Abstract
The objective of this study was to systematically assess the bifidogenic effect of three commonly used prebiotic products using in vitro cultures of infant fecal samples. Fresh stool samples collected from six term infants, each exclusively fed human milk (n = 3) or infant formula (n = 3), at 28 days of age were used as inocula. The following prebiotic products were added at concentrations applicable to infant formula: Vivinal GOS 15 (containing 28.5% galacto-oligosaccharide [GOS]) at 7.2 g/liter, Beneo HP (99.5% long-chain inulin [IN]) at 0.8 g/liter, Beneo Synergy 1 (enriched oligofructose and inulin [OF-IN]) at 4 g/liter, and a combination of Vivinal GOS 15 (7.2 g/liter) and Beneo HP (0.8 g/liter) (GOS-IN). The growth of total bacteria, Bifidobacterium, Bacteroides, Bifidobacterium longum, and Escherichia coli was quantified using specific quantitative PCR (qPCR). Bifidobacterium was also enumerated on selective Beerens agar plates, with representative colonies identified by sequencing of their 16S rRNA genes. Volatile fatty acids (VFA) and pH in the cultures were also determined. Irrespective of the feeding methods, the GOS product, either alone or in combination with Beneo HP, resulted in substantially higher growth of total bifidobacteria, and much of this growth was attributed to growth of B. longum. Beneo Synergy 1 also increased the abundance of total bifidobacteria and B. longum. Corresponding to the increases in these two bacterial groups, acetic acid concentrations were higher, while there was a trend of lower E. coli levels and pH. The lower pH and higher acetic acid concentration might be directly responsible for the lower E. coli population. At the concentrations studied, the GOS product was more bifidogenic and potent in inhibiting E. coli than the other products tested. These results suggest that supplementation of infant formula with GOS may increase intestinal bifidobacteria and benefit infant health.
INTRODUCTION
It is generally accepted that human milk-fed infants and formula-fed infants can have different gut microbiomes. The demonstrated differences in the gut microbiomes include a greater abundance of Bifidobacterium and a lower abundance of clostridia and enteric bacteria in human milk-fed infants than in formula-fed infants (1, 2). Such differences in gut microbiomes are believed to contribute to the benefits associated with human milk feeding, such as protection against infection and allergy (3–5), as well as long-term health and neurodevelopment (5–7). Human milk contains high levels of more than 200 structures of nondigestible oligosaccharides (8, 9), whereas cow milk contains virtually no oligosaccharides (10). Therefore, without supplementation, oligosaccharides are nearly absent in cow milk-based infant formulas. The difference in nondigestible oligosaccharides between human milk and infant formula is believed to be a main reason for the observed differences in intestinal microbiomes between infants receiving these two types of feeding (9, 11).
Nondigestible oligosaccharides can be added to infant formula as prebiotics to increase its oligosaccharide content (12). However, the prebiotics commercially available for inclusion in infant formula are limited in variety, and the nondigestible oligosaccharides contained in most prebiotic products are much simpler in structure than most of those found in human milk (13). Clinical feeding trials have been conducted to determine the effect of nondigestible oligosaccharides added to infant formula, which include fructo-oligosaccharides, galacto-oligosaccharides, and inulin (14). The conclusions of these investigations regarding changes in microbiome are conflicting (15). Indeed, some of these studies demonstrated a significant increase in beneficial bacteria (i.e., Bifidobacterium and Lactobacillus) and a decrease in harmful bacteria (i.e., clostridia and Escherichia coli) (16–18), whereas other studies showed little or no measurable effect (19–21). This discrepancy may be attributable to differences in laboratory methodology, formula composition, infant populations studied, and their associated individualized intestinal microbiomes (15). Indeed, tremendous variations in intestinal microbiomes were reported among infants in a number of studies (22–24), and such variations often exceed the treatment effects, thus making it difficult to ascertain the actual impact of prebiotic supplementation.
Evaluation of prebiotics for their effects using in vitro cultures can overcome the limitations posed by some of the uncontrollable variations in clinical trials, such as variations in feeding regimens and intestinal microbiomes. Additionally, in vitro cultures overcome the difficulties in comparing the efficacy of different prebiotics owing to differences in methodology, form of prebiotic products, dose, duration, number of subjects, and measurements taken. Several researchers have used in vitro cultures to evaluate the effect of prebiotics on individual strains of intestinal bacteria (25–27). Although these studies have advanced our knowledge on the characteristics (e.g., the ability to grow on the test prebiotics and the fermentation products) of these bacterial strains, the conclusions drawn probably do not apply to the complex intestinal microbiomes of infants due to the absence of other intestinal bacteria. Although several studies have compared the prebiotic or bifidogenic effects of multiple prebiotics using in vitro cultures of adult fecal samples (28–30), no study has been reported that used in vitro cultures of infant stool samples. Because the intestinal microbiomes differ greatly between adults and infants (31), different effects of prebiotics on intestinal microbiome are expected. The objective of this study was to comparatively evaluate the effects of prebiotics commonly added to infant formula (32) on the growth of select populations of intestinal bacteria and their fermentation using in vitro cultures of infant fecal samples. Fresh stool samples from both human milk-fed and infant formula-fed infants were used as the inocula in an attempt to encompass the gut microbiome supported by both feeding types. In addition, the prebiotic oligosaccharides were compared at the concentrations representative of those added to infant formula.
MATERIALS AND METHODS
Study design and sample collection.
This was a nonrandomized, single-center, unblinded study. An independent ethics committee and institutional review board reviewed and approved the protocol and consent forms. Informed consent was obtained from the legally authorized representatives of the subjects prior to the study. The study was designed to enroll infants between the ages of 0 (date of birth) and 5 days to collect one fecal sample from each subject at approximately 28 days of age. The study was designed to enroll 5 infants into each group. Infants who were fed exclusively human milk were enrolled into the human milk-fed (HM) group, while infants who were consuming exclusively a milk-based infant formula, with no added prebiotics, were enrolled into the formula-fed (FF) group. The parent(s) of each infant were instructed to use the designated feeding ad libitum as the sole source of nutrition for the duration of the study. Infants were excluded from further analyses if they (or the mothers of HM-fed infants) received any medication that could affect gut bacteria. There were 3 study visits during this study (day 0, 15 ± 3, and 28 ± 3) to gather information on the infants and protocol compliance.
Fresh fecal samples were collected at the last visit (day 28 ± 3). Detailed sample collection procedures were provided to the study coordinator at the clinical site. Within 60 min of a bowel movement, the diaper was removed and placed in a plastic bag, then placed in an insulated cooler bag containing frozen ice packs, and delivered to the study coordinator. The fecal sample was immediately transferred into a sterile 15-ml preweighed glass bead-containing serum vial using a sterile tongue depressor. The vial was then immediately filled completely with an anaerobic buffer (0.1% peptone, 0.85% NaCl, and 0.5% cysteine-HCl [pH 7.0]) of known volume, capped with a rubber stopper, and sealed with an aluminum crimp cap. The sample vial was placed in an insulated cooler containing ice packs and brought to the laboratory within 3.4 to 5.3 h from the time the stool was passed. Upon receipt of each sample in the laboratory, the sample vial, which contained the fresh stool sample and the anaerobic buffer added, was weighed (total sample vial weight). The amount of the anaerobic buffer transferred into each sample vial was calculated as the difference between the initial volume of the anaerobic buffer and the volume left in the original buffer vial. The amount of each fresh stool sample collected was determined as the difference between the total weight of each sample vial and the amount of buffer transferred to the sample vial from the buffer vial. The dilution factor for each of the fresh stool samples was determined by dividing the amount of each stool sample (in grams) by the total weight of each sample vial (in grams). Of the infants enrolled in each feeding group, 3 in each group met the enrollment criteria and their fresh stool samples were collected and used as the inocula.
In vitro fermentation, enumeration, and isolation of Bifidobacterium organisms.
The prebiotic products used in this study were selected based on their market availability, potential as prebiotics, and suitability to be included in infant formulas. The tested products included Vivinal GOS 15 (FrieslandCampina Domo, Rolling Meadows, IL), Beneo HP (Beneo, Inc., Morris Plains, NJ), and Beneo Synergy 1 (Beneo, Inc.). Vivinal GOS 15 contained (percent dry matter) polysaccharides (45.6%), GOS (28.5%), lactose (10.1%), and glucose (9.7%) as main components. Beneo HP contained chicory inulin (>99.5% purity) with the small oligofructose (degree of polymerization < 5) removed, while Beneo Synergy 1 was a combination of chicory inulin and oligofructose produced by partial enzymatic hydrolysis of chicory inulin (90 to 94% enriched oligofructose and inulin). These products were evaluated in the following treatments: Vivinal GOS 15 alone at 7.2 g/liter (referred to as GOS), Beneo HP alone at 0.8 g/liter (referred to as IN), Beneo Synergy 1 alone at 4.0 g/liter (referred to as OF-IN), and a combination of Vivinal GOS 15 (7.2 g/liter) and Beneo HP (0.8 g/liter) (referred to as GOS-IN). These doses were selected based on those tested in clinical studies and on tolerance information available in the literature of clinical studies conducted with young infants (20, 33). A no-prebiotics control was also included.
The basal medium contained (in g/liter) peptone, 2.0; yeast extract, 2.0; NaCl, 0.1; K2HPO4, 0.04; KH2PO4, 0.04; MgSO4·7H2O, 0.01; CaCl2·2H2O, 0.01; NaHCO3, 2.0; cysteine-HCl, 0.5; and bile salts, 0.5, as well as Tween 80 at 2.0 ml, phylloquinone at 10 μl, hemin solution at 1.0 ml, and sodium resazurin (27, 29, 34). The medium was made anaerobic as described previously (29) and dispensed into Hungate tubes (9 ml per tube). Each product was added (0.5 ml) from a sterile stock solution to the final concentration intended. The control cultures received no prebiotics but received 0.5 ml of phosphate-buffered saline (PBS). Each fresh stool sample, diluted 2.9- to 15.8-fold in the anaerobic buffer in the sampling vials, was mixed well by vortexing vigorously, and 0.5 ml of each was inoculated into each of the 3 replicate Hungate tubes for each treatment. These culture tubes were incubated at 37°C for 24 h. Three subsamples (3.5 ml) were collected at 12 and 24 h postincubation from each culture tube and aliquoted into 3 microtubes. One subsample was processed and frozen at −80°C for volatile fatty acid (VFA) analysis using gas chromatography (35); the second subsample was centrifuged at 4°C for 10 min at 16,000 × g to harvest the bacterial biomass, which was frozen immediately at −80°C; and the third subsample was used to enumerate bifidobacteria on selective Beerens agar plates immediately.
Each of the sampled cultures was diluted in 1:10 series in an anaerobic chamber using the same anaerobic buffer as used in stool sample collection. An aliquot of 0.1 ml of 3 dilutions each was plated on selective Beerens agar plates in triplicate (29); the plates were then incubated at 37°C for 24 h inside an anaerobic chamber that was filled with 90% N2, 5% H2, and 5% CO2. The colonies formed were counted from the plates that had 30 to 300 colonies. The number of CFU/ml of culture for each time point and each treatment was calculated based on the number of colonies and the corresponding dilution factor.
Analysis of Bifidobacterium isolates by ERIC-PCR and DNA sequencing.
To identify the bifidobacteria grown on the Beerens plates, 96 random colonies were picked for each treatment and each time point. They were inoculated into 1 ml of Columbia broth dispensed into deep-well plates (96 wells per plate and 1 ml of medium per well). These plates were incubated in an anaerobic incubator at 37°C for 24 h. Then 100-μl quantities of each culture were transferred to the wells of 96-well PCR plates, which were sealed with a thermal adhesive film (Fisher Scientific) and incubated at 95°C for 5 min to lyse the cells, using a thermocycler. Following centrifugation, the cell lysate was used directly as the template to perform enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR) to identify unique colonies (36, 37). The banding patterns of ERIC-PCR were clustered using BioNumerics (v.5.1; Applied Maths, Inc., Austin, TX). The 16S rRNA gene (rrs) of each representative ERIC-PCR phylotype, rather than the 16S rRNA gene of all the colonies, was amplified by PCR using the cell lysate as the template and universal bacterial primers (38). Following confirmation of the expected band by agarose gel (1.5%) electrophoresis, the PCR products were purified using QIAquick PCR purification kits (Qiagen, Inc., Valencia, CA) and sequenced using a 3730 DNA analyzer (Applied Biosystems, Inc., Foster City, CA). After manual verification of base calling, the sequences were compared to GenBank sequences using BLASTn to identify the most similar sequences for each isolate.
Microbial community DNA extraction.
Community DNA was extracted from each of the cultures using the repeated bead-beating and column purification method as previously described (39). The resultant DNA quality was visually assessed by agarose gel (1.0%) electrophoresis. The DNA concentration was quantified using a Quant-it kit (Invitrogen Corporation, Carlsbad, CA) and an Mx3000P real-time PCR system (Stratagene, La Jolla, CA).
Quantification of bacterial groups by specific real-time PCR.
The primers used in the real-time PCR assays are listed in Table 1. The abundances of the following bacterial groups were quantified using respective specific real-time PCR as described previously: total bacteria (40), Bifidobacterium (40), Bacteroides (41), Bifidobacterium longum (42), Clostridium difficile (43), and E. coli (44). Lactobacilli were not quantified because initial endpoint PCR analysis of the samples showed either no detection or very low occurrence of this group of bacteria (data not shown). One sample-derived standard was prepared for each of the real-time PCR assays for total bacteria, Bifidobacterium, and Bacteroides as described previously (40, 45). The rrs genes of B. longum ATCC 15708 and E. coli wild-type MG1655 were PCR amplified using bacterial cells of each strain and universal bacterial primers 63f/1389r (46, 47), and the PCR products were used as respective real-time PCR standards for these two species. All the real-time PCR assays were performed using an Mx3000P real-time PCR system (Stratagene) and 25-μl reaction volumes. All samples were analyzed in triplicate together with respective standards (also in triplicate) using the same master mix on the same PCR plate. A no-template control in triplicate was always included in parallel.
TABLE 1.
PCR primers and probe used in this study
| Primer | Sequence (5′→3′) | Annealing positiona | Target | Annealing temp (°C) | Amplicon length (bp)b | Reference |
|---|---|---|---|---|---|---|
| 27f | AGA GTT TGA TCM TGG CTC AG | 8–27 | Most bacteria | 54 | 1,535 | 38 |
| 1525r | AAG GAG GTG WTC CAR CC | 1,525–1,542 | ||||
| 340f | TCC TAC GGG AGG CAG CAG T | 340–258 | Most bacteria | 60 | 467 | 76 |
| 806r | GGA CTA CCA GGG TAT CTA ATC CTG TT | 781–806 | ||||
| TaqMan probe | 6-FAM-5′-CGT ATT ACC GCG GCT GCT GGC AC-3′-TAMRA | 515–537 | ||||
| Bac303f | GAA GGT CCC CCA CAT TG | 302–318 | Bacteroides | 56 | 418 | 41 |
| Bac708r | CAA TCG GAG TTC TTC GTG | 708–725 | 77 | |||
| Bif164f | GGG TGG TAA TGC CGG ATG | 164–181 | Bifidobacterium | 60 | 530 | 78 |
| Bif662rc | CCA CCG TTA CAC CGG GAA | 676–693 | ||||
| ECA75f | GGA AGA AGC TTG CTT CTT TGCT GAC | 75–99 | E. coli | 56 | 545 | 44 |
| ECR619r | AGC CCG GGG ATT TCA CAT CTG ACT TA | 594–619 | ||||
| Cdif-706f | ATT AGG AGG AAC ACC AGT TG | 164–181 | C. difficile | 54 | 307 | 43 |
| Cdif-994r | AGG AGA TGT CAT TGG GAT GT | 994–1,012 | ||||
| BiLON-1 | TTC CAG TTG ATC GCA TGG TC | 186–207 | B. longum | 63 | 831 | 42 |
| BiLON-2 | GGG AAG CCG TAT CTC TAC GA | 1,009–1,028 | ||||
| ERIC 1 | ATG TAA GCT CCT GGG GAT TCA C | Variable | Bacteria | 52 | Variable | 37 |
| ERIC 2 | AAG TAA GTG ACT GGG GTG AGC G |
Based on numbering of the rrs gene of Bacteroides.
Calculated based on the rrs gene of Bacteroides.
When used in PCR-DGGE, a 40-bp GC clamp (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG G-3′) was attached to the primer at the 5′ end.
Profiling of Bifidobacterium by PCR-DGGE.
The population of Bifidobacterium was profiled using genus-specific PCR-denaturing gradient gel electrophoresis (PCR-DGGE) as described previously (40). Distinct DGGE bands were excised from the DGGE gels and the DNA reamplified using the PCR primers used in the DGGE but without the GC clamp (48). Successful reamplification was confirmed as a single band upon agarose gel (1.5%) electrophoresis. The PCR products were purified using QIAquick PCR purification kits (Qiagen) and sequenced as mentioned above. After manual confirmation of base calling and chimera checking, the sequences were compared with the GenBank sequences using BLASTn to identify the most similar database sequences.
Analysis of volatile fatty acids.
All the culture samples were subjected to analysis for volatile fatty acids using gas chromatography as described by Knol et al. (35). Lactic acid, which is not a VFA, was not analyzed.
Statistical analysis.
To compare the prebiotic effects among the treatments (GOS, IN, GOS-IN, OF-IN, and control), the PROC MIXED procedure of SAS (SAS Institute, Inc., Cary, NC) was used to fit a randomized complete-block analysis of variance (ANOVA) to the data obtained at 12 or 24 h for each measurement. The model contained a fixed effect for the treatment and a random effect for the infant. For those measurements with a statistically significant effect of the prebiotic treatments, the least-squares means (LSM) were compared pairwise with a Tukey-Kramer adjustment made to the P values. Only the measurements for which most of the infant stool samples (at least 5 of the 6 samples for each treatment) resulted in complete data (values at 12 or 24 h for all 5 treatments) were analyzed. The bacterial plate counts and real-time PCR data were first log10 transformed to improve normality before analysis. Statistical analysis was not performed on the abundance of C. difficile because many of the samples had no detectable C. difficile organisms. The time-zero data (calculated from the data of the inocula) were obtained from the cultures prior to incubation. Thus, the five treatments had the same beginning values for a particular measurement. Statistical significance was declared at a P value of ≤0.05. All values are presented as means (± standard errors of the means [SEM]).
RESULTS
Demographics of the enrolled infant subjects.
Three infant subjects from each feeding group met the requirements of the study protocol. The subjects enrolled in the two feeding groups were similar in age, gender, birth weight, and birth method (Table 2). No serious adverse events were reported for any of the infants during the study. Fresh stool samples from each of these infants were used as the inocula to evaluate all the test prebiotic products.
TABLE 2.
Demographic information for the enrolled infants
| Variable | Feeding groupa |
|
|---|---|---|
| FF | HM | |
| No. of subjects | 3 | 3 |
| Gender | 2 males, 1 female | 2 males, 1 female |
| Ethnicity | All non-Hispanic | All non-Hispanic |
| Race | All white | All white |
| Method of delivery | All vaginal | All vaginal |
| Birth wt, g (mean ± SEM) | 3,534.2 ± 223.4 | 3,411.4 ± 125.0 |
| Age at study enrollment, days (mean ± SEM) | 1.0 ± 0.0 | 1.3 ± 0.3 |
FF, formula fed; HM, human milk fed.
Abundance of total bacteria and Bacteroides.
In an attempt to encompass the gut microbiome supported by both feeding types, the abundance of total bacteria, Bacteroides, total bifidobacteria, B. longum, and E. coli was determined for each inoculum collected from both the HM and the FF feeding groups, with all six fresh stool samples (three replicates per sample) used to determine the effects of the different prebiotics. In all the cultures, including the control, the average abundance of total bacteria increased by about 3 log rrs copies/ml culture over the first 12 h of incubation (data not shown). However, no further growth of total bacteria was observed thereafter. The abundances of total bacteria did not differ significantly among the treatments at either 12 or 24 h. The abundance of Bacteroides increased from approximately 4 to 7 log rrs copies/ml by 12 h in all cultures, but the cultures that received the GOS product (Vivinal GOS 15) increased the least, to 6.9 (±0.87) log rrs copies/ml. From 12 to 24 h, Bacteroides continued to increase in abundance by another log in the control, IN, and OF-IN cultures but decreased slightly in the GOS and the GOS-IN cultures (data not shown).
Abundances of total bifidobacteria and B. longum.
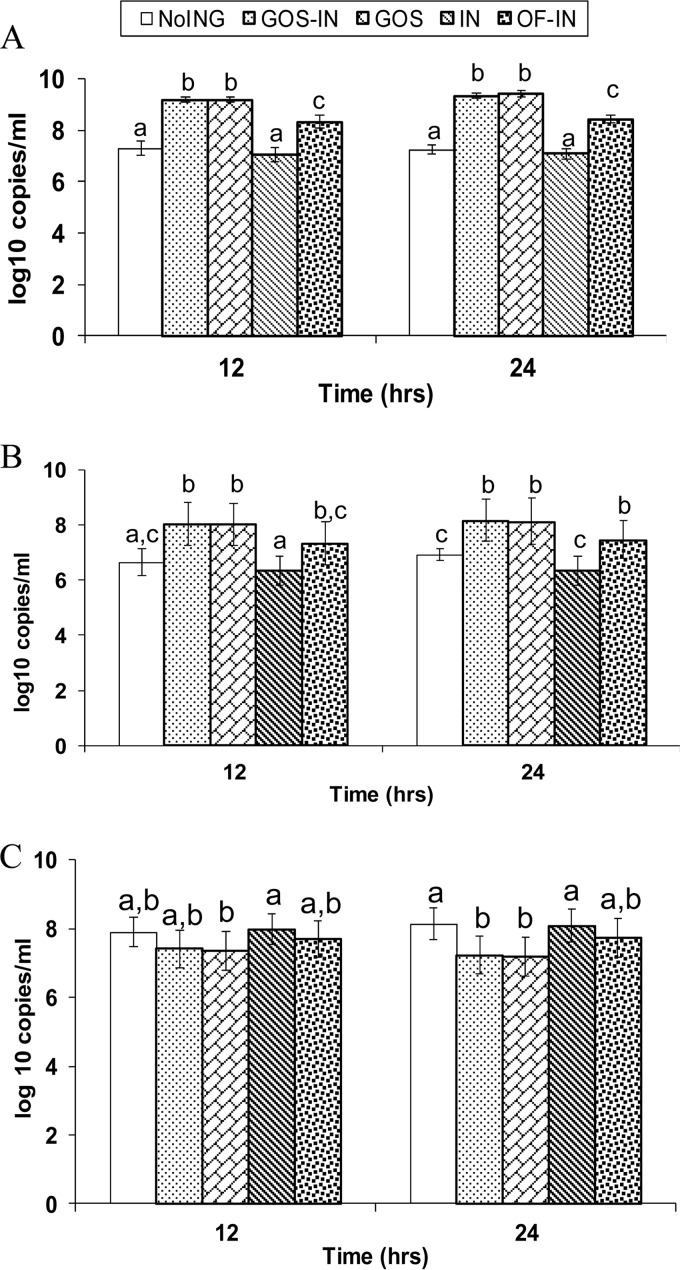
The in vitro cultures had a bifidobacterial abundance of 5.12 (±0.17) log rrs copies/ml prior to incubation. At 12 h the bifidobacterial population increased by about 4 log rrs copies/ml in both the GOS and the GOS-IN cultures and about 3 log rrs copies/ml in the OF-IN cultures (Fig. 1A). Over the same period, bifidobacteria in the control and the IN cultures increased only by approximately 1.5 logs. From 12 to 24 h, the bifidobacterial population did not increase further, irrespective of the prebiotics added.
FIG 1.
Abundances of bifidobacteria (A), B. longum (B), and E. coli (C) determined in in vitro cultures. The time-zero values were 5.12 (±0.17) log 16S rRNA gene copies/ml for total bifidobacteria, 3.87 (±0.67) for B. longum, and 3.23 (±0.51) for E. coli. Different letters indicate significant differences at P values of <0.01 (A) and <0.05 (B and C). NoING, the control containing no ingredient; GOS-IN, GOS and inulin; IN, inulin; OF-IN, oligofructose and inulin.
The abundance of B. longum approximated 3.87 (±0.67) log rrs copies/ml of culture prior to the incubation, which increased by various magnitudes in all the cultures, including the control (Fig. 1B). At both 12 and 24 h, the B. longum population in the GOS and the GOS-IN cultures was greater (P ≤ 0.001) than that in the IN and the control cultures. The OF-IN cultures had a significantly greater abundance of B. longum than the IN cultures at both 12 and 24 h (P ≤ 0.02) and the control cultures at 24 h (P = 0.001), while the IN and the control cultures had similar B. longum population sizes.
The relative abundance of B. longum was calculated by dividing the abundance of B. longum by that of total bifidobacteria. The relative abundance of B. longum varied among the cultures at time zero and accounted for about 81% of total bifidobacteria. During the in vitro incubation, the relative abundance of B. longum increased numerically in all the cultures. Overall, B. longum accounted for 87% to 91% of total bifidobacteria at 12 h and from 86% to 95% at 24 h. The predominance of B. longum numerically increased to a lesser extent in the GOS and the GOS-IN cultures than in other cultures. The relative abundance was greatest in the control cultures at 24 h, reaching 95%.
Abundances of Escherichia coli and C. difficile.
The abundance of E. coli in the cultures prior to incubation was 3.23 (± 0.51) log rrs copies/ml and increased approximately by 4 logs during the first 12 h of incubation (Fig. 1C). The E. coli population size stabilized from 12 to 24 h. The addition of the prebiotics affected the growth of E. coli differently. Pairwise comparisons showed that the GOS cultures had a significantly lower abundance of E. coli organisms than the IN cultures at 12 h. The GOS and the GOS-IN cultures also had significantly fewer (P < 0.05) E. coli organisms than the control and the IN cultures at 24 h. The E. coli population in the OF-IN cultures was numerically smaller only than that in the control cultures (P = 0.835 and 0.249 at 12 and 24 h, respectively) and the IN cultures (P = 0.581 and 0.355 at 12 and 24 h, respectively) but numerically greater than that in the GOS cultures (P = 0.438 and 0.058 at 12 and 24 h, respectively) and the GOS-IN cultures (P = 0.616 and 0.098 at 12 and 24 h, respectively).
The abundance of C. difficile organisms averaged about 2.70 log rrs copies/ml prior to the in vitro incubation, increased to 5.35 to 5.38 log rrs copies/ml by 12 h among the treatments, and thereafter increased or decreased slightly by 24 h. Because two or three of the six inocula did not result in detectable C. difficile growth in the in vitro cultures, no statistical analysis was done on the qPCR data for C. difficile. However, there were numerical differences in the abundance of C. difficile among the treatments (data not shown).
Cultured bifidobacteria.
Beerens agar plates were used to enumerate and recover bifidobacteria from each of the cultures (for ease of reference, referred to as “cultured bifidobacteria,” though Beerens plates permit growth of some bacteria other than bifidobacteria). Based on the selective plating, the time-zero cultures had about 6 log CFU/ml of culture on average. By 12 h, the control and the IN cultures had approximately 7.5 log CFU/ml, while the other 3 cultures had about 8 log CFU/ml. By 24 h all the cultures had slightly decreased CFU, but not the OF-IN cultures.
Some differences in species distribution were seen between the two feeding groups, and thus the data for cultured bifidobacteria were examined separately for the two feeding groups. In the initial time-zero samples, most of the isolates (81.0%) from the HM infants were identified, through sequencing of 16S rRNA genes, as Bifidobacterium longum subsp. infantis, with the remaining isolates as B. breve (Table 3). The initial time-zero samples of the FF group were also dominated by B. longum subsp. infantis (83.7%), but Bifidobacterium pseudocatenulatum, Bifidobacterium breve, and Enterococcus faecalis were also found (Table 4). Over the course of the in vitro incubation, B. longum subsp. infantis remained predominant in all the cultures. The GOS cultures of the HM group and the GOS and GOS-IN cultures of the FF group had a numerically greater prevalence of B. breve than the other cultures. The GOS cultures of the HM group also had fewer Enterococcus faecalis organisms. No such trend was observed in the GOS cultures within the FF group.
TABLE 3.
Abundance of total cultured bacteria and prevalence of bacterial species recovered from the Beerens plates inoculated from the samples of the HM group
| Culture | Total bacteria (log10 CFU/ml [mean ± SEM]) | % prevalence (mean ± SEM) |
||||||
|---|---|---|---|---|---|---|---|---|
| B. longum subsp. infantis | B. breve | B. bifiduma | E. faecalis | B. licheniformisa | Clostridium sp.a | Collinsella aerofaciensa | ||
| Time zero (0 h) | 6.07 (0.82) | 81.0 (33.0) | 19.0 (33.0) | |||||
| Control | ||||||||
| 12 h | 7.56 (0.51) | 75.0 (43.3) | 12.5 (21.6) | 12.5 (21.6) | ||||
| 24 h | 7.14 (0.41) | 40.3 (32.3) | 7.2 (9.1) | 28.5 (26.4) | 15.3 (26.5) | 8.7 (15.1) | ||
| IN | ||||||||
| 12 h | 7.34 (0.36) | 73.8 (19.0) | 4.3 (7.5) | 19.0 (28.8) | 2.9 (5.0) | |||
| 24 h | 7.17 (0.62) | 64.0 (40.8) | 10.1 (17.6) | 2.4 (4.1) | 23.4 (26.5) | |||
| OF-IN | ||||||||
| 12 h | 8.09 (0.29) | 88.8 (15.7) | 2.8 (4.8) | 6.9 (12.0) | 1.4 (2.5) | |||
| 24 h | 7.81 (0.41) | 90.9 (15.7) | 1.5 (2.6) | 6.1 (10.5) | 1.5 (2.6) | |||
| GOS | ||||||||
| 12 h | 8.15 (0.59) | 71.8 (44.9) | 22.2 (38.5) | 4.4 (7.7) | 1.5 (2.6) | |||
| 24 h | 7.1 (0.38) | 77.1 (39.7) | 20.8 (36.1) | 2.1 (3.6) | ||||
| GOS-IN | ||||||||
| 12 h | 8.27 (0.65) | 87.7 (14.2) | 3.7 (6.4) | 5.6 (9.6) | 3.0 (5.2) | |||
| 24 h | 7.16 (0.46) | 74.2 (44.6) | 6.1 (10.5) | 18.2 (31.5) | 1.5 (2.6) | |||
Found in the cultures of only one infant.
TABLE 4.
Abundance of total cultured bacteria and prevalence of bacterial species recovered from the Beerens plates inoculated from the samples of the FF group
| Culture | Total bacteria (log10 CFU/ml [mean ± SEM]) | % prevalence (mean ± SEM) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| B. longum subsp. infantis | B. brevea | B. bifidum | E. faecalis | B. licheniformis | Clostridium sp. | Collinsella aerofaciensa | B. pseudocatenulatuma | ||
| Time zero (0 h) | 6.7 (0.23) | 83.7 (24.5) | 1.4 (2.5) | 9.3 (16.0) | 5.6 (9.6) | ||||
| Control | |||||||||
| 12 h | 7.5 (0.14) | 79.1 (18.1) | 4.5 (7.9) | 5.1 (8.9) | 8.6 (9.1) | 2.6 (4.4) | |||
| 24 h | 7.7 (0.36) | 81.1 (12.4) | 3.0 (5.2) | 2.4 (4.1) | 6.1 (10.5) | 1.5 (2.6) | 5.9 (10.2) | ||
| IN | |||||||||
| 12 h | 7.6 (0.29) | 79.3 (32.2) | 1.4 (2.5) | 19.3 (33.4) | |||||
| 24 h | 7.4 (0.42) | 66.5 (30.1) | 2.0 (3.4) | 17.8 (13.3) | 11.8 (20.4) | 2.0 (3.4) | |||
| OF-IN | |||||||||
| 12 h | 8.0 (1.0) | 74.4 (35.9) | 3.3 (5.8) | 22.2 (38.5) | |||||
| 24 h | 8.1 (0.30) | 75.0 (43.3) | 8.3 (14.4) | 8.3 (14.4) | 8.3 (14.4) | ||||
| GOS | |||||||||
| 12 h | 7.9 (0.57) | 52.1 (21.9) | 12.5 (21.6) | 5.6 (9.6) | 13.2 (17.7) | 8.3 (14.4) | 8.3 (14.4) | ||
| 24 h | 7.7 (0.09) | 77.1 (39.7) | 6.25 (10.8) | 2.1 (3.6) | 10.4 (18.0) | 4.2 (7.2) | |||
| GOS-IN | |||||||||
| 12 h | 7.7 (0.63) | 71.0 (27.4) | 11.8 (20.4) | 7.8 (13.6) | 1.6 (2.7) | 7.8 (13.6) | |||
| 24 h | 7.6 (0.14) | 59.8 (42.7) | 28.3 (49.1) | 2.4 (4.1) | 9.5 (16.5) | ||||
Found in the cultures of only one infant.
Culture pH and VFA production.
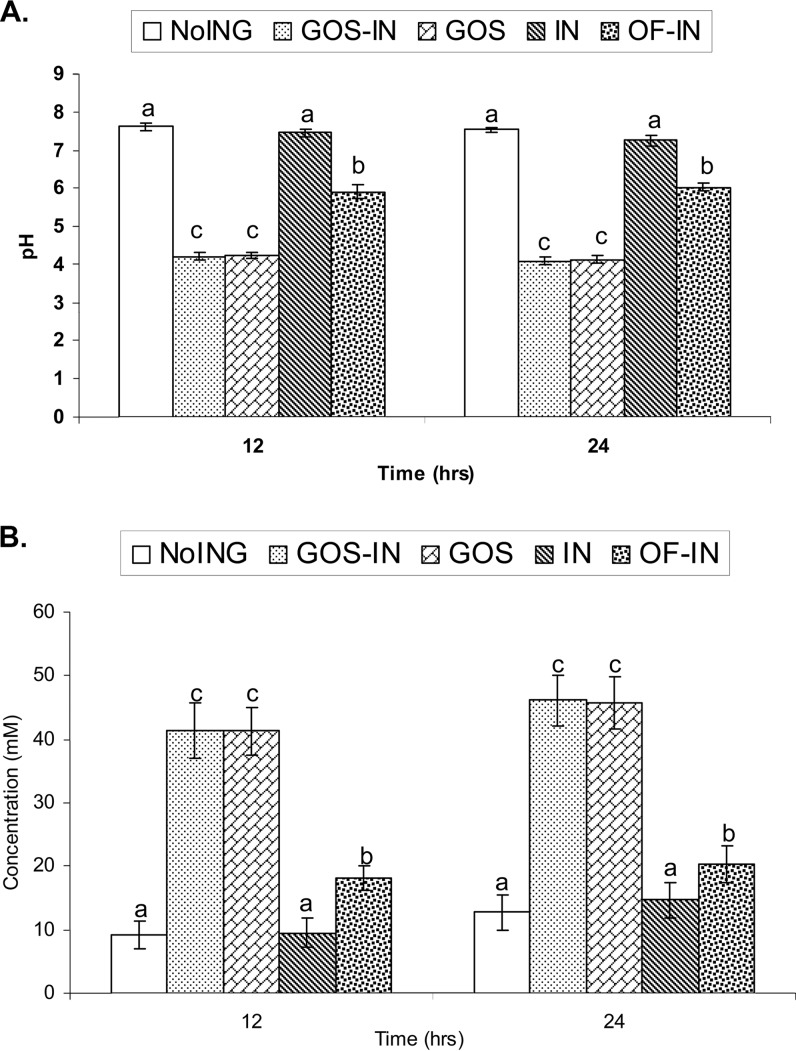
The pH of the initial cultures prior to incubation was approximately 6.4 (Fig. 2A). The addition of the GOS product or both the GOS product and the IN product (Beneo HP) decreased the culture pH by >2 pH units after 12 h of incubation. The inclusion of OF-IN product (Beneo Synergy 1) also decreased the culture pH at 12 h but to a smaller magnitude (approximately 0.5 pH unit). The pH in both the control and the IN cultures increased at 12 h. No appreciable change in pH was observed in any of the cultures after the initial 12 h of incubation.
FIG 2.
pHs (A) and acetic acid concentrations (B) determined in in vitro cultures. The time-zero value for pH was 6.36 ± 0.38 (mean ± SEM), and that for acetic acid concentration was 0.06 ± 0.02 mM (mean ± SEM). Different letters indicate significant differences at P values of <0.01 (A) and <0.05 (B).
The concentrations of acetic, butyric, propionic, isobutyric, valeric, and isovaleric acids were analyzed for all the culture samples. Very little acetic acid was detected in the cultures prior to the incubation, but after 12 h of incubation, more than 40 mM acetic acid was detected in both the GOS and the GOS-IN cultures (Fig. 2B). The OF-IN cultures also had an increased acetic acid concentration relative to the control and the IN cultures. Only small increases in acetic acid production were seen after 12 h. Almost no butyric acid was detected in the cultures prior to the incubation (data not shown). Although the concentrations remained very low in all of the cultures, a treatment effect was observed (not statistically analyzed) on butyric acid concentrations at both 12 and 24 h (data not shown). The addition of the GOS product, either alone or in combination with the IN product, resulted in the lowest concentration of butyric acid. Little propionic acid was detected in the cultures prior to incubation, but the different prebiotic products resulted in different increases in propionic acid concentration (data not shown) at 12 and 24 h. The largest increase was seen for the OF-IN cultures, followed by the IN and the control cultures. The addition of the GOS product or the GOS product and the IN product together had very little effect on propionic acid production at both 12 and 24 h. The concentrations of isobutyric, valeric, and isovaleric acids averaged below 1 mM in all the cultures, except for isovaleric acid in the control (1.5 mM) and the IN cultures (1.7 mM) of one formula-fed infant.
DGGE profiling and identification of bifidobacteria from the excised DGGE bands.
There were differences in intensities of some DGGE bands but no visual differences in the presence and absence of DGGE bands between the two feeding groups or among the cultures supplemented with the different prebiotics (data not shown). The two most common species recovered from the DGGE bands were B. longum (the short 16S rRNA gene region did not allow for identification to subspecies level) and B. breve. No obvious effect of the added prebiotics was found on the bifidobacterial species identified from the excised DGGE bands (data not shown).
DISCUSSION
This study is among the few studies that have comparatively evaluated the common prebiotics that have been added to infant formula using an in vitro model and fresh infant stool samples from breast-fed and formula-fed infants as inocula. As shown in this study, fresh stool samples are difficult to collect from multiple young infants within a narrow time window. It is also a challenge to maintain anaerobiosis of the samples and viability of the microbes. However, compared to frozen stool samples, fresh stool samples would allow for more applicable conclusions. From a microbiological perspective, it is logical to use the same concentration for all the prebiotic products to compare their prebiotic effect and fermentation profiles. However, because the tested prebiotics resulted in differences in several aspects, such as tolerance and stool consistency (49–52), they were evaluated at the concentrations that can be practically used in infant formula. It should be noted that 7.2 g/liter of the GOS product (Vivinal GOS 15; 28.5% GOS content) corresponded to 2.05 g/liter of pure GOS in the cultures, with the remaining being polysaccharides (3.28 g/liter) and lactose and glucose (about 0.7 g/liter each). The differences observed between the GOS product and the other products (Beneo HP and Beneo Synergy 1) thus might not be solely attributable to GOS itself.
The quantification of the major groups of bacteria that are important to infant health and the analysis for the major fermentation products (i.e., VFA) and pH allow for evaluation of these prebiotic products with respect to their bifidogenic effect and the effect on fermentation. Because it is rather difficult to maintain sample anaerobiosis and viability of the fecal microbes in samples collected from young infants from different geographic regions, a relatively small number of infants were sampled. However, the evaluation of each of the tested prebiotic products using each of the fresh stool samples, collected from both human milk-fed and formula-fed infants, allows for comparison of these prebiotic products using multiple replicates in the laboratory setting. Results of in vitro studies cannot be directly extrapolated to in vivo studies. However, this in vitro study allowed us to evaluate four different prebiotics, and such in vitro studies using fresh fecal samples may be used to test other promising prebiotic ingredients and their combinations simultaneously. The findings of this study may help in designing future clinical trials to further evaluate these prebiotics.
Overall, the prebiotics at the tested levels had various effects on different bacterial groups analyzed and on the in vitro fermentation by infants' fecal microbiomes. The GOS product exhibited the most stimulatory effect on proliferation of total bifidobacteria and B. longum. This observation is consistent with the ability of bifidobacteria, including B. longum, to grow on a variety of oligosaccharides (53, 54) and their preference for GOS (27, 55) even though this product also contains polysaccharides, lactose, and glucose. Even though the combination of the GOS product and the inulin product (0.8 g/liter) promoted bifidobacteria to a level comparable to that of the GOS product, the bifidogenic effect observed almost certainly stemmed from the stimulatory effect of Vivinal GOS 15, because Beneo HP alone at the tested level did not increase the population of bifidobacteria. The Beneo HP concentration used might have been too low to produce a significant bifidogenic effect. In addition, a few studies showed that inulin with long chains was fermented slower and could only be fermented by a slightly lower number of bifidobacteria than short-chain oligofructose (56, 57), and long-chain inulin did not exhibit a bifidogenic effect in humanized rats (58). Thus, the long chain lengths of the inulin tested in the present study might be another reason for the lack of a significant bifidogenic effect.
The oligofructose-enriched inulin (Beneo Synergy 1) increased bifidobacteria to a smaller (P < 0.05) magnitude than the GOS product, which may be largely attributed to the oligofructose present in the mixture. It is interesting that the total population of Bifidobacterium and that of B. longum had a similar trend (Fig. 1A and B), suggesting that B. longum was the primary bifidobacterial species that was stimulated by the added prebiotics in the cultures. This observation is in line with the finding that B. longum was one of the major bifidobacterial species of adults' fecal microbiomes that were stimulated by GOS (59). In addition, the relative abundance of B. longum as quantified by the species-specific qPCR was similar to that of infantis subsp. infantis recovered on Beerens plates, suggesting that the majority of the B. longum organisms in the cultures were probably B. longum subsp. infantis. It should be noted that there was very little increase in the abundance of bifidobacteria after 12 h of incubation. The ceased population growth could be attributed to depletion of the added products or another nutrient(s) or to accumulation of metabolites (including decreased pH due to accumulation of VFA) to inhibitory levels.
Most of the cultured bifidobacteria recovered from the fecal samples of the HM group were B. infantis, while B. pseudocatenulatum and E. faecalis were also recovered from the fecal samples of the FF group. Such differential distributions of bifidobacteria between formula- and human milk-fed infants are consistent with the findings of previous studies (60, 61). Collectively, more species were recovered from the in vitro cultures than from the initial fecal samples, including nonbifidobacterial species. The HM and the FF groups also exhibited differences in the prevalences of the cultured species. As shown recently (62), the prevalence of B. breve increased at the expense of that of B. longum in all the cultures, especially in the GOS culture of the HM group and the GOS and the GOS-IN cultures of the FF group. The dynamic successions of bifidobacterial species in the cultures were also incubation time and prebiotics independent. Different bifidobacterial species have various ability to utilize different prebiotics (63–65), which may explain the observed differences in population shifts during the cultivation. The results of this study and a previous study (66) suggest the occurrence of different “types” (a set of specific bifidobacterial species) of bifidobacteria under different conditions (nutrient, environmental, and microbial). More studies using cocultures or other specific analyses are needed to verify this premise.
The prevalences of the recovered bifidobacterial species differed from some of the earlier studies (67, 68) in which Bifidobacterium adolescentis and Bifidobacterium dentium were also found to be prevalent. Differences in methods used (including media) and host gut microbiomes might be major contributing factors affecting the species captured on agar plates. It should also be noted that a few nonbifidobacterial species were also recovered from the Beerens agar plates, indicating that this type of selective plate is not exclusively specific for bifidobacteria. This has been observed in a previous study, and the use of plate count to enumerate bifidobacteria is hindered by the lack of selectivity of media (69). Caution should be taken when quantitative data for bifidobacteria from cultivation-based and molecular methods are compared.
The Bacteroides population increased in all the cultures within the first 12 h of incubation but increased the least in the GOS and the OF-IN cultures. Additionally, the Bacteroides population continued to increase by another log, irrespective of feeding groups, in the control, the IN, and the OF-IN cultures from 12 to 24 h, while it decreased slightly in the GOS and the GOS-IN cultures during the same period (data not shown). These results suggest possible inhibition of Bacteroides from the increased bifidobacterial population, including the pH decline caused by fermentation of the GOS product by bifidobacteria. The total bacterial populations were similar among all the cultures irrespective of the prebiotics added, suggesting that the prebiotics added to the basal medium was relatively small or can be utilized only by a few selected groups of bacteria present in the initial inocula.
The concentrations of acetic acid appeared to be inversely associated with the pH and the E. coli population in the cultures. The addition of the GOS or the GOS-IN product had the most profound effects on these 3 parameters. Most bifidobacterial species produce both acetic acid and lactic acid at a characteristic 3:2 ratio through the bifid shunt pathway during carbohydrate fermentation (70), and they are also acid tolerant and able to grow well at low pH (71). E. coli is known to be inhibited at low pH (72). It can be concluded that the addition of the GOS product in the GOS and the GOS-IN cultures stimulated the growth of bifidobacteria and subsequent production of acetic acid and lactic acid (not analyzed in this study) by primarily this group of bacteria, resulting in a lowered culture pH (approximately 4.0), which inhibited E. coli in the GOS and the GOS-IN cultures. This premise is consistent with the lower fecal pH and E. coli abundance in breast-fed infants than in formula-fed infants (73, 74). The lower concentration of acetic acid and higher pH and E. coli population in both the control and the IN cultures seem to corroborate the above conclusion and the limited bifidogenic effect of the inulin at the tested level. The addition of the OF-IN product also stimulated acetate production and lowered the culture pH, but not to the magnitudes observed in the GOS or the GOS-IN cultures. This observation is also consistent with the relatively lower (P < 0.05) abundance of bifidobacteria in the OF-IN cultures than in the GOS or the GOS-IN cultures. Although not statistically analyzed, the OF-IN cultures had the highest concentrations of propionic and butyric acids (2.7 and 1.25 mM, respectively, at 24 h), followed by the IN (1.9 and 1.0 mM, respectively) and the control cultures (1.3 and 0.98 mM, respectively). On the other hand, the GOS and the GOS-IN cultures had the lowest concentrations of these 2 acids (approximately 0.56 and 0.09 mM, respectively). These results suggest that inulin, but not GOS, may stimulate the growth of butyrate- or propionate-producing bacteria, which is consistent with the findings of previous studies (57, 75). Nevertheless, relatively low concentrations of acetic acid corresponded with relatively higher concentrations of both butyric and propionic acids in the in vitro cultures. Detailed studies of the bacteria present in such microbiomes will help determine the bacteria that are likely involved in the production of both butyric and propionic acids.
ACKNOWLEDGMENT
This study was partially supported by Abbott Nutrition.
Footnotes
Published ahead of print 19 September 2014
REFERENCES
- 1.Fanaro S, Chierici R, Guenini P, Vigi V. 2003. Intestinal microflora in early infancy: composition and development. Acta Paediatr. 92:S48–S55. 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 2.Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30:61–67. 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 3.Cebra JJ. 1999. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69:1046S–1051S. [DOI] [PubMed] [Google Scholar]
- 4.Chierici R, Fanaro S, Saccomandi D, Vigi V. 2003. Advances in the modulation of the microbial ecology of the gut in early infancy. Acta Paediatr. 92:56–63. 10.1111/j.1651-2227.2003.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 5.Lönnerdal B. 2004. Human milk proteins: key components for the biological activity of human milk. Adv. Exp. Med. Biol. 554:11–25. 10.1007/978-1-4757-4242-8_4. [DOI] [PubMed] [Google Scholar]
- 6.Horwood LJ, Darlow BA, Mogridge N. 2001. Breast milk feeding and cognitive ability at 7–8 years. Arch. Dis. Child. Fetal Neonatal Ed. 84:F23–F27. 10.1136/fn.84.1.F23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savino F, Lupica MM. 2006. Breast milk: biological constituents for health and well-being in infancy. Recenti Prog. Med. 97:519–527 (In Italian.) 10.1701/183.1934. [DOI] [PubMed] [Google Scholar]
- 8.Barile D, Rastall RA. 2013. Human milk and related oligosaccharides as prebiotics. Curr. Opin. Biotechnol. 24:214–219. 10.1016/j.copbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Coppa GV, Zampini L, Galeazzi T, Gabrielli O. 2006. Prebiotics in human milk: a review. Dig. Liver Dis. 38(Suppl 2):S291–S294. 10.1016/S1590-8658(07)60013-9. [DOI] [PubMed] [Google Scholar]
- 10.Boehm G, Stahl B. 2003. Oligosaccharides, p 203–243 In Mattila-Sandholm T. (ed), Functional dairy products. Woodhead Publishing Limited, Cambridge, England. [Google Scholar]
- 11.Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25:37–58. 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 12.Vandenplas Y. 2002. Oligosaccharides in infant formula. Br. J. Nutr. 87:293–296. 10.1079/BJNBJN/2002551. [DOI] [PubMed] [Google Scholar]
- 13.Roberfroid M. 2007. Prebiotics: the concept revisited. J. Nutr. 137:830S–837S. [DOI] [PubMed] [Google Scholar]
- 14.Boehm G, Moro G. 2008. Structural and functional aspects of prebiotics used in infant nutrition. J. Nutr. 138:1818S–1828S. [DOI] [PubMed] [Google Scholar]
- 15.Williams TA. 2009. A DNA-based investigation of intestinal microbiota of infants and the impact of prebiotics and maternal intestinal microbiota. Ph.D. dissertation The Ohio State University, Columbus, OH. [Google Scholar]
- 16.Ben XM, Zhou XY, Zhao WH, Yu WL, Pan W, Zhang WL, Wu SM, Van Beusekom CM, Schaafsma A. 2004. Supplementation of milk formula with galacto-oligosaccharides improves intestinal micro-flora and fermentation in term infants. Chin. Med. J. (Engl.) 117:927–931. [PubMed] [Google Scholar]
- 17.Boehm G, Lidestri M, Casetta P, Jelinek J, Negretti F, Stahl B, Marini A. 2002. Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Arch. Dis. Child. Fetal Neonatal Ed. 86:F178–F181. 10.1136/fn.86.3.F178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, Lee DH, Meyer D. 2007. Supplementation of baby formula with native inulin has a prebiotic effect in formula-fed babies. Asia Pac. J. Clin. Nutr. 16:172–177. [PubMed] [Google Scholar]
- 19.Brunser O, Gotteland M, Cruchet S, Figueroa G, Garrido D, Steenhout P. 2006. Effect of a milk formula with prebiotics on the intestinal microbiota of infants after an antibiotic treatment. Pediatr. Res. 59:451–456. 10.1203/01.pdr.0000198773.40937.61. [DOI] [PubMed] [Google Scholar]
- 20.Euler AR, Mitchell DK, Kline R, Pickering LK. 2005. Prebiotic effect of fructo-oligosaccharide supplemented term infant formula at two concentrations compared with unsupplemented formula and human milk. J. Pediatr. Gastroenterol. Nutr. 40:157–164. 10.1097/00005176-200502000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura N, Gaskins HR, Collier CT, Nava GM, Rai D, Petschow B, Russell WM, Harris C, Mackie RI, Wampler JL, Walker DC. 2009. Molecular ecological analysis of fecal bacterial populations from term infants fed formula supplemented with selected blends of prebiotics. Appl. Environ. Microbiol. 75:1121–1128. 10.1128/AEM.02359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14:169–181. 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magne F, Hachelaf W, Suau A, Boudraa G, Mangin I, Touhami M, Bouziane-Nedjadi K, Pochart P. 2006. A longitudinal study of infant faecal microbiota during weaning. FEMS Microbiol. Ecol. 58:563–571. 10.1111/j.1574-6941.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- 24.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaretti A, Bernardi T, Tamburini E, Zanoni S, Lomma M, Matteuzzi D, Rossi M. 2007. Kinetics and metabolism of Bifidobacterium adolescentis MB 239 growing on glucose, galactose, lactose, and galactooligosaccharides. Appl. Environ. Microbiol. 73:3637–3644. 10.1128/AEM.02914-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada V, Nevoral J, Trojanová I, Tománková E, Smehilová M, Killer J. 2008. Growth of infant faecal bifidobacteria and clostridia on prebiotic oligosaccharides in in vitro conditions. Anaerobe 14:205–208. 10.1016/j.anaerobe.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Vernazza CL, Gibson GR, Rastall RA. 2006. Carbohydrate preference, acid tolerance and bile tolerance in five strains of Bifidobacterium. J. Appl. Microbiol. 100:846–853. 10.1111/j.1365-2672.2006.02832.x. [DOI] [PubMed] [Google Scholar]
- 28.Olano-Martin E, Gibson GR, Rastell RA. 2002. Comparison of the in vitro bifidogenic properties of pectins and pectic-oligosaccharides. J. Appl. Microbiol. 93:505–511. 10.1046/j.1365-2672.2002.01719.x. [DOI] [PubMed] [Google Scholar]
- 29.Olano-Martin E, Mountzouris KC, Gibson GR, Rastall RA. 2000. In vitro fermentability of dextran, oligodextran and maltodextrin by human gut bacteria. Br. J. Nutr. 83:247–255. [DOI] [PubMed] [Google Scholar]
- 30.Rycroft CE, Jones MR, Gibson GR, Rastall RA. 2001. A comparative in vitro evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91:878–887. 10.1046/j.1365-2672.2001.01446.x. [DOI] [PubMed] [Google Scholar]
- 31.Yu Z, Morrison M. 2009. The gut microbiome: current understanding and future perspective, p 19–40 In Jaykus L-A, Wang H, Schlesinger LS. (ed), Food-borne microbes: shaping the host ecosystem. ASM Press, Washington, DC. [Google Scholar]
- 32.Veereman-Wauters G, Staelens S, Van de Broek H, Plaskie K, Wesling F, Roger L, McCartney A, Assam P. 2011. Physiological and bifidogenic effects of prebiotic supplements in infant formulae. J. Pediatr. Gastroenterol. Nutr. 52:763–771. 10.1097/MPG.0b013e3182139f39. [DOI] [PubMed] [Google Scholar]
- 33.Moro G, Minoli I, Mosca M, Fanaro S, Jelinek J, Stahl B, Boehm G. 2002. Dosage-related bifidogenic effects of galacto- and fructo-oligosaccharides in formula-fed term infants. J. Pediatr. Gastroenterol. Nutr. 34:291–295. 10.1097/00005176-200203000-00014. [DOI] [PubMed] [Google Scholar]
- 34.Sanz ML, Gibson GR, Rastall RA. 2005. Influence of disaccharide structure on prebiotic selectivity in vitro. J. Agric. Food Chem. 53:5192–5199. 10.1021/jf050276w. [DOI] [PubMed] [Google Scholar]
- 35.Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, Klarczyk M, Schöpfer H, Böckler H-M, Wells J. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 40:36–42. 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Shuhaimi M, Ali AM, Saleh NM, Yazid AM. 2001. Utilisation of enterobacterial repetitive intergenic consensus (ERIC) sequence-based PCR to fingerprint the genomes of Bifidobacterium isolates and other probiotic bacteria. Biotechnol. Lett. 23:731–736. 10.1023/A:1010355003674. [DOI] [Google Scholar]
- 37.Ventura M, Meylan V, Zink R. 2003. Identification and tracing of Bifidobacterium species by use of enterobacterial repetitive intergenic consensus sequences. Appl. Environ. Microbiol. 69:4296–4301. 10.1128/AEM.69.7.4296-4301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow MD. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY. [Google Scholar]
- 39.Yu Z, Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812. [DOI] [PubMed] [Google Scholar]
- 40.Anderson K, Yu Z, Chen J, Jenkins J, Courtney P, Morrison M. 2008. Analyses of Bifidobacterium, Lactobacillus, and total bacterial populations in healthy volunteers consuming calcium gluconate by denaturing gradient gel electrophoresis and real-time PCR. Int. J. Probiotics Prebiotics 3:31–36. [Google Scholar]
- 41.Bartosch S, Fite A, Macfarlane GT, McMurdo MET. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70:3575–3581. 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuki T, Watanabe K, Fujimoto J, Kado Y, Takada T, Matsumoto K, Tanaka R. 2004. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 70:167–173. 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia Q, Williams T, Hustead D, Price P, Morrison M, Yu Z. 2012. Quantitative analysis of intestinal bacterial populations from term infants fed formula supplemented with fructo-oligosaccharides. J. Pediatr. Gastroenterol. Nutr. 55(3):314–320. 10.1097/MPG.0b013e3182523254. [DOI] [PubMed] [Google Scholar]
- 44.Sabat G, Rose P, Hickey WJ, Harkin JM. 2000. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 66:844–849. 10.1128/AEM.66.2.844-849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Yu Z, Michel FC, Jr, Wittum T, Morrison M. 2007. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73:4407–4416. 10.1128/AEM.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osborn AM, Moore ER, Timmis KN. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39–50. 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 48.Yu Z, Garcia-Gonzalez R, Schanbacher FL, Morrison M. 2008. Evaluations of different hypervariable regions of archaeal 16S rRNA genes in profiling of methanogens by archaea-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 74:889–893. 10.1128/AEM.00684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marteau P, Seksik P. 2004. Tolerance of probiotics and prebiotics. J. Clin. Gastroenterol. 38:S67–S69. 10.1097/01.mcg.0000128929.37156.a7. [DOI] [PubMed] [Google Scholar]
- 50.Mugambi MN, Musekiwa A, Lombard M, Young T, Blaauw R. 2012. Synbiotics, probiotics or prebiotics in infant formula for full term infants: a systematic review. Nutr. J. 11:81. 10.1186/1475-2891-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao S, Srinivasjois R, Patole S. 2009. Prebiotic supplementation in full-term neonates: a systematic review of randomized controlled trials. Arch. Pediatr. Adolesc. Med. 163:755–764. 10.1001/archpediatrics.2009.94. [DOI] [PubMed] [Google Scholar]
- 52.Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, Wolvers D, Watzl B, Szajewska H, Stahl B, Guarner F, Respondek F, Whelan K, Coxam V, Davicco MJ, Leotoing L, Wittrant Y, Delzenne NM, Cani PD, Neyrinck AM, Meheust A. 2010. Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 104(Suppl 2):S1–S63. 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 53.Macfarlane GT, Steed H, Macfarlane S. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305–344. 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- 54.Zivkovic AM, German JB, Lebrilla CB, Mills DA. 2 August 2010. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garrido D, Ruiz-Moyano S, Jimenez-Espinoza R, Eom HJ, Block DE, Mills DA. 2013. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol. 33:262–270. 10.1016/j.fm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pompei A, Cordisco L, Raimondi S, Amaretti A, Pagnoni UM, Matteuzzi D, Rossi M. 2008. In vitro comparison of the prebiotic effects of two inulin-type fructans. Anaerobe 14:280–286. 10.1016/j.anaerobe.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 57.Rossi M, Corradini C, Amaretti A, Nicolini M, Pompei A, Zanoni S, Matteuzzi D. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 71:6150–6158. 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kleessen B, Hartmann L, Blaut M. 2001. Oligofructose and long-chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291–300. 10.1079/BJN2001403. [DOI] [PubMed] [Google Scholar]
- 59.Maathuis AJ, van den Heuvel EG, Schoterman MH, Venema K. 2012. Galacto-oligosaccharides have prebiotic activity in a dynamic in vitro colon model using a 13C-labeling technique. J. Nutr. 142:1205–1212. 10.3945/jn.111.157420. [DOI] [PubMed] [Google Scholar]
- 60.Kleessen B, Bunke H, Tovar K, Noack J, Sawatzki G. 1995. Influence of two infant formulas and human milk on the development of faecal flora in newborn infants. Acta Paediatr. 84:1347–1356. 10.1111/j.1651-2227.1995.tb13567.x. [DOI] [PubMed] [Google Scholar]
- 61.Sakata S, Tonooka T, Ishizeki S, Takada M, Sakamoto M, Fukuyama M, Benno Y. 2005. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 243:417–423. 10.1016/j.femsle.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Avershina E, Storro O, Oien T, Johnsen R, Wilson R, Egeland T, Rudi K. 2013. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl. Environ. Microbiol. 79:497–507. 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 286:34583–34592. 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garrido D, Dallas DC, Mills DA. 2013. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology 159:649–664. 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruiz-Moyano S, Totten SM, Garrido D, Smilowitz JT, German JB, Lebrilla CB, Mills DA. 2013. Variation in consumption of human milk oligosaccharides by infant-gut associated strains of Bifidobacterium breve. Appl. Environ. Microbiol. 79:6040–6049. 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 67.Rinne MM, Gueimonde M, Kalliomäki M, Hoppu U, Salminen SJ, Isolauri E. 2005. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol. Med. Microbiol. 43:59–65. 10.1016/j.femsim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 68.Vlkova E, Rada V, Bujnakova D, Kmet V. 2004. Enumeration, isolation, and identification of bifidobacteria from infant feces. Folia Microbiol. (Praha) 49:209–212. 10.1007/BF02931404. [DOI] [PubMed] [Google Scholar]
- 69.Harmsen HJ, Gibson GR, Elfferich P, Raangs GC, Wildeboer-Veloo AC, Argaiz A, Roberfroid MB, Welling GW. 2000. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125–129. 10.1111/j.1574-6968.2000.tb08945.x. [DOI] [PubMed] [Google Scholar]
- 70.Fushinobu S. 2010. Unique sugar metabolic pathways of bifidobacteria. Biosci. Biotechnol. Biochem. 74:2374–2384. 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- 71.Gomes AMP, Malcata FX. 1999. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci. Technol. 10:139–157. 10.1016/S0924-2244(99)00033-3. [DOI] [Google Scholar]
- 72.Wilson M. 2005. The gastrointestinal tract and its indigenous microbiota, p 251–317 In Wilson M. (ed), Microbial inhabitants of humans. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 73.Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernandez L. 1992. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J. Pediatr. Gastroenterol. Nutr. 15:248–252. 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 74.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. 2005. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 243:141–147. 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 75.Scott KP, Martin JC, Duncan SH, Flint HJ. 28 August 2013. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 76.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257–266. [DOI] [PubMed] [Google Scholar]
- 77.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574. 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satokari RM, Vaughan EE, Akkermans ADL, Saarela M, de Vos WM. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504–513. 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]