Abstract
2,4-Dinitroanisole (DNAN) is an insensitive munition ingredient used in explosive formulations as a replacement for 2,4,6-trinitrotoluene (TNT). Little is known about the environmental behavior of DNAN. There are reports of microbial transformation to dead-end products, but no bacteria with complete biodegradation capability have been reported. Nocardioides sp. strain JS1661 was isolated from activated sludge based on its ability to grow on DNAN as the sole source of carbon and energy. Enzyme assays indicated that the first reaction involves hydrolytic release of methanol to form 2,4-dinitrophenol (2,4-DNP). Growth yield and enzyme assays indicated that 2,4-DNP underwent subsequent degradation by a previously established pathway involving formation of a hydride-Meisenheimer complex and release of nitrite. Identification of the genes encoding the key enzymes suggested recent evolution of the pathway by recruitment of a novel hydrolase to extend the well-characterized 2,4-DNP pathway.
INTRODUCTION
DNAN (2,4-dinitroanisole) is one of the insensitive nitroaromatic ingredients increasingly used as a replacement for 2,4,6-trinitrotoluene (TNT) in munitions. DNAN or its metabolites can be toxic to earthworms, bacteria, algae, and plants (1, 2). Therefore, the release of DNAN to the environment could pose ecological and health risks. There is little information about the environmental behavior of DNAN (2), and no bacteria capable of complete biodegradation have been reported.
The initial reaction in the biotransformation of DNAN by bacteria and in abiotic transformation with zero valent iron is the reduction of the nitro group in the ortho position to yield 2-amino-4-nitroanisole (3–5). Under anoxic conditions, DNAN is biotransformed to toxic metabolites such as diaminoanisole (3, 5–8). A Bacillus strain was reported to transform DNAN slowly under aerobic conditions to 2-amino-4-nitroanisole as a predominant dead-end product (4). A recent investigation revealed substantial aerobic biodegradation of DNAN by enrichment cultures derived from activated sludge, but the responsible bacteria were not isolated (9). During alkaline hydrolysis, DNAN is converted to 2,4-dinitrophenolate via an unstable hydride-Meisenheimer complex (10). Phototransformation of DNAN resulted in 2-hydroxy-4-nitroanisole and 2,4-dinitrophenol (2,4-DNP) as major and minor products, respectively (5, 11). The pathway of 2,4-DNP biodegradation under aerobic conditions is well-known, and the genes involved have been characterized for Rhodococcus erythropolis and Nocardia (12, 13). A Rhodococcus sp. has been reported to degrade 4-nitroanisole by a pathway involving removal of the methyl group and subsequent degradation of the resulting 4-nitrophenol via 4-nitrocatechol and 1,2,4-trihydroxybenzene (14). Biotransformation of DNAN to 2,4-DNP has been reported in mammals (15).
We isolated bacteria able to grow on DNAN as the sole carbon source under aerobic conditions and elucidated the catabolic pathway. An initial O-demethylation catalyzed by a hydrolase was followed by degradation of the resultant 2,4-dinitrophenol by a pathway involving formation of a hydride-Meisenheimer complex (16, 17).
MATERIALS AND METHODS
Isolation of DNAN-degrading bacteria.
An activated sludge sample from Holston Army Ammunition Plant was inoculated (20% [vol/vol]) into 1/4-strength minimal salts medium (MSB) (18) (pH 6.5) containing 2,4-dinitroanisole (DNAN) (100 μM), and the suspension was incubated at 30°C with shaking. Following the disappearance of DNAN as monitored by high-performance liquid chromatography (HPLC) (see below), samples (20% [vol/vol]) were repeatedly transferred into fresh medium and then spread on MSB agar (1.5%) plates containing DNAN (100 μM). Individual colonies that appeared after 4 days of incubation were tested for the ability to degrade DNAN in carbon- and nitrogen-free MSB. Isolates that used DNAN as the sole source of carbon, nitrogen, and energy were selected for further study. 16S rRNA gene analysis was used for identification of the strains from the draft genome sequences obtained by using Illumina sequencing technologies (Oregon State University). The genomes were assembled using the A5 pipeline (19) and annotated by Rapid Annotation using Subsystem Technology (RAST) (20).
Growth of DNAN-degrading isolate.
The JS1661 isolate was grown with shaking at 30°C in MSB liquid medium (pH 6.5) containing ammonium sulfate (1.9 mM) as an additional nitrogen source. When large amounts of biomass were required, the strain was grown in MSB containing DNAN (100 μM) and sodium acetate (10 mM) or in 1/4-strength Trypticase soy broth. To determine growth yields, the MSB medium was supplemented with either DNAN (200 μM) or 2,4-dinitrophenol (2,4-DNP) (200 μM) and inoculated to an optical density at 600 nm (OD600) of 0.004 with cultures pregrown on DNAN (100 μM). Biomass was quantified by measuring protein 6 h after the disappearance of 2,4-DNP. Controls with DNAN or 2,4-DNP were inoculated with autoclaved cells. One-milliliter samples were centrifuged, and the cell pellets were suspended in NaOH (0.1 N), lysed by heating at 95°C for 10 min, and assayed for protein as indicated below. Specific activities of cells grown on DNAN or 2,4-DNP were determined using induced and noninduced cells. Induced cells were grown in MSB supplemented with DNAN or 2,4-DNP (100 μM) as the sole carbon and energy source. Noninduced cells were grown in MSB supplemented with sodium acetate (10 mM). Cells were harvested during logarithmic growth, washed twice with MSB, and suspended to an OD600 of 0.004 (corresponding to a protein concentration of about 6 and 8.5 μg ml−1 for cells grown in acetate and DNAN or 2,4-DNP, respectively) in MSB supplemented with DNAN (100 μM) or 2,4-DNP (100 μM) and incubated at 30°C. Disappearance of DNAN or 2,4-DNP was monitored at appropriate intervals by HPLC.
Enzyme assays.
Cells were grown for 48 h in MSB containing DNAN (100 μM) or 2,4-DNP (100 μM) supplemented with sodium acetate (10 mM). Additional DNAN or 2,4-DNP was added after complete degradation of the initial nitro compound. Immediately following disappearance of the second addition of DNAN or 2,4-DNP, cells were harvested by centrifugation, washed twice with phosphate buffer (pH 7.0, 20 mM), and suspended in the same buffer. Cells were broken by two passages through a French pressure cell at 10,000 lb/in2. The lysates were clarified by centrifugation at 100,000 × g for 30 min at 4°C. For some of the experiments, the resulting cell lysates were either ultrafiltered using Microcon centrifugal filters (10 kDa) (Millipore, MA, USA) or dialyzed overnight using Slide-A-Lyzer dialysis membranes (10 kDa) (Pierce, IL, USA) against potassium phosphate buffer (pH 7.0, 20 mM) at 4°C to remove the cofactors.
Demethylase assays were performed in 3-ml reaction mixtures in phosphate buffer (pH 7.0, 20 mM) containing DNAN (100 μM) and cell extract (0.3 to 0.5 mg of protein). At appropriate intervals, samples were removed, and the reactions were stopped by addition of trifluoroacetic acid (TFA) (0.5%). Precipitated proteins were removed by centrifugation, and the supernatant was analyzed by HPLC as indicated below. Oxygen uptake was measured in 1.8-ml reaction mixtures in phosphate buffer (pH 7.0, 20 mM) containing DNAN (100 μM) and cell extract (0.3 to 0.5 mg of protein). Boiled lysates were used in controls.
Concentrations of methanol were determined in 1-ml reaction mixtures by an indirect method. Briefly, after complete transformation of DNAN to 2,4-DNP by dialyzed cells in 2-ml sealed HPLC vials, methanol was converted to formaldehyde by incubation with alcohol oxidase (0.5 U) for 15 min as described by Klavons and Bennett (21). The reactions were stopped by the addition of TFA (0.5%) to avoid further conversion of formaldehyde to formic acid by alcohol oxidase. Formaldehyde was then derivatized with 2,4-pentanedione for 15 min at 58°C in the presence of ammonium acetate and glacial acetic acid as described by Summers (22). The derivative was analyzed by HPLC as indicated below.
Hydride transferase activity was assayed in 3-ml reaction mixtures as described by Behrend and Heesche-Wagner (17). Briefly, the reaction mixtures contained potassium phosphate (pH 7.0, 20 mM), 2,4-DNP (100 μM), NADPH (200 μM), and cell extract (0.3 to 0.5 mg of protein). The reaction mixtures were incubated at 30°C. At appropriate time intervals, samples were removed, and the reactions were stopped by the addition of TFA (0.5%). The acidified reaction mixtures were clarified by centrifugation and depletion of 2,4-DNP analyzed by HPLC.
Purification of DNAN demethylase.
JS1661 cells were grown in Trypticase soy broth, harvested by centrifugation, washed with 0.02 M phosphate buffer (pH 7), and lysed with a Micro-Fluidics M-110P homogenizer at 20,000 lb/in2. After centrifugation to remove unbroken cells, proteins were precipitated with ammonium sulfate. The 30 to 45% ammonium sulfate fraction was dissolved in phosphate buffer and desalted on a Hi Prep 26/10 desalting column (GE Healthcare), the resulting mixture was applied to a Fractogel DEAE column, and proteins were eluted with a 0 to 1 M NaCl gradient in phosphate buffer (pH 7). Active fractions were combined, concentrated by membrane filtration (10-kDa cutoff), applied to a Superdex 200 gel filtration column (130 ml), and eluted with 20 mM Tris (pH 7.5, 150 mM NaCl). Active fractions were combined and applied to a Source 15Q column, and proteins were eluted with a 0 to 1 M NaCl gradient in 20 mM Tris (pH 7.5). Pooled active fractions were mixed with ammonium sulfate to a final concentration of 2.0 mM, applied to a Source 15 PHE column and eluted with a 3 to 0 M gradient of ammonium sulfate. All purification steps were carried out at 4°C. During purification, DNAN hydrolase activity was measured by monitoring the release of 2,4-DNP spectrophotometrically at 400 nm in the reaction mixture described above.
Purified proteins were separated by SDS-PAGE, and bands were excised from the gel. After digestion with trypsin, peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an Agilent 1100 capillary liquid chromatography system with an Agilent XCT ion trap mass spectrometer. Mass spectral data resulting from the tryptic digests were matched with a custom sequence database of translations of all open reading frames (ORFs) (>150 bp) in the genome of strain JS1661 using Agilent's Spectrum Mill software (revision A.03.02.060). In-gel digestions, peptide sequencing, and database matching were performed as described previously (23).
Analytical methods.
Concentrations of DNAN, 2,4-DNP, and the hydride-Meisenheimer complex of 2,4-DNP were determined using an Agilent 1100 HPLC system with a Merck Chromolith C18 reverse-phase column (4.6 mm by 100 mm; 5 μm). The mobile phase for DNAN or 2,4-DNP consisted of 95% water and 5% acetonitrile with TFA (12.7 mM), delivered at a flow rate of 1.5 ml min−1 over a period of 8 min. The hydride-Meisenheimer complex of 2,4-DNP was analyzed as described by Blasco et al. (24) using the same HPLC column as described above but with a mobile phase of 98% phosphate buffer (pH 7.0, 20 mM) without TFA and 2% acetonitrile with TFA (0.13 mM) delivered at a flow rate of 1.5 ml min−1. The formaldehyde derivative (3,5-diacetyl-1,4-dihydrolutidine) was analyzed on a Zorbax ODS C18 reverse-phase column (4.6 mm by 100 mm; 5 μm) with a mobile phase of 60% water and 40% acetonitrile with TFA (10.3 mM) at a flow rate of 0.5 ml min−1. DNAN was monitored at 298 nm (retention time [RT], 5.7 min), 2,4-DNP at 260 nm (RT, 4 min in water-acetonitrile and 3.1 min in phosphate buffer), hydride-Meisenheimer complex at 400 nm (RT, 1.5 min), and 3,5-diacetyl-1,4-dihydrolutidine at 410 nm (RT, 2.1 min). Oxygen uptake was measured polarographically using a Clark-type oxygen electrode and a YSI model 5300 oxygen monitor. Nitrite concentrations were determined colorimetrically as described previously (25). Protein assays were done using a bicinchoninic acid (BCA) assay reagent kit (Rockford, IL, USA). Samples were centrifuged for 5 min at 16,000 × g after the addition of BCA reagents and then analyzed spectrophotometrically. All experiments were performed in biological duplicate.
Chemicals.
DNAN was from Alfa Aesar (Ward Hill, MA, USA), and 2,4-DNP, formaldehyde, and methanol were from Sigma-Aldrich (St, Louis, MO, USA). The hydride-Meisenheimer complex of 2,4-DNP was chemically synthesized as described by Behrend and Heesche-Wagner (17).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of Nocardioides sp. strain JS1661 and JS1660 were deposited in GenBank with accession numbers KM026539 and KM026540, respectively. Catabolic gene clusters involved in DNAN demethylase and 2,4-DNP degradation were deposited in GenBank with accession numbers KM213001 and KM189438, respectively.
RESULTS
Isolation and identification of DNAN-degrading strains.
Enrichment cultures yielded two bacterial isolates able to grow on DNAN as the sole source of carbon, nitrogen, and energy. 16S rRNA gene sequence analysis indicated that one of the isolates, designated Nocardioides sp. strain JS1661, was most closely related (98% sequence identity) to Nocardioides nitrophenolicus (26). The second isolate, designated Nocardioides sp. strain JS1660 was most closely related (98% sequence identity) to Nocardioides oleivorans (27). The two strains behaved similarly in growth experiments except that strain JS1660 was more sensitive to high DNAN concentrations (>200 μM). Therefore, subsequent experiments were conducted with Nocardioides sp. strain JS1661.
Growth of strain JS1661.
During growth on DNAN, 2,4-DNP accumulated transiently, and its disappearance was accompanied by the release of nitrite (Fig. 1). About 90% of the theoretically expected nitrite accumulated in the culture fluid. In a separate but similar experiment, stoichiometric accumulation of methanol (110 ± 2 μM) was observed after complete biodegradation of DNAN (100 ± 9 μM). Methanol (up to 5 mM) did not support growth when provided as the sole carbon and energy source. Growth yield was 18 ± 2 and 17 ± 0.5 g of protein mol−1 of DNAN and 2,4-DNP, respectively. The modest yield was expected, because most previous reports of 2,4-DNP biodegradation either involved use of supplemental carbon sources or provided no quantitative results of growth yields. The JS1661 strain did not grow on or transform 4-nitroanisole or 2-nitroanisole (100 μM) under conditions identical to those used for growth on DNAN.
FIG 1.
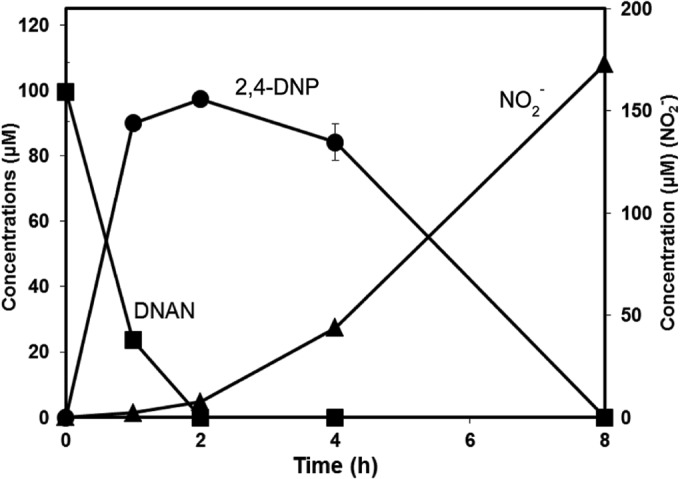
Biodegradation of DNAN by Nocardioides sp. strain JS1661.
Experiments were done to determine whether the enzymes involved in DNAN and 2,4-DNP metabolism are inducible or constitutive. Cells pregrown in either DNAN or acetate and transferred to MSB supplemented with DNAN exhibited no lag period prior to DNAN transformation. The initial specific activity was 153 ± 22 nmol min−1 mg−1 protein for DNAN-grown cells and 141 ± 8 nmol min−1 mg protein−1 for acetate-grown cells. In contrast, cells grown on acetate did not transform 2,4-DNP at a detectable rate, whereas 2,4-DNP-grown cells degraded 2,4-DNP immediately (86 ± 1 nmol min−1 mg protein−1). 2,4-DNP degradation activity became evident after 30 h for noninduced cells (see Fig. S1 in the supplemental material). These results indicated clearly that the enzymes involved in 2,4-DNP biodegradation are inducible, whereas the demethylase activity is constitutive.
Enzyme assays.
Soluble enzymes in dialyzed extracts of DNAN-grown cells of Nocardioides sp. JS1661 catalyzed stoichiometric transformation of DNAN to 2,4-DNP and methanol with an initial specific activity of 170 ± 2 nmol min−1 mg protein−1 (Fig. 2). No attempt was made to optimize the assay conditions. Transformation of DNAN (101 ± 2 μM) to 2,4-DNP (103 ± 7 μM) was accompanied by stoichiometric release of methanol (104 ± 4 μM). Formaldehyde was not detected in the absence of alcohol oxidase. Addition of cofactors, such as NADPH (200 μM), NADH (200 μM), or tetrahydrofolate (200 μM) did not stimulate the transformation of DNAN to 2,4-DNP, and oxygen consumption was not required for the reaction (data not shown). No activity was detected with boiled crude lysate. These results indicated that the initial reaction in biodegradation of DNAN is hydrolytic removal of the methyl group to yield 2,4-DNP without a requirement for added cofactors.
FIG 2.
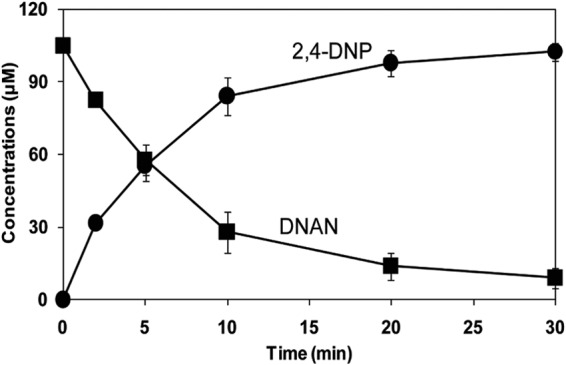
Transformation of DNAN catalyzed by soluble enzymes from Nocardioides sp. JS1661.
2,4-DNP hydride transferase activity in extracts prepared from cells grown on 2,4-DNP was 35 nmol min−1 mg protein−1. The disappearance of 2,4-DNP was accompanied by formation of the hydride-Meisenheimer complex of 2,4-DNP which is consistent with the well-established pathway (16, 17). The identity of the product was verified by HPLC comparison with the chemically synthesized standard (Fig. 3). The hydride-Meisenheimer complex of 2,4-DNP is not commercially available, and the synthetic standard was not sufficiently pure to allow quantification, as noted previously (16, 17, 28). The hydride-Meisenheimer complex disappeared from the reaction mixtures after a few minutes, and release of nitrite or accumulation of other metabolites was not detected during the reaction catalyzed by crude or dialyzed cell extracts. This could be due to the instability of the hydride-Meisenheimer complex that has been shown to spontaneously regenerate 2,4-DNP (16, 17) or to the sensitivity of the hydride transferase enzymes to oxygen and light (29–31). 2,4-DNP was not transformed when NADPH was substituted for NADH. The results indicated that the initial reactions in 2,4-DNP biodegradation involved the transfer of hydride ions to form hydride-Meisenheimer complexes of 2,4-DNP as established previously in Rhodococcus (28, 32) and Nocardia (16, 17, 31).
FIG 3.
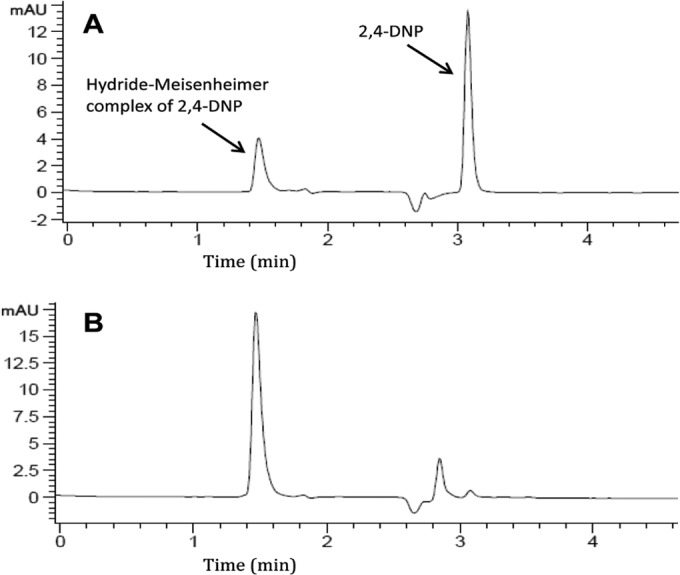
HPLC analysis of the reaction mixture during 2,4-DNP biotransformation by Nocardioides sp. JS1661. The results are from a sample analyzed after incubation with 2,4-DNP for 2 min (A) and the hydride-Meisenheimer complex synthetic standard (B).
Genes encoding DNAN demethylase.
DNAN demethylase was purified from JS1661 cells in order to determine the protein sequence and identify the gene(s) responsible. The preliminary purification described here yielded a single active band with an estimated molecular mass of 265 kDa on a native gel and a single peak with an approximate molecular mass of 234 kDa on size exclusion chromatography. When the major band was cut from the native gel and suspended in 300 μl of 500 μM DNAN in 50 mM HEPES buffer (pH 8.5), the yellow color due to DNAN hydrolysis appeared immediately, whereas there was no color appearance in identical reaction mixtures incubated with gel slices from elsewhere in the gel. The purified protein yielded two bands of approximately 36 and 34 kDa on SDS-polyacrylamide gels (see Fig. S2 in the supplemental material), which suggests that the active enzyme is a multimer of two subunits. The specific activity of the purified enzyme was 6.4 μmol min−1 mg protein−1. Detailed characterization of the enzyme will require optimization and scale-up of the purification or heterologous expression of the genes and will be reported elsewhere.
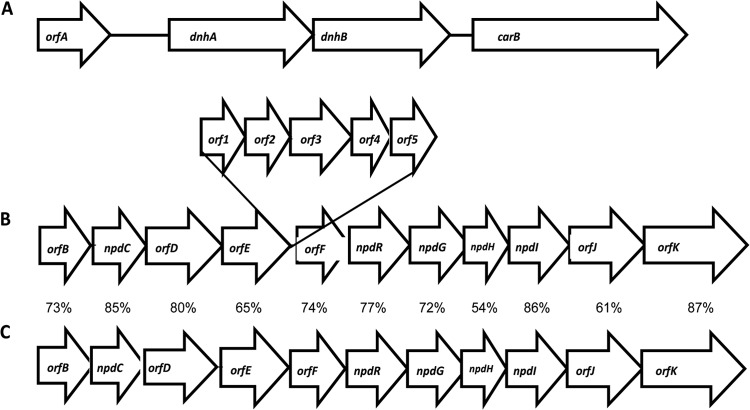
Based on peptide sequences obtained from the purified proteins and annotation of the contig, two adjacent genes were identified on a 5-kb contig in the draft genome of strain JS1661. A homolog search based on protein family (Pfam) revealed that one of the genes, designated dnhA (328 amino acids), had only 22% amino acid identity to a metallo-beta-lactamase protein (PF00753) of Bacillus cereus (33), while no Pfam identity was identified for the second gene designated dnhB (318 amino acids). dnhA has a start codon of ATG, while dnhB appears to start immediately at the stop codon of dnhA with GTG as the start codon. The sequences of the peptides and locations of the ribosomal binding site and putative start/stop codons are shown in the supplemental material (see Fig. S3 in the supplemental material). The nearest upstream ORF encodes a hypothetical protein, and the downstream ORF is most closely related (25% amino acid identity) to a beta phytoene dehydrogenase from Myxococcus xanthus and appears to have no involvement with the hydrolase genes (Fig. 4A).
FIG 4.
(A and B) Organization of genes encoding enzymes involved in DNAN demethylation (A) and 2,4-DNP degradation (B) in Nocardioides sp. JS1661. (C) 2,4-DNP catabolic genes were compared with homologs from Rhodococcus opacus (12), and the percentage amino acid identity is indicated. Protein-coding genes include the following genes: orfA, gene encoding hypothetical protein; dnhA, upstream hydrolase; dnhB, downstream hydrolase; carB, gene encoding putative phytoene dehydrogenase; orfB, l-carnitine dehydratase; npdC, hydride transferase I; orfD, aldehyde dehydrogenase; orfE, acetyl coenzyme A (acetyl-CoA) synthetase; orf1, ABC transporter; orf2, amino acid transport system permease; orf3, ABC transporter permease; orf4, ABC transporter ATP-binding protein; orf5, amino acid transport ATP-binding protein; orfF, lyase; npdR, transcriptional regulator; npdG, NADPH-dependent F420 oxidoreductase; npdH, protein converting the dihydride-picric acid complex (2H−-PA) to product X; npdI, hydride transferase II; orfJ, enoyl-CoA hydratase; orfK, acyl-CoA dehydrogenase.
Genes encoding 2,4-DNP catabolic pathways.
Nocardioides sp. JS1661 contains genes very similar in sequence and organization to the genes involved in picric acid and 2,4-DNP degradation in Rhodococcus opacus (12, 13), except for insertion of several genes that appear to be involved mainly in transport functions in Nocardioides sp. strain JS1661 (Fig. 4B and C). Homologs of the inserted genes were not found in Rhodococcus (Table 1). The genes that encode the 2,4-DNP catabolic pathway and the DNAN demethylase genes were not located on the same contig in the draft genome.
TABLE 1.
Genes encoding DNAN hydrolase and 2,4-DNP catabolic enzymes in comparison with the closest amino acid matches from the NCBI database
| Genea | Enzyme or protein size (no. of aa)b | Proposed/confirmed function | Identity (%) | Closest organism | Accession no. of closest match |
|---|---|---|---|---|---|
| orfA | 107 | Hypothetical protein | 59 | Achromobacter xylosoxidans | WP_006384465 |
| dnhA | 328 | DNAN hydrolase α-subunitc (metallo-beta-lactamase) | 22 | Bacillus cereus | GI-157836766 |
| dnhB | 318 | DNAN hydrolase β-subunitc (hypothetical protein) | 26 | Marinobacterium jannaschii | WP_027858191 |
| carB | 541 | Phytoene dehydrogenase | 26 | Myxococcus xanthus | WP_011551016 |
| orfB | 394 | l-Carnitine dehydratase | 73 | Rhodococcus opacus | WP_005257508 |
| npdC | 295 | Hydride transferase | 85 | Rhodococcus opacus | AAK29142d |
| orfD | 483 | Aldehyde dehydrogenase | 80 | Rhodococcus opacus | WP_005257514 |
| orfE | 532 | Acetyl-CoA synthetase | 65 | Rhodococcus opacus | AAK38099 |
| orf1 | 386 | ABC transporter | 35 | Sphaerobacter thermophilus | WP_012873153 |
| orf2 | 287 | Amino acid transport system permease | 40 | Actinotalea ferrariae CF5-4 | EYR64100 |
| orf3 | 355 | ABC transporter permease | 35 | Bradyrhizobium sp. strain ORS 375 | WP_009030206 |
| orf4 | 222 | ABC transporter ATP-binding protein | 43 | Geobacillus thermocatenulatus | WP_025950112 |
| orf5 | 231 | Amino acid transport ATP-binding protein | 45 | Amphimedon queenslandica | XP_003390336 |
| orfF | 160 | Lyase | 74 | Rhodococcus opacus | AAK38100 |
| npdR | 268 | Transcriptional regulator | 77 | Rhodococcus opacus | AAK38101 |
| npdG | 220 | NADPH-dependent F420 oxidoreductase | 72 | Rhodococcus opacus | AAK29135d |
| npdH | 95 | Protein converting 2H−-PA to product X | 54 | Rhodococcus opacus | WP_005257523 |
| npdI | 349 | Hydride transferase II | 86 | Rhodococcus opacus | AAK29131d |
| orfJ | 232 | Enoyl-CoA hydratase | 61 | Rhodococcus opacus | AAK38105 |
| orfK | 384 | Acyl-CoA dehydrogenase | 87 | Rhodococcus opacus | WP_005257529 |
The first 4 genes in the table are on a separate contig from the genes below.
aa, amino acids.
Functions established in this study.
Biochemically characterized previously.
DISCUSSION
Growth of bacteria on methoxy-substituted aromatic compounds is common, and the pathways including O-demethylation have been established. Three types of O-demethylation mechanisms have been reported. They include the Rieske nonheme iron oxygenases (34–36) and the cytochrome P-450-dependent O-demethylases (37, 38), both of which require oxygen and NAD(P)H, and the tetrahydrofolate-dependent O-demethylases (39, 40). DNAN biodegradation in Nocardioides sp. strain JS1661 is also initiated by removal of the methyl group to yield 2,4-DNP and methanol. The release of methanol and formation of 2,4-DNP without a requirement for cofactors or oxygen indicates that cleavage of the ether bond of DNAN in Nocardioides involves a novel hydrolytic demethylase (Fig. 5). Atrazine hydrolase (TrzN) from Arthrobacter has been reported to catalyze the cleavage of the methoxy group of the heterocyclic pesticide atratone at a low rate (41, 42). However, TrzN had no amino acid similarity to DNAN hydrolase.
FIG 5.
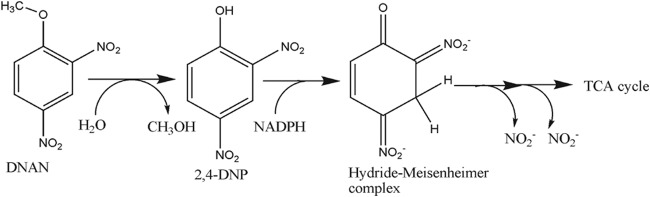
Proposed pathway of DNAN biodegradation by Nocardioides sp. JS1661. TCA, tricarboxylic acid.
The biodegradation pathway of 2,4-DNP is well established for Gram-positive bacteria such as Rhodococcus erythropolis (28, 32), Nocardioides simplex FJ2-1A (16, 31), and Nocardioides sp. strain CB 22-2 (17). The pathway is initiated by the formation of the hydride-Meisenheimer complex of 2,4-DNP by the action of an NADPH-dependent F420 reductase and hydride transferases (16) (Fig. 5). In contrast, it was suggested that the initial step of 2,4-DNP biodegradation in Gram-negative bacteria such as Burkholderia sp. strain KU-46, is removal of nitrite to form 4-nitrophenol (43). Neither nitrite nor 4-nitrophenol was detected during transformation of 2,4-DNP by cell extracts from Nocardioides sp. strain JS1661, and the strain did not grow on 4-nitrophenol. Taken with the presence of the genes closely related to those encoding the pathway for 2,4-DNP degradation, the results indicate clearly that the pathway of 2,4-DNP biodegradation in Nocardioides JS1661 is similar to the pathway previously established for the Rhodococcus and Nocardia species.
The presence of genes involved in 2,4-DNP biodegradation, constitutive demethylase activity, and the inducible 2,4-DNP degradation suggest that recent acquisition of the demethylase genes by the parental 2,4-DNP-degrading strain enabled growth on DNAN. Evolution of degradation pathways for novel organic compounds often involves recruitment of genes encoding catabolic enzymes to extend existing pathways (44–47). The early stages of pathway assembly often involve loss of regulatory functions that results in constitutive expression of key enzymes (44).
In conclusion, the robust biodegradation of DNAN by strain JS1661 suggests that the isolates would be good candidates for waste treatment or biodegradation applications. The fact that they were isolated from a DNAN manufacturing plant suggests that they are involved in DNAN degradation in the waste treatment system at the site. We are currently investigating their activity and distribution in other ecosystems and further characterizing the hydrolase enzyme and its evolutionary origins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bob Winstead at BAE Systems for providing samples from Holston Army Ammunition Plant and John Oakeshott for insightful discussions. We also thank Jian-Wei Liu and Peter Campbell for assistance in enzyme purification and peptide sequencing.
This work was funded in part by CSIRO's Land and Water Flagship, the Defense Threat Reduction Agency, and U.S. Army Research Office grant W911NF-07-1-0077.
Footnotes
Published ahead of print 3 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02752-14.
REFERENCES
- 1.Liang J, Olivares C, Field JA, Sierra-Alvarez R. 2013. Microbial toxicity of the insensitive munitions compound, 2,4-dinitroanisole (DNAN), and its aromatic amine metabolites. J. Hazard. Mater. 262:281–287. 10.1016/j.jhazmat.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 2.Dodard SG, Sarrazin M, Hawari J, Paquet L, Ampleman G, Thiboutot S, Sunahara GI. 2013. Ecotoxicological assessment of a high energetic and insensitive munitions compound: 2,4-dinitroanisole (DNAN). J. Hazard. Mater. 262:143–150. 10.1016/j.jhazmat.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 3.Olivares C, Liang J, Abrell L, Sierra-Alvarez R, Field JA. 2013. Pathways of reductive 2,4-dinitroanisole (DNAN) biotransformation in sludge. Biotechnol. Bioeng. 110:1595–1604. 10.1002/bit.24820. [DOI] [PubMed] [Google Scholar]
- 4.Perreault NN, Manno D, Halasz A, Thiboutot S, Ampleman G, Hawari J. 2012. Aerobic biotransformation of 2,4-dinitroanisole in soil and soil Bacillus sp. Biodegradation 23:287–295. 10.1007/s10532-011-9508-7. [DOI] [PubMed] [Google Scholar]
- 5.Hawari J, Monteil-Rivera F, Perreault NN, Halasz A, Paquet L, Radovic-Hrapovic Z, Deschamps S, Thiboutot S, Ampleman G. 2015. Environmental fate of 2,4-dinitroanisole (DNAN) and its reduced products. Chemosphere 119:16–23. 10.1016/j.chemosphere.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 6.Platten WE, III, Bailey D, Suidan MT, Maloney SW. 2010. Biological transformation pathways of 2,4-dinitroanisole and N-methyl paranitroaniline in anaerobic fluidized-bed bioreactors. Chemosphere 81:1131–1136. 10.1016/j.chemosphere.2010.08.044. [DOI] [PubMed] [Google Scholar]
- 7.Niedzwiecka J, Millerick K, Galloway S, Schlautman M, Finneran K. 2014. Microbially mediated 2,4-dinitroanisole (DNAN) and nitroguanidine (NQ) degradation, abstr Q-383 Abstr. 114th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 8.Arnett CM, Rodriguez G, Maloney SW. 2009. Analysis of bacterial community diversity in anaerobic fluidized bed bioreactors treating 2,4-dinitroanisole (DNAN) and n-methyl-4-nitroaniline (MNA) using 16S rRNA gene clone libraries. Microbes Environ. 24:72–75. 10.1264/jsme2.ME08556. [DOI] [PubMed] [Google Scholar]
- 9.Richard T, Weidhaas J. 2014. Biodegradation of IMX-101 explosive formulation constituents: 2,4-dinitroanisole (DNAN), 3-nitro-1,2,4-triazol-5-one (NTO), and nitroguanidine. J. Hazard. Mater. 280:372–379. 10.1016/j.jhazmat.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Salter-Blanc AJ, Bylaska EJ, Ritchie JJ, Tratnyek PG. 2013. Mechanisms and kinetics of alkaline hydrolysis of the energetic nitroaromatic compounds 2,4,6-trinitrotoluene (TNT) and 2,4-dinitroanisole (DNAN). Environ. Sci. Technol. 47:6790–6798. 10.1021/es304461t. [DOI] [PubMed] [Google Scholar]
- 11.Rao B, Wang W, Cai Q, Anderson T, Gu B. 2013. Photochemical transformation of the insensitive munitions compound 2,4-dinitroanisole. Sci. Total Environ. 443:692–699. 10.1016/j.scitotenv.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 12.Heiss G, Hofmann KW, Trachtmann N, Walters DM, Rouviere P, Knackmuss HJ. 2002. npd gene functions of Rhodococcus (opacus) erythropolis HL PM-1 in the initial steps of 2,4,6-trinitrophenol degradation. Microbiology 148:799–806. [DOI] [PubMed] [Google Scholar]
- 13.Walters DM, Russ R, Knackmuss HJ, Rouviere PE. 2001. High-density sampling of a bacterial operon using mRNA differential display. Gene 273:305–315. 10.1016/S0378-1119(01)00597-2. [DOI] [PubMed] [Google Scholar]
- 14.Schäfer A, Harms H, Zehnder AB. 1996. Biodegradation of 4-nitroanisole by two Rhodococcus spp. Biodegradation 7:249–255. 10.1007/BF00058184. [DOI] [PubMed] [Google Scholar]
- 15.Hoyt N, Brunell M, Kroeck K, Hable M, Crouse L, O'Neill A, Bannon DI. 2013. Biomarkers of oral exposure to 3-nitro-1,2,4-triazol-5-one (NTO) and 2,4-dinitroanisole (DNAN) in blood and urine of rhesus macaques (Macaca mulatta). Biomarkers 18:587–594. 10.3109/1354750X.2013.829522. [DOI] [PubMed] [Google Scholar]
- 16.Ebert S, Fischer P, Knackmuss HJ. 2001. Converging catabolism of 2,4,6-trinitrophenol (picric acid) and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. Biodegradation 12:367–376. 10.1023/A:1014447700775. [DOI] [PubMed] [Google Scholar]
- 17.Behrend C, Heesche-Wagner K. 1999. Formation of hydride-Meisenheimer complexes of picric acid (2,4,6-trinitrophenol) and 2,4-dinitrophenol during mineralization of picric acid by Nocardioides sp. strain CB 22-2. Appl. Environ. Microbiol. 65:1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271. 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 19.Tritt A, Eisen JA, Facciotti MT, Darling AE. 2012. An integrated pipeline for de novo assembly of microbial genomes. PLoS One 7:e42304. 10.1371/journal.pone.0042304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42:D206–D214. 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klavons JA, Bennett RD. 1986. Determination of methanol using alcohol oxidase and its application to methyl ester content of pectins. J. Agric. Food Chem. 34:597–599. 10.1021/jf00070a004. [DOI] [Google Scholar]
- 22.Summers WR. 1990. Characterization of formaldehyde and formaldehyde-releasing preservatives by combined reversed-phase cation-exchange high-performance liquid chromatography with postcolumn derivatization using Nash's reagent. Anal. Chem. 62:1397–1402. 10.1021/ac00213a010. [DOI] [Google Scholar]
- 23.Campbell PM, Cao AT, Hines ER, East PD, Gordon KHJ. 2008. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. 38:950–958. 10.1016/j.ibmb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Blasco R, Moore E, Wray V, Pieper D, Timmis K, Castillo F. 1999. 3-Nitroadipate, a metabolic intermediate for mineralization of 2,4-dinitrophenol by a new strain of a Rhodococcus species. J. Bacteriol. 181:149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu Y, Spain JC. 2010. Biodegradation of 5-nitroanthranilic acid by Bradyrhizobium sp. strain JS329. Appl. Environ. Microbiol. 76:1417–1422. 10.1128/AEM.02816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon J-H, Cho Y-G, Lee ST, Suzuki K-I, Nakase T, Park Y-H. 1999. Nocardioides nitrophenolicus sp. nov., a p-nitrophenol-degrading bacterium. Int. J. Syst. Bacteriol. 49:675–680. 10.1099/00207713-49-2-675. [DOI] [PubMed] [Google Scholar]
- 27.Schippers A, Schumann P, Spröer C. 2005. Nocardioides oleivorans sp. nov., a novel crude-oil-degrading bacterium. Int. J. Syst. Evol. Microbiol. 55:1501–1504. 10.1099/ijs.0.63500-0. [DOI] [PubMed] [Google Scholar]
- 28.Rieger PG, Sinnwell V, Preuss A, Francke W, Knackmuss HJ. 1999. Hydride-Meisenheimer complex formation and protonation as key reactions of 2,4,6-trinitrophenol biodegradation by Rhodococcus erythropolis. J. Bacteriol. 181:1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schönheit P, Keweloh H, Thauer RK. 1981. Factor F420 degradation in Methanobacterium thermoautotrophicum during exposure to oxygen. FEMS Microbiol. Lett. 12:347–349. 10.1111/j.1574-6968.1981.tb07671.x. [DOI] [Google Scholar]
- 30.Cheeseman P, Toms-Wood A, Wolfe RS. 1972. Isolation and properties of a fluorescent compound, factor 420, from Methanobacterium strain M.o.H. J. Bacteriol. 112:527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert S, Rieger PG, Knackmuss HJ. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J. Bacteriol. 181:2669–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenke H, Pieper DH, Bruhn C, Knackmuss HJ. 1992. Degradation of 2,4-dinitrophenol by two Rhodococcus erythropolis strains, HL 24-1 and HL 24-2. Appl. Environ. Microbiol. 58:2928–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. 1995. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 14:4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunel F, Davison J. 1988. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J. Bacteriol. 170:4924–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Civolani C, Barghini P, Roncetti AR, Ruzzi M, Schiesser A. 2000. Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13. Appl. Environ. Microbiol. 66:2311–2317. 10.1128/AEM.66.6.2311-2317.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudtachat N, Ito N, Itakura M, Masuda S, Eda S, Mitsui H, Kawaharada Y, Minamisawa K. 2009. Aerobic vanillate degradation and C1 compound metabolism in Bradyrhizobium japonicum. Appl. Environ. Microbiol. 75:5012–5017. 10.1128/AEM.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werck-Reichhart D, Hehn A, Didierjean L. 2000. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 5:116–123. 10.1016/S1360-1385(00)01567-3. [DOI] [PubMed] [Google Scholar]
- 38.Snyder R. 2000. Cytochrome P450, the oxygen-activating enzyme in xenobiotic metabolism. Toxicol. Sci. 58:3–4. 10.1093/toxsci/58.1.3. [DOI] [PubMed] [Google Scholar]
- 39.Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M. 2005. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 187:2030–2037. 10.1128/JB.187.6.2030-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, Katayama Y, Fukuda M. 2004. A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J. Bacteriol. 186:2757–2765. 10.1128/JB.186.9.2757-2765.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seffernick JL, Reynolds E, Fedorov AA, Fedorov E, Almo SC, Sadowsky MJ, Wackett LP. 2010. X-ray structure and mutational analysis of the atrazine chlorohydrolase TrzN. J. Biol. Chem. 285:30606–30614. 10.1074/jbc.M110.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapir N, Rosendahl C, Johnson G, Andreina M, Sadowsky MJ, Wackett LP. 2005. Substrate specificity and colorimetric assay for recombinant TrzN derived from Arthrobacter aurescens TC1. Appl. Environ. Microbiol. 71:2214–2220. 10.1128/AEM.71.5.2214-2220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwaki H, Abe K, Hasegawa Y. 2007. Isolation and characterization of a new 2,4-dinitrophenol-degrading bacterium Burkholderia sp. strain KU-46 and its degradation pathway. FEMS Microbiol. Lett. 274:112–117. 10.1111/j.1574-6968.2007.00816.x. [DOI] [PubMed] [Google Scholar]
- 44.Cases I, de Lorenzo V. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1–11. 10.1093/emboj/20.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson GR, Spain JC. 2003. Evolution of catabolic pathways for synthetic compounds: bacterial pathways for degradation of 2,4-dinitrotoluene and nitrobenzene. Appl. Microbiol. Biotechnol. 62:110–123. 10.1007/s00253-003-1341-4. [DOI] [PubMed] [Google Scholar]
- 46.Kolvenbach BA, Helbling DE, Kohler H-PE, Corvini PFX. 2014. Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria. Curr. Opin. Biotechnol. 27:8–14. 10.1016/j.copbio.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Schulenburg C, Miller BG. 2014. Enzyme recruitment and its role in metabolic expansion. Biochemistry 53:836–845. 10.1021/bi401667f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.