-size filter membranes (polycarbonate) were aseptically placed on each LB agar plate. Next, a 7- to 10-μl volume of the prey lysate was placed on each filter membrane. The plate was incubated at room temperature for 12 to 48 h to investigate the young film and for 7 to 21 days to study the mature film. The film also forms robustly on bare LB agar plates.
Imaging techniques. (i) FISH.
Before performing fluorescence in situ hybridization (FISH) microscopy, we prepared prey lysates of both B. bacteriovorus 109J and the environmental isolate. Once the coculture cleared, the prey lysate was deposited on a sterile filtration membrane over an LB plate, allowed to grow for 1 to 2 days for young film analysis and for 1 to 3 weeks for mature film analysis, and processed according to the methods described by Mahmoud and colleagues using a Cy3-labeled 16S rRNA probe for B. bacteriovorus 109J (33). Hybridization and wash buffers were made as described previously using 35% formamide in the hybridization buffer and 450 mM NaCl in the wash solution (34). A genus-specific Bdellovibrio probe (probe BDE525, 5′-GATCCCTCGTCTTACCGC-3′; identifies B. bacteriovorus 109J 16S rRNA sequence 3′-CTAGGGAGCAGAATGGCG-5′) (33) was labeled with Cy3 (for young-film FISH) or Alexa Fluor 555 (for mature-film FISH). A different label for Bdellovibrio was used for mature films due to supplier constraints; the Alexa Fluor 555 dye fluoresces in a similar wavelength range as the Cy3 dye. Hybridization processing times were 3 to 15 h, followed by washes of 15 min (34). For the young-film FISH studies, cells were counterstained with a dilute 4′,6′-diamidino-2-phenylindole (DAPI) solution (5 mg/ml) for 1 min. For the mature-film samples, a gammaproteobacterial probe (probe GAM42a, 5′-GCCTTCCCACATCGTTT-3′; identifies E. coli) (35) or the universal bacterial probe (probe EUB338, 5′-GCTGCCTCCCGTAGGAGT-3′; identifies most bacteria) (36) was conjugated to Alexa Fluor 488 dye. In each case, samples were examined by epifluorescence microscopy using a Nikon E80i epifluorescence research microscope with a Nikon DS-Qi1Mc high-sensitivity monochrome digital camera.
(ii) AFM.
The young film at 12 to 18 h after deposition was still clear and translucent, so filter membranes with young films were removed from the medium plate, adhered to a stainless steel support, and imaged directly with atomic force microscopy (AFM). Since the mature film had grown too thick and soft to image directly, we applied a horizontal lift technique reminiscent of the method of Langmuir and Schaefer (37), in which the top layers of the mature film were peeled off to a fresh new filter surface and subsequently imaged by AFM. Constant-force contact mode AFM images under ambient conditions were acquired using a Digital Instruments MultiMode scanning probe microscope with a NanoScope IV controller. Standard silicon nitride tips were used for these measurements. Many details of the procedures and instrumental parameters for imaging bacterial cells by AFM are provided in our previous publications (1, 27, 29).
Predator phenotype assay.
To determine the phenotypic behavior of cells from different regions of the mature film, cells from each region were picked with a sterile toothpick or micropipette and spread on double agar YPSC plates containing E. coli ML-35 in the top layer. The plates were incubated for 1 to 2 weeks at room temperature and monitored for the formation of cleared plaques.
Sanger sequencing of PCR products.
The kits, protocols, and associated experimental details to isolate the PCR products for Sanger sequencing are discussed in the supplemental material. The hit gene (Bd0108) of an HD prey lysate as well as cells from both regions of a mature film were sequenced; the 16S rRNA partial gene of our environmental isolate was also examined. These isolated nucleic acid product samples were sent to Alpha Biolabs, Inc., in Burlingame, CA, for Sanger sequencing.
RESULTS
We prepared bacterial films of mixed composition from unfiltered prey lysates, allowing us to observe the interactions between B. bacteriovorus and E. coli cells in complex communities living on a surface. A small aliquot of this cleared coculture was deposited on a filter membrane over an LB agar plate. Within about 2 days, the young bacterial film appeared thin and light yellow. However, as the film grew radially outward and grew in height over days, it changed appearance. The mature film appeared as an inner yellow region and an outer white region, resembling a fried egg with an approximate 1- to 2-cm diameter. Figure 1 shows a typical spatially organized film with concentric inner and outer regions grown over 1 week at room temperature. The time frame for development of the two regions in a film varied from several days to about 1 week. All measurements presented here were made on mature films aged 1 to 3 weeks and grown on porous membranes over LB agar.
FIG 1.
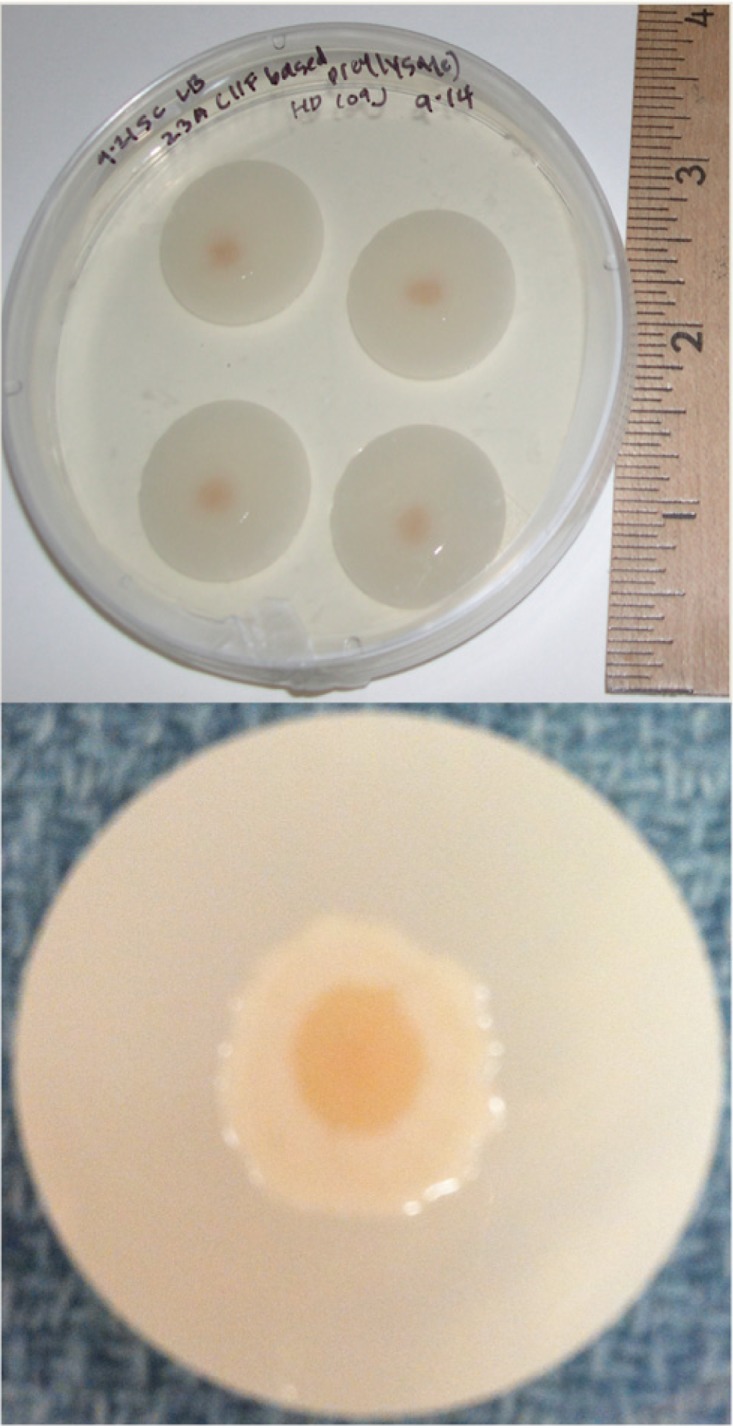
The spatially organized film on a porous membrane. At top is a digital photograph of a petri dish with four individual porous filter membranes over LB agar. Each membrane supports a mature, spatially organized film begun from an initial deposition of a tiny volume of a B. bacteriovorus prey lysate. At bottom is a magnified digital image of a mature film on a filter membrane.
The young film (∼1 to 2 days after deposition of prey lysate). (i) FISH.
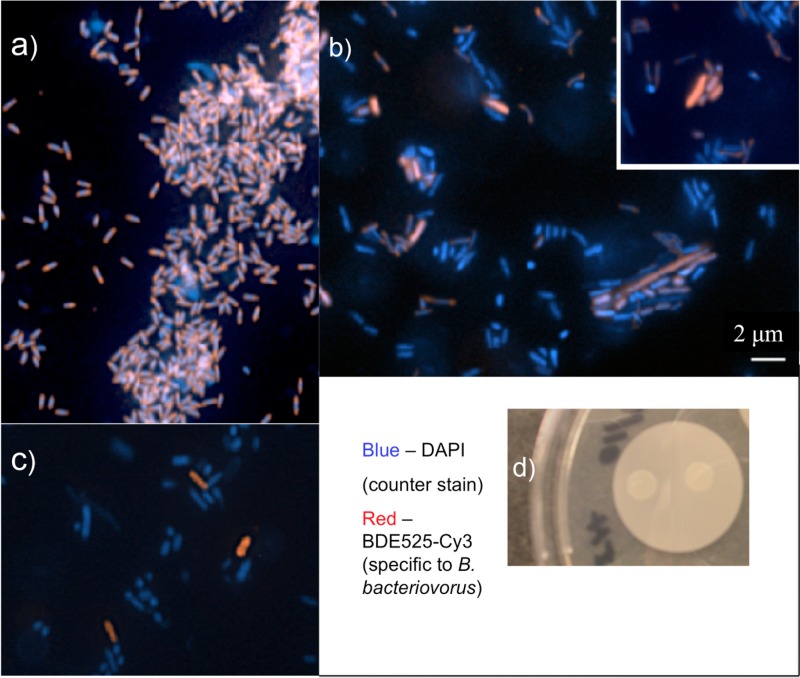
Fluorescence in situ hybridization (FISH) was used to characterize the composition of the lysates and the bacterial film shortly after deposition onto the filter. Bdellovibrio-specific probes targeted to the 16S rRNA and conjugated to Cy3 were designed and characterized by Mahmoud and colleagues (33). In a fresh prey lysate coculture of Bdellovibrio 109J and E. coli ML-35, a profusion of small, vibroid predators was observed stained red by the probe (Fig. 2a). About 1 μm in length and narrow and oblong in shape, these classic predator cells have clearly multiplied vigorously in the culture. Few E. coli cells remain (stained blue by the DAPI counterstain). In contrast, in a young film the small, narrow Bdellovibrio predator cells and blue E. coli prey cells are joined by a third type of cell (Fig. 2b). Identified as Bdellovibrio by the bright red stain, these cells are long and/or wide, and some appear to be actively dividing into progeny cells. This pleomorphism is characteristic of HI bdellovibrios or perhaps unusually shaped bdelloplasts. An analogous film prepared from a prey lysate of an environmental B. bacteriovorus isolate also shows a mixture of large, robustly stained red cells and blue cells, providing the molecular evidence that our purified environmental samples contain B. bacteriovorus (Fig. 2c). The shapes and sizes of these cells are consistent with the morphologies seen for B. bacteriovorus 109J cells from young films in Fig. 2b. Note also that in Fig. 2a to c, the fluorescent blue cells include both E. coli and nonactive cells of either species. In Fig. 2b and c, blue cells of dimensions consistent with both HD B. bacteriovorus and E. coli are visible. Figure 2d is a digital camera image of the young film of B. bacteriovorus 109J as observed on a filter membrane over LB agar. To identify species in the young film, PCR of extracted DNA and gel electrophoresis experiments were also conducted (see the supplemental material).
FIG 2.
Fluorescence in situ hybridization (FISH) images of a young film. Populations of bacterial cells in young films were examined using rRNA-targeted oligonucleotide probes specific to B. bacteriovorus 109J and tethered to a Cy3 dye (red). DAPI was used as a counterstain (blue). (a) An abundance of predatory B. bacteriovorus 109J cells are seen in a fresh prey lysate (small red/blue cells), but only a few E. coli prey remain (large, blue). (b) In a young film, small B. bacteriovorus predators are identified along with probable E. coli prey. In addition, long, wide B. bacteriovorus cells appear, strongly stained red. (c) In a young film from our B. bacteriovorus environmental isolate, the specific probe recognizes Bdellovibrio cells. (d) A digital photograph of the young film from a B. bacteriovorus 109J prey lysate. The scale bar in panel b also applies to panels a and c as well as to the inset in panel b, which was relocated for ease of viewing.
(ii) AFM.
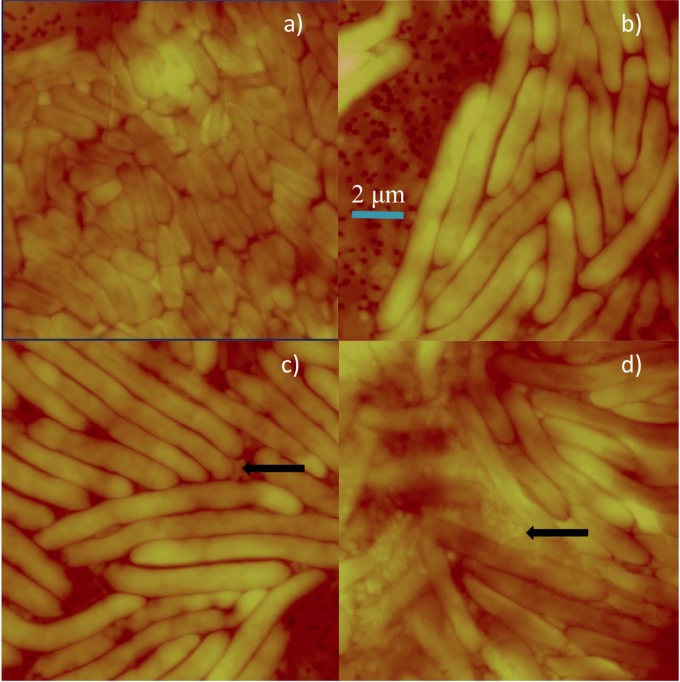
We employed atomic force microscopy (AFM) imaging to visualize the development of the young films, and representative images of films at 12 h postdeposition are displayed in Fig. 3. Unlike the FISH images shown in Fig. 2, AFM requires no fixation or staining, and thus the cells are entirely native; however, AFM alone is insufficient to distinguish the identity of cells with similar morphologies. The AFM tip was moved to three regions of the translucent but nascent film. Many cells were wide and elongated, with lengths ranging from about 4 to 10 μm, but small (∼1 μm), narrow cells of classic Bdellovibrio predator morphology are also present (Fig. 3, arrows). In contrast, deposition of E. coli ML-35 control yields a 10- to 18-h film, shown in Fig. 3a, with more uniform cells of lengths of 1 to 3 μm, suggesting that the elongated cells shown in Fig. 3b to d may be B. bacteriovorus cells with HI morphology. Deposits of this same prey lysate developed into spatially organized films within 1 to 2 weeks. Therefore, the AFM images in Fig. 3b to d give us evidence of significant cell morphology changes preceding the development of the characteristics of the mature film, consistent with the FISH results.
FIG 3.
Atomic force microscopy (AFM) of a young film. Shown are contact AFM images (constant-force mode) in air for films produced by deposition of bacteria on a 0.2-μm-pore-size filter membrane over LB agar. Images were obtained 10 to 18 h (a) or 12 h (b to d) after deposition. Panel a, depicting the single-species film resulting from deposition of E. coli ML-35, serves as a control image. Typical cell dimensions are about 1 μm wide by 2 to 3 μm long. Panels b to d show different locations on the young film resulting from an unfiltered prey lysate. All panels have the same x and y dimensions (12.9 μm; the scale bar in panel b applies to all images) and a similar z scale of about 1 μm. In panels a and b, the filter holes of the membrane are visible. Black arrows indicate small, vibroid-shaped cells consistent with HD B. bacteriovorus hunter cells with dimensions of 400 nm wide by 800 nm long.
The mature, spatially organized film (>3 days to ∼3 weeks). (i) Predation phenotype assay.
The mature film's two differently colored concentric regions were assayed for predatory ability (Table 1). A sterile micropipette tip or toothpick was used to collect cell mass from the film's inner and outer regions. This material was spread directly on the top agar of double-layer YPSC plates containing E. coli ML-35 prey. Cells from the film's inner region induced clearing of the E. coli lawn in from 1 to several days, whereas cells from the outer region did not, even after extended incubation. On a very few occasions, this subculturing of the outer region resulted in small yellow colonies on the top agar, but still no clearing of the top agar was observed. These results were reproduced many times over 2 years in different films from prey lysates.
TABLE 1.
Phenotype assay for predation of E. coli prey by Bdellovibrio bacteriovorus sampled in mature films
| Origin of B. bacteriovorus | Region of organized film sampled | Phenotype of cells from organized filma | No. and type(s) of replicates |
|---|---|---|---|
| ATCC 109J | Inner | + | 3 PFU, 2 non-PFU |
| Outer | − | ||
| Environmental isolate | Inner | + | 3 non-PFU |
| Outer | − |
Biofilm material was spread on the top agar of double-layer YPSC plates containing E. coli ML-35 prey. +, cells clear opaque top agar containing E. coli ML-35; −, cells do not clear opaque top agar.
(ii) Optical microscopy.
Phase-contrast optical microscopy was used to examine the size, shape, and motility of unstained cells from the film's inner and outer regions. Cells were suspended in HM buffer and imaged under 60× oil and, occasionally, 100× oil objectives. Inner-region cells swam frenetically and similarly to HD prey lysate bdellovibrios, qualitatively indicating that they have flagella, and appeared tiny (<1 μm). Some appeared vibroid shaped if attached to an apparent E. coli prey cell. Qualitatively, the outer-region cells displayed noticeably less frenetic motility and were quite pleomorphic, with some notably long cells.
(iii) Atomic force microscopy.
Because B. bacteriovorus cells are too small to resolve well by optical microscopy, we turned to AFM imaging. In Fig. 4, AFM constant-force images are displayed for the topmost layers of mature films (∼3 weeks old). Figure 4a illustrates the outermost edge of a mature, organized film, with the bare filter membrane surface at top right and many large cells (1 to 3 μm in length) packed closely together. Cell morphologies in this region of the film are most similar to those of E. coli cells shown in Fig. 3a. Images obtained near the border of the inner and outer regions reveal several long, thin cells as well as many smaller cells, as indicated in Fig. 4b and c. Though these long, thin cells could be E. coli cells, Bdellovibrio variants, or possibly abnormally shaped bdelloplasts, they are too large to be predatory B. bacteriovorus cells. Figure 4d shows an inner region primarily packed with cells averaging 800 nm in length, consistent with classic predatory B. bacteriovorus. Somewhat longer cells than these are also present in the inner region but as a minor population. As a control comparison, Fig. 4e shows the top layer of E. coli cells grown as a film under similar conditions as a mature B. bacteriovorus film. We emphasize that this two-member spatially organized biofilm is a three-dimensional entity and that these AFM data reveal the cell morphologies of the biofilm surface only.
FIG 4.
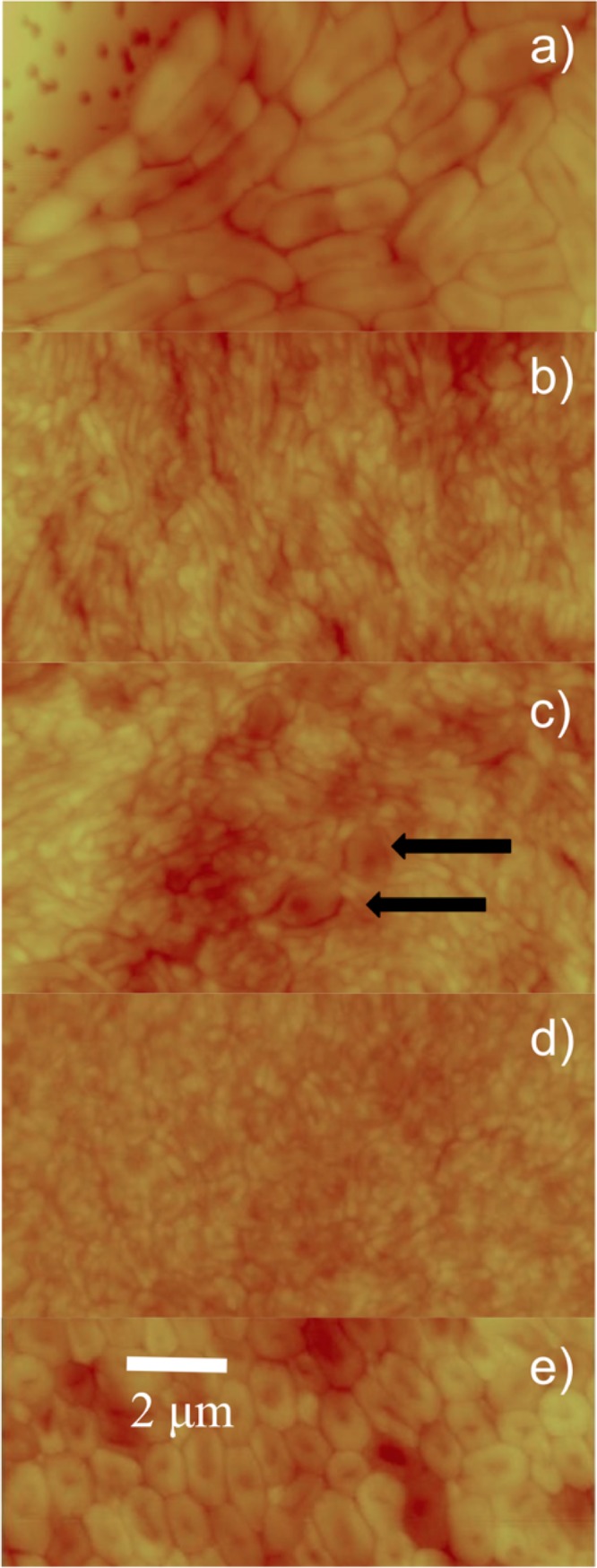
AFM images of the mature film. Contact AFM images (constant-force mode) in air were collected from the top layer of cells from a mature, spatially resolved film. The film was sampled at the outermost edge (a), the area where inner and outer regions meet (b and c), and the inner region (d). A control image of a film of E. coli ML-35 is presented in panel e. The scale bar in panel e applies to all images; panels a to d have a z scale of 1 μm, while panel e has a 0.5-μm z scale. In panel a, the filter holes of the membrane are visible. Black arrows in panel c indicate bdelloplasts.
(iv) FISH.
Fluorescence in situ hybridization (FISH) was also used to characterize the inner and outer regions of the mature film. A Bdellovibrio-specific probe targeted to the 16S rRNA was linked to the red Alexa Fluor 555 dye. To differentiate E. coli prey, the green Alexa Fluor 488 dye was conjugated to a 23S rRNA oligonucleotide specific to Gammaproteobacteria (GAM42a). Because the inner region is packed with HD bdellovibrios and sometimes E. coli is not present in a field of view, we alternatively employed the EUB338 oligonucleotide (16S rRNA) linked to the green Alexa Fluor 488 dye as a universal bacterial probe. To confirm that there was minimal cross-hybridization of Bdellovibrio-specific and Gammaproteobacteria-specific probes with our protocol, we performed controls with pure E. coli cultures (see Fig. S1 in the supplemental material) and a cleared B. bacteriovorus prey lysate (see Fig. S2).
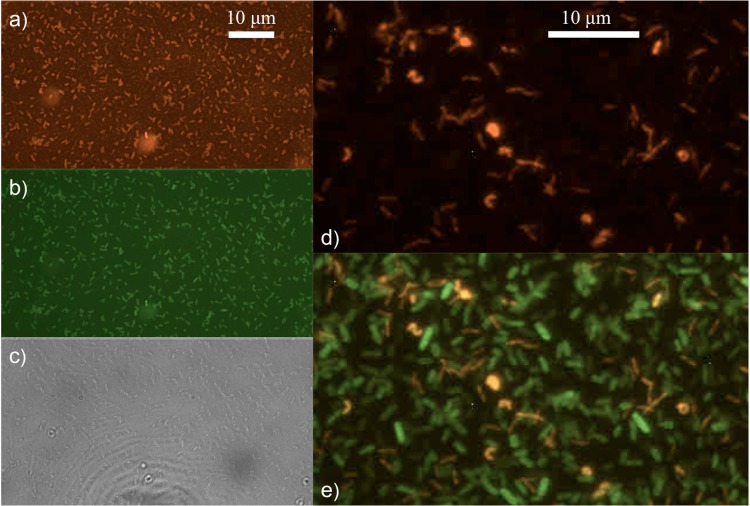
The inner region of non-PFU mature films contains primarily short, small bdellovibrios (<1 μm). In the representative images shown in Fig. 5a to c, a plethora of cells with morphologies consistent with HD bdellovibrios stain with both the red Bdellovibrio-specific probe, BDE525 (Fig. 5a), and the green, general bacterial probe EUB338 (Fig. 5b). In the dense mat of cells shown in Fig. 5d, small, slender bdellovibrios are revealed by the Bdellovibrio-specific red probe. The overlay with the green, general bacterial probe in Fig. 5e illustrates the coexistence of bdellovibrios (small, slender yellow cells), bdelloplasts (round, intense yellow cells), and E. coli prey (green).
FIG 5.
Fluorescence in situ hybridization (FISH) images of the inner region of the mature film. Cells from the inner region of a non-PFU mature film (aged 7 to 17 days) were examined using rRNA-targeted oligonucleotide probes specific to B. bacteriovorus 109J and tethered to an Alexa Fluor 555 dye (orange/red). The EUB338 probe was used as a universal bacterial counterstain (green), allowing us to identify most bacterial cells. An abundance of predatory B. bacteriovorus 109J cells are seen from the inner region with the BDE525 probe (a) and EUB338 probe (b) and in bright-field images (c). The scale bar in panel a applies to the images in b and c. Bdellovibrio cells are observed in panel d with the BDE525 probe and with E. coli prey in panel e as an overlay of panel d with same view using the EUB338 probe. The scale bar in panel d applies to the image in panel e.
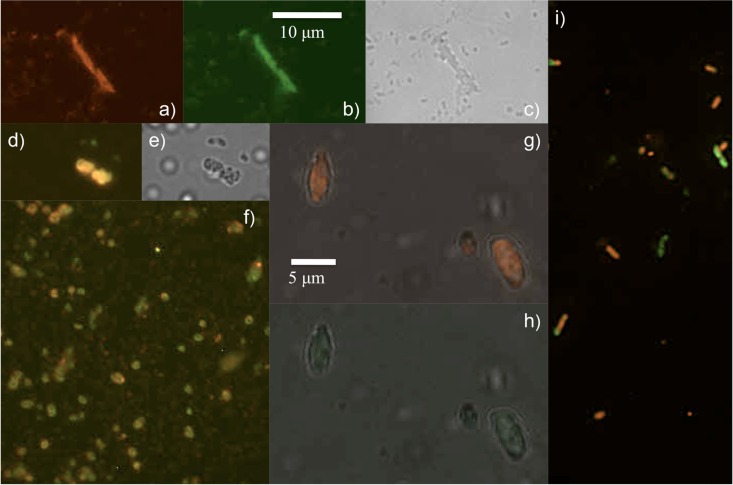
In contrast, the outer region probed by FISH (Fig. 6) contains much larger cells, including pleomorphic bdellovibrios and bdelloplasts and E. coli prey. A nearly 10-μm-long bdellovibrio variant or long bdelloplast is observed in Fig. 6a to c, with the red Bdellovibrio probe and the green, general bacterial (EUB338) probe and in bright-field images. (E. coli cells can be long as shown in Fig. S1 in the supplemental material, and previous work with Aquaspirillum has yielded long bdelloplasts [29]). Two large bdelloplasts observed in overlay (with E. coli in green from the GAM42a probe) and bright-field images, respectively, are displayed in panels d and e. In Fig. 6f, an array of small bdelloplasts and some small bdellovibrios are observed in the overlay (where EUB338 stains both prey and predator cells green). Another view of larger bdelloplasts is observed in overlays of the bright-field image and BDE525 (panel g) and the bright-field image and GAM42a (panel h). As noted previously, bdelloplast fluorescence intensity is much higher than the fluorescence intensity of bdellovibrios (33). Nonetheless, while our methods of fixation and hybridizations performed well for FISH imaging of planktonic E. coli and routine prey lysates, we noticed from our experiments that FISH imaging of bdelloplasts from biofilms either provided bright images (presumably newly formed bdelloplasts that are metabolically active) or dim images (perhaps older, metabolically quiescent bdelloplasts). Bdellovibrio cells of various sizes, long bdelloplasts, and a few E. coli prey cells are observed as an overlay image of BDE525 and GAM42a in Fig. 6i. In addition, PCR and gel electrophoresis experiments, including Sanger sequencing of the hit gene, characterize both inner and outer regions of the mature film (see the supplemental material).
FIG 6.
Fluorescence in situ hybridization (FISH) images of the outer region of the mature film. Cells imaged are taken from the outer region of a non-PFU mature film (aged 7 to 17 days). Pleomorphic Bdellovibrio cells, bdelloplasts, and a few E. coli cells are observed. A long narrow cell consistent with a Bdellovibrio variant, or perhaps a bdelloplast, is observed with the BDE525 probe (a) and EUB338 probe (b) and in the bright-field image (c). Rounded, large bdelloplasts are displayed in panel d as overlays of BDE525 and GAM42a probes; the latter probe identified E. coli prey as gammaproteobacteria (green), with its bright-field counterpart shown in panel e. Many small bdelloplasts are seen in panel f as an overlay image of BDE525 and EUB338 views. Large bdelloplasts are observed in overlays of the bright-field image and the BDE525 probe (g) and of the bright-field image and the GAM42a probe (h). E. coli cells and different morphologies of bdellovibrios and bdelloplasts are observed in panel i as an overlay of BDE525 and GAM42a views. The scale bar in panel b applies to panels a and c to f; the scale bar in panel g applies to panels h and i.
(v) The spatially organized film on a bare LB agar surface.
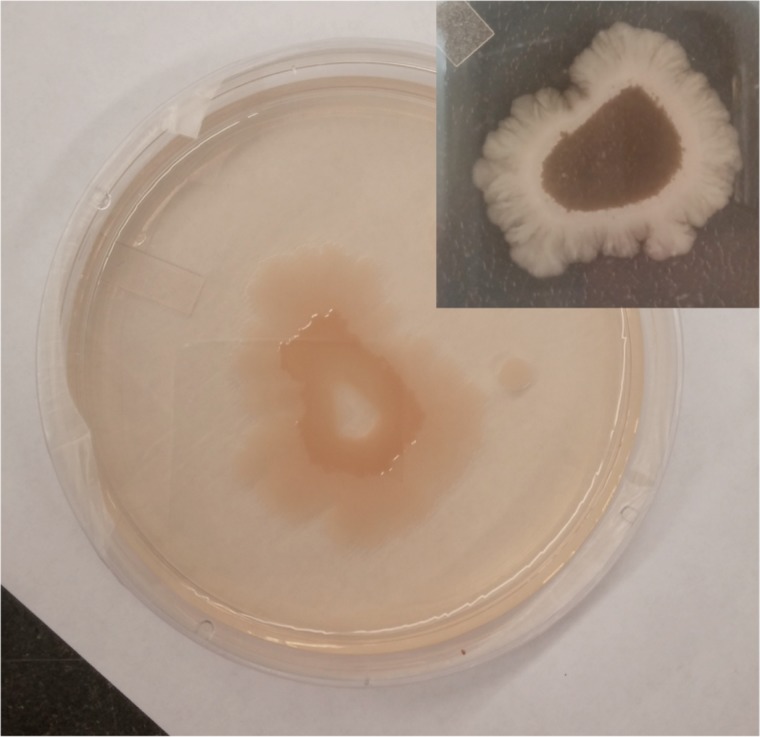
Spatially organized films also developed on the surface of LB agar plates without a filter, and these displayed the same striking segregation into radial domains as those characterized here on a porous membrane (Fig. 7). Notably, the films grown on bare LB agar expanded to a larger surface area, consistent with some motility restrictions being lifted without the membrane. There are fine structural features in the outermost region.
FIG 7.
The spatially organized film on agar. Films develop on a bare LB agar surface from a small volume of prey lysate, with the same general features as those investigated on a filter membrane. However, as seen in the digital image, the film expands to a much larger surface area. Also, while the inset shows a contiguous inner yellow region, the main image shows concentric rings of inner and outer regions with an uncovered center where the initial deposition was spotted. The outer region displays fine structural features.
DISCUSSION
Our experimental results show that a self-organized Bdellovibrio community emerges from a population of predatory B. bacteriovorus and E. coli confined to an interface. FISH and AFM data together show that on a surface, cells from a prey lysate quickly begin to exhibit pleomorphism, even as E. coli cells continue to be subject to invasion by predatory bdellovibrios. Predation assays, DNA sequencing, AFM, and FISH of the mature film clearly show that the inner region is primarily composed of short, narrow cells characteristic of classic, predatory B. bacteriovorus cells with some bdelloplasts and E. coli cells while the outer region contains a complex mixture of cells.
This film is novel in that a complex community of nonhunter Bdellovibrio variants, bdelloplasts, and prey cells separates but coexists in direct proximity to their predatory counterparts. The film formation process appears to be general as it occurs for both the laboratory strain 109J as well as a newly isolated B. bacteriovorus strain from human sewage. Furthermore, this phenomenon occurs whether or not the HD B. bacteriovorus cells are sampled from a coculture generated from a single plaque or from a multiply passaged HD B. bacteriovorus prey lysate. Given that each instance of prey lysate deposition on a surface will likely sample a different set of random point mutations in B. bacteriovorus, our findings indicate that the selective pressure induced by a low-prey, high-nutrient, confined surface setting leads to the population separation.
These results encourage us to consider how this macroscopic spatial segregation of phenotypes occurs. Given that a tiny fraction of HI B. bacteriovorus cells are likely to arise in an HD prey lysate as spontaneous mutants (12), then it may not be too surprising that both classic HD and HI morphologies develop together within 12 to 48 h on a surface: the HD B. bacteriovorus cells can continue to prey upon E. coli cells, while any HI cells can grow and replicate with available nutrients. It is also possible that E. coli responds in different ways to this environment and grows quite long. What is most interesting is that the hunter/nonhunter phenotypes eventually spatially separate instead of either clumping randomly or growing homogeneously as a biofilm of interspersed hunter and nonhunter B. bacteriovorus variants. Also notable is that not all of the E. coli cells are devoured by the classic predators early in the film formation. Finally, the cells in the outer region of the mature film are particularly pleomorphic and include a significant population of bdelloplasts with some E. coli prey.
One possible explanation for the radial location preferences of B. bacteriovorus phenotypes is the difference in the motilities of nonhunter variants and bdelloplasts compared to the motility of classic hunters. Although the polar flagellum that grants bdellovibrios such distinctive mobility in solution is less effective on a surface, HD hunters on surfaces can use gliding to locate prey (38). Yet rather than all HD hunters moving outward to hunt for prey, we find a reservoir of hunters in the center. Some of these hunters may indeed move outward, as seen in Fig. 7, but some remain segregated in an inner region. We note that B. bacteriovorus cells of the inner mature film retain their flagella and utilize them to swim vigorously when transferred to a buffer solution, as viewed under an optical microscope. The presence of functional flagella also suggests that these cells are not simply confusing the filter surface with the periplasmic space.
Conversely, the cells that we encounter in the outer region of the film do not exhibit the frenetic motility of classic hunters when removed to a buffer solution and observed by light microscopy, but this does not preclude effective surface-associated movements for both. On the contrary, the hydrated surface may provide an optimal environment for gliding motility. Gliding motility has been directly observed recently in both predatory and HI Bdellovibrio cells by Lambert et al. (38), and the Bdellovibrio genome contains at least 16 gliding and 2 twitching genes putatively related to nonflagellar motility (17).
The published transcriptional profiles of hunting HD B. bacteriovorus, early bdelloplast B. bacteriovorus, and axenic HI B. bacteriovorus (39) as well as the RNA sequencing of HD B. bacteriovorus and a late bdelloplast B. bacteriovorus each provide guidance in considering the role of motility in spatial segregation of phenotypes (40). Lambert et al. monitored the expression of the four operons in the B. bacteriovorus genome for tol- and ton-like genes. These genes are homologous to tol and ton adventurous gliding genes in Myxococcus xanthus, with a possible connection to outer membrane transport (secretion and uptake) due to contact or mechanical sensing (39, 41). In Bdellovibrio these four operons were linked to specific growth phases based on their up- or downregulation. Although the precise functional roles of these genes in each Bdellovibrio growth phase have not yet been elucidated, we hypothesize that these and other motility genes are likely to play a role in film formation.
We observed that bdelloplasts in the outer region are pleomorphic. Finding many bdelloplasts in the outer region is consistent with the results of our 2003 study (29) that showed an advancing tide of bdelloplasts when HD bdellovibrios were deposited on a extant, flat monolayer of E. coli prey. Flannagan et al. (6) downregulated an motA motility gene, resulting in variously shaped bdelloplasts, reduction in the motility of progeny bdellovibrios, and a much delayed lysis release. Another group also observed delayed bdelloplast lysis when Bdellovibrio motility genes were altered (7). Some of the outer-region bdelloplasts in our film show the elongating, enclosed growing bdellovibrios, but these cells do not appear to be septating into progeny. Sanchez-Amat and Torella (5) observed several marine Bdellovibrio strains creating “stable” or quiescent bdelloplasts that were less metabolically active yet remained viable over several months. The researchers could manipulate the proportion of stable bdelloplast formation by altering nutrient concentrations and predator-prey ratios although they were not able to pinpoint the chemical signals that induced either stable bdelloplast formation or lysis. Under the experimental conditions of this study, the best chance of survival as prey numbers diminish may involve B. bacteriovorus bacteria remaining in bdelloplasts until more favorable prey conditions occur.
A second contributing factor in the radial localization of the phenotypes rests on the idea that chemical communication in a consortium of sessile bacteria will lead to biofilm formation and dispersal. The radial distribution is consistent with the diffusion of a small signaling molecule, concentrated at the area of initial deposition of the prey lysate. These molecules are unlikely to be acyl-homoserine lactones as the B. bacteriovorus genome contains no genes homologous to known genes coding for synthesis or detection of these molecules (17). This signaling molecule may be linked to the synthesis of cyclic-di-GMP, a component of the well-known signal transduction system widely utilized in many bacterial processes, including switching between planktonic and sessile lifestyles in Proteobacteria (42). Both predatory and axenic B. bacteriovorus bacteria utilize specific cyclic-di-GMP signaling pathways, and these are linked to gliding and flagellar motility (43). If secreted by B. bacteriovorus or prey cells, cyclic-di-GMP would be expected to concentrate where many cells are confined. Establishment of a high concentration of signal molecule could set in motion gene expression related to surface attachment and motility.
We have been unsuccessful in repeated attempts to induce the nonhunter variants from the outer region of the mature film to revert to classic hunters. Even more intriguingly, when putative nonhunter cells from the mature film's outer radii are streaked on bare LB agar, HI colonies are rarely observed though we would expect true HI mutants to grow on agar when they are dispersed as well as they do when crowded on a surface. By removing the nonhunting Bdellovibrio cells from a high-cell-density environment of the film to a low-cell-density environment (liquid medium or agar plate), we hypothesize that we also removed the cells from critical signaling molecules that encouraged the nonhunting phenotype and growth. The observation that Bdellovibrio variants rarely grow prolifically when diluted or streaked out has been known for decades though previously it was attributed to a low frequency of HI mutants (12, 44). Unexpectedly, even with bdelloplasts in the outer region, no hunting phenotype is observed. Since persistent bdelloplasts were previously demonstrated to lyse only under certain conditions, most likely as a result of an unidentified chemical signal (5), the conditions under which the bdelloplasts were placed in our film may not have delivered such a signal.
Phenotypic differentiation in a biofilm has been reported for many diverse bacterial species, including M. xanthus and Pseudomonas aeruginosa (23, 45). Films of both species organize with high-motility cells in the center and lower-motility cells in the outer regions. A fruiting body dispersal strategy, in which the inner cells are signaled to disperse at some advantageous moment, has been verified for M. xanthus and implicated for P. aeruginosa (17, 45, 46). It appears that this type of biofilm eventuality may be general as it, too, has a life cycle (42, 47). In our film, chemical communication between the B. bacteriovorus phenotypes (and perhaps even with bdelloplasts) seems plausible although detection of signaling molecules from the prey cells may also be important, as it can be for surface-bound M. xanthus (23, 43). Any small-molecule secretions by E. coli prey will decrease as these cells are consumed by surface-bound classic hunters; this biofilm may develop from a Bdellovibrio prey starvation mechanism not unlike the predation, swarming, and fruiting body patterns exhibited by M. xanthus under low-prey conditions (12, 44, 48, 49, 50).
This discussion of signaling and motility does not negate the role of genotypic change in the transition between predatory and HI phenotypes. Sanger sequencing of the hit locus in the mature film confirmed that many of the Bdellovibrio cells were still wild type (see the supplemental material), and several groups reported saprophytic bdellovibrios with no mutation in the hit gene (18–20). The hit gene is known to be critical for host interaction, mainly host invasion and penetration, but the entire hit locus may have many ways to impact change from the HD to HI phenotype (44), particularly in the sharply constrained environment of a surface and the resulting film growth.
In conclusion, a spatially organized, mature bacterial community develops from a largely predatory Bdellovibrio population. This segregated community is formed reproducibly by the long-cultured 109J strain and our recent environmental isolate, indicating the general nature of this phenomenon. This investigation suggests that B. bacteriovorus populations at surfaces including the mammalian gut (51) as well as relevant agricultural settings may also form segregated regions with different functions. Our laboratories are working to further elucidate the interfacial coexistence of B. bacteriovorus lifestyles with the bacteria's prey and the transduced signals that allow the B. bacteriovorus variants to emerge, segregate, and coexist with prey at a surface. This surface behavior may reflect a persistence strategy in interfacial biofilm communities, serving to sequester predatory cells, multiply any nonhunter cells, and retain bdelloplasts in a nutrient-rich, low-prey environment.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply grateful to the National Science Foundation (CHE-0910578) for support of this work. E.M.S. could not have completed this work without support through the ACS-PRF UFS grant program (49377-UFS) and the Environmental Science and Engineering program at the California Institute for Technology, particularly the laboratory of Jared Leadbetter, as well as the Chemistry Department and Undergraduate Research Center of Occidental College. The work was also supported in part by a 2008 grant to Occidental College from Howard Hughes Medical Institute through the Undergraduate and Graduate Science Education Programs. M.E.N. acknowledges the Camille and Henry Dreyfus Foundation and the Clare Boothe Luce Foundation for their support.
E.M.S. thanks Aram Nersissian and Chris Craney for generously allowing the use of lab equipment as well as for their expertise.
Footnotes
Published ahead of print 19 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02423-14.
REFERENCES
- 1.Núñez ME, Martin MO, Chan PH, Spain EM. 2005. Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloids Surf. B Biointerfaces 42:263–271. 10.1016/j.colsurfb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Ruby EG. 1992. The genus Bdellovibrio, p 3400–3415 In Balows A, Starr MP, Stolp H, Truper HG, Schlegel HG. (ed), The prokaryotes. Springer, New York, NY. [Google Scholar]
- 3.Sockett RE. 2009. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 63:523–539. 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 4.Martin MO. 2002. Predatory prokaryotes: an emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4:467–477. [PubMed] [Google Scholar]
- 5.Sanchez-Amat A, Torella F. 1990. Formation of stable bdelloplasts as a starvation-survival strategy of marine bdellovibrios. Appl. Environ. Microbiol. 56:2717–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flannagan RS, Valvano MA, Koval SF. 2004. Downregulation of the motA gene delays the escape of the obligate predator Bdellovibrio bacteriovorus 109J from bdelloplasts of bacterial prey cells. Microbiology 150:649–656. 10.1099/mic.0.26761-0. [DOI] [PubMed] [Google Scholar]
- 7.Morehouse KA, Hobley L, Capeness M, Sockett RE. 2011. Three motAB stator gene products in Bdellovibrio bacteriovorus contribute to motility of a single flagellum during predatory and prey-independent growth. J. Bacteriol. 193:932–943. 10.1128/JB.00941-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenton AK, Lambert C, Wagstaff PC, Sockett RE. 2010. Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J. Bacteriol. 192:1299–1311. 10.1128/JB.01157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert C, Sockett RE. 2008. Laboratory maintenance of Bdellovibrio. Curr. Protoc. Microbiol. Chapter 7:Unit 7B.2. 10.1002/9780471729259.mc07b02s9. [DOI] [PubMed] [Google Scholar]
- 10.Varon M, Seijffers J. 1975. Symbiosis-independent and symbiosis-incompetent mutants of Bdellovibrio bacteriovorus 109J. J. Bacteriol. 124:1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishiguro EE. 1973. A growth initiation factor for host-independent derivatives of Bdellovibrio bacteriovorus. J. Bacteriol. 115:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidler RJ, Starr MP. 1969. Isolation and characterization of host-independent bdellovibrios. J. Bacteriol. 100:769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horowitz AT, Kessel M, Shilo M. 1974. Growth cycle of predacious bdellovibrios in a host-free extract system and some properties of the host extract. J. Bacteriol. 117:270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg D. 1978. Growth of host dependent Bdellovibrio in host cell free system. Arch. Microbiol. 116:185–190. 10.1007/BF00406035. [DOI] [PubMed] [Google Scholar]
- 15.Diedrich DL, Denny CF, Hashimoto T, Conti SF. 1970. Facultatively parasitic strain of Bdellovibrio bacteriovorus. J. Bacteriol. 101:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotter T, Thomashow MF. 1992. Identification of a Bdellovibrio bacteriovorus genetic locus, hit, associated with the host-independent phenotype. J. Bacteriol. 174:6018–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Barr C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 18.Roschanski N, Klages S, Reinhardt R, Linscheid M, Strauch E. 2011. Identification of genes essential for prey-independent growth of Bdellovibrio bacteriovorus HD100. J. Bacteriol. 193:1745–1756. 10.1128/JB.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wurtzel O, Dori-Bachash M, Pietrokovski S, Jurkevitch E, Sorek R. 2010. Mutation detection with next-generation resequencing through a mediator genome. PLoS One 5:e15628. 10.1371/journal.pone.0015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barel G, Jurkevitch E. 2001. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch. Microbiol. 176:211–216. 10.1007/s002030100312. [DOI] [PubMed] [Google Scholar]
- 21.Medina AA, Kadouri DE. 2009. Biofilm formation of Bdellovibrio bacteriovorus host-independent derivatives. Res. Microbiol. 160:224–231. 10.1016/j.resmic.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Stoodley P, Sauer K, Davies DG, Costerton JW. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187–209. 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 23.Straight PD, Kolter R. 2009. Interspecies chemical communication in bacterial development. Annu. Rev. Microbiol. 63:99–118. 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- 24.Lopez D, Vlamakis H, Kolter R. 2010. Biofilms. Cold Spring Harb. Perspect. Biol. 2:a000398. 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watnick P, Kolter R. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675–2679. 10.1128/JB.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert C, Sockett RE. 2013. Nucleases in Bdellovibrio bacteriovorus contribute towards efficient self-biofilm formation and eradication of preformed prey biofilms. FEMS Microbiol. Lett. 340:109–116. 10.1111/1574-6968.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Núñez ME, Martin MO, Chan PH, Duong LK, Sindhurakar AR, Spain EM. 2005. Atomic force microscopy of bacterial communities. Methods Enzymol. 397:256–268. 10.1016/S0076-6879(05)97015-8. [DOI] [PubMed] [Google Scholar]
- 28.Kadouri D, O'Toole GA. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71:4044–4051. 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Núñez ME, Martin MO, Duong LK, Ly E, Spain EM. 2003. Investigations into the life cycle of the bacterial predatory Bdellovibrio bacteriovorus 109J at an interface by atomic force microscopy. Biophys. J. 84:3379–3388. 10.1016/S0006-3495(03)70061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferguson MA, Schmitt JL, Sindhurakar AR, Volle C, Núñez ME, Spain EM. 2008. Rapid isolation of host-independent Bdellovibrio bacteriovorus. J. Microbiol. Methods 73:279–281. 10.1016/j.mimet.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Hobley L, Lerner TR, Williams LE, Lambert C, Till R, Milner DS, Basford SM, Capeness MJ, Fenton AK, Atterbury RJ, Harris MATS, Sockett RE. 2012. Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13:670. 10.1186/1471-2164-13-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolcott R, Costerton JW, Raoult D, Cutler SJ. 2013. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 19:107–112. 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 33.Mahmoud KK, McNeely D, Elwood C, Koval SF. 2007. Design and performance of a 16S rRNA-targeted oligonucleotide probe for detection of members of the genus Bdellovibrio by fluorescence in situ hybridization. Appl. Environ. Microbiol. 73:7488–7493. 10.1128/AEM.01112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303–2309. 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600. 10.1016/S0723-2020(11)80121-9. [DOI] [Google Scholar]
- 36.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmuir I, Schaefer VJ. 1938. Activities of urease and pepsin monolayers. J. Am. Chem. Soc. 60:1351–1360. 10.1021/ja01273a023. [DOI] [Google Scholar]
- 38.Lambert C, Fenton AK, Hobley L, Sockett RE. 2011. Predatory Bdellovibrio bacteria use gliding motility to scout for prey on surfaces. J. Bacteriol. 193:3139–3141. 10.1128/JB.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lambert C, Chang C-Y, Capeness MJ, Sockett RE. 2010. The first bite—profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One 5:e8599. 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karunker I, Rotem O, Dori-Bachash M, Jurkevitch E, Sorek R. 2013. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One 8:e61850. 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nan B, Zusman DR. 2011. Uncovering the mystery of gliding motility in the myxobacteria. Annu. Rev. Genet. 45:21–39. 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hobley L, Fung RKY, Lambert C, Harris MATS, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S-I, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 8:e1002493. 10.1371/journal.ppat.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, Chaudhuri R, Alderwick LJ, Lee DJ, Swarbreck D, Liddell S, Aizawa S-I, Sockett RE. 2013. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS One 8:e79759. 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purevdorj-Gage B, Costerton WJ, Stoodley P. 2005. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology 151:1569–1576. 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- 46.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell. Microbiol. 11:1034–1043. 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 47.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2012. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10:39–50. 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Plamann L. 1996. Purification and in vitro phosphorylation of Myxococcus xanthus AsgA protein. J. Bacteriol. 178:289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berleman JE, Kirby JR. 2007. Multicellular development in Myxococcus xanthus is stimulated by predator-prey interactions. J. Bacteriol. 189:5675–5682. 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, Athar R, Zheng G, Williams HN. 2011. Prey bacteria shape the community structure of their predators. ISME J. 5:1314–1322. 10.1038/ismej.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iebba V, Santangelo F, Totino V, Nicoletti M, Gagliardi A, De Biase RV, Cucchiara S, Nencioni L, Conte MP, Schippa S. 2013. Higher prevalence and abundance of Bdellovibrio bacteriovorus in the human gut of healthy subjects. PLoS One 8:e61608. 10.1371/journal.pone.0061608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.