Abstract
Harmful algal blooms, caused by massive and exceptional overgrowth of microalgae and cyanobacteria, are a serious environmental problem worldwide. In the present study, we looked for Bacillus strains with sufficiently strong anticyanobacterial activity to be used as biocontrol agents. Among 24 strains, Bacillus amyloliquefaciens FZB42 showed the strongest bactericidal activity against Microcystis aeruginosa, with a kill rate of 98.78%. The synthesis of the anticyanobacterial substance did not depend on Sfp, an enzyme that catalyzes a necessary processing step in the nonribosomal synthesis of lipopeptides and polyketides, but was associated with the aro gene cluster that is involved in the synthesis of the sfp-independent antibiotic bacilysin. Disruption of bacB, the gene in the cluster responsible for synthesizing bacilysin, or supplementation with the antagonist N-acetylglucosamine abolished the inhibitory effect, but this was restored when bacilysin synthesis was complemented. Bacilysin caused apparent changes in the algal cell wall and cell organelle membranes, and this resulted in cell lysis. Meanwhile, there was downregulated expression of glmS, psbA1, mcyB, and ftsZ—genes involved in peptidoglycan synthesis, photosynthesis, microcystin synthesis, and cell division, respectively. In addition, bacilysin suppressed the growth of other harmful algal species. In summary, bacilysin produced by B. amyloliquefaciens FZB42 has anticyanobacterial activity and thus could be developed as a biocontrol agent to mitigate the effects of harmful algal blooms.
INTRODUCTION
Eutrophication of surface waters has many undesirable effects and can lead to major water quality issues in freshwater and coastal systems (1). This phenomenon results in blooms of harmful algal species in freshwater lakes and brackish waters throughout the world. Moreover, the excessive growth of harmful algae, such as microalgae and cyanobacteria, often increases the production of inherent toxins such as microcystins and nodularins that cause acute poisonings of fish, birds, and mammals, including humans (2). For example, dogs died after they were exposed to a cyanobacterial bloom of Microcystis aeruginosa in Lake Amstelmeer (The Netherlands), and the concentration of microcystin in this lake was up to 5.27 × 103 μg g−1 of dry weight (3). In recent years, harmful algal blooms of eutrophic water in China have occurred frequently, including in Lake Taihu and Lake Chaohu, and Microcystis is thought to be the dominant bloom genera (4).
Many control techniques have been used to prevent and mitigate bloom problems, including yellow loess (5), clay (6), and chemical agents such as copper sulfate and hydrogen peroxide (7). However, each of the physical and chemical methods available to remediate eutrophic water is associated with certain disadvantages (8). Therefore, there is still a pressing need for environmentally friendly, cost-effective, and convenient bactericidal agents directed against cyanobacterial blooms in eutrophic lakes. Biological control agents such as bacteria, viruses, and protozoa are of particular interest (9).
A growing body of evidence suggests that some bacteria can inhibit the growth of red-tide algae effectively through direct or indirect attack. Myxobacter spp. (10), Cytophaga spp. (11, 12), and Saprospira spp. (13) can invade through cell walls into the interior of algal cells. Bacteria that act indirectly exert killing activity through the production of extracellular algicidal substances, such as the phenazine pigments and 1-methyl-β-carboline secreted by certain Pseudomonas spp. (14, 15), β-cyano-l-alanine from Vibrio spp. (16), and lactones produced by Ruegeria pomeroyi (17). Recent studies have demonstrated that Bacillus spp. can suppress the growth of harmful algal bloom species (9, 18, 19). In a previous study, Ahn et al. (18) revealed that the culture broth of Bacillus subtilis C1 containing 10 mg liter−1 surfactin completely inhibited the growth of M. aeruginosa, although these researchers did not further isolate the surfactin from the culture broth or construct a mutant to verify its inhibitory activity. As such, the active compound(s) and mechanisms of action remain to be identified.
Some species from the genus Bacillus, such as B. subtilis and B. amyloliquefaciens, are plant-growth promoting bacteria (PGPR), and these species have been developed as biocontrol agents due to their ability to form heat- and desiccation-resistant spores. Several Bacillus-based commercial products are available, such as Quantum-400 (B. subtilis GB03), Serenade (B. subtilis QST713), and Rhizovital (B. amyloliquefaciens FZB42) (20, 21). Bacillus spp. produce a variety of bioactive metabolites that exert antagonistic actions against pathogens. Prominent classes of such compounds are the sfp-dependent lipopeptides and polyketides which need Sfp, a 4′-phosphopantetheinyl transferase to transfer 4′-phosphopantetheine from coenzyme A onto peptidyl carrier proteins in the nonribosomal peptide synthesis pathway (22). Lipopeptides consist of a lipid connected to a peptide, and these are largely amphiphilic membrane-active biosurfactants and also peptide antibiotics with mainly antifungal activity (23). Polyketides are biosynthesized through decarboxylative condensation, and these form a large family of secondary metabolites with antibacterial, immunosuppressive, antitumor, and other physiologically relevant bioactivities (24).
The rhizosphere-colonizing B. amyloliquefaciens FZB42 is an environmental strain that has the impressive ability to stimulate plant growth while suppressing the growth of plant-pathogenic organisms. Genome analysis reveals that B. amyloliquefaciens FZB42 harbors an array of giant gene clusters involved in ribosome-dependent and nonribosomal peptide synthesis (25). The non-ribosomally synthesized lipopeptides (e.g., surfactin, fengycin, and bacillomycin D) and polyketides (e.g., bacillaene, difficidin, and macrolactin) exhibit potent antifungal, hemolytic, and antibacterial activities (24–26). Meanwhile, the ribosomally synthesized peptide antibiotics plantazolicin A and B show moderate nematicidal activity (27, 28). In the present study, we report that B. amyloliquefaciens FZB42 displays high inhibitory activity against M. aeruginosa. We managed to identify the anticyanobacterial substance present in the culture filtrates and clarify the underlying mechanisms responsible for the specific bactericidal activity against harmful algal bloom species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Twenty-four Bacillus strains of different origins were used in a first round of screening. B. amyloliquefaciens FZB42T was kindly supplied by R. Borriss (ABiTEP GmbH, Berlin, Germany). Other strains were isolated from soil samples in Tibet, China. Escherichia coli DH5α (TaKaRa Bio, Inc., Dalian, China) was used as the host for all plasmids. S. aureus ATCC 9144 (29) was used as the assay organism in bacilysin determinations. Luria broth (30) was used for growing S. aureus and E. coli. Landy medium (31) was used to ferment all Bacillus isolates and mutants. For general bacilysin production, B. amyloliquefaciens FZB42 and associated mutants were grown in Perry and Abraham (PA) medium (32). When required, antibiotics were added to the following final concentrations: ampicillin (Amp) at 100 μg ml−1, chloramphenicol (Cm) at 5 μg ml−1, erythromycin (Erm) at 10 μg ml−1, and kanamycin (Km) at 5 μg ml−1.
Cyanobacterial culture.
M. aeruginosa NIES-843, A. flos-aquae FACHB-1040, Nostoc sp. strain FACHB-1135, and Anabaena sp. strain FACHB-1383 were purchased from the Freshwater Algae Culture Collection of the Institute of Hydrobiology (China). All cultures were grown at 25 ± 1°C under white fluorescent lamps (60 μM m−2 s−1, 16:8 h light-dark cycle). The cyanobacteria were cultured in sterilized BG11 medium (33) and transferred once a week to ensure that the experiments were always conducted with cultures during exponential growth phase.
Transformation, DNA manipulation, and transposon mutagenesis of B. amyloliquefaciens FZB42.
The isolation and manipulation of recombinant DNA were performed using standard techniques. E. coli and B. amyloliquefaciens were transformed as described by Sambrook and Russell (34) and Spizizen (35), respectively. The transposon mutagenesis library was constructed using pMarA and Southern blotting was used to analyze the insertion copies of the pMarA plasmid into the selected transposon mutants of B. amyloliquefaciens FZB42, as described previously (36). All enzymes used in the present study were purchased from TaKaRa Bio.
Generation of B. amyloliquefaciens FZB42 mutant and complementation strains.
The mutant B. amyloliquefaciens strain FZB42ΔbacB (devoid of bacilysin production) was obtained as follows. About 700-bp genomic regions upstream and downstream of the bacB gene were amplified from B. amyloliquefaciens FZB42 chromosomal DNA, respectively. The two gel-purified double-stranded DNA fragments were linked by a Cmr resistance cassette and then ligated into pMD-18. The linearized plasmid was integrated into the genome of B. amyloliquefaciens FZB42 by double-crossover recombination, yielding the knockout mutant B. amyloliquefaciens FZB42ΔbacB.
For complementation, the entire bacB gene and two homologous recombination arms obtained from the amyE gene were amplified from the chromosomal DNA of B. amyloliquefaciens FZB42. Fragments were linked with a Kmr resistance cassette by overlap PCR, and then the sequence was ligated into pMD-18. Finally, the vector was transformed into the B. amyloliquefaciens FZB42ΔbacB mutant and selected on solid LB agar medium supplemented with 5 μg ml−1 Cm and 50 μg ml−1 Kan. The specific primers used above are listed in Table 1.
TABLE 1.
Oligonucleotide DNA primers used in this study
| Function and primer | Sequence (5′-3′) | Purpose |
|---|---|---|
| Construction of mutants | ||
| bacB1-F | CCTTGTTCCAATCGCTCAG | Construction of the site-directed mutant FZB42ΔbacB |
| bacB1-R | GTCGGAGATGTCACAAGAAA | Construction of the site-directed mutant FZB42ΔbacB |
| bacB2-F | AGAAAGCAGAACTTCCGTAT | Construction of the site-directed mutant FZB42ΔbacB |
| bacB2-R | CCTGAAGGGACAAGTAGTGAG | Construction of the site-directed mutant FZB42ΔbacB |
| amyE1-F | CCTCTTTACTGCCGTTATT | Complementation of the mutant FZB42ΔbacB |
| amyE1-R | ATGCCCGTAGTTAGAAGC | Complementation of the mutant FZB42ΔbacB |
| amyE2-F | ACAAGTTAGTCACATGGGTG | Complementation of the mutant FZB42ΔbacB |
| amyE2-R | TGCGGAAGATAACCATTCAAAC | Complementation of the mutant FZB42ΔbacB |
| bacB-F | GTCGGGAATGTCAATGCT | Complementation of the mutant FZB42ΔbacB |
| bacB-R | GTGACGACGTTGGAAGAT | Complementation of the mutant FZB42ΔbacB |
| Real-time PCR analysis | ||
| 16S-F | GGACGGGTGAGTAACGCGTA | Internal reference |
| 16S-R | CCCATTGCGGAAAATTCCCC | Internal reference |
| glmS-F | TGTGCCTCCGATGTCAGT | Detection of the expression of glmS |
| glmS-R | ATGAAGTGACGATAACCCT | Detection of the expression of glmS |
| psbA1-F | GGTCAAGARGAAGAAACCTACAAT | Detection of the expression of psbA1 |
| psbA1-R | GTTGAAACCGTTGAGGTTGAA | Detection of the expression of psbA1 |
| mcyB-F | CCTACCGAGCGCTTGGG | Detection of the expression of mcyB |
| mcyB-R | GAAAATCCCCTAAAGATTCCTGAGT | Detection of the expression of mcyB |
| ftsZ-F | TCGCTGCTATTTCCTCGC | Detection of the expression of ftsZ |
| ftsZ-R | TGACTTCTCCCTGCATTTTCT | Detection of the expression of ftsZ |
Preparation and assay of bacilysin activity.
To obtain the pure bacilysin, culture filtrates of B. amyloliquefaciens FZB42 in PA medium were extracted twice in ice-cold ethanol. These extracts were subjected to Dowex 50WX8-200 separation (Sigma, USA) on a column equilibrated with 50/50 ethanol-water and eluted (after washing with water) by the application of 4% aqueous ammonium hydroxide (32). The eluate was immediately lyophilized to dryness and resuspended in 1 ml of water for loading onto the high-performance liquid chromatography-mass spectrometry (HPLC-MS). The sample was injected onto a ZoRBX Eclipse XDB-C18 column at a flow rate of 1 ml min−1. A gradient of solvent A (0.1% [vol/vol] HCOOH) and solvent B (CH3CN) was prepared; 100% solvent B was reached after 10 min, and this was held for 2 min (26). The retention time of bacilysin was 4.087 min, as detected by the absorbance at 230 nm and the expected molecular mass of 271 Da (26, 32). The eluate at the corresponding retention time was collected and rerun three times as described above. After lyophilization of the eluates, we obtained 1.81 g liter−1 of pure bacilysin (Fig. 1). Bacilysin in culture broths was determined by the paper-disc agar diffusion assay, and the antibiotic activity was estimated as previously described (37). N-Acetylglucosamine (Sigma), a known specific antagonist of bacilysin/anticapsin activity (38), was used to verify bacilysin activity on bioassay plates.
FIG 1.
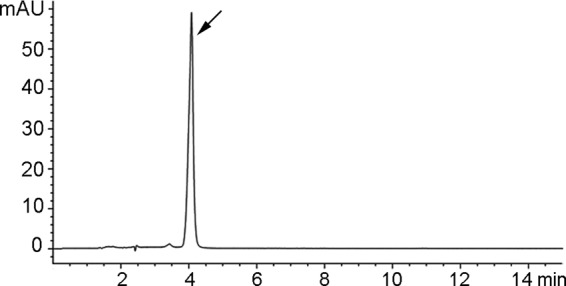
HPLC analysis of bacilysin from B. amyloliquefaciens FZB42. The retention time of bacilysin is 4.087 min.
Bactericidal activity.
Bactericidal activity of Bacillus spp. on the cyanobacteria was investigated as previously reported (39). To obtain culture filtrates, the fermentation broth and PA culture medium of each strain was centrifuged at 12,000 × g for 20 min at 4°C and then filtered through 0.22-μm-pore-size Millipore membranes. Filtrates or bacilysin were inoculated into 30-ml algae cultures at the desired concentrations.
The chlorophyll a content was determined as previously described (40). The cyanobacterial cells incubated in the absence or presence of filtrates or bacilysin were harvested at the indicated times. Cell pellets were resuspended and extracted in 90% acetone for 24 h at 4°C. The samples were centrifuged at 10,000 × g for 10 min to remove cell debris, and then the chlorophyll a concentrations were determined by using the following equation: chlorophyll a concentration (μg liter−1) = (11.47 × OD664) − (0.40 × OD630) (40), where OD664 is the optical density at 664 nm. The bactericidal activity was calculated by using the following equation: bactericidal activity (%) = (1 − T/C) ×100, where T (treatment) and C (control) are the chlorophyll a contents of M. aeruginosa with and without treatment, respectively (41). Small amounts of bacilysin were added into cyanobacterium cultures to obtain five concentration groups (1 to 10 mg liter−1). The median effective concentration (EC50) was calculated by the probit unit method using SPSS 16.0 software (39).
SEM and TEM studies.
To investigate the changes in cell shape and ultrastructure of M. aeruginosa, cells treated with 15 mg liter−1 bacilysin for 2 h were centrifuged at 10,000 × g for 10 min before being washed twice with sodium phosphate buffer (50 mM, pH 7.2). The samples were then prefixed with 2.5% glutaraldehyde. For scanning electron microscopy (SEM) observation, samples were mounted on copper grids, sputter-coated with gold–palladium and examined with a Hitachi S-3000N scanning electron microscope (Hitachi, Tokyo, Japan). For transmission electron microscopy (TEM) observations, prefixed samples were washed three times with phosphate buffer, postfixed with 1% osmium tetroxide for 1 h, dehydrated in a graded series of ethanol solution, embedded in Epon 812, sectioned with an ultramicrotome (LKB-V, Sweden), and observed by using a Hitachi H-600 transmission electron microscope.
Real-time PCR analysis.
For the determination of gene expression, M. aeruginosa was exposed to 4 mg liter−1 bacilysin for 1, 2, and 3 days. After incubation, 10 ml of algal culture was centrifuged at 10,000 × g for 10 min at 4°C to collect the algal cells. Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions. First-strand cDNA was obtained by using reverse transcriptase (TaKaRa Bio) with oligo(T) primer. Real-time PCR was performed with SYBR Premix Ex Taq (TaKaRa Bio) using a 7500 Fast Real-Time PCR detection system. The 16S rRNA gene was used as the internal reference for normalization. Primers for these genes are listed in Table 1.
Statistical analysis.
Each experiment was conducted in at least three independent replications. The data were statistically evaluated by using analysis of variance, followed by a Fisher least significant difference test (P ≤ 0.05) using the SPSS v16.0 software (SPSS, Chicago, IL).
RESULTS
Screening of Bacillus strains for anti-M. aeruginosa activity.
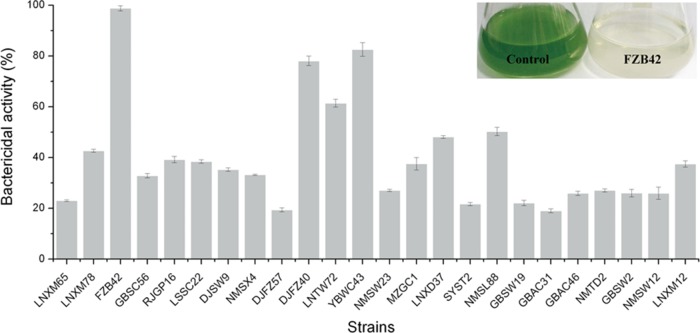
Twenty-four strains of Bacillus spp. were tested in competitive mixed-culture experiments for the ability to suppress the growth of M. aeruginosa. The anticyanobacterial ability was evaluated by measuring the content of chlorophyll a at 7 days. As shown in Fig. 2, all Bacillus strains inhibited the growth of M. aeruginosa to various extents. B. amyloliquefaciens FZB42, Bacillus sp. strain YBWC43 and B. amyloliquefaciens DJFZ40 showed most potent activities, but it was B. amyloliquefaciens FZB42 that displayed the greatest anti-M. aeruginosa activity (killing rate of 98.78%). After treatment with B. amyloliquefaciens FZB42, the water quality was restored (Fig. 2). Thus, these data suggest that metabolites may exist in the culture filtrates that exert anticyanobacterial activities.
FIG 2.
Suppressive activity of Bacillus strains against M. aeruginosa. The inset shows the effect of B. amyloliquefaciens FZB42 against an M. aeruginosa culture after 7 days (right). The control (left) is without B. amyloliquefaciens FZB42. The bactericidal activity was determined as described in Materials and Methods. All error bars represent the standard deviations.
The bactericidal substances against M. aeruginosa are Sfp independent.
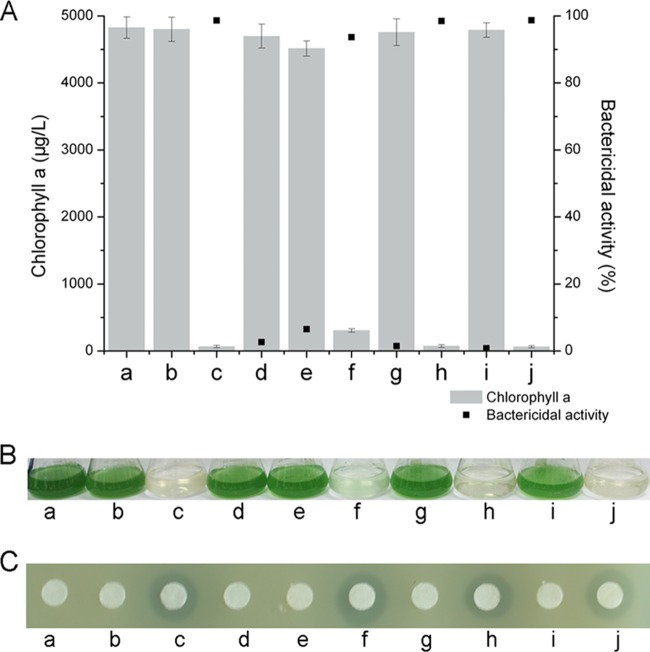
B. amyloliquefaciens FZB42 and other plant-associated strains of this species produce a broad spectrum of non-ribosomally synthesized antimicrobial lipopeptides and polyketides (25, 30). Sfp, a 4′-phosphopantetheinyl transferase, acts as the peptidyl carrier protein, and it is essential for the production of non-ribosomally synthesized lipopeptides and polyketides (22). Therefore, in order to confirm whether non-ribosomally synthesized lipopeptides and polyketides from B. amyloliquefaciens FZB42 were involved in suppressing the growth M. aeruginosa, we used a mutant deficient in the synthesis of Sfp (B. amyloliquefaciens CH03 [30]) to investigate whether this strain would suppress the growth of M. aeruginosa. At 7 days after the M. aeruginosa culture had been treated with B. amyloliquefaciens CH03 culture filtrate, the concentration of chlorophyll a was 245.8 μg liter−1, which corresponded to an inhibitory effect of 94.39%. Thus, there was no difference in growth inhibition caused by B. amyloliquefaciens FZB42 and the CH03 strain. This indicates that nonribosomal lipopeptides and polyketides synthesized through the sfp-dependent pathway are not involved in the suppression of M. aeruginosa growth that and the anticyanobacterial effect of B. amyloliquefaciens FZB42 culture filtrate must be due to other metabolites (Fig. 3A and B).
FIG 3.
Detection of antagonistic action against M. aeruginosa (A and B) and S. aureus by paper disc agar diffusion assay (C). Columns: a, control; b, N-acetylglucosamine (10 mM); c, B. amyloliquefaciens FZB42; d, M436 (random mutant FZB42aroA::TnYLB-1); e, M1125 (random mutant FZB42aroE::TnYLB-1); f, CH03 (site-directed mutant FZB42Δsfp); g, site-directed mutant FZB42ΔbacB; h, complemented FZB42ΔbacBΔamyE::bacB; i, bacilysin supplemented with 10 mM N-acetylglucosamine; j, bacilysin. All error bars represent the standard deviations.
Screening of mutant libraries and identification of anticyanobacterial related genes.
To identify the anticyanobacterial agent, 2,000 TnYLB-1 transposon-inserted mutants were screened after B. amyloliquefaciens FZB42 was transformed with transposon-carrying pMarA. Two mutants, M436 (bactericidal activity 2.65%) and M1125 (bactericidal activity, 6.48%), were unable to inhibit the growth of M. aeruginosa (Fig. 3A and B). Insertion copy analysis by Southern blotting demonstrated that both mutants contained single insertions (data not shown). To identify the insertion sites, the inserted transposon and its flanking regions were cloned by inverse PCR and sequenced. Sequence analysis revealed that the aroA gene of M436 and aroE gene of M1125 had been disrupted by the TnYLB-1 transposon. The aroA gene encodes a bifunctional enzyme consisting of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase and chorismate mutase (42), while the aroE gene encodes 5-enolpyruvylshikimate-3-phosphate synthase (43, 44). Both genes are responsible for aromatic acid biosynthesis in many Bacillus spp. (42). All aro mutants are deficient in bacilysin biosynthesis, and it is known that the bacilysin pathway branches off the aromatic amino acid pathway at prephenate (29, 45, 46). Thus, we hypothesize that it is bacilysin produced by B. amyloliquefaciens FZB42 that has specific bactericidal activity against M. aeruginosa.
Construction of mutants and supplementation with an antagonist.
Bacilysin, a dipeptide consisting of nonproteinogenic l-anticapsin and N-terminal l-alanine, is one of the simplest known peptide antibiotics, and it exhibits antifungal and antibacterial activities (38). In order to confirm the role of bacilysin in the anticyanobacterial activity, we constructed B. amyloliquefaciens FZB42ΔbacB that was deficient in bacilysin synthesis. bacB, the second gene in the bacilysin biosynthetic pathway, encodes an isomerase that catalyzes an allylic isomerization to generate a conjugated dienone (47–49). Furthermore, the B. amyloliquefaciens FZB42ΔbacB mutant was complemented with the entire bacB gene fused to the amyE gene. First, we investigated bacilysin production in the mutants and Staphylococcus aureus was used as an indicator strain because it is sensitive to this antibiotic (29). As shown in Fig. 3C, B. amyloliquefaciens FZB42ΔbacB did not cause an inhibition zone, and M436 and M1125 also showed no antagonistic action, thus indicating that all of these strains were devoid of bacilysin production. The level of bacilysin produced by the complemented transformant corresponded to that of the B. amyloliquefaciens FZB42 wild type. Second, we tested the anticyanobacterial activity of the mutants. Treatment of M. aeruginosa with the complemented transformant resulted in a bactericidal effect similar to that of the wild type, whereas B. amyloliquefaciens FZB42ΔbacB had no inhibitory effect since the bactericidal activity was just 1.45%, and the chlorophyll a content at 7 days was 4,756.5 μg liter−1, which was not significantly different from the control (Fig. 3A and B). On the other hand, bacilysin obtained from B. amyloliquefaciens FZB42 wild-type culture filtrates showed inhibitory effects against S. aureus and M. aeruginosa, while supplementation with N-acetylglucosamine (10 mM), a known specific antagonist of bacilysin, abrogated the growth-inhibitory activities (Fig. 3). These data suggest that bacilysin produced by B. amyloliquefaciens FZB42 has specific bactericidal activity against M. aeruginosa.
Micro- and ultrastructural changes of M. aeruginosa caused by bacilysin.
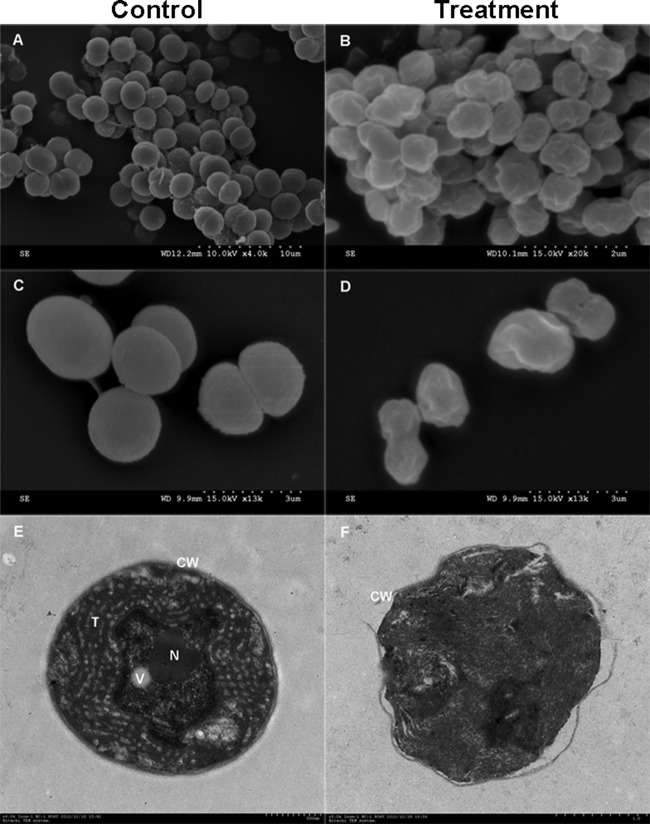
To determine the mechanism of inhibition of bacilysin, M. aeruginosa cells were examined before and after exposure by bacilysin by SEM. As shown in Fig. 4A and C, normal cells were intact, plump, and spherical with smooth exteriors; some cells showed segmentation. After treatment with bacilysin, the majority of cells were obviously depressed or distorted from their normal spherical shape and the integrity of cell wall was damaged (Fig. 4B and D). Moreover, TEM was used to observe changes at the ultrastructural level. In the absence of bacilysin, M. aeruginosa cells possessed an intact and very distinct cell wall and the space between the cell membrane and cell wall was uniform. The cytoplasm enveloped by the plasma membrane contained a large number of thylakoids with regularly scattered phycobilisomes, and there was a distinct nuclear area, vesicles, and other cell organelles (Fig. 4E). Cell damage caused by exposure to bacilysin is illustrated in Fig. 4F. In comparison to the untreated control, there was severe cell damage. The cell wall was partly ruptured, and the cytoplasm was condensed, resulting in slight plasmolysis. Furthermore, there were no thylakoids, and the cells had lost their basic structure.
FIG 4.
Micro- and ultrastructural changes of M. aeruginosa in the presence of 15 mg liter−1 bacilysin for 2 h. (A) Normal M. aeruginosa cells at 10 kV × 4.0 k; (B) damaged M. aeruginosa cells at 15 kV (magnification, 20,000×); (C) normal cells at 15 kV (magnification, 13,000×): (D) damaged cells at 15 kV (magnification, 13,000×); (E) a control M. aeruginosa cell at (magnification, 6,000×); (F) a damaged M. aeruginosa cell (magnification, 5,000×). CW, cell wall; N, nuclear area; T, thylakoid; V, gas vesicle.
Effect on M. aeruginosa gene expression after exposure to bacilysin.
To explore the effects of bacilysin on cyanobacterial gene expression, we assessed the expression of glmS, psbA1, mcyB, and ftsZ in M. aeruginosa at 1, 2, and 3 days. glmS encodes l-glutamine: d-fructose-6-phosphate amidotransferase (known as glucosamine-6-phosphate synthase) that is important for the biosynthesis of peptidoglycan, a component of the bacterial cell wall (50). psbA encodes for the integral membrane protein D1 of photosystem II (51). mcyB encodes McyB, a protein involved in the synthesis of microcystins (52), while ftsZ encodes FtsZ, which is involved in cell division (53).
Real-time PCR analysis revealed that glmS showed a significant decrease in expression at all exposure times (35.3, 19.1, and 9.7% of the control values at day 1, 2, and 3, respectively). Meanwhile, transcript levels of psbA1, mcyB, and ftsZ were slightly downregulated after 1 day, while expression was reduced significantly after 2- and 3-day exposures (Fig. 5).
FIG 5.
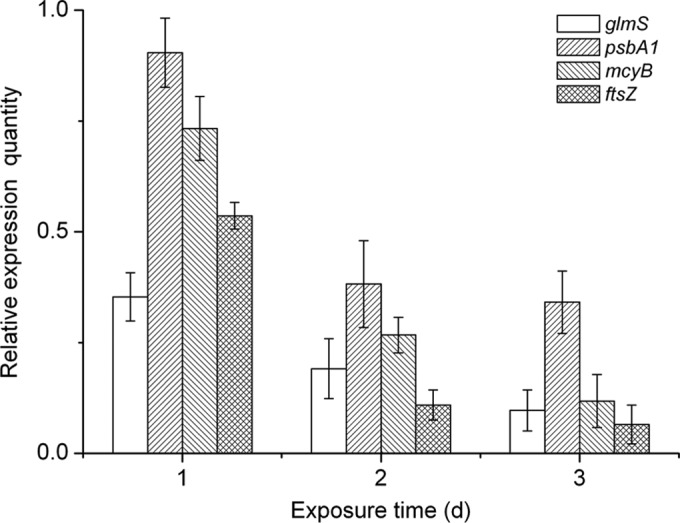
Real-time PCR analysis of expression of glmS, psbA1, mcyB, and ftsZ in M. aeruginosa cells in response to 4 mg liter−1 bacilysin for 1, 2 and 3 days. Values were normalized to the levels of the 16S rRNA gene, an internal reference gene. The y axis values represent the mean expression ± the standard deviations (n = 3) relative to the control.
Bacilysin has potent bactericidal activity against other harmful algal species.
To investigate whether bacilysin showed bactericidal activity against other harmful algal species, we examined effects against Aphanizomenon flos-aquae, Nostoc sp., and Anabaena sp. As shown in Table 2, bacilysin had potent growth inhibitory effects against each of these species. The chlorophyll a contents for treated cultures and controls at 7 days ranged from 103.5 to 591.3 μg liter−1 and 2.58 to 13.97%, respectively. The color of the cultures changed from blue-green to turbid white or pale green, and the bactericidal activity was >85%, with an EC50 of <5.0 mg liter−1.
TABLE 2.
Bactericidal activity of bacilysin against various harmful algal bloom speciesa
| Algal species | Mean amt of chlorophyll a (μg liter−1) ± SD at day 7 |
Bactericidal activity (%) | EC50 (mg liter−1) | |
|---|---|---|---|---|
| Control | Treatment | |||
| Anabaena sp. | 4,234.1 ± 154.8 | 591.3 ± 34.7 | 86.04 | 5.01 |
| A. flos-aquae | 4,012.4 ± 201.3 | 103.5 ± 15.3 | 97.43 | 4.29 |
| Nostoc sp. | 4,893.5 ± 161.7 | 229.4 ± 21.4 | 95.32 | 4.51 |
Values are means from five independent experiments.
DISCUSSION
Harmful algal blooms have increased throughout the world, and these have caused serious problems in recent decades, such as the loss of aquaculture industries, environmental pollution, and damage to human health (54). Previous reports have demonstrated that many bacteria have a significant algicidal effect on several harmful algal bloom species (10, 11, 14, 17). Most algicidal bacteria isolated from the environment are characterized as belonging to the genera Cytophaga, Saprospira, Pseudoalteromonas, and Alteromonas (55). Recently, other studies have confirmed that some species of Bacillus, such as B. subtilis and B. fusiformis, can inhibit the growth of M. aeruginosa (18, 19). In the present study, we showed that 24 Bacillus strains exerted bactericidal activity against M. aeruginosa. Among these, B. amyloliquefaciens FZB42 exhibited the strongest suppressive effect, with a killing rate of 98.78%.
In B. amyloliquefaciens FZB42, numerous gene clusters are devoted to the nonribosomal synthesis of secondary metabolites that are dependent on the Sfp phosphopantetheinyl transferase (25). Four types of these metabolites, surfactin and polyketides (difficidin, macrolactin, and bacillaene), are known for their antibacterial activity (24, 30). So, as a first step to determine whether these compounds were involved in the antagonistic effects on cyanobacteria, we used an Sfp mutant strain of B. amyloliquefaciens FZB42 (CH03), which is unable to synthesize antibacterial surfactin and polyketides. Surprisingly, the suppressive activity against M. aeruginosa was almost unaffected, suggesting that metabolites other than surfactin and polyketides were involved in the antagonistic activity. To further identify the substance, we prepared a mutant library and performed site-specific mutagenesis. The results demonstrated that bacilysin, produced independently of Sfp, was the bactericidal substance produced by B. amyloliquefaciens FZB42 that acted against harmful algal bloom species.
Bacilysin (l-alanyl-[2,3-epoxycyclohexanone-4]-l-alanine) is a dipeptide antibiotic that contains an l-alanine residue and the nonproteinogenic amino acid l-anticapsin. This nonribosomal dipeptide is synthesized by the bacABCDEFG gene cluster and generated independently of the Sfp pathway (32, 47–49). Many studies have demonstrated that bacilysin is active against a wide range of bacteria (38). Bacilysin, together with difficidin produced by plant-associated B. amyloliquefaciens, is efficient for controlling fire blight disease caused by Erwinia amylovora (26). Bacilysin also exhibits a certain antifungal activity against the yeast (38). Although bacilysin is antimicrobial, its inhibitory activity against harmful algal bloom species has not been reported previously. In the present study, we demonstrated that bacilysin significantly inhibited the growth of M. aeruginosa with an EC50 of 4.13 mg liter−1. Moreover, bacilysin was shown to exert specific bactericidal activity against A. flos-aquae, Nostoc sp., and Anabaena sp. Compared to other algicidal compounds, bacilysin exerts similar potency of anti-algal effects (Table 3). Thus, our results suggest that bacilysin not only acts as a bactericide but also possesses significant inhibitory effects against cyanobacteria and microalgae.
TABLE 3.
Comparison of the activity of bacilysin to the activities of other algicidal compounds
| Algicidal compound | Target species | Initial algae density | EC50 (mg liter−1) | Source or reference |
|---|---|---|---|---|
| Bacilysin | M. aeruginosa | 106 | 4.13 | This study |
| Artemisinin | M. aeruginosa | 106 | 3.2 | 39 |
| Sinapic acid | M. aeruginosa | 104–105 | 4.9 | 56 |
| Copper sulfate | M. aeruginosa | 106 | 0.3 | 57 |
| Ginkgolic acids | M. aeruginosa | 107 | 2.03 | 58 |
| Rhamnolipid | P. dentatum | 104–105 | 5.00 | 59 |
| HPA3 | P. minimum | 104 | 16 μM | 60 |
Numerous studies on the mode of action of bacilysin have demonstrated that its antibacterial activity depends on the anticapsin moiety, which is released by an intracellular peptidase (61) after bacilysin is taken up into susceptible cells by a distinct peptide permease system (62). Anticapsin behaves as a glutamine analogue, and reaction of its epoxide group with a thiol group of glucosamine synthase results in its covalent linkage to the enzyme, thus blocking its function and therefore bacterial peptidoglycan or fungal mannoprotein biosynthesis. This inhibition leads to cell protoplasting and lysis (49, 61, 63–65). In the present study, the mechanism of action of bacilysin against M. aeruginosa was clarified. We found that bacilysin produced by B. amyloliquefaciens FZB42 primarily affected the cell wall, as evidenced by microscopic and ultramicroscopic observations. The fast breakdown of the cell wall and plasma membrane led to increased cell permeability and the efflux of intracellular components (see Fig. S1 and S2 in the supplemental material). In addition, the transcript levels in M. aeruginosa of four target genes (glmS, psbA1, mcyB, and ftsZ) involved in the synthesis of peptidoglycan, the photosynthesis system, microcystins, and cell division were downregulated, which suggests that the metabolism of M. aeruginosa was significantly inhibited by bacilysin. A similar phenomenon was observed that ginkgolic acids extracted from Ginkgo biloba exocarp cause pleiotropic effects on M. aeruginosa such as destruction of the cellular structure, induction of oxidative damage, and reduced photosynthesis (58).
In conclusion, the present study provides direct evidence for the remarkable anticyanobacterial effects of bacilysin produced by B. amyloliquefaciens FZB42 and the mechanism by which this compound acts against algal cell walls. These results might provide an alternative method for managing harmful algal blooms, although further research has to confirm whether bacilysin could be applied safely to eutrophic lakes and reservoirs.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from Agro-Scientific Research in the Public Interest (20130315), the Special Fund for the Fundamental Research Funds for the Central Universities (KYZ201404), the National Natural Science Foundation of China (31100056 and 31471811), the Doctoral Fund of Ministry of Education of China (20100097120011), and the National High-Tech R&D Program of China (2012AA101504).
Footnotes
Published ahead of print 26 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02605-14.
REFERENCES
- 1.Smith VH, Schindler DW. 2009. Eutrophication science: where do we go from here? Trends Ecol. Evol. 24:201–207. 10.1016/j.tree.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Dittmann E, Wiegand C. 2006. Cyanobacterial toxins: occurrence, biosynthesis, and impact on human affairs. Mol. Nutr. Food Res. 50:7–17. 10.1002/mnfr.200500162. [DOI] [PubMed] [Google Scholar]
- 3.Lürling M, Faassen EJ. 2013. Dog poisoning associated with a Microcystis aeruginosa bloom in the Netherlands. Toxins 5:556–567. 10.3390/toxins5030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan X, Kong FX, Yu Y, Zhang M. 2009. Spatio-temporal variations of phytoplankton community composition assayed by morphological observation and photosynthetic pigment analyses in Lake Taihu (China). Afr. J. Biotechnol. 8:4977–4982. [Google Scholar]
- 5.Na GH, Choi WJ, Chun YY. 1996. A study on red tide control with loess suspension. Kor. J. Aquacult. 9:239–245. [Google Scholar]
- 6.Sun XX, Choi JK, Kim EK. 2004. A preliminary study on the mechanism of harmful algal bloom mitigation by use of sophorolipid treatment. J. Exp. Mar. Biol. Ecol. 304:35–49. 10.1016/j.jembe.2003.11.020. [DOI] [Google Scholar]
- 7.Steidinger KA. 1983. A re-evaluation of toxic dinoflagellate biology and ecology. Prog. Phycol. Res. 2:147–188. [Google Scholar]
- 8.Ferrier MD, Butler BR, Terlizzi DE, Lacouture RV. 2005. The effects of barley straw (Hordeum vulgare) on the growth of freshwater algae. Bioresour. Technol. 96:1788–1795. 10.1016/j.biortech.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Kim YS, Lee DS, Jeong SY, Lee WJ, Lee MS. 2009. Isolation and characterization of a marine algicidal bacterium against the harmful Raphidophyceae Chattonella marina. J. Microbiol. 47:9–18. 10.1007/s12275-008-0141-z. [DOI] [PubMed] [Google Scholar]
- 10.Shilo M. 1970. Lysis of blue-green algae by Myxobacter. J. Bacteriol. 104:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai I, Ishida Y, Hata Y. 1993. Killing of marine phytoplankton by a gliding bacterium Cytophaga sp., isolated from the coastal sea of Japan. Mar. Biol. 116:527–532. 10.1007/BF00355470. [DOI] [Google Scholar]
- 12.Mitsutani A, Takesue K, Kirita M, Ishida Y. 1992. Lysis of Skeletonema costatum by Cytophaga sp. isolated from the coastal water of the Ariake Sea. Nippon Suisan Gakkaishi 58:2159–2169. 10.2331/suisan.58.2159. [DOI] [Google Scholar]
- 13.Furusawa G, Yoshikawa T, Yasuda A, Sakata T. 2003. Algicidal activity and gliding motility of Saprospira sp. SS98-5. Can. J. Microbiol. 49:92–100. 10.1139/w03-017. [DOI] [PubMed] [Google Scholar]
- 14.Dakhama A, De la Noüe J, Lavoie MC. 1993. Isolation and identification of antialgal substances produced by Pseudomonas aeruginosa. J. Appl. Phycol. 5:297–306. 10.1007/BF02186232. [DOI] [Google Scholar]
- 15.Kodani S, Imoto A, Mitsutani A, Murakami M. 2002. Isolation and identification of the antialgal compound, harmane (1-methyl-β-carboline), produced by the algicidal bacterium, Pseudomonas sp. K44-1. J. Appl. Phycol. 14:109–114. 10.1023/A:1019533414018. [DOI] [Google Scholar]
- 16.Yoshikawa K, Adachi K, Nishijima M, Takadera T, Tamaki S, Harada KI. 2000. β-Cyanoalanine production by marine bacteria on cyanide-free medium and its specific inhibitory activity toward cyanobacteria. Appl. Environ. Microbiol. 66:718–722. 10.1128/AEM.66.2.718-722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riclea R, Gleitzmann J, Bruns H, Junker C, Schulz B, Dickschat JS. 2012. Algicidal lactones from the marine Roseobacter clade bacterium Ruegeria pomeroyi. Beilstein J. Org Chem. 8:941–950. 10.3762/bjoc.8.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn CY, Joung SH, Jeon JW, Kim HS, Yoon BD, Oh HM. 2003. Selective control of cyanobacteria by surfactin-containing culture broth of Bacillus subtilis C1. Biotechnol. Lett. 25:1137–1142. 10.1023/A:1024508927361. [DOI] [PubMed] [Google Scholar]
- 19.Mu RM, Fan ZQ, Pei HY, Yuan XL, Liu SX, Wang XR. 2007. Isolation and algae-lysing characteristics of the algicidal bacterium B5. J. Environ. Sci. 19:1336–1340. 10.1016/S1001-0742(07)60218-6. [DOI] [PubMed] [Google Scholar]
- 20.Brannen P, Kenney DS. 1997. Kodiak®—a successful biological-control product for suppression of soil-borne plant pathogens of cotton. J. Ind. Microbiol. Biotechnol. 19:169–171. 10.1038/sj.jim.2900439. [DOI] [Google Scholar]
- 21.Ngugi H, Dedej S, Delaplane K, Savelle A, Scherm H. 2005. Effect of flower-applied Serenade biofungicide (Bacillus subtilis) on pollination related variables in rabbiteye blueberry. Biol. Control 33:32–38. 10.1016/j.biocontrol.2005.01.002. [DOI] [Google Scholar]
- 22.Mootz HD, Finking R, Marahiel MA. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289–37298. 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- 23.Thimon L, Peypoux F, Maget-Dana R, Roux B, Michel G. 1992. Interactions of bioactive lipopeptides, iturin A and surfactin from Bacillus subtilis. Biotechnol. Appl. Biochem. 16:144–151. [PubMed] [Google Scholar]
- 24.Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084–1096. 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R. 2009. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J. Biotechnol. 140:27–37. 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Chen XH, Scholz R, Borriss M, Junge H, Mögel G, Kunz S, Borriss R. 2009. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 140:38–44. 10.1016/j.jbiotec.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Budiharjo A, Wang P, Shi H, Fang J, Borriss R, Zhang K, Huang X. 2013. The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Appl. Microbiol. Biotechnol. 97:10081–10090. 10.1007/s00253-013-5247-5. [DOI] [PubMed] [Google Scholar]
- 28.Scholz R, Molohon KJ, Nachtigall J, Vater J, Markley AL, Süssmuth RD, Mitchell DA, Borriss R. 2011. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J. Bacteriol. 193:215–224. 10.1128/JB.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilton MD, Alaeddinoglu NG, Demain AL. 1988. Bacillus subtilis mutant deficient in the ability to produce the dipeptide antibiotic bacilysin: isolation and mapping of the mutation. J. Bacteriol. 170:1018–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, Gottschalk G, Süssmuth RD, Borriss R. 2006. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB42. J. Bacteriol. 188:4024–4036. 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landy M, Warren GH, Rosenman SB, Colio LG. 1948. Bacillomycin: an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67:539–541. 10.3181/00379727-67-16367. [DOI] [PubMed] [Google Scholar]
- 32.Parker JB, Walsh CT. 2013. Action and timing of BacC and BacD in the late stages of biosynthesis of the dipeptide antibiotic bacilysin. Biochemistry 52:889–901. 10.1021/bi3016229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong Y, Hu HY, Xie X, Li FM. 2008. Responses of enzymatic antioxidants and non-enzymatic antioxidants in the cyanobacterium Microcystis aeruginosa to the allelochemical ethyl 2-methyl acetoacetate (EMA) isolated from reed (Phragmites communis). J. Plant Physiol. 165:1264–1273. 10.1016/j.jplph.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 35.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:1072–1078. 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breton YL, Mohapatra NP, Haldenwang WG. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl. Environ. Microbiol. 72:327–333. 10.1128/AEM.72.1.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Özcengiz G, Alaeddinoglu NG. 1991. Bacilysin production and sporulation in Bacillus subtilis. Curr. Microbiol. 23:61–64. 10.1007/BF02092250. [DOI] [PubMed] [Google Scholar]
- 38.Kenig M, Abraham E. 1976. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 94:37–45. 10.1099/00221287-94-1-37. [DOI] [PubMed] [Google Scholar]
- 39.Ni L, Acharya K, Hao X, Li S. 2012. Isolation and identification of an anti-algal compound from Artemisia annua and mechanisms of inhibitory effect on algae. Chemosphere 24:387–392. 10.1016/j.chemosphere.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Jeffrey SW, Humphrey GF. 1975. New spectrophotometric equations for determining chlorophylls a, b, c1, and c2 in higher plants, algae, and natural phytoplankton. Biochem. Physiol. Pflanzen. 167:191–194. [Google Scholar]
- 41.Kim JD, Kim B, Lee CG. 2007. Alga-lytic of Pseudomonas fluorescens against the red tide causing marine alga Heterosigma akashiwo (Raphidophyceae). Biol. Control 41:296–303. 10.1016/j.biocontrol.2007.02.010. [DOI] [Google Scholar]
- 42.Hoch JA, Nester EW. 1973. Gene-enzyme relationships of aromatic acid biosynthesis in Bacillus subtilis. J. Bacteriol. 116:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian YS, Xu J, Han J, Zhao W, Fu XY, Peng RH, Yao QH. 2013. Complementary screening, identification, and application of a novel class II 5-enopyruvylshikimate-3-phosphate synthase from Bacillus cereus. World J. Microbiol. Biotechnol. 29:549–557. 10.1007/s11274-012-1209-9. [DOI] [PubMed] [Google Scholar]
- 44.Vallier H, Welker NE. 1990. Genetic map of the Bacillus stearothermophilus NUB36 chromosome. J. Bacteriol. 172:793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilton MD, Alaeddinoglu NG, Demain AL. 1988. Synthesis of bacilysin by Bacillus subtilis branches from prephenate of the aromatic amino acid pathway. J. Bacteriol. 170:482–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roscoe J, Abraham EP. 1966. Experiments relating to the biosynthesis of bacilysin. Biochem. J. 99:793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker JB, Walsh CT. 2012. Olefin isomerization regiochemistries during tandem action of BacA and BacB on prephenate in bacilysin biosynthesis. Biochemistry 51:3241–3251. 10.1021/bi300254u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker JB, Walsh CT. 2012. Stereochemical outcome at four stereogenic centers during conversion of prephenate totetrahydrotyrosine by BacABGF in the bacilysin pathway. Biochemistry 51:5622–5632. 10.1021/bi3006362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinborn G, Hajirezaei M, Hofemeister J. 2005. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 183:71–79. 10.1007/s00203-004-0743-8. [DOI] [PubMed] [Google Scholar]
- 50.Wojciechowski M, Milewski S, Mazerski J, Borowski E. 2005. Glucosamine-6-phosphate synthase, a novel target for antifungal agents: molecular modeling studies in drug design. Acta Biochim. Pol. 52:647–653. [PubMed] [Google Scholar]
- 51.Aro EM, Virgin I, Andersson B. 1993. Photoinhibition of photosystem. II. Inactivation, protein damage, and turnover. Biochim. Biophys. Acta 1143:113–134. [DOI] [PubMed] [Google Scholar]
- 52.Pearson LA, Neilan BA. 2008. The molecular genetics of cyanobacterial toxicity as a basis for monitoring water quality and public health risk. Curr. Opin. Biotechnol. 19:281–288. 10.1016/j.copbio.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Mazouni K, Domain F, Cassier-Chauvat C, Chauvat F. 2004. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN, and MinCDE. Mol. Microbiol. 52:1145–1158. 10.1111/j.1365-2958.2004.04042.x. [DOI] [PubMed] [Google Scholar]
- 54.Edvardsen B, Imai I. 2006. The ecology of harmful flagellates within Prymnesiophyceae and Raphidophyceae. Ecol. Harmful Algae 189:67–79. 10.1007/978-3-540-32210-8_6. [DOI] [Google Scholar]
- 55.Wang B, Yang XR, Lu J, Zhou Y, Su J, Tian Y, Zhang J, Wang GZ, Zheng TL. 2012. A marine bacterium producing protein with algicidal activity against Alexandrium tamarense. Harmful Algae 13:83–88. 10.1016/j.hal.2011.10.006. [DOI] [Google Scholar]
- 56.Nakai S, Inoue Y, Hosomi M. 2001. Algal growth inhibition effects and inducement modes by plant-producing phenols. Water Res. 35:1855–1859. 10.1016/S0043-1354(00)00444-9. [DOI] [PubMed] [Google Scholar]
- 57.Hadjoudja S, Vignoles C, Deluchat V, Lenain JF, Le Jeune AH, Baudu M. 2009. Short-term copper toxicity on Microcystis aeruginosa and Chlorella vulgaris using flow cytometry. Aquat. Toxicol. 94:255–264. 10.1016/j.aquatox.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 58.Zhang C, Ling F, Yi YL, Zhang HY, Wang GX. 2014. Algicidal activity and potential mechanisms of ginkgolic acids isolated from Ginkgo biloba exocarp on Microcystis aeruginosa. J. Appl. Phycol. 26:323–332. 10.1007/s10811-013-0057-9. [DOI] [Google Scholar]
- 59.Wang XL, Gong LY, Liang SK, Han XR, Zhu CJ, Li YB. 2005. Algicidal activity of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa. Harmful Algae 4:433–443. 10.1016/j.hal.2004.06.001. [DOI] [Google Scholar]
- 60.Park SC, Lee JK, Kim SW, Park Y. 2011. Selective algicidal action of peptides against harmful algal bloom species. PLoS One 6:26727–26733. 10.1371/journal.pone.0026733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenig M, Vandamme E, Abraham EP. 1976. The mode of action of bacilysin and anticapsin and biochemical properties of bacilysin-resistant mutants. J. Gen. Microbiol. 94:46–54. 10.1099/00221287-94-1-46. [DOI] [PubMed] [Google Scholar]
- 62.Perry D, Abraham EP. 1979. Transport and metabolism of bacilysin and other peptides by suspensions of Staphylococcus aureus. J. Gen. Microbiol. 115:213–221. 10.1099/00221287-115-1-213. [DOI] [PubMed] [Google Scholar]
- 63.Chmara H. 1985. Inhibition of glucosamine synthase by bacilysin and anticapsin. J. Gen. Microbiol. 131:265–271. [DOI] [PubMed] [Google Scholar]
- 64.Milewski S. 1993. Chemical modification studies of the active site of glucosamine-6-phosphate synthase from baker's yeast. Biochim. Biophys. Acta 1161:279–284. 10.1016/0167-4838(93)90225-G. [DOI] [PubMed] [Google Scholar]
- 65.Whitney JG, Funderburk SS, Westhead JE, Lively DH, Solenberg JM, Denney JW. 1972. Anticapsin, a new biologically active metabolite: screening and assay procedures. Appl. Microbiol. 24:907–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.