Abstract
Gut microbiota has been recognized as an important environmental factor in health, as well as in metabolic and immunological diseases, in which perturbation of the host gut microbiota is often observed in the diseased state. However, little is known on the role of gut microbiota in systemic lupus erythematosus. We investigated the effects of host genetics, sex, age, and dietary intervention on the gut microbiome in a murine lupus model. In young, female lupus-prone mice resembling women at childbearing age, a population with the highest risk for lupus, we found marked depletion of lactobacilli, and increases in Lachnospiraceae and overall diversity compared to age-matched healthy controls. The predicted metagenomic profile in lupus-prone mice showed a significant enrichment of bacterial motility- and sporulation-related pathways. Retinoic acid as a dietary intervention restored lactobacilli that were downregulated in lupus-prone mice, and this correlated with improved symptoms. The predicted metagenomes also showed that retinoic acid reversed many lupus-associated changes in microbial functions that deviated from the control. In addition, gut microbiota of lupus-prone mice were different between sexes, and an overrepresentation of Lachnospiraceae in females was associated with an earlier onset of and/or more severe lupus symptoms. Clostridiaceae and Lachnospiraceae, both harboring butyrate-producing genera, were more abundant in the gut of lupus-prone mice at specific time points during lupus progression. Together, our results demonstrate the dynamics of gut microbiota in murine lupus and provide evidence to suggest the use of probiotic lactobacilli and retinoic acid as dietary supplements to relieve inflammatory flares in lupus patients.
INTRODUCTION
The mammalian gut harbors trillions of microorganisms known as the microbiota (1). Increasing evidence in recent years suggests that the host microbiota and immune system interact to maintain tissue homeostasis in healthy individuals (2–6). The importance of microbiota for the host is highlighted by altered immune responses in the absence of commensal bacteria, where both systemic and gut-specific lymphoid tissues are generally smaller and less developed (7). Higher susceptibility to infectious pathogens (8) and, in some cases, attenuated symptoms in autoimmune disorders (2), have been observed in mice raised under germfree conditions. Indeed, perturbation of the host microbiota, especially that in the gut, is shown to be associated with many diseases. These include inflammatory bowel disease (9, 10), metabolic syndrome (e.g., obesity [11–15] and type 2 diabetes [16], and autoimmune diseases, e.g., type 1 diabetes [17–20], rheumatoid arthritis [5, 21–23], and multiple sclerosis [24, 25]).
However, whether gut microbiota differ in systemic lupus erythematosus (lupus) versus healthy individuals has not been reported. Lupus is an autoimmune disorder characterized by the generation of autoantibodies, recruitment of autoreactive or inflammatory T cells, and abnormal production of proinflammatory cytokines (26, 27). The hallmark of the disease is severe and persistent inflammation that leads to tissue damage in multiple organs, including kidneys, lungs, joints, heart, and brain (26). According to the Lupus Foundation of America, about 2 million Americans currently live with the disease. The prevalence ranges from 20 to 200 cases per 100,000 persons, with higher prevalence for people of African, Hispanic, or Asian ancestry. Although the disease affects both males and females, women of childbearing age are diagnosed nine times more often than men. African-American women suffer from more severe symptoms and a higher mortality rate. The cause of lupus is unclear and there is currently no cure. To date, treatment and prevention of disease flares have relied on long-term use of immunosuppressants where side effects, such as susceptibility to infections, are of particular concern (28). There is a need for better understanding of the disease and for better approaches in lupus treatment and management.
Here, we report the dynamics of gut microbiota in a murine lupus model. Disease-, sex-, and time-dependent differences were identified. Using vitamin A as a potential intervention to reduce lupus nephritis (29–32), we also showed that specific changes of the gut microbiota strongly correlated with disease attenuation. Predicted metagenomes associated with murine lupus and vitamin A treatments were also described.
MATERIALS AND METHODS
Mice.
MRL/Mp (MRL), MRL/Mp-Faslpr (MRL/lpr, stock number 000485), C56BL/6 (B6), and B6.MRL-Faslpr (B6/lpr) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and bred and maintained in a specific-pathogen-free facility according to the requirements of Institutional Animal Care and Use Committee at Virginia Polytechnic Institute and State University. All four strains had been housed in the same room under the same condition for at least 12 months prior to the initiation of the study. The breeding rotations were 3 months for MRL and MRL/lpr due to the short life span of MRL/lpr and 6 months for mice of B6 background. All-trans-retinoic acid (RA) and all-trans-retinyl palmitate (RP) were purchased from Sigma (St. Louis, MO) and prepared and used in the dark to avoid exposure to light. Both retinoids were dissolved in canola oil (vehicle) and administered orally, directly into the mouth, to mice from 6 to 14 weeks of age. For RA treatment, 6 mg RA/kg of body weight was given per day. For the daily VARA (vitamin A-retinoic acid) treatment, 11.2 mg RP/kg of body weight (equivalent to 6 mg retinol/kg) was mixed with 0.6 mg RA/kg of body weight (10% of the amount of retinol) before being given to the mice. Mice were weighed twice weekly, and the retinoid doses were adjusted accordingly.
Microbiota sampling, DNA extraction, and PCR.
Fecal microbiota samples were obtained by taking individual mice out of their cage and collecting a fecal pellet. Colonic microbiota samples were collected within 20 min after euthanasia. To avoid cross-contamination, each microbiota sample was collected by using a new pair of sterile tweezers. All samples were stored at −80°C till being processed at the same time. Sample homogenization and cell lysis was aided by mixing 0.1-g portions of samples with 0.1-mm sterile zirconia beads using a bead-beater (BioSpec Mini-Beadbeater-16) for 45 s. DNA was extracted by a phenol-chloroform method (14). The V4 region (ca. 252 bp) of 16S rRNA gene was PCR amplified with 515F and 12-base GoLay barcoded 806R primers (33). The purified amplicons were sequenced bidirectionally (paired-end 150 bp) on an Illumina MiSeq at Argonne National Laboratory.
Taxonomy assignments and diversity analyses.
Contigs were made from the paired-end reads by using the fastq-join program with a minimum overlap of 10 bp and minimum percent identity of 95% (34). High-quality reads with Phred score of ≥20 were obtained by using QIIME (Quantitative Insights Into Microbial Ecology) (35). Chimeric sequences were identified with usearch (36) and removed from further analysis. A total of 2,021,399 nonchimeric sequences were mapped into 1,005 operational taxonomic units (OTU) in the Greengene reference database (version 13.5, gg13.5) with the program uclust (36). Sequences without a match to gg13.5 were clustered into new OTU at 97% similarity. Taxonomy was assigned by using a naive Bayes classifier (37) trained with the Greengenes taxonomy with more than 200,000 reference sequences (38). A phylogenetic tree was constructed from aligned sequences representing each OTU. For beta diversity calculations, microbiota used for comparison were randomly sampled to the same depth coverage. Principal coordinates were calculated from unweighted UniFrac metrics (39). Alpha diversity metrics, including phylogenetic diversity (40) and the Shannon index, were calculated by using QIIME.
Renal function.
Proteinuria was measured weekly with Chemstrip 2GP from 12 to 14 weeks of age (Roche, Indianapolis, IN). A scale of 0 to 4 was used for proteinuria, corresponding to no protein, trace amounts of protein (5 to 20 mg/dl), and 30, 100, and ≥500 mg/dl total protein, respectively. When mice were euthanized at 14 weeks of age, kidneys were fixed in formalin for 24 h, paraffin embedded, sectioned, and stained with hematoxylin and eosin at the Histopathology Laboratory at Virginia-Maryland Regional College of Veterinary Medicine. Slides were read with an Olympus BX43 microscope. All slides were scored in a blinded fashion by a certified veterinary pathologist. Glomerular lesions were graded on a scale of 0 to 3 for each of the following five categories: increased cellularity, increased mesangial matrix, necrosis, the percentage of sclerotic glomeruli, and the presence of crescents. Renal function was calculated as follows: (the composite glomerular score ÷ 5) + the average proteinuria score in the last 3 weeks.
Metagenomic prediction.
Bacterial metagenomes were predicted using PICRUSt (41) by comparing 16S to database gg13.5 and then to Integrated Microbial Genomes (IMG). The OTU were mapped to the gg13.5 database at 97% similarity by using the QIIME's pick_closed_otus script. OTU abundance was normalized by using the 16S rRNA gene copy numbers from known bacterial genomes. The normalized OTU were used for metagenomes prediction in PICRUSt. The predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs (42) was summarized to level 3 functional categories and compared among groups by using the Statistical Analysis of Metagenomic Profile package (43). Differentially represented gene families were identified by two-sided Welch's t test with Storey's false-discovery-rate correction.
Statistics.
Two-sample comparison was performed by unpaired Student t test and Mann-Whitney test. For >2 groups, one-way analysis of variance (ANOVA) was performed. If the overall effect is significant, pairwise comparisons were made by using unpaired t test with P values adjusted for Tukey's honest significance differences (TukeyHSD). Two-way ANOVA was used to determine the effect of sex and strain. Correlation analysis (Pearson for continuous variables or Spearman for categorical variables) was performed. Principal coordinate analysis was tested for significance by a permutational multivariate method PERMANOVA (44). All statistics were calculated by using R version 3.0.2.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences from the present study were deposited in the Metagenomics RAST server under the accession number 4557629.3. The original sequences were also deposited in the Sequence Read Archive under accession number SRP045474.
RESULTS
Lupus-associated gut microbial composition and diversity.
We have used the classical lupus-prone MRL/Mp-Faslpr (MRL/lpr) mice to model systemic lupus erythematosus (45). Mice homozygous for the mutation Faslpr show systemic autoimmunity, lymphadenopathy, and glomerulonephritis (46), which are similar to lupus-associated manifestations in humans. Female MRL/lpr mice die at an average age of 18 weeks. Starting at about 12 weeks of age, the levels of circulating autoantibodies and immune complexes increase greatly in MRL/lpr mice. The control mouse strain, MRL/Mp (MRL), does not show lupus-like symptoms before 18 weeks of age (47).
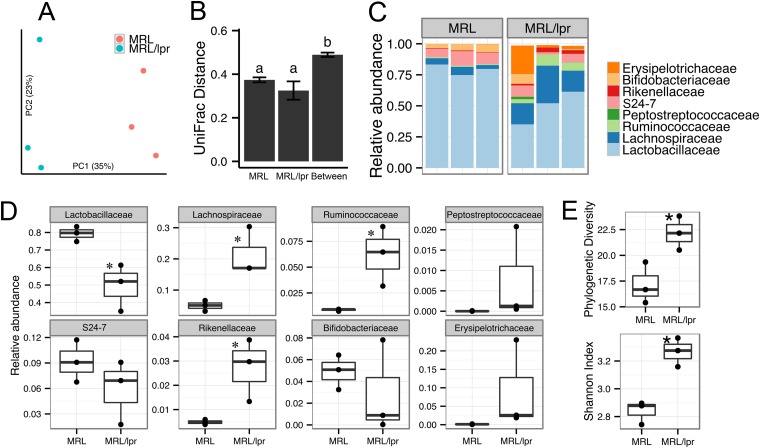
Women of childbearing age have the highest risk for lupus (26). We thus compared the gut microbiota of female MRL versus MRL/lpr mice at 5 weeks of age that mimic this population (n = 3 per strain). We found a clear distinction in the microbiota revealed by principal coordinate analysis (Fig. 1A). UniFrac distances between mouse strains were significantly higher than distances within strains (TukeyHSD test, P < 0.005, Fig. 1B), indicating an overall phylogenetic difference. The gut microbiota of lupus-prone MRL/lpr mice were characterized by a significant reduction in the family Lactobacillaceae (under “Firmicutes_Bacilli”) and a concurrent increase in the family Lachnospiraceae (under “Firmicutes_Clostridia”), which together accounted for more than 65% of all bacteria in the distal gut (Fig. 1C and D). A total of 105 operational taxonomic units (OTU) representing all Lactobacillaceae sequences were further classified to genus Lactobacillus, with the majority (92 of the 105 OTU) being uncultured species of this genus (see Table S1, highlighted rows, in the supplemental material). The Lachnospiraceae family comprises a phylogenetically diverse group of anaerobes defined as Clostridium cluster XIVa (48) that includes butyrate-producing genera such as Butyrivibrio and Roseburia (49). The predominant Lachnospiraceae-affiliated OTU were uncultured (see Table S2, highlighted rows, in the supplemental material). Other lupus-enriched families included Ruminococcaceae (uncultured) and Rikenellaceae (genus Alistipes) (Fig. 1D). Rare OTU (defined as <0.1% of total bacteria from all samples) accounted for <3.5% of individual microbiota (see Fig. S1 in the supplemental material). Among them, the RF9 group (“Tenericutes_Mollicutes”), Clostridiales family XIII, and the Streptococcaceae were significantly enriched in lupus-prone mice (see Fig. S1 in the supplemental material). Overall, gut microbiota in lupus-prone mice had a higher diversity measured as Faith's phylogenetic diversity and Shannon's entropy (Fig. 1E). These results show that lupus-prone mice have distinct bacterial composition and diversity compared to healthy controls, suggesting that lupus could alter gut microbiota.
FIG 1.
Differences in gut microbiota between healthy and lupus-prone mice. (A) Microbiota separation on the first two principal coordinates calculated from unweighted UniFrac distances. MRL, healthy control mice; MRL/lpr, lupus-prone mice. Fecal samples from 5-week-old female mice were used. The number of sequences in each of the six samples was 7,800. (B) UniFrac distances within and between mouse strains. Error bars indicate the standard errors of the mean for n = 3 (within MRL), n = 3 (within MRL/lpr), and n = 9 (between the two strains). Statistical significance was determined by one-way ANOVA, followed by pairwise t test with TukeyHSD-adjusted P values. (C) Taxonomy breakdown at the family level for abundant (>0.1%) bacterial OTU. (D) Bacterial abundance comparison between MRL and MRL/lpr microbiota. (E) Diversity indices. Statistical comparison was based on an unpaired Student t test (n = 3 in each group; *, P < 0.05; **, P < 0.01).
Microbiota dynamics associated with sex and age in murine lupus.
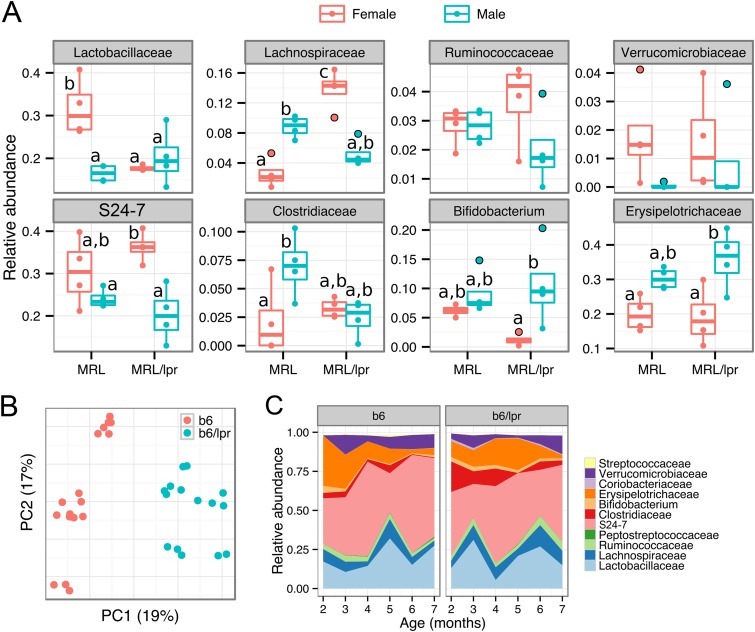
Lupus is a sex-specific disease affecting more women than men (26). In the MRL/lpr model, while lupus develops spontaneously in both females and males, female mice exhibit accelerated disease (50). We thus analyzed the gut microbiota separately in female and male mice (Fig. 2A and see Fig. S2A in the supplemental material). To try to minimize the cage effect as a confounding factor, we randomly selected one mouse of each sex from four different litters for each mouse strain (MRL or MRL/lpr, n = 4 per strain). In 14-week-old females, comparing lupus-prone MRL/lpr mice to MRL controls, significantly lower levels of Lactobacillaceae and higher levels of Lachnospiraceae were noted (Fig. 2A), a finding consistent with our observations in 5-week-old females (Fig. 1D). In males, however, the two strains of mice were not significantly different from each other in their gut microbial composition (Fig. 2A). Since sex and disease status appeared to interact, we also compared female and male mice within each mouse strain. In MRL mice, females had significantly higher Lactobacillaceae and Streptococcaceae levels and lower Lachnospiraceae and Clostridiaceae levels than did male mice (Fig. 2A and see Fig. S2B in the supplemental material). In contrast, in the lupus-prone MRL/lpr group, female mice had higher levels of Lachnospiraceae and Bacteroidetes S24-7, and lower levels of Bifidobacterium and Erysipelotrichaceae, whereas the levels of Lactobacillaceae were comparable between female and male mice (Fig. 2A). Principal coordinate analysis showed distinct microbiota between sexes, albeit the difference depended on mouse strains (see Fig. S2C in the supplemental material). Although no sex-dependent difference in bacterial diversity was observed in control mice, female lupus-prone mice had significantly higher diversity than their male counterparts (see Fig. S2D in the supplemental material). These results suggest that the gut microbiota of lupus-prone mice are different between sexes and that female-specific changes in certain bacterial phylotypes (such as the higher abundance of Lachnospiraceae) may be associated with an earlier onset of and/or more severe lupus symptoms in female MRL/lpr mice.
FIG 2.
Microbiota dynamics associated with sex and age. (A) Sex-associated microbiota differences. Colonic contents from 14-week-old MRL and MRL/lpr mice were used. Statistical significance was determined by two-way ANOVA, followed by pairwise t test with TukeyHSD-adjusted significance (n = 4 in each group). (B and C) Age-associated microbiota differences. (B) Fecal microbiota separation on the first two principal coordinates calculated from unweighted UniFrac distances. b6, female healthy control mice; b6/lpr, female lupus-prone mice. (C) Time-dependent changes of fecal microbiota in b6 and b6/lpr strains. Abundant bacterial OTU (>0.1%) were summarized (n = 3 per group).
We next determined time-dependent changes of gut microbiota in murine lupus. Because MRL/lpr mice have a short life span of approximately 18 weeks (47), we chose to investigate the time course in another lupus-prone model with the same Faslpr mutation. B6.MRL-Faslpr (B6/lpr) mice, which are of C57BL/6 (B6) background, have a life span of at least 7 months. Both MRL/lpr and B6/lpr mice start to develop lupus-like symptoms at about 2 months of age, but disease progression is slower in B6/lpr mice (51–53). We found that the gut microbiota of female B6/lpr mice was distinct from healthy B6 mice, as shown by principal coordinate analysis (Fig. 2B, n = 3 per strain). In this pair of mouse strains we observed dynamic changes in microbiota composition over a 5-month time period (Fig. 2C). Notably, B6/lpr mice had higher levels of Clostridiaceae than did B6 mice from 2 to 5 months (P < 0.05, unpaired t test), a time frame wherein B6/lpr mice have just started to show lupus-like symptoms. The family Clostridiaceae includes the butyrate producers in Clostridium cluster I (48). In addition, the abundance of Lactobacillaceae oscillated in the guts of both B6/lpr and B6 mice, whereas Lachnospiraceae appeared to be more abundant in B6/lpr mice during the last month (P = 0.003, Fig. 2C). These results suggest that the composition of gut microbiota change over time in both healthy and lupus-prone mice. Certain bacterial phylotypes, such as Clostridiaceae and Lachnospiraceae (both butyrate producers), are more abundant in the guts of B6/lpr mice at specific time points during lupus progression.
Restoration of lactobacilli and improvement of lupus symptoms with RA.
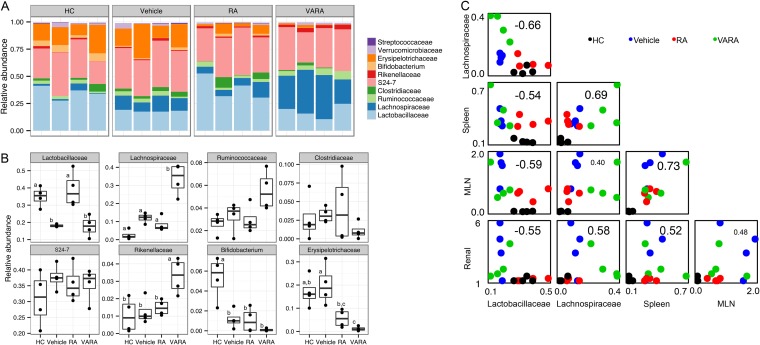
Vitamin A has been shown to reduce lupus nephritis (29–32). However, while orally administered vitamin A may come directly in contact with commensal bacteria in the intestinal tract during absorption, little is known about whether and how this nutrient affects gut microbiota. We thus investigated the effects of vitamin A (retinol) and a major physiological active metabolite of vitamin A, retinoic acid (RA), on gut microbiota in the MRL/lpr mouse model. We treated female MRL/lpr mice, orally and daily, with RA at 6 mg/kg of body weight (suspended in canola oil as the vehicle) from 6 to 14 weeks of age. RA of comparable doses and with similar treatment time frames was previously reported to reduce spleen and lymph node sizes and improve renal pathology in murine lupus models (29, 30). To mimic natural vitamin A supplementation, where retinol (retinyl palmitate) is the primary ingredient, we also treated female MRL/lpr mice daily with 11.2 mg retinyl palmitate/kg of body weight (equivalent to 6 mg retinol/kg) mixed with 0.6 mg RA/kg of body weight (10% of the amount of retinol) from 6 to 14 weeks of age. The small amount of RA in this formulation, which we refer to as VARA, has been shown to enhance the storage of retinol in the body (54). Because the vehicle itself (canola oil, which contains polyunsaturated fatty acids) may affect the gut microbiota (55–57), MRL/lpr mice treated with canola oil were used as the diseased control, whereas MRL mice treated with canola oil were used as the healthy control. To try to minimize the cage effect as a confounding factor, we randomly selected female MRL/lpr mice from four different litters and housed them in four cages. Each cage had one mouse randomly assigned to vehicle treatment, one with RA, and a third with VARA. The MRL mice were from two different litters. Colonic contents were collected after treatment and the microbiota were analyzed.
Gut microbiota after vitamin A treatments were not significantly different from the vehicle control among MRL/lpr groups based on principal coordinate analysis (see Fig. S3A in the supplemental material, n = 4 per group). However, we noted a number of significant differences in the taxonomy breakdown at the family level for abundant bacterial OTU. Strikingly, we found that RA restored Lactobacillaceae (Fig. 3A and B) that was downregulated in MRL/lpr mice (Fig. 1D, Fig. 2A, and Fig. 3B). VARA, on the other hand, did not have such an effect. Rather, it significantly increased the levels of Lachnospiraceae and Rikenellaceae (Fig. 3B), two bacterial families that were already higher in MRL/lpr than MRL mice, at least for 5-week-old mice (Fig. 1D). Importantly, we observed strong correlations between the abundance of two bacterial families, Lactobacillaceae and Lachnospiraceae, and lupus disease indices (Fig. 3C and see Table S3 in the supplemental material). Lupus disease parameters, lymphadenopathy (increased spleen and mesenteric lymph node weights) and glomerulonephritis (impaired renal function indicated by increasing scores), correlated negatively with the abundance of Lactobacillaceae and positively with Lachnospiraceae in the gut (Fig. 3C; P < 0.05, except for lymph node weight versus Lachnospiraceae, where P = 0.124). This suggests that RA improved lupus symptoms while restoring intestinal colonization of Lactobacillaceae, whereas VARA worsened the symptoms while increasing the abundance of Lachnospiraceae that were already higher in lupus-prone mice. Whether retinoid-mediated changes of gut microbiota directly affected lupus symptoms or they were by-products of the effects of vitamin A on lupus requires further investigation.
FIG 3.
Changes of bacterial abundance with vitamin A treatments. (A) Taxonomy breakdown at the family level abundant (>0.1%) bacterial OTU. HC, MRL healthy control treated with canola oil; vehicle, lupus-prone MRL/lpr treated with canola oil; RA, MRL/lpr treated with retinoic acid (6 mg/kg of body weight); VARA, MRL/lpr treated with retinol (6 mg retinol/kg of body weight) and retinoic acid (0.6 mg/kg of body weight, which facilitates retinol storage). All mice were female and treated orally from 6 to 14 weeks of age. Mice were sacrificed at 14 weeks of age and colonic contents were collected (n = 4 per group). (B) Comparison of individual families shown in panel A. Statistical significance was determined by one-way ANOVA, followed by a pairwise t test with TukeyHSD-adjusted significance. (C) Correlation between bacterial abundance and lupus disease indices. Spleen, spleen weight in grams; MLN, mesenteric lymph node weight in grams; renal, renal function estimated from proteinuria and glomerular scores as described in Materials and Methods. Correlation coefficients are shown on each individual plot. For example, the correlation between Lactobacillaceae and Lachnospiraceae was −0.66. Spearman's correlation tests were used for pairs involving ranked renal function data; all other correlations were calculated by using the Pearson method. Raw data can be found in the supplemental material.
Both RA and VARA also decreased the levels of Erysipelotrichaceae compared to the vehicle control in MRL/lpr mice (Fig. 3B). In addition, lupus-prone mice had significantly lower levels of Bifidobacterium compared to the healthy control, and neither RA nor retinol restored the abundance of this bacterial family (Fig. 3B). Lower representation of Erysipelotrichaceae and Bifidobacterium could potentially make available more substrates for other commensal bacteria, such as Lactobacillaceae in the case of RA treatments and Lachnospiraceae in the case of VARA treatments (Fig. 3B). Consistent with prior observations (Fig. 1E), vehicle-treated MRL/lpr microbiota had higher diversity than vehicle-treated MRL microbiota based on Shannon's entropy analysis, whereas neither RA nor VARA treatments changed this phenomenon (see Fig. S3B in the supplemental material) despite their effects on lupus symptoms, suggesting that microbial diversity in the gut may depend on genetic predisposition of the mouse strains but not their phenotype.
Changes of microbiome metabolic functions with lupus and vitamin A treatments.
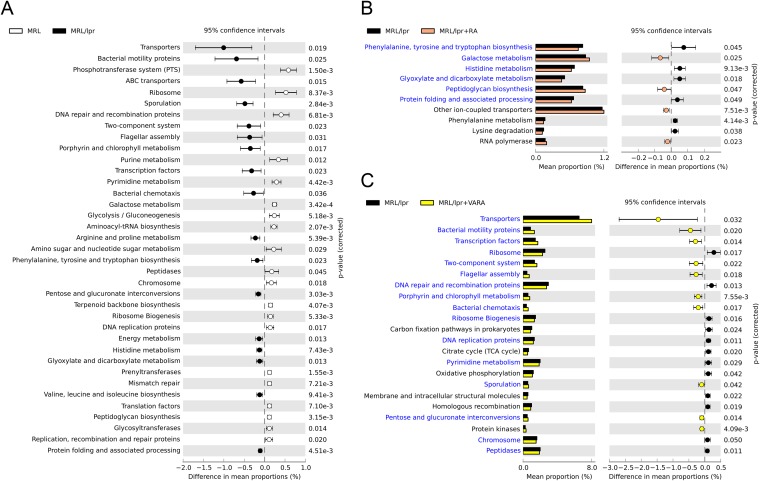
To investigate microbiome functions associated with lupus, we used PICRUSt to infer putative metagenomes from 16S rRNA gene profiles of 14-week-old female MRL and MRL/lpr mice (41). Comparing the predicted gut microbiome of lupus-prone mice to that of the control, categories representing genes important for genetic information processing (e.g., DNA replication and repair, protein synthesis) and nucleotide metabolism were significantly lower in MRL/lpr microbiome (Fig. 4A), indicating reduced turnover of bacterial DNA, RNA, and protein synthesis (58). This was accompanied by a significant increase in cell motility genes (e.g., bacterial chemotaxis and flagellar assembly) in the gut microbiome of lupus-prone mice (Fig. 4A), suggesting that the bacteria may be actively accessing substrates and/or adjusting their locations (59). Sporulation genes also increased with lupus disease, possibly due to the increase of spore-forming clostridia (Fig. 2A). In the carbohydrate metabolism category, galactose metabolism and glycolysis decreased, while glyoxylate and dicarboxylate metabolism increased with lupus disease (Fig. 4A). Lupus-associated increases in membrane transport (e.g., ATP-binding cassette transporters), signal transduction (two-component system), and amino acid metabolism were also observed (Fig. 4A).
FIG 4.
Bacterial metagenomes predicted from microbial community identities. Gene functional categories were from level 3 of KEGG pathways. Gene functions with a significant difference are shown (P < 0.05). (A) Differences between MRL control and lupus-prone MRL/lpr mice. The effect size was 0.10%. ABC transporters, ATP-binding cassette transport systems. (B) Effect of RA treatment on lupus-prone bacterial metagenomes. The effect size was 0.02%. (C) Effect of retinol treatment (VARA) on lupus-prone bacterial metagenomes. The effect size was 0.08%.
To assess whether vitamin A treatments could affect gut microbiome functions in lupus-prone mice, we performed a similar analysis between vehicle- and RA-treated MRL/lpr mice (Fig. 4B), as well as between vehicle- and VARA-treated MRL/lpr mice (Fig. 4C). Metabolic functions that overlapped with those in Fig. 4A were identified and color-coded in blue. Interestingly, we found that RA reversed many lupus-associated changes in microbial functions that deviated from the MRL control (Fig. 4B). These included genes in carbohydrate metabolism (e.g., galactose metabolism, glyoxylate and dicarboxylate metabolism) and amino acid metabolism (e.g., histidine metabolism, and phenylalanine, tyrosine and tryptophan biosynthesis). In contrast, VARA worsened lupus-associated changes on all overlapped functional categories, including cell motility, membrane transport, DNA replication and repair, protein synthesis, signal transduction, and sporulation (Fig. 4C). Together, these results suggest that microbiome metabolic functions in the gut could change with lupus disease. RA treatment, which attenuated symptoms, reversed lupus-associated changes in microbial gene functions. VARA, on the other hand, exacerbated the changes of metagenomic profiles in lupus-prone mice while worsening the disease, suggesting a possible correlation between gut microbiome and disease symptoms in lupus.
DISCUSSION
In this study, we reported the dynamics of gut microbiota in murine lupus. By using lupus-prone mice carrying the Faslpr mutation, we showed that the composition and diversity of gut microbiota differed between control and diseased mice (Fig. 1). Many differences were sex specific and varied over time during lupus progression (Fig. 2). In addition, we found that the use of different vitamin A treatments that increased abundance of Lactobacillaceae in the intestine correlated with improved lupus symptoms, whereas greater colonization of Lachnospiraceae correlated with disease deterioration (Fig. 3). Through prediction analysis of metagenomes (Fig. 4), we also showed that certain functions, such as bacterial motility, increased with lupus disease in mice. In addition, gut microbiome metabolic functions appeared to correlate with disease severity in lupus.
Members of the Lactobacillaceae family have been recommended as probiotics with anti-inflammatory functions (60). Lactobacillus rhamnosus GG, for example, suppresses proinflammatory responses by increasing interleukin-10 and regulatory T (Treg) cells (61, 62). Several other members of Lactobacillaceae, including L. acidophilus and L. reuteri, were also found to induce anti-inflammatory Treg cells (63, 64). Although colonization of Lactobacillaceae in the guts of both humans and mice has been shown to suppress inflammation (65, 66), a direct relationship between intestinal Lactobacillaceae and autoimmune lupus has not been established. In the present study we show that lower colonization of lactobacilli in the gut is associated with disease state in lupus, an autoimmune disease characterized by chronic inflammation.
Several groups have recently reported that short-chain fatty acids produced by gut microbiota, especially butyrate produced by Clostridia, promote the differentiation of Treg cells to suppress inflammation (67–70). However, in the present study, we found that both young (5-week-old) and old (14-week-old) female MRL/lpr mice had higher levels of Lachnospiraceae, where butyrate-producing Clostridium XIVa belongs, than their MRL counterparts (Fig. 1D and Fig. 2A). In the B6/lpr model, the levels of Lachnospiraceae, as well as of another butyrate-producing family, Clostridiaceae, were also higher in lupus-prone mice compared to B6 controls (Fig. 2C). Moreover, we found strong positive correlations between the abundance of Lachnospiraceae and lupus disease parameters, including lymphadenopathy and renal pathology (Fig. 3C). All of these results suggest that Lachnospiraceae, or butyrate-producing bacteria in general, may not be able to suppress inflammation in the lupus-prone MRL/lpr and B6/lpr models. Indeed, a recent report has shown that in mice that are deficient in Fas signaling (e.g., Faslpr mutation), butyrate-induced apoptosis of T cells is abolished (71). Thus, even though Lachnospiraceae accumulated in MRL/lpr and B6/lpr mice, the butyrate that they produced would lack the mechanism required for T-cell apoptosis. This would likely lead to uncontrolled proinflammatory T-cell responses and thereby lupus progression. The notion is supported by our observation that further changes of the abundance of Lachnospiraceae through vitamin A intervention of MRL/lpr mice failed to alter the percentages of different T-cell populations in the periphery (data not shown).
Many autoimmune disorders are more prevalent in females (72). Sex-specific differences in gut microbiota have not been reported until recently, where microbiota was found to influence sex bias in type 1 diabetes (19, 20). In nonobese diabetic (NOD) mice, females were found to possess greater microbial diversity than male counterparts (19). In addition, it was found that the levels of Alistipes (family Erysipelotrichaceae) were higher in male NOD mice, although the difference was not statistically significant (20). Our data from the MRL/lpr model agreed with the above reports that female mice had significantly higher microbial diversity (see Fig. S2D in the supplemental material), and Erysipelotrichaceae was significantly enriched in the gut microbiota of male MRL/lpr mice (Fig. 2A). In addition to these differences, we found that in lupus-prone mice, females had significantly higher levels of Lachnospiraceae, whereas males had significantly higher levels of Bifidobacterium (Fig. 2A). Bifidobacterium, like lactobacilli, has been suggested to exert anti-inflammatory functions (73). Thus, a greater abundance of Bifidobacterium in the gut may be associated with attenuated lupus symptoms in males. It was also noted that the variation of the abundance of some bacteria appeared to be greater in females than in males (see, for example, Verrucomicrobiaceae in Fig. 2A). This could be due to the lack of synchronization of the estrus cycle, since microbiota may change according to the female reproductive cycle (74).
Important roles of vitamin A and its derivatives, including RA, in regulating antiviral and/or antibacterial immune responses have been recognized since the 1920s (75). However, whether and how RA affects lupus is not clearly understood. We found here that treatment with RA attenuated lupus-like symptoms while increasing the Lactobacillaceae family of commensal bacteria in the colons of lupus-prone MRL/lpr mice (Fig. 3C). RA can sustain the stability and function of natural Treg cells (76) and promote the differentiation of inducible Treg cells in the periphery (77–79), which may in turn attenuate lupus. Lactobacillus spp. can also suppress proinflammatory responses by increasing the number of inducible Treg cells (61, 62). Thus, it is likely that RA and Lactobacillaceae that RA can enrich in the gut of lupus-prone mice (Fig. 3A and B) could work in concert to induce Treg cells and attenuate inflammatory flares in lupus.
Retinol is the primary ingredient in vitamin A supplements. In this report we found that retinol, represented by VARA treatment, increased the fraction of Lachnospiraceae in the gut of MRL/lpr mice (Fig. 3B) that was already higher than that in MRL mice (Fig. 1D). It did not attenuate lupus (Fig. 3C) or affect members of the Lactobacillaceae family (Fig. 3B). This suggests that unlike RA, retinol may promote intestinal colonization of Lachnospiraceae that does not attenuate lupus pathogenesis. In fact, the percentage of Lachnospiraceae in MRL/lpr mice negatively correlated with that of Lactobacillaceae and positively correlated with lymphadenopathy of the spleen and impaired renal function (Fig. 3C). Therefore, our results suggest that RA, given as a direct dose, may be better than retinol in suppressing inflammation in lupus. Metagenomic predication of microbial metabolic functions, where RA was shown to reverse, while retinol (VARA) exacerbated, lupus-associated changes (Fig. 4), supports this hypothesis.
The composition of gut microbiota can be greatly altered by the housing condition. In the present study, all mice were maintained in the same room under the same specific-pathogen-free housing condition. In addition, mice in each experimental group were randomly selected from different litters to eliminate the cage effect as a potential confounding factor. We kept control and lupus animals in different cages to avoid the possible effect of coprophagy on disease progression. It has been noted, however, after the completion of the study, that the combination of housing condition adaptation and dam cohousing between control and lupus strains prior to their respective breeding could have further equilibrated the baseline microbiota.
Altogether, our study showed several important lupus-associated changes in gut microbiota. Intestinal colonization of Lactobacillaceae, in particular, was found to negatively correlate with lupus activity. This suggests that probiotics containing lactobacilli may be able to decrease the occurrence and/or severity of inflammatory flares suffered by lupus patients, but future studies are required to address whether a causal relationship between lactobacilli and lupus exists. In addition, the potential therapeutic benefit of probiotics against lupus may be enhanced when combined with RA, a dietary supplement that can increase the levels of lactobacilli in the gut and attenuate lupus symptoms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sarah Owens for assistance on sequencing and Thomas Cecere for scoring the histopathological slides.
This study was supported by X. Luo and H. Zhang's startup funds provided by Virginia Tech College of Veterinary Medicine and College of Engineering, respectively.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 26 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02676-14.
REFERENCES
- 1.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Seo SU, Chen GY, Nunez G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13:321–335. 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn KA, Stappenbeck TS. 2013. Peripheral education of the immune system by the colonic microbiota. Semin. Immunol. 25:364–369. 10.1016/j.smim.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strober W. 2013. Impact of the gut microbiome on mucosal inflammation. Trends Immunol. 34:423–430. 10.1016/j.it.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. 2013. The role of the microbiome in rheumatic diseases. Curr. Rheumatol. Rep. 15:314. 10.1007/s11926-012-0314-y. [DOI] [PubMed] [Google Scholar]
- 6.Brown EM, Sadarangani M, Finlay BB. 2013. The role of the immune system in governing host-microbe interactions in the intestine. Nat. Immunol. 14:660–667. 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 7.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313–323. 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inagaki H, Suzuki T, Nomoto K, Yoshikai Y. 1996. Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of l-selectin+ CD44+ T cells in sites of inflammation. Infect. Immun. 64:3280–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8:292–300. 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104:13780–13785. 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez J-P, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482:179–185. 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1131. 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 110:9066–9071. 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102:11070–11075. 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. 2009. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U. S. A. 106:2365–2370. 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 17.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109–1113. 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathis D, Benoist C. 2012. The influence of the microbiota on type-1 diabetes: on the threshold of a leap forward in our understanding. Immunol. Rev. 245:239–249. 10.1111/j.1600-065X.2011.01084.x. [DOI] [PubMed] [Google Scholar]
- 19.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity 39:400–412. 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339:1084–1088. 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 21.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. 2008. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 35:1500–1505. [PubMed] [Google Scholar]
- 22.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, Heuvelmans-Jacobs M, Akira S, Nicklin MJ, Ribeiro-Dias F, van den Berg WB. 2008. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 118:205–216. 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32:815–827. 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):S4615–S4622. 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G. 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479:538–541. 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 26.Tsokos GC. 2011. Systemic lupus erythematosus. N. Engl. J. Med. 365:2110–2121. 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 27.Ohl K, Tenbrock K. 2011. Inflammatory cytokines in systemic lupus erythematosus. J. Biomed. Biotechnol. 2011:432595. 10.1155/2011/432595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houssiau F. 2007. Thirty years of cyclophosphamide: assessing the evidence. Lupus 16:212–216. [DOI] [PubMed] [Google Scholar]
- 29.Perez de Lema G, Lucio-Cazana FJ, Molina A, Luckow B, Schmid H, de Wit C, Moreno-Manzano V, Banas B, Mampaso F, Schlondorff D. 2004. Retinoic acid treatment protects MRL/lpr lupus mice from the development of glomerular disease. Kidney Int. 66:1018–1028. 10.1111/j.1523-1755.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita K, Yoo BS, Nozaki Y, Sugiyama M, Ikoma S, Ohno M, Funauchi M, Kanamaru A. 2003. Retinoic acid reduces autoimmune renal injury and increases survival in NZB/W F1 mice. J. Immunol. 170:5793–5798. 10.4049/jimmunol.170.11.5793. [DOI] [PubMed] [Google Scholar]
- 31.Nozaki Y, Yamagata T, Yoo BS, Sugiyama M, Ikoma S, Kinoshita K, Funauchi M, Kanamaru A. 2005. The beneficial effects of treatment with all-trans-retinoic acid plus corticosteroid on autoimmune nephritis in NZB/WF mice. Clin. Exp. Immunol. 139:74–83. 10.1111/j.1365-2249.2005.02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinoshita K, Kishimoto K, Shimazu H, Nozaki Y, Sugiyama M, Ikoma S, Funauchi M. 2010. Successful treatment with retinoids in patients with lupus nephritis. Am. J. Kidney Dis. 55:344–347. 10.1053/j.ajkd.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinformatics J. 7:1–8. 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 35.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conservation 61:1–10. 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 41.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31:814–821. 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parks DH, Beiko RG. 2010. Identifying biologically relevant differences between metagenomic communities. Bioinformatics (Oxford, England) 26:715–721. 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 44.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26:32–46. 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 45.Perry D, Sang A, Yin Y, Zheng YY, Morel L. 2011. Murine models of systemic lupus erythematosus. J. Biomed. Biotechnol. 2011:271694. 10.1155/2011/271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrews BS, Eisenberg RA, Theofilopoulos AN, Izui S, Wilson CB, McConahey PJ, Murphy ED, Roths JB, Dixon FJ. 1978. Spontaneous murine lupus-like syndromes: clinical and immunopathological manifestations in several strains. J. Exp. Med. 148:1198–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hewicker M, Kromschroder E, Trautwein G. 1990. Detection of circulating immune complexes in MRL mice with different forms of glomerulonephritis. Z. Versuchstierkd. 33:149–156. [PubMed] [Google Scholar]
- 48.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812–826. 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 49.Cotta M, Forster R. 2006. The family Lachnospiraceae, including the genera Butyrivibrio, Lachnospira, and Roseburia, p 1002–1021 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer US, New York, NY. [Google Scholar]
- 50.Smathers PA, Santoro TJ, Chused TM, Reeves JP, Steinberg AD. 1984. Studies of lymphoproliferation in MRL-lpr/lpr mice. J. Immunol. 133:1955–1961. [PubMed] [Google Scholar]
- 51.Wofsy D, Murphy ED, Roths JB, Dauphinee MJ, Kipper SB, Talal N. 1981. Deficient interleukin 2 activity in MRL/Mp and C57BL/6J mice bearing the lpr gene. J. Exp. Med. 154:1671–1680. 10.1084/jem.154.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morse HC, III, Roths JB, Davidson WF, Langdon WY, Fredrickson TN, Hartley JW. 1985. Abnormalities induced by the mutant gene, lpr. Patterns of disease and expression of murine leukemia viruses in SJL/J. mice homozygous and heterozygous for lpr. J. Exp. Med. 161:602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izui S, Kelley VE, Masuda K, Yoshida H, Roths JB, Murphy ED. 1984. Induction of various autoantibodies by mutant gene lpr in several strains of mice. J. Immunol. 133:227–233. [PubMed] [Google Scholar]
- 54.Ross AC, Li NQ, Wu L. 2006. The components of VARA, a nutrient-metabolite combination of vitamin A and retinoic acid, act efficiently together and separately to increase retinyl esters in the lungs of neonatal rats. J. Nutr. 136:2803–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q, Zhang Q, Wang C, Tang C, Zhang Y, Li N, Li J. 2011. Fish oil enhances recovery of intestinal microbiota and epithelial integrity in chronic rejection of intestinal transplant. PLoS One 6:e20460. 10.1371/journal.pone.0020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Druart C, Neyrinck AM, Vlaeminck B, Fievez V, Cani PD, Delzenne NM. 2014. Role of the lower and upper intestine in the production and absorption of gut microbiota-derived PUFA metabolites. PLoS One 9:e87560. 10.1371/journal.pone.0087560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu HN, Zhu J, Pan WS, Shen SR, Shan WG, Das UN. 2014. Effects of fish oil with a high content of n-3 polyunsaturated fatty acids on mouse gut microbiota. Arch. Med. Res. 45:195–202. 10.1016/j.arcmed.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 58.Klaassens ES, Ben-Amor K, Vriesema A, Vaughan EE, de Vos W. 2011. The fecal bifidobacterial transcriptome of adults: a microarray approach. Gut Microbes 2:217–226. 10.4161/gmic.2.4.16798. [DOI] [PubMed] [Google Scholar]
- 59.He S, Ivanova N, Kirton E, Allgaier M, Bergin C, Scheffrahn RH, Kyrpides NC, Warnecke F, Tringe SG, Hugenholtz P. 2013. Comparative metagenomic and metatranscriptomic analysis of hindgut paunch microbiota in wood- and dung-feeding higher termites. PLoS One 8:e61126. 10.1371/journal.pone.0061126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. 2012. The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12:728–734. 10.1038/nri3312. [DOI] [PubMed] [Google Scholar]
- 61.So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, Hwang KC, Jeon YH, Im SH. 2008. Lactobacillus casei suppresses experimental arthritis by downregulating T helper 1 effector functions. Mol. Immunol. 45:2690–2699. 10.1016/j.molimm.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 62.Khailova L, Baird CH, Rush AA, McNamee EN, Wischmeyer PE. 2013. Lactobacillus rhamnosus GG improves outcome in experimental Pseudomonas aeruginosa pneumonia: potential role of regulatory T cells. Shock 40:496–503. 10.1097/SHK.0000000000000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khazaie K, Zadeh M, Khan MW, Bere P, Gounari F, Dennis K, Blatner NR, Owen JL, Klaenhammer TR, Mohamadzadeh M. 2012. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. U. S. A. 109:10462–10467. 10.1073/pnas.1207230109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sims IM, Frese SA, Walter J, Loach D, Wilson M, Appleyard K, Eason J, Livingston M, Baird M, Cook G, Tannock GW. 2011. Structure and functions of exopolysaccharide produced by gut commensal Lactobacillus reuteri 100-23. ISME J. 5:1115–1124. 10.1038/ismej.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O'Sullivan GC, Kiely B, Collins JK, Shanahan F, Quigley EM. 2005. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 128:541–551. 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 66.von Schillde MA, Hormannsperger G, Weiher M, Alpert CA, Hahne H, Bauerl C, van Huynegem K, Steidler L, Hrncir T, Perez-Martinez G, Kuster B, Haller D. 2012. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11:387–396. 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 67.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 70.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40:128–139. 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. 2012. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am. J. Physiol. Gastrointest. Liver Physiol. 302:G1405–G1415. 10.1152/ajpgi.00543.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whitacre CC. 2001. Sex differences in autoimmune disease. Nat. Immunol. 2:777–780. 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 73.Preising J, Philippe D, Gleinser M, Wei H, Blum S, Eikmanns BJ, Niess J-H, Riedel CU. 2010. Selection of bifidobacteria based on adhesion and anti-inflammatory capacity in vitro for amelioration of murine colitis. Appl. Environ. Microbiol. 76:3048–3051. 10.1128/AEM.03127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mulak A, Tache Y, Larauche M. 2014. Sex hormones in the modulation of irritable bowel syndrome. World J. Gastroenterol. 20:2433–2448. 10.3748/wjg.v20.i10.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Green HN, Mellanby E. 1928. Vitamin A as an anti-infective agent. BMJ 2:691–696. 10.1136/bmj.2.3537.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X, Kong N, Wang J, Fan H, Zou H, Horwitz D, Brand D, Liu Z, Zheng SG. 2010. Cutting edge: all-trans retinoic acid sustains the stability and function of natural regulatory T cells in an inflammatory milieu. J. Immunol. 185:2675–2679. 10.4049/jimmunol.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B, Kuchroo VK. 2008. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-β-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J. Immunol. 181:2277–2284. 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317:256–260. 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 79.Hall JA, Grainger JR, Spencer SP, Belkaid Y. 2011. The role of retinoic acid in tolerance and immunity. Immunity 35:13–22. 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.